Abstract
Remarkable clinical benefits in several advanced cancers are observed under the treatment of immune checkpoint inhibitor (ICI) agents. However, only a smaller proportion of patients respond to the treatments. Reelin (RELN) is frequently mutated in the cancer genome. In this study, the RELN mutation association with ICI treatment efficacy in melanoma and non-small cell lung cancer (NSCLC) was elucidated. Data from 631 melanoma and 109 NSCLC patients with both ICI treatment data and pre-treatment mutational profiles were collected. In addition, from the Cancer Genome Atlas (TCGA) project, we also obtained both tumors to explore the immunologic features behind RELN mutations. Melanoma patients with RELN mutations exhibited a favorable ICI survival benefit when compared with wild-type patients (HR: 0.66, 95% CI: 0.51–0.87, p = 0.003). A higher response rate was also noticed in RELN-mutated patients (38.9% vs. 28.3%, p = 0.017). The association of RELN mutations with a preferable immunotherapy outcome and response was further confirmed in NSCLC. Further exploration demonstrated that favorable immunocyte infiltration and immune response signaling pathways were found in patients with RELN mutations. In this study, RELN mutations were identified to connect with a better immune microenvironment and an improved ICI efficacy in melanoma and NSCLC, which provides a potential biomarker for immunological feature evaluation and immunotherapeutic outcome prediction at the molecular level.
Keywords: RELN mutations, immune checkpoint inhibitor, melanoma, non-small cell lung cancer, molecular determinant
1. Introduction
Recently, mutations in a single gene were reported to act as potential indicators for tumor immunogenicity and immunotherapeutic response. Li et al. leveraged the somatic mutational profiles of gastric cancer (GC) patients and found that MUC16 mutations were linked with an elevated TMB and immunogenicity-related signaling pathways [1]. Further analysis showed that MUC16-mutated GC patients exhibited an improved survival outcome, which confirms the clinical immunotherapy significance of MUC16 mutations [1]. Consistent with Li et al.’s results, two recent studies [2,3] validated that MUC16 mutations were predictive of a favorable ICI treatment response and outcome. Mutations in POLE [4], FAT1 [5], TP53 [6], COL3A1 [7], and HSPG2 [8] were also identified to be associated with a better ICI prognosis or response rate. In addition, ICI treatment resistance was observed in patients with mutations of JAK1/2 [9,10] or B2M [11].
Immune checkpoint inhibitors (ICIs) directing the programmed cell death (ligand) 1 [PD-(L)1] and cytotoxic T lymphocyte antigen 4 (CTLA-4) signals have become the routine clinical treatment regimens for multiple cancer types [12,13]. However, durable clinical response to ICI treatments has been observed only in a few patients [14]. Therefore, multiple biomarkers are determined to assist in selecting patients who will respond effectively to ICI treatments. Several biomarkers have been identified for predicting immunotherapeutic efficacy, such as tumor mutation burden (TMB) [15], PD-L1 protein expression on tumor cells [16], and neoantigen burden [17,18]. Nevertheless, many shortcomings have emerged regarding the above biomarkers in clinical practice; for instance, the uncertain cutoff value, the cost of genomic sequencing is too high, and the predictive rate is unsatisfactory, limiting the wide utilization of these markers in cancer immunotherapy [19]. Therefore, more effective immunotherapeutic determinants are urgently necessary for guiding treatment efficacy.
Reelin (RELN) encodes a large secreted extracellular matrix protein, and it is critical for cell positioning. Studies conducted in the null reeler mouse have indicated that the serine protease activity of RELN is critical for developing the brain [19]. Pathways triggered by RELN depend on the recruitment of distinct cell surface receptors, i.e., very low density lipoprotein receptor (VLDLR) [20], apolipoprotein E receptor 2 (ApoER2 or LRP8) [20,21], α3β1 integrin [22], and members of the cadherin-related neuronal receptor (CNR) family [23]. A number of studies have revealed changes in the expression of RELN in different cancer types [24]. RELN expression has been observed to reduce in breast [25], colorectal [26], and pancreatic cancers [27], while it has been noticed to be increased in retinoblastoma [28], myelomas [29], and prostate cancers [30]. To our knowledge, the immunological and cancer immune treatment implications of RELN mutations have not been reported in clinical practice.
Since melanoma and NSCLC are two cancer types commonly used immunotherapies, in this genomic association analysis, we retrospectively integrated a total of 631 melanoma and 119 NSCLC samples; their pre-treatment mutational data and clinical ICI therapy information were also obtained. We discovered that RELN mutations may be a potential biomarker for cancer immunotherapeutic efficacy prediction.
2. Materials and Methods
2.1. Sample Collection and Genomic Data Processing
From previously published melanoma [10,31,32,33,34,35,36,37] and NSCLC [38,39] immunogenomic studies, we retrospectively collected a total of 631 and 109 samples, with both somatic mutational data and ICI treatment information (i.e., response and survival information). All the above samples were treated with checkpoint blockade treatments of CTLA-4, PD-1/PD-L1, or a combination drug. Taking into account that the mutational data were obtained from distinct sequencing platforms, we therefore uniformly unscrambled them with the Oncotator [40]. In this work, non-synonymous mutations were included for the subsequent analyses. Detailed clinical data and immunotherapy response information for curated melanoma and NSCLC samples are exhibited in Tables S1 and S2, respectively.
A total of 457 melanoma and 995 NSCLC cases with mutational data, gene expression profiles, and clinicopathological information in the Cancer Genome Atlas (TCGA) cohort were acquired from Genome Data Commons (https://gdc.cancer.gov, accessed on 1 September 2022). In particular, the gene expression profiles of both tumors in the TCGA were employed for the immunological mechanism exploration of RELN mutations.
2.2. Detection of Tumor Mutational Signatures
Tumor mutational signatures were determined by using a nonnegative matrix factorization-based algorithm proposed by a recent study [41], which could divide the somatic mutational matrix A into 2 nonnegative matrices W and H (i.e., A ≈ W × H). Of these, W indicates the determined mutational signatures and H indicates the mutational activities for each signature. All identified signatures were then compared with 30 well-annotated signatures reserved in the COSMIC (version 2, https://cancer.sanger.ac.uk/cosmic, accessed on 1 September 2022) based on the cosine similarity.
