Abstract
Physical mutagens are a powerful tool used for genetic research and breeding for over eight decades. Yet, when compared to chemical mutagens, data sets on the effect of different mutagens and dosages on the spectrum and density of induced mutations remain lacking. To address this, we investigated the landscape of mutations induced by gamma and X-ray radiation in the most widely cultivated crop species: rice. A mutant population of a tropical upland rice, Oryza sativa L., was generated and propagated via self-fertilization for seven generations. Five dosages ranging from 75 Gy to 600 Gy in both X-ray and gamma-irradiated material were applied. In the process of a forward genetic screens, 11 unique rice mutant lines showing phenotypic variation were selected for mutation analysis via whole-genome sequencing. Thousands of candidate mutations were recovered in each mutant with single base substitutions being the most common, followed by small indels and structural variants. Higher dosages resulted in a higher accumulation of mutations in gamma-irradiated material, but not in X-ray-treated plants. The in vivo role of all annotated rice genes is yet to be directly investigated. The ability to induce a high density of single nucleotide and structural variants through mutagenesis will likely remain an important approach for functional genomics and breeding.
Keywords: mutation breeding, structural variants, forward genetics, whole-genome sequencing, transposable elements
1. Introduction
Crop biodiversity plays a key role in overcoming existing and emerging climate-related challenges that threaten world food security. Yet, domestication and thousands of years of human selection resulted in bottlenecks that greatly reduced genetic diversity [1,2,3]. While large collections of diverse germplasm are being created and are being utilized to address food insecurity, limitations including linkage drag can hamper the timely introgression of desired traits into elite cultivars [4,5]. An alternative approach to using existing diversity for crop improvement is to generate new genetic variation. While genome editing represents the latest technological iteration, the concept is not new. Scientists continue to create novel variation in plants as they have since the 1920s [6]. The first mutagenic treatments were performed by irradiating cells with X-rays. Pioneering work was carried out in the insect Drosophila melanogaster and shortly thereafter in plants [7,8]. X-ray irradiation is an important tool for genetic research. In D. melanogaster and C. elegans, X-ray irradiation was used to create balancer chromosomes that facilitated stock management and continue to be used as a tool for genome research [9,10]. In crops, the first “mutant” variety was created using X-ray irradiation of tobacco and released in the 1930s [11]. In the decades following, gamma irradiation became the predominant method for crop mutation breeding, with the earliest variety found in the IAEA’s Mutant Variety Database being the “Pink Hat” rose released in 1960 (https://mvd.iaea.org/, accessed 15 September 2022). Today there are more than 3360 officially registered mutant varieties in the IAEA’s Mutant Variety Database, with approximately 50% of varieties developed from direct or indirect use of gamma- and 17% from X-ray irradiation. This compares to approximately 11% of varieties derived from chemical mutagenesis and less than 1% listed as being derived from ion beam irradiation (https://mvd.iaea.org/, accessed 15 September 2022). The exact number of unique mutation events that contributed to varieties is difficult to estimate owing to the fact that some founder varieties were used to introgress traits in other backgrounds [12]. However, the total value of mutant varieties is estimated to be in the billions of dollars, showing the effectiveness of random mutagenesis in crop improvement [13].
The causative mutations leading to improved mutant varieties and their underlying mechanisms remain largely unknown. In addition to furthering our understanding of gene function, knowledge of the effect of mutagen and dosage on the number and type of heritable induced mutations will facilitate optimizing the mutation breeding projects so that the maximum probability of achieving the desired trait can be obtained with the smallest population size. Of the more than 240 species where mutations were used to create new varieties, rice (Oryza sativa L.) is the most prominent, representing approximately 25% of all registered varieties (https://mvd.iaea.org/, accessed 15 September 2022) [14]. Indeed, rice is a staple for more than half of the world’s population and a future with sustainable food security must include approaches to make rice more climate resilient [5].
The availability of relatively low-cost DNA sequencing enabled a genome-wide view of the effect of mutagens on plant genomes. Large data sets exist for chemically mutagenized plants. Decades of research using the chemical mutagen ethyl methane sulfonate (EMS) revealed that the mutagen produces primarily G:C to A:T point mutations with limited positional bias [15,16,17]. The effect of physical mutagens on plant genomes is more complex and less clear. Whole-genome sequencing of 1504 rice mutants (variety Kitaake) treated with fast neutron mutagenesis revealed a broad spectrum of induced mutations, including insertions, duplications, and single-base substitutions [18]. Whole-genome sequencing of seven rice mutants (variety Hitomebore) treated with C-ion and seven treated with gamma-rays showed both mutagens producing single-base substitutions, indels, and larger structural variations with more structural variants recovered in C-ion mutants [19]. An additional study of six gamma-irradiated lines (cultivar Nipponbare), also showed a predominance of single-base substitutions with indels and structural variants accumulating at a lower frequency [20]. More recently, a larger-scale study of 123 gamma-irradiated rice (subsp. japonica cv.) mutant lines created for TILLING assays focused on the recovery of single-base and small indel mutations and found a higher percentage of indels compared to SNVs [21]. Larger insertion, deletion and structural variations were not evaluated in this study. In contrast, while numerous reports were published on the effect of X-ray irradiation on rice phenotype, there is limited information on the effect of X-ray treatment at the sequence level in plants. When considering mutation breeding, a bulk of the seeds from a single cultivar are typically irradiated. In addition, mutation breeding often involves the choice of locally adapted cultivars where information is limited on the effect that genotype may have on the accumulation of induced mutations.
To address the limited knowledge of the effect of X-rays on the rice genome, and to expand knowledge on gamma irradiation, and the spectrum and density of induced mutations in a different rice cultivar, mutant populations of a Malagasy rice variety were developed and evaluated. The effect of dosage showed gamma-irradiated material to have more survivability over a broader range. The highest density of mutations (8816) was observed in the highest dosage (450 Gy). This trend was not observed in X-ray-irradiated material where one of four mutant lines treated with 75 Gy had 3-fold more accumulated mutations than the mutant with the lowest number at the same dosage.
2. Results
2.1. Generation of Mutant Population and Mutant Selection
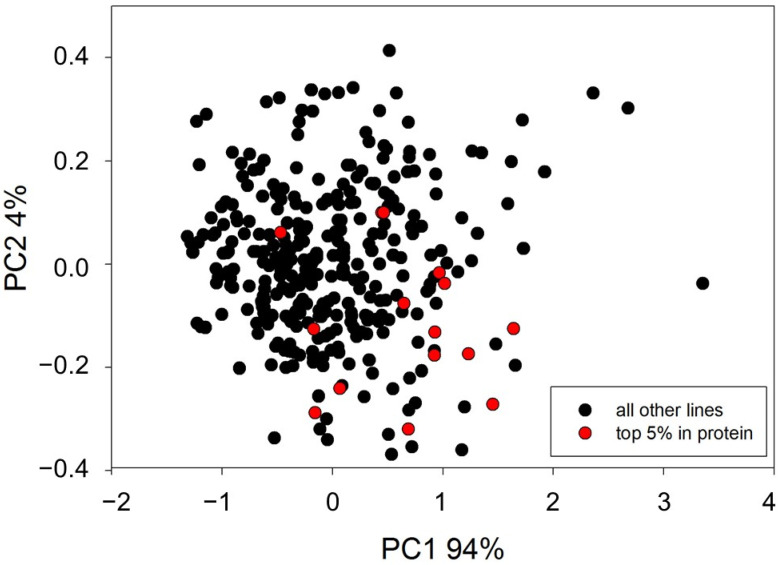
To evaluate the effect of gamma and X-ray irradiation on Oryza sativa L. ‘Marotia’, seeds were treated with one of six selected dosages of gamma irradiation or one of six dosages of X-ray irradiation. The survival rate of M1 plants negatively correlated with dosage starting at 150 Gy (Table 1). Prior to DNA sequencing, plants were evaluated for phenotypic variation with the rational that variation in plant phenotype may indicate the presence of novel nucleotide variation. Phenotypic variation between mutated and non-mutated control material was observed for survival rate, flowering date, plant height, panicle length, and number of seed per plant (Table 2). In addition, a subset of 329 mutant lines were subjected to qualitative and quantitative near-infrared reflectance spectroscopy (NIRS), with data collected for ash, fat, fibre, protein, and moisture. Principle component analysis (PCA) resulted in a clustering of samples with 14 lines as statistical outliers within two or more standard deviations from the mean (Figure 1). While the small sample size used in NIRS screening did not uncover mutants with higher seed protein content, the variation observed in NIRS and the other phenotypic traits measured suggested that genome sequencing may uncover novel induced mutations. Data from phenotypic analyses was therefore. used to select 11 mutant lines for genotypic evaluation. (Table 2 and Figure 2).
Table 1.
