Abstract
Simple Summary
Phylogenomics is an effective method to resolve phylogenetic problems. Herein, the phylogenetic relationships among 12 Sciaenidae species were reconstructed using genome-wide data. Two major phylogenetic groups were distinguished in our study, namely, the Western Atlantic species group, which contained four species, and the Eastern Atlantic, Indian Ocean, and West Pacific species group, which contained eight species. The Western Atlantic species were indicted as the more ancient group in the studied species. Meanwhile, the Eastern Atlantic, Indian Ocean, and West Pacific species were the younger group. However, a close phylogenetic relationship between the Collichthys and Larmichthys genera was not supported.
Abstract
Uncertainty and controversy exist in the phylogenetic status of the Sciaenidae family because of the limited genetic data availability. In this study, a data set of 69,098 bp, covering 309 shared orthologous genes, was extracted from 18 genomes and 5 transcriptomes of 12 species belonging to the Sciaenidae family and used for phylogenetic analysis. The maximum likelihood (ML) and Bayesian approach (BA) methods were used to reconstruct the phylogenetic trees. The resolved ML and BA trees showed similar topology, thus revealing two major evolutionary lineages within the Sciaenidae family, namely, Western Atlantic (WA) and Eastern Atlantic–Indo–West Pacific (EIP). The WA group included four species belonging to four genera: Cynoscion nebulosus, Equetus punctatus, Sciaenops ocellatus, and Micropogonias undulatus. Meanwhile, the EIP group formed one monophyletic clade, harboring eight species (Argyrosomus regius, A. japonicus, Pennahia anea, Nibea albiflora, Miichthys miiuy, Collichthys lucidus, Larimichthys polyactis, and L. crocea) from six genera. Our results indicated that the Western Atlantic (WA) group was more ancient in the studied species, while the Eastern Atlantic–Indo–West Pacific (EIP) group was a younger group. Within the studied species, the genera Collichthys and Larmichthys were the youngest lineages, and we do not suggest that Collichthys and Larmichthys should be considered as one genus. However, the origin of the Sciaenidae family and problems concerning the basal genus were not resolved because of the lack of genomes. Therefore, further sampling and sequencing efforts are needed.
Keywords: phylogenomics, orthologous, Sciaenidae, genome-wide, phylogeny
1. Introduction
The Sciaenidae family, commonly known as croakers or drums, is a unique group of fish that produce characteristic sounds through the sonic muscles and swim bladder. It is considered that the generated sounds may be used by those fish for communication and sex identification during reproduction [1,2]. In addition, the Sciaenidae family includes 68 genera that comprise approximately 293 extant species [3,4]. The members of this family are distributed across temperate and tropical seas worldwide, thus displaying a high level of biodiversity in the Atlantic, Indian, and Western Pacific regions. Moreover, this family harbors some commercially important species in China, such as the large yellow croaker, Larimichthys crocea, and the small yellow croaker, Larimichthys polyactis [5]. Their large economic and scientific values have always attracted great interest from biogeographers in studying the origins, patterns, and phylogeny of Sciaenidae fish [6]. Nevertheless, uncertainty still exists regarding the phylogenetic relationships of the family, primarily because of limitations in the available morphological and genetic data [7].
Thus far, Chao proposed an initial phylogenetic classification of the Sciaenidae family based on the anatomical features of the swim bladder [8]. In turn, Sasaki was the first to discuss the origin of the Sciaenidae family and proposed its monophyly based on morphology. The author also postulated the hypothesis that the origin of Sciaenidae occurred in the New World and diversified via an eastward and/or westward expansion path [9]. However, that hypothesis was merely based on morphological data, thus lacking strong evidence.
Phylogenetic studies based on molecular data have become a hot topic since the 21st century. At first, a limited set of mitochondrial and nuclear genes were used as markers in the phylogeny of Sciaenidae fish [10,11,12,13,14,15,16,17]. However, they may not be reliable because of the introgression, saturation, or selection of mitochondrial genes [18]. In addition, the phylogenetic analysis based on the different genetic markers often results in different evolutionary inferences. For example, Lo et al. performed a comprehensive study based on the cyt b, COⅠ, RAG1, RH, EGR1, and EGR2B genes of Sciaenidae and revealed that the genus Argyrosomus was the basal genus of the Western Pacific species [6]. However, the study based on mtDNA genomes supported that the genus Argyrosomus and the genus Sciaenops formed a sister lineage, whereas the genus Miichthys occupied the basal position within the West Pacific lineage [17]. Therefore, to date, the phylogenetic status of the Sciaenidae family has not been ambiguously resolved, mainly because of the application of the limited amount of genetic data for the study. Therefore, in this study, we summarized three main problems of the Sciaenidae phylogeny that still require solutions: (1) Which genus occupies the basal position within the family? (2) Which genus is the most ancestral within the high-diversified Indo–Western Pacific Sciaenidae group? (3) Should the Collichthys and Larimichthys genera be merged into one genus? Thus far, none of the listed questions have been answered by phylogenomic analysis.