2.3. Infiltration Abundance of Immune Cell Subtypes
We used the CIBERSORT algorithm [42] to conclude the distinct infiltration levels of 22 immunocytes in RELN-mutant and wild-type subgroups. A total of 547 feature genes for the above immune cells, termed LM22 signature within the CIBERSORT, were employed to evaluate infiltration levels.
2.4. IFNγ-Related Gene Signature
Interferon γ (IFNγ) signature [43] includes immune genes (i.e., GBP1, IFI16, IFI30, IFNG, IRF1, STAT1, TAP1, TAP2, PSMB9, IL15RA, GZMA, GZMB, CXCL10, CXCL9, and TBX21) associated with antigen presentation, cytotoxic activity, and adaptive immune response. A previous study has demonstrated that this T cell-inflamed gene expression signature could serve as an indicator for quantifying tumor microenvironment and is predictive of the clinical response to anti-PD-1 therapies.
2.5. GSVA and GSEA
We performed differential expression analysis of the whole genome between RELN-mutated and wild-type subgroups by using the DESeq2 R package [44]. All genes with their corresponding t values obtained from differential analysis were put into fgsea functions embedded in R fgsea packages to conduct gene set enrichment analysis (GSEA). Signaling pathways in the Hallmark database were utilized to infer dysregulated pathways. In addition, in order to calculate the enrichment scores of IFNγ signature for each sample with specific feature genes, a single sample GSEA method in R GSVA package [45] was used.
2.6. Statistical Analysis
R software (version 4.2.1) was used in this study to complete related analyses and plots. Mutational patterns for specific genes were illustrated with a waterfall plot under R maftools package [46]. In this analysis, TMB was defined as the log2 transformation of total non-synonymous mutations per megabase in both tumors. The Kaplan-Meier method was used to achieve survival curves and the log-rank test was used to compare the survival difference significance. Multivariable regression analyses (i.e., logistic and Cox regression) with multiple confounding factors taken into account were performed with R forestmodel package. RELN mutation associations with continuous and categorical variables were evaluated with Wilcoxon rank-sum test and Fisher exact test, respectively.
3. Results
3.1. ICI Response Information for Melanoma Patients and RELN Mutations
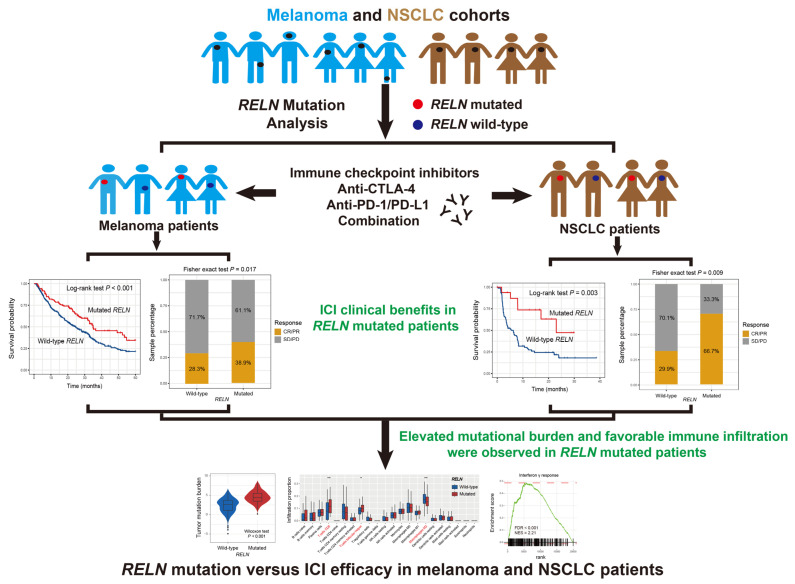
The detailed workflow of this study is shown in Figure 1. A total of 631 melanoma patients were included in this study, of which 193 (30.6%) exhibited the ICI response statuses (i.e., complete response or partial response), 430 (68.1%) were non-responders (i.e., stable disease or progressive disease), and the rest (1.3%) were unavailable. The mutational waterfall plot showed that C > T mutations were the primary base substitution pattern in the melanoma cohort (Figure S1). Mutational patterns of RELN and frequently mutated driver genes in melanoma are illustrated in Figure S1. A total of 160 of the 631 patients (25.4%) harbored RELN mutations and RELN mutation-induced amino acid changes are exhibited using a lollipop plot in Figure S2.
Figure 1.
The detailed workflow operating in this work to explore the clinical ICI treatment implications of RELN mutations based on the genomic data and immunotherapy information.
3.2. RELN Mutations in Predicting ICI Treatment Efficacy in Melanoma
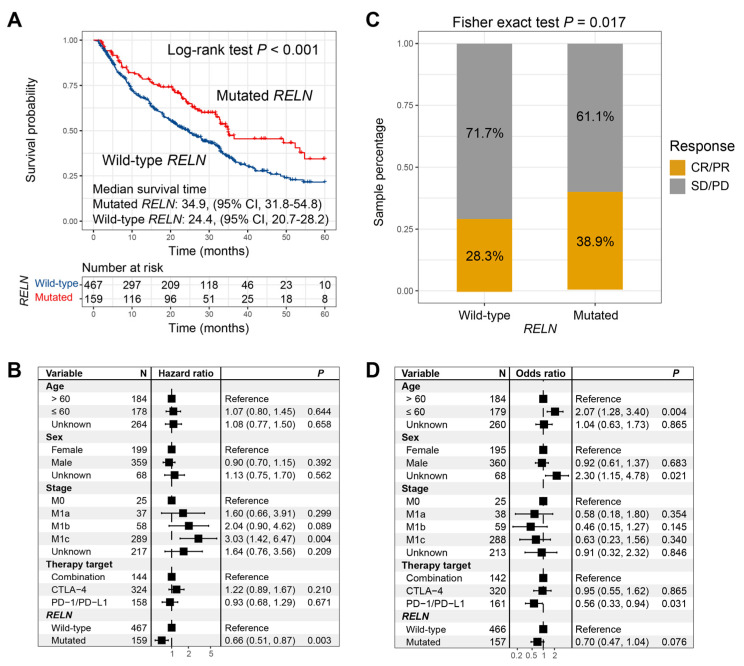
Significant ICI survival benefits were observed in melanoma patients who harbored RELN mutations (median survival time: 34.9 vs. 24.4 months, Log-rank test p < 0.001; Figure 2A). We further incorporated multiple confounding factors (e.g., age, sex, stage, and therapy type) into a multivariable Cox regression analysis, and the association between RELN mutations and favorable ICI survival was still noticed (HR: 0.66, 95% CI: 0.51–0.87, p = 0.003; Figure 2B). Roles of RELN mutations in evaluating ICI treatment prognosis in included single cohorts, and distinct treatment types are illustrated in Figures S3 and S4. Further analysis demonstrated that RELN mutations were also connected with a significantly elevated ICI response rate (38.9% vs. 28.3%, Fisher exact test p = 0.017; Figure 2C). a multivariable logistic regression analysis with the confounding variables taken into account still revealed a positive association (OR: 0.70, 95% CI: 0.47–1.04, p = 0.076; Figure 2D).
