Abstract
We report the cloning of the gene encoding the 32-kDa lipoprotein, designated LipL32, the most prominent protein in the leptospiral protein profile. We obtained the N-terminal amino acid sequence of a staphylococcal V8 proteolytic-digest fragment to design an oligonucleotide probe. A Lambda-Zap II library containing EcoRI fragments of Leptospira kirschneri DNA was screened, and a 5.0-kb DNA fragment which contained the entire structural lipL32 gene was identified. Several lines of evidence indicate that LipL32 is lipid modified in a manner similar to that of other procaryotic lipoproteins. The deduced amino acid sequence of LipL32 would encode a 272-amino-acid polypeptide with a 19-amino-acid signal peptide, followed by a lipoprotein signal peptidase cleavage site. LipL32 is intrinsically labeled during incubation of L. kirschneri in media containing [3H]palmitate. The linkage of palmitate and the amino-terminal cysteine of LipL32 is acid labile. LipL32 is completely solubilized by Triton X-114 extraction of L. kirschneri; phase separation results in partitioning of LipL32 exclusively into the hydrophobic, detergent phase, indicating that it is a component of the leptospiral outer membrane. CaCl2 (20 mM) must be present during phase separation for recovery of LipL32. LipL32 is expressed not only during cultivation but also during mammalian infection. Immunohistochemistry demonstrated intense LipL32 reactivity with L. kirschneri infecting proximal tubules of hamster kidneys. LipL32 is also a prominent immunogen during human leptospirosis. The sequence and expression of LipL32 is highly conserved among pathogenic Leptospira species. These findings indicate that LipL32 may be important in the pathogenesis, diagnosis, and prevention of leptospirosis.
Leptospirosis is considered to be the most widespread zoonosis in the world (Centers for Disease Control, http://www .cdc.gov/ncidod/dbmd/diseaseinfo/leptospirosis_t.htm, 1998). Renal tubular infection with pathogenic Leptospira species has been documented for a wide variety of wild and domestic animals. Infections are typically transmitted to humans through contact with water or soil contaminated with the urine of infected animals. It is likely that the wide distribution of Leptospira species reflects the ability to survive in diverse environmental conditions combined with genetic adaptation, which is reflected in the plasticity of the leptospiral genome (40). Pathogenic Leptospira species are capable of surviving as free-living bacteria for extended periods of time after urinary shedding. Inactivated whole-cell leptospiral vaccines have been in use for many years in an effort to control disease in animals and exposure to humans (35). However, currently available leptospiral vaccines are ineffective at preventing either disease or infection in cattle (5–7) and produce only short-term immunity in other animals (35). The immunity provided by these vaccines is based on the lipopolysaccharide (LPS) carbohydrate serovar determinant; there is little cross-protection against infection with the more than 250 different leptospiral serovars that are now known to exist (A.F. Kaufmann et al., http://www.pasteur.fr/recherche/Leptospira/Strains.html, 1999).
Because of the deficiencies of LPS-based vaccines, protein components of the leptospiral outer membrane represent an important approach to the development of alternative immunoprotection strategies. Studies suggest that the leptospiral outer membrane has a relatively complex protein profile (10, 18, 25, 41). To date, only a few leptospiral outer membrane proteins have been characterized in detail, including a porin, OmpL1 (15, 29), and two lipoproteins, LipL36 (16) and LipL41 (31). The most prominent band in the leptospiral total protein profile is a protein with a molecular mass of approximately 32 kDa. Zuerner et al. have shown that this major outer membrane protein (MOMP) could be solubilized by extraction of the outer membrane with the nonionic detergents Triton X-100 and Triton X-114 and was the most prominent antigen identified by infection-derived swine sera in the Triton X-114 detergent phase (41).
Many of the most abundant proteins in spirochetes are lipoproteins, including TpN47 of Treponema pallidum (12, 13), OspA of Borrelia burgdorferi (4, 8), the Vmp proteins of the relapsing Borrelia fever (11, 30), and SmpA of Brachyspira hyodysenteriae (37). In this report we describe LipL32, the MOMP of pathogenic Leptospira spp. Evidence is presented that LipL32 is a lipoprotein covalently modified at its amino-terminal cysteine by fatty acid(s). We also examined infected hamster kidney tissue and clinical leptospirosis sera to provide documentation that LipL32 is expressed not only during cultivation but also during mammalian infection.
(Portions of this work were presented at the 99th General Meeting of the American Society for Microbiology in Chicago, Ill., May 30 to June 3, 1999.)
MATERIALS AND METHODS
Bacterial strains, media, and plasmids.
Leptospira kirschneri RM52 (36) and other leptospiral strains were obtained from the National Leptospirosis Reference Center (National Animal Disease Center, Agricultural Research Service, U.S. Department of Agriculture, Ames, Iowa). Most of the leptospiral strains used in this publication are described in a recent DNA relatedness study (9). Leptospires were cultivated in Johnson-Harris bovine serum albumin-Tween 80 medium (Bovuminar PLM-5 Microbiological Media; Intergen) (21). Escherichia coli DH5α [supE44 ΔlacU169 (φ80 lacZ ΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1] was used as the host strain for transformations of recombinant DNA. E. coli PLK-F′ (recA lac mcrA mcrB hsdR gal supE [F′ proAB lacIqZΔM15 Tn10 (Tetr)]) was used as the host strain for infection with the Lambda-Zap II vector (Stratagene). E. coli PLK-F′ and the ExAssist helper phage were used for in vivo excision of the pBluescript phagemid (Stratagene). E. coli SOLR (e14−[mcrA], Δ[mcrCB-hsdSMR-mrr)171 sbcC recB recJ umuC::Tn5[Kanr] uvrC lac gyrA96 relA1 thi-1 endA1 λr [F′ proAB lacIqZΔM15], Su− [nonsuppressing]) was used as the host strain for replication of the excised pBluescript phagemid from the Lambda-Zap II vector (Stratagene). E. coli BLR(DE3)/pLysS [F− ompT hsdSB (rB−mB) gal dcm Δ(srl-recA)306::Tn10(TcR) (DE3) pLysS(CmR)] (Novagen) was used as the host strain for the pRSET expression vector (Invitrogen). The DE3 lysogen of E. coli JM109 (Promega) was used as the host strain for pET-15b (Novagen). E. coli cells were routinely grown in Luria-Bertani broth or on Luria-Bertani agar, unless otherwise mentioned (27).
Gel electrophoresis and immunoblotting.
Samples for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) were solubilized in final sample buffer (FSB) composed of 62.5 mM Tris hydrochloride (pH 6.8), 10% glycerol, 5% 2-mercaptoethanol, and 2% SDS. Proteins were separated on a 12% gel with a discontinuous buffer system (22) and stained with Coomassie brilliant blue or were transferred to nitrocellulose (Schleicher & Schuell) for immunoblotting. For antigenic detection on immunoblots, the nitrocellulose was blocked with 5% nonfat dry milk in 0.1 M phosphate-buffered saline (PBS; pH 7.4)–0.1% Tween 20 (PBS-T), incubated for 1h with antiserum diluted 1:5,000 (unless otherwise noted) in PBS-T, and probed with donkey anti-rabbit antiserum conjugated to horseradish peroxidase (Amersham). Samples containing E. coli expressing full-length LipL32 were separated in 10% Tris-tricine gels (28), and the proteins were transferred to Immobilon-P (Millipore Corp., Bedford, Mass.) for immunoblotting (performed as described above except that the secondary antibody was horseradish peroxidase-conjugated anti-rabbit antiserum obtained from Kirkegaard and Perry Laboratories, Gaithersburg, Md.). Antigen-antibody binding was detected by using the Enhanced Chemiluminescence System (ECL; Amersham). Blots were incubated in ECL reagents for 1 min and then exposed to XAR-5 film (Kodak).