Recently, phylogenomic analysis has become the primary trend to solve problems in phylogenetic studies [19]. The cost of genome sequencing has dramatically reduced along with the development of next-generation sequencing technologies, thereby enabling robust phylogenetic reconstructions based on genome-wide data [20]. Within the past two decades, almost a thousand fish genomes have been sequenced, thus providing vast usable data for large-scale phylogenetic studies [21]. Among plenty of available genomic data, exon genetic information is widely used because of the easy alignment and high level of conservation among species, which makes it particularly robust in phylogenetic studies [22]. For example, Malmstrøm et al. initially established the firm phylogenetic relationships of the teleost and gadiform using a data set of 567 homologous exons from 111 genes; thus, this genomic analysis revealed the evolution of immune genes in the teleost fish [23]. Hughes et al. successfully reconstructed the life-tree of 303 ray-finned fish by applying a total of 1105 single-copy orthologous data sets [7].
Moreover, we applied the available genomic and transcriptomic data to perform a phylogenomic analysis and reconstruct more reliable and comprehensive phylogenetic relationships within the Sciaenidae family. Our phylogenomic study provides a reference to resolve the controversy and origin of the Sciaenidae fish.
2. Materials and Methods
2.1. Data Collection and Transcriptome De Novo Assembly
The genomic data of 12 species from 10 genera of the Sciaenidae family and two outgroup species were collected from the NCBI GeneBank database (SRR5903997, SRR10001351, SRR5997754, SRR12344258, and SRR13555243) (Table 1). Coreoperca whiteheadi (from the GeneBank database) and Sillago sinica [24] were used as the outgroup. The Trimmomatic software (version 0.36) was used to clean the raw RNA-seq data, removing reads with sequencing adapters, unknown nucleotides (N ratio > 10%), and low-quality reads (quality scores < 20) [25]. Meanwhile, the software Trinity (version 2.8.5) [26] was used for the transcriptome de novo assembly of each selected species, and the parameters were as follows: -genome_guided_max_intron 10,000. Then, we used a Perl script (namely “extract_longest_isoforms_from_TrinityFasta.pl”, which can be found in Supplementary Materials) with the default to extract the longest unigene from the resulting Trinity file. The longest unigene will be used for the next phylogenetic study.
Table 1.
Genomic and transcriptome data information for the species examined in this study.
| Scientific Name | Distribution * | Data Type | Total Sequence (Mb) | Contig N50 (bp) | Source |
|---|---|---|---|---|---|
| 1 Sciaenops | |||||
| Sciaenops ocellatus | Western Atlantic | Genome | 648,634 | 40,922 | GCA_14183145.1 |
| 2 Micropogonias | |||||
| Micropogonias undulatus | Western Atlantic | Transcriptome | 157,291 | 1304 | SRR10001351 |
| 3 Equetus | |||||
| Equetus punctatus | Western Atlantic | Transcriptome | 90,036 | 1976 | SRR5997754 |
| 4 Cynoscion | |||||
| Cynoscion nebulosus | Western Atlantic | Transcriptome | 46,823 | 953 | SRR12344258 |
| 5 Argyrosomus | |||||
| Argyrosomus regius | Eastern Atlantic | Transcriptome | 812,965 | 3012 | SRR5903997 |
| Argyrosomus japonicus | Indo–West Pacific | Genome | 79,196 | 1984 | GCA_15710095.1 |
| 6 Pennahia | |||||
