Abstract
Background:
miRNA 223 /125a and Cordon-bleu Protein Like 1 (COBLL1) are novel biomarkers that can predict prognosis and guide treatment decisions in patients with chronic lymphocytic leukemia (CLL). Also, there is a growing interest in CLL monitoring based on flow cytometry of receptor tyrosine kinase-like orphan receptor-1 (ROR-1).
Objective:
This study aimed to evaluate the relationship between miRNA 223 /125a and COBLL1 expressions and ROR-1 expression in patients with CLL. Also, the study evaluated the relationship between the expression of these biomarkers with tumor staging and cancer progression.
Methods:
Our study included 40 patients newly diagnosed with B-CLL. In peripheral blood (PB), miRNA 223/125a and COBLL1 expressions were detected by real-time polymerase chain reaction (real-time PCR) and ROR-1 percentage was detected by flow cytometry before and after treatment.
Results:
High level of COBLL1 expression was statistically significantly associated with high ROR-1 percentage expression (P= 0.03). However, a high level of miRNA 223/125a expression was statistically significantly associated with low ROR-1 percentage expression (P=0.002). The sensitivity and specificity of ROR-1 as a predictor of high WBCs count after treatment were 96.6 and 81.1%, respectively. There was a statistically significant reduction of ROR-1 percentage after treatment compared to before treatment (P <0.001).
Conclusion:
ROR-1 percentage expression can be considered a possible prognostic predictor in CLL along with miRNA 223/125a and COBLL1 expressions. This can be explained by the significant correlation between ROR-1 and the studied molecular biomarkers; miRNA 223/125a and COBLL1. In addition, there was a significantly higher ROR-1 percentage in patients with higher WBC counts. Moreover, there was a significant reduction in ROR-1 percentage after treatment.
Key Words: miRNA 223/125a, COBLL1, ROR-1, CLL
Introduction
Chronic lymphocytic leukemia (CLL) is characterized by an overabundance of lymphocytes (mostly B lymphocytes) in the bone marrow, lymph nodes, and peripheral blood (PB) (Schulz et al., 2007). The etiology of CLL is unknown, and several factors have been investigated to better understand the pathogenesis (Strati et al., 2018). Several genetic and molecular features have been discovered in CLL that may have a substantial impact on the disease’s prognosis.
Transmembrane receptor tyrosine kinase-like orphan receptor-1 (ROR-1) is expressed on CLL patients’ neoplastic B cells but not on normal CD5 B-cells (Cui et al., 2016). ROR1 is highly expressed during embryonic development but is undetectable in adults. It is a member of the Wnt/ planar cell polarity (PCP) pathway which regulates various processes during embryonic development like cell polarity, survival, and migration (Fukuda et al., 2008). It is a Wnt5a receptor that promotes cell survival, proliferation, and migration in CLL (Yu et al., 2016). ROR-1 is expressed in all cases of CLL where low-level ROR-1 expression in leukemia cells is linked to more indolent illness (Cui et al., 2016). ROR-1 is upregulated in CLL patients, although its activity is dependent on posttranslational modifications and the availability of specialized ligands (Janovska et al., 2016).
Cordon-bleu Protein Like 1 (COBLL1) is a ROR-1 binding partner. It is required for neural tube closure, a process regulated by the Wnt/PCP pathway. COBLL1 levels in CLL are variable and sometimes do not correlate with ROR1. So, its expression can be an independent molecular marker in CLL (Plešingerová et al., 2018).
MicroRNAs (miRNAs) are a family of non-coding RNAs that contain 20–22 nucleotides. They regulate gene transcription and expression (Shahjahani et al., 2020). They control the gene expression after transcription by inhibiting the translation of mRNA or inducing its degradation (Kim et al., 2011). Dysregulation of the expression of some microRNAs leads to carcinogenesis by influencing cell growth by interfering with cell cycle regulators (Sethi et al., 2014; Nazarian et al., 2019). In CLL, dysregulation of some miRNAs disrupts apoptosis of malignant cells enhancing cell proliferation and disease progression and decreasing patient survival (Davari et al., 2021).
Tumor suppressors and oncogenes are the two main types of miRNAs target genes. In CLL, targeting tumor suppressor genes is frequently impaired and targeting oncogenes is elevated (Rezaeeyan et al., 2017). miRNA 223 and miRNA 125a as tumor suppressor genes target multiple transcription factors like STAT3 (Chen et al., 2012; Fan et al., 2015). Davari et al. (2021) found that miRNA-125a and miRNA-223 expression decreased in the patients with CLL compared to the control group. In addition, upregulation of their target genes, BCL-2 and STAT3, was observed in CLL compared to the control subjects. On the other hand, Bader El-Din et al. (2021) found that miRNA 223 was significantly upregulated in both colorectal carcinoma tissues and serum samples with a highly significant prognostic value.
Iorio et al., (2005) used a miRNA microarray to evaluate the miRNA expression in cancer breast tissues. They found that miRNA 125a was reduced in breast cancer. Also, Ahmadvand et al., (2019) reported decreased miRNA 125 expression in CLL patients. Moreover, Rigolin et al., (2014) found that reduced miRNA 125a expression was associated with more genetic abnormalities in CLL patients.
