Abstract
Microsporidia are intracellular organisms of increasing importance as opportunistic pathogens in immunocompromised patients. Host cells are infected by the extrusion and injection of polar tubes located within spores. The spore is surrounded by a rigid spore wall which, in addition to providing mechanical resistance, might be involved in host cell recognition and initiation of the infection process. A 51-kDa outer spore wall protein was identified in Encephalitozoon cuniculi with the aid of a monoclonal antibody, and the corresponding gene, SWP1, was cloned by immunoscreening of a cDNA expression library. The cDNA encodes a protein of 450 amino acids which displays no significant similarities to known proteins in databases. The carboxy-terminal region consists of five tandemly arranged glycine- and serine-rich repetitive elements. SWP1 is a single-copy gene that is also present in the genomes of Encephalitozoon intestinalis and Encephalitozoon hellem as demonstrated by Southern analysis. Indirect immunofluorescence and immunoelectron microscopy revealed that SWP1 is differentially expressed during the infection cycle. The protein is absent in replicative meronts until 24 h postinfection, and its expression is first induced in early sporonts at a time when organisms translocate from the periphery to the center of the parasitophorous vacuole. Expression of SWP1 appears to be regulated at the mRNA level, as was shown by reverse transcriptase PCR analysis. Further identification and characterization of stage-specific genes might help to unravel the complex intracellular differentiation process of microsporidia.
Microsporidia are obligately intracellular organisms that are able to infect numerous vertebrate and invertebrate species (3). Several genera of the phylum Microsporidia (i.e., Encephalitozoon, Enterocytozoon, and Septata) have been recognized as important opportunistic pathogens in humans, particularly in patients infected with the human immunodeficiency virus (22). Recently performed phylogenetic analysis suggested that microsporidia are related to fungi rather than being early eucaryotes (5, 9, 10, 12, 13). The infective stage of microsporidia is the spore, which has an unique, phylum-specific mechanism for infecting host cells. A filamentous tube, coiled within the interior of the spore and attached to a structure termed the anterior disk, is explosively extruded after an appropriate stimulus (7). If the tube penetrates the plasma membrane of an adjacent host cell, the sporoplasm is translocated through the hollow tube into the host cell while the spore wall and tube remain extracellular (15, 23). The intracellular life cycle can be divided into two phases: merogony and sporogony. Meronts are morphologically simple cells which are limited by a unit membrane and replicate by binary or multiple fission. Spore wall formation is initiated early in sporogony by deposition of electron-dense material at the plasma membrane and continues while the organisms differentiate into sporoblasts and finally into mature spores (3). The rigid spore wall protects the sporoplasm of mature spores against environmental stress and permits long-term survival after release from host cells. In addition, the rigid spore wall prevents the sporoplasm from expanding when the internal pressure necessary for extrusion of the polar tube is generated (7). Two layers of the spore wall can be distinguished ultrastructurally: an outer electron-dense layer of 25 to 30 nm (the exospore) and an inner electron-lucent layer of 30 to 35 nm (the endospore), which is believed to contain polysaccharides, particularly chitin (6, 19). Using electron microscopy of thin sections and freeze-fracture techniques, the exospore of Encephalitozoon hellem was recently described to be composed of an outer spinny layer, an electron-lucent intermediate lamina, and an inner fibrous layer (2). There are hints that the spore surface, besides providing mechanical protection, is involved in the initiation of polar tube extrusion and that modifications of the spore wall architecture occur during this activation (24, 25). Electron microscopy revealed that the outer spore envelope of Spraguea lophii and Thelohania sp. completely disassembles at the time of spore activation (24, 25). Using antikeratin antibodies it was demonstrated that the outer spore wall of Thelohania sp. consists in part of keratin-like proteins that form 10-nm intermediate filaments which become phosphorylated and disassemble during spore activation (25). It has been possible to distinguish subcompartments within the spore wall using polyclonal antisera against partially purified microsporidial proteins. A 30-kDa antigen was found to be located on the outer spore wall, while a 33-kDa protein was found in a region close to the plasma membrane (4). In addition, several monoclonal antibodies (MAbs) have been reported in previous studies to recognize spore wall antigens (1, 16, 21). However, identification of the corresponding genes has not been reported so far. In this work we describe for the first time the cloning and characterization of a gene encoding a spore wall protein and demonstrate that it is differentially regulated during the intracellular life cycle of Encephalitozoon cuniculi.
MATERIALS AND METHODS
Cultivation of microsporidia.
E. cuniculi, E. hellem, and Encephalitozoon intestinalis were routinely propagated in tissue culture with human foreskin fibroblasts (HFF) as host cells. The culture medium consisted of Dulbecco's modified Eagle medium (DMEM) supplemented with 1% fetal calf serum and penicillin-streptomycin. Microsporidia were passaged by scraping off an infected monolayer 4 to 8 days postinfection and passing the suspension through a 26-gauge needle in order to disrupt intact host cells and release microsporidia from the parasitophorous vacuoles. The microsporidial suspension was then used to infect a new HFF monolayer.
MAbs.
A panel of mouse anti-E. cuniculi MAbs was obtained by immunization of BALB/c mice with E. cuniculi lysate and subsequent fusion of spleen cells with Ag 8.653 myeloma cells. Hybridoma supernatants were tested on methanol-fixed E. cuniculi spores by indirect immunofluorescence and in parallel by immunoblotting on E. cuniculi lysate. Positive hybridomas were subcloned in 96-well plates. The isotype of MAb 11A1 is immunoglobulin G1 (IgG1).
cDNA cloning.
Total RNA was isolated, using the RNeasy Kit (Qiagen), from two partly lysed T-75 HFF tissue culture flasks that were heavily infected with E. cuniculi. RNA was treated with DNase, phenol-chloroform extracted, and ethanol precipitated. Poly(A) RNA was isolated with Dynabeads (Dynal) according to the manufacturer's protocol. cDNA synthesis and cloning into the ZAP Express vector (Stratagene) were performed according to the instruction manual. A total of 2.5 × 105 PFU was obtained. Ten randomly picked clones had insert sizes between 0.4 and 2 kb. The cDNA library was amplified and immunoscreened with a hybridoma supernatant of MAb 11A1, diluted 1:10 in Tris-buffered saline–1% bovine serum albumin (BSA). Positive clones were subjected to “in vivo excision” according to the manufacturer's protocol (Stratagene).