Figure 2.
ICI treatment prognosis and response rate analyses of RELN mutations in melanoma. (A) Survival curves of RELN-mutated and wild-type patients. (B) Multivariable Cox regression analysis of RELN mutations was performed with clinical confounders taken into consideration. (C) Bar plot representation of ICI response rates of RELN-mutated and wild-type patients. (D) Multivariable logistic regression analysis of RELN mutations was achieved.
3.3. RELN Mutations in Predicting ICI Treatment Efficacy in NSCLC
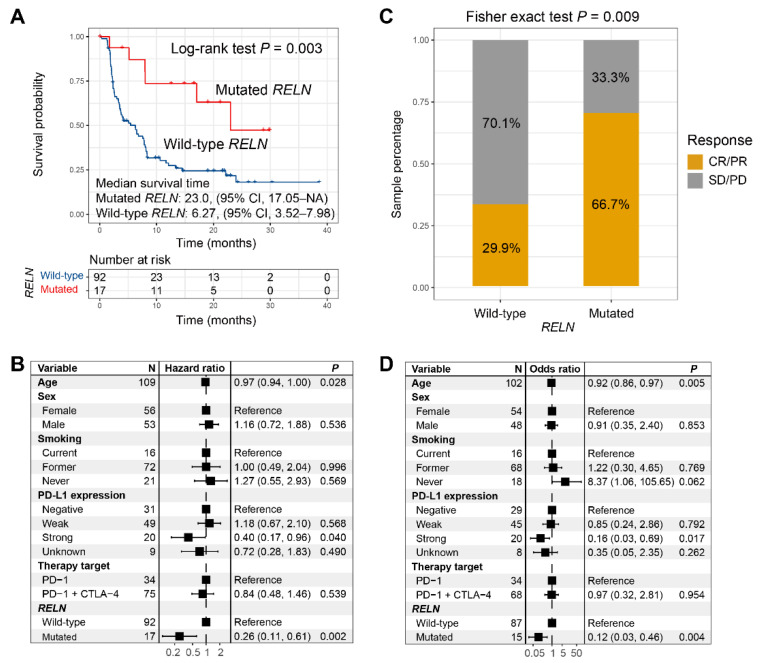
A total of 36 (33.0%) of the 109 included NSCLC patients exhibited the ICI complete response or partial response statuses. RELN mutated in 17 (15.6%) of the above NSCLC patients. Survival analysis revealed that a significantly improved ICI survival benefit was found in NSCLC patients with RELN mutations (median survival time: 23.0 vs. 6.27 months, Log-rank test p = 0.003; Figure 3A). We incorporated multiple clinical confounding factors into a multivariable Cox regression analysis, and the association of RELN mutations with preferable ICI prognosis was still observed (HR: 0.26, 95% CI: 0.11–0.61, p = 0.002; Figure 3B). RELN mutation associations with ICI prognosis in diverse NSCLC ICI types are shown in Figure S5. Subsequent exploration indicated that an enhanced immunotherapeutic response rate was also found in RELN-mutated NSCLC patients (66.7% vs. 29.9%, Fisher exact test p = 0.009; Figure 3C). Multivariable adjusted analysis still confirmed this connection between RELN mutations and elevated ICI response rate (OR: 0.12, 95% CI: 0.03–0.46, p = 0.004; Figure 3D).
Figure 3.
ICI treatment prognosis and response rate analyses of RELN mutations in NSCLC. (A) Survival curves of RELN-mutated and wild-type NSCLC patients. (B) Multivariable Cox regression analysis of RELN mutations was performed with clinical confounders taken into consideration. (C) Bar plot representation of ICI response rates of RELN-mutated and wild-type patients. (D) Multivariable logistic regression analysis of RELN mutations was achieved with multiple confounding variables adjusted.
3.4. RELN Mutation Association with TMB
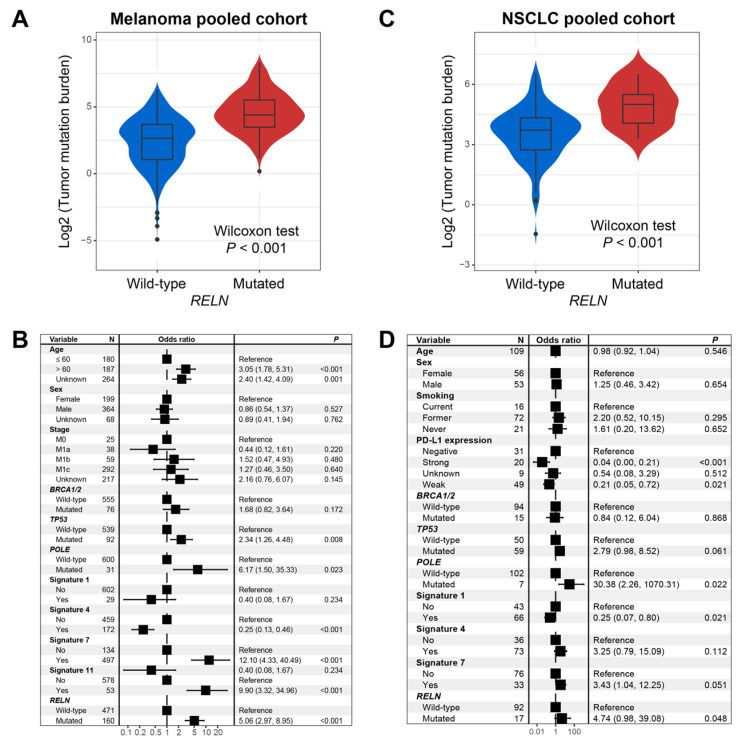
In melanoma, genomic mutational analysis showed that patients with RELN mutations had a markedly higher TMB than RELN wild-type patients (Wilcoxon rank-sum test, p < 0.001; Figure 4A). Several recent studies have demonstrated that mutational signatures are linked with genomic instability and mutation rate. We therefore extracted four mutational signatures from melanoma mutation profiles; these were signatures 1, 4, 7, and 11 (Table S3). Subsequently, in order to adjust confounding factors and obtain a real association between RELN mutations and TMB, we conducted a multivariable logistic regression model with clinical variables, alterations in DNA repair genes, and four mutational signatures taken into account. The association of RELN mutations with elevated TMB was still significant (OR: 5.06, 95% CI: 2.97–8.95, p < 0.001; Figure 4B).