Several studies evaluated the individual expression of these biomarkers. However, the relationship between all these indicators and their impact on CLL prognosis was poorly investigated. So, this study aimed to evaluate the relationship between ROR-1 percentage and miRNA 223/125a and COBLL1 expressions in B-CLL and to evaluate the relationship between the expression of these biomarkers with tumor staging and cancer progression.
Materials and Methods
This study was conducted in the Oncology, Clinical Pathology, and Medical Biochemistry & Molecular Biology Departments, Faculty of Medicine, Zagazig University, Zagazig, Egypt during the period from January 2020 to December 2021.
This study included 40 newly diagnosed B-CLL patients (22 males and 18 females) who were diagnosed according to the WHO criteria, 2016. All patients were subjected to medical history taking, full clinical examination, and routine laboratory investigations. The clinical staging was determined based on the Rai staging which is one of the two currently used staging systems for the assessment of CLL. It includes stages from 0 to IV and classifies CLL into low, intermediate, and high-risk categories (Rai et al., 1975). miRNA 223 /125a and COBLL1 expression were detected by real-time PCR. ROR-1 percentage was detected by flow cytometry before and after treatment.
Therapeutic regimen
The patients with the early disease were eligible for the watch and wait strategy until they become symptomatic. Thirty one (77.5%) of patients received the chemotherapeutic regimen such as Fludarabine/ Cyclophosphamide (Fludarabine 25mg/m2 IV on days 1, 2, 3 and Cyclophosphamide 250 mg/ m2 IV on days 1, 2, 3 for each cycle to be repeated every 4 weeks) (Catovsky et al., 2007), Cyclophosphamide/ Vincristine/ Prednisone (Cyclophosphamide 750 mg/m2 IV on day 1, Vincristine 1.4 mg/m2 (max 2 mg), Prednisone 40 mg/m2/d on days1, 2, 3, 4, 5 for each cycle to be repeated every 3 weeks) (Hallek, 2019).
RNA extraction and cDNA synthesize
Total RNA was extracted from 5 mL peripheral blood with EDTA. The extraction procedure was performed within 24 hours of the sample collection to avoid RNA degradation using PAXgene Blood miRNA Kit (Qiagen, Germany). To assess the quality and the quantity of the RNA, it was run on the gel electrophoresis and measured on Nanodrop spectrophotometry (ND 1000-NanoDrop®). Then, the cDNA was synthesized with high capacity cDNA reverse transcription kit from 1 µg RNA according to the instructions of the manufacturer (Applied Biosystem) with a total reaction volume of 20 µL (10 µL containing 1 µg RNA, 2 µL 10x RT Buffer, 0.8 µL 25X dNTP mix (100 mM), 2.0 µL 10x RT random primers, 1.0 µL multiscribe reverse transcriptase, 1.0 µL RNase inhibitors, and 3.2 µL nuclease-free water) in MicroAmp™ fast 96-well reaction plate thermal cycler (Applied Biosystem) with cycling condition of; 25oC for 10 minutes, 37oC for 120 minutes, and 85oC for 5 minutes for enzyme deactivation. cDNA was diluted at 1:5 and stored at -20oC for further use.
Reverse transcription of the miRNA
Fifty ng of the total extracted RNA was reverse transcribed in a final volume of 20 µL (50 ng dissolved in 5µL nuclease-free water, 4µL of 5× miRCURY RT reaction buffer, 2.5 µl of 10x miRCURY RT Enzyme Mix, 1.2 µL of a predesigned stem-loop primer (Table 1) and 10µL of RNase-free water) with a cycling condition of 42oC for 60 minutes for the reverse transcription stage and 95oC for the inactivation of the enzyme according to the instructions of the manufacturer (Qiagen, Germany) then the cDNA was aliquoted and stored at -20oC until used.
Table 1.
Primer Sequence of the Studied Genes
| Gene | Forward | Reverse | Stem loop |
|---|---|---|---|
| COBLL1 | CGGGGAAGGGAAGAGTAGGA | CTCAGCTGGAGGAAGTGGTG | |
| PGK1 housekeeping | CCACTGTGGCTTCTGGCATA | ATGAGAGCTTTGGTTCCCCG | |
| has-mir-125a | GGTGTCCCTGAGACCCTTTAA | GTGCAGGGTCCGAGGT | GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACTCACAG |
| has-mir-223 | GTGTTTTTTTTCGTGTATTTGACAAGC | GTGCAGGGTCCGAGGT | GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAAC AACTCA |
Real-time PCR protocol
The real-time PCR was done by the thermal cycler Rotor-Gene Q 2 plex (Qiagen, Germany) in a final volume reaction of 20 µL using 10 µL of TOPreal syberGreen (Enzynomics, Korea), 1 µL of each forward and reverse primers synthesized by Sangon Biotech (Beijing, China) (Table1), 1 µL of cDNA and up to 20 µL of Nuclease free water. The cycling conditions were initial denaturation at 95oC for 10 minutes, 40 cycles of (denaturation at 95oC for 10 seconds, annealing at 60 oC for 15 seconds, and extension at 72oC for 15 seconds). The gene expression was measured as the relative fold change to an internal control reference gene (PGK1). Δ ct was calculated as the ct difference between the target gene and the reference gene. Then, ΔΔ ct was calculated as the difference between Δ ct of the sample and the average Δ ct of the control. Finally, the fold gene expression was calculated as 2-ΔΔct.