Computational analysis.
The LaserGene Software Package (DNASTAR) was used for sequence data alignment and cDNA analysis. Searches for DNA and protein homologies were performed with BLAST (http://www.ncbi.nlm.nih.gov/). A search for protein motifs was done in PROSITE. The SignalP program (http://www.cbs.dtu.dk/services/SignalP/) was used for prediction of the peptide leader cleavage site. Scanning for transmembrane regions was performed with TMpred (http://www.isrec.isb-sib.ch/software/TMPRED_form.html).
5′ rapid amplification of cDNA ends (5′ RACE).
Total RNA was isolated, using the RNeasy Kit (Qiagen), from two partly lysed T-75 HFF tissue culture flasks that had been heavily infected with E. cuniculi 3 days before. SWP1 mRNA was reverse transcribed with the SWP1 antisense primer 11A1-P5 (5′-TTGGTTTGCAGTCAGAAGAGGAA-3′), located 213 nucleotides (nt) downstream of the ATG start codon, and 200 U of Superscript reverse transcriptase (RT) (BRL) in a total volume of 20 μl for 15 min at 37°C. QuickSpin (Qiagen)-purified cDNA was dA tailed with 30 U of terminal transferase (Boehringer) for 5 min at 37°C in the presence of 0.2 mM dATP. The reaction was stopped by heating at 65°C for 5 min, followed by QuickSpin purification. Double-stranded cDNA was generated from 20% of the eluate utilizing a poly(T) adapter (8) followed by 30 PCR cycles with the nested SWP1 antisense primer 11A1-P6 (5′-ACATCACTGCTCATTCCGTCAC-3′) located 164 nt downstream of the ATG start codon and an adapter primer (8). PCR conditions were as follows: 94°C for 30 s, 55°C for 1 min, and 72°C for 1 min. A PCR product of 250 nt was obtained and cloned into the T/A cloning vector (Invitrogen). Eight clones were sequenced with the vector-specific primer M13.
RT-PCR analysis.
Extracellular spores were obtained from the supernatant of partly lysed E. cuniculi cultures and passed through a 26-gauge needle to rupture remaining intact host cells. A synchronous infection of HFF was obtained by allowing the spores to infect an HFF monolayer for 3 h, followed by extensive washing with DMEM in order to remove residual extracellular spores. Total RNA was isolated from two T-25 flasks at 24, 48, and 72 h postinfection. RNA was treated with 10 U of RNase-free DNase for 1 h at 37°C, followed by phenol-chloroform extraction and ethanol precipitation. Poly(A) RNA was isolated with Dynabeads (Dynal) according to the manufacturer's protocol and reverse transcribed with 200 U of Superscript RT (BRL) for 1 h at 37°C. A control sample without RT was incubated in parallel. Samples were adjusted to 100 μl with H2O and used for PCR in fivefold dilutions starting with 0.5 μl/PCR. Tubulin was amplified using primer pairs E.c.Tub+ (5′-CTACAGGGGTTTCAGATTACACAT-3′) and E.c.Tub− (5′-ACAAGGGAGACAAGGTGGTTC-3′). Volumes of cDNA samples were adjusted to give tubulin bands of comparable intensities, and normalized samples were subjected to PCR with SWP1-specific primer pairs sense-4+ (5′-TCGCCAGAGTCAACACAATG-3′) and antisense-10− (5′-TAGCATGTGGGTCGTACATCTAACCAC-3′). PCR conditions for tubulin and SWP1 were as follows: 25 cycles of 94°C for 30 s, 55°C for 1 min, and 72°C for 1 min. No PCR products were obtained in the control sample lacking RT, demonstrating the absence of contaminating genomic DNA. To demonstrate specificity, PCR products were blotted onto Hybond-N+ membranes (Amersham) and hybridized with digoxigenin-dUTP (Boehringer)-labeled tubulin and SWP1 cDNA, respectively. Signals were detected by chemifluorescence on Hyperfilm (Amersham) using CSPD (Boehringer) as a substrate.
Invasion assay.
A total of 106 freshly harvested E. cuniculi spores were incubated with protein G-Sepharose-purified MAb 11A1 in a volume of 0.5 ml of DMEM under permanent rotation for 3 h at room temperature. Final antibody concentrations were 1, 10, and 100 μg/ml. Parasites were centrifuged, washed twice in DMEM, and used to infect an HFF monolayer on cover slides in 24-well plates. Samples were fixed 3 days postinfection and immunostained with a polyclonal rabbit anti-E. cuniculi antiserum as described below under “Immunolabeling.” Numbers of parasitophorous vacuoles per field were determined with an immunofluorescent microscope.
Protein analysis.
Pellets of 7 × 106 E. cuniculi organisms were solubilized in denaturing buffer, separated in by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and blotted onto nitrocellulose membranes. After blocking for 1 h in phosphate-buffered saline (PBS)–1% BSA, the membrane was incubated for 3 h with MAb 11A1 supernatant (1:50 in PBS–1% BSA), followed by three washes for 10 min each. A 1:5,000-diluted alkaline phosphatase-conjugated goat anti-mouse IgG (Dianova) served as a secondary antibody. Reactive bands were visualized using the substrates 5-bromo-4-chloro-3-indolylphosphate and nitroblue tetrazolium. Alkaline hydrolysis of O-linked glycosaminoglycans was performed as previously described (14a). Briefly, the pellet of 7 × 106 E. cuniculi organisms was incubated for 12 h in 0.3 M NaBH4 in 0.4 M NaOH; then 60 μl of 2 M HCl was added to eliminate excess borohydride by converting it to hydrogen gas. The pH was adjusted to pH 6 to 8 with NaOH, and the sample was subjected to immunoblot analysis. For enzymatic digestion of glycosaminoglycan residues, the pellets of freshly harvested E. cuniculi were washed in the appropriate enzyme incubation buffer (14a) and incubated in a volume of 50 μl for 15 h at 37°C with either (i) 5 U of heparinase I, (ii) 0.5 U of heparinase III, or (iii) 0.5 U of chondroitase ABC. All enzymes were purchased from Sigma. A 10-μl aliquot was subjected to immunoblot analysis.
