Abstract
The period (PER) and cryptochrome (CRY) families play critical roles in circadian rhythms. The imbalance of circadian factors may lead to the occurrence of cancer. Expressions of PER and CRY family members decrease in various cancers. Nevertheless, expression levels, genetic variations, and molecular mechanisms of PER and CRY family members in lung adenocarcinoma (LUAD) and their correlations with prognoses and immune infiltration in LUAD patients are still unclear. In this study, to identify their biological functions in LUAD development, comprehensive high-throughput techniques were applied to analyze the relationships of expressions of PER and CRY family members with genetic variations, molecular mechanisms, and immune infiltration. The present results showed that transcription levels of PER1 and CRY2 in LUAD were significantly downregulated. High expression levels of PER2, PER3, CRY1, and CRY2 indicated longer overall survival. Some cancer signaling pathways were related to PER and CRY family members, such as cell-cycle, histidine metabolism, and progesterone-mediated oocyte maturation pathways. Expressions of PER and CRY family members significantly affected the infiltration of different immune cells. In conclusion, our findings may help better understand the molecular basis of LUAD, and provide new perspectives of PER and CRY family members as novel biomarkers for LUAD.
Keywords: circadian rhythm, lung adenocarcinoma, biomarker, PER, CRY
INTRODUCTION
According to a report provided by cancer statistics 2022, the most commonly diagnosed cancer is lung cancer, which is also the leading cause of cancer deaths in the world [1]. Lung cancer is divided into two categories, namely small-cell lung cancer (SCLC) and non-SCLC (NSCLC). NSCLC accounts for more than 85% of all lung cancers, and it is subdivided into lung adenocarcinoma (LUAD), squamous cell lung cancer, and large-cell lung cancer. LUAD accounts for the highest proportion of NSCLC cases, and finding efficacious treatments for LUAD is one of the main research goals of researchers. In recent years, although the pathogenesis of LUAD and new treatment strategies have been discovered, LUAD is still one of the most aggressive and fatal types of lung cancer, with low 5-year overall survival (OS) rates. Therefore, finding novel biomarkers for LUAD is desperately in demand [2–6]. Recently, it was proven that circadian rhythms act as a crucial factor causing cancer, as an abnormal lifestyle may disrupt and break natural circadian rhythms [7–9]. Circadian rhythms can regulate cell proliferation, cell death, DNA repair, and metabolic functions [10, 11]. Changes in circadian rhythms may lead to loss of these regulatory functions and further lead to the development of cancer. The suprachiasmatic nucleus (SCN) plays an important role in the circadian rhythms of mammals [12], and it uses a molecular oscillator to maintain clock oscillation at a normal pace [13]. The molecular oscillator consists of interacting molecular loops, composed of positive elements including circadian locomotor output cycles kaput (CLOCK) and brain and muscle ARNT-like 1 (BMAL1), and negative elements including period (PER) circadian regulators and cryptochrome (CRY) circadian regulators [14]. In addition, the core oscillatory mechanism of the SCN begins from the heterodimer CLOCK/BMAL1 complex binding to E-box elements in their regulatory regions and activating target genes to initiate transcription of PER and CRY [15], which are translated into proteins and accumulate in the cytoplasm during the daytime. After the proteins have accumulated to a certain level, the PER and CRY proteins form a complex that is then translocated into nuclei to inhibit their own transcription at night [16]. Afterward, the PER and CRY proteins are gradually phosphorylated at night, and degraded by the proteasome after being ubiquitinated by a specific E3 ligase [17–19]. The SCN can adjust the circadian rhythm stage by receiving photic and non-photic signals [20].
PER and CRY are important negative regulators of circadian rhythms [21]. Studies showed that CRY-deficient mice produce angiopoietin-like protein 2 expression [22], and CRY was proven to be related to insulin-like growth factor (IGF), which plays important roles in cell proliferation, growth, and cancer [23]. Similarly, according to clinicopathological features, PER1, PER2, and PER3 are obviously methylated in breast cancer patients [24]. PER expression in colorectal cancer cells is also significantly lower than that of normal colorectal mucosal cells [25]. Recent studies discovered the mechanisms by which circadian factors affect certain cancers. For example, melatonin can inhibit the activity and the growth of prostate cancer cells by upregulating PER2 [26]. CLOCK/BMAL1/PER/CRY were also found to alter the c-Myc/p21 and Wnt/β-catenin pathways to varying degrees to affect DNA damage [27]. In addition, circadian rhythms are thought to be related to the immune system, as CRY can affect some key inflammatory pathways such as nuclear factor (NF)-κB [28]. Despite the fact that there is research on PER and CRY in various cancers, current studies have not fully elucidated expression levels, gene variations, molecular functions, or their relationships with prognoses and immune infiltrations in LUAD.
Few previous studies reported the roles of PER and CRY in lung cancer. In particular, interactions and pathways between all PER and CRY family members and related molecules in tumorigenesis are still unclear. Multiple microarray and sequencing technologies have enhanced the ability of robust computational algorithms to rapidly analyze biomedical data [29–34]. Examining gene expressions and employing appropriate algorithms are thought to be able to help us understand the respective functions of PER and CRY in lung cancer development. In this study, different bioinformatics databases were incorporated to analyze various PER and CRY family members in LUAD to understand expressions of these factors, molecular functions such as proliferation or tumorigenesis, and their impacts on OS, genetic changes, immune infiltration, and immune checkpoints in LUAD, which would help identify whether PER and CRY are suitable biomarkers for precision treatment and detection of LUAD.
RESULTS
Transcriptional levels of PER and CRY family members in LUAD patients
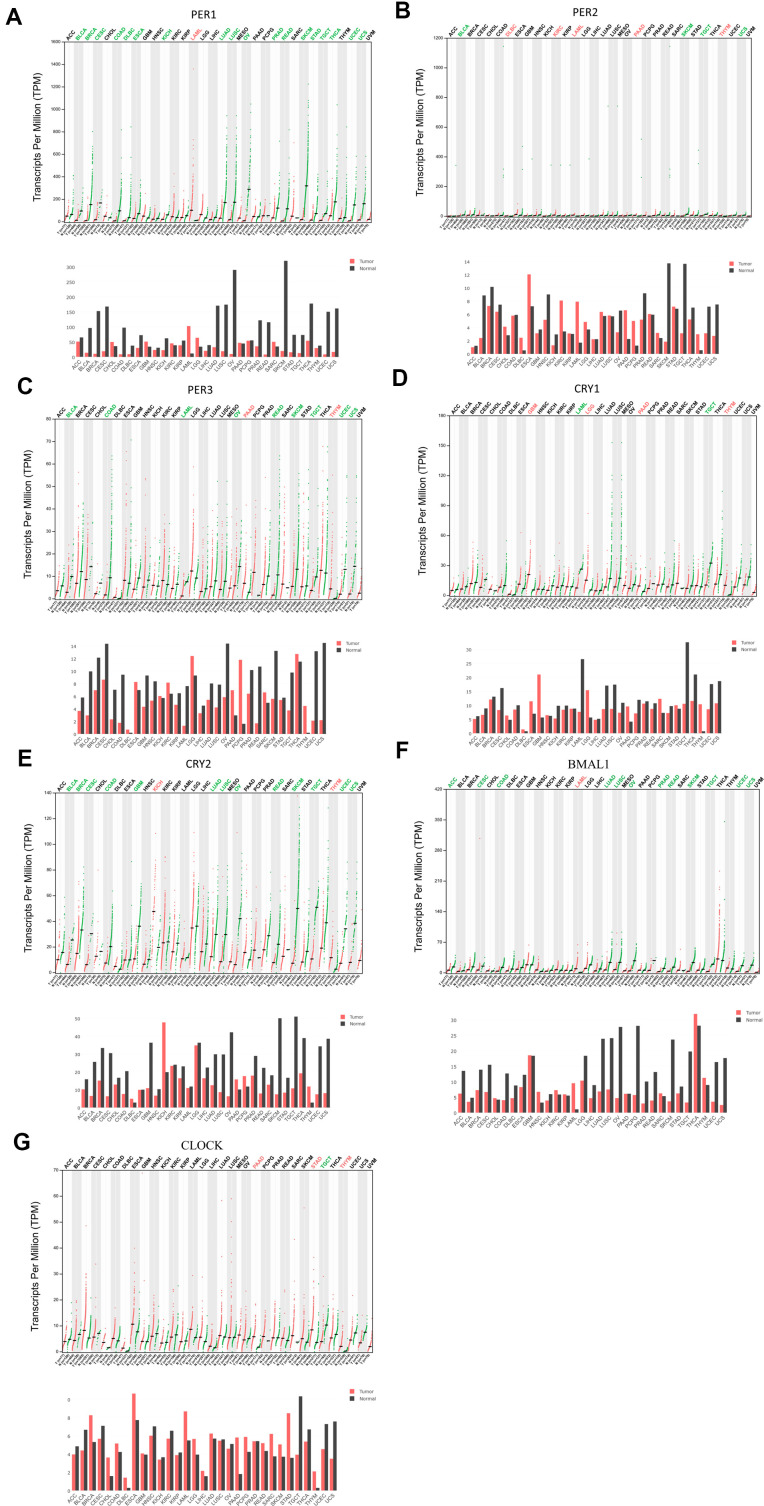
Circadian factors are widely expressed by mammalian cells, but have different expression levels in different tumor tissues. The UALCAN database showed expressions of circadian factors, including PER and CRY family members, BMAL1 (ARNTL), and CLOCK, with differential expression levels between different types of cancers and normal tissues (Supplementary Figure 1). GEPIA was used to understand the messenger (m)RNA expressions of circadian rhythm-related factors in the PER family (PER1, PER2, and PER3), CRY family (CRY1 and CRY2), BMAL1, and CLOCK in different types of cancer (Figure 1).
Figure 1.
Expression levels of period (PER) family (PER1, PER2, and PER3), cryptochrome (CRY) family (CRY1 and CRY2), and other circadian factors such as BMAL1 and CLOCK in different types of cancer (GEPIA). (A–G) This figure shows mRNA expression levels of the PER and CRY families of circadian factors in different cancer tissues. If a cancer had significant overexpression of a gene, the name of the cancer is shown in red. Conversely, if a cancer had a significantly low expression of the gene, green color indicates the name of the cancer.
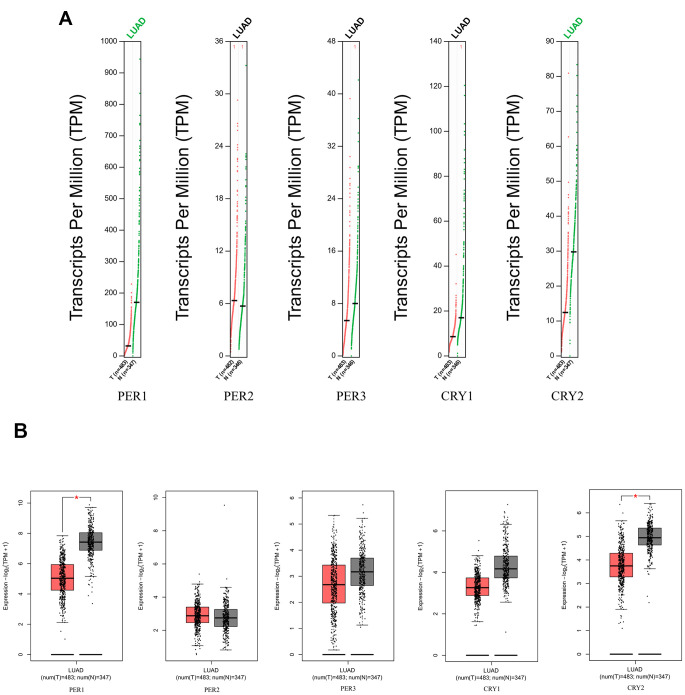
The Oncomine database displays mRNA expression levels of PER and CRY family members in different types of tumors and normal tissue samples (Supplementary Figure 2). The Oncomine analysis showed that transcription levels of both PER and CRY family members were downregulated in lung cancer patients. Transcription levels of all members of the PER and CRY families were significantly lower than those in normal tissues (Supplementary Table 1). In the LUAD dataset of Bhattacharjee Lung [35], Stearman Lung [36], Landi Lung [37] and Okayama Lung [38], transcriptional levels of PER1 in tumors were lower than those in normal samples with -5.555- (p = 3.35E-5), -1.717- (p = 1.38E-7), -2.125- (p = 8.29E-19), and -2.148-fold changes (p = 1.77E-8), respectively. In the Su Lung dataset [39], expressions of PER1, PER2, CRY1, and CRY2 were -1.861- (p = 6.23E-7), -1.803- (p = 1.29E-7), -2.058- (p = 1.39E-6), and -3.450-fold lower (p = 2.47E-5) in LUAD than in normal samples. In the Hou Lung [40] dataset, PER3, CRY1, and CRY2 were significantly lower than in normal tissues in LUAD patients with respective fold changes of -2.024 (p = 1.05E-9), -1.702 (p = 5.10E-13), and -1.836 (p = 9.60E-13). In GEPIA2, PER1 and CRY2 were also found to have higher expressions in normal lung tissues than in LUAD tissues (Figure 2).