Amino acid sequencing of an internal polypeptide fragment.
L. kirschneri strain RM52 cells (4 × 1011) were washed two times in PBS and resuspended in 30 ml of ice-cold TENP buffer (50 mM Tris, pH 8.0; 1 mM EDTA; 100 mM NaCl; 0.1 mM phenylmethylsulfonyl fluoride [PMSF]). The cells were disrupted by tip sonication. The membrane fraction was recovered by centrifugation for 20 min at 12,000 × g and washed once in TENP buffer. The membrane proteins were separated on a 12.5% SDS-PAGE gel. A test strip was stained with Coomassie brilliant blue in order to locate the 32-kDa band, which was cut out of the remainder of the gel and loaded onto a second SDS-PAGE gel in the presence of staphylococcal V8 protease at a concentration of 100 μg ml−1 (Sigma). The proteins were allowed to migrate into the stacking gel by electrophoresis, and the current was disconnected for 45 min, followed by completion of electrophoresis. The polypeptide fragments were subjected to SDS-PAGE, transferred to Trans-Blot PVDF Protein Sequencing Membrane (Bio-Rad, Richmond, Calif.) and submitted to the UCLA Protein Microsequencing Facility. N-terminal amino acid sequence analysis was performed on a Porton 1090-E gas-phase sequenator with on-line detection of PTH amino acids.
Southern blot analysis.
L. kirschneri DNA was prepared by the method of Yelton and Charon (39). Leptospiral DNA was digested with EcoRI and electrophoresed in a 1.0% agarose gel. After depurination, denaturation, and neutralization, the DNA was transferred to a nylon filter (Zeta-Probe; Bio-Rad) by the method of Southern (27). Filters were prehybridized for 3 h at 37°C in buffer containing 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 1× Denhardt's solution, 0.05% sodium PPi, 0.5% SDS, and 100 μg of denatured salmon sperm DNA per ml. Two degenerate oligonucleotides, 32A-1 and 32A-2, were designed based upon the first 8 and last 8 amino acids, respectively, of a 32-amino-acid sequence of a proteolytic fragment of LipL32. The sequence of oligonucleotide 32A-1 was GDCAYAAYAARTAYAAYWSYYT. The sequence of oligonucleotide 32A-2 was AARAAYATHGAYACNAARAARYT. Oligonucleotides 32A-1 and 32A-2 were prepared commercially (Gibco Life Technologies) and used to amplify the intervening region of the lipL32 gene. The 96-bp amplification product was radiolabeled by PCR amplification in the presence of [32P]dCTP. The filters were then hybridized overnight at 42°C with radiolabeled probe. After hybridization, the filters were washed at 42°C in 2× SSC–0.5% SDS.
Cloning and sequencing of the lipL32 gene.
Standard recombinant DNA procedures were performed as described earlier (27). Restriction endonuclease digests were performed as recommended by the suppliers (New England Biolabs and Promega). EcoRI fragments of L. kirschneri genomic DNA were ligated into the Lambda-Zap II vector (Stratagene). The ligated DNA was packaged with Gigapack II Gold packaging extract (Stratagene) and stored in 0.3% chloroform at 4°C. The plaque titer was determined by infecting E. coli PLK F′ (Stratagene). Plaques were plated, transferred to filters in duplicate, and processed as previously described (15). Filters were probed with the 96-bp amplified lipL32 gene fragment described above using the same hybridization and washing conditions as in the Southern hybridization. Recombinant pBluescript SK(−) clones were recovered from phage producing positive plaques by in vivo excision according to the manufacturer. After restriction mapping, appropriate DNA fragments were subcloned into pBluescript KS and sequenced at the UCLA Core DNA Sequencing Facility by the dideoxy chain termination method with fluorescein-labeled dideoxy nucleotides (Applied Biosystems).
The lipL32 genes of L. borgpetersenii serovar hardjo strain 203, L. interrogans serovar pomona strain RZ11, L. noguchii serovar fortbragg strain Fort Bragg, and L. santarosai serovar tropica strain CA299 were amplified by PCR with primers P662 (5′-CTA-AGT-TCA-TAC-CGT-GAT-TT-3′), starting 39 bp upstream from the lipL32 start codon, and P663 (5′-ATT-ACT-TAG-TCG-CGT-CAG-AA-3′), starting 1 bp from the end of termination codon on the complementary strand of DNA. Amplification was done with Tth polymerase (Clontech, Palo Alto, Calif.) as follows: 94°C for 2 s, 60°C for 1.5 min for 40 cycles, and then 67°C for 1 h. The amplified lipL32 genes of L. santarosai serovar tropica strain CA299 and L. borgpetersenii serovar hardjo strain 203 were sequenced directly, while the amplified lipL32 genes of L. interrogans serovar pomona strain RZ11 and L. noguchii serovar fortbragg strain Fort Bragg were cloned into pCRII (Invitrogen, La Jolla, Calif.) prior to sequencing. DNA sequencing was performed by using dye chain termination reactions separated in a Prism 377 DNA Sequencer (Applied Biosystems, Foster City, Calif.) at the Iowa State Nucleic Acid Facility. Sequence data were analyzed using Sequencher version 4.0.2 (Genecodes, Ann Arbor, Mich.). The sequences were aligned using Clustal_X (37a).
Antisera.
Antisera to OmpL1 and LipL41 were prepared as previously described (15, 31). Briefly, New Zealand White rabbits were immunized with purified His6 fusion proteins, expressed by E. coli JM109 (Invitrogen) transformed with the pRSET plasmid (Invitrogen) containing either the ompL1 gene or the lipL41 gene (15, 31). Antiserum prepared to a 2% Triton X-114 outer membrane extract of L. interrogans serovar pomona strain RZ11 was used in an immunoblot analysis of E. coli harboring pRZ1132K.
Antiserum to LipL32 was prepared as follows. The PCR was used to amplify the portion of the lipL32 gene encoding the mature protein beginning with the first residue after the amino-terminal cysteine. The 5′ oligonucleotide contained the nucleotide sequence coding for the six amino acids following the amino-terminal cysteine of mature LipL32, including a XhoI restriction endonuclease site (underlined): 5′-TTA CCG CTC GAG GTG CTT TCG GTG GTC TGC-3′. The 3′ oligonucleotide contained the nucleotide sequence coding for the five carboxy-terminal amino acids and the lipL32 stop codon, including a SmaI restriction endonuclease site (underlined): 5′-TGT TAA CCC GGG TTA CTT AGT CGC GTC AGA-3′. L. kirschneri genomic DNA was used as template. The 782-bp amplified lipL32 gene was digested with XhoI and SmaI and ligated into pRSETc (Invitrogen) digested with XhoI and PvuII. The resulting construct, pRSETc-LipL32, was transformed into E. coli BLR(DE3)/pLysS (Novagen). Expression of the His6-LipL32 fusion protein was achieved by isopropyl-β-d-thiogalactopyranoside (IPTG; Sigma) induction. The His6-LipL32 fusion protein was solubilized in 6 M guanidine and purified by affinity chromatography using Ni2+-nitrilotriacetic acid-agarose (Qiagen) and dialyzed in PBS containing 10% glycerol, 0.3% Triton X-100, and 0.025% sodium azide. The purified His6-LipL32 fusion protein was loaded onto an SDS-PAGE gel. After electrophoresis, the His6-LipL32 band containing approximately 150 μg of protein was cut out of the acrylamide gel, dessicated, ground to powder, mixed with Freund's complete adjuvant, and inoculated subcutaneously and intramuscularly into a New Zealand White male rabbit. Additional immunizations with approximately 150 μg of His6-LipL32 fusion protein in Freund's incomplete adjuvant were given at 4 and 8 weeks after primary immunization. The rabbit was bled 10 weeks after the primary immunization.