Figure 4.
Association of RELN mutations with TMB in melanoma and NSCLC. (A) Univariate analysis between RELN mutations and TMB in melanoma. (B) Multivariable logistic analysis of RELN mutations was achieved with multiple confounding factors adjusted. (C) Univariate analysis between RELN mutations and TMB in NSCLC. (D) Multivariable logistic analysis of RELN mutations was achieved with multiple confounding factors controlled to acquire a real association.
In NSCLC, we validated the association between RELN mutations and higher TMB (Wilcoxon rank-sum test, p < 0.001; Figure 4C). We also extracted three mutational signatures (i.e., signatures 1, 4, and 7) from NSCLC mutational profiles (Table S4). Consistently, a multivariable logistic regression model with confounders still confirmed that RELN mutations were linked with a significantly enhanced TMB (OR: 4.74, 95% CI: 0.98–39.08, p = 0.048; Figure 4D).
3.5. Immune Infiltration and Signaling Pathways Associated with RELN Mutations
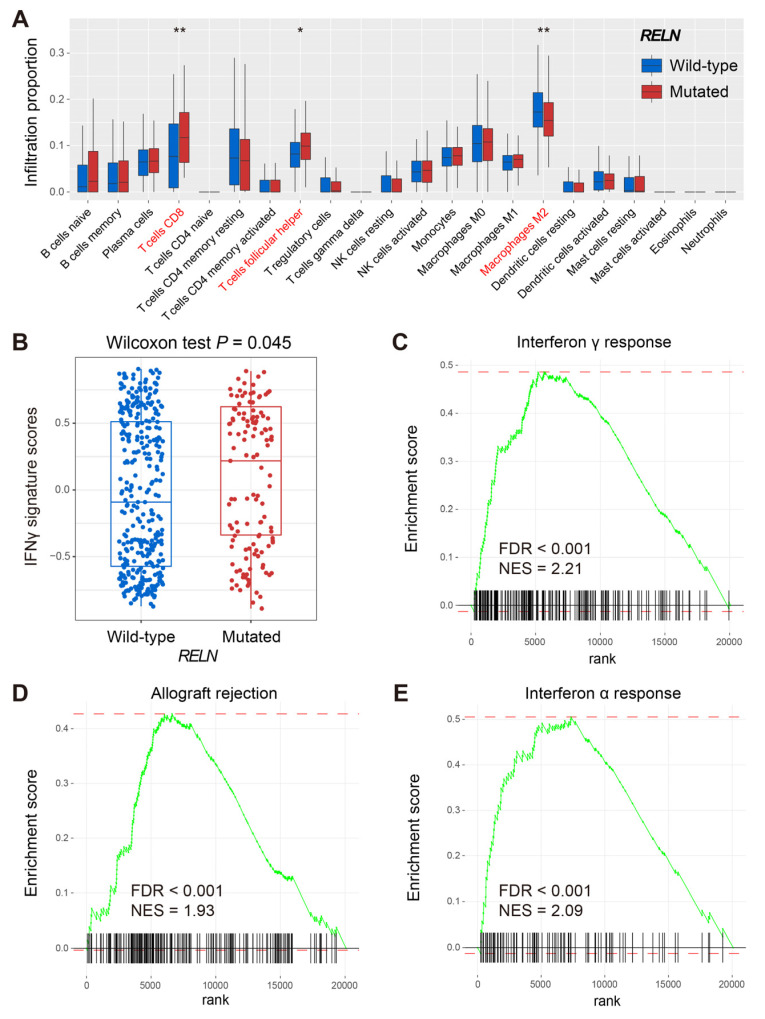
The CIBERSORT algorithm was used to evaluate distinct immunocyte infiltration levels between RELN subgroups in melanoma (Figure 5A). Results showed that pro-inflammatory immunocytes (e.g., CD8 T cells and T follicular helper cells) were significantly enriched in RELN-mutated melanoma patients (Wilcoxon rank-sum test, both p < 0.05). However, the infiltration abundance of immune-suppressive cells (e.g., M2 macrophages) was decreased in the subgroup mutated in this way (Wilcoxon rank-sum test, p < 0.01). The subsequent ssGSEA method revealed that patients with RELN mutations harbored significantly higher enrichment scores of IFNγ signature when compared with wild-type patients (Wilcoxon rank-sum test, p = 0.045; Figure 5B). GSEA results indicated that immunogenicity-related signaling pathways of interferon γ/α response and allograft rejection were observed in RELN-mutated melanoma patients (all FDR < 0.001; Figure 5C–E).
Figure 5.
Immune infiltration and signaling pathways behind RELN mutations in melanoma. (A) CIBERSORT method revealed the distinct immunocyte infiltration in RELN two subgroups. (B) Distinct enrichment scores of IFNγ signature in RELN two subgroups. Immunogenicity-related signaling pathways of (C) interferon γ response, (D) allograft rejection, and (E) interferon α response were enriched in patients with RELN mutations. * p < 0.05, ** p < 0.01.
We also calculated the immunocyte infiltration level differences between two RELN subgroups in NSCLC. Consistently, favorable immunocyte infiltration was observed in patients with RELN mutations (Figure S6).