Figure 2.
Expression levels of PER (period) and CRY (cryptochrome) family members in lung adenocarcinoma (LUAD) patients (GEPIA2), with the q-value cutoff set to 0.01. (A) mRNA expressions of PER1, PER2, PER3, CRY1, and CRY2 in LUAD (red) and normal lung tissues (green). The name of a cancer in green indicates that the mRNA expression of the gene in normal tissues was significantly higher than in cancer tissues. (B) mRNA expressions of PER1, PER2, PER3, CRY1, and CRY2 in LUAD (red) and normal lung tissues (gray). Red stars indicate significant differences in the data.
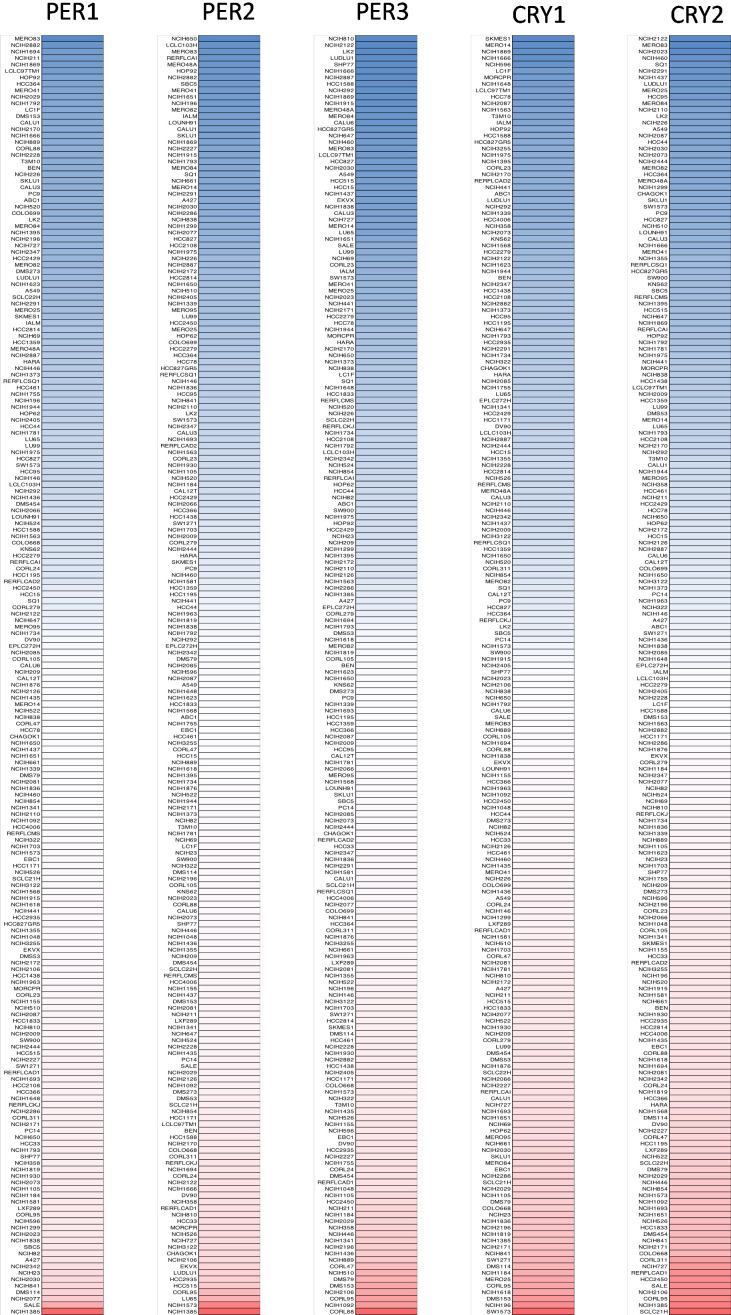
Furthermore, we used the CCLE to analyze mRNA expression levels of PER1, PER2, PER3, CRY1, and CRY2 in lung cancer cell lines in current lung cancer research. Then, the CCLE analysis was presented to reveal transcriptomic levels of PER and CRY family members in 198 lung cancer cell lines (Figure 3). Comprehensive results are clearly described in Figure 3 and show mRNA expression levels of PER1, PER2, PER3, CRY1, and CRY2 in different lung cancer cell lines. Blue represents a lowly expressed gene in a cell line, while red indicates a highly expressed gene in a cell line. The shade of the color represents the degree of high or low expression.
Figure 3.
Gene expression levels of PER (period) and CRY (cryptochrome) family members when screening 198 lung cancer cell lines (CCLE database). PER1, PER2, PER3, CRY1, and CRY2 expression levels were differentiated into five columns. The blue blocks indicated under-expression, whereas the red blocks represented overexpression.
Prognostic values of PER and CRY family members in LUAD patients
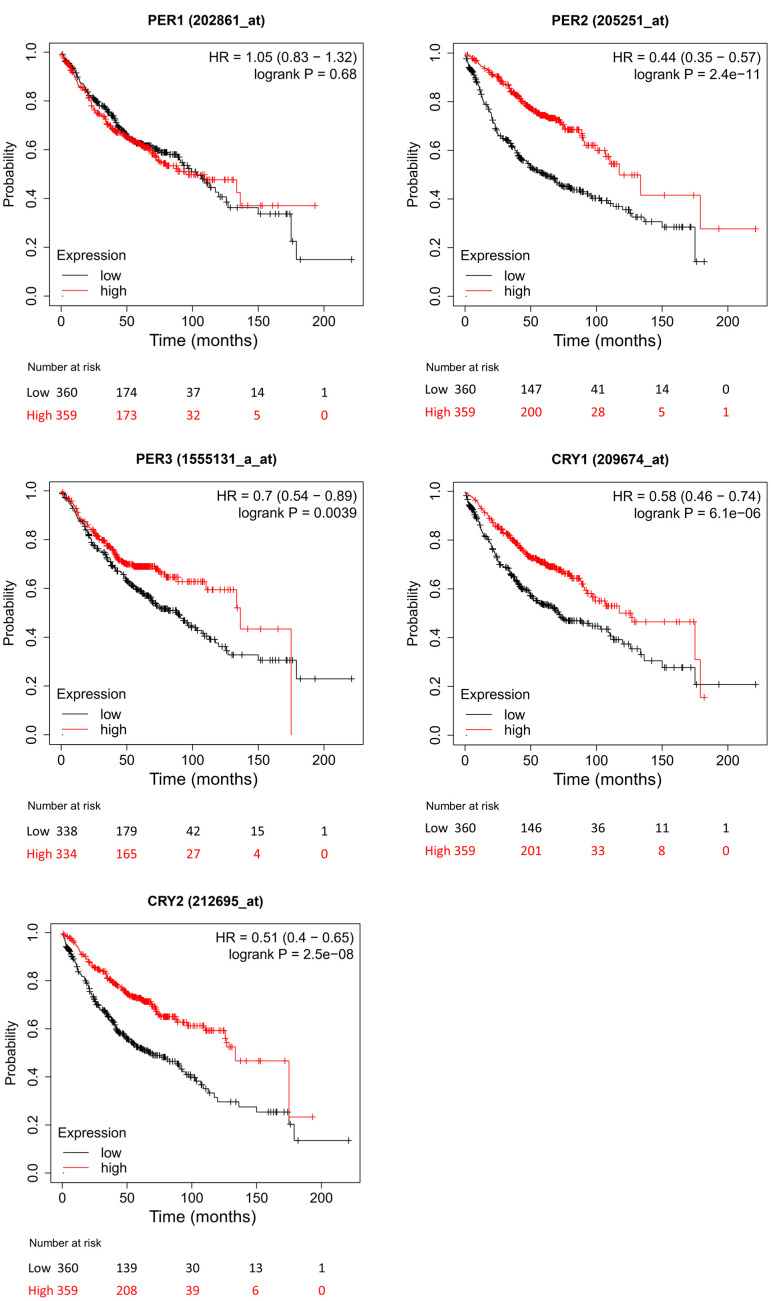
To evaluate different transcription levels of PER and CRY family members in the LUAD, we used the KM plotter to analyze correlations of PER and CRY family members with clinical results. OS results showed that high expression levels of PER2, PER3, CRY1, and CRY2 were significantly related to longer OS in LUAD patients (Figure 4). In contrast, PER1 expression in LUAD patients was not significantly related to OS.
Figure 4.
Different expressions of PER (period) and CRY (cryptochrome) family members in lung adenocarcinoma (LUAD) patients in the overall survival (OS) curve (using the Kaplan-Meier plotter). The red line represents the survival rate curve of patients with LUAD who expressed the gene, and the black line represents the survival rate curve of LUAD patients who did not express the gene.
Analysis of genetic changes and coexpressions of PER and CRY family proteins in LUAD patients
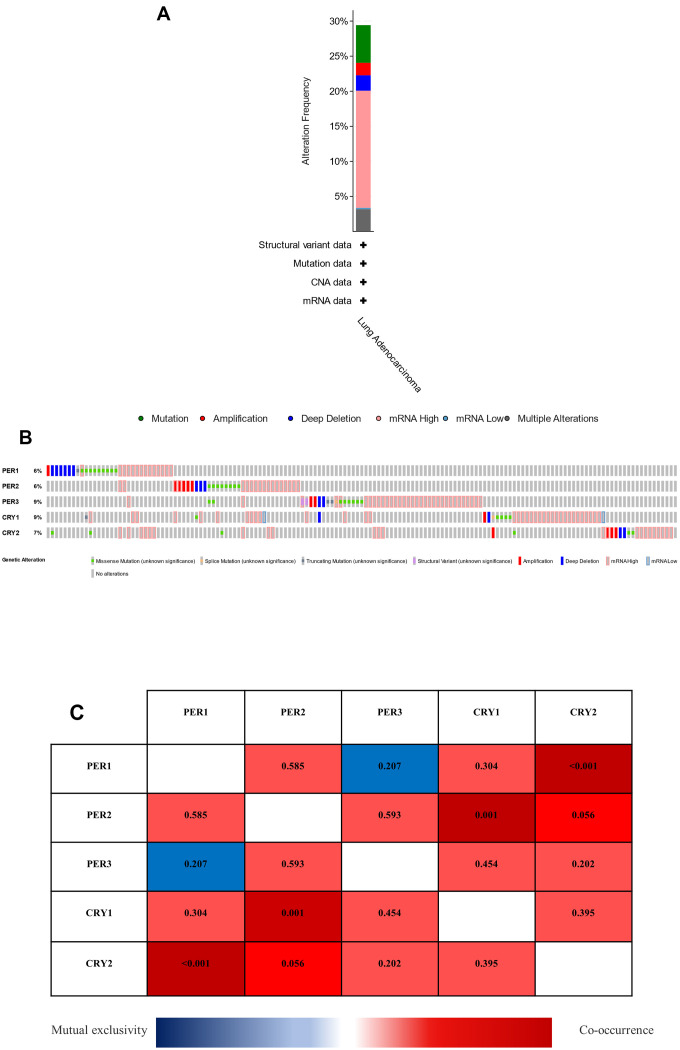
The cBioPortal web tool was used to analyze changes in PER and CRY family proteins in LUAD patients. Among 503 cases, 148 cases (29.42%) of LUAD patients had genetic changes in PER and CRY circadian rhythm-related factors (Figure 5A and Supplementary Figure 3). TCGA dataset showed that among circadian rhythm-related factors, mutation rates were highest in PER3 and CRY1 (9%), followed by CRY2 (7%), and mutation rates were lowest in PER1 and PER2 (6%) (Figure 5B). In addition, results showed the coexpression and mutually exclusive relationships among these genes, with only PER1 and CRY2, and PER2 and CRY1 exhibiting statistically significant coexpression relationships (p<0.05). Others showed coexpression and mutual exclusion without statistical significance (p>0.05) possibly due to insufficient sample sizes (Figure 5C).