Flow cytometry
Fresh leukemic cells from PB were stained with a combination of monoclonal antibodies (mAbs). They were analyzed using the FACS Canto II flow cytometry (Becton Dickinson (BD, San Jose, CA, USA). CLL panel included CD45 V500, CD19 PEcy7, CD5 PerCP, CD20 V450, CD23 FITC, CD38 APC-H7, CD10 APC, (Kappa FITC/ Lambda PE/CD19 PerCP in Tripler markers), and CD79 APC. ROR1 PE antibodies were used (Becton Dickinson). 100 µ of EDTA blood was incubated with 10µ antibody combinations detecting specific surface markers. The expression of ROR1 was assessed for CD19, CD5 positive cells. The acquisition was carried out by collecting at least 50 000 events per tube on diagnostic samples (Ozturk et al., 2021).
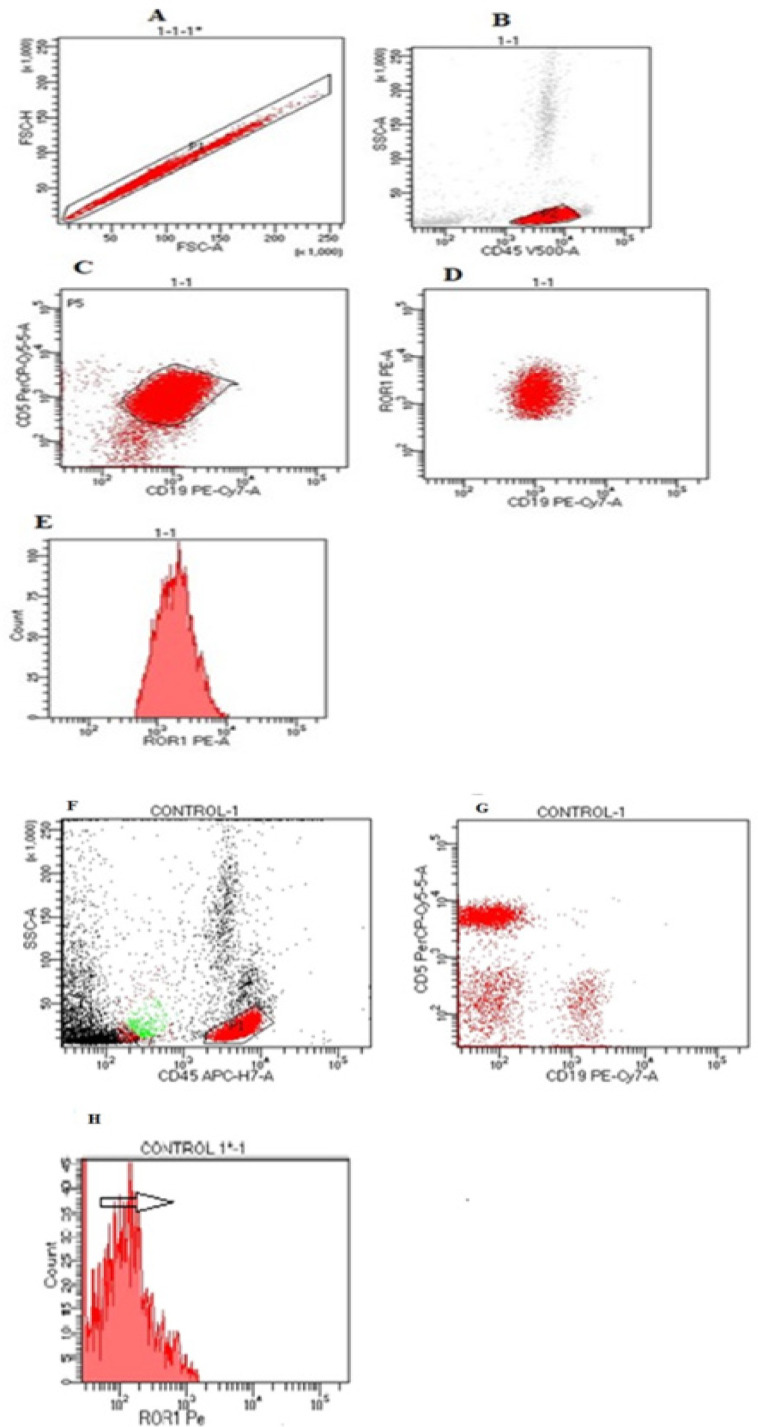
In each run, the CLL cell population was identified by the following gating strategy: (1) Identification of CD45-positive leucocytes. (2) Lymphocyte identification by determination of CD45/CD19 positive population. (3) CLL cell identification by displaying CD19 versus CD5. (4) ROR1 expression was analyzed in the CD19/CD5 double positive cell population. In healthy volunteers, ROR1 expression was analyzed in normal lymphocytes subpopulations using the following antibody/fluorochrome panel: CD3 FITC, CD19 PeCY7, CD45 APCH-7, CD56 PecP5.5, and ROR1 PE (all from Becton Dickinson (Fig.1) It was used to determine a positive result with any antibody using a cut-off limit of 30% of lymphoid cells, as recommended by the British Committee for Standards in Hematology (BCSH) guideline (Oscier et al., 2012; Pochtar et al., 2022)
Figure 1.
