Abstract
High-risk human papillomavirus (HPV) persistent infection is the major tumorigenesis factor for cervical cancer (CC). However, the incidence of HPV-negative CC is 5% to 30% with different HPV detection methods. High-risk HPV E6/E7 mRNA in situ hybridization (RISH) can detect HPV-driven tumors. Our study aimed to explore whether HPV typing-negative CC was caused by HPV infection. The tissues of CC patients with HPV typing results, collected from cervical biopsies, conization, or hysterectomies, were submitted to RISH using RNAscope chromogenicin. Immunohistochemistry was performed to evaluate the expression of p16INK4a and Ki-67. A total of 308 women with HPV typing results were enrolled, and 30 (9.74%) cases of HPV typing were negative. In HPV typing-negative CCs, 28/30 (93.3%) were positive for RISH, which contained 22/22 (100%) squamous cell carcinomas and 6/8 (75%) adenocarcinomas. RISH was positive in 278/278 (100%) HPV typing-positive CCs, which included 232/232 (100%) squamous cell carcinomas and 46/46 (100%) adenocarcinomas. Positive RISH in HPV typing-negative CC was significantly lower than in the HPV typing-positive group (P=0.002, 95% confidence interval: 0.848–1.027). However, this significant difference only existed in adenocarcinoma. No significant differences were seen in the expression of p16INK4a and Ki-67 (all P>0.05). HPV typing may cause misdiagnosis in 9.74% of CC patients, and HPV E6/E7 mRNA can detect HPV in CC with HPV typing-negative patients. This approach could provide a novel option to accurately detect high-risk HPVs in cervical tumors and help to eliminate the percentage of misdiagnosed HPV-related cases.
Key Words: Cervical cancer, HPV typing-negative, HPV E6/E7 mRNA in situ hybridization, HPV detection
A causal link between persistent infection with high-risk human papillomaviruses (HR-HPVs) and the development of cervical cancer (CC) is well established and is the cause of 99.7% of CC and >75% of cervical adenocarcinoma 1,2. Therefore, testing for HR-HPV has been proven to be effective in screening for CC 3–5. However, in the literature, the incidence of HPV-negative CC is ∼5% to 30% and may be related to differences in HPV testing methodology resulting in a high number of false-negative cases 6–8. Therefore, accurate detection of HR-HPVs in cervical lesions is crucial. Current HPV detection methods, which include hybrid capture-II (HC-II), HPV DNA in situ hybridization, polymerase chain reaction (PCR), and HPV mRNA in situ hybridization (RISH), have varying levels of sensitivity and specificity. The HC-II test, which was a homogeneous hybridization assay using unmarked single-strand RNA probes compatible with the targeted sequences, was practical, but it could not identify the various genotypes. HPV typing assay (PCR) is the most commonly used HPV DNA detection method for persistent HPV infection, which is the key cause of CC. HPV typing tests have been classified according to the expression of HPV L1 capsid proteins, which are one of the main targets of the cellular immune response and are influenced by HPV DNA integration into the human genome. A reduction or loss of capsid antigen production can result in a reduction in the cellular immune response, and previous research revealed that the reduction in L1 capsid antigen was higher in high-grade squamous intraepithelial lesions than in low-grade squamous intraepithelial lesions. HPV typing negativity in CC may be associated with HPV L1 capsid protein loss. Moreover, it cannot distinguish HPV transcriptionally active infections from those defined as “passenger” HPV.
The accumulated data indicated that the viral E6 and E7 proteins are the key factors that maintain the malignant phenotype of HPV-positive cancer cells 9. It is worth pointing out that E6 and E7 are the only viral genes that are always retained and expressed in HPV-positive cancer cells. HPV-positive cancer cells are oncogene addicted in that their growth is dependent on sustained E6/E7 expression. The overexpression of HR-HPV E6/E7 genes is the key causative factor for cervical intraepithelial neoplasias and CC. Quantitative reverse transcriptase PCR to detect HPV E6/E7 mRNA would seem to be an ideal HPV testing method since it reveals that HPV is not merely present but is transcriptionally active. The Food and Drug Administration (FDA) approved PCR to detect E6/E7 mRNA as the “gold standard” for the detection and typing of HPV. However, this assay can be performed exclusively in fresh-frozen tumor tissue and lacks the capacity to specify the detected genotypes; as a result, its use is limited in clinical practice. RISH for detecting HPV E6/E7 mRNA transcripts represents an advance in HPV testing because of the ability to detect the virus in its active transcriptional status in formalin-fixed paraffin-embedded (FFPE) tumor tissues. Leading research has shown that RISH is a highly specific and sensitive method for detecting HPV in oropharyngeal squamous cell carcinoma (SCC) 10. In addition, our previous research proved the accuracy of RISH in diagnosing cervical lesions 11.