4. Discussion
Immunotherapies are clinically confirmed as promising cancer treatment strategies, especially for advanced or metastatic cancers. Although remarkable clinical benefits are observed, only a subset of patients is responsive. Therefore, newly identified biomarkers for evaluating ICI efficacy are needed immediately. In this work, we uncovered that RELN mutations were predictive of a better ICI treatment outcome and response in melanoma and NSCLC. Moreover, an elevated TMB and a favorable immune infiltration were also observed in patients with RELN mutations in both tumors. The above findings suggest that RELN mutations may be regarded as a possible indicator for assessing immunotherapeutic efficacy and used for selecting cancer patients to receive immune checkpoint-based therapies.
In our analysis, RELN mutations were found to be connected with the preferable ICI therapy outcome and response in both melanoma and NSCLC patients, which suggests the immunotherapeutic significance of RELN mutations in clinical practice. To explore whether RELN mutations play a role in other therapeutic types, we acquired mutational profiles and clinical features data of melanoma and NSCLC samples from the TCGA. Survival analysis demonstrated that no significant survival differences were noticed between RELN-mutated and wild-type subgroups in both tumors (Log-rank test p = 0.852 and 0.136, respectively; Figure S7). The above findings indicate that RELN mutations may play an efficacy predictive role in immunotherapeutic settings, rather than a prognostic role. Further analyses are necessary to elucidate the roles of RELN mutations in other treatment types.
Tumor mutation burden (TMB) has recently been reported as a promising molecular biomarker for evaluating ICI treatment outcome and response in several cancers [7,47,48,49]. Its high level is always correlated with favorable clinical ICI benefits. Nevertheless, the determination of TMB requires the performance of whole-exome sequencing, which is costly. On the other hand, the cut-off values for stratifying high and low TMB in diverse cancer types are distinct [50]. Several recent studies have revealed that mutations in a single gene, such as POLE [51], TP53 [52], and FAT1 [5], may be the potential surrogates for TMB. In this analysis, we observed that RELN mutations were related to an elevated TMB in both tumors. Taking into account that some confounding factors may influence the real association, we conducted multivariable-adjusted analyses to verify the association of RELN mutations with high TMB. The above findings suggest that RELN mutations may be regarded as a surrogate for TMB to evaluate immune treatment efficacy.
A favorable tumor microenvironment is important for the immune response and treatment efficacy [53]. Tumor-infiltrating immunocytes are vital elements in the microenvironment for regulating a series of biological processes [54]. We therefore explored the distinct immunocyte infiltration levels and signaling pathway distributions in RELN-mutated and wild-type groups. We observed that the higher infiltration abundance of CD8 T cells and the lower abundance of immune-suppressive M2 macrophages were enriched in melanoma patients with RELN mutations. Moreover, the immunogenicity-relevant pathways were also noticed in the group thus mutated. Consistently, in NSCLC, a preferable immunocyte infiltration and immune microenvironment were found in patients with RELN mutations. The above evidence showed that RELN mutations are predictive of better immune infiltration, which further supports the observed relationship between RELN mutation and favorable ICI treatment efficacy.
TMB and neoantigen burden have emerged as promising indicators for assessing ICI efficacy, and previous evidence has demonstrated their positive connection with the immunotherapy response rate and outcome via multiple clinical trials [15,47,55]. Nevertheless, a few studies concluded controversial results; that is, high TMB could not always accurately predict ICI response [47]. Immune checkpoints, such as PD-L1 expression, are another widely used biomarker linked with ICI therapies’ efficacy. Similarly, it may not work in some trials [56]. In view of the current situation, novel and more effective indicators are needed to distinguish subpopulations that are likely to be sensitive to ICI treatment.
A recent study has reported that FAT1 mutations were associated with favorable ICI treatment efficacy in melanoma and NSCLC patients. To elucidate, RELN and FAT1 mutations were two independent biomarkers for evaluating immunotherapeutic efficacy. We performed multivariable Cox regression models in melanoma and NSCLC cohorts with multiple confounding factors, including RELN and FAT1 mutations, taken into account. We observed that both mutations exhibited preferable ICI treatment prognoses in melanoma and NSCLC patients after mutually adjusting (all HR < 1, all p < 0.05; Figures S8 and S9), which suggests that RELN and FAT1 mutations are two independent biomarkers for predicting ICI response.
Some shortcomings exist in this study. First, the melanoma and NSCLC samples used in this study were acquired from publicly available databases and lacked in-house result validation. Second, the integrated immunogenomic cohorts were obtained based on several single cohorts, thus some biases may be introduced during data processing. Third, analyses on the transcriptomic level were performed by using gene expression data from TCGA cohorts, with no corresponding expression data for integrated cohorts.
5. Conclusions
Collectively, by leveraging genomic profiles and clinical information, RELN mutations were determined as a potential biomarker for ICI treatment efficacy prediction, which may provide some clues for selecting cancer patients to receive immunotherapies.
Acknowledgments
The authors thank the contributors in the TCGA project for their selfless spirit in the conduct of this scientific research.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cells11233841/s1, Figure S1: mutational patterns of RELN and common melanoma driver genes exhibited with waterfall plot; Figure S2: detailed amino acid changes induced by RELN mutations in the integrated melanoma cohort; Figure S3: Kaplan-Meier survival analyses of RELN mutations in individual ICI-treated melanoma cohorts; Figure S4: Kaplan-Meier survival analyses of RELN mutations in distinct ICI treatment types in melanoma; Figure S5: Kaplan-Meier survival analyses of RELN mutations in individual ICI-treated NSCLC cohorts; Figure S6: distinct infiltration of 22 immunocytes of RELN-mutated and wild-type groups evaluated with CIBERSORT algorithm in NSCLC. Immunocytes highlighted with red are significantly differentially infiltrated; Figure S7: prognostic capacities of RELN mutations in (A) melanoma and (B) NSCLC patients derived from the TCGA project; Figure S8: Multivariable Cox regression analysis of RELN mutations was performedwith multiple clinical confounding factors taken into consideration in melanoma; Figure S9: Multivariable Cox regression analysis of RELN mutations was performed with multiple clinical confounding factors taken into consideration in NSCLC; Table S1: detailed clinical data and immunotherapy response information for 631 pooled melanoma patients; Table S2: detailed clinical data and immunotherapy response information for 109 pooled NSCLC patients; Table S3: the detected 4 mutational signatures with detailed mutational activities in the pooled melanoma cohort; Table S4: the detected 3 mutational signatures with detailed mutational activities in the pooled NSCLC cohort.