Figure 5.
Analysis of genetic changes, coexpressions of PER (period) and CRY (cryptochrome) in lung adenocarcinoma (LUAD) patients. (A) Summary of changes in PER and CRY in LUAD patients. (B) Summary of OncoPrint query changes to PER and CRY family members. (C) Heat map of expressions of different PER and CRY family members in LUAD patients. The value in each color represents the p value, while red represents coexpression, and blue represents mutual exclusion.
Analysis of gene interactions among PER1, PER2, PER3, CRY1, and CRY2 in LUAD patients
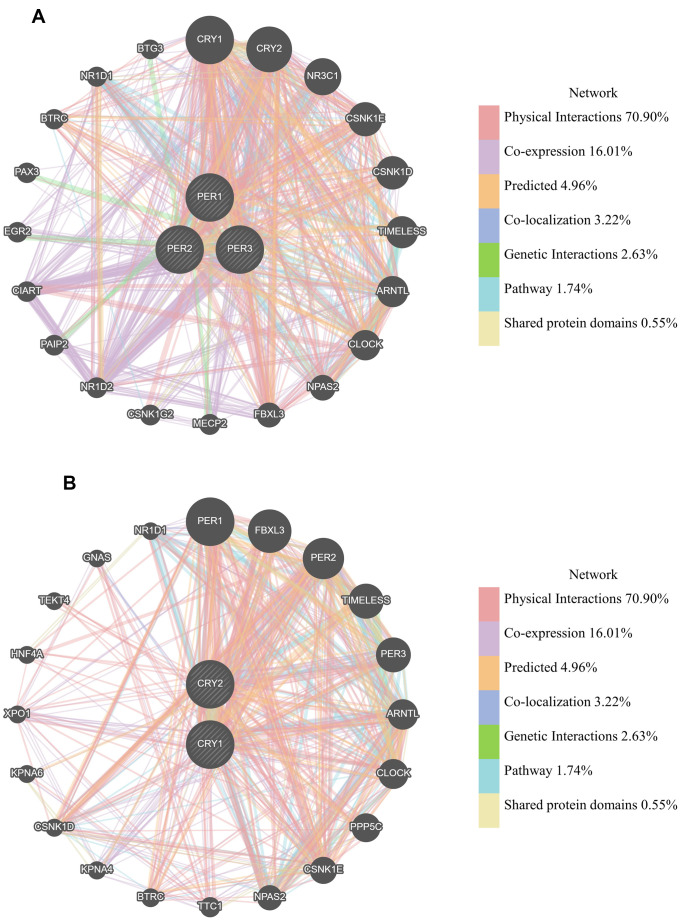
From previous studies, we know that the functions of these genes are often related to regulating circadian rhythms. For instance, CLOCK and ARNTL (aryl hydrocarbon receptor nuclear translocator-like protein 1, also known as BMAL1) are positive mediators of circadian rhythms and mediate CRY and PER transcription [14]. GeneMANIA was used to construct a gene-gene interaction (GGI) network composed of PER1, PER2, PER3, CRY1, and CRY2, and analyze the functions that may be related to networks composed of PER and CRY family members. They were all surrounded by 20 nodes, representing genes that may have physical interactions, coexpressions, predictions, co-localizations, pathways, gene interactions, and shared protein domains with PER and CRY family members (Figure 6). In the PER family network (Figure 6A), the most relevant genes were CRY1, CRY2, NR3C1 (nuclear receptor subfamily 3 group C member 1), CSNK1E (casein kinase 1 epsilon), CSNK1D (casein kinase 1 delta), TIMELESS (timeless circadian regulator), ARNTL, and CLOCK. In the CRY family network (Figure 6B), the most relevant genes were PER1, FBXL3 (F-box and leucine-rich repeat protein 3), PER2, TIMELESS, PER3, ARNTL, CLOCK, and PPP5C (protein phosphatase 5 catalytic subunit).
Figure 6.
Gene interactions among PER1, PER2, PER3, CRY1, and CRY2 in lung adenocarcinoma (LUAD) patients (GeneMANIA). (A) PER (period) family network constructed by GeneMANIA. (B) CRY (cryptochrome) family network constructed by GeneMANIA. Each node in the figure represents a gene, and the size of the node represents the intensity of the interaction. Connecting lines between nodes represent gene-gene interactions. The color of the connecting line represents the type of interaction.
PPIs and functional analysis of PER and CRY family members in LUAD patients
DAVID was utilized to analyze molecular functions and Online Mendelian inheritance in man (OMIM) diseases of PER1-, PER2-, PER3-, CRY1-, and CRY2-related genes (Supplementary Table 2). The first five molecular functions of PER1 (Supplementary Table 2A) were protein binding, ATP binding, microtubule-binding, single-stranded DNA binding, and microtubule motor activity. As to OMIM diseases, PER1 was related to breast cancer and colorectal cancer (Supplementary Table 2B). The first five molecular functions of PER2 (Supplementary Table 2C) were protein binding, poly(A) RNA binding, single-stranded DNA binding, microtubule binding, and chromatin binding. As to OMIM diseases, PER2 was related to skin/hair/eye pigmentation 1, blond/brown hair, skin/hair/eye pigmentation 1, blue/non-blue eyes (Supplementary Table 2D). The first five molecular functions of PER3 (Supplementary Table 2E) were protein binding, poly (A) RNA binding, MHC class II receptor activity, metal ion binding, and structural constituents of ribosomes. As to OMIM diseases, PER3 was related to congenital dysfibrinogenemia and congenital afibrinogenemia (Supplementary Table 2F). The first five molecular functions of the CRY family (Supplementary Table 2G) were protein binding, single-stranded DNA-dependent ATPase activity, protein kinase binding, DNA clamp loader activity, and ATP binding [41–45]. Results demonstrated the proportions of related genes with protein-binding function were the highest with statistical significance among these five genes. Moreover, we used STRING database to separately analyze PPIs of PER and CRY family members. Supplementary Figure 4 shows the protein networks closely related to the PER (Supplementary Figure 4A) and CRY families (Supplementary Figure 4B).
In addition, to verify the detail pathway relative to PER and CRY, the database of DAVID was used to analyze and select KEGG pathways with the highest correlations with PER1, PER2, PER3, and CRY family members (Supplementary Figure 5). The most closely related KEGG pathway to PER2 (Supplementary Figure 5A) was histidine metabolism. The KEGG pathway of progesterone-mediated oocyte maturation was the most relevant to PER3 (Supplementary Figure 5B). The most relevant KEGG pathway to CRY family members (Supplementary Figure 5C) was the cell cycle like PER1, but the associated genes were not the same.
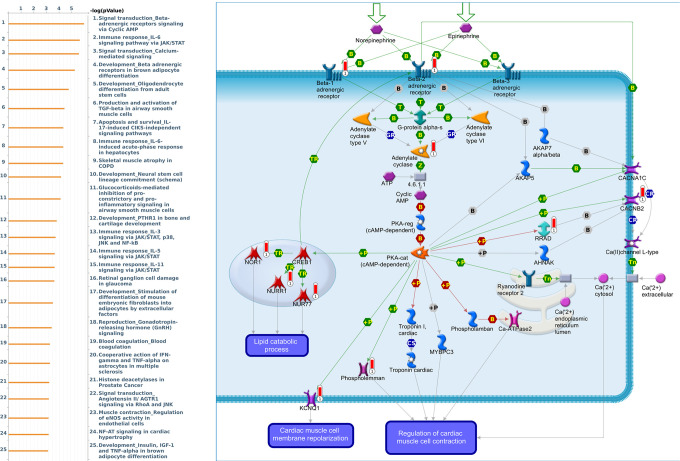
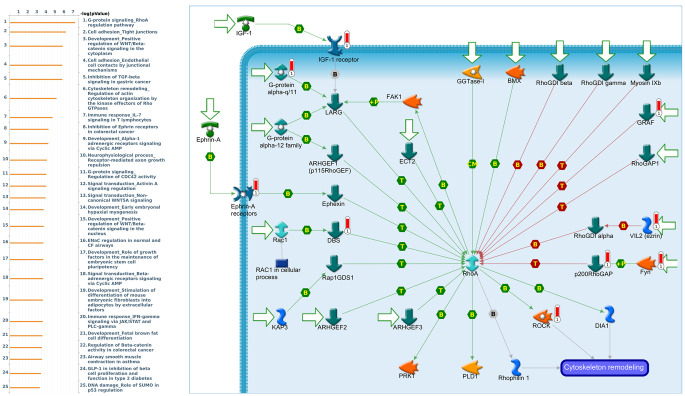
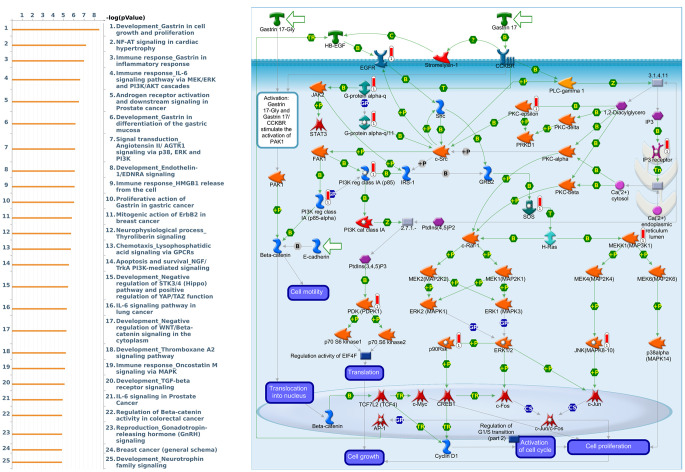
Genes coexpressed with PER1 were correlated with “Signal transduction_Beta-adrenergic receptors signaling via cyclic AMP”, “Immune response_IL-6 signaling pathway via JAK/STAT”, and “Signal transduction_Calcium-mediated signaling” (Figure 7 and Supplementary Table 3). Genes coexpressed with PER2 were correlated with “G-protein signaling_RhoA regulation pathway”, “Cell adhesion_Tight junctions”, and “Development_Positive regulation of WNT/Beta-catenin signaling in the cytoplasm” (Figure 8 and Supplementary Table 4). Genes coexpressed with PER3 were correlated with “Development_Gastrin in cell growth and proliferation”, “NF-AT signaling in cardiac hypertrophy”, and “Immune response_Gastrin in inflammatory response” (Figure 9 and Supplementary Table 5).
Figure 7.
Expression of the PER1 signaling pathway in lung cancer (using MetaCore). The functional analysis of “Signal transduction_Beta-adrenergic receptors signaling via cyclic AMP” was correlated with lung cancer development.
Figure 8.
Expression of the PER2 signaling pathway in lung cancer (using MetaCore). The functional analysis of the “G-protein signaling_RhoA regulation pathway” was correlated with lung cancer development.
Figure 9.
Expression of the PER3 signaling pathway in lung cancer (using MetaCore). The functional analysis of “Development_Gastrin in cell growth and proliferation” was correlated with lung cancer development.