Herein, we describe a cohort of CC patients with negative HPV typing on FFPE material and explore the expression of HPV E6/E7 RISH. RNAscope technology is a new generation of single-molecule RISH analysis technology. A novel probe design strategy and a hybridization-based signal amplification system to simultaneously amplify signals and suppress the background are applied during this procedure. mRNA expression was observed at the single-cell level under a standard bright-field microscope.
MATERIALS AND METHODS
Study Population and Selection
Patients with CC detected by cervical biopsy, conization, or hysterectomies were retrospectively collected from April 2018 to September 2021 at the Affiliated Hospital of Weifang Medical University in China. The exclusion criteria were as follows: (1) not tested for HPV typing; (2) history of cervical treatment, regardless of physical therapy or excision; (3) pregnancy status; (4) chemotherapy and/or radiotherapy; and (5) resistant to anti-HPV treatment. In the cohort, CCs with negative HPV typing were included; CC patients with positive HPV typing in the same period were selected as controls (Fig. 1).
FIG. 1.
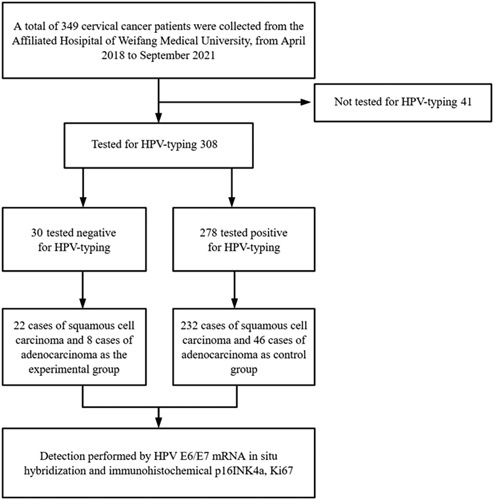
Flowchart of patient inclusion in the study. HPV indicates human papillomavirus.
The patients’ clinical data, which included age at diagnosis, smoking history, drinking history, fertility status, menopausal status, surgical method, and pathologic diagnosis, were collected and evaluated. Subjects who smoked at least 1 cigarette a day and continuously for >6 mo were considered positive for smoking history. Drinking at least once a month, including social engagements, was considered positive for drinking history.
Depending on the procedure, samples included cervical biopsies, cervical conization, and hysterectomies. The tissue blocks were cut into 5 μm sections on positively charged glass slides for the following assays: (1) hematoxylin and eosin (H&E) staining for morphologic identification; (2) p16INK4a immunohistochemistry (IHC); (3) Ki-67 IHC; (4) RISH; and (5) Hs-PPIB RISH (housekeeping/positive control).
Morphologic Evaluation
Two senior pathologists of gynecology who were blind to the results reviewed the H&E-stained slides independently, and any ambiguity was resolved by coexamination using a multihead microscope. Based on the World Health Organization (WHO) 2014 criteria, the research subjects contained 254 cases of SCC and 54 cases of adenocarcinoma, which contained usual, mucinous, serous, clear cell, and minimal deviation adenocarcinoma.
IHC
IHC was performed on FFPE tissue as per standard protocols, and antibodies against p16INK4a (clone: G175-405; ZSGB-BIO, China) and Ki-67 (clone: MIB1; ZSGB-BIO) were used. Phosphate-buffered saline was used in lieu of the primary antibody as a negative control. A positive control slide was used to ensure the validity of the staining procedure. According to the manufacturer’s instructions, the sections were deparaffinized and dehydrated, and antigen retrieval was performed by boiling the slides with EDTA antigen retrieval solution (pH 9.0) in a pressure cooker for 15 min. After blocking endogenous peroxidase with H2O2, the sections were incubated with antibodies, followed by incubation with the secondary antibody. The reaction was detected by diaminobenzidine and counterstained with H&E. The IHC results were analyzed independently by 2 pathologists blinded to the samples.
The p16INK4a staining pattern was classified as negative (no staining), patchy (patchy+, focal and uneven staining in the nuclei and cytoplasm), and block-like (block+, diffuse and even staining in the nuclei and cytoplasm in 100% of the tumor cells). p16INK4a block-like was considered positive 12. For Ki-67, the cells with nuclear staining were counted in at least 10 fields per slide, and the average was calculated. In the cancer cell nuclear staining only, continuous staining was considered positive.