| Pennahia anea | Indo–West Pacific | Transcriptome | 812,965 | 2223 | SRR13555243 |
| 7 Nibea | |||||
| Nibea albiflora 1 | Northwest Pacific | Genome | 619,301 | 1212 | GCA_014281875.1 |
| Nibea albiflora 2 | Northwest Pacific | Genome | 574,466 | 34,769 | GCA_900327885.1 |
| Nibea albiflora 3 | Northwest Pacific | Genome | 595,677 | 3712 | GCA_902410095.1 |
| 8 Miichthys | |||||
| Miichthys miiuy | Northwest Pacific | Genome | 627,579 | 20,386 | GCA_001593715.1 |
| 9 Collichthys | |||||
| Collichthys lucidus 1 | Western Atlantic | Genome | 877,615 | 2911 | GCA_004119905.2 |
| Collichthys lucidus 2 | Western Atlantic | Genome | 671,995 | 137,747 | GCA_009852395.1 |
| 10 Larimichthys | |||||
| Larimichthys crocea 1 | Northwest Pacific | Genome | 65,794 | 16,979 | GCA_000972845.1 |
| Larimichthys crocea 2 | Northwest Pacific | Genome | 648,391 | 51,577 | GCA_000742935.1 |
| Larimichthys crocea 3 | Northwest Pacific | Genome | 744,270 | 4085 | GCA_003711585.2 |
| Larimichthys crocea 4 | Northwest Pacific | Genome | 721,257 | 1576 | GCA_003845795.1 |
| Larimichthys crocea 5 | Northwest Pacific | Genome | 7,441,011 | 3905 | GCA_004352675.2 |
| Larimichthys crocea 6 | Northwest Pacific | Genome | 689,173 | 9930 | GCA_90024615.1 |
| Larimichthys polyactis 1 | Northwest Pacific | Genome | 649,447 | 172,294 | GCA_10119295.1 |
| Larimichthys polyactis 2 | Northwest Pacific | Genome | 706,148,513 | 1,208,045 | GCA_18985215.1 |
| Outgroup | |||||
| Sillago sinica | Genome | 536,576 | 2662 | Xu et al. (2018) [25] | |
| Coreoperca whiteheadi | Genome | 712,457 | 76,587 | GCA_011952105.1 |
* Distribution data derived from FishBase: https://www.fishbase.org (accessed on 30 September 2022).
2.2. Exon Captures and Phylogenetic Inference
Sixteen Sciaenidae genomes, five transcriptomes (longest extracted unigene files), and two outgroup genomes were included for the phylogenomic analysis. A data set of 1105 single-copy orthologs (conserved exon markers > 200 bp) by comparing eight well-annotated fish genomes was obtained from Hughes et al. [7]. The gathered data set was subjected to paralogy filtering through gene genealogy interrogation to confirm the orthologous state of each locus. Meanwhile, the collected genomes and unigene files were used to search against this 1105 single-copy orthologous data set using the nHMMER program within the HMMER (v.3.3.2) through the hidden Markov model (HMM) [27]. Subsequently, the identified exons were aligned and concatenated by Perl script (File S1). Gblocks v0.91b was used to eliminate unaligned loci [28]. Furthermore, the ProtTest software (v.3.4.2; Oxford University Press, New York, NY, USA) was used to obtain the best-fitting model for gene alignment, where the best model was BLOSUM62+I+G+F based on both AIC and BIC [29]. For the maximum likelihood (ML) analysis, a total of 1000 bootstrap replicates were performed for all of the concatenated nucleotide sequences GTRGAMMAI model implemented in the RAxML software (v.8.2.12; Oxford University Press, New York, NY, USA) [30]. Then, the Bayesian analysis (BA) was also carried out by Mr. Bayes 3.2.7 under the BLOSUM62+I+G+F model [31]. Hence, 100,000 metropolis-coupled Markov chain Monte Carlo (MCMCMC) generations were applied for this purpose, where every 100th generation was sampled and then the first 25% of the sampled generations were discarded as burn-in.