Author Contributions
Conceptualization, Q.W. and S.W.; methodology, Z.L. and Q.W.; software, X.W. and Y.Y.; validation, Z.L., X.W. and Y.Y.; resources, Q.W.; data curation, Q.W., S.W., F.S. and W.Z.; writing—original draft preparation, Q.W. and Z.L.; writing—review and editing, S.W.; visualization, Q.W.; supervision, Q.W. and S.W.; project administration, Q.W.; funding acquisition, Q.W. and S.W. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All samples included in this work were publicly obtained and can be acquired by contacting the corresponding author under reasonable requests.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study, in the collection, analyses, or interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
Funding Statement
This research was funded by the Medicine and Health Science and Technology Development Plan Project of Shandong Province (grant number 202112050480), National Natural Science Foundation of China (grant number 32200512 and 32000495), and the APC was funded by National Natural Science Foundation of China (grant number 32200512).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Li X., Pasche B., Zhang W., Chen K. Association of MUC16 Mutation With Tumor Mutation Load and Outcomes in Patients With Gastric Cancer. JAMA Oncol. 2018;4:1691–1698. doi: 10.1001/jamaoncol.2018.2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Q., Yang Y., Yang M., Li X., Chen K. High mutation load, immune-activated microenvironment, favorable outcome, and better immunotherapeutic efficacy in melanoma patients harboring MUC16/CA125 mutations. Aging. 2020;12:10827–10843. doi: 10.18632/aging.103296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang L., Han X., Shi Y. Association of MUC16 Mutation With Response to Immune Checkpoint Inhibitors in Solid Tumors. JAMA Netw. Open. 2020;3:e2013201. doi: 10.1001/jamanetworkopen.2020.13201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma X., Dong L., Liu X., Ou K., Yang L. POLE/POLD1 mutation and tumor immunotherapy. J. Exp. Clin. Cancer Res. 2022;41:216. doi: 10.1186/s13046-022-02422-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang W., Tang Y., Guo Y., Kong Y., Shi F., Sheng C., Wang S., Wang Q. Favorable immune checkpoint inhibitor outcome of patients with melanoma and NSCLC harboring FAT1 mutations. NPJ Precis. Oncol. 2022;6:46. doi: 10.1038/s41698-022-00292-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong Z.Y., Zhong W.Z., Zhang X.C., Su J., Xie Z., Liu S.Y., Tu H.Y., Chen H.J., Sun Y.L., Zhou Q., et al. Potential Predictive Value of TP53 and KRAS Mutation Status for Response to PD-1 Blockade Immunotherapy in Lung Adenocarcinoma. Clin. Cancer Res. 2017;23:3012–3024. doi: 10.1158/1078-0432.CCR-16-2554. [DOI] [PubMed] [Google Scholar]
- 7.Zhang W., Kong Y., Li Y., Shi F., Lyu J., Sheng C., Wang S., Wang Q. Novel Molecular Determinants of Response or Resistance to Immune Checkpoint Inhibitor Therapies in Melanoma. Front. Immunol. 2021;12:798474. doi: 10.3389/fimmu.2021.798474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang W., Lin Z., Shi F., Wang Q., Kong Y., Ren Y., Lyu J., Sheng C., Li Y., Qin H., et al. HSPG2 Mutation Association with Immune Checkpoint Inhibitor Outcome in Melanoma and Non-Small Cell Lung Cancer. Cancers. 2022;14:3495. doi: 10.3390/cancers14143495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shin D.S., Zaretsky J.M., Escuin-Ordinas H., Garcia-Diaz A., Hu-Lieskovan S., Kalbasi A., Grasso C.S., Hugo W., Sandoval S., Torrejon D.Y., et al. Primary Resistance to PD-1 Blockade Mediated by JAK1/2 Mutations. Cancer Discov. 2017;7:188–201. doi: 10.1158/2159-8290.CD-16-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zaretsky J.M., Garcia-Diaz A., Shin D.S., Escuin-Ordinas H., Hugo W., Hu-Lieskovan S., Torrejon D.Y., Abril-Rodriguez G., Sandoval S., Barthly L., et al. Mutations Associated with Acquired Resistance to PD-1 Blockade in Melanoma. N. Engl. J. Med. 2016;375:819–829. doi: 10.1056/NEJMoa1604958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sade-Feldman M., Jiao Y.J., Chen J.H., Rooney M.S., Barzily-Rokni M., Eliane J.P., Bjorgaard S.L., Hammond M.R., Vitzthum H., Blackmon S.M., et al. Resistance to checkpoint blockade therapy through inactivation of antigen presentation. Nat. Commun. 2017;8:1136. doi: 10.1038/s41467-017-01062-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Long J., Lin J., Wang A., Wu L., Zheng Y., Yang X., Wan X., Xu H., Chen S., Zhao H. PD-1/PD-L blockade in gastrointestinal cancers: Lessons learned and the road toward precision immunotherapy. J. Hematol. Oncol. 2017;10:146. doi: 10.1186/s13045-017-0511-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Z., Wu H.X., Lin W.H., Wang Z.X., Yang L.P., Zeng Z.L., Luo H.Y. EPHA7 mutation as a predictive biomarker for immune checkpoint inhibitors in multiple cancers. BMC Med. 2021;19:26. doi: 10.1186/s12916-020-01899-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang W., Li Y., Lyu J., Shi F., Kong Y., Sheng C., Wang S., Wang Q. An aging-related signature predicts favorable outcome and immunogenicity in lung adenocarcinoma. Cancer Sci. 2022;113:891–903. doi: 10.1111/cas.15254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan T.A., Yarchoan M., Jaffee E., Swanton C., Quezada S.A., Stenzinger A., Peters S. Development of tumor mutation burden as an immunotherapy biomarker: Utility for the oncology clinic. Ann. Oncol. 2019;30:44–56. doi: 10.1093/annonc/mdy495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teng F., Meng X., Kong L., Yu J. Progress and challenges of predictive biomarkers of anti PD-1/PD-L1 immunotherapy: A systematic review. Cancer Lett. 2018;414:166–173. doi: 10.1016/j.canlet.2017.11.014. [DOI] [PubMed] [Google Scholar]