Survival rates (%) of irradiated material after one month of hydroponic growth.
| Treatment | 0 Gy | 75 Gy | 150 Gy | 300 Gy | 450 Gy | 600 Gy |
|---|---|---|---|---|---|---|
| gamma | 74 | 78 | 80 | 76 | 28 | 2 |
| X-ray | 72 | 82 | 70 | 6 | 0 | 0 |
Table 2.
Phenotypic characteristics of 11 mutant lines and the control selected for sequencing.
| Sample | Treatment | Days to Flowering | Plant Height | Panicle Length | Total No of Spikelets | Empty Spikelets | Fertile Spikelets | 1000 Seed Weight |
|---|---|---|---|---|---|---|---|---|
| Control | N/A | 117 | 80 | 8 | 26 | 18 | 8 | 27.19 |
| M149 | gamma 150 Gy | 90 | 112 | 14 | 67 | 28 | 40 | 30.07 |
| M153 | gamma 150 Gy | 97 | 125 | 23 | 202 | 75 | 127 | 33.43 |
| M166 | gamma 300 Gy | 103 | 100 | 16 | 49 | 20 | 30 | 31.94 |
| M193 | gamma 450 Gy | 94 | 120 | 14 | 68 | 26 | 42 | 30.8 |
| M225 | X-ray 75 Gy | 107 | 75 | 13 | 38 | 18 | 20 | 25.52 |
| M232 | X-ray 75 Gy | 90 | 72 | 10 | 45 | 19 | 26 | 19.64 |
| M238 | X-ray 75 Gy | 87 | 105 | 17 | 102 | 25 | 76 | 29.64 |
| M242 | X-ray 75 Gy | 108 | 52 | 6 | 19 | 8 | 11 | 21.35 |
| M244 | X-ray 75 Gy | 101 | 75 | 13 | 50 | 26 | 24 | 20.94 |
| M314 | X-ray 150 Gy | 97 | 80 | 12 | 38 | 17 | 21 | 26.22 |
| M317 | X-ray 150 Gy | 89 | 80 | 10 | 25 | 9 | 16 | 24.49 |
Figure 1.
PCA of NIRS data on 329 mutant rice lines. Lines with seed protein in the top 5% of the total are marked in red.
Figure 2.
Grain length and color variation in rice mutant lines (scale used cm).
2.2. Genome Sequencing
Sequencing was performed using an Illumina HiSeq2500 system and 2 × 125 PE reads on genomic DNA from two biological replicates of each of the 11 selected mutants and non-mutated control. Between 125,509,494 and 182,922,162 reads were produced, resulting in a mean coverage between 26.4 and 37.0 for all samples sequenced (Supplemental Tables S1 and S2, Supplemental Figure S1).
2.3. Single Nucleotide and Insertion/Deletion Variants Detected in Rice Mutant Lines
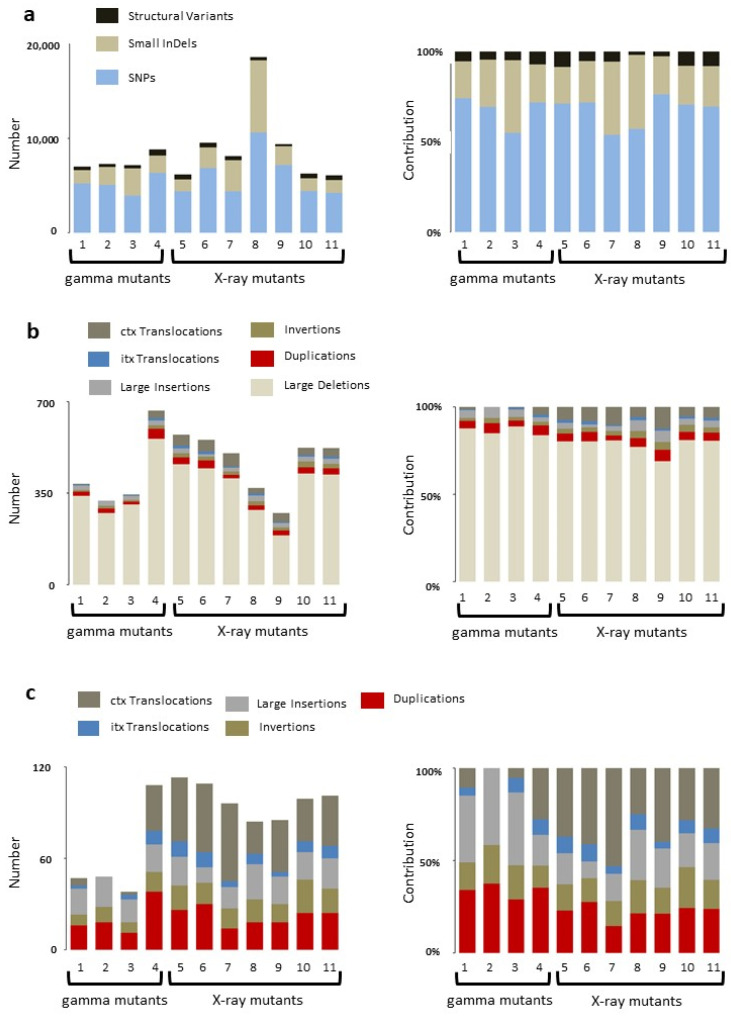
SNV and small indel variants were identified using GATK HaplotypeCaller. Variants found in the non-mutagenized control material, variants previously identified by resequencing non-mutagenized plants, and variants common in more than one mutant line were considered natural variations and not reported as induced mutations (Supplemental Table S3). This resulted in between 18,612 and 6069 total mutations per plant, with more than 90% being SNV and small indels in all irradiated material. This represents an estimated mutation frequency between 1 mutation/23 kb and 1/71 kb (Table 3). No clear trend between dosage and mutation frequency was observed. Line M242 (treated with 75 Gy X-rays) had the highest number of SNV and indel variants. Other lines treated with 75 Gy showed slightly higher accumulation of indels compared to 150 Gy X-ray-irradiated lines, with SNV variants sometimes higher and sometimes lower. In gamma-irradiated material, plants from 150 Gy-irradiated seeds accumulated more SNVs than 300 Gy gamma-irradiated material and more indel mutations than either 300 or 450 Gy gamma-irradiated material.
Table 3.
Distribution of all predicted induced mutations identified in mutant rice.
| Mutant Line | Treatment Type | Total Mutations * | SNVs | Indels | Total SNV/InDel Mutations | SNV/indel Mutation Frequency (bp/Event) ** | Mutation Frequency All Variants bp/Event) ** | SVs | Large Deletions | Duplications | Inversions | Insertions | Itx *** | Ctx *** | SV Frequency (bp/Event) ** |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 43,801 (control) * | None | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 43,802_control | None | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| M149 | gamma 150 Gy | 7016 | 5205 | 1432 | 6637 | 64,788.31 | 61,288.48 | 379 | 339 | 16 | 7 | 17 | 2 | 5 | 1,134,564.64 |
| M153 | gamma 150 Gy | 7294 | 5054 | 1919 | 6973 | 61,666.43 | 58,952.56 | 321 | 273 | 18 | 10 | 20 | 0 | 0 | 1,339,563.86 |
| M166 | gamma 300 Gy | 7147 | 3924 | 2884 | 6808 | 63,160.99 | 60,165.10 | 339 | 306 | 11 | 7 | 15 | 3 | 2 | 1,268,436.58 |
| M193 | gamma 450 Gy | 8816 | 6328 | 1861 | 8189 | 52,509.46 | 48,774.95 | 627 | 558 | 38 | 13 | 18 | 9 | 30 | 685,805.42 |
| M225 | X-ray 75 Gy | 6158 | 4389 | 1248 | 5637 | 76,281.71 | 69,827.87 | 521 | 460 | 26 | 16 | 19 | 10 | 42 | 825,335.89 |
| M232 | X-ray 75 Gy | 9546 | 6859 | 2188 | 9047 | 47,529.57 | 45,045.05 | 499 | 445 | 30 | 14 | 10 | 10 | 45 | 861,723.45 |
| M238 | X-ray 75 Gy | 8128 | 4378 | 3303 | 7681 | 55,982.29 | 52,903.54 | 447 | 406 | 14 | 13 | 14 | 4 | 51 | 961,968.68 |
| M242 | X-ray 75 Gy | 18,612 | 10,619 | 7652 | 18,271 | 23,534.56 | 23,103.37 | 341 | 285 | 18 | 15 | 23 | 7 | 21 | 1,260,997.07 |
| M244 | X-ray 75 Gy | 9392 | 7172 | 1984 | 9156 | 46,963.74 | 45,783.65 | 236 | 188 | 18 | 12 | 18 | 3 | 34 | 1,822,033.90 |
| M314 | X-ray 150 Gy | 6271 | 4425 | 1357 | 5782 | 74,368.73 | 68,569.61 | 489 | 425 | 24 | 22 | 18 | 7 | 28 | 879,345.60 |
| M317 | X-ray 150 Gy | 6069 | 4216 | 1372 | 5588 | 76,950.61 | 70,851.87 | 481 | 421 | 24 | 16 | 20 | 8 | 33 | 893,970.89 |
* Nucleotide variation identified in non-mutated control material is considered to be natural variation. Natural variants found in mutated material are removed prior to mutation counting. ** Mutation frequency calculated as (genome size = 430 Mbp)/# observed mutations. *** Translocations are also recorded as deletions from the original location. This is reflected in the total SV count.