Immunolabeling.
For immunoelectron microscopy, cell cultures infected with E. cuniculi for 1, 4, 7, and 11 days were fixed in 2% paraformaldehyde in 0.1 M phosphate buffer. The cells were then scraped off and centrifuged. The cell pellets were dehydrated and embedded in L R White resin. Thin sections were cut and placed on Formvar-coated nickel grids. For immunolabeling, grids were placed on drops of 1% BSA in Tris buffer to block nonspecific staining, followed by the primary antibody, 11A1 (or 4A5 as a negative control), in Tris buffer, and incubated overnight at 4°C. The grids were then washed and placed on drops of goat anti-mouse IgG conjugated to 10 nm of colloidal gold. After washing, the grids were stained with uranyl acetate prior to examination with a JEOL 1200EX electron microscope.
Indirect immunofluorescence analysis was performed as follows: a synchronous infection with E. cuniculi was obtained by incubation of HFF monolayers with freshly harvested spores for 1 h only, followed by extensive washing in order to remove residual extracellular spores. Samples were fixed with 4% paraformaldehyde–PBS for 10 min and permeabilized with 0.25% Triton X-100–PBS for 15 min. The kinetics of the spore wall antigen expression were analyzed from day 1 to day 4 postinfection by double-immunofluorescence staining using MAb 11A1 and a polyclonal rabbit anti-E. cuniculi antiserum for counterstaining. Samples were incubated sequentially for 1 h with (i) MAb 11A1 hybridoma supernatant, (ii) fluorescein isothiocyanate (FITC)-conjugated anti-mouse IgG (diluted 1:50 in PBS), (iii) polyclonal rabbit anti-E. cuniculi serum (kindly provided by Louis M. Weiss), and (iv) tetramethyl rhodamine isocyanate (TRITC)-conjugated anti-rabbit IgG (diluted 1:50 in PBS). For immunostaining of intact spores, freshly harvested E. cuniculi spores were incubated sequentially for 1 h with (i) MAb 11A1 hybridoma supernatant (1:10 in PBS–1% BSA) and (ii) FITC-conjugated anti-mouse IgG (diluted 1:50 in PBS–1% BSA). A non-E. cuniculi-related IgG of the same isotype served as a control.
Nucleotide sequence accession number.
Sequence data reported in this paper have been submitted to the DDBJ/EMBL/GenBank database under accession number AJ133745.
RESULTS
Identification of a spore wall antigen in E. cuniculi.
A panel of anti-E. cuniculi MAbs was tested by indirect immunofluorescence for reactivity with the outer spore wall of extracellular E. cuniculi spores. MAb 11A1 exhibited strong surface staining on unfixed and paraformaldehyde-fixed spores, suggesting that a spore wall antigen located on the surface was detected. The polar tube of discharged spores was unlabeled, indicating that MAb 11A1 does not cross-react with polar tube proteins (PTPs) (Fig. 1). The MAb 11A1-reactive antigen has an apparent molecular size of 51 kDA in immunoblot analysis under reducing conditions (see Fig. 6).
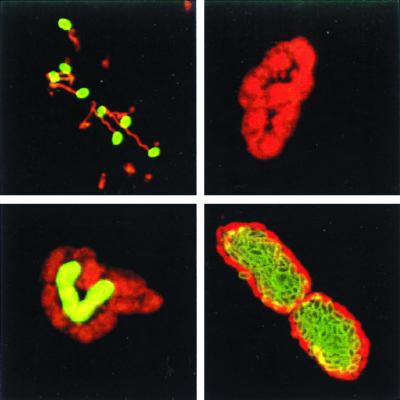
FIG. 1.
MAb 11A1 detects a differentially expressed spore wall antigen. HFF monolayers were infected with E. cuniculi spores and fixed 2 h (top left), 24 h (top right and bottom left), and 48 h (bottom right) postinfection. Double immunostaining was performed with MAb 11A1 detected with an FITC conjugate (green fluorescence), followed by staining with a polyclonal rabbit anti-microsporidia antiserum detected with a rhodamine conjugate (red fluorescence). MAb 11A1 stains a spore wall antigen located on the surfaces of extracellular spores. Expression of the spore wall antigen is first induced after 24 h in a small fraction of organisms located in the center of the vacuole. Most parasitophorous vacuoles contained MAb 11A1-positive E. cuniculi after 48 h. Note that a monolayer of negative organisms is located at the periphery of the vacuole.
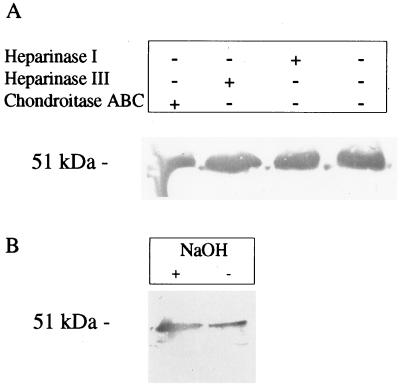
FIG. 6.
SWP1 is not modified by glycosaminoglycans. (A) A total of 7 × 106 freshly harvested E. cuniculi organisms were digested with heparinase I, heparinase III, or chondroitase ABC and subjected to immunoblot analysis using MAb 11A1. (B) The pellet of 7 × 106 E. cuniculi organisms was incubated for 12 h with 0.3 M NaBH4 in 0.4 M NaOH in order to release O-linked glycosaminoglycans by alkaline hydrolysis. Samples were subjected to immunoblot analysis using MAb 11A1. The molecular size of SWP1 from treated samples is identical to that from untreated samples, indicating that glycosaminoglycan residues are not attached to the protein.
Since MAb 11A1 recognizes a surface molecule of E. cuniculi, we tested its ability to inhibit invasion by mature spores. Freshly harvested extracellular spores were incubated with MAb 11A1 at a concentration of 1, 10, or 100 μg/ml for 3 h and used to infect an HFF monolayer. No difference in infectivity was found compared to untreated controls, indicating that incubation of spores with MAb 11A1 has no influence on the infection process.