- 17.Xu P., Luo H., Kong Y., Lai W.F., Cui L., Zhu X. Cancer neoantigen: Boosting immunotherapy. Biomed. Pharmacother. 2020;131:110640. doi: 10.1016/j.biopha.2020.110640. [DOI] [PubMed] [Google Scholar]
- 18.Alban T.J., Chan T.A. Immunotherapy biomarkers: The long and winding road. Nat. Rev. Clin. Oncol. 2021;18:323–324. doi: 10.1038/s41571-021-00498-w. [DOI] [PubMed] [Google Scholar]
- 19.Quattrocchi C.C., Wannenes F., Persico A.M., Ciafre S.A., D’Arcangelo G., Farace M.G., Keller F. Reelin is a serine protease of the extracellular matrix. J. Biol. Chem. 2002;277:303–309. doi: 10.1074/jbc.M106996200. [DOI] [PubMed] [Google Scholar]
- 20.Trommsdorff M., Gotthardt M., Hiesberger T., Shelton J., Stockinger W., Nimpf J., Hammer R.E., Richardson J.A., Herz J. Reeler/Disabled-like disruption of neuronal migration in knockout mice lacking the VLDL receptor and ApoE receptor 2. Cell. 1999;97:689–701. doi: 10.1016/S0092-8674(00)80782-5. [DOI] [PubMed] [Google Scholar]
- 21.Dlugosz P., Nimpf J. The Reelin Receptors Apolipoprotein E receptor 2 (ApoER2) and VLDL Receptor. Int. J. Mol. Sci. 2018;19:3090. doi: 10.3390/ijms19103090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dulabon L., Olson E.C., Taglienti M.G., Eisenhuth S., McGrath B., Walsh C.A., Kreidberg J.A., Anton E.S. Reelin binds alpha3beta1 integrin and inhibits neuronal migration. Neuron. 2000;27:33–44. doi: 10.1016/S0896-6273(00)00007-6. [DOI] [PubMed] [Google Scholar]
- 23.Senzaki K., Ogawa M., Yagi T. Proteins of the CNR family are multiple receptors for Reelin. Cell. 1999;99:635–647. doi: 10.1016/S0092-8674(00)81552-4. [DOI] [PubMed] [Google Scholar]
- 24.Khialeeva E., Carpenter E.M. Nonneuronal roles for the reelin signaling pathway. Dev. Dyn. 2017;246:217–226. doi: 10.1002/dvdy.24462. [DOI] [PubMed] [Google Scholar]
- 25.Ndoye A., Miskin R.P., DiPersio C.M. Integrin alpha3beta1 Represses Reelin Expression in Breast Cancer Cells to Promote Invasion. Cancers. 2021;13:344. doi: 10.3390/cancers13020344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vignot S., Lefebvre C., Frampton G.M., Meurice G., Yelensky R., Palmer G., Capron F., Lazar V., Hannoun L., Miller V.A., et al. Comparative analysis of primary tumour and matched metastases in colorectal cancer patients: Evaluation of concordance between genomic and transcriptional profiles. Eur. J. Cancer. 2015;51:791–799. doi: 10.1016/j.ejca.2015.02.012. [DOI] [PubMed] [Google Scholar]
- 27.Sato N., Fukushima N., Chang R., Matsubayashi H., Goggins M. Differential and epigenetic gene expression profiling identifies frequent disruption of the RELN pathway in pancreatic cancers. Gastroenterology. 2006;130:548–565. doi: 10.1053/j.gastro.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 28.Seigel G.M., Hackam A.S., Ganguly A., Mandell L.M., Gonzalez-Fernandez F. Human embryonic and neuronal stem cell markers in retinoblastoma. Mol. Vis. 2007;13:823–832. [PMC free article] [PubMed] [Google Scholar]
- 29.Qin X., Lin L., Cao L., Zhang X., Song X., Hao J., Zhang Y., Wei R., Huang X., Lu J., et al. Extracellular matrix protein Reelin promotes myeloma progression by facilitating tumor cell proliferation and glycolysis. Sci. Rep. 2017;7:45305. doi: 10.1038/srep45305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stein T., Cosimo E., Yu X., Smith P.R., Simon R., Cottrell L., Pringle M.A., Bell A.K., Lattanzio L., Sauter G., et al. Loss of reelin expression in breast cancer is epigenetically controlled and associated with poor prognosis. Am. J. Pathol. 2010;177:2323–2333. doi: 10.2353/ajpath.2010.100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Snyder A., Makarov V., Merghoub T., Yuan J., Zaretsky J.M., Desrichard A., Walsh L.A., Postow M.A., Wong P., Ho T.S., et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N. Engl. J. Med. 2014;371:2189–2199. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Allen E.M., Miao D., Schilling B., Shukla S.A., Blank C., Zimmer L., Sucker A., Hillen U., Foppen M.H.G., Goldinger S.M., et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350:207–211. doi: 10.1126/science.aad0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hugo W., Zaretsky J.M., Sun L., Song C., Moreno B.H., Hu-Lieskovan S., Berent-Maoz B., Pang J., Chmielowski B., Cherry G., et al. Genomic and Transcriptomic Features of Response to Anti-PD-1 Therapy in Metastatic Melanoma. Cell. 2016;165:35–44. doi: 10.1016/j.cell.2016.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Riaz N., Havel J.J., Makarov V., Desrichard A., Urba W.J., Sims J.S., Hodi F.S., Martin-Algarra S., Mandal R., Sharfman W.H., et al. Tumor and Microenvironment Evolution during Immunotherapy with Nivolumab. Cell. 2017;171:934–949.e16. doi: 10.1016/j.cell.2017.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roh W., Chen P.L., Reuben A., Spencer C.N., Prieto P.A., Miller J.P., Gopalakrishnan V., Wang F., Cooper Z.A., Reddy S.M., et al. Integrated molecular analysis of tumor biopsies on sequential CTLA-4 and PD-1 blockade reveals markers of response and resistance. Sci. Transl. Med. 2017;9:eaah3560. doi: 10.1126/scitranslmed.aah3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miao D., Margolis C.A., Vokes N.I., Liu D., Taylor-Weiner A., Wankowicz S.M., Adeegbe D., Keliher D., Schilling B., Tracy A., et al. Genomic correlates of response to immune checkpoint blockade in microsatellite-stable solid tumors. Nat. Genet. 