[3H]palmitate radiolabeling and immunoprecipitation of native LipL32.
A 35-ml culture containing 5 × 107 L. kirschneri cells per ml in the log phase of growth was intrinsically labeled by addition of [9,10(n)-3H]palmitate (250 μCi; 60 Ci/mmol; Amersham), followed by further incubation in a shaker incubator at 30°C for 48 h until the bacterial concentration reached 109/ml. Organisms were washed in 5 mM MgCl2 in PBS. A sample for immunoprecipitation containing 8 × 109 L. kirschneri cells was resuspended in 1.25 ml of 10 mM Tris HCl (pH 8.0)–10 mM EDTA–1 mM PMSF. To this suspension was added 12.5 μl of 10% protein-grade Triton X-100 (Calbiochem), followed by gentle agitation for 30 min at 4°C. The insoluble material was removed by centrifugation at 16,000 × g for 10 min. To the supernatant was added 0.2 ml of LipL32 rabbit antiserum and 0.2 ml of a slurry of staphylococcal protein. A-Sepharose CL-4B (Sepharose-SpA) (Sigma). The suspension was gently agitated for 1 h. The Sepharose-SpA-antibody-antigen complexes were washed twice in 0.1% Triton X-100 in 10 mM Tris HCl (pH 8.0)–20 mM CaCl2–150 mM NaCl and then resuspended in FSB. After SDS-PAGE, gels were prepared for autoradiography by being soaked in Amplify (Amersham), dried, and exposed to Tritium film (Amersham). In some cases, gels were stained with Coomassie brilliant blue and destained in 10% acetic acid–45% methanol prior to being soaked in Amplify (Amersham), dried, and exposed to Tritium film (Amersham).
Triton X-114 extraction of L. kirschneri.
L. kirschneri was extracted with 1% Triton X-114 by a modification of the method described previously (18). In brief, culture-attenuated L. kirschneri cells were washed in PBS–5 mM MgCl2 and then extracted in the presence of 1% protein grade Triton X-114 (Calbiochem)–150 mM NaCl–10 mM Tris (pH 8)–1 mM EDTA at 4°C. The insoluble material was removed by centrifugation at 17,000 × g for 10 min. After centrifugation, 20 mM CaCl2 was added to half of the supernatant. Phase separation was performed by warming the supernatant to 37°C and subjecting it to centrifugation for 10 min at 1,000 × g. The detergent and aqueous phases were separated and precipitated with acetone.
Expression of full-length LipL32 in E. coli.
The lipL32 amplicon was generated with primers P662 and P663 by using template DNA from strain RZ11 and cloned into the pCRII plasmid vector (Invitrogen), resulting in plasmid pRZ1132K. In pRZ1132K, lipL32 is oriented downstream of the lacZ promoter. E. coli INV?′ cells harboring pRZ1132K or pCRII were grown to stationary phase in DYT broth (24). Cells were harvested by centrifugation, and membrane and cytoplasmic fractions were prepared according to the method of Weigel et al. (38).
Immunohistochemistry.
The methods used to perform immunohistochemistry and to obtain L. kirschneri-infected hamster kidney tissue have been previously described (3). In brief, 5-week-old Golden Syrian hamsters (Harlan Sprague-Dawley), in groups of three, were inoculated intraperitoneally with 105 virulent L. kirschneri RM52. Moribund hamsters were euthanized; liver and kidney tissues were removed, fixed in formalin, and paraffin embedded. Hamsters surviving 28 days after challenge were euthanized, and the liver and kidney tissues were removed, fixed in formalin, and paraffin embedded. Tissue sections were stained with hematoxylin and eosin or silver stain by the technique of Steiner and Steiner (32).
Serial 5-μm sections of kidney taken 10 and 28 days after infection with L. kirschneri were cut. Tissue sections were placed on Pro-Bond Plus slides. Paraffin was removed from sections with xylene and ethanol by using standard procedures. Tissues were pretreated with 0.1% trypsin in 0.1 M Tris HCl (pH 7.6) with 0.1% CaCl2 for 5 min at 37°C. Nonspecific staining of tissue sections was blocked by using 10% normal goat serum with incubation at room temperature for 20 min prior to incubation overnight at 4°C with primary antibody. Anti-LipL32 antiserum was used at a 1:6,000 dilution. Controls included “negative” primary antibody, normal rabbit serum, and hyperimmune serum on kidney sections from uninfected hamsters. Unbound primary antibody was removed, and tissues were incubated at room temperature for 30 min with biotinylated goat anti-rabbit immunoglobulin (Vector). After being washed, sections were incubated for 20 min at room temperature with supersensitive streptavidin-alkaline phosphatase (Biogenex). Enzyme reactions were developed by using new Fuchsin (Biogenex). All slides were counterstained with hematoxylin before dehydration in alcohols and propar (xylene substitute), and coverslips were then mounted. Smears of organisms from actively growing cultures were processed as tissue sections without the removal of paraffin.
ELISA.
Convalescent sera from five patients with culture-proven infection due to L. kirschneri serovar bim were used to compare the enzyme-linked immunosorbent assay (ELISA) reactivity with five leptospiral His6 fusion proteins. Immulon microtiter plates (Dynatech) were coated at 37°C overnight with 125 ng of purified His6 fusion proteins in 0.05 M sodium carbonate buffer (pH 9.6). The plates were washed three times with PBS containing 0.05% Tween 20, followed by the addition of 200 μl of blocking buffer (PBS containing 0.05% Tween 20 and 1% nonfat dried milk), and were then incubated at 37°C for 1 h. After removal of the blocking buffer, 100 μl of antisera diluted 1:100 with blocking buffer was added, and the plates were incubated at 37°C for 2 h. After washing the plates three times, 100 μl of 1:5,000-diluted sheep anti-human immunoglobulin G conjugated to horseradish peroxidase (Amersham) was added, and the plates were incubated at 37°C for 2 h. After three more washes, 100 μl of 3,3′,5,5′-tetramethylbenzidine substrate was added, and the plates were incubated at 37°C for 20 min on an orbital shaker. Then, 100 μl of a 1 M solution of sulfuric acid was added to stop the reaction. The absorbance was read at a wavelength of 450 nm with a microplate reader (model 550; Bio-Rad). Each serum sample was tested four separate times. Absorbance readings were normalized by subtracting the value obtained from antigen-negative control wells. Statistical analysis was performed by using the two-tailed Student's t test for samples with unequal variance.
Nucleotide sequence accession numbers.
The nucleotide sequence of the lipL32 gene from L. kirschneri serovar grippotyphosa strain RM52 has been deposited in the GenBank database under the accession number AF121192. The nucleotide sequences of the lipL32 genes of L. interrogans serovar pomona strain RZ11, L. borgpetersenii serovar hardjo strain 203, L. santarosai serovar tropica strain CA299, and L. noguchii serovar fortbragg strain Fort Bragg have been deposited in the GenBank database under accession numbers AF181553, AF181554, AF181555, and AF181556, respectively. The nucleotide sequence of the lipL32 gene from L. interrogans serovar lai had previously been deposited in the GenBank database under accession number LIU89708.
RESULTS
Cloning and sequence analysis of the lipL32 gene.