2.4. Structural Variants Detected in Rice Mutant Lines
Discovery of larger structural variants (SVs) was carried out using the programs Manta, Lumpy, Breakdancer, and bin-by-sam. Large deletions, insertions, inversions, duplications, and translocations unique to mutant lines were identified. A comparison of SNV and indel variations versus SVs showed that at least over 90% of induced mutations are SNVs and indels with structural variants making up between 1.8% and 8.5% of total variation (Table 3 and Figure 3a). The largest number of structural variants was identified in the highest (450 Gy) dosage gamma-irradiated line, with the second largest recovered in a line treated with the lowest dosage of X-rays (75 Gy). Evaluation of structural variants revealed large deletions to be the predominant SV, comprising more than 80% of all SVs (Figure 3b). The remaining SV types were present at varying ratios in mutants with intra-chromosomal translocations (itx) being the least common (Figure 3c). While the overall number of SV events is low, the percentage of the genome affected is high. For example, evaluation of mutant M149 revealed 340 deletion events covering 49.5 Mbp (Supplemental Table S4). Of these, 126 (37%) are within intergenic regions, and 213 (63%) span genes with 141 deletions (42%) spanning regions annotated as containing transposons or retrotransposons.
Figure 3.
Distribution of variants in mutant lines: (a) total variation identified in gamma and X-ray mutagenized rice; (b) distribution of SVs; and (c) comparison of the least represented SVs in rice mutants.
Interestingly, the total number of translocations predicted within X-ray-irradiated material is higher than that identified in gamma treated samples, with less of an observable trend based on dosage (Supplemental Figures S2 and S3). A large number of translocations are also predicted between non-mutagenized parental genotype and the reference genome (Supplemental Figure S3). Intrachromosomal translocations are predicted at a lower frequency in mutated material.
2.5. Validation of Predicted Mutations
Twenty-four small variants (SNVs and short indels) unique to a single mutant line and six non-unique variants were selected for validation by Sanger sequencing. To test for the possibility of false negative errors caused by true mutations being removed during the data filtering step, four putative variants that did not pass filtering parameters were also sequenced. Three of the four were removed from the data because of the allele frequency threshold, and one predicted variant was removed due to low coverage. All 30 variants predicted by GATK and passing downstream filtration were confirmed by Sanger sequencing. None of the GATK-predicted variants removed due to allele ratio or coverage could be identified by Sanger sequencing (Table 4). In addition, 27 predicted larger structural variants ranging in size from 179 to 7732 were all confirmed by PCR size polymorphism (Table 5).
Table 4.
Validation of variants by Sanger sequencing.
| Mutant Line | Mutation Type | Mutation Name | Position Call | Mutation | Type | Sanger Confirmed |
|---|---|---|---|---|---|---|
| M149 | indel | 149_IND1 | 1:23,789,397 | GAT to G | Unique in mutant | Y |
| M153 | indel | 153_IND4 | 4:15,522,985 | C to CTCAGCCG | Unique in mutant | Y |
| M166 | indel | 166_IND1 | 1:18,131,046 | AGCGCATGTGCCATG to A | Unique in mutant | Y |
| M166 | indel | 166_IND3 | 6:5,694,121 | CA to C | Unique in mutant | Y |
| M193 | SNP | 193_SNP1 | 2:32,736,082 | A to ACCAACG | Unique in mutant | Y |
| M193 | SNP | 193_SNP4 | 6:28,952,090 | A to G | Unique in mutant | Y |
| M193 | SNP | 193_SNP5 | 7:10,033,642 | T to C | Unique in mutant | Y |
| M193 | SNP | 193_SNP6 | 12:24,819,926 | C to A | Filtered for allele ratio * | N |
| M193 | indel | 193_IND6 | 12:1,851,681 Identified in IRRI set |
AC to A | Unique in mutant | Y |
| M225 | SNP | 225_SNP2 | 3:30,121,299 | T to G | Unique in mutant | Y |
| M225 | SNP | 225_SNP3 | 5:18,773,517 | G to A | Not unique in mutant | Y |
| M232 | indel | 232_IND1 | 1:38,864,179 | AGG to A | Unique in mutant | Y |
| M238 | SNP | 238_SNP4 | 8:24,636,417 | G to A | Unique in mutant | Y |
| M238 | SNP | 238_SNP5 | 9:19,013,812 | G to C | Unique in mutant | Y |
| M238 | indel | 238_IND1 | 1:2,382,470 | CG to C | Unique in mutant | Y |
| M238 | indel | 238_IND2 | 1:3,324.566 | GGTGGT to G | Unique in mutant | Y |
| M238 | indel | 238_IND4 | 6:12,475,810 | C to CAAGT | Unique in mutant | Y |
| M238 | indel | 238_IND5 | 6:16,993,055 | TA to A | Unique in mutant | Y |
| M238 | indel | 238_IND6 | 6:17,139,752 | A to AGATGCTCTAGGACAGTTTGTTGG | Not unique in Mutant | Y |
| M242 | SNP | 242_SNP1 | 1:14,877,967 | G to T | Not unique in mutant | Y |
| M242 | SNP | 242_SNP3 | 3:8,704,769 | G to A | Not unique in mutant | Y |
| M242 | SNP | 242_SNP5 | 8:8,951,950 | G to T | Unique in mutant | Y |
| M242 | indel | 242_IND1 | 1:13,935,043 | A to AT | Low coverage | N |
| M242 | indel | 242_IND4 | 7:29,013,042 | CAAGG to C | Unique in mutant | Y |
| M242 | indel | 242_IND4 | 7:29,013,050 | G to GC | Unique in mutant | Y |
| M244 | SNP | 244_SNP3 | 7:11,432,559 | C to T | Filtered for allele ratio ** | N |
| M244 | SNP | 244_SNP4 | 11:22,433,103 | C to T | Filtered for allele ratio *** | N |
| M244 | indel | 244_IND1 | 8:24,737,886 | A to ACCAACG | Not unique in mutant | Y |
| M314 | SNP | 314_SNP2 | 4:28,342,904 | G to T | Unique in mutant | Y |
| M314 | indel | 314_IND1 | 4:3,682,688 | G to GAT | Unique in mutant | Y |
| M314 | indel | 314_IND1 | 4:3,682,689 | G to GA | Not unique in mutant | Y |
| M314 | indel | 314_IND1 | 4:3,682,697 | GGT to G | Unique in mutant | Y |
| M314 | indel | 314_IND1 | 4:3,682,701 | CAG to C | Unique in mutant | Y |
| M314 | indel | 314_IND2 | 7:15,991,122 | AC to A | Unique in mutant | Y |
* 3 of 29 reads supporting alt allele, GATK called as heterozygous. ** 2 of 21 reads supporting alt allele. *** 2 of 25 reads supporting alt allele.
Table 5.
Locus affected, and function of confirmed SV mutations.