The spore wall antigen is differentially expressed during intracellular development.
The kinetics of the spore wall antigen expression were monitored during the intracellular development of E. cuniculi by indirect immunofluorescence using MAb 11A1. At 24 h postinfection, host cells contained parasitophorous vacuoles harboring four to eight organisms. At this time >98% of E. cuniculi organisms were MAb 11A1 negative, based on immunofluorescence assay (IFA) analysis; however, a small number of parasitophorous vacuoles harbored a heterogenous population with MAb 11A1-positive and -negative organisms within the same vacuole (Fig. 1). Positively stained E. cuniculi organisms were located in the centers of these vacuoles, while microsporidia at the peripheries were MAb 11A1 negative. At 48 h postinfection, the number of organisms within parasitophorous vacuoles had increased to >20 and most vacuoles (>95%) harbored MAb 11A1-positive organisms. The centers of these vacuoles were completely filled with positively labeled E. cuniculi organisms; however, a unicellular ring of MAb 11A1-negative E. cuniculi organisms was still located at the periphery (Fig. 1). The same staining pattern was observed at 72 h postinfection in parasitophorous vacuoles, which had increased in size and harbored more than 50 organisms.
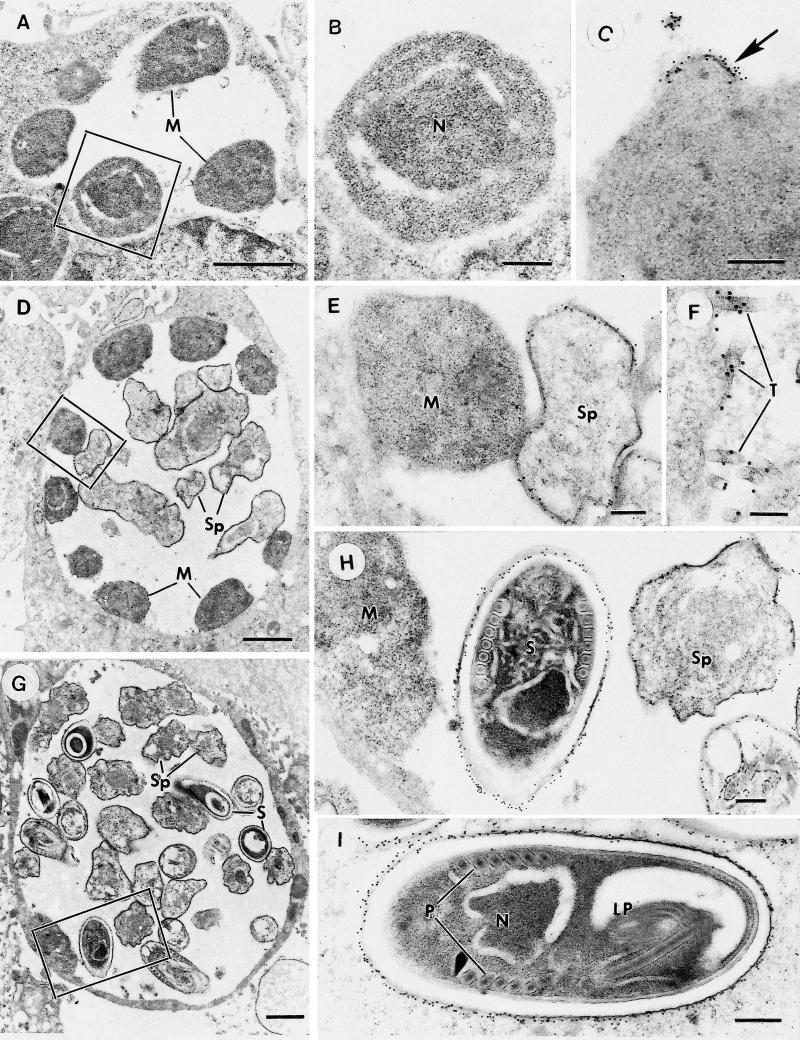
Immunoelectron microscopy was performed on cultures infected with E. cuniculi at 1, 4, 7, and 11 days postinfection. At day 1, vacuoles containing low numbers (one to eight) of meronts, which closely adhered to the membrane of the parasitophorous vacuole and were negative for MAb 11A1, were observed (Fig. 2A and B). At 4 days postinfection, there were numerous parasitophorous vacuoles containing organisms at various stages of development. Some parasitophorous vacuoles contained meronts and early sporonts, while others contained all stages from meronts to mature spores, facilitating the examination of the stage-specific expression of the protein recognized by MAb 11A1. Meronts were still not labeled with 11A1, but at the stage when E. cuniculi detached from the parasitophorous vacuole membrane and the outline of the organisms became more distinct (very early sporonts), there was labeling of the thickened surface membrane (Fig. 2D and E). This was initially localized to certain areas of the plasma membrane which appeared thickened (Fig. 2C). This may represent the earliest deposition of spore wall material, which was recognized by MAb 11A1. The level of surface labeling increased with spore maturation and spore wall formation (Fig. 2G and H). In addition, within the parasitophorous vacuoles a few tubular structures were also stained with MAb 11A1 (Fig. 2F). These tubules were distinct from extruded polar tubes, which were unstained by MAb 11A1. At 7 and 11 days postinfection, the size of the parasitophorous vacuoles and the number of organisms contained was increased. The proportion of mature spores also increased, and by 11 days postinfection, meronts were rarely seen. At both time points, the staining pattern with MAb 11A1 was similar to that described for 4 days postinfection. In addition, increasing numbers of extracellular spores were observed, a proportion of which had discharged their polar tube and contents. No labeling of the extruded polar tube was observed in discharged spores (data not shown). Labeling of mature spores was restricted to the electron-dense exospore, while the electron-lucent endospore remained negative (Fig. 2I).
FIG. 2.