Staphylococcal V8 protease digestion of LipL32 resulted in two fragments which comigrated by SDS-PAGE with an apparent molecular mass of 20 kDa. Because the two fragments were present in unequal amounts, N-terminal amino acid sequence analysis of the 20-kDa band revealed two sequences: AFKAATPEEKSM and RHNKYNSLTRIKIPNPPKSFDDLKNIDTKKLL. A PCR-amplified 96-bp probe based on the longer sequence bound to a single 5.0-kb band in EcoRI digests of total genomic L. kirschneri serovar grippotyphosa strain RM52 DNA. Restriction mapping, Southern blot analysis, and DNA sequencing revealed that the entire lipL32 gene is encoded by the 5.0-kb EcoRI fragment (Fig. 1). A BLAST search of the GenBank database revealed a nearly 99% identical gene of L. interrogans serovar lai (GenBank accession number LIU89708) (1; BLAST Sequence Similarity Searching, http://www.ncbi.nlm.nih.gov/BLAST/, 1998). The L. interrogans lipL32 homolog was isolated and sequenced by Kim et al. on a DNA fragment containing the sphH sphingomyelinase gene by screening a library of leptospiral DNA fragments using the leptospiral sphA gene as a probe (M. J. Kim et al., Meet. Int. Leptospirosis Soc., 1996). Besides sphH, two other genes were found in proximity to lipL32. A 398-bp open reading frame (ORF), ORF-1, is located 232 bp upstream of lipL32. Although ORF-1 and lipL32 are in the same orientation, ORF-1 is followed by a 15-bp inverted repeat, which may function as a rho-independent transcription terminator, suggesting that ORF-1 and lipL32 are not cotranscribed. However, E. coli ς70 consensus −35 and −10 promoter regions were not found upstream of lipL32. Upstream of ORF-1 and oriented in the opposite direction is a gene which is highly homologous to the S-adenosylmethionine synthetase genes of a variety of gram-negative and gram-positive bacteria.
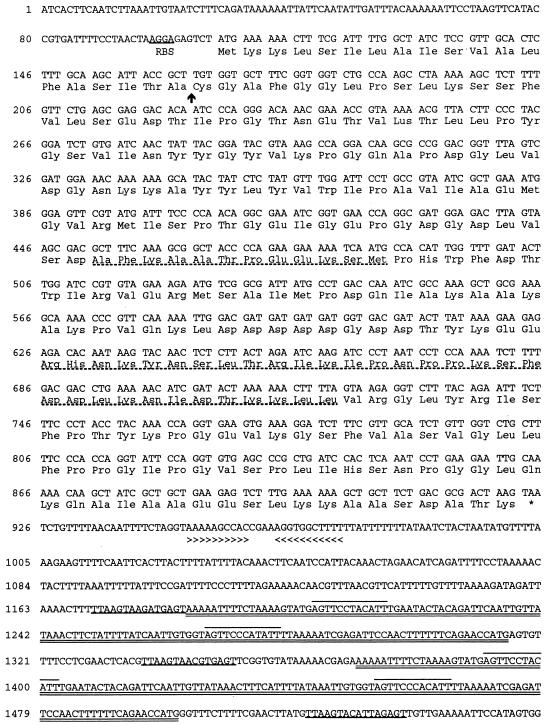
FIG. 1.
Nucleotide sequence and deduced amino acid sequence of lipL32. The putative ribosome-binding site (RBS) is shown. The putative signal peptidase cleavage site is indicated by an arrow. The amino acid sequences obtained from the staphylococcal V8 protease digestion of the native protein are shown (broken underline). The location of the TAA stop codon is indicated by an asterisk. An inverted repeat is indicated by arrowheads. Repeated sequences following the lipL32 gene include a shorter sequence repeated three times (underlined), flanking a longer sequence repeated twice (double underlined). The longer sequences contain another short sequence which is repeated four times (overlined).
The lipL32 structural gene consists of 816 bases encoding a protein of 272 amino acids. A consensus ribosome-binding site (AAGGA) is present upstream of the initiation codon. An inverted repeat located 30 bp downstream from the termination codon may function as a rho-independent transcription terminator. This inverted repeat is followed by a region of reiterated elements: a 12-bp sequence (AGTTCCCACATT) is repeated four times after the L. kirschneri lipL32 gene and six times after the L. interrogans lipL32 gene, as well as near the L. interrogans heat shock operon (GenBank accession number AF007813) and within the L. interrogans insertion element IS1501a (GenBank accession number AF038933).
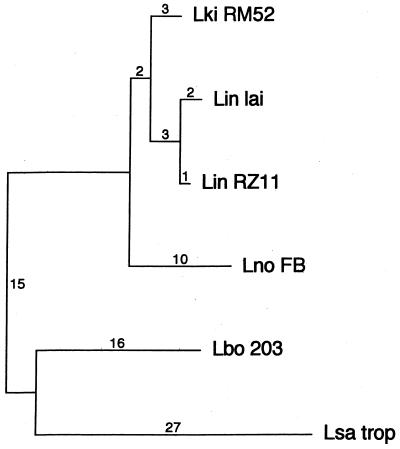
The lipL32 sequence is highly conserved among Leptospira species. In addition to the lipL32 genes of L. kirschneri serovar grippotyphosa strain RM52 and L. interrogans serovar lai, the lipL32 genes from four additional strains were sequenced: L. interrogans serovar pomona strain RZ11, L. noguchii serovar fortbragg strain Fort Bragg, L. borgpetersenii serovar hardjo strain 203, and L. santarosai serovar tropica. Comparison of the lipL32 DNA sequences of the six leptospiral strains, representing five of the seven pathogenic species, reveals a high degree of sequence conservation, with an average DNA sequence identity of 96.4% (range, 93.5 to 99.6%) (alignment available upon request). Most of the sequence polymorphisms detected (50 of 73) were silent, suggesting that there is evolutionary pressure to maintain the primary sequence of this protein. Comparison of the deduced amino acid sequences of the six LipL32 variants reveals an average amino acid sequence identity of 97.8% (range, 95.2 to 99.6%) (alignment available upon request). The six lipL32 DNA sequences were used to perform phylogenetic analysis (Fig. 2). The segregation pattern found using lipL32 sequences is remarkably similar to that resulting from analysis of 16S RNA sequences, which determined that the leptospiral pathogens formed three groups: group I was represented by L. interrogans and L. kirschneri species, group II included L. borgpetersenii and L. santarosai, while group III consisted of L. noguchii and L. meyeri (20). The similarity of the lipL32 DNA and 16S rDNA phylogenetic trees indicates that the observed sequence differences are a result of genetic drift.
FIG. 2.
Phylogenetic tree based on lipL32 nucleotide sequence differences. L. kirschneri serovar grippotyphosa, L. interrogans serovar pomona, and L. interrogans serovar lai formed one monophyletic group; a second group was formed by L. borgpetersenii serovar hardjo and L. santarosai serovar tropica, while L. noguchii serovar fortbragg segregated separately. Sequences were analyzed by using the PAUP software package (version 3.1; D. Swafford, Smithsonian Institution, Washington, D.C.). Horizontal lengths are proportional to nucleotide step differences (indicated above the lines).
As expected for a lipoprotein, the deduced amino acid sequence begins with a 19-residue signal peptide, represented by the N-terminal peak of 2.8 on the Kyte-Doolittle hydrophobicity plot (data not shown). The LipL32 sequence conforms to the rules established for procaryotic lipoprotein signal peptides (19, 26). The LipL32 signal peptide has a basic amino-terminal region (including lysines at positions 2 and 3), a hydrophobic core (amino acids 4 through 19), and a carboxy-terminal I-T-A-C signal peptidase II cleavage site, which is similar to the L-T-A-C lipoprotein signal peptidase cleavage site of LipL36. An isoleucine at the −3 position relative to cysteine is less common than leucine in signal peptidase II cleavage sites, but there are at least eight other examples of isoleucine occurring at this position in bacterial lipoproteins, including OspE, OspF, ErpG, BBA59, and BBA60 of B. burgdorferi, EnvC and ExcC of E. coli, and Lpp20 of Helicobacter pylori (alignment available upon request). When LipL32 is expressed in E. coli, processing is incomplete, and the recombinant LipL32 is equally distributed between the membrane and cytoplasmic fractions of E. coli (data not shown). This result suggests either that LipL32 secretion was inefficient in E. coli, that the level of LipL32 synthesis using pRZ1132K had overwhelmed the E. coli secretory apparatus, or there was a combination of both effects.