| Mutant Line | Mutation Name | Mutation Call | Call Size | Verified Mutation Size | Locus Affected | Function |
|---|---|---|---|---|---|---|
| M149 | 149_DEL3 | 7:42,391–44,131 | 1740 | 1743 bp | LOC_Os07g01070.1 | Membrane transport activity |
| M153 | 153_DEL3 | 2:30,638,481–30,639,067 | 586 | 592 bp | Intron after LOC_Os02g50150 | |
| M232 | 232_DEL3_2 | 3:15,993,208–15,994,132 | 924 | 952 bp | Intron after LOC_Os03g27840 | |
| M242 | 242_DUP3 | 5:1,790,511–1,790,511 | 179 | 179 bp | LOC_Os05g03972.1 | Plant protein, unknown function |
| M317 | 317_DEL2 | 2:30,048,905–30,049,501 | 596 | 603 bp | Intron | |
| M317 | 317_DEL3 | 5:86,454–87,418 | 964 | 960 bp | Intron region bordering upstream of LOC_Os05g01080 | Uncharacterised protein |
| M166 | 166_DEL3 | 3:14,816,703–14,817,352 | 649 | 654 bp | Intron | |
| M225 | 225_DEL3 | 3:14,816,703–14,817,352 | 649 | 654 bp | Intron | |
| M193 | 193_DEL1 | 2:35,131,943–35,132,357 | 414 | 413 bp | Intron after LOC_Os02g57350 | |
| M232 | 232_DEL1 | 2:35,131,943–35,132,357 | 414 | 413 bp | Intron after LOC_Os02g57350 | |
| M238 | 238_DEL2 | 2:35,131,943–35,132,357 | 414 | 413 bp | Intron after LOC_Os02g57350 | |
| M242 | 242_DEL1 | 2:35,131,943–35,132,357 | 414 | 415 bp | Intron after LOC_Os02g57350 | |
| M244 | 244_DEL1 | 2:35,131,943–35,132,357 | 414 | 413 bp | Intron after LOC_Os02g57350 | |
| M225 | 225_DEL2 | 7:606,112–607,134 | 1022 | 1000 bp | LOC_Os07g01990.1 | Uncharacterised |
| M193 | 193_DEL3 | 7:606,112–607,134 | 1022 | 1000 bp | LOC_Os07g01990.1 | Uncharacterised |
| M314 | 314_DEL2 | 7:606,112–607,134 | 1022 | 1022 bp | LOC_Os07g01990.1 | Uncharacterised |
| M149 | 149 DEL2 | 2:34,566,631–34,570,008 | 3377 | Primer pair designed in deletion, WT (1200), MUT (0) | Intron | |
| M149 | 149 DEL3 | 3:34,303,046–34,303,733 | 687 | WT (750), MUT (50) | Intron | |
| M232 | 232 DUP3 | 4:2,243,338–2,244,529 | 1191 | WT (0), MUT (bands at 1200, 550) | LOC_Os04g04660.1 | Expressed protein |
| M238 | 238 DEL3_2 | 5:4,851,233–4,852,248 | 1015 | WT (1000), MUT (100) | Intron | |
| M242 | 242 DEL2 | 6:712,560–713,948 | 1388 | WT (400), MUT (200) | Intron | |
| M242 | 242 DEL3 | 10:23,140,818–23,144,340 | 3522 | WT (4200), MUT (450) | LOC_Os10g42910.1, LOC_Os10g42920.1 LOC_Os10g42930.1 | Transposon protein, conserved hypothetical protein, expressed protein |
| M317 | 317 DEL1_2 | 7:547,127–550,124 | 2534 | WT (600), MUT (2200), Primer pair designed in deletion, expected WT (609), MUT (0) | LOC_Os07g01904.1 | Expressed protein |
| M244 | 244 DUP2 | 3:35,152,931–35,153,261 | 330 | WT (550), MUT (850,550) | LOC_Os03g62040.1 | Retrotransposon protein |
| M232 | 232 DUP2 | 3:35,152,931–35,153,261 | 330 | WT (650), (1100,650) | LOC_Os03g62040.1 | Retrotransposon protein |
| M153 | 153 DEL1_4 | 10:23,131,290–23,139,022 | 7732 | WT (~8000), MUT (220) | LOC_Os10g42900.1; LOC_Os10g42910.1 | Peptide transporter PTR2, transposon protein |
| M314 | 314 DEL3_4 | 10:23,131,290–23,139,022 | 7732 | WT (~8000), MUT (220) | LOC_Os10g42900.1; LOC_Os10g42910.1 | Peptide transporter PTR2, transposon protein |
2.6. Predicted Effect of Induced Mutations
The potential effect of SNV and indel mutations on gene function was evaluated using SNPeff. The frequency of nonsense changes ranged between 0.38 and 1.99% (Table 6). Similarly, high impact SNVs represented the lowest frequency with the majority of variation found in intergenic regions. The distribution of indels is similarly highest in intergenic regions. In contrast to SNVs, predicted high-impact indels predominate over low and moderate ones (Table 6). Larger variants were also recovered that affect coding regions (Table 5).
Table 6.
Predicted effect of small variants.
| Mutant Line | Missense | Nonsense | Silent | High | Low | Moderate | Modifier | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Count | % | Count | % | Count | % | Count | % | Count | % | Count | % | Count | % | |
| M149 | 131 | 60.93% | 1 | 0.465% | 83 | 38.605% | 42 | 0.261% | 91 | 0.565% | 147 | 0.913% | 15,823 | 98.261% |
| M153 | 204 | 55.135% | 3 | 0.811% | 163 | 44.054% | 54 | 0.286% | 209 | 1.108% | 223 | 1.183% | 18,371 | 97.423% |
| M166 | 422 | 57.415% | 4 | 0.544% | 309 | 42.041% | 48 | 0.129% | 374 | 1.006% | 468 | 1.259% | 36,281 | 97.606% |
| M193 | 148 | 56.705% | 1 | 0.383% | 112 | 42.912% | 52 | 0.253% | 141 | 0.685% | 162 | 0.787% | 20,238 | 98.276% |
| M225 | 87 | 55.769% | 0 | 0.00% | 69 | 44.231% | 28 | 0.221% | 82 | 0.646% | 95 | 0.749% | 12,481 | 98.384% |
| M232 | 204 | 58.621% | 4 | 1.149% | 140 | 40.23% | 64 | 0.273% | 164 | 0.7% | 231 | 0.986% | 22,966 | 98.041% |
| M238 | 419 | 60.201% | 4 | 0.575% | 273 | 39.224% | 74 | 0.173% | 353 | 0.827% | 458 | 1.073% | 41,798 | 97.927% |
| M242 | 1363 | 58.198% | 26 | 1.11% | 953 | 40.692% | 264 | 0.245% | 1294 | 1.203% | 1479 | 1.375% | 104,546 | 97.177% |
| M244 | 158 | 55.052% | 5 | 1.742% | 124 | 43.206% | 58 | 0.287% | 159 | 0.787% | 167 | 0.826% | 19,829 | 98.1% |
| M314 | 107 | 52.709% | 2 | 0.985% | 94 | 46.305% | 46 | 0.363% | 107 | 0.845% | 118 | 0.931% | 12,398 | 97.861% |
| M317 | 100 | 49.751% | 4 | 1.99% | 97 | 48.259% | 35 | 0.292% | 111 | 0.925% | 108 | 0.9% | 11,751 | 97.884% |
3. Discussion
When inducing novel mutations, a balance must be struck whereby a sufficient type and number of variants accumulate that are transmissible to the next generation, while at the same time limiting plant death and sterility. Phenotypic measurements, such as hypocotyl length and survivability, are typically conducted in the first (M1) generation to estimate the effect of mutagen dosage [19]. When using seed mutagenesis, the first generation is chimeric, making the link between early observed phenotypes and heritable mutations difficult, as plants undergo a broad response to irradiation-induced DNA damage that includes cell cycle arrest [22]. In the current study, survival rates increased slightly at lower dosages compared to control (0–150 Gy for gamma and 0–75 Gy in X-ray) and then dropped as dosage increased. While small variations may represent experimental stochasticity, low dosages of ionizing radiation are reported to have a stimulating effect on plant growth, a process known as hormesis [23]. Further studies are required to evaluate if low dosage irradiation has an effect on seed germination and plant growth.
Genome sequencing of rice and other species revealed that gamma irradiation induces a broad spectrum of heritable mutations ranging from single nucleotide variants to large chromosomal aberrations [19,24,25]. In Arabidopsis thaliana, studies were undertaken to evaluate the transmissibility of mutations in gamma and carbon ion-irradiated pollen, suggesting a link between non-transmissible large deletions and semisterility [26]. This phenomenon is likely also occurring in plants where seed is mutagenized. In the current study, increasing gamma irradiation dosage from 150 Gy to 600 Gy resulted in survival rates dropping from 80% to 2%. The highest dosage evaluated at the DNA sequence level, 450 Gy, resulted in the highest accumulation of mutations (8816), including the highest overall number of structural variants (637). This represents 19% more mutations than 300 Gy treated material, but also a drop in survivability from 76% to 28%. In rice seed (cv. Nipponbare) treated with gamma irradiation (a 137 Cs source versus 60 Co used in the current study) more mutations were reported in lower dosages (4698 at 165 Gy) compared to the highest dosage (3326 at 389 Gy). In addition, the highest dosage produced the lowest number of structural variants [20]. This is in contrast with the present study, where the number of structural variants were highest in 450 Gy-treated material, followed by the next highest in plants from 150 Gy-treated seed. These differences likely occur due to a combination of variation in treatment conditions such as seed moisture content, source of gamma rays, and also genotype-specific DNA and epigenomic variation, and differences in other chromosome features that are shown to affect spontaneous mutations in plants [27,28]. In addition, care must be taken when interpreting results, as the number of mutant lines subjected to whole-genome sequencing can be low, and it may be difficult to remove all natural or spontaneous mutations prior to estimating the frequency of induced mutations. For example, Li et al. sequenced six gamma-irradiated plants and found the total number of SNV and small indel mutations to range between 3135 and 4698 [20]. In the present study, four lines selected based on high phenotypic variability showed a range of between 8816 and 7016. When considering that random mutagenesis might produce a distribution of mutation frequencies in a collection of treated seed, such observed differences may be explained as a sampling bias in the present study towards highly mutagenized material. However, while efforts were made to remove natural genetic variation and Sanger sequencing revealed no false positive or false negative errors, an inflation of mutation frequency due to the presence of natural variants cannot be ruled out in the current data set. Bulk seed was chosen for irradiation to mimic the standard practice used in mutation breeding. The observed frequency of small induced mutations (1/23 kb to 1/71 kb) is much higher than mutation rates reported in rice (~1/135 kb) and more similar to rates reported in diploid, tetraploid, and hexaploid wheat (1/92 kb, 1/51 kb, and 1/24 kb, respectively) [6]. To control for natural variation, each biological replicate from the same mutant line was compared to two non-irradiated controls and 20 other plants from different mutant lines, and only mutations unique to the mutant line and also present in both replicates were considered induced mutations. The level of segregation distortion at the sequence level is unknown for Oryza sativa L. ‘Marotia’, and therefore it remains possible that sequencing of more plants from the same bulk seed is required to ensure removal of all potential natural variants present in the population. In addition, spontaneous mutations occurring during the propagation of mutated plants would be indistinguishable from irradiation-induced mutations. Studies in Arabidopsis suggest that plants may experience increased rates of spontaneous mutations after multigenerational growth in elevated temperatures [29]. For example, in mutation accumulation populations grown in high heat, a mean of 36.6 total novel SNV and indel spontaneous mutations were reported. This represents less than 1% of predicted induced mutations of the same class in any of the mutant lines described in the present study. Less is known about sequence variation due to X-ray irradiation in plants [30]. In the present study, the survival rate peaked at 82% in 75 Gy-treated material with a reduction to 70% in 150 Gy, followed by a sharp drop off to 6% at 300 Gy. Total mutation accumulation was highly variable with a threefold variation observed at 75 Gy. Interestingly, the ratio of structural variants to total mutations was highest in X-ray-irradiated material with the lowest number of total accumulated mutations. In addition, more translocations were predicted within X-ray-irradiated material. This may indicate an increase in the generation of double strand breaks, variation in response of the DNA repair machinery, or a combination of both [31]. Whole-genome sequencing of many more samples will be required to determine the extent to which dosage and environmental conditions influence the spectrum and density of induced mutations in both gamma and X-ray-irradiated material. This will become more amenable as sequencing prices continue to drop.