Immunoelectron microscopy of sections through cells infected with E. cuniculi at various stages of development which have been immunostained with MAb 11A1 and labeled with 10-nm gold particles. (A) Low-power micrograph of an early stage of intracellular development showing a parasitophorous vacuole containing peripherally located meronts (M). Bar, 1 μm. (B) Detail of the enclosed area in panel A. Note that the surface of the meront is unlabeled. Bar, 200 nm. (C) Part of a meront-like organism in which a focal area of the surface is strongly labeled (arrow). Note that the label is associated with an area of membrane thickening. Bar, 200 nm. (D) Low-power micrograph of a stage in development slightly later than that shown in panel A. The parasitophorous vacuole contains a few centrally located early sporonts (Sp) plus peripherally located meronts (M). Bar, 1 μm. (E) Enlargement of the enclosed area in panel D, contrasting the strong labeling of the surface of the early sporont (Sp) to the absence of staining of the adjacent meront (M). Bar, 200 nm. (F) Detail of the parasitophorous vacuole showing a number of labeled tubules (T) adjacent to the surface of a sporont. Bar, 100 nm. (G) Low-power micrograph of a cell containing E. cuniculi organisms at a late stage in development, in which the vacuole contains various stages in sporont development (Sp), including mature spores (S) with few remaining meronts. Bar, 1 μm. (H) Enlargement of the enclosed area in panel G, showing the absence of label on the meront (M) surface, while there is strong labeling of the surfaces of the sporont (Sp) and the mature spore (S). Bar, 200 nm. (I) Longitudinal section through a mature spore showing the strong labeling of the outer surface of the spore wall. N, nucleus; LP, lamellar polaroplast; P, polar tube. Bar, 200 nm.
Molecular characterization of the gene encoding the spore wall antigen (SWP1).
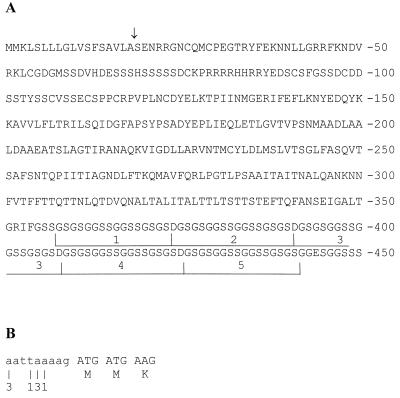
Immunoscreening of an E. cuniculi cDNA library using MAb 11A1 resulted in the isolation of five positive clones with insert sizes ranging from 1.4 to 1.6 kb. Protein lysates of these clones were obtained from isopropyl-1-thio-β-d-galactopyranoside (IPTG)-induced Escherichia coli cultures, separated by SDS-PAGE, and submitted to immunoblotting using MAb 11A1. Depending on the clone analyzed, the molecular sizes of the recombinant LacZ fusion proteins were 46 to 52 kDa, while control lysates did not react with MAb 11A1. DNA sequencing revealed that all clones had identical 3′ ends, including the poly(A) tail, but overlapping 5′ ends of different sizes. Starting with the first in-frame ATG, codon an open reading frame of 1,350 nt predicts a 450-amino-acid (aa) protein of 46 kDa. The coding sequence for SWP1 was submitted to the EMBL database under accession number AJ133745. The N-terminal region displays the typical characteristics of a signal peptide for endoplasmic reticulum (ER) translocation, and based on the “von Heijne” algorithm (17) the cleavage site was predicted between aa 18 (alanine) and aa 19 (serine). SWP1 is rich in glycine (14%) and serine (19%), due to five tandemly arranged repeats located at the carboxy-terminal region with the sequence GSGSGGSSGGSSGSGSD (Fig. 3A). No significant homology (P > 0.14) was found in the GenBank database by using the SWP1 amino acid sequence without the repetitive region (aa 1 to 357) in a BLASTP search. Due to the high glycine content of the carboxy-terminal region, a search with the complete amino acid sequence displayed similarities to glycine-rich proteins such as keratins and loricrins. The hydrophobicity pattern of the mature protein is triphasic: a hydrophilic amino-terminal region (aa 19 to 150) is linked to a moderately hydrophobic core region (aa 151 to 357) followed by the weakly hydrophilic carboxy-terminal region, which consists of the repetitive elements (aa 358 to 450). The cysteine residues are unequally distributed within the primary sequence; eight of the nine cysteines are located near the hydrophilic amino-terminal region within the first 130 aa. A further two in-frame ATG codons are located 81 and 171 nt downstream of the first ATG. However, these are unlikely to represent the start codon because (i) only the first ATG is followed by a hydrophobic von Heijne signal peptide as expected for a secreted protein which is located on the spore surface and (ii) only the first ATG is preceded by an A in the −3 position as is typical for 5′ untranslated regions of protozoa (26). In order to determine the transcription start site of SWP1, 5′ RACE was performed as described in Materials and Methods. Sequencing of eight clones revealed that the start site is located within a stretch of 6 nt immediately upstream from the putative start codon, resulting in an unusual short 5′ untranslated region of 3 to 8 nt (Fig. 3B). A poly(A) signal (AATAAA) was present in the 3′ untranslated region 165 nt downstream of the stop codon. The poly(A) tails of all five cDNA clones started 10 nt downstream of this poly(A) signal.
FIG. 3.
(A) Amino acid sequence of SWP1. The five tandemly arranged glycine- and serine-rich repeats at the carboxy-terminal region are underlined. The putative cleavage site of the signal peptide is indicated by an arrow. (B) 5′ untranslated region of SWP1 (lowercase letters) as determined by 5′ RACE. The start nucleotides of eight sequenced clones are indicated. The number of clones starting with each is given below the nucleotide.
SWP1 homologs in E. intestinalis and E. hellem.