After cleavage of the 19-amino-acid signal peptide by leptospiral signal peptidase II, the mature polypeptide would have a predicted molecular mass of 27.6 kDa. Staphylococcal V8 protease is known to cleave peptides following acidic amino acids. The predicted amino acid sequences following aspartate residue 115 and glutamate residue 173 are identical to those obtained from N-terminal amino acid sequencing of proteolytic fragments of the native protein. Beginning at residue 161 there is an unusual cluster of seven aspartate residues in a span of eight amino acids, a finding reminiscent of the six-aspartate cluster of LipL36 (16). The secondary structure of LipL32 is predicted to be 24% alpha-helix and 24% beta-sheet by using the GOR secondary structure prediction method version IV (14; P. B.-I., Lyonnais, GOR IV Secondary Structure, http://pbil.ibcp.fr/cgi-bin/npsa-automat.pl?page=gor4.html, 1999). The relatively low amount of alpha-helical structure is consistent with the finding of only 10 potential helix-stabilizing salt bridges conforming to the “N+4” rule (23). This is in contrast to LipL41, which has 22 potential salt bridges and is predicted to be 60% alpha-helix (31).
Expression of LipL32 in Leptospira species.
To address the level and distribution of LipL32 expression, immunoblot analysis was performed on a panel of Leptospira species, using antiserum from a rabbit immunized with purified His6-LipL32 (Fig. 3). The LipL32 antiserum is reactive with a single band with a molecular mass of 32 kDa, demonstrating the specificity of the LipL32 antiserum. The molecular mass and amount of LipL32 expressed among all seven pathogenic Leptospira species is highly conserved. LipL32 is expressed in relatively the same amount by all leptospiral pathogens tested. There was a positive correlation between leptospiral pathogenicity and reactivity with antiserum to LipL32. No LipL32 was detected in the nonpathogenic Leptospira species L. biflexa, L. meyeri, and L. wolbachii or in the related nonpathogen L. illini.
FIG. 3.
Immunoblot of a panel of Leptospira species obtained by using LipL32 antiserum. LipL32 antiserum detected a single band in each of the lanes containing pathogenic Leptospira species. LipL32 expression is highly conserved among the pathogenic Leptospira species, L. interrogans, L. kirschneri, L. borgpetersenii, L. inadai, L. noguchii, L. santarosai, and L. weilii. L. biflexa, L. meyeri, and L. wolbachii are nonpathogenic species, as is the related organism Leptonema illini.
Behavior of LipL32 during Triton X-114 extraction and phase partitioning.
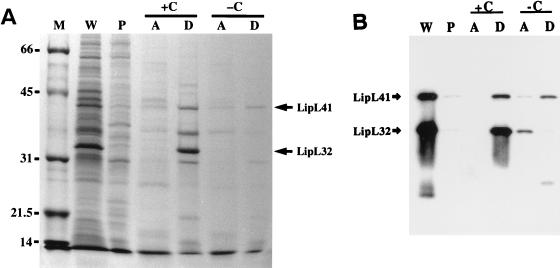
We analyzed the behavior of L. kirschneri LipL32 in the nonionic detergent Triton X-114 (Fig. 4). We found that extraction of the leptospiral outer membrane with Triton X-114 resulted in complete solubilization of LipL32. No LipL32 was seen in the detergent-insoluble pellet. When 20 mM CaCl2 was added to the detergent-solubilized material prior to phase partitioning, LipL32 was found to partition exclusively into the detergent phase. Lipoproteins characteristically partition into the Triton X-114 detergent phase because of the hydrophobicity of the fatty acids. In contrast to LipL41, LipL32 was found to be completely absent from the detergent phase when phase partitioning was performed without addition of CaCl2. Comparison of the Coomassie brilliant blue-stained gel (Fig. 4A) with the immunoblot probed with LipL32 and LipL41 antisera (Fig. 4B) demonstrates that LipL32 is the most prominent protein in the leptospiral protein profile. The results presented here for L. kirschneri LipL32 mirror those found with the previously described L. interrogans 31-kDa MOMP, strongly indicating that they are the same protein (41).
FIG. 4.
Behavior of LipL32 and other leptospiral proteins in Triton X-114. Triton X-114 fractions of L. kirschneri organisms were separated by SDS-PAGE and stained with Coomassie brilliant blue (A) or probed with either a combination of LipL32 and LipL41 antisera (B). Fractions analyzed were the whole organism (W), Triton X-114-insoluble pellet (P), and aqueous-phase (A) or detergent-phase (D) material with (+C) or without (−C) 20 mM CaCl2. LipL32, but not LipL41, requires CaCl2 to avoid proteolytic degradation during Triton X-114 phase partitioning.
Specificity of LipL32 antiserum.
The specificity of the LipL32 antiserum has been demonstrated both by one-dimensional (Fig. 3) and two-dimensional (data not shown) immunoblots. Preimmune serum (prior to rabbit immunization with purified, recombinant protein) was tested and found not to be reactive with any leptospiral antigens (data not shown). The LipL32 antiserum was prepared by the same method and with the same adjuvant as that used in the preparation of antisera specific for several other leptospiral outer membrane proteins, including OmpL1 (15) and the lipoproteins LipL36 (16) and LipL41 (31). Our prior studies show that each of these antisera react only with the unique protein used in the immunization and not with the 32-kDa antigen identified by the LipL32 antiserum. Prior immunoprecipitation experiments showed that LipL36 and LipL41 antisera selectively immunoprecipitate proteins of the correct molecular mass; in neither case did immunoprecipitation of a 32-kDa protein occur (17, 31). In addition, immunohistochemistry studies with L. kirschneri-infected hamster kidney sections demonstrated differential reactivity; OmpL1 and LipL41 antisera were positive, while LipL36 antiserum was negative (3). These findings confirm the specificity of the antisera used in our immunoprecipitation and immunohistochemistry experiments.
L. kirschneri acylates LipL32.
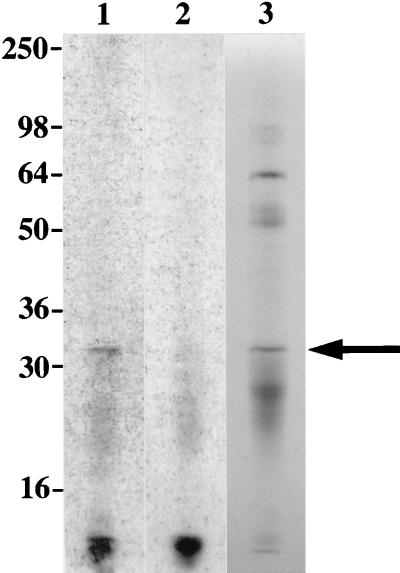
Intrinsic labeling of L. kirschneri with [3H]palmitate resulted in the incorporation of label in leptospiral LipL32 immunoprecipitated with LipL32 antiserum (Fig. 5, lane 1). When an SDS-PAGE gel containing the same material was stained with Coomassie brilliant blue in a solution containing 10% acetic acid, the immunoprecipitated LipL32 could be visualized (Fig. 5, lane 3), but it was no longer labeled (Fig. 5, lane 2), indicating that the chemical bond between [3H]palmitate and the N-terminal cysteine of LipL32 is acid labile. This experiment also demonstrates that intrinsic labeling of leptospiral LipL32 with [3H]palmitate does not involve the incorporation of tritium into amino acid biosynthetic pathways.
FIG. 5.