3. Results
Phylogenetic Analysis of the Studied Species
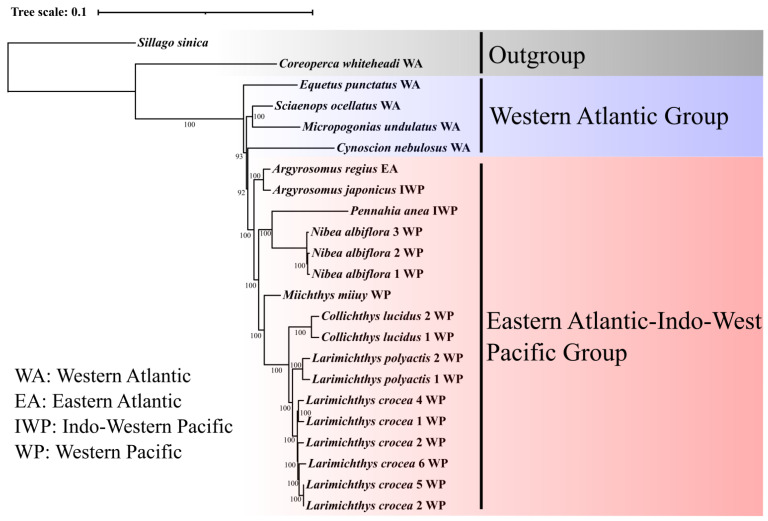
Herein, a filtered, concatenated, and aligned data set of 69,098 bp, covering 309 shared single-copy orthologs, was used for the phylogenetic analyses. The reconstructed ML and BA trees were highly congruent and well supported by high bootstrap pseudo-replicate scores and significant posterior probabilities (>95%) for most nodes (Figure 1 and Figure 2).
Figure 1.
ML tree based on concatenated nucleotide data. Numbers at each node indicate the degree of support (only bootstrap values that are >70% are shown).
Figure 2.
BA tree based on concatenated nucleotide data. Numbers at each node indicate the degree of support (only bootstrap values that are >70% are shown).
In our study, the reconstructed phylogenetic trees indicated the presence of two main evolutionary lineages within the Sciaenidae family, namely: (1) West Atlantic (WA) and (2) Eastern Atlantic–Indo–West Pacific (EIP) cluster, and the former was more ancient on both trees. The WA cluster included four species belonging to four genera: C. nebulosus, E. punctatus, S. ocellatus, and M. undulatus. Within the WA group, the ML tree resolved that a lineage comprising M. undulatus and S. ocellatus has a closer relationship with the genus Equetus. In turn, the BA tree displayed a closer relationship between this lineage and the genus Cynoscion. Both trees indicated that EIP represents a monophyletic lineage that includes congruent phylogenetic topology within the EIP lineage, which was monophyletic and included eight species representing six genera: A. regius, A. japonicus, P. anea, N. albiflora, M. miiuy, C. lucidus, L. polyactis, and L. crocea. Both trees supported close evolutionary relationships between M. undulatus and S. ocellatus. Six L. crocea individuals were well separated as a single distinct lineage in the topology of both resolved trees.
4. Discussion
4.1. Representativeness of Studied Species
Herein, we collected all available genome-wide data on Sciaenidae to reconstruct reliable and comprehensive phylogenetic relationships within the Sciaenidae family. Although only 12 species that belong to 10 genera were included, our sampling strategy well represented the high diversity of Sciaenidae. First, our taxa sampling covered the main habitat of Sciaenidae as the studied species were distributed from tropical to subfrigid zones, including the Atlantic, Indian Ocean, and Western Pacific (Table 1). Widely distributed species such as A. regius and regional species such as L. crocea examined in our study were also included [32,33]. Finally, the studied species represent large behavioral and morphological diversities regardless of living in a homogeneous habitat. For example, two sibling species in the Western Pacific, L. crocea and L. polyactis, had large differences in their behavior such as spawning season and migratory route [33,34]. Although most representative species of the Sciaenidae family were included in our study, more genomes are still required for further investigation.
4.2. Phylogeny of Sciaenidae
For the first time in this study, we reconstructed the phylogenetic relationship within the Sciaenidae family based on genome-wide data. The results of our study indicate that the Sciaenidae family is represented by two main phylogenetic lineages, namely: (1) Western Atlantic (WA) and (2) Eastern Atlantic–Indo–West Pacific (EIP) clusters. The WA phylogenetic lineage resolved in our study was represented by four species belonging to four genera, namely: C. nebulosus, E. punctatus, S. ocellatus, and M. undulates. Herein, the sister relationship between the genera Sciaenops and Micropogonias was revealed, which is concordant with the previous results reported by Lo et al. based on the analysis of the multi-gene data set of several Sciaenidae fish species [6]. However, our study did not provide an answer as to which genus represents the basal genus within the WA cluster because of the non-correspondence in the resolved ML and BA topologies, which is most probably caused by the limited taxa sampling. In addition, the Western Atlantic Sciaenidae species were supported as the more ancient group, which conformed to the fossil evidence [35,36]. However, we did not resolve the problems concerning the origin of Sciaenidae due to the lack of samples. Thus, further studies should focus on the Western Atlantic species with the application of a larger number of genes for the comprehensive phylogeny reconstruction of this lineage.