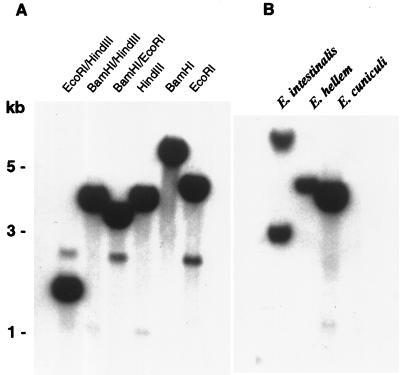
BamHI/HindIII-digested genomic DNA from three Encephalitozoon species (E. cuniculi, E. intestinalis, and E. hellem) was subjected to Southern analysis and hybridized with a labeled SWP1 cDNA fragment from E. cuniculi. Specific bands of 3 to 8 kb were detected in all three species (Fig. 4B), suggesting that SWP1 homologs are also present in the E. intestinalis and E. hellem genomes. However, MAb 11A1 reacted with E. cuniculi in immunofluorescence and in immunoblotting but not with E. intestinalis or E. hellem; thus, the epitope recognized by this MAb appears to be species specific. The hybridization pattern obtained by Southern analysis of E. cuniculi genomic DNA is in accordance with SWP1 being a single-copy gene (Fig. 4A). Weak bands at 1 kb for HindIII and 2.5 kb for EcoRI are due to internal restriction sites within the SWP1 locus. Primers flanking the complete cDNA sequence were used to amplify E. cuniculi genomic DNA and resulted in PCR products of sizes identical to that of the cDNA clone, indicating that introns are absent in SWP1.
FIG. 4.
SWP1 is a single-copy gene present in at least three Encephalitozoon species. (A) Genomic DNA from 1.5 × 108 E. cuniculi organisms was digested with a panel of different restriction enzymes, separated on a 0.7% agarose gel, blotted onto nylon membranes, hybridized with a 32P-labeled SWP1 cDNA fragment, and washed under high-stringency conditions (0.2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]–0.1% SDS at 65°C). The restriction pattern is in accordance with SWP1 being a single-copy gene. (B) Genomic DNA from E. cuniculi, E. hellem, and E. intestinalis was digested with BamHI/HindIII and hybridized as described in panel A, followed by washing under moderate stringency (0.2× SSC–0.1% SDS at 45°C).
The mRNA level of SWP1 is upregulated during differentiation from meronts to sporonts.
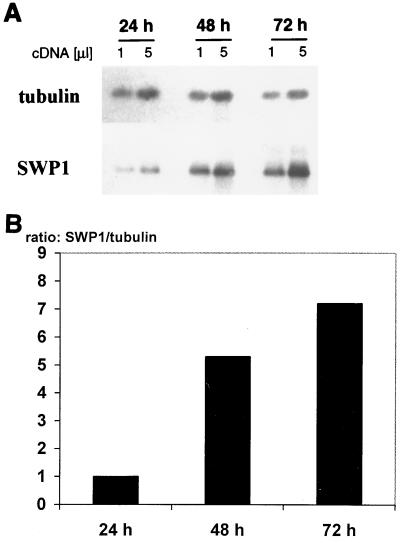
Poly(A) RNA was isolated from HFF cultures infected with E. cuniculi at 24, 48, and 72 h postinfection and analyzed by RT-PCR with gene-specific primers for E. cuniculi beta-tubulin and SWP1 transcripts. Two dilutions were analyzed for each sample in order to demonstrate the linearity of the PCR. Samples normalized for beta-tubulin transcripts showed a fivefold increase in the SWP1 transcript level from 24 to 48 h and a 1.5-fold increase from 48 to 72 h (Fig. 5). The specificity of the SWP1 PCR products was demonstrated by blotting the PCR products onto Hybond membranes and hybridizing with a digoxigenin-labeled SWP1 cDNA fragment. Control PCRs on RNA samples treated in parallel without RT did not reveal any bands, demonstrating the absence of contaminating genomic DNA. These results indicate that the steady-state level of SWP1 mRNA is upregulated during the intracellular life cycle of E. cuniculi.
FIG. 5.
(A) SWP1 mRNA is upregulated during intracellular development. HFF monolayers were infected with E. cuniculi, and mRNA was isolated 24, 48, and 72 h postinfection. The volume of the 48-h sample was diluted 8-fold, and that of the 72-h sample was diluted 20-fold, to normalize for tubulin transcript. RT-PCR was performed with tubulin and SWP1-specific primer pairs on two cDNA amounts. PCR products were blotted onto nylon membranes and hybridized with digoxigenin-labeled tubulin or SWP1 cDNA fragments. The level of SWP1 mRNA was strongly upregulated from 24 to 48 h and moderately increased from 48 to 72 h postinfection. (B) Densitometric quantification of SWP1 RT-PCR products. Densitometric analysis of RT-PCR film exposures was performed using the BioDocII video documentation system (Biometra). Peak areas for SWP1 and tubulin were determined with the ScanPack 3.0 software (Biometra). The ratio of the SWP1 to the tubulin peak area was calculated for each time point of the kinetics; these ratios are expressed in arbitrary units. The amount of SWP1 mRNA, normalized to that of tubulin, increases fivefold from 24 to 48 h and more than sevenfold from 24 to 72 h postinfection.
SWP1 is not modified with O-glycosidic linked glycosaminoglycans.
The glycosaminoglycan attachment motif SGXG (where X represents any amino acid) is present 10 times within the glycine- and serine-rich repetitive region of SWP1. Since modifications with glycosaminoglycan residues are frequently found in extracellular proteins with structural functions, the possible linking of glycosaminoglycans to SWP1 was examined. E. cuniculi cell lysates were either subjected to, alkaline treatment, which hydrolyzes the O-specific glycosidic linkages of carbohydrates, or incubated with the glycosaminoglycan-degrading enzymes heparinase I, heparinase III, and chondroitase ABC. Samples were separated by SDS-PAGE, and the SWP1 antigen was detected with MAb 11A1 after immunoblotting. No difference in size was observed for treated and untreated samples (Fig. 6), indicating that the glycine and serine repeats do not serve as attachment sites for glycosaminoglycans.