Naturally occurring structural variation was implicated in plant phenotype variation, adaptation, and domestication [32,33]. In rice, a tandem duplication of the GL7 locus was shown to lead to an increase in grain length [34]. Large-scale structural variant analysis in 3000 genomes resulted in the discovery of 63 million variants, suggesting an important role for natural SVs in gene function and phenotypic diversity in rice [35]. Thus, it is expected that mutation-induced structural variants will have a large impact on plant phenotype. Indeed, this may explain the popularity of physical irradiation compared to chemical mutagenesis in plant mutation breeding. Large single-loci events are easier to genetically fix and maintain in a population as compared to traits that require multiple small mutations in unlinked genomic regions. More studies are needed to understand the mechanisms of SV accumulation in mutated plants. For example, transposons were implicated as drivers of plant genome plasticity and can function synergistically with DNA repair mechanisms to generate structural variation [36]. Irradiation of plant cells induces double strand breaks, affects DNA methylation state, which can change transposon activity, and activates DNA repair machinery [37,38]. Consequently, numerous processes, along with plant genotype, can contribute to the accumulation of germ-line mutations, making a priori predictions regarding spectrum and density of mutations from different irradiation dosages difficult.
The use of chemical mutagens or ionizing radiation can produce thousands of novel induced mutations per mutant line. This was exploited for high-throughput reverse genetic screens, such as TILLING [6]. While precision genome editing tools were developed since the advent of TILLING, new screening approaches promise continued value for reverse genetics with random mutagenesis. Automated phenomics can more efficiently and accurately link genotype to phenotype.
In addition, by combining high-density mutagenesis and large population sizes, DNA libraries can be prepared where there is a high probability of novel mutations at all base pair positions that are the target of the applied mutagen. This allows the application of genotypic screens for a specific desired base pair change, rather than discovery of all mutations within a PCR amplicon that is common for typical TILLING by sequencing screens. Genotypic screening can provide increased throughput at reduced costs. A proof of principle of this approach was described by Knudsen et al. who used digital PCR to identify EMS-induced mutations in barley in a method known as fast identification of nucleotide variants by DigITal PCR (FIND-IT) [39]. Thus, mutant populations can approach the precision of genome editing. Nevertheless, when utilizing a mutagen that produces thousands of heritable mutations per line, causative versus background mutations must be considered. Successive rounds of self-fertilization and single-seed descent can be used to fix a desired trait without genotypic evaluation so long as background mutations do not have a pleiotropic effect. Indeed, of the 872 mutant rice varieties listed in the Mutant Variety Database, 462 (53%) are listed as being directly released without crossing to another genotype. Alternatively, the desired trait can be introgressed into an elite line. This can be advantageous, as background mutations may reduce fitness. Mutation load can be reduced through repeated rounds of backcrossing or through applying genomic background selection where a small number of molecular markers can be used to select progeny with a higher percentage of the elite genome [12]. In the present study, phenotypic variation in mutants was observed throughout several generations in a number of traits, including fat, fibre, and protein content in seed, seed morphology, days to flowering, and 1000 seed weight. The extent to which these are controlled by single or multiple genes is unknown. The high percentage of direct-release rice varieties may reflect a combination of the ease of fixing mutations in self-fertile plants and the need to maintain mutations in multiple loci for the expression of the desired trait (s). Tools to rapidly map mutations can be used to address the gap in knowledge regarding the causative genes involved in many economically important mutant varieties [40].
While next generation sequencing became more routine, challenges still exist for the discovery and analysis of induced mutations using short-read sequencing. This includes the accurate assignment of heterozygous SNV mutation calls. Evaluation of carbon ion and gamma-ray-induced mutations in rice showed a high correlation between the variant allele frequency and genotype call accuracy when using GATK Haplotype caller [19]. In the current study, applying an 80% ratio to support homozygous calls allowed accurate recovery of heterozygous SNVs when evaluated using Sanger sequencing. Recovery of larger variants from short-read data is more challenging, as the estimating extent of false negative errors from different tools remains difficult. Use of longer-read sequencing will enable a more comprehensive view of mutation-induced structural variants in plants. Indeed, genomic tools are allowing a deeper understanding of plant genome plasticity. The rate of spontaneous mutations was reported to vary between different tissues [41]. In the context of mutation breeding, this highlights the utility of including biological or technical replicates so that spontaneous mutations can be differentiated from induced mutations.
Researchers and plant breeders now have a powerful suite of tools to understand gene function and improve crops as compared to 80 years ago when inducing mutations in a plant was first described. While targeted genome editing approaches, such as CRISPR, promise to revolutionize agriculture, the in vivo function of the majority of annotated plant genes remains yet to be established. This limits target selection in reverse genetic methods. Thus, it is expected that random mutagenesis and forward genetics will remain a useful approach for establishing gene function, and also for crop breeding.