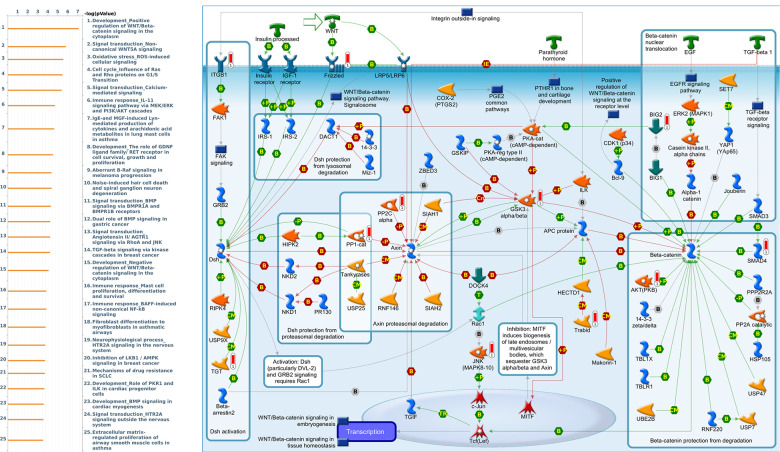
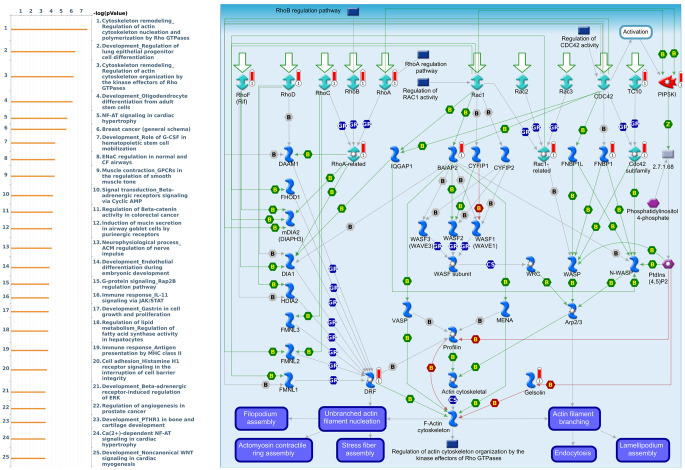
Genes coexpressed with CRY1 were correlated with “Development_Positive regulation of WNT/Beta-catenin signaling in the cytoplasm”, “Signal transduction_Non-canonical WNT5A Signaling”, and “Oxidative stress_ROS-Induced cellular signaling” (Figure 10 and Supplementary Table 6). Genes coexpressed with CRY2 were correlated with “Cytoskeleton remodeling_Regulation of actin cytoskeleton nucleation and polymerization by Rho GTPases”, “Development_Regulation of lung epithelial progenitor cell differentiation”, and “Cytoskeleton remodeling_Regulation of actin cytoskeleton organization by the kinase effectors of Rho GTPases” (Figure 11 and Supplementary Table 7).
Figure 10.
Expression of the CRY1 signaling pathway in lung cancer (using MetaCore). The functional analysis of “Development_Positive regulation of WNT/Beta-catenin signaling in the cytoplasm” was correlated with lung cancer development.
Figure 11.
Expression of the CRY2 signaling pathway in lung cancer (using MetaCore). The functional analysis of “Cytoskeleton remodeling_Regulation of actin cytoskeleton nucleation and polymerization by Rho GTPases” was correlated with lung cancer development.
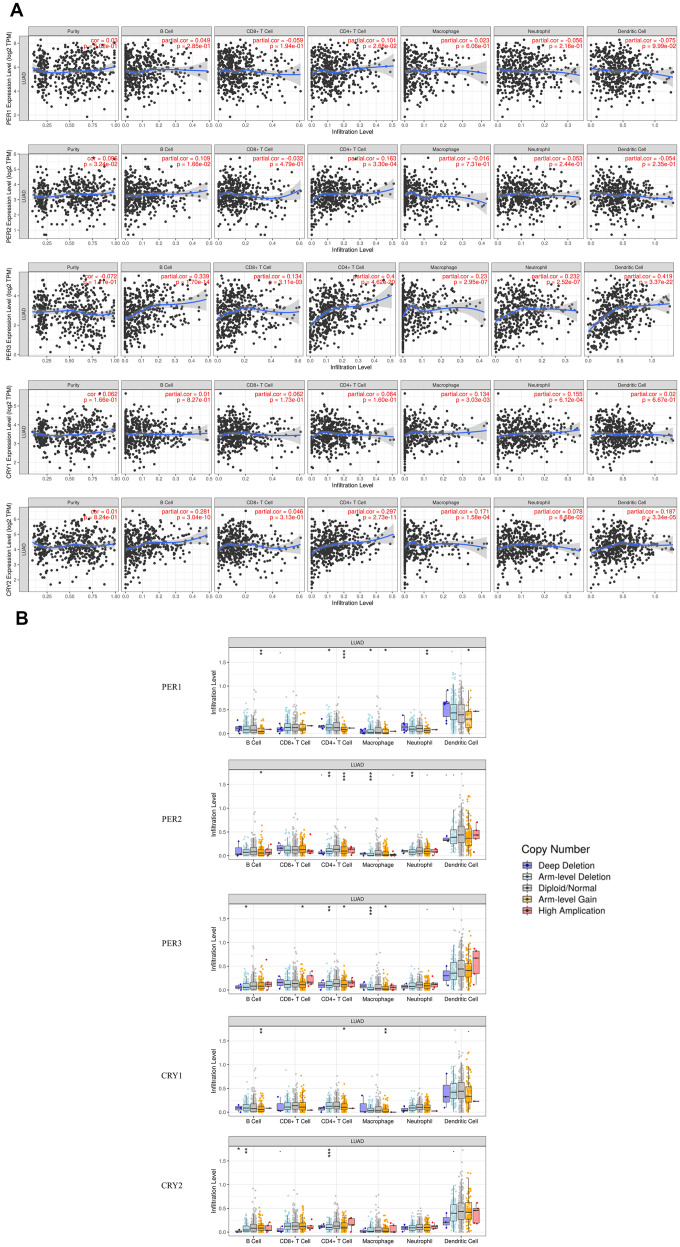
Relationships between expressions of circadian factors in the PER and CRY families with immune infiltration in LUAD
In the current research, some relationships between the immune system and circadian rhythms were discovered. For example, it is believed that PER and CRY are related to the inflammasome [28]. It is, however, unclear whether PER and CRY are related to immune cell infiltration in LUAD patients. TIMER was utilized to understand relationships between immune infiltration and circadian factor expressions in LUAD (Figure 12A). The analysis showed that expressions of PER1 and PER2 were only positively related to infiltration of cluster of differentiation 4-positive (CD4+) T cells. PER3 expression was positively related to the infiltration of B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells (DCs). CRY1 expression was positively related to the infiltration of macrophages and neutrophils. CRY2 expression was positively related to the infiltration of B cells, CD4+ T cells, macrophages, and DCs. These results demonstrated that the circadian-related factors of the PER and CRY families were related to immune infiltration in LUAD patients.
Figure 12.
Relationships of expressions and copy number alteration (CNA) effects of PER (period) and CRY (cryptochrome) family members with immune infiltration-related cells, including B cells, cluster of differentiation 4-positive (CD4+) T cells, CD8+ T cells, macrophages, neutrophils, and dendritic cells (using TIMER). (A) Correlations of immune infiltration-related cells and expressions of PER1, PER2, PER3, CRY1, and CRY2. (B) CNAs of different circadian factors affect infiltration levels in LUAD. * p<0.05, ** p<0.01, and *** p<0.001, compared to arm-level deletions, the diploid/normal group, and arm-level gain.
In addition, somatic copy number alterations (CNAs) of circadian factors were significantly associated with infiltration levels (Figure 12B). Among them, CNAs of PER1 affected the infiltration level of B cells, CD4+ T cells, macrophage, neutrophil, and DCs; CNAs of PER2 affected the infiltration levels of B cells, CD4+ T cells, macrophage, and neutrophils; CNAs of PER3 affected the infiltration levels of B cells, CD8+ T cells, CD4+ T cells, and macrophages; CNAs of CRY1 affected the infiltration levels of B cells, CD4+ T cells, and macrophages; and CNAs of CRY2 affected the infiltration levels of B cells and CD4+ T cells. These results demonstrated that genetic alterations of the PER and CRY families in LUAD led to changes in immune infiltration levels.
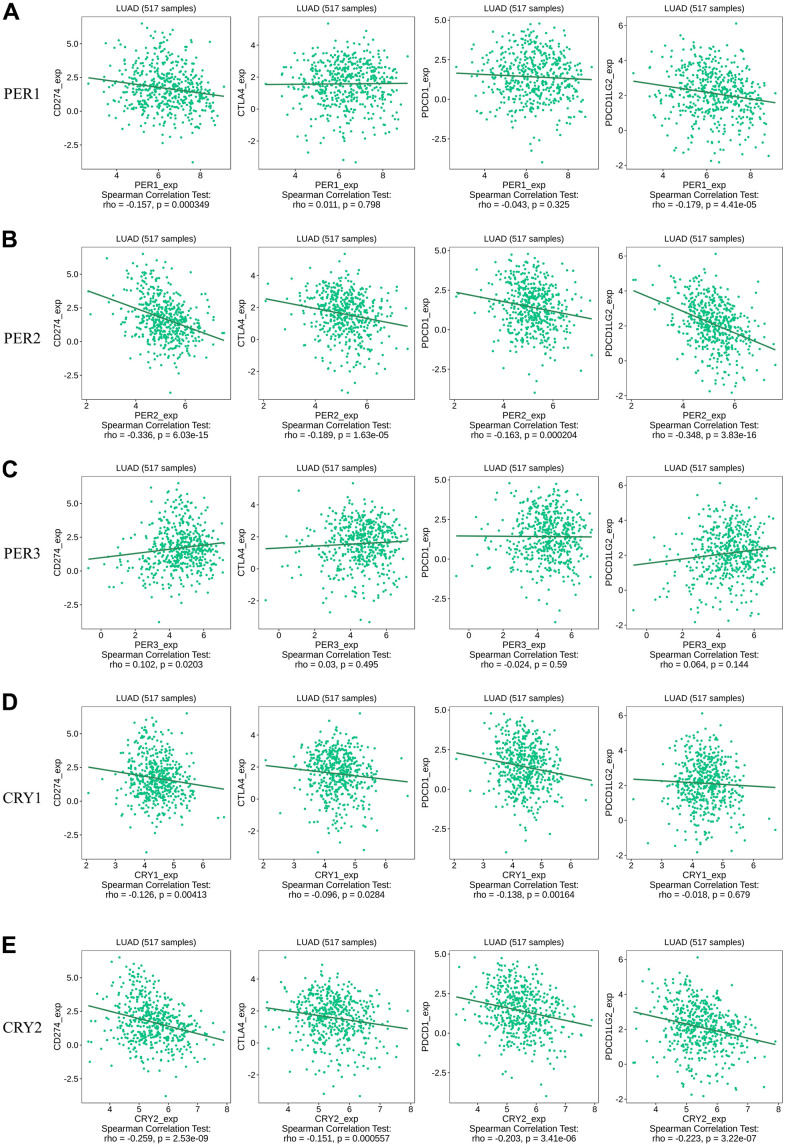
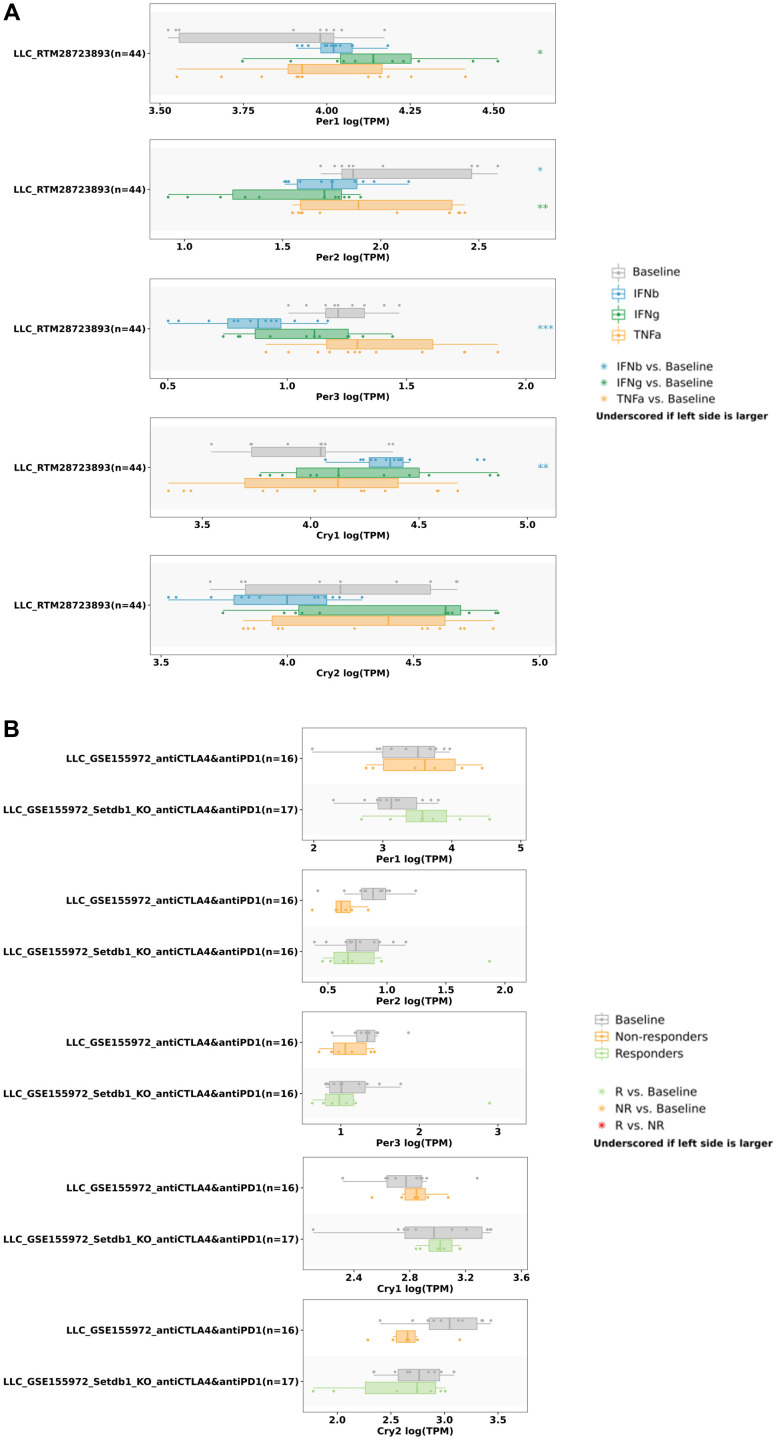
We also utilized the TISIDB database to understand relationships between various immune checkpoints and circadian factors (Figure 13). The results showed that the expression of PER1 was correlated with CD274 (rho = -0.157, p = 0.000349) and PDCD1LG2 (rho = -0.179, p = 4.41E-05) (Figure 13A); the expression of PER2 was correlated with CD274 (rho = -0.336, p = 6.03E-15), CTLA4 (rho = -0.189, p = 1.63e-05), PDCD1 (rho = -0.163, p = 0.000204), and PDCD1LG2 (rho = -0.348, p = 3.83e-16) (Figure 13B); the expression of PER3 was correlated with CD274 (rho = 0.102, p = 0.0203) (Figure 13C); the expression of CRY1 was correlated with CD274 (rho = -0.126, p = 0.00413), CTLA4 (rho = -0.096, p = 0.0284), and PDCD1 (rho = -0.138, p = 0.00164) (Figure 13D); and the expression of CRY2 was correlated with CD274 (rho = -0.259, p = 2.53E-09), CTLA4 (rho = -0.151, p = 0.000557), PDCD1 (rho = -0.203, p = 3.41E-06), and PDCD1LG2 (rho = -0.223, p = 3.22e-07) (Figure 13E). Finally, we used the TISMO database to recognize whether the expressions of circadian factors were affected by different immunotherapies (Figure 14). Expressions of circadian rhythm factors in an LLC (lung carcinoma) cancer model changed under stimulation with different cytokines (Figure 14A). However, the expressions of these circadian factors were not significantly affected under different immune checkpoint blockade treatments (Figure 14B).