LipL32 is acylated by L. kirschneri. LipL32 was isolated from L. kirschneri organisms intrinsically labeled with [3H]palmitate by extraction with Triton X-100 and immunoprecipitation with LipL32 antiserum. Lanes 1, autoradiogram of immunoprecipitated LipL32 without treatment of SDS-PAGE gel with acetic acid; 2, autoradiogram of immunoprecipitated LipL32 after treatment of SDS-PAGE gel with acetic acid; 3, immunoprecipitated LipL32 stained with Coomassie brilliant blue. Acetic acid treatment of SDS-PAGE gel results in a loss of 3H label, a finding consistent with an acid-labile bond between the labeled palmitate and the N-terminal cysteine of LipL32. Locations of molecular size standards are shown (in kilodaltons) on the left.
Immunohistochemistry with antiserum specific for LipL32.
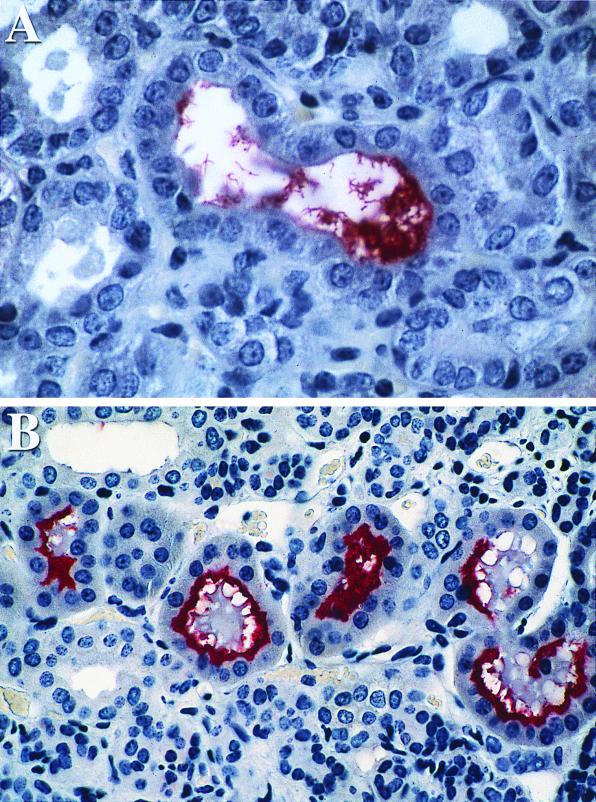
Leptospires within the proximal tubules of kidney sections obtained 10 and 28 days after infection with virulent L. kirschneri stained positive with LipL32 antiserum. Immunohistochemical visualization of discrete organisms is possible at 10 days after infection (Fig. 6A). Staining was more intense at 28 days than at 10 days, in correlation with the greater burden of organisms present at the later time point (3). At 28 days after infection, adherent organisms were detected as a continuous rim of intense staining lining the luminal surface of the tubular epithelial cells (Fig. 6B). In contrast to the distribution of leptospiral LPS or OmpL1 (3), little LipL32 antigen was observed at the sites of interstitial inflammatory cell infiltrates. In control experiments, normal rabbit serum did not show reactivity to infected hamster kidney sections. Uninfected hamster kidney sections were used as controls in a previous study using antisera to three other leptospiral proteins (OmpL1, LipL36, and LipL41), none of which showed any staining of uninfected kidney (3).
FIG. 6.
Immunohistochemistry of kidney tissue obtained at 10 days (A) and 28 days (B) postinfection with virulent L. kirschneri by using LipL32 antiserum. Antigen was detected on leptospires within the renal tubular lumen at both time points. The increased reactivity at 28 days after challenge is consistent with a higher burden of organisms.
Humoral immune response to leptospiral proteins in patients with leptospirosis.
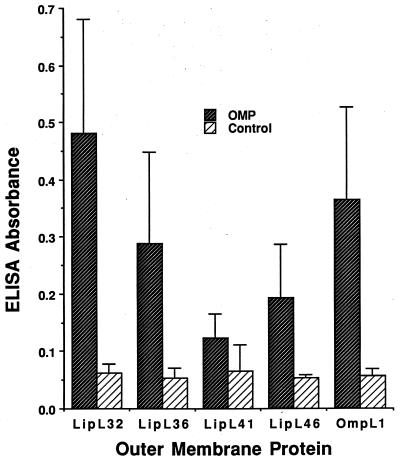
Convalescent sera from five patients with culture-proven infection due to L. kirschneri serovar bim were used to compare the ELISA reactivity with five leptospiral His6 fusion proteins. As shown in Fig. 7, LipL32 and the porin OmpL1 were found to be stronger leptospiral immunogens during naturally occurring infection than the three other leptospiral lipoproteins LipL36, LipL41, or LipL46 (P < 0.05). Control experiments with 62 normal human sera found no significant ELISA reactivity with LipL32 (A. Ko, personal communication).
FIG. 7.
Antibodies to L. kirschneri outer membrane proteins (OMPs) in humans with leptospirosis. Mean ELISA readings to leptospiral outer membrane proteins in the convalescent sera from five leptospirosis patients from Barbados are shown. Control readings are from wells with sera but without antigen. Standard deviations are shown by the error bars. Differences between wells with or without antigen were significant for all five membrane proteins (P < 0.01), but there was a greater degree of statistical significance for OmpL1 (P < 0.001) and LipL32 (P < 0.0001).
DISCUSSION
In this study we have characterized LipL32, the most prominent protein in the leptospiral total protein profile, and the major Triton X-114 detergent-phase protein. Extraction of L. kirschneri with the nonionic detergents Triton X-100 or Triton X-114 solubilizes the leptospiral outer membrane, including the LPS, the porin OmpL1, and the lipoproteins LipL36 and Lip41 (15, 16, 18, 31). The specificity of Triton detergent extraction for the outer membrane has been assessed by demonstrating that the leptospiral cell wall and morphology remain intact after extraction and that the control proteins endoflagella, penicillin-binding proteins, and GroEL are not extractable under these conditions. The conclusion that LipL32 is the leptospiral MOMP is consistent with previous findings of a prominent band of approximately the same molecular mass in the protein profiles of leptospiral outer membrane fractions isolated using the technique of Auran et al. (2, 10, 25).
The results of the studies presented here differ in significant ways from previously published reports analyzing leptospiral outer membrane proteins by detergent extraction. Although, in an earlier study, a 32-kDa major protein was apparent in the total protein profile of L. kirschneri, it was absent from the Triton X-114 detergent phase because CaCl2 was not included during the phase-partitioning step (18). In a study published at approximately the same time on the closely related organism L. interrogans, Zuerner et al. demonstrated that CaCl2, ZnCl2, or CuCl2 must be present during the phase-partitioning step to prevent the loss of LipL32 from the Triton X-114 detergent phase (41). In the current study, we have demonstrated that these are properties of LipL32 by using antiserum specific for this protein (Fig. 4), which was made possible by the isolation of the lipL32 gene and the generation of purified recombinant protein used to immunize rabbits.
The CaCl2 requirement for stability of LipL32 is not understood. Zuerner et al. proposed that the loss of LipL32 during Triton X-114 phase partitioning is due to degradation by an endogenous protease (41). In this study, we found that CaCl2 is not required until the temperature is raised to 37°C to separate the Triton X-114 extract into aqueous and detergent phases. Neither temperature nor Triton X-114 are sufficient alone to degrade LipL32. Presumably, the combination of heat and detergent result in denaturation of LipL32, which in turn exposes sites sensitive to proteolytic degradation. We were unable to determine whether or not LipL32 was autocatalytic, since attempts to reproduce LipL32 degradation using an in vitro-synthesized protein were unsuccessful. The protective effect of CaCl2 could operate in several ways. It is possible that Ca2+ binding is required for conformational stability of LipL32. On the other hand, the finding that several Triton X-114 detergent-phase proteins other than LipL32 are also partially or completely lost from the Triton X-114 extract during phase partitioning without CaCl2 (Fig. 4) suggests that the mechanism of Ca2+ stabilization is indirect. For example, either the putative protease is inhibited by Ca2+ or leptospiral phospholipids and/or LPS associated with leptospiral outer membrane proteins require CaCl2 for their stability. This latter explanation would be consistent with the role of divalent cations in maintaining the integrity of the E. coli outer membrane. Based on this idea, it would be anticipated that calcium chelators, such as EDTA, would destabilize the leptospiral outer membrane. Further studies to elucidate the protective mechanism of CaCl2 would shed light on leptospiral outer membrane structure and the interaction of leptospiral outer membrane proteins with other outer membrane components.