4. Materials and Methods
4.1. Plant Material and Generation of a Mutant Population
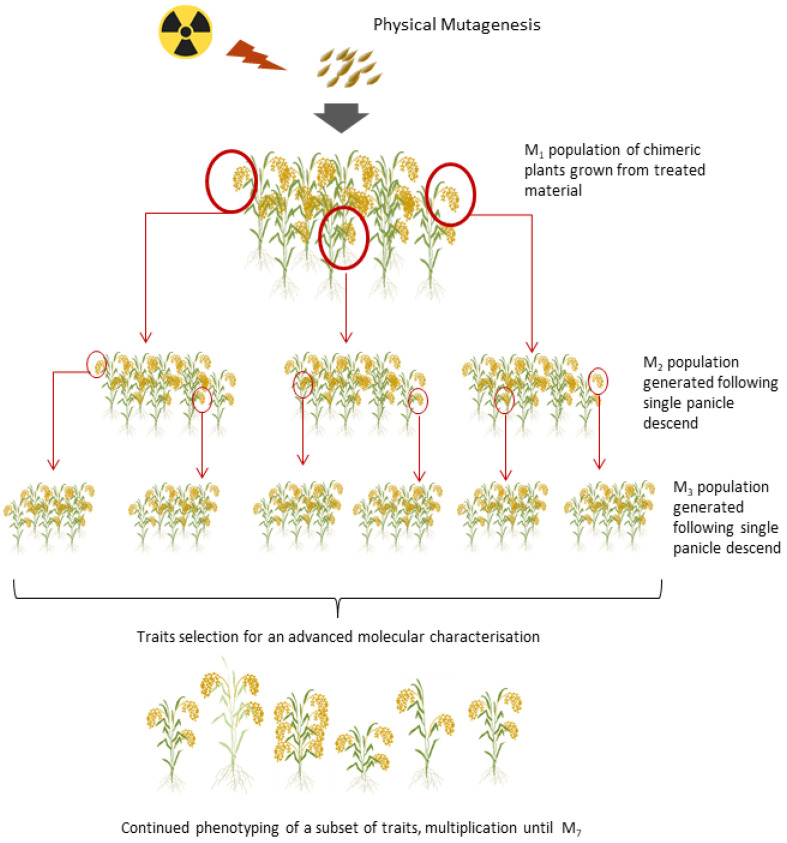
An upland, local rice cultivar, Marotia (synonym CNA4136, collection number 3729), was obtained from the Antananarivo University, Antananarivo, Madagascar. Reported phenotypic characteristics are a growth cycle of 115–120 days, plant height of 120–130 cm, semi-erect plant architecture, and semi-long seed (paddy length 9.6 mm, caryopsis length 7.2 mm) with the 1000 seed weight of 33.1 g. The cultivar is characterized as lodging-sensitive. A bulk of 600 grains was mutagenized with gamma or X-rays at 0, 75, 150, 300, 450, and 600 grays of irradiation using Co-60 source located at the Plant Breeding and Genetics Laboratory in Seibersdorf, Austria (https://www.iaea.org/topics/plant-breeding/laboratory, (accessed 15 September 2022)). Seeds were pre-germinated in petri plates and upon germination transplanted into a hydroponic growth system. Survivability was determined by evaluating 50 seeds per dose per treatment type (X-ray or gamma irradiation), and carried out according to [27]. Cultures were maintained following procedures described in [42]. Non-irradiated seeds served as control material during the screening process. Upon flowering, every M1 panicle was bagged in order to avoid cross-contamination and to ensure a pureness of the resulting line. Over 4600 M2 seeds were harvested from mutagenized material. Harvested seeds were labeled following the designed nomenclatures reflecting the origin of the seed, irradiation mode, and dose applied. This nomenclature was maintained over the next propagation cycles with the addition of information on the mutant generation. The mutant population was maintained and multiplied following the principle of single-panicle descent by self-fertilization to the seventh generation (M7) [27]. Plants were maintained in the greenhouse with the temperature set to 28 ± 3 °C and humidity to 80 ± 5%. From November to April, artificial lights were supplemented to maintain the light intensity and the day/night period to 14/10.
4.2. Glasshouse Trials and Agronomic Traits Measurement for Forward Genetics
All glasshouse experiments were performed in hydroponic systems using a randomized complete block design [27]. Each mutant line was grown with a planting density of 4 cm × 4 cm using the test platforms as described in Bado et al. [43]. As of M2 (Figure 4, Supplemental Figure S4), agronomic traits were measured for every single plant. These included flowering date, plant height, panicle length, number of panicles per plant, number of tillers per plant, number of empty and fertile spikelets, 1000-grain weight, and total number of seed. Phenotypic characterization was repeated for every mutant generation until the M4, at which stage 329 independent lines were identified for near infrared reflectance spectroscopy (NIRS) [44]. Seed weight and seed number are important considerations when selecting the generation to screen with the use of NIRS; therefore, obtained yield was the main criterion in the identification of lines to be analyzed.
Figure 4.
A mutant population was generated with the use of gamma or X-ray at different doses covering the range between 75 and 600 Gy plus a 0 Gy control. Plants were grown in glasshouse conditions with a hydroponic system. Single panicle descent was used for maintenance of mutant lines and for subsequent mutant selection.
Qualitative and quantitative NIRS analyses were performed as outlined in Vollmann and Jankowicz-Cieslak [44]. Spectral data obtained during measurements were subjected to principal component analysis (PCA). PCA scores for samples were calculated and further used in score plots to visualize classification results. In the rice mutant population, spectroscopic outliers were subsequently detected based on the distance to untreated control genotypes of the same genetic background. For quantitative analyses, external calibration was used for prediction of analyte values, which availed the measurement of traits such as ash, fat, fibre, protein, and moisture content in the subset of 329 rice mutant lines.
4.3. DNA Sequencing and Read Mapping
Five seeds per selected M7 mutant line and a control were planted (germination rate was 100%). All germinated seedlings were transplanted into a hydroponic system and allowed to grow until a second flag leaf appeared. Two healthy seedlings per line and two from the control were randomly selected for tissue collection. Young leaf tissue was harvested into Eppendorf tubes, frozen in liquid nitrogen, and stored in a −80 °C freezer until further use. High-quality genomic DNA was isolated using the Qiagen DNeasy kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. To determine stock concentrations of each sample, DNA was quantified using a NanoDrop spectrophotometer (Thermo Scientific, Waltham, MA, USA) and Qubit® 2.0 Fluorimeter (QubitTM Assays, Invitrogen, Waltham, MA, USA) following manufacturer’s instructions. DNA was then diluted to the working concentration of 1 ng/µL in the final volume of 100 µL. In order to produce sequencing libraries, 60 ng of each DNA in 50 µL volume was fragmented in a microTUBE AFA Fiber (Pre-Slit-Snap-Cap 6 × 16 mm; Covaris, Brighton, UK) to a size ranging of 550 bp using Covaris M220 Focused ultrasonicator (Covaris, Brighton, UK). Fragmented, double-stranded DNA was quality checked with the use of a Bioanalyzer instrument (Agilent, Santa Clara, CA, USA) and libraries were prepared using the NEB DNA Ultra kit. The libraries were sequenced on an Illumina HiSeq2500 instrument using paired-end mode and 125 bp read length. Sequencing was targeted to a sequencing depth of 30-fold for each rice sample. The quality of the raw sequence reads was analyzed with FastQC [45]. The 2 × 125 bp paired-end sequence reads were mapped to the Oryza sativa subsp. japonica reference genome (IRGSP 1.0.23 build) using the mapping tool Burrows–Wheeler Aligner-MEM (version 0.7.17-r1188) [46]. Coverage analysis was carried out on BAM files using SAMtools depth [47].
4.4. Detection and Analysis of Point Mutations and Small Indels
Variant calling for SNVs and small indels was performed with GATK HaplotypeCaller (Version 4.1.8.1, Broad Institute, Cambridge, MA, USA) using default settings and following best practices, including marking and removing duplicates (Picard MarkDuplicates), and producing a multi-sample genomic variant call format file (gVCF) [48]. The multi-sample gVCF was split by sample and filtered for a minimum coverage depth of 20× using SAMtools. Zygosity calls were recalibrated such that heterozygous calls with less than 20% or more than 80% of reads supporting the alternative allele were re-scored as homozygous reference or homozygous alternative, respectively, based on previous work studying the effect of allele ratios and false positive errors in mutagenized rice [19]. Identification of variants unique to a specific mutant line was carried out using pairwise analysis with bcftools isec (version 1.10.2, Sanger Institute, Hinxton, England, Broad Institute, Cambridge, Massachusetts, United States of America) [47]. Four filtering steps were applied to remove natural nucleotide variation not induced by irradiation. First, variants were cataloged from 2 non-irradiated plants grown from the bulk seed for mutagenesis, with the first (#43801) having 1,810,784 variants and the second (#43802) having 1,814,721, representing 2,147,350 unique variants. When present, these variants were removed from the mutated material (see Supplemental Table S3). Second, variation present in more than one mutant line was considered to be natural and was removed. Third, variation not present in both biological replicates from the same line was removed. Finally, putative natural nucleic variation present in the SNP-Seek database from resequencing 3000 rice genomes found in the mutant lines was also removed [49]. The effect of the remaining small, predicted induced mutations was evaluated using SNPeff (version 5.0e, Wayne State University; Detroit, Michigan, United States of America; McGill University; Quebec, QC, Canada) [50]. A subset of called variants were selected for Sanger sequencing validation, including variants specific to a single mutant line, those common to more than one line, and also negative controls consisting of called variants that were filtered from the data set due to low coverage or allele ratio (see Table 5). Primer pairs were designed using the online tool ‘Primer3′ with the following parameters; primer size: 18–24 (opt: 22) and primer Tm 58–62 (opt: 60) [51].
4.5. Genomic Variant Detection: Structural Variants
BreakDancer [52], Lumpy [53], bin-by-sam [54], and Manta [55] were used to detect structural variants (SVs). Parameters for BreakDancer (version 1.1.2, Washin1gton University School of Medicine, St. Louis, MO, USA) were the default—except for −r 50—minimum number of read pairs required to establish a connection. Only translocations, which were supported by more than 100 reads, were used for further analysis. Overlapping translocations within sample replicates were merged into one if they shared similar start and end coordinates (±2000 bp) in the targeted chromosome. Default parameters were used for Manta (version 0.2.13, Illumina, San Diego, CA, USA). For Lumpy (version 1.0.2, University of Virginia, Charlottesville, VA, USA) default parameters were used, followed by filtering for the number of reads supporting one SV using both 10 and 50 reads. Bin sizes of 1, 5, 10, and 100 kb were used in bin-by-sam (version 2.0, University of California, Davis, CA, USA) analysis. The effect of the resulting structural variants unique to a specific mutant line were annotated using intansv [36].