Figure 13.
Spearman correlations between expressions of different circadian factors with CD274/CTLA4/PDCD1/PDCD1LG2. (A) Correlations of expressions of immune checkpoint factors CD274/CTLA4/PDCD1/PDCD1LG2 with PER1. (B) Correlations of expressions of immune checkpoint factors CD274/CTLA4/PDCD1/PDCD1LG2 with PER2. (C) Correlations of expressions of immune checkpoint factors CD274/CTLA4/PDCD1/PDCD1LG2 with PER3. (D) Correlations of expressions of immune checkpoint factors CD274/CTLA4/PDCD1/PDCD1LG2 with CRY1. (E) Correlations of expressions of immune checkpoint factors CD274/CTLA4/PDCD1/PDCD1LG2 with CRY2.
Figure 14.
Correlations of PER and CRY family members expressions with different cytokine and immune checkpoint blockade treatments. (A) PER and CRY family member expressions in the LLC lung cancer model stimulated by different cytokines. (B) PER and CRY family member expressions in the LLC lung cancer model were not significantly related to immune checkpoint blockade treatment. * p<0.05, ** p<0.01, *** p<0.001; comparison results are summarized in boxplots.
DISCUSSION
The physiological behavior of animals often exhibits periodic changes to adapt to repeated environmental changes. The most typical one is the sleep-wake cycle, and others include neurological, metabolic, endocrine, cardiovascular, and immune functions [46]. There are very close links between circadian rhythms and sleep. Both circadian rhythms and sleep play important roles in disease and health, which complicates the process of finding links between circadian rhythms and diseases. This study is the first attempt to elucidate possible links between some gene families associated with circadian rhythms and LUAD.
Previous studies showed that an imbalance of circadian rhythm-related factors may lead to the occurrence of cancer. At present, circadian rhythm-related factors have been discovered, and the basic operation mode of circadian rhythms has been established. PER and CRY family members were found to be basic factors in modulating circadian rhythms [47], and their relationships with many types of cancer have been explored. Previous studies showed that circadian rhythm-related factors such as the ARNTL, CLOCK, RORA, RORB, CRY1, CRY2, and PER3 genes were associated with a higher risk of lung cancer [48]. Moreover, some genes of numerous circadian factors can affect the prognosis of NSCLC and changes in immune infiltration and cell functions. Many circadian factors are involved in numerous biological processes, such as inhibiting levels of immune cell infiltration. However, no study has discussed whether the PER and CRY families have the same effect on lung cancer development [49].
Thus, this is the first study to use bioinformatics to analyze and discuss different PER and CRY transcription levels, genetic variations, molecular functions, diseases, and their relationships with prognoses and immune infiltration in LUAD patients. We conducted an Oncomine database analysis and found that compared to normal tissues, expressions of circadian rhythm-related factors of PER1, PER2, PER3, CRY1, and CRY2 in LUAD tissues were relatively low. Consistently, expressions of PER1 and CRY2 were found in a GEPIA2 analysis to be lower than in normal tissues. Moreover, the KM analysis showed that high expression levels of PER2, PER3, CRY1, and CRY2 in LUAD patients were related to a better OS.
In addition, PER and CRY family members were found to have higher mutation rates (29.42%) in LUAD patients. Mutually exclusive coexpressions were found between differentially expressed PER and CRY family members (mainly coexpression), which meant that LUAD might be induced by the co-inhibition of these gene family members. Furthermore, a molecular mechanism pathway analysis demonstrated that the functions of PER- and CRY-related genes were mainly involved protein-binding, cell-cycle, histidine-metabolism, and progesterone-mediated oocyte-maturation pathways. In particular, progesterone-mediated oocyte-maturation pathways and the cell cycle were previously demonstrated to be correlated with LUAD [50]. Histidine metabolism was also related to the oncogenic function of FAM83A in LUAD [51]. Therefore, the development and inhibition of these pathways may respectively be related to the occurrence and development of LUAD. Results indicated that differentially expressed PER and CRY family members in LUAD have the potential to become crucial genes for targeted therapy.
We also analyzed MetaCore and found genes that are coexpressed with PER and CRY family members and the functions associated with those genes. We found that there were many signaling pathways involved in immune evasion, cancer migration and proliferation, and other functions. Among them, PER1 was related to the Immune response_IL-6 signaling pathway via JAK/STAT. Interleukin (IL)-6 can be found in all human inflammatory diseases and cancers due to its dysregulation and overexpression [52]. It is also known that IL-6-activated Janus kinase 1 (JAK1) might lead to the phosphorylation of Y112 of programmed death ligand 1 (PD-L1) and consequently induce cancer immune evasion [53]. The top three pathways related to PER2 included a G-protein signaling RhoA regulatory pathway, cell adhesion tight junctions, and development positive regulation of WNT/Beta-catenin signaling in the cytoplasm, which are all related to the function of cell migration [54–56]. Therefore, overexpression of PER2 may allow cancer cells to easily migrate and spread. Signaling pathways associated with PER3 were more related to gastrin, including the development of gastrin in cell growth and proliferation, and immune response of gastrin in the inflammatory response. Gastrin is currently considered to be related to cancer development, proliferation, and anti-apoptosis in addition to digestion-related functions [57]. The CRY1-related pathway was the WNT signaling pathway. The canonical WNT signaling pathway is associated with cell migration, and the WNT5A non-canonical signaling pathway was also found to be associated with a variety of human cancers [58]. Finally, CRY2 was mainly related to the cytoskeletal remodeling regulation signaling pathway. The abnormality of cytoskeletal remodeling was related to the invasion and metastasis of cancer cells in previous research [59]. These findings revealed possible related cancer pathways of PER and CRY family members and provide insights into why dysregulation of circadian rhythms may contribute to cancer development.
Previous studies demonstrated that deregulation of circadian clock genes was indicated in the development of cancers. Melatonin can resynchronize rhythmic patterns of gene expressions, correcting defects in various circadian rhythm oncogenes. Melatonin also inhibits myeloperoxidase catalytic activity [60], which is crucial for tumorigenesis. The action of melatonin requires two receptors known as MT1 and MT2, and these two receptors are present in high densities in the SCN and other organ parts, which may indicate that melatonin affects other organ systems in addition to the SCN [61]. For instance, the mean nocturnal melatonin level ratio and melatonin nocturnal levels decrease in patients with untreated LUAD [62]. Expression levels of the CRY1 and BMAL1 core-clock genes were correlated with clinical parameters in epithelial ovarian cancer [63, 64]. Melatonin can inhibit the development of breast cancer by interfering with estrogen [65], and has a certain degree of benefit in colorectal cancer in the elderly [66]. In addition, melatonin has functions of stimulating cell apoptosis, regulating cell survival and tumor-related metabolism, and inhibiting angiogenesis [67]. In the cytokine signaling pathway, PER1 expression is suppressed by tumor necrosis factor (TNF)-α, and knockdown of PER1 decreases the proliferation of pancreatic carcinoma cells [68]. These findings correspond to our results that PER and CRY family members and related circadian clock genes interfere with melatonin secretion and circadian rhythms, which have effects on pathogeneses of malignancy.
Circadian rhythms can directly interact with components of the immune system, thereby affecting aspects of the immune system such as inflammation. Recent studies also indicated that phagocytosis, migration of inflammatory or infected tissues, cytolytic activity, and proliferative responses to antigens are closely related to circadian rhythms [69]. Furthermore, our data demonstrated that immune cell infiltration in LUAD patients was related to expressions of PER and CRY family members. In LUAD, expressions of PER1 and PER2 were positively correlated with the immune infiltration of cluster of differentiation 4-positive (CD4+) T and natural killer (NK) cells. CRY1 expression was positively correlated with the infiltration of NK cells, macrophages, and neutrophils, and CRY2 was correlated with the infiltration of NK cells, B cells, CD4+ T cells, macrophages, and DCs. PER3 was connected to the immune infiltration of NK cells, B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and DCs. Additionally, it was found that circadian rhythm factors were negatively correlated with the expressions of immune checkpoint-related genes. Further analysis of TISMO found that circadian rhythm factors had no significant relationships with immune checkpoint blockades, but were more related to stimulation of cytokines. These results suggest that circadian rhythm factors may contribute to increases in levels of partial immune infiltration by downregulating expressions of immune checkpoint genes [70–73].
Our results correspond to other research in immunotherapy [74]. PER1 and CRY2 were correlated with the expression of CD4+ T cells, and the expression of PD-1 exhibited a robust circadian rhythm in normal lung tissues [75], which supported the results that circadian rhythm factors might downregulate expressions of immune checkpoint factors and thus enhance the effects of immunotherapy in LUAD. Although there is a lack of further evident clinical data to prove our hypothesis, we propose a positive association between circadian rhythm factors and immunity.
Taken together, the results indicated that PER1 and CRY2 are significantly downregulated in LUAD. Except for PER1, high expressions of PER2, PER3, CRY1, and CRY2 lead to better OS in LUAD patients. In the functional analysis of these circadian coexpressed genes, many factors related to cancer development were also found. In addition, PER1, PER2, PER3, CRY1, and CRY2 were related to six different immune cells to varying degrees, which may be related to the downregulation of different immune checkpoints. Given the above results, these circadian rhythm factors may be involved in tumor immunity of LUAD. PER and CRY family members could be novel and promising prognostic biomarkers of LUAD.