DISCUSSION
The electron-dense exospore of microsporidia is the outermost layer of the infective spore stage and provides protection and resistance to environmental stress. In addition, surface molecules of the exospore might be involved in the initiation of the infection process (24, 25), and it is conceivable that the spore surface harbors molecules which mediate host and tissue specificity. In this report we describe for the first time the cloning and molecular characterization of a gene encoding a spore wall protein of microsporidia. The SWP1 gene codes for a protein of 450 aa and was cloned by immunoscreening of a cDNA library with MAb 11A1. This MAb revealed strong surface staining in immunofluorescence analysis on unfixed E. cuniculi spores, indicating that the recognized epitope is located at the surface of the exospore. Immunoelectron microscopy confirmed that the electron-dense exospore was labeled while the endospore and the interior of the spore were negative. The surface location of SWP1 makes it unlikely that the hydrophobic region from aa 230 to 253, which is predicted by several computer programs (e.g., TMpred) as a transmembrane region, is indeed membrane spanning. The distance of 60 nm between the plasma membrane and the spore wall surface would be too large to be crossed by a 51-kDa protein. The presence of a typical von Heijne signal sequence (17) suggests that SWP1 is translocated into the ER, as expected for a secreted protein. Two recently cloned genes encoding PTPs also possess hydrophobic signal sequences and are probably imported into the ER (4, 14). However, while PTPs remain intracellular, SWP1 as a spore wall component most likely enters a secretory pathway.
Electron microscopic analysis revealed that deposition of electron-dense material during spore wall synthesis in several microsporidian species starts at a few surface foci and subsequently spreads over wider surface areas (20). MAb 11A1 stained focal areas which were observed by immunoelectron microscopy at the plasma membranes of some organisms most likely represent these areas of early spore wall deposition. Later in sporogony, the continuous layer of electron-dense material of several microsporidian species forms tubular or fibrillar expansions which protrude from the surface of the sporoblast into the parasitophorous vacuole (20). The observed MAb 11A1-positive extracellular tubules might thus be a result of a budding process of these protrusions, resulting in the secretion of material into the parasitophorous vacuole.
SWP1 consists of an unusual carboxy-terminal region of five tandemly arranged glycine- and serine-rich repeats. Repetitive stretches of these amino acids were previously described as structural elements in three distinct protein families: intermediate filaments (i.e., keratins), loricrins, and single-stranded RNA binding proteins (18). These regions have been proposed to form a structural motif termed the “glycine loop” which might interact with similar structures on neighboring molecules, thereby increasing adhesion (18). It is proposed that the loop structure is stabilized by hydrophobic interaction of two aliphatic amino acid residues flanking the loop (18). However, the glycine and serine repeats of SWP1 are flanked by aspartates, an amino acid which is unlikely to form hydrophobic interactions. Thus, it is unclear whether the repetitive elements of SWP1 have the capacity to form loop structures. Although SWP1 shares the glycine-rich repetitive region with keratins, other structural elements conserved in keratins are absent. Thus, SWP1 does not belong to the keratin family, and in fact, the SWP1 sequence without the repetitive region does not display any significant similarities to proteins in public databases. Several putative attachment sites for glycosaminoglycans with the sequence SGXG are present within the repetitive domains. Neither alkaline treatment, which hydrolyzes O-glycosidic linkages, nor treatment with heparinases and chondroitases, which digest glycosaminoglycans, resulted in a size shift of SWP1, suggesting the absence of O-glycosidic linked carbohydrates. In addition, SWP1 lacks N-glycosylation sites. Thus, SWP1 does not appear to be modified by carbohydrates.
Immunofluorescence and immunoelectron microscopic analysis revealed that SWP1 is differentially expressed during the intracellular life cycle of E. cuniculi. SWP1 is absent in meronts and is first induced in early sporonts, which display a thickened plasma membrane due to the deposition of electron-dense material. Expression of SWP1 is associated with a translocation of organisms from the periphery to the center of the parasitophorous vacuole. A unicellular layer of organisms at the periphery remained SWP1 negative throughout the life cycle, even in late stages of development, when vacuoles harbored more than 100 E. cuniculi organisms. This morphology of the parasitophorous vacuole with sporonts inside and meronts outside is typical for Encephalitozoon species, as determined by electron microscopy (20). SWP1 expression can thus serve as a molecular marker to determine the differentiation status of organisms by IFA.
The intracellular life cycle of microsporidia is a highly coordinated differentiation process in which the organisms undergo morphological changes and progressively acquire spore-specific structures such as the spore wall or the polar tube. RT-PCR analysis demonstrated that the amount of SWP1-specific mRNA increases with infection time, suggesting that regulation of SWP1 expression occurs at the mRNA level. Investigations of the regulation of gene expression have not been reported for microsporidia so far, and SWP1 appears to be the first microsporidian gene for which regulation at the transcript level is demonstrated. An interesting feature of the SWP1 mRNA is the short AT-rich 5′ untranslated region of 8 nt, which is otherwise typical for amitochondrial protozoa of the genera Entamoeba, Trichomonas, and Giardia (11). Future studies on the intracellular development of microsporidia will certainly benefit from the characterization of additional stage-specific markers, each induced at specific time points during spore differentiation.
ACKNOWLEDGMENTS
We thank Anne Wirsing for excellent technical assistance; Peter Deplazes, Zürich, Switzerland, for providing E. cuniculi organisms; and Louis M. Weiss for providing a polyclonal anti-E. cuniculi antiserum.
W.B. is supported by an AIDS-Stipendium from the Deutsches Krebsforschungszentrum. D.J.P.F. is supported by an equipment grant from the Wellcome Trust.