(A, B, C& D) ROR1 antigen evaluated on the lymphocyte gate in combination with CD5 and CD19 at the time of CLL diagnosis. (E) Histogram analysis revealed positive expression of ROR1 that was evaluated on the malignant tumor cells CD5/CD19 co-expression. (F,G&H): Flow cytometry analysis of ROR1 expression in healthy B-cell population as negative control
Statistical analysis
IBM SPSS 23.0 for Windows (SPSS Inc., Chicago, IL, USA) and NCSS 11 for Windows (NCSS LCC., Kaysville, UT, USA) were used to analyze the data. Quantitative data were expressed as mean ± standard deviation (SD). Qualitative data were expressed as frequency and percentage. Independent sample t-test, Mann-Whitney test for not normally distributed data, Chi-square and Fisher exact for qualitative data analysis, and ROC curve analysis for validity data were all used. All tests were two-tailed. P values of less than 0.05 were considered significant, P values of less than 0.001 were considered highly significant, and P values of more than 0.05 were considered insignificant.
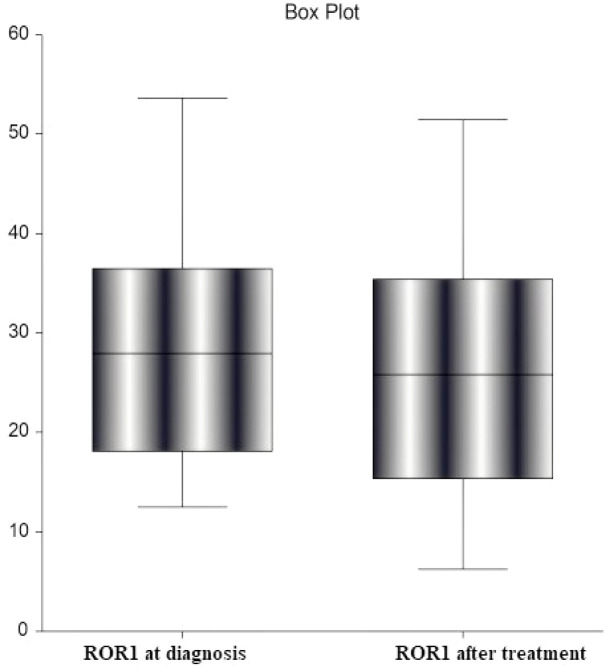
Figure 3.
Reduction in ROR-1 after Treatment (P ˂ 0.001)
Results
Demographic and basic characteristics of the studied participants
The mean age of the participants was 65.7 ± 7.13 years. They were 22 males and 18 females. They had a mean hemoglobin level of 11.2 ± 1.15 gm/dL, and a mean WBC count of 48.4 ±33.1 x103/mm3. The mean platelet count was 170.2 ± 97.98 x103/mm3. The mean percentage of ROR-1 was 30.1 ± 12.13 in B-CLL patients. According to Ria classification risk categories, 22.5% of our patients were low risk, 35% were intermediate-risk and 42.5% of them were high-risk. High miRNA 223/125a expression was detected in 47.5% of patients while high COBLL1 expression was detected in 72.5% of patients (Table 2).
Table 2.
The Relation between COBLL 1 Expression and the Basic Characteristics in the Studied Cases
| Characteristics | CLL Cases N=40 |
High COBLL 1 expression N=26 |
Low COBLL 1 expression N=14 |
P$ |
|---|---|---|---|---|
| Age (years)& | 65.7 ± 7.13 | 64.7 ± 7.23 | 68.4 ± 6.42 | 0.14 |
| WBCs (103/mm3)& | 48.4 ±33.1 | 48 (17.7-111.6) | 21.7 (16.5-59.6) | 0.002* |
| Hemoglobin (gm/dL)& | 11.2 ± 1.15 | 11.1 ± 1.28 | 11.2 ± 0.75 | 0.84 |
| Platelets (103/mm3)& | 170.2 ± 97.98 | 165.1 (46.2-450.5) | 165.3 (69.1-360.1) | 0.96 |
| ROR-1 percentage expression& | 30.1 ± 12.13 | 32.2 (12.5-53.6) | 18 (18.1-53) | 0.03* |
| Gender# | ||||
| Male | 22 (55%) | 14 (53.8%) | 8 (57.1%) | 0.84 |
| Female | 18 (45%) | 12 (46.2%) | 6 (42.9) | |
| Ria classification risk categories# | ||||
| Low | 9 (22.5%) | 2 (7.7%) | 7 (50%) | 0.41 |
| Intermediate | 14 (35%) | 8 (30.8%) | 6 (42.9%) | |
| High | 17(42.5%) | 16 (61.5%) | 1 (7.1%) | |
| miRNA 223/125a expression# | ||||
| High | 19 (47.5%) | 14 (53.8%) | 5 (35.7%) | 0.87 |
| Low | 21(52.5%) | 12 (46.2 %) | 9 (64.3) | |
| COBLL1 expression# | ||||
| High | 29 (72.5%) | |||
| Low | 11(27.5%) |
&, values are expressed in mean± SD while non-parametric data are presented as median and range compared with Mann-Whitney test; #, values are expressed in number (%); $, the difference between high and low expression groups; P-value>0.05 is not significant; *, P-value<0.05 is significant; **, P-value <0.001 is highly significant
Relation of COBLL1 expression with basic characteristics of the studied cases
Patients with high COBLL1 expression showed significantly higher WBC count and ROR-1 percentage compared to those with low expression (P=0.002 and 0.03 respectively) (Table 2). However, no significant relationships between COBLL1 expression and other studied demographic or clinical data were found.