In summary, we used several high-throughput bioinformatics databases to analyze and investigate gene expressions of PER and CRY family members and their influences. The present study may help us better understand the molecular functions of circadian rhythm-related factors in LUAD and may provide possible molecular targets for LUAD in chemotherapy and immunotherapy.
MATERIALS AND METHODS
UALCAN database analysis
UALCAN (http://ualcan.path.uab.edu/) is an online tool for analyzing cancer OMICS data and was used to analyze the relationship between gene expressions and various cancers. Its functions include (1) analysis of the relative expressions of genes in tumor and normal samples, (2) analysis of the effects of gene expressions on survival rates of different cancer patients, and (3) analysis of high and low expressions of genes in different cancer samples. These helped us understand expression levels of circadian factors in different cancer types [76].
GeneMANIA
GeneMANIA (https://genemania.org/), an online server for prediction, is used to prioritize genes and predict gene function biological networks [77]. This tool was used to understand relationships between circadian rhythm-related genes in the PER and CRY families and other genes, and to establish a network.
Cancer cell line encyclopedia (CCLE) analysis
Cancer cell lines are the most relevant approach in cancer biology research to verify targets and determine drug efficacies (https://portals.broadinstitute.org/ccle). This platform was established with multiple human cancer cell lines (n = 1457) and plenty of unique datasets (n = 136,488) [78]. Of interest, we obtained gene expression levels in 198 lung cancer cell lines and visualized the data with default settings as in our previous studies [79–82].
Gene expression profiling interactive analysis dataset analysis 2 (GEPIA2)
GEPIA2 (http://gepia2.cancer-pku.cn/#index), an upgraded version of GEPIA, is a web-based data platform that can be used to compare tumor tissues and normal tissues, and provides 60,498 genes and 198,619 isoforms for querying. Like the older version of GEPIA, functions include differential expression analyses, spectrogram drawing, correlation analyses, patient survival analyses, similar gene detections, and dimensionality reduction analyses. In addition, some of the original older functions have been upgraded, and there are also new functions such as survival maps, isoform use profiling, uploaded expression data comparisons, and cancer-subtype classifiers [83, 84].
The Kaplan-Meier (KM) plotter analysis
The KM plotter (https://kmplot.com/analysis/), an online database with gene expression and clinical data, can be used to analyze relationships between gene expressions and cancer survival rates. Types of cancer that can be analyzed include lung cancer [85], breast cancer [86], ovarian cancer [87], gastric cancer [88], liver cancer [89] and pan-cancer [90]. We used this tool to understand prognostic values of expression levels of the circadian rhythm-related PER1, PER2, PER3, CRY1, and CRY2 genes in lung cancer patients and analyzed the OS of lung cancer patients under expressions of related genes, as well as the number of patients, median values of messenger (m)RNA expressions, 95% confidence intervals (CIs), hazard ratios (HRs), p values, and other related information.
The cancer genome atlas (TCGA) data and cBioPortal
TCGA is an open database with genome sequencing and related pathological data of more than 30 human tumors [91]. We selected LUAD data (TCGA, PanCancer Atlas) containing 503 pathological reports, and further used the cBioPortal (https://www.cbioportal.org/) to analyze expression levels, coexpressions, and network analyses of circadian rhythm-related genes of PER and CRY family members [92–94].
STRING analysis
STRING (https://string-db.org/) is a biological database and web resource for searching and predicting protein-protein interactions (PPIs) [95]. In this study, we used this tool to understand proteins related to the PER and CRY families, and establish a relationship network.
DAVID analysis
DAVID (https://david.ncifcrf.gov/) is a database that aims to provide functional explanations for a large number of genes from genome research. DAVID has four analytical modules, namely Annotation Tool, GoCharts, KeggCharts, and DomainCharts [96, 97]. In the study, we used this tool to understand gene functions of the PER1, PER2, PER3, CRY1, and CRY2 gene lists after cross-comparisons of different databases to evaluate how PER1, PER2, PER3, CRY1, and CRY2 affect molecular functions and may be related to various diseases.
Tumor immune estimation resource (TIMER) analysis
TIMER (cistrome.shinyapps.io/timer) and its upgraded version TIMER2.0 (http://timer.cistrome.org/) were established to study interactions between malignant cells and host immune systems. It can be used to understand relationships between genes and tumor-infiltrating immune cells and evaluate their clinical impacts [98–101]. This analytical website was used to evaluate the impacts of PER and CRY family gene expressions on tumor-infiltrating immune cells.
TISIDB
TISIDB (http://cis.hku.hk/TISIDB/) is an integrated repository portal for tumor-immune system interactions, which integrates multiple heterogeneous data types including the PubMed database, genomics, transcriptomics, and clinical data of 30 cancer types from TCGA, high-throughput screening data, exome and RNA sequencing datasets of patients, and other public databases including UniProt, GO, DrugBank, etc. It can be used to analyze correlations between immune checkpoint factors and circadian factors [102].
TISMO database analysis
TISMO (http://tismo.cistrome.org/) is a database for hosting and analyzing an extensive collection of syngeneic mouse model data. The entire repository contains raw sequencing data from 1518 mouse samples, including 68 cell lines and 19 cancer types, which can be used to analyze relationships between cancers receiving different treatments (e.g., cytokines and immune checkpoint blockade) and gene expressions of circadian factors [103].
Functional enrichment analysis
The MetaCore platform was used to identify cancer risk pathways and tumorigenesis in enrichment pathways as we previously described. Expression profiles of TCGA dataset on PER1, PER2, PER3, CRY1, and CRY2 gene expressions were pooled and in-depth integrated to describe potential key candidate genes and pathways in lung cancer [104–107].
Statistical analysis
We utilized TCGA Pan-Cancer Atlas, a dataset from cBioPortal, to obtain patient data and query the effects of the expressions of different PER and CRY family members on overall survival (OS). For the survival analysis, a KM plotter was applied, with all default settings, and recurrence-free survival (RFS) was preferred, with the auto-best cutoff values and J best probe set. All possible cutoff values between the lower and upper quartiles were determined, and the best presenting threshold was subsequently used as the cutoff. A log-rank p value of <0.05 was considered statistically significant [84, 108, 109].
Data availability statement
CBioPortal: https://cbioportal.org; The Human Protein Atlas: https://www.proteinatlas.org; Kaplan-Meier plot database https://kmplot.com; MetaCore analysis https://portal.genego.com. The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Supplementary Material
ACKNOWLEDGMENTS
The authors acknowledge the (statistical/computational/technical) support of the Clinical Data Center, Office of Data Science, Taipei Medical University, Taiwan. The authors also give special thanks to Daniel P. Chamberlin for his professional English editing from the Office of Research and Development at Taipei Medical University.
Footnotes
AUTHOR CONTRIBUTIONS: Study concept and design: C.-C.W., W.-H.L., S.-C.K., C.-Y.W., and W.-J.W.; Literature search and data extraction: W.-J.S., F.-W.L, G.A., and H.D.K.T.; Data analysis: C.-F.S., S.-H.W., and C.-C.Y.; Manuscript drafting: C.-Y.W. and W.-J.W. All authors approved the final version for publication.
CONFLICTS OF INTEREST: The authors declare that no conflicts of interest exists.
FUNDING: This research was supported by grants from the Ministry of Science and Technology (MOST) of Taiwan (MOST-110-2320-B-039-068 to W-J.W. and 109-2320-B-038-009-MY2 to C-Y.W.), China Medical University (CMU110-MF-47 to W-J.W.), Taipei Medical University (TMU-108-AE1-B16 to C-Y.W.), Kaohsiung Chang Gung Memorial Hospital (CMRPG8E1661-3, CMRPG8F1441, CMRPG8F1351, CMRPG8H1201, CORPG8F1491-3, CMRPG8K0481-2, and CMRPG8L0521 to C-C.W.), and the TMU Research Center of Cancer Translational Medicine from The Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan (DP2-111-21121-01-N-10).
REFERENCES
- 1.Nasim F, Sabath BF, Eapen GA. Lung Cancer. Med Clin North Am. 2019; 103:463–73. 10.1016/j.mcna.2018.12.006 [DOI] [PubMed] [Google Scholar]
- 2.Chung CC, Huang TY, Chu HR, De Luca R, Candelotti E, Huang CH, Yang YSH, Incerpi S, Pedersen JZ, Lin CY, Huang HM, Lee SY, Li ZL, et al. Heteronemin and tetrac derivatives suppress non-small cell lung cancer growth via ERK1/2 inhibition. Food Chem Toxicol. 2022; 161:112850. 10.1016/j.fct.2022.112850 [DOI] [PubMed] [Google Scholar]
- 3.Hsiao SH, Chen WT, Chung CL, Chou YT, Lin SE, Hong SY, Chang JH, Chang TH, Chien LN. Comparative survival analysis of platinum-based adjuvant chemotherapy for early-stage squamous cell carcinoma and adenocarcinoma of the lung. Cancer Med. 2022; 11:2067–78. 10.1002/cam4.4570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuo KT, Lin CH, Wang CH, Pikatan NW, Yadav VK, Fong IH, Yeh CT, Lee WH, Huang WC. HNMT Upregulation Induces Cancer Stem Cell Formation and Confers Protection against Oxidative Stress through Interaction with HER2 in Non-Small-Cell Lung Cancer. Int J Mol Sci. 2022; 23:1663. 10.3390/ijms23031663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tseng PC, Chen CL, Lee KY, Feng PH, Wang YC, Satria RD, Lin CF. Epithelial-to-mesenchymal transition hinders interferon-γ-dependent immunosurveillance in lung cancer cells. Cancer Lett. 2022; 539:215712. 10.1016/j.canlet.2022.215712 [DOI] [PubMed] [Google Scholar]
- 6.Lee HC, Lu YH, Huang YL, Huang SL, Chuang HC. Air Pollution Effects to the Subtype and Severity of Lung Cancers. Front Med (Lausanne). 2022; 9:835026. 10.3389/fmed.2022.835026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farhud D, Aryan Z. Circadian Rhythm, Lifestyle and Health: A Narrative Review. Iran J Public Health. 2018; 47:1068–76. [PMC free article] [PubMed] [Google Scholar]
- 8.Fu L, Kettner NM. The circadian clock in cancer development and therapy. Prog Mol Biol Transl Sci. 2013; 119:221–82. 10.1016/B978-0-12-396971-2.00009-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Papagiannakopoulos T, Bauer MR, Davidson SM, Heimann M, Subbaraj L, Bhutkar A, Bartlebaugh J, Vander Heiden MG, Jacks T. Circadian Rhythm Disruption Promotes Lung Tumorigenesis. Cell Metab. 2016; 24:324–31. 10.1016/j.cmet.2016.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shafi AA, Knudsen KE. Cancer and the Circadian Clock. Cancer Res. 2019; 79:3806–14. 10.1158/0008-5472.CAN-19-0566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang WJ, Lai HY, Zhang F, Shen WJ, Chu PY, Liang HY, Liu YB, Wang JM. MCL1 participates in leptin-promoted mitochondrial fusion and contributes to drug resistance in gallbladder cancer. JCI Insight. 2021; 6:e135438. 10.1172/jci.insight.135438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morse D, Sassone-Corsi P. Time after time: inputs to and outputs from the mammalian circadian oscillators. Trends Neurosci. 2002; 25:632–7. 10.1016/s0166-2236(02)02274-9 [DOI] [PubMed] [Google Scholar]