The EIP phylogenetic lineage was represented by eight species belonging to six genera, namely: Argyrosomus regius, A. japonicus, Pennahia anea, Nibea albiflora, Miichthys miiuy, Collichthys lucidus, Larimichthys polyactis, and L. crocea. ML and Bayesian trees suggested the same phylogenetic relationships. Based on our results, the genus Argyrosomus was more ancient within the EIP clade. However, our results did not resolve the lack of samples for the problem of the basal genus of the EIP group. The study based on the multi-gene dataset indicated that the genus Totoaba was more ancestral to this clade, and the genus Argyrosomus branched after it [6]. Moreover, our study also indicated the close evolutionary relationships between Miichthys, Collichthys, and Larimichthys genera, wherein all included species from those genera formed one monophyletic lineage within the EIP cluster. However, the phylogenetic analysis we carried out did not support previous studies based on analysis of mitochondrial COI, cyt b, and 16S rRNA genes, which suggested that Collichthys and Larmichthys should be considered as one genus [37]. Nevertheless, other species from the genus Collichthys (like C. niveatus) were not included in our study because of the lack of genomes. Therefore, more sequencing efforts are needed in further phylogenetic analysis to confirm this conclusion and resolve the remaining problems.
5. Conclusions
In this study, genome-wide data were used in the phylogenetic analysis of the Sciaenidae family. The results indicated that the Sciaenidae family harbors two major evolutionary lineages: (1) the Western Atlantic (WA) and (2) the Eastern Atlantic and Indo–Western Pacific (EIP) lineages. Thus, the results of our phylogenetic study provide a reference to resolve the existing controversy of the Sciaenidae family: (1) the Western Atlantic species is the more ancient group of the family Sciaenidae; however, the origin and the specific basal genus were not confirmed. (2) The genus Argyrosomus is the more ancient genus of the studied Indo–Western Pacific Sciaenidae species; however, the problem concerning the basal genus was not resolved because of the lack of samples. (3) The genera Collichthys and Larimichthys should not be merged into one genus. Despite our study unraveling phylogenetic relationships among the fish belonging to the Sciaenidae family, some shortages remain in the overall evolutionary picture of the family because of the gaps in the available data. Therefore, further taxonomic sampling and sequencing efforts are needed to determine more comprehensive phylogenetic relationships within the Sciaenidae family.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ani12233386/s1. File S1. Alignment and concatenation Perl script, namely, fastas2aligned_by_mafft.txt and orthomcl_findSingleCopyOrtholog.txt. File S2. A detailed protocol of using the 1105 single-copy orthologous data set to reconstruct the phylogenetic relationships of fish, namely, detailed_protocol.txt.
Author Contributions
X.H. performed the data analysis and article writing. S.J. performed the article writing. T.G. reviewed and made corrections to the manuscript. Z.H. designed the study and reviewed the manuscript. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This study is supported by the National Natural Science Foundation of China (No. 41976083), the Zhejiang Provincial Natural Science Foundation of China (No. LR21D060003) and the National Key Research and Development Program of China (No. 2017YFA0604904).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ramcharitar J., Gannon D.P., Popper A.N. Bioacoustics of fishes of the family Sciaenidae (croakers and drums) Trans. Am. Fish. Soc. 2006;135:1409–1431. doi: 10.1577/T05-207.1. [DOI] [Google Scholar]
- 2.Mok H.K., Yu H., Ueng J.P., Wei R. Characterization of sounds of the blackspotted croaker Protonibea diacanthus (Sciaenidae) and localization of its spawning sites in estuarine coastal waters of Taiwan. Zool. Stud. 2009;48:325–333. [Google Scholar]
- 3.Nelson J.S. Fishes of the World. 4th ed. John Wiley and Sons; New York, NY, USA: 2006. [Google Scholar]
- 4.Froese R., Pauly D. FishBase. World Wide Web Electronic Publication. 2022. [(accessed on 30 September 2022)]. Available online: http://www.fishbase.org.