To validate candidate SVs, regions with a large (>2 kbp), medium (~1 kbp), and smaller (<1 kbp) deletion were chosen for analysis. Each region was visualized using IGV and PCR primers designed to flanking sequence (Supplemental Figure S4). Depending on the size of the region and the extent of the deletion in the region, primers were designed for PCR products of size ranging between 526 and 7932 bp. Primer pairs were designed using the online tool ‘Primer3′ using the same parameters as for Sanger sequencing validation. To detect the presence/absence of molecular weight variations due to insertions or deletions, 5 µL of the PCR product and 2µL of orange G loading dye were loaded into wells on a 1.5% agarose gel with ethidium bromide.
Acknowledgments
Authors wish to thank Lydia Razafinirina, Kamila Kozak-Stankiewicz and Harimialimalala Jhonny Rabefiraisana for support provided in development and maintenance of mutant population used in this study. Authors would also like to thank Johann Vollmann for the analyses of NIRS results.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/plants11233232/s1. Supplemental Figure S1. Sequencing coverage per chromosome. Supplemental Figure S2. Cross-chromosomal translocations detected in each sample. Supplemental Figure S3. Summary of translocations in all mutants. Supplemental Figure S4. Generation of mutagenic population and selection of candidate mutant lines. Supplemental Table S1. Summary of alignment statistics. Supplemental Table S2. Mean coverage per sample. Supplemental Table S3. Summary of filtering natural point mutations and small indels. Supplemental Table S4. Deletion events called in mutant line M149.
Author Contributions
Conceptualization: J.J.-C. and B.J.T. Investigation: J.J.-C., S.J., B.J.H., B.G., L.J., A.G., I.L.I. and B.J.T. Review and editing: J.J.-C., I.L.I. and B.J.T. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Raw sequencing reads have been uploaded to the NCBI database under BioProject ID PRJNA826906 (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA826906, accessed 14 April 2022).
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
Funding for the work was provided by the Food and Agriculture Organization of the United Nations and the International Atomic Energy Agency through their Joint FAO/IAEA Program of Nuclear Techniques in Food and Agriculture.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Purugganan M.D., Fuller D.Q. The nature of selection during plant domestication. Nature. 2009;457:843–848. doi: 10.1038/nature07895. [DOI] [PubMed] [Google Scholar]
- 2.Joukhadar R., Daetwyler H.D., Bansal U.K., Gendall A.R., Hayden M.J. Genetic Diversity, Population Structure and Ancestral Origin of Australian Wheat. Front. Plant Sci. 2017;8:2115. doi: 10.3389/fpls.2017.02115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mondal S., Rutkoski J.E., Velu G., Singh P.K., Crespo-Herrera L.A., Guzmán C., Bhavani S., Lan C., He X., Singh R.P. Harnessing Diversity in Wheat to Enhance Grain Yield, Climate Resilience, Disease and Insect Pest Resistance and Nutrition Through Conventional and Modern Breeding Approaches. Front. Plant Sci. 2016;7:991. doi: 10.3389/fpls.2016.00991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCouch S., Baute G.J., Bradeen J., Bramel P., Bretting P.K., Buckler E., Burke J.M., Charest D., Cloutier S., Cole G., et al. Feeding the future. Nature. 2013;499:23–24. doi: 10.1038/499023a. [DOI] [PubMed] [Google Scholar]
- 5.Bailey-Serres J., Parker J.E., Ainsworth E.A., Oldroyd G.E.D., Schroeder J.I. Genetic strategies for improving crop yields. Nature. 2019;575:109–118. doi: 10.1038/s41586-019-1679-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Till B.J., Datta S., Jankowicz-Cieslak J. TILLING: The Next Generation. In: Varshney R.K., Pandey M.K., Chitikineni A., editors. Plant Genetics and Molecular Biology. Springer International Publishing; Cham, Switzerland: 2018. pp. 139–160. [Google Scholar]
- 7.Muller H.J. Artificial Transmutation of the Gene. Science. 1927;66:84–87. doi: 10.1126/science.66.1699.84. [DOI] [PubMed] [Google Scholar]
- 8.Stadler L.J. Genetic Effects of X-Rays in Maize. Proc. Natl. Acad. Sci. USA. 1928;14:69–75. doi: 10.1073/pnas.14.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edgley M.L., Baillie D.L., Riddle D.L., Rose A.M. Genetic balancers. WormBook; Pasadena, CA, USA: 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghavi-Helm Y., Jankowski A., Meiers S., Viales R.R., Korbel J.O., Furlong E.E.M. Highly rearranged chromosomes reveal uncoupling between genome topology and gene expression. Nat. Genet. 2019;51:1272–1282. doi: 10.1038/s41588-019-0462-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jankowicz-Cieslak J., Till B.J. Advances in Plant Breeding Strategies: Breeding, Biotechnology and Molecular Tools. Springer International Publishing; Cham, Switzerland: 2015. Forward and Reverse Genetics in Crop Breeding; pp. 215–240. [Google Scholar]
- 12.Jung C., Till B. Mutagenesis and genome editing in crop improvement: Perspectives for the global regulatory landscape. Trends Plant Sci. 2021;26:1258–1269. doi: 10.1016/j.tplants.2021.08.002. [DOI] [PubMed] [Google Scholar]
- 13.Ahloowalia B.S., Maluszynski M., Nichterlein K. Global impact of mutation-derived varieties. Euphytica. 2004;135:187–204. doi: 10.1023/B:EUPH.0000014914.85465.4f. [DOI] [Google Scholar]
- 14.Jankowicz-Cieslak J., Mba C., Till B.J. Mutagenesis for Crop Breeding and Functional Genomics. In: Jankowicz-Cieslak J., Tai T.H., Kumlehn J., Till B.J., editors. Biotechnologies for Plant Mutation Breeding: Protocols. Springer International Publishing; Cham, Switzerland: 2017. pp. 3–18. [Google Scholar]
- 15.Greene E.A., Codomo C.A., Taylor N.E., Henikoff J.G., Till B.J., Reynolds S.H., Enns L.C., Burtner C., Johnson J.E., Odden A.R., et al. Spectrum of chemically induced mutations from a large-scale reverse-genetic screen in Arabidopsis. Genetics. 2003;164:731–740. doi: 10.1093/genetics/164.2.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henry I.M., Nagalakshmi U., Lieberman M.C., Ngo K.J., Krasileva K.V., Vasquez-Gross H., Akhunova A., Akhunov E., Dubcovsky J., Tai T.H., et al. Efficient Genome-Wide Detection and Cataloging of EMS-Induced Mutations Using Exome Capture and Next-Generation Sequencing. Plant Cell. 2014;26:1382–1397. doi: 10.1105/tpc.113.121590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan W., Deng X.W., Yang C., Tang X. The Genome-Wide EMS Mutagenesis Bias Correlates With Sequence Context and Chromatin Structure in Rice. Front. Plant Sci. 2021;12:579675. doi: 10.3389/fpls.2021.579675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li G., Jain R., Chern M., Pham N.T., Martin J.A., Wei T., Schackwitz W.S., Lipzen A.M., Duong P.Q., Jones K.C., et al. The Sequences of 1504 Mutants in the Model Rice Variety Kitaake Facilitate Rapid Functional Genomic Studies. Plant Cell. 2017;29:1218–1231. doi: 10.1105/tpc.17.00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li F., Shimizu A., Nishio T., Tsutsumi N., Kato H. Comparison and Characterization of Mutations Induced by Gamma-Ray and Carbon-Ion Irradiation in Rice (Oryza sativa L.) Using Whole-Genome Resequencing. G3 Genes Genomes Genet. 2019;9:3743–3751. doi: 10.1534/g3.119.400555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li S., Zheng Y., Cui H., Fu H., Shu Q., Huang J. Frequency and type of inheritable mutations induced by γ rays in rice as revealed by whole genome sequencing. J. Zhejiang Univ. Sci. B. 2016;17:905–915. doi: 10.1631/jzus.B1600125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hwang S.-G., Lee S.C., Lee J., Lee J.W., Kim J.-H., Choi S.Y., Kim J.B., Choi H.I., Jang C.S. Resequencing of a Core Rice Mutant Population Induced by Gamma-Ray Irradiation and Its Application in a Genome-Wide Association Study. J. Plant Biol. 2020;63:463–472. doi: 10.1007/s12374-020-09266-2. [DOI] [Google Scholar]
- 22.Hefner E., Huefner N., Britt A.B. Tissue-specific regulation of cell-cycle responses to DNA damage in Arabidopsis seedlings. DNA Repair. 2006;5:102–110. doi: 10.1016/j.dnarep.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 23.Song K.E., Lee S.H., Jung J.G., Choi J.E., Jun W., Chung J.-W., Hong S.H., Shim S. Hormesis effects of gamma radiation on growth of quinoa (Chenopodium quinoa) Int. J. Radiat. Biol. 2021;97:906–915. doi: 10.1080/09553002.2021.1919783. [DOI] [PubMed] [Google Scholar]