- 13.Bollinger T, Schibler U. Circadian rhythms - from genes to physiology and disease. Swiss Med Wkly. 2014; 144:w13984. 10.4414/smw.2014.13984 [DOI] [PubMed] [Google Scholar]
- 14.Shearman LP, Sriram S, Weaver DR, Maywood ES, Chaves I, Zheng B, Kume K, Lee CC, van der Horst GT, Hastings MH, Reppert SM. Interacting molecular loops in the mammalian circadian clock. Science. 2000; 288:1013–9. 10.1126/science.288.5468.1013 [DOI] [PubMed] [Google Scholar]
- 15.Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet. 2006; 15 Spec No 2:R271–7. 10.1093/hmg/ddl207 [DOI] [PubMed] [Google Scholar]
- 16.Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, Takahashi JS. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012; 338:349–54. 10.1126/science.1226339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shirogane T, Jin J, Ang XL, Harper JW. SCFbeta-TRCP controls clock-dependent transcription via casein kinase 1-dependent degradation of the mammalian period-1 (Per1) protein. J Biol Chem. 2005; 280:26863–72. 10.1074/jbc.M502862200 [DOI] [PubMed] [Google Scholar]
- 18.Blau J. PERspective on PER phosphorylation. Genes Dev. 2008; 22:1737–40. 10.1101/gad.1696408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gossan N, Boot-Handford R, Meng QJ. Ageing and osteoarthritis: a circadian rhythm connection. Biogerontology. 2015; 16:209–19. 10.1007/s10522-014-9522-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reghunandanan V, Reghunandanan R. Neurotransmitters of the suprachiasmatic nuclei. J Circadian Rhythms. 2006; 4:2. 10.1186/1740-3391-4-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Langmesser S, Tallone T, Bordon A, Rusconi S, Albrecht U. Interaction of circadian clock proteins PER2 and CRY with BMAL1 and CLOCK. BMC Mol Biol. 2008; 9:41. 10.1186/1471-2199-9-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kadomatsu T, Uragami S, Akashi M, Tsuchiya Y, Nakajima H, Nakashima Y, Endo M, Miyata K, Terada K, Todo T, Node K, Oike Y. A molecular clock regulates angiopoietin-like protein 2 expression. PLoS One. 2013; 8:e57921. 10.1371/journal.pone.0057921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chaudhari A, Gupta R, Patel S, Velingkaar N, Kondratov R. Cryptochromes regulate IGF-1 production and signaling through control of JAK2-dependent STAT5B phosphorylation. Mol Biol Cell. 2017; 28:834–42. 10.1091/mbc.E16-08-0624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lesicka M, Jabłońska E, Wieczorek E, Seroczyńska B, Kalinowski L, Skokowski J, Reszka E. A different methylation profile of circadian genes promoter in breast cancer patients according to clinicopathological features. Chronobiol Int. 2019; 36:1103–14. 10.1080/07420528.2019.1617732 [DOI] [PubMed] [Google Scholar]
- 25.Fonnes S, Donatsky AM, Gögenur I. Expression of core clock genes in colorectal tumour cells compared with normal mucosa: a systematic review of clinical trials. Colorectal Dis. 2015; 17:290–7. 10.1111/codi.12847 [DOI] [PubMed] [Google Scholar]
- 26.Jung-Hynes B, Huang W, Reiter RJ, Ahmad N. Melatonin resynchronizes dysregulated circadian rhythm circuitry in human prostate cancer cells. J Pineal Res. 2010; 49:60–8. 10.1111/j.1600-079X.2010.00767.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karantanos T, Theodoropoulos G, Pektasides D, Gazouli M. Clock genes: their role in colorectal cancer. World J Gastroenterol. 2014; 20:1986–92. 10.3748/wjg.v20.i8.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hergenhan S, Holtkamp S, Scheiermann C. Molecular Interactions Between Components of the Circadian Clock and the Immune System. J Mol Biol. 2020; 432:3700–13. 10.1016/j.jmb.2019.12.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, Yefanov A, Lee H, Zhang N, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013; 41:D991–5. 10.1093/nar/gks1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin JC, Liu TP, Yang PM. CDKN2A-Inactivated Pancreatic Ductal Adenocarcinoma Exhibits Therapeutic Sensitivity to Paclitaxel: A Bioinformatics Study. J Clin Med. 2020; 9:4019. 10.3390/jcm9124019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin TY, Wang PW, Huang CH, Yang PM, Pan TL. Characterizing the Relapse Potential in Different Luminal Subtypes of Breast Cancers with Functional Proteomics. Int J Mol Sci. 2020; 21:6077. 10.3390/ijms21176077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu LW, Hsieh YY, Yang PM. Bioinformatics Data Mining Repurposes the JAK2 (Janus Kinase 2) Inhibitor Fedratinib for Treating Pancreatic Ductal Adenocarcinoma by Reversing the KRAS (Kirsten Rat Sarcoma 2 Viral Oncogene Homolog)-Driven Gene Signature. J Pers Med. 2020; 10:130. 10.3390/jpm10030130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang PM, Hsieh YY, Du JL, Yen SC, Hung CF. Sequential Interferon β-Cisplatin Treatment Enhances the Surface Exposure of Calreticulin in Cancer Cells via an Interferon Regulatory Factor 1-Dependent Manner. Biomolecules. 2020; 10:643. 10.3390/biom10040643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang PM, Lin LS, Liu TP. Sorafenib Inhibits Ribonucleotide Reductase Regulatory Subunit M2 (RRM2) in Hepatocellular Carcinoma Cells. Biomolecules. 2020; 10:117. 10.3390/biom10010117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M, Loda M, Weber G, Mark EJ, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci USA. 2001; 98:13790–5. 10.1073/pnas.191502998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stearman RS, Dwyer-Nield L, Zerbe L, Blaine SA, Chan Z, Bunn PA Jr, Johnson GL, Hirsch FR, Merrick DT, Franklin WA, Baron AE, Keith RL, Nemenoff RA, et al. Analysis of orthologous gene expression between human pulmonary adenocarcinoma and a carcinogen-induced murine model. Am J Pathol. 2005; 167:1763–75. 10.1016/S0002-9440(10)61257-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Landi MT, Dracheva T, Rotunno M, Figueroa JD, Liu H, Dasgupta A, Mann FE, Fukuoka J, Hames M, Bergen AW, Murphy SE, Yang P, Pesatori AC, et al. Gene expression signature of cigarette smoking and its role in lung adenocarcinoma development and survival. PLoS One. 2008; 3:e1651. 10.1371/journal.pone.0001651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okayama H, Kohno T, Ishii Y, Shimada Y, Shiraishi K, Iwakawa R, Furuta K, Tsuta K, Shibata T, Yamamoto S, Watanabe S, Sakamoto H, Kumamoto K, et al. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res. 2012; 72:100–11. 10.1158/0008-5472.CAN-11-1403 [DOI] [PubMed] [Google Scholar]
- 39.Su LJ, Chang CW, Wu YC, Chen KC, Lin CJ, Liang SC, Lin CH, Whang-Peng J, Hsu SL, Chen CH, Huang CY. Selection of DDX5 as a novel internal control for Q-RT-PCR from microarray data using a block bootstrap re-sampling scheme. BMC Genomics. 2007; 8:140. 10.1186/1471-2164-8-140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hou J, Aerts J, den Hamer B, van Ijcken W, den Bakker M, Riegman P, van der Leest C, van der Spek P, Foekens JA, Hoogsteden HC, Grosveld F, Philipsen S. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS One. 2010; 5:e10312. 10.1371/journal.pone.0010312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thorat MA, Balasubramanian R. Breast cancer prevention in high-risk women. Best Pract Res Clin Obstet Gynaecol. 2020; 65:18–31. 10.1016/j.bpobgyn.2019.11.006 [DOI] [PubMed] [Google Scholar]
- 42.Lin CY, Lee CH, Chuang YH, Lee JY, Chiu YY, Wu Lee YH, Jong YJ, Hwang JK, Huang SH, Chen LC, Wu CH, Tu SH, Ho YS, Yang JM. Membrane protein-regulated networks across human cancers. Nat Commun. 2019; 10:3131. 10.1038/s41467-019-10920-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsai HT, Huang CS, Tu CC, Liu CY, Huang CJ, Ho YS, Tu SH, Tseng LM, Huang CC. Multi-gene signature of microcalcification and risk prediction among Taiwanese breast cancer. Sci Rep. 2020; 10:18276. 10.1038/s41598-020-74982-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nguyen HD, Liao YC, Ho YS, Chen LC, Chang HW, Cheng TC, Liu D, Lee WR, Shen SC, Wu CH, Tu SH. The α9 Nicotinic Acetylcholine Receptor Mediates Nicotine-Induced PD-L1 Expression and Regulates Melanoma Cell Proliferation and Migration. Cancers (Basel). 2019; 11:1991. 10.3390/cancers11121991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee KL, Kuo YC, Ho YS, Huang YH. Triple-Negative Breast Cancer: Current Understanding and Future Therapeutic Breakthrough Targeting Cancer Stemness. Cancers (Basel). 2019; 11:1334. 10.3390/cancers11091334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patke A, Young MW, Axelrod S. Molecular mechanisms and physiological importance of circadian rhythms. Nat Rev Mol Cell Biol. 2020; 21:67–84. 10.1038/s41580-019-0179-2 [DOI] [PubMed] [Google Scholar]
- 47.Takahashi JS. Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet. 2017; 18:164–79. 10.1038/nrg.2016.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mocellin S, Tropea S, Benna C, Rossi CR. Circadian pathway genetic variation and cancer risk: evidence from genome-wide association studies. BMC Med. 2018; 16:20. 10.1186/s12916-018-1010-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li M, Chen Z, Jiang T, Yang X, Du Y, Liang J, Wang L, Xi J, Lin M, Feng M. Circadian rhythm-associated clinical relevance and Tumor Microenvironment of Non-small Cell Lung Cancer. J Cancer. 2021; 12:2582–97. 10.7150/jca.52454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu B, Li T, Chen J, Wang FQ, Fu JH, Liu SM, Wang Y, Zhang X, Yang HT. Identification of activated pathways in lung adenocarcinoma based on network strategy. J Cancer Res Ther. 2020; 16:793–9. 10.4103/0973-1482.199458 [DOI] [PubMed] [Google Scholar]
- 51.Yu J, Hou M, Pei T. FAM83A Is a Prognosis Signature and Potential Oncogene of Lung Adenocarcinoma. DNA Cell Biol. 2020; 39:890–9. 10.1089/dna.2019.4970 [DOI] [PubMed] [Google Scholar]
- 52.Lokau J, Garbers C. Activating mutations of the gp130/JAK/STAT pathway in human diseases. Adv Protein Chem Struct Biol. 2019; 116:283–309. 10.1016/bs.apcsb.2018.11.007 [DOI] [PubMed] [Google Scholar]
- 53.Chan LC, Li CW, Xia W, Hsu JM, Lee HH, Cha JH, Wang HL, Yang WH, Yen EY, Chang WC, Zha Z, Lim SO, Lai YJ, et al. IL-6/JAK1 pathway drives PD-L1 Y112 phosphorylation to promote cancer immune evasion. J Clin Invest. 2019; 129:3324–38. 10.1172/JCI126022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haga RB, Ridley AJ. Rho GTPases: Regulation and roles in cancer cell biology. Small GTPases. 2016; 7:207–21. 10.1080/21541248.2016.1232583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trepat X, Chen Z, Jacobson K. Cell migration. Compr Physiol. 2012; 2:2369–92. 10.1002/cphy.c110012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Katoh M. Multi-layered prevention and treatment of chronic inflammation, organ fibrosis and cancer associated with canonical WNT/β-catenin signaling activation (Review). Int J Mol Med. 2018; 42:713–25. 10.3892/ijmm.2018.3689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zeng Q, Ou L, Wang W, Guo DY. Gastrin, Cholecystokinin, Signaling, and Biological Activities in Cellular Processes. Front Endocrinol (Lausanne). 2020; 11:112. 10.3389/fendo.2020.00112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Remtisch L, Wiltberger G, Schierle K, Yousef M, Thieme R, Jansen Winkeln B, Wittekind C, Gockel I, Lyros O. The WNT5A/ROR2 signaling pathway in pancreatic ductal adenocarcinoma (PDAC). J BUON. 2021; 26:1595–606. [PubMed] [Google Scholar]
- 59.Li X, Wang J. Mechanical tumor microenvironment and transduction: cytoskeleton mediates cancer cell invasion and metastasis. Int J Biol Sci. 2020; 16:2014–28. 10.7150/ijbs.44943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Galijasevic S, Abdulhamid I, Abu-Soud HM. Melatonin is a potent inhibitor for myeloperoxidase. Biochemistry. 2008; 47:2668–77. 10.1021/bi702016q [DOI] [PubMed] [Google Scholar]