2018;50:1271–1281. doi: 10.1038/s41588-018-0200-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu D., Schilling B., Liu D., Sucker A., Livingstone E., Jerby-Arnon L., Zimmer L., Gutzmer R., Satzger I., Loquai C., et al. Integrative molecular and clinical modeling of clinical outcomes to PD1 blockade in patients with metastatic melanoma. Nat. Med. 2019;25:1916–1927. doi: 10.1038/s41591-019-0654-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rizvi N.A., Hellmann M.D., Snyder A., Kvistborg P., Makarov V., Havel J.J., Lee W., Yuan J., Wong P., Ho T.S., et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hellmann M.D., Nathanson T., Rizvi H., Creelan B.C., Sanchez-Vega F., Ahuja A., Ni A., Novik J.B., Mangarin L.M.B., Abu-Akeel M., et al. Genomic Features of Response to Combination Immunotherapy in Patients with Advanced Non-Small-Cell Lung Cancer. Cancer Cell. 2018;33:843–852.e844. doi: 10.1016/j.ccell.2018.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramos A.H., Lichtenstein L., Gupta M., Lawrence M.S., Pugh T.J., Saksena G., Meyerson M., Getz G. Oncotator: Cancer variant annotation tool. Hum. Mutat. 2015;36:E2423–E2429. doi: 10.1002/humu.22771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim J., Mouw K.W., Polak P., Braunstein L.Z., Kamburov A., Kwiatkowski D.J., Rosenberg J.E., Van Allen E.M., D’Andrea A., Getz G. Somatic ERCC2 mutations are associated with a distinct genomic signature in urothelial tumors. Nat. Genet. 2016;48:600–606. doi: 10.1038/ng.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Newman A.M., Liu C.L., Green M.R., Gentles A.J., Feng W., Xu Y., Hoang C.D., Diehn M., Alizadeh A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ayers M., Lunceford J., Nebozhyn M., Murphy E., Loboda A., Kaufman D.R., Albright A., Cheng J.D., Kang S.P., Shankaran V., et al. IFN-gamma-related mRNA profile predicts clinical response to PD-1 blockade. J. Clin. Investig. 2017;127:2930–2940. doi: 10.1172/JCI91190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hanzelmann S., Castelo R., Guinney J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mayakonda A., Lin D.C., Assenov Y., Plass C., Koeffler H.P. Maftools: Efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28:1747–1756. doi: 10.1101/gr.239244.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Samstein R.M., Lee C.H., Shoushtari A.N., Hellmann M.D., Shen R., Janjigian Y.Y., Barron D.A., Zehir A., Jordan E.J., Omuro A., et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat. Genet. 2019;51:202–206. doi: 10.1038/s41588-018-0312-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klempner S.J., Fabrizio D., Bane S., Reinhart M., Peoples T., Ali S.M., Sokol E.S., Frampton G., Schrock A.B., Anhorn R., et al. Tumor Mutational Burden as a Predictive Biomarker for Response to Immune Checkpoint Inhibitors: A Review of Current Evidence. Oncologist. 2020;25:e147–e159. doi: 10.1634/theoncologist.2019-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shi F., Zhang W., Yang Y., Yang Y., Zhao J., Xie M., Sheng C., Wang S., Wang Q. Sex Disparities of Genomic Determinants in Response to Immune Checkpoint Inhibitors in Melanoma. Front. Immunol. 2021;12:721409. doi: 10.3389/fimmu.2021.721409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang W., Shi F., Kong Y., Li Y., Sheng C., Wang S., Wang Q. Association of PTPRT mutations with immune checkpoint inhibitors response and outcome in melanoma and non-small cell lung cancer. Cancer Med. 2022;11:676–691. doi: 10.1002/cam4.4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang F., Zhao Q., Wang Y.N., Jin Y., He M.M., Liu Z.X., Xu R.H. Evaluation of POLE and POLD1 Mutations as Biomarkers for Immunotherapy Outcomes Across Multiple Cancer Types. JAMA Oncol. 2019;5:1504–1506. doi: 10.1001/jamaoncol.2019.2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Assoun S., Theou-Anton N., Nguenang M., Cazes A., Danel C., Abbar B., Pluvy J., Gounant V., Khalil A., Namour C., et al. Association of TP53 mutations with response and longer survival under immune checkpoint inhibitors in advanced non-small-cell lung cancer. Lung Cancer. 2019;132:65–71. doi: 10.1016/j.lungcan.2019.04.005. [DOI] [PubMed] [Google Scholar]
- 53.Hinshaw D.C., Shevde L.A. The Tumor Microenvironment Innately Modulates Cancer Progression. Cancer Res. 2019;79:4557–4566. doi: 10.1158/0008-5472.CAN-18-3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lei X., Lei Y., Li J.K., Du W.X., Li R.G., Yang J., Li J., Li F., Tan H.B. Immune cells within the tumor microenvironment: Biological functions and roles in cancer immunotherapy. Cancer Lett. 2020;470:126–133. doi: 10.1016/j.canlet.2019.11.009. [DOI] [PubMed] [Google Scholar]
- 55.Yarchoan M., Hopkins A., Jaffee E.M. Tumor Mutational Burden and Response Rate to PD-1 Inhibition. N. Engl. J. Med. 2017;377:2500–2501. doi: 10.1056/NEJMc1713444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sharma P., Callahan M.K., Bono P., Kim J., Spiliopoulou P., Calvo E., Pillai R.N., Ott P.A., de Braud F., Morse M., et al. Nivolumab monotherapy in recurrent metastatic urothelial carcinoma (CheckMate 032): A multicentre, open-label, two-stage, multi-arm, phase 1/2 trial. Lancet Oncol. 2016;17:1590–1598. doi: 10.1016/S1470-2045(16)30496-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All samples included in this work were publicly obtained and can be acquired by contacting the corresponding author under reasonable requests.