Relation of miRNA 223/125a expression with basic characteristics of the studied cases
Patients with high miRNA 223/125a expression showed significantly lower WBCs count and ROR-1 percentage (P<0.001, 0.002 respectively), while they showed higher hemoglobin level and platelet count (P=0.02, 0.001 respectively) compared to those with low miRNA 223/125a expression. Ria classification risk showed a significant difference between the two groups (P<0.001) (Table 3).
Table 3.
The Relation between miRNA 223/125a Expression and Basic Characteristics of the Studied Cases
| Characteristics | High miRNA 223/125a expression N=19 |
Low miRNA 223/125a expression N=21 |
P$ |
|---|---|---|---|
| Age (years)& | 64.2 ± 7.55 | 67.1 ± 6.59 | 0.19 |
| WBCs (103/mm3)□ | 19.8 (16.5-30.1) | 81.2 (35.3-111.6) | <0.001** |
| Hemoglobin (gm/dL)& | 11.6 ± 0.79 | 10.8 ± 1.28 | 0.02* |
| Platelets (103/mm3)□ | 255.2 (69.1-450.5) | 78.2 (46.2-282.1) | 0.001* |
| ROR-1 percentage expression□ | 21.2 (12.5-46.5) | 34.5 (18.1-53.6) | 0.002* |
| Gender# | |||
| Male | 10 (52.6%) | 12 (57.1%) | 0.78 |
| Female | 9 (46.4%) | 9 (42.9 %) | |
| Ria classification risk categories# | |||
| Low | 9 (46.4%) | 0 (0.0%) | <0.001** |
| Intermediate | 7 (36.8%) | 7 (33.3%) | |
| High | 3 (15.8%) | 14 (66.7 %) | |
| COBLL1 expression# | |||
| High | 14 (73.7%) | 15 (71.4%) | 0.89 |
| Low | 5 (26.3%) | 6 (28.6%) |
&, values are expressed in mean± SD; #, values are expressed in number and percentage (%); $, the difference between high and low expression groups; □, non-parametric data presented as median & range and compared with Mann-Whitney test; P-value>0.05 is not significant; *, P-value<0.05 is significant; **, P-value <0.001 is highly significant.
Relation of ROR-1 percentage with WBC count among the studied cases
ROR-1 percentage was significantly higher in patients with WBC count > 50,000/mm3 compared to those with WBC count <20,000/mm3 (P= 0.005) (Table 4).
Table 4.
The Relation between the Count of WBCs and ROR-1 Expression Percentage among the Studied Cases
| High WBCs | Low WBCs | P | |
|---|---|---|---|
| >50,000/mm3 | <20,000/mm3 | ||
| ROR-1 percentage in Peripheral blood (mean± SD) | 38.9 ± 9.91 | 23.2 ± 12.4 | 0.005* |
| Median | 36.3 | 16.1 | |
| Range | 21 – 53.6 | 12.5 – 46.5 |
*, P-value<0.05 is significant
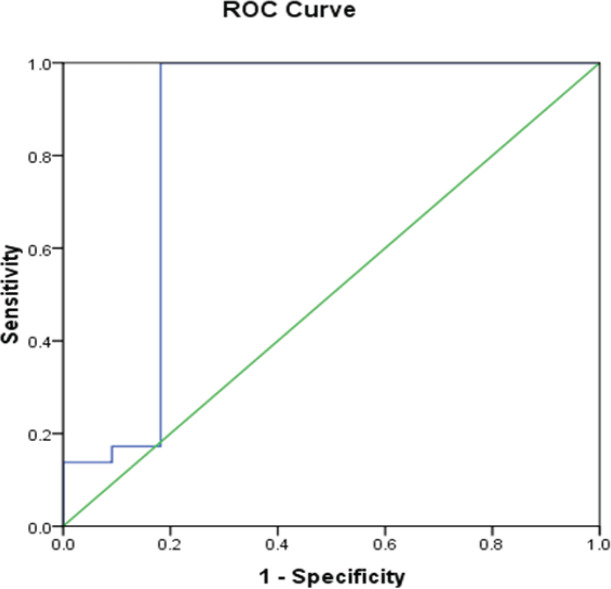
Validity data of ROR-1 percentage as a predictor of high WBC count after treatment
The sensitivity and specificity of ROR-1 overexpression as a predictor of persistent high WBC count even after treatment were 96.6% and 81.1%, respectively. The positive predictive value (PPV) was 93.3% and the negative predictive value (NPV) was 90%. The accuracy was 92.5% at a cut-off of 16.6%. Area under the curve (AUC) was 0.846 (0.654-1.04) (P< 0.001) (Figure 1).