- 61.Khullar A. The role of melatonin in the circadian rhythm sleep-wake cycle: a review of endogenous and exogenous melatonin. Psychiatric Times. 2012; 26. [Google Scholar]
- 62.Hu S, Shen G, Yin S, Xu W, Hu B. Melatonin and tryptophan circadian profiles in patients with advanced non-small cell lung cancer. Adv Ther. 2009; 26:886–92. 10.1007/s12325-009-0068-8 [DOI] [PubMed] [Google Scholar]
- 63.Savvidis C, Koutsilieris M. Circadian rhythm disruption in cancer biology. Mol Med. 2012; 18:1249–60. 10.2119/molmed.2012.00077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tokunaga H, Takebayashi Y, Utsunomiya H, Akahira J, Higashimoto M, Mashiko M, Ito K, Niikura H, Takenoshita S, Yaegashi N. Clinicopathological significance of circadian rhythm-related gene expression levels in patients with epithelial ovarian cancer. Acta Obstet Gynecol Scand. 2008; 87:1060–70. 10.1080/00016340802348286 [DOI] [PubMed] [Google Scholar]
- 65.González-González A, Mediavilla MD, Sánchez-Barceló EJ. Melatonin: A Molecule for Reducing Breast Cancer Risk. Molecules. 2018; 23:336. 10.3390/molecules23020336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mirza-Aghazadeh-Attari M, Mohammadzadeh A, Mostavafi S, Mihanfar A, Ghazizadeh S, Sadighparvar S, Gholamzadeh S, Majidinia M, Yousefi B. Melatonin: An important anticancer agent in colorectal cancer. J Cell Physiol. 2020; 235:804–17. 10.1002/jcp.29049 [DOI] [PubMed] [Google Scholar]
- 67.Talib WH. Melatonin and Cancer Hallmarks. Molecules. 2018; 23:518. 10.3390/molecules23030518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Suzuki T, Sato F, Kondo J, Liu Y, Kusumi T, Fujimoto K, Kato Y, Sato T, Kijima H. Period is involved in the proliferation of human pancreatic MIA-PaCa2 cancer cells by TNF-alpha. Biomed Res. 2008; 29:99–103. 10.2220/biomedres.29.99 [DOI] [PubMed] [Google Scholar]
- 69.Labrecque N, Cermakian N. Circadian Clocks in the Immune System. J Biol Rhythms. 2015; 30:277–90. 10.1177/0748730415577723 [DOI] [PubMed] [Google Scholar]
- 70.Chan DV, Gibson HM, Aufiero BM, Wilson AJ, Hafner MS, Mi QS, Wong HK. Differential CTLA-4 expression in human CD4+ versus CD8+ T cells is associated with increased NFAT1 and inhibition of CD4+ proliferation. Genes Immun. 2014; 15:25–32. 10.1038/gene.2013.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Konkel JE, Frommer F, Leech MD, Yagita H, Waisman A, Anderton SM. PD-1 signalling in CD4(+) T cells restrains their clonal expansion to an immunogenic stimulus, but is not critically required for peptide-induced tolerance. Immunology. 2010; 130:92–102. 10.1111/j.1365-2567.2009.03216.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Linsley PS, Brady W, Urnes M, Grosmaire LS, Damle NK, Ledbetter JA. CTLA-4 is a second receptor for the B cell activation antigen B7. J Exp Med. 1991; 174:561–9. 10.1084/jem.174.3.561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang XB, Fan ZZ, Anton D, Vollenhoven AV, Ni ZH, Chen XF, Lefvert AK. CTLA4 is expressed on mature dendritic cells derived from human monocytes and influences their maturation and antigen presentation. BMC Immunol. 2011; 12:21. 10.1186/1471-2172-12-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang Z, Zeng P, Gao W, Zhou Q, Feng T, Tian X. Circadian clock: a regulator of the immunity in cancer. Cell Commun Signal. 2021; 19:37. 10.1186/s12964-021-00721-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang Y, Yuan G, Xie H, Wei T, Zhu D, Cui J, Liu X, Shen R, Zhu Y, Yang X. Circadian clock associates with tumor microenvironment in thoracic cancers. Aging (Albany NY). 2019; 11:11814–28. 10.18632/aging.102450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVS, Varambally S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia. 2017; 19:649–58. 10.1016/j.neo.2017.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, Maitland A, Mostafavi S, Montojo J, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010; 38:W214–20. 10.1093/nar/gkq537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nusinow DP, Szpyt J, Ghandi M, Rose CM, McDonald ER 3rd, Kalocsay M, Jané-Valbuena J, Gelfand E, Schweppe DK, Jedrychowski M, Golji J, Porter DA, Rejtar T, et al. Quantitative Proteomics of the Cancer Cell Line Encyclopedia. Cell. 2020; 180:387–402.e16. 10.1016/j.cell.2019.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Choy TK, Wang CY, Phan NN, Khoa Ta HD, Anuraga G, Liu YH, Wu YF, Lee KH, Chuang JY, Kao TJ. Identification of Dipeptidyl Peptidase (DPP) Family Genes in Clinical Breast Cancer Patients via an Integrated Bioinformatics Approach. Diagnostics (Basel). 2021; 11:1204. 10.3390/diagnostics11071204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Phan NN, Liu S, Wang CY, Hsu HP, Lai MD, Li CY, Chen CF, Chiao CC, Yen MC, Sun Z, Jiang JZ. Overexpressed gene signature of EPH receptor A/B family in cancer patients-comprehensive analyses from the public high-throughput database. Int J Clin Exp Pathol. 2020; 13:1220–42. [PMC free article] [PubMed] [Google Scholar]
- 81.Kao TJ, Wu CC, Phan NN, Liu YH, Ta HDK, Anuraga G, Wu YF, Lee KH, Chuang JY, Wang CY. Prognoses and genomic analyses of proteasome 26S subunit, ATPase (PSMC) family genes in clinical breast cancer. Aging (Albany NY). 2021; 13:17970. 10.18632/aging.203345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang CY, Chiao CC, Phan NN, Li CY, Sun ZD, Jiang JZ, Hung JH, Chen YL, Yen MC, Weng TY, Chen WC, Hsu HP, Lai MD. Gene signatures and potential therapeutic targets of amino acid metabolism in estrogen receptor-positive breast cancer. Am J Cancer Res. 2020; 10:95–113. [PMC free article] [PubMed] [Google Scholar]
- 83.Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019; 47:W556–60. 10.1093/nar/gkz430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chou CW, Hsieh YH, Ku SC, Shen WJ, Anuraga G, Khoa Ta HD, Lee KH, Lee YC, Lin CH, Wang CY, Wang WJ. Potential Prognostic Biomarkers of OSBPL Family Genes in Patients with Pancreatic Ductal Adenocarcinoma. Biomedicines. 2021; 9:1601. 10.3390/biomedicines9111601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Győrffy B, Surowiak P, Budczies J, Lánczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013; 8:e82241. 10.1371/journal.pone.0082241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Györffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010; 123:725–31. 10.1007/s10549-009-0674-9 [DOI] [PubMed] [Google Scholar]
- 87.Gyorffy B, Lánczky A, Szállási Z. Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr Relat Cancer. 2012; 19:197–208. 10.1530/ERC-11-0329 [DOI] [PubMed] [Google Scholar]
- 88.Szász AM, Lánczky A, Nagy Á, Förster S, Hark K, Green JE, Boussioutas A, Busuttil R, Szabó A, Győrffy B. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016; 7:49322–33. 10.18632/oncotarget.10337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Menyhárt O, Nagy Á, Győrffy B. Determining consistent prognostic biomarkers of overall survival and vascular invasion in hepatocellular carcinoma. R Soc Open Sci. 2018; 5:181006. 10.1098/rsos.181006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nagy Á, Munkácsy G, Győrffy B. Pancancer survival analysis of cancer hallmark genes. Sci Rep. 2021; 11:6047. 10.1038/s41598-021-84787-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tomczak K, Czerwińska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn). 2015; 19:A68–77. 10.5114/wo.2014.47136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012; 2:401–4. 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013; 6:pl1. 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ramezani M, Baharzadeh F, Almasi A, Sadeghi M. A Systematic Review and Meta-Analysis: Evaluation of the β-Human Papillomavirus in Immunosuppressed Individuals with Cutaneous Squamous Cell Carcinoma. Biomedicine (Taipei). 2020; 10:1–10. 10.37796/2211-8039.1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, Mering CV. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019; 47:D607–13. 10.1093/nar/gky1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003; 4:P3. [PubMed] [Google Scholar]
- 97.Huang DW, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC, Lempicki RA. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007; 35:W169–75. 10.1093/nar/gkm415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li B, Severson E, Pignon JC, Zhao H, Li T, Novak J, Jiang P, Shen H, Aster JC, Rodig S, Signoretti S, Liu JS, Liu XS. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 2016; 17:174. 10.1186/s13059-016-1028-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, Li B, Liu XS. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017; 77:e108–10. 10.1158/0008-5472.CAN-17-0307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, Li B, Liu XS. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020; 48:W509–14. 10.1093/nar/gkaa407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Xuan DTM, Wu CC, Kao TJ, Ta HDK, Anuraga G, Andriani V, Athoillah M, Chiao CC, Wu YF, Lee KH, Wang CY, Chuang JY. Prognostic and immune infiltration signatures of proteasome 26S subunit, non-ATPase (PSMD) family genes in breast cancer patients. Aging (Albany NY). 2021; 13:24882–913. 10.18632/aging.203722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ru B, Wong CN, Tong Y, Zhong JY, Zhong SSW, Wu WC, Chu KC, Wong CY, Lau CY, Chen I, Chan NW, Zhang J. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics. 2019; 35:4200–2. 10.1093/bioinformatics/btz210 [DOI] [PubMed] [Google Scholar]
- 103.Zeng Z, Wong CJ, Yang L, Ouardaoui N, Li D, Zhang W, Gu S, Zhang Y, Liu Y, Wang X, Fu J, Zhou L, Zhang B, et al. TISMO: syngeneic mouse tumor database to model tumor immunity and immunotherapy response. Nucleic Acids Res. 2022; 50:D1391–7. 10.1093/nar/gkab804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Anuraga G, Wang WJ, Phan NN, An Ton NT, Ta HDK, Berenice Prayugo F, Minh Xuan DT, Ku SC, Wu YF, Andriani V, Athoillah M, Lee KH, Wang CY. Potential Prognostic Biomarkers of NIMA (Never in Mitosis, Gene A)-Related Kinase (NEK) Family Members in Breast Cancer. J Pers Med. 2021; 11:1089. 10.3390/jpm11111089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Liu HL, Yeh IJ, Phan NN, Wu YH, Yen MC, Hung JH, Chiao CC, Chen CF, Sun Z, Jiang JZ, Hsu HP, Wang CY, Lai MD. Gene signatures of SARS-CoV/SARS-CoV-2-infected ferret lungs in short- and long-term models. Infect Genet Evol. 2020; 85:104438. 10.1016/j.meegid.2020.104438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang CY, Chao YJ, Chen YL, Wang TW, Phan NN, Hsu HP, Shan YS, Lai MD. Upregulation of peroxisome proliferator-activated receptor-α and the lipid metabolism pathway promotes carcinogenesis of ampullary cancer. Int J Med Sci. 2021; 18:256–69. 10.7150/ijms.48123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang CY, Chang YC, Kuo YL, Lee KT, Chen PS, Cheung CHA, Chang CP, Phan NN, Shen MR, Hsu HP. Mutation of the PTCH1 gene predicts recurrence of breast cancer. Sci Rep. 2019; 9:16359. 10.1038/s41598-019-52617-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Weng TY, Huang SS, Yen MC, Lin CC, Chen YL, Lin CM, Chen WC, Wang CY, Chang JY, Lai MD. A novel cancer therapeutic using thrombospondin 1 in dendritic cells. Mol Ther. 2014; 22:292–302. 10.1038/mt.2013.236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chen PS, Hsu HP, Phan NN, Yen MC, Chen FW, Liu YW, Lin FP, Feng SY, Cheng TL, Yeh PH, Omar HA, Sun Z, Jiang JZ, et al. CCDC167 as a potential therapeutic target and regulator of cell cycle-related networks in breast cancer. Aging (Albany NY). 2021; 13:4157–81. 10.18632/aging.202382 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
CBioPortal: https://cbioportal.org; The Human Protein Atlas: https://www.proteinatlas.org; Kaplan-Meier plot database https://kmplot.com; MetaCore analysis https://portal.genego.com. The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.