- 5.National Bureau of Statistics of China, editor. China Fishery Statistical Yearbook. Ministry of Agriculture and Rural Affairs of the People’s Republic of China, National Fisheries Technology Extension Center, China Society of Fisheries; Beijing, China: 2020. [Google Scholar]
- 6.Lo P., Liu S., Chao N., Nunoo F.K.E., Mok H.K., Chen W. A multi-gene dataset reveals a tropical New World origin and early miocene diversification of croakers (Perciformes: Sciaenidae) Mol. Phylogenet. Evol. 2015;88:132–143. doi: 10.1016/j.ympev.2015.03.025. [DOI] [PubMed] [Google Scholar]
- 7.Hughes L.C., Ortí G., Huang Y., Sun Y., Baldwin C.C., Thompson A.W., Arcila D., Beetancur-R R., Li C., Becker L., et al. Comprehensive phylogeny of ray-finned fishes (Actinopterygii) based on transcriptomes and genomic Data. Proc. Natl. Acad. Sci. USA. 2018;115:6249–6254. doi: 10.1073/pnas.1719358115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chao N.L. A Basis for Classifying Western Atlantic Sciaendiae (Teleostei: Perciformes) Department of Commerce, National Oceanic and Atmospheric Administration, National Marine Fisheries Service; Silver Spring, MD, USA: 1978. pp. 1–65. NOAA Technical Report Circular-415. [Google Scholar]
- 9.Sasaki K. Phylogeny of the family Sciaenidae, with notes on its zoogeography (Teleostei, Perciformes) Mem. Fac. Fish. Hokkaido Univ. 1989;36:1–137. [Google Scholar]
- 10.Vinson C., Gomes G., Schneider H., Sampaio I. Sciaenidae fish of the Caeté River estuary, northern Brazil: Mitochondrial DNA suggests explosive radiation for the western Atlantic assemblage. Genet. Mol. Biol. 2004;27:174–180. doi: 10.1590/S1415-47572004000200008. [DOI] [Google Scholar]
- 11.Vergara-Chen C., Aguirre W.E., González-Wangüemert M., Bermingham E. A mitochondrial DNA based phylogeny of weakfish species of the Cynoscion group (Pisces: Sciaenidae) Mol. Phylogenet. Evol. 2009;53:602–607. doi: 10.1016/j.ympev.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 12.Cooke G.M., Chao N.L., Beheregaray L.B. Marine incursions, cryptic species and ecological diversification in Amazonia: The biogeographic history of the croaker Genus Plagioscion (Sciaenidae) J. Biogeogr. 2012;39:724–738. doi: 10.1111/j.1365-2699.2011.02635.x. [DOI] [Google Scholar]
- 13.Santos I., Gomes M., Ferreira A., Sampaio I., Schneider H. Molecular phylogeny of the western south Atlantic Sciaenidae based on mitochondrial and nuclear data. Mol. Phylogenet. Evol. 2013;66:423–428. doi: 10.1016/j.ympev.2012.09.020. [DOI] [PubMed] [Google Scholar]
- 14.Barbosa A.J.B., Sampai I., Schneider H., Santos S. Molecular phylogeny of weakfish species of the Stellifer group (Sciaenidae, Perciformes) of the Western South Atlantic based on mitochondrial and nuclear Data. PLoS ONE. 2014;9:e102250. doi: 10.1371/journal.pone.0102250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boeger W.A., Flávio M., Marteleto L.Z., Braga M.P. Tracking the history of an invasion: The freshwater croakers (Teleostei: Sciaenidae) in south America. Zool. Scr. 2014;44:250–262. doi: 10.1111/zsc.12098. [DOI] [Google Scholar]
- 16.Li W., Huang H., Lin X., Chen B. Molecular phylogeny of the Sciaenidae based on the complete mitochondrial sequences. Mitochondrial DNA B. 2018;3:1180–1182. doi: 10.1080/23802359.2018.1524719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wen H., Ma X., Xu P., Cao Z., Zheng B., Hua D., Jin W., Sun G.M., Gu R. Structure and evolution of the complete mitochondrial genome of the freshwater drum, Aplodinotus grunniens (Actinopterygii: Perciformes: Sciaenidae) Acta Ichthyol. Piscat. 2020;50:23–35. doi: 10.3750/AIEP/02701. [DOI] [Google Scholar]
- 18.Funk D.J., Omland K.E. Species-level paraphyly and polyphyly: Frequency, causes, and consequences, with insights from animal mitochondrial DNA. Annu. Rev. Ecol. Evol. Syst. 2003;34:397–423. doi: 10.1146/annurev.ecolsys.34.011802.132421. [DOI] [Google Scholar]