Reduction of ROR-1 percentage after treatment among the studied cases
There was a statistically significant decrease in ROR-1 percentage post-treatment compared to before treatment among B-CLL cases (P <0.001) (Figure 2).
Figure 2.
Receiver Operating Curve (ROC) for ROR-1 as a Prognostic Predictor of High WBCs Count after Treatment
Discussion
During the last decades, researchers have searched for new biomarkers that can predict prognosis and guide treatment decisions in CLL. COBLL1 expression could serve as an independent molecular marker in CLL patients with a poor prognosis by inhibiting apoptosis (Plesingerova et al., 2018). Besides, miRNAs play an important role in regulating hematopoiesis and the production of various blood cell lines (Haybar et al., 2018). So, some miRNAs can be used as predictive factors in hematological diseases because they target numerous genes in the cell cycle and apoptosis (Balatti et al., 2018). Furthermore, CLL cells express high levels of ROR-1 while it is not expressed in normal B cells (Uhrmacher et al., 2011).
In our study, we reported that miRNA 223/125a expression was inversely correlated to WBC count and platelet count while it was directly correlated with hemoglobin (Hb) concentration. These findings agreed with Davari et al., (2021). They stated that these miRNAs may have regulatory effects by controlling WBC production based on the inverse correlation between WBC count and Hb concentration. However, miRNA 223 expression was not correlated to any of the demographic data in their study except smoking. On the other hand, Zhou et al., (2012) did not observe a significant relationship between miRNA 223 and leukocytic count. However, they found no relationship between miRNA 223 and age, similar to our result.
Our study revealed a significant difference between miRNA 223 expression and Ria classification risk categories. However, no significant relationship was detected between miRNA 223 and gender. Similarly, Stamatopoulos et al., (2009) and Zhou et al., (2012) reported a significant relationship between miRNA 223 and Binet staging system of CLL with no significant relationship between miR-223 and gender. Rodríguez-Vicente et al., (2015) found that miRNA 223 expression was reduced in CLL patients lacking the immunoglobulin heavy chain mutation, which was associated with poor prognosis and disease progression. Also, Zhou et al., (2012) declared that reduced miRNA 223 expression in CLL patients was associated with aggressive disease and decreased response to treatment.
In agreement with our study, Plešingerová et al., (2018) detected that higher COBLL1 expression was detected in patients with high ROR-1 expression. However, they found no significant difference in WBC count regarding COBLL1 expression while we found a significant direct correlation between WBC count and COBLL1 expression.
There was a controversy regarding the prognostic role of COBLL1 expression in cancer. Bilous et al., (2019) and Plešingerová et al., (2018) indicated that COBLL1 expression resulted in shorter overall survival (OS) and time to second treatment which indicates poor CLL prognosis. Besides, COBLL1 upregulation in patients with chronic myeloid leukemia (CML) was associated with a reduction in apoptosis, disease progression, and shorter OS (Han et al., 2017). On the contrary, COBLL1 upregulation was associated with a better prognosis after surgery in malignant pleural mesothelioma, where it acts as a negative regulator of apoptosis (Gordon et al., 2003). In our study, no significant correlation between COBLL1 expression with Ria classification was detected
Our study detected that higher expression of ROR-1 was associated with higher circulating leukemic cells. This agrees with Stefania et al., (2020). Furthermore, our study noticed a statistically significant decrease in the level of ROR-1 expression after treatment compared to before treatment, but this disagreed with Stefania et al., (2020) who reported no significant changes in ROR-1 before and after treatments.
In conclusion, ROR-1 expression can be considered a possible marker for CLL along with miRNA 223/125a and COBLL1 expressions. This can be explained by the significant correlation between ROR-1 and the studied molecular biomarkers; miRNA 223/125a and COBLL1. In addition, there was a significantly higher ROR-1 percentage in patients with higher WBC counts. Moreover, there was a significant reduction in ROR-1 percentage after treatment.
Author Contribution Statement
Conception: Huda F Ebian and Samia Hussein; Interpretation or analysis of data: Abdallah S. Abdelazem, AL-Shabrawy M. Abdelnabi, Tarek Khamis, Ahmed Ali Obaya, Huda F Ebian, and Samia Hussein; Preparation of the manuscript: Huda F Ebian, Samia Hussein and Shimaa Abdelmoneem; Revision for important intellectual content: All authors; Supervision: Huda F Ebian and Samia Hussein
Acknowledgments
Recommendation
Prospective broader studies in the future are highly recommended to estimate the correlation of these biomarkers with other prognostic factors and to confirm ROR-1 accuracy and reliability as a marker for CLL.
Ethical Approval
The experimental protocol was approved by the Faculty of Medicine, Zagazig University, Zagazig, Egypt.
Availability of data
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Compliance with Ethical Standards Ethical approval
All procedures performed in studies involving human participants were following the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Conflict of interests
The authors declare no conflict of interest.