REFERENCES
- 1.Beckers P J, Derks G J, Gool T, Rietveld F J, Sauerwein R W. Encephalocytozoon intestinalis-specific monoclonal antibodies for laboratory diagnosis of microsporidiosis. J Clin Microbiol. 1996;34:282–285. doi: 10.1128/jcm.34.2.282-285.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bigliardi E, Selmi M G, Lupetti P, Corona S, Gatti S, Scaglia M, Sacchi L. Microsporidian spore wall: ultrastructural findings on Encephalitozoon hellem exospore. J Eukaryot Microbiol. 1996;43:181–186. doi: 10.1111/j.1550-7408.1996.tb01388.x. [DOI] [PubMed] [Google Scholar]
- 3.Canning E U. Microsporidia. In: Kreier J P, Baker J R, editors. Parasitic Protozoa. 2nd ed. Vol. 6. New York, N.Y: Academic Press; 1993. pp. 299–385. [Google Scholar]
- 4.Delbac F, Duffieux F, David D, Metenier G, Vivares C P. Immunocytochemical identification of spore proteins in two microsporidia, with emphasis on extrusion apparatus. J Eukaryot Microbiol. 1998;45:224–231. doi: 10.1111/j.1550-7408.1998.tb04529.x. [DOI] [PubMed] [Google Scholar]
- 5.Edlind T D, Li J, Visvesvara G S, Vodkin M H, McLaughlin G L, Katiyar S K. Phylogenetic analysis of beta-tubulin sequences from amitochondrial protozoa. Mol Phylogenet Evol. 1996;5:359–367. doi: 10.1006/mpev.1996.0031. [DOI] [PubMed] [Google Scholar]
- 6.Erickson B W, Blanquet R S. The occurrence of chitin in the spore wall of Glugea weissenbergi. J Invertebr Pathol. 1969;14:358–364. doi: 10.1016/0022-2011(69)90162-1. [DOI] [PubMed] [Google Scholar]
- 7.Frixione E, Ruiz L, De Vargas L V, Tejero J M, Undeen A H. Dynamics of polar filament discharge and sporoplasm expulsion by microsporidian spores. Cell Motil Cytoskelet. 1992;22:38–50. [Google Scholar]
- 8.Frohman M A, Dush M K, Martin G R. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific primer. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Germot A, Philippe H, Le Guyader H. Evidence for loss of mitochondria in Microsporidia from a mitochondrial-type HSP70 in Nosema locustae. Mol Biochem Parasitol. 1997;87:159–168. doi: 10.1016/s0166-6851(97)00064-9. [DOI] [PubMed] [Google Scholar]
- 10.Hirt R P, Logsdon J M, Jr, Healy B, Dorey M W, Doolittle W F, Embley T M. Microsporidia are related to Fungi: evidence from the largest subunit of RNA polymerase II and other proteins. Proc Natl Acad Sci USA. 1999;96:580–585. doi: 10.1073/pnas.96.2.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katiyar S K, Visvesvara G S, Edlind T D. Comparisons of ribosomal RNA sequences from amitochondrial protozoa: implications for processing, mRNA binding and paromomycin susceptibility. Gene. 1995;152:27–33. doi: 10.1016/0378-1119(94)00677-k. [DOI] [PubMed] [Google Scholar]
- 12.Keeling P J, Doolittle W F. Alpha-tubulin from early-diverging eucaryotic lineages and the evolution of the tubulin family. Mol Biol Evol. 1996;13:1297–1305. doi: 10.1093/oxfordjournals.molbev.a025576. [DOI] [PubMed] [Google Scholar]
- 13.Keeling P J, McFadden G I. Origins of microsporidia. Trends Microbiol. 1998;6:19–23. doi: 10.1016/S0966-842X(97)01185-2. [DOI] [PubMed] [Google Scholar]
- 14.Keohane E M, Orr G A, Zhang H S, Takvorian P M, Cali A, Tanowitz H B, Wittner M, Weiss L M. The molecular characterization of the major polar tube protein gene from Encephalitozoon hellem, a microsporidian parasite of humans. Mol Biochem Parasitol. 1998;94:227–236. doi: 10.1016/s0166-6851(98)00071-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14a.Linhardt R J. Analysis of glycosaminoglycans with polysaccharide lyases. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1997. p. 17.13B. [DOI] [PubMed] [Google Scholar]
- 15.Lom J, Vavra J. The mode of sporoplasm extrusion in microsporidian spores. Acta Protozool. 1963;1:81–92. [Google Scholar]
- 16.Lujan H D, Conrad J T, Clark C G, Touz M C, Delbac F, Vivares C P, Nash T E. Detection of microsporidia spore-specific antigens by monoclonal antibodies. Hybridoma. 1998;17:237–243. doi: 10.1089/hyb.1998.17.237. [DOI] [PubMed] [Google Scholar]
- 17.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 18.Steinert P M, Mack J W, Korge B P, Gan S Q, Haynes S R, Steven A C. Glycine loops in proteins: their occurrence in certain intermediate filament chains, loricrins and single-stranded RNA binding proteins. Int J Biol Macromol. 1991;13:130–139. doi: 10.1016/0141-8130(91)90037-u. [DOI] [PubMed] [Google Scholar]
- 19.van Gool T, Snijders F, Reiss P, Eeftinck Schattenkerk J K, van den Bergh Weerman M A, Bartelsman J F, Bruins J J, Canning E U, Dankert J. Diagnosis of intestinal and disseminated microsporidial infections in patients with HIV by a new rapid fluorescence technique. J Clin Pathol. 1993;46:694–699. doi: 10.1136/jcp.46.8.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vavra J, Larsson J I R. Structure of the microsporidia. In: Wittner M, Weiss L M, editors. The Microsporidia and microsporidiosis. Washington, D.C.: American Society for Microbiology; 1999. pp. 7–84. [Google Scholar]
- 21.Visvesvara G S, Leitch G J, Da Silva A J, Croppo G P, Moura H, Wallace S, Slemenda S B, Schwartz D A, Moss D, Bryan R T, Pieniazek N J. Polyclonal and monoclonal antibody and PCR-amplified small-subunit rRNA identification of a microsporidian, Encephalitozoon hellem, isolated from an AIDS patient with disseminated infection. J Clin Microbiol. 1994;32:2760–2768. doi: 10.1128/jcm.32.11.2760-2768.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weber R, Bryan R T, Schwartz D A, Owen R L. Human microsporidial infections. Clin Microbiol Rev. 1994;7:426–461. doi: 10.1128/cmr.7.4.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weidner E, Byrd W, Scarborough A, Pleshinger J, Sibley D. Microsporidian spore discharge and the transfer of polaroplast organelle membrane into the plasma membrane. J Protozool. 1984;31:195–198. [Google Scholar]
- 24.Weidner E. Cytoskeletal proteins expressed by microsporidian parasites. Subcell Biochem. 1992;18:385–399. doi: 10.1007/978-1-4899-1651-8_12. [DOI] [PubMed] [Google Scholar]
- 25.Weidner E, Halonen S K. Microsporidian spore envelope keratins phosphorylate and disassemble during spore activation. J Eukaryot Microbiol. 1993;40:783–788. [Google Scholar]
- 26.Yamauchi K. The sequence flanking translational initiation site in protozoa. Nucleic Acids Res. 1991;19:2715–2720. doi: 10.1093/nar/19.10.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]