- 19.Betancur-R R., Arcila D., Vari R.P., Hughes L.C., Oliveira C., Sabaj M.H., Ortí G. Phylogenomic incongruence, hypothesis testing, and taxonomic sampling: The monophyly of Characiform Fishes. Evolution. 2019;73:329–345. doi: 10.1111/evo.13649. [DOI] [PubMed] [Google Scholar]
- 20.Jarvis E.D., Mirarab S., Aberer A.J., Li B., Houde P., Li C., Ho S., Faircloth B.C., Nabholz B., Howard J.T., et al. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science. 2014;346:1320–1331. doi: 10.1126/science.1253451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Faircloth B.C. Ultraconserved elements anchor thousands of genetic markers spanning multiple evolutionary timescales. Syst. Biol. 2012;6:717–726. doi: 10.1093/sysbio/sys004. [DOI] [PubMed] [Google Scholar]
- 22.Jiang J., Yuan H., Zheng X., Wang Q., Kuang T., Li J., Liu J., Song S., Wang W., Cheng F., et al. Gene markers for exon capture and phylogenomics in ray-finned fishes. Ecol. Evol. 2019;9:3973–3983. doi: 10.1002/ece3.5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malmstrøm M., Matschiner M., Tørresen O.K., Star B., Snipen L.G., Hansen T.F., Baalsrud H.T., Nederbragt A.J., Hanel R., Salzburger W., et al. Evolution of the immune system influences speciation rates in Teleost fishes. Nat. Genet. 2016;48:1204–1210. doi: 10.1038/ng.3645. [DOI] [PubMed] [Google Scholar]
- 24.Xu S., Xiao S., Zhu S., Zeng X., Jing L., Liu J., Gao T., Chen N. A draft genome assembly of the Chinese sillago (Sillago sinica), the first reference genome for Sillaginidae fishes. GigaScience. 2018;7:giy108. doi: 10.1093/gigascience/giy108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anthony M.B., Marc L., Bjoern U. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grabherr M.G., Haas B.J., Yassour M., Levin J.Z., Thompson D.A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q., et al. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jaina M., Finn R.D., Eddy S.R., Bateman A., Punta M. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 2013;41:e121. doi: 10.1093/nar/gkt263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 29.Abascal F., Zardoya R., Posada D. Prottest: Selection of best-fit models of orotein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- 30.Alexandros S. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ronquist F., Huelsenbeck J.P. MRBAYES: Bayesian inference of phylogeny. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 32.Klimogianni A., Pagoulatou M.T., Maria H., George N. Investigation on early development, the feeding ability and larval survival under starvation in common meagre, Argyrosomus regius (Asso, 1801) J. Aquat. Sci. 2013;1:1–6. [Google Scholar]
- 33.Xu Z., Chen J. Analysis on migratory routine of Larimichthy crocea in the East China Sea and Yellow Sea. J. Fish. Sci. China. 2011;35:429–437. [Google Scholar]
- 34.Xu Z., Chen J. Analysis on migratory routine of Larimichthy polyactis. J. Fish. Sci. China. 2009;16:931–940. [Google Scholar]
- 35.Nolf D. Studies on Fossil OTOLITHS—The State of the Art. In: Secor D.H., Dean J.M., Campana S.E., editors. Recent Developments in Fish Otolith Research. University of South Carolina; Columbia, SC, USA: 1995. pp. 513–544. [Google Scholar]
- 36.Bannikov A.F., Carnevale G., Landini W. A new early Miocene genus of the family Sciaenidae (Teleostei, Perciformes) from the eastern paratethys. C. R. Palevol. 2009;8:535–544. doi: 10.1016/j.crpv.2009.03.001. [DOI] [Google Scholar]
- 37.Xu T., Cheng Y., Sun Y., Shi G., Wang R. The complete mitochondrial genome of bighead croaker, Collichthys niveatus (Perciformes, Sciaenidae): Structure of control region and phylogenetic considerations. Mol. Biol. Rep. 2011;38:4673–4685. doi: 10.1007/s11033-010-0602-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.