References
- Ahmadvand M, Eskandari M, Khakpour G, et al. Identifcation of MiR-125a as a novel plasma diagnostic biomarker for chronic lymphoblastic leukemia. Clin Lab. 2019;65:10. doi: 10.7754/Clin.Lab.2018.180815. [DOI] [PubMed] [Google Scholar]
- Bader El Din NG, Farouk S, Abdel-Salam LO, Khairy A. The potential value of miRNA-223 as a diagnostic biomarker for Egyptian colorectal patients. Eur J Gastroenterol Hepatol. 2021;33:25–31. doi: 10.1097/MEG.0000000000001961. [DOI] [PubMed] [Google Scholar]
- Balatti V, Tomasello L, Rassenti LZ, et al. MiR-125a and MiR-34a expression predicts Richter syndrome in chronic lymphocytic leukemia patients. Blood. 2018;132:2179–82. doi: 10.1182/blood-2018-04-845115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilous N, Abramenko I, Chumak A, et al. Analysis of LPL gene expression in patients with chronic lymphocytic leukemia. Exp Oncol. 2019;41:39–45. [PubMed] [Google Scholar]
- Catovsky D, Richards S, Matutes E, et al. Assessment of fludarabine plus cyclophosphamide for patients with chronic lymphocytic leukaemia (the LRF CLL4 Trial): a randomised controlled trial. Lancet. 2007;370:230–9. doi: 10.1016/S0140-6736(07)61125-8. [DOI] [PubMed] [Google Scholar]
- Chen Q, Wang H, Liu Y, et al. Inducible microRNA-223 down-regulation promotes TLR-triggered IL-6 and IL-1β production in macrophages by targeting STAT3. PLoS One. 2012;7:42971. doi: 10.1371/journal.pone.0042971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiorazzi N. Implications of new prognostic markers in chronic lymphocytic leukemia. Hematol Am Soc Hematol Educ Program. 2012;1:76–87. doi: 10.1182/asheducation-2012.1.76. [DOI] [PubMed] [Google Scholar]
- Cui B, Ghia E, Chen L, et al. High-level ROR1 associates with accelerated disease progression in chronic lymphocytic leukemia. BLOOD. 2016;128:2931–40. doi: 10.1182/blood-2016-04-712562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davari N, Ahmadpour F, Kiani A, et al. Evaluation of microRNA-223 and microRNA-125a expression association with STAT3 and Bcl2 genes in blood leukocytes of CLL patients: a case-control study. BMC Res Notes. 2021;14:21. doi: 10.1186/s13104-020-05428-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Z, Cui H, Xu X, et al. MiR-125a suppresses tumor growth, invasion and metastasis in cervical cancer by targeting STAT3. Oncotarget. 2015;6:25266. doi: 10.18632/oncotarget.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda T, Chen L, Endo T, et al. Antisera induced by infusions of autologous AdCD154-leukemia B cells identify ROR1 as an oncofetal antigen and receptor for Wnt5a. Proc Natl Acad Sci U S A. 2008;105:3047–52. doi: 10.1073/pnas.0712148105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon GJ, Jensen R V, Hsiao LL, et al. Using gene expression ratios to predict outcome among patients with mesothelioma. J Natl Cancer Inst. 2003;95:598–5. doi: 10.1093/jnci/95.8.598. [DOI] [PubMed] [Google Scholar]
- Hallek , M Chronic lymphocytic leukemia: 2020 update on diagnosis, risk stratification and treatment. Am J Hematol. 2019;94:1266–87. doi: 10.1002/ajh.25595. [DOI] [PubMed] [Google Scholar]
- Han SH, Kim SH, Kim HJ, et al. Cobll1 is linked to drug resistance and blastic transformation in chronic myeloid leukemia. Leukemia. 2017;31:1532–9. doi: 10.1038/leu.2017.72. [DOI] [PubMed] [Google Scholar]
- Haybar H, Jalali MT, Zayeri ZD. What genetics tells us about cardiovascular disease in diabetic patients? Cardiovasc Haematol Disord Drug Targets. 2018;18:147–52. doi: 10.2174/1871529X18666180212114305. [DOI] [PubMed] [Google Scholar]
- Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–70. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- Janovska P, Poppova L, Plevova K, et al. Autocrine signaling by Wnt-5a deregulates chemotaxis of leukemic cells and predicts clinical outcome in chronic lymphocytic leukemia. Clin Cancer Res. 2016;22:459–69. doi: 10.1158/1078-0432.CCR-15-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Kasinski AL, Slack FJ. MicroRNA therapeutics in preclinical cancer models. Lancet Oncol. 2011;12:319–21. doi: 10.1016/S1470-2045(11)70067-5. [DOI] [PubMed] [Google Scholar]
- Nazarian A, Mohamadnia A, Danaee E, Bahrami N. Examining the Expression of miR-205 and CEA mRNA in Peripheral Blood of Patients with OSCC (Oral Squamous Cell Carcinomas) and Comparing them with Healthy People. Asian Pac J Cancer Biol. 2019;4:65–8. [Google Scholar]