Another important aspect of the method described here is the inclusion of 150 mM NaCl in the buffer used in the initial extraction at 4°C with Triton X-114. Previous studies that did not include 150 mM NaCl in the extraction buffer found that Triton detergent extraction of proteins such as OmpL1 and LipL41 was incomplete (15, 16, 31). NaCl is probably important in the solubility of these and potentially other leptospiral outer membrane proteins. On the basis of the findings in this study, we believe that the method of Triton X-114 extraction and phase partitioning described here and in the study by Zuerner et al. (41) results in isolation of fractions which most accurately represent the components of the leptospiral outer membrane. We have not excluded the possibility that some cytoplasmic membrane proteins are extractable under these conditions. Confirmation of the Triton X-114 solubilization and phase-partitioning approach will require the development of better markers for the cytoplasmic membrane and independent approaches for the separation of the cytoplasmic membrane from the outer membrane.
In this study, we have presented several lines of evidence to support the conclusion that LipL32 is a lipoprotein, covalently modified by fatty acid(s) at its amino-terminal cysteine residue. The atypical lipoprotein signal peptidase cleavage site of LipL32 and the inefficient cleavage of LipL32 expressed in E. coli (data not shown) are consistent with the idea that the leptospiral lipoprotein signal peptidase and/or other components of the leptospiral membrane protein export pathway differ in their specificity from those of E. coli (D. A. Haake, submitted for publication). Even though the two other lipoproteins that have been described in Leptospira species, LipL36 and LipL41, have an L-X-Y-C lipoprotein signal peptidase cleavage site, they are also inefficiently processed in E. coli unless the amino acids between leucine and cysteine are altered to conform to those found in the E. coli murein lipoprotein (17). Like LipL36 and LipL41, LipL32 is labeled by intrinsic labeling of L. kirschneri by [3H]palmitate. Previous [3H]palmitate labeling studies of leptospiral lipoproteins were subject to the criticism that the tritium label could have been incorporated into amino acids through beta-oxidation of [3H]palmitate. In this study, we have shown that in the process of staining LipL32 with Coomassie brilliant blue and treatment with 10% acetic acid, the tritium label is lost. This is consistent with an acid-labile bond between palmitate and the N-terminal cysteine, a pattern found in other spirochetal lipoproteins (8, 33). Another form of evidence that LipL32 is a lipoprotein is its behavior during Triton X-114 phase partitioning. Native LipL32 partitions selectively into the hydrophobic detergent phase, a result consistent with modification by hydrophobic fatty acids, while the recombinant His6-LipL32 fusion protein lacking N-terminal lipidation partitions into the hydrophilic aqueous phase (data not shown).
Although there is a discrepancy of >4 kDa between the observed mobility of LipL32 and its calculated molecular mass of 27.6 kDa, we believe that the lipL32 sequence data we have presented are correct. The >99% nucleotide sequence identity between the L. kirschneri LipL32 and other versions of LipL32 make it unlikely that there were internal deletions during the cloning process. The location of the TAA stop codon is confirmed by the inverted repeat located 23 bp downstream. A likely explanation for the slower-than-predicted electrophoretic mobility of LipL32 is the high percentage of acidic residues (34 of 253 [13.4%]), which could reduce SDS binding. Leptospiral LipL41 (31) and the T. pallidum lipoprotein TpN34 (34) are two other examples of lipoproteins with high percentages of acidic residues associated with discrepancies between the observed electrophoretic mobility and the calculated molecular mass.
Our findings indicate that LipL32 is expressed at high levels both during cultivation and during infection. Some proteins found in organisms grown in leptospiral culture conditions are not expressed during infection. For example, LipL36 is expressed at high levels by cultivated L. kirschneri but is not detectable either within infected hamster kidneys by immunohistochemistry or by sera from hamsters infected with host-derived organisms (3, 16). In contrast, OmpL1 and LipL41 were detectable both by immunohistochemistry and by immunoblot (3). The immunohistochemistry results presented here indicate that, like OmpL1 and LipL41, LipL32 is expressed by L. kirschneri within renal tubules (Fig. 6). In addition, sera from humans with leptospirosis had a stronger antibody response to LipL32 than to the four other leptospiral membrane proteins (Fig. 7). These results suggest that LipL32 may be useful in the development of new approaches for the serological diagnosis of leptospirosis. Further studies are needed to define the time course of the humoral immune response to LipL32 during the course of leptospirosis. Research on outer membrane proteins expressed by Leptospira spp. during infection is essential both to an understanding of leptospiral pathogenesis and for development of subunit vaccines. It is anticipated that the reagents and findings developed during the course of this investigation will make it possible to elucidate the role of LipL32 in the interaction of pathogenic Leptospira spp. with the mammalian host.
ACKNOWLEDGMENTS
This work was supported by funding from a Public Health Service grant AI-34431 (to D.A.H.).
We thank Tonia McNunn and Annette Olson for excellent technical assistance.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Auran N E, Johnson R C, Ritzi D M. Isolation of the outer sheath of Leptospira and its immunogenic properties in hamsters. Infect Immun. 1972;5:968–975. doi: 10.1128/iai.5.6.968-975.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnett J K, Barnett D, Bolin C A, Summers T A, Wagar E A, Cheville N F, Hartskeerl R A, Haake D A. Expression and distribution of leptospiral outer membrane components during renal infection of hamsters. Infect Immun. 1999;67:853–861. doi: 10.1128/iai.67.2.853-861.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergstrom S, Bundoc V G, Barbour A G. Molecular analysis of linear plasmid-encoded major surface proteins, OspA and OspB, of the Lyme disease spirochaete Borrelia burgdorferi. Mol Microbiol. 1989;3:479–486. doi: 10.1111/j.1365-2958.1989.tb00194.x. [DOI] [PubMed] [Google Scholar]
- 5.Bolin C A, Cassells J A, Zuerner R L, Trueba G. Effect of vaccination with a monovalent Leptospira interrogans serovar hardjo type hardjo-bovis vaccine on type hardjo-bovis infection of cattle. Am J Vet Res. 1991;52:1639–1643. [PubMed] [Google Scholar]
- 6.Bolin C A, Thiermann A B, Handsaker A L, Foley J W. Effect of vaccination with a pentavalent leptospiral vaccine on Leptospira interrogans serovar hardjo type hardjo-bovis infection of pregnant cattle. Am J Vet Res. 1989;50:161–165. [PubMed] [Google Scholar]
- 7.Bolin C A, Zuerner R L, Trueba G. Effect of vaccination with a pentavalent leptospiral vaccine containing Leptospira interrogans serovar hardjo type hardjo-bovis on type hardjo-bovis infection of cattle. Am J Vet Res. 1989;50:2004–2008. [PubMed] [Google Scholar]