- 24.Komura S., Jinno H., Sonoda T., Oono Y., Handa H., Takumi S., Yoshida K., Kobayashi F. Genome sequencing-based coverage analyses facilitate high-resolution detection of deletions linked to phenotypes of gamma-irradiated wheat mutants. BMC Genomic. 2022;23:111. doi: 10.1186/s12864-022-08344-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hase Y., Satoh K., Seito H., Oono Y. Genetic Consequences of Acute/Chronic Gamma and Carbon Ion Irradiation of Arabidopsis thaliana. Front. Plant Sci. 2020;11:336. doi: 10.3389/fpls.2020.00336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Naito K., Kusaba M., Shikazono N., Takano T., Tanaka A., Tanisaka T., Nishimura M. Transmissible and Nontransmissible Mutations Induced by Irradiating Arabidopsis thaliana Pollen With γ-Rays and Carbon Ions. Genetics. 2005;169:881–889. doi: 10.1534/genetics.104.033654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spencer-Lopes M.M., Forster B.P., Jankuloski L., editors. Manual on Mutation Breeding. FAO; Vienna, Austria: IAEA; Vienna, Austria: 2018. [Google Scholar]
- 28.Monroe J.G., Srikant T., Carbonell-Bejerano P., Becker C., Lensink M., Exposito-Alonso M., Klein M., Hildebrandt J., Neumann M., Kliebenstein D., et al. Mutation bias reflects natural selection in Arabidopsis thaliana. Nature. 2022;602:101–105. doi: 10.1038/s41586-021-04269-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu Z., Cui J., Wang L., Teng N., Zhang S., Lam H.-M., Zhu Y., Xiao S., Ke W., Lin J., et al. Genome-wide DNA mutations in Arabidopsis plants after multigenerational exposure to high temperatures. Genome Biol. 2021;22:160. doi: 10.1186/s13059-021-02381-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jo Y.D., Kim J.-B. Frequency and Spectrum of Radiation-Induced Mutations Revealed by Whole-Genome Sequencing Analyses of Plants. Quantum Beam Sci. 2019;3:7. doi: 10.3390/qubs3020007. [DOI] [Google Scholar]
- 31.Ramsden D.A., Nussenzweig A. Mechanisms driving chromosomal translocations: Lost in time and space. Oncogene. 2021;40:4263–4270. doi: 10.1038/s41388-021-01856-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liao Z., Zhang X., Zhang S., Lin Z., Zhang X., Ming R. Structural variations in papaya genomes. BMC Genomic. 2021;22:335. doi: 10.1186/s12864-021-07665-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Songsomboon K., Brenton Z., Heuser J., Kresovich S., Shakoor N., Mockler T., Cooper E.A. Genomic patterns of structural variation among diverse genotypes of Sorghum bicolor and a potential role for deletions in local adaptation. G3 Genes Genomes Genet. 2021;11:jkab154. doi: 10.1093/g3journal/jkab154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y., Xiong G., Hu J., Jiang L., Yu H., Xu J., Fang Y., Zeng L., Xu E., Xu J., et al. Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nat. Genet. 2015;47:944–948. doi: 10.1038/ng.3346. [DOI] [PubMed] [Google Scholar]
- 35.Fuentes R.R., Chebotarov D., Duitama J., Smith S., De la Hoz J.F., Mohiyuddin M., Wing R.A., McNally K.L., Tatarinova T., Grigoriev A., et al. Structural variants in 3000 rice genomes. Genome Res. 2019;29:870–880. doi: 10.1101/gr.241240.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krasileva K.V. The role of transposable elements and DNA damage repair mechanisms in gene duplications and gene fusions in plant genomes. Curr. Opin. Plant Biol. 2019;48:18–25. doi: 10.1016/j.pbi.2019.01.004. [DOI] [PubMed] [Google Scholar]
- 37.Laanen P., Saenen E., Mysara M., Van de Walle J., Van Hees M., Nauts R., van Nieuwerburgh F., Voorspoels S., Jacobs G., Cuypers A., et al. Changes in DNA Methylation in Arabidopsis thaliana Plants Exposed Over Multiple Generations to Gamma Radiation. Front. Plant Sci. 2021;12:611783. doi: 10.3389/fpls.2021.611783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.West C.E., Waterworth W.M., Jiang Q., Bray C.M. Arabidopsis DNA ligase IV is induced by gamma-irradiation and interacts with an Arabidopsis homologue of the double strand break repair protein XRCC4. Plant J. 2000;24:67–78. doi: 10.1046/j.1365-313x.2000.00856.x. [DOI] [PubMed] [Google Scholar]
- 39.Knudsen S., Wendt T., Dockter C., Thomsen H.C., Rasmussen M., Jørgensen M.E., Lu Q., Voss C., Murozuka E., Østerberg J.T., et al. FIND-IT: Ultrafast mining of genome diversity. bioRxiv. 2021:bioRxiv:444969. doi: 10.1101/2021.05.20.444969. [DOI] [Google Scholar]
- 40.Abe A., Kosugi S., Yoshida K., Natsume S., Takagi H., Kanzaki H., Matsumura H., Yoshida K., Mitsuoka C., Tamiru M., et al. Genome sequencing reveals agronomically important loci in rice using MutMap. Nat. Biotechnol. 2012;30:174–178. doi: 10.1038/nbt.2095. [DOI] [PubMed] [Google Scholar]
- 41.Wang L., Ji Y., Hu Y., Hu H., Jia X., Jiang M., Zhang X., Zhao L., Zhang Y., Jia Y., et al. The architecture of intra-organism mutation rate variation in plants. PLoS Biol. 2019;17:e3000191. doi: 10.1371/journal.pbio.3000191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bado S., Forster B.P., Nielen S., Ali A.M., Lagoda P.J.L., Till B.J., Laimer M. Plant mutation breeding: Current progress and future assessment. Plant Breed. Rev. 2015;39:23–88. [Google Scholar]
- 43.Bado S., Forster B.P., Ghanim A.M.A., Jankowicz-Cieslak J., Berthold G., Luxiang L. Protocol for Screening for Salt Tolerance in Rice. In: Bado S., Forster B.P., Ghanim A.M.A., Jankowicz-Cieslak J., Berthold G., Luxiang L., editors. Protocols for Pre-Field Screening of Mutants for Salt Tolerance in Rice, Wheat and Barley. Springer International Publishing; Cham, Switzerland: 2016. pp. 21–31. [Google Scholar]
- 44.Utilising NIRS for Qualitative and Non-Destructive Identification of Seed Mutants in Large Populations. 2021. [(accessed on 23 March 2022)]. Available online: https://www.iaea.org/resources/book/utilising-nirs-for-qualitative-and-non-destructive-identification-of-seed-mutants-in-large-populations.
- 45.FASTQC A Quality Control Tool for High Throughput Sequence Data. BibSonomy. [(accessed on 17 March 2022)]. Available online: https://www.bibsonomy.org/bibtex/f230a919c34360709aa298734d63dca3.
- 46.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scaling Accurate Genetic Variant Discovery to Tens of Thousands of Samples. bioRxiv. [(accessed on 17 March 2022)]. Available online: https://www.biorxiv.org/content/10.1101/201178v3.
- 49.Alexandrov N., Tai S., Wang W., Mansueto L., Palis K., Fuentes R.R., Ulat V.J., Chebotarov D., Zhang G., Li Z., et al. SNP-Seek database of SNPs derived from 3000 rice genomes. Nucleic Acids Res. 2015;43:D1023–D1027. doi: 10.1093/nar/gku1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cingolani P., Platts A., Wang L.L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Untergasser A., Cutcutache I., Koressaar T., Ye J., Faircloth B.C., Remm M., Rozen S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fan X., Abbott T.E., Larson D., Chen K. BreakDancer-Identification of Genomic Structural Variation from Paired-End Read Mapping. Curr. Protoc. Bioinforma. 2014;45:bi1506s45. doi: 10.1002/0471250953.bi1506s45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Layer R.M., Chiang C., Quinlan A.R., Hall I.M. LUMPY: A probabilistic framework for structural variant discovery. Genome Biol. 2014;15:R84. doi: 10.1186/gb-2014-15-6-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Henry I.M., Zinkgraf M.S., Groover A.T., Comai L. A System for Dosage-Based Functional Genomics in Poplar. Plant Cell. 2015;27:2370–2383. doi: 10.1105/tpc.15.00349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen X., Schulz-Trieglaff O., Shaw R., Barnes B., Schlesinger F., Källberg M., Cox A.J., Kruglyak S., Saunders C.T. Manta: Rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics. 2016;32:1220–1222. doi: 10.1093/bioinformatics/btv710. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing reads have been uploaded to the NCBI database under BioProject ID PRJNA826906 (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA826906, accessed 14 April 2022).