- Oscier D, Dearden C, Eren E, et al. British Committee for Standards in Haematology Guidelines on the diagnosis, investigation and management of chronic lymphocytic leukaemia. Br J Haematol. 2012;159:541–64. doi: 10.1111/bjh.12067. [DOI] [PubMed] [Google Scholar]
- Ozturk HBA, Albayrak M, Yildiz A, et al. The Importance of Tyrosine Kinase-Like Orphan Receptor 1 (ROR-1) in Chronic Lymphocytic Leukemia. UHOD. 2021;4:199–204. [Google Scholar]
- Pochtar EV, Lugovskaya SA, Naumova EV, Dmitrieva EA, Dolgov VV. ROR-1 Expression in the Diagnosis and Monitoring of Minimal Residual Disease in Chronic Lymphocytic Leukemia. Clinl Oncol. 2022;2:148–55. [Google Scholar]
- Plesingerova H, Janovska P, Mishra A, et al. Expression of COBLL1 encoding novel ROR1 binding partner is robust predictor of survival in chronic lymphocytic leukemia. Haematologica. 2018;103:313–24. doi: 10.3324/haematol.2017.178699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plešingerová H, Janovská P, Mishra A, et al. Expression of COBLL1 encoding novel ROR1binding partner is robust predictor of survival in chronic lymphocytic leukemia. Haematologica. 2018;103:313–24. doi: 10.3324/haematol.2017.178699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai KR, Sawitsky A, Cronkite EP, et al. Clinical staging of chronic lymphocytic leukemia. Blood. 1975;46:219–34. [PubMed] [Google Scholar]
- Rezaeeyan H, Hassani S N, Barati M, et al. PD-1/PD-L1 as a 3-prognostic factor in leukemia. J Hematopathol. 2017;10:17–24. [Google Scholar]
- Rigolin GM, Saccenti E, Rizzotto L, et al. Genetic subclonal complexity and miR125a-5p down-regulatio identify a subset of patients with inferior outcome in low-ris CLL patients. Oncotarget. 2014;5:140. doi: 10.18632/oncotarget.1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Vicente AE, Quwaider D, Benito R, et al. MicroRNA-223 is a novel negative regulator of HSP90B1 in CLL. BMC Cancer. 2015;15:238. doi: 10.1186/s12885-015-1212-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz H, Bohlius J F, Trelle S, et al. Immunochemotherapy with rituximab and overall survival in patients with indolent or mantle cell lymphoma: a systematic review and meta-analysis. J Natl Cancer Inst. 2007;99:706–14. doi: 10.1093/jnci/djk152. [DOI] [PubMed] [Google Scholar]
- Sethi N, Wright A, Wood H, Rabbitts P. MicroRNAs and head and neck cancer: reviewing the first decade of research. Eur J Cancer. 2014;50:2619–35. doi: 10.1016/j.ejca.2014.07.012. [DOI] [PubMed] [Google Scholar]
- Shahjahani M, Abroun A, Saki N, et al. STAT5 from pathogenesis mechanism to therapeutic approach in acute leukemia. Lab Med. 2020;51:345–51. doi: 10.1093/labmed/lmz074. [DOI] [PubMed] [Google Scholar]
- Stamatopoulos B, Meuleman N, Haibe-Kains B, et al. microRNA-29c and microRNA-223 down-regulation has in vivo signifcance in chronic lymphocytic leukemia and improves disease risk stratifcation. Blood. 2009;113:5237–45. doi: 10.1182/blood-2008-11-189407. [DOI] [PubMed] [Google Scholar]
- Stefania M, Intoppa S, Laura M M, et al. ROR1 is an accurate and reliable marker of minimal residual disease in chronic lymphocytic leukemia. Br J Haematol. 2020;190:346–49. doi: 10.1111/bjh.16910. [DOI] [PubMed] [Google Scholar]
- Strati P, Jain N, O’Brien S. Chronic lymphocytic leukemia: diagnosis and treatment. Mayo Clinic Proceedings. 2018;93: 651–64. doi: 10.1016/j.mayocp.2018.03.002. [DOI] [PubMed] [Google Scholar]
- Uhrmacher S, Schmidt C, Erdfelder F, et al. Use of the receptor tyrosine kinase-like orphan receptor 1(ROR1) as a diagnostic tool in chronic lymphocytic leukemia (CLL) Leukemia Res. 2011;35:1360–6. doi: 10.1016/j.leukres.2011.04.006. [DOI] [PubMed] [Google Scholar]
- Yu J, Chen L, Cui B, et al. Wnt5a induces ROR1/ ROR2 heterooligomerization to enhance leukemia chemotaxis and proliferation. J Clin Invest. 2016;126:585–98. doi: 10.1172/JCI83535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Chen L, Cui B, et al. Wnt5a induces ROR1/ROR2 heterooligomerization to enhance leukemia chemotaxis and proliferation. J Clin Investig. 2016;126:585–98. doi: 10.1172/JCI83535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K, Yi S, Yu Z, et al. MicroRNA-223 expression is uniformly down-regulated in B cell lymphoproliferative disorders and is associated with poor survival in patients with chronic lymphocytic leukemia. Leuk Lymph. 2012;53:1155–61. doi: 10.3109/10428194.2011.642303. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.