- 8.Brandt M E, Riley B S, Radolf J D, Norgard M V. Immunogenic integral membrane proteins of Borrelia burgdorferi are lipoproteins. Infect Immun. 1990;58:983–991. doi: 10.1128/iai.58.4.983-991.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brenner D J, Kaufmann A F, Sulzer K R, Steigerwalt A G, Rogers F C, Weyant R S. Further determination of DNA relatedness between serogroups and serovars in the family Leptospiraceae with a proposal for Leptospira alexanderi sp. nov. and four new Leptospira genomospecies. Int J Syst Bacteriol. 1999;49(Pt. 2):839–858. doi: 10.1099/00207713-49-2-839. [DOI] [PubMed] [Google Scholar]
- 10.Brown J A, LeFebvre R B, Pan M J. Protein and antigen profiles of prevalent serovars of Leptospira interrogans. Infect Immun. 1991;59:1772–1777. doi: 10.1128/iai.59.5.1772-1777.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burman N, Bergstrom S, Restrepo B I, Barbour A G. The variable antigens Vmp7 and Vmp21 of the relapsing fever bacterium Borrelia hermsii are structurally analogous to the VSG proteins of the African trypanosome. Mol Microbiol. 1990;4:1715–1726. doi: 10.1111/j.1365-2958.1990.tb00549.x. [DOI] [PubMed] [Google Scholar]
- 12.Chamberlain N R, Brandt M E, Erwin A L, Radolf J D, Norgard M V. Major integral membrane protein immunogens of Treponema pallidum are proteolipids. Infect Immun. 1989;57:2872–2877. doi: 10.1128/iai.57.9.2872-2877.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chamberlain N R, DeOgny L, Slaughter C, Radolf J D, Norgard M V. Acylation of the 47-kilodalton major membrane immunogen of Treponema pallidum determines its hydrophobicity. Infect Immun. 1989;57:2878–2885. doi: 10.1128/iai.57.9.2878-2885.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garnier J, Gibrat J-F, Robson B. GOR secondary structure prediction method version IV. Methods Enzymol. 1996;266:540–553. doi: 10.1016/s0076-6879(96)66034-0. [DOI] [PubMed] [Google Scholar]
- 15.Haake D A, Champion C I, Martinich C, Shang E S, Blanco D R, Miller J N, Lovett M A. Molecular cloning and sequence analysis of the gene encoding OmpL1, a transmembrane outer membrane protein of pathogenic Leptospira spp. J Bacteriol. 1993;175:4225–4234. doi: 10.1128/jb.175.13.4225-4234.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haake D A, Martinich C, Summers T A, Shang E S, Pruetz J D, McCoy A M, Mazel M K, Bolin C A. Characterization of leptospiral outer membrane lipoprotein LipL36: downregulation associated with late-log-phase growth and mammalian infection. Infect Immun. 1998;66:1579–1587. doi: 10.1128/iai.66.4.1579-1587.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haake D A, Mazel M K, McCoy A M, Milward F, Chao G, Matsunaga J, Wagar E A. Leptospiral outer membrane proteins OmpL1 and LipL41 exhibit synergistic immunoprotection. Infect Immun. 1999;67:6572–6582. doi: 10.1128/iai.67.12.6572-6582.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haake D A, Walker E M, Blanco D R, Bolin C A, Miller M N, Lovett M A. Changes in the surface of Leptospira interrogans serovar grippotyphosa during in vitro cultivation. Infect Immun. 1991;59:1131–1140. doi: 10.1128/iai.59.3.1131-1140.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayashi S, Wu H C. Lipoproteins in bacteria. J Bioenerg Biomembr. 1990;22:451–471. doi: 10.1007/BF00763177. [DOI] [PubMed] [Google Scholar]
- 20.Hookey J V, Bryden J, Gatehouse L. The use of 16S rDNA sequence analysis to investigate the phylogeny of Leptospiaceae and related spirochaetes. J Gen Microbiol. 1993;139(Pt. 11):2585–2590. doi: 10.1099/00221287-139-11-2585. [DOI] [PubMed] [Google Scholar]
- 21.Johnson R C, Harris V G. Differentiation of pathogenic and saprophytic letospires. I. Growth at low temperatures. J Bacteriol. 1967;94:27–31. doi: 10.1128/jb.94.1.27-31.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.Marqusee S, Baldwin R L. Helix stabilization by Glu−…Lys+ salt bridges in short peptides of de novo design. Proc Natl Acad Sci USA. 1987;84:8898–8902. doi: 10.1073/pnas.84.24.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller J H. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 25.Nunes-Edwards P L, Thiermann A B, Bassford P J, Jr, Stamm L V. Identification and characterization of the protein antigens of Leptospira interrogans serovar hardjo. Infect Immun. 1985;48:492–497. doi: 10.1128/iai.48.2.492-497.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pugsley A P. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 28.Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 29.Shang E S, Exner M M, Summers T A, Martinich C, Champion C I, Hancock R E W, Haake D A. The rare outer membrane protein, OmpL1, of pathogenic Leptospira species is a heat-modifiable porin. Infect Immun. 1995;63:3174–3181. doi: 10.1128/iai.63.8.3174-3181.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shang E S, Skare J T, Exner M M, Blanco D R, Kagan B L, Miller J N, Lovett M A. Isolation and characterization of the outer membrane of Borrelia hermsii. Infect Immun. 1998;66:1082–1091. doi: 10.1128/iai.66.3.1082-1091.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shang E S, Summers T A, Haake D A. Molecular cloning and sequence analysis of the gene encoding LipL41, a surface-exposed lipoprotein of pathogenic Leptospira species. Infect Immun. 1996;64:2322–2330. doi: 10.1128/iai.64.6.2322-2330.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steiner G, Steiner G. New simple silver stain for demonstration of bacteria, spirochetes, and fungi in sections of paraffin-embedded tissue blocks. J Lab Clin Med. 1944;29:868–871. [Google Scholar]
- 33.Swancutt M A, Radolf J D, Norgard M V. The 34-kilodalton membrane immunogen of Treponema pallidum is a lipoprotein. Infect Immun. 1990;58:384–392. doi: 10.1128/iai.58.2.384-392.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swancutt M A, Riley B S, Radolf J D, Norgard M V. Molecular characterization of the pathogen-specific, 34-kilodalton membrane immunogen of Treponema pallidum. Infect Immun. 1989;57:3314–3323. doi: 10.1128/iai.57.11.3314-3323.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thiermann A B. Leptospirosis: current developments and trends. J Am Vet Med Assoc. 1984;184:722–725. [PubMed] [Google Scholar]
- 36.Thiermann A B, McClellan R D, Hill H T. Improved techniques for the isolation of leptospires from swine abortion cases. Ann Proc Am Assoc Vet Lab Diagn. 1984;27:233–244. [Google Scholar]
- 37.Thomas W, Sellwood R. Molecular cloning, expression, and DNA sequence analysis of the gene that encodes the 16-kilodalton outer membrane lipoprotein of Serpulina hyodysenteriae. Infect Immun. 1993;61:1136–1140. doi: 10.1128/iai.61.3.1136-1140.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37a.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weigel L M, Belisle J T, Radolf J D, Norgard M V. Digoxigenin-ampicillin conjugate for detection of penicillin-binding proteins by chemiluminescence. Antimicrob Agents Chemother. 1994;38:330–336. doi: 10.1128/aac.38.2.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yelton D B, Charon N W. Cloning of a gene required for tryptophan biosynthesis from Leptospira biflexa serovar patoc into Escherichia coli. Gene. 1984;28:147–152. doi: 10.1016/0378-1119(84)90251-8. [DOI] [PubMed] [Google Scholar]
- 40.Zuerner R L, Herrmann J L, Saint Girons I. Comparison of genetic maps for two Leptospira interrogans serovars provides evidence for two chromosomes and intraspecies heterogeneity. J Bacteriol. 1993;175:5445–5451. doi: 10.1128/jb.175.17.5445-5451.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zuerner R L, Knudtson W, Bolin C A, Trueba G. Characterization of outer membrane and secreted proteins of Leptospira interrogans serovar pomona. Microb Pathog. 1991;10:311–322. doi: 10.1016/0882-4010(91)90014-2. [DOI] [PubMed] [Google Scholar]