HPV E6/E7 RISH
RISH was performed using the RNAscope 2.5 HD Detection Reagent-BROWN and the HR-HPV 18 probe cocktail (Advanced Cell Diagnostics, Hayward, CA) with the HybEZ hybridization system (Advanced Cell Diagnostics) according to the manufacturer’s instructions. Target-specific probes were used to detect the E6 and E7 genes of HR-HPV 18 genotypes (HPV 16, 18, 26, 31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66, 68, 73, and 82). Hs-PPIB was used as the positive control, while normal epithelial cells in the FFPE block were used as the internal negative control. A positive control that showed effective positive staining in the batch of slides effective was used to ensure that the slide processing was successful. The assay was performed according to the supplier’s instructions (Advanced Cell Diagnostics). Pretreatment: After being deparaffinized and dehydrated, these sections were serially treated with pretreatment 1 and pretreatment 2 solutions, followed by incubation with pretreatment 3 solution overnight. Hybridization: The sections were hybridized in HR-HPV 18 hybridization solution without a coverslip in a HybEZ Oven (Advanced Cell Diagnostics). Signal amplification: The hybridized probe was performed through the serial application of Amp 1 to 6. Visualization: Diaminobenzidine was used to demonstrate the amplified signal. The sections were counterstained with H&E, dehydrated with graded ethanol and xylene, and mounted with Cytoseal.
Images of the RISH slides were taken at 20× magnification using a Motic light microscope (Motic BA600 Mot, Germany). A positive RISH test result was defined as positive if any of the malignant cells showed brown punctuate dot-like nuclear and/or cytoplasmic positivity 13,14.
Following identification and screening by H&E staining, the effective nucleus was taken as the core while eliminating the identified oversized adherent nuclei and undersized cell fragments to identify the areas with diaminobenzidine staining signals within the cell range and to record the positive patterns. To exclude any possible morphologic influences, the RISH slides were evaluated by 2 pathologists who were blinded to the morphologic diagnoses.
Statistical Analysis
Statistical analyses were performed using SPSS 25.0 (SPSS; IBM). The rate and percentage of expression were used to describe the general situation of the study subject. The χ2 analysis was used to test for differences in expression rate. A P-value <0.05 was considered statistically significant.
RESULTS
In the total cohort, 308 women with HPV typing results were enrolled, and most CC patients (300/308, 97.5%) had clinical symptoms of contact bleeding, irregular vaginal bleeding, or/and abnormal discharge; conversely, some patients (8/308, 2.5%) had no abnormal symptoms and were diagnosed by the health examination. The clinical characteristics of these patients are presented in Table 1. The mean age at diagnosis was 47.0±9.7 yr old, with a range of 26 to 73 yr old. The median age was 46 yr old, while the age of most patients at diagnosis was 47 yr old. Furthermore, 56 women (56/308, 18.2%) were postmenopausal, and no women were nulliparous. All the patients had a reproductive history of birth at least once, and women of minimum age were included.
TABLE 1.
The clinical characteristics of the cervical cancer patients included in the study
| Parameters | n (%) |
|---|---|
| Age [mean±SD (range)] | 47.0±9.7 (26–73) |
| Smoking history | |
| Yes | 0 (0) |
| No | 308 (100) |
| Drinking | |
| Yes | 0 (0) |
| No | 308 (100) |
| Fertility status | |
| Nulliparous | 0 (0) |
| Pluriparous | 308 (100) |
| Menopausal status | |
| Premenopausal | 252 (81.8) |
| Postmenopausal | 56 (18.2) |
| Presentation | |
| Contact bleeding | 300 (97.5) |
| Symptomless | 8 (2.5) |
| HR-HPV infection | |
| Yes | 278 (90.3) |
| No | 30 (9.7) |
| Pathologic diagnosis | |
| SCC | 254 (82.5) |
| Adenocarcinoma | 54 (17.5) |
HR-HPV indicates high-risk human papilloma virus; SCC, squamous cell carcinoma.
HPV typing was negative in 30 cases (9.74%), which included 22/30 (73.3%) cases of SCC and 8/30 (26.7%) cases of adenocarcinoma. The details are presented in Table 2. In the CC patients who were HPV typing-negative, 28/30 (93.3%) were positive for RISH and p16INK4a block+ staining, which included 22/22 (100%) SCCs and 6/8 (75%) adenocarcinomas. RISH was positive in 278/278 (100%) HPV typing-positive CCs, which included 232/232 (100%) SCCs and 46/46 (100%) adenocarcinomas. While 273/278 (98.2%) showed p16INK4a block+ staining, the 5 negative cases were SCC. Positive RISH in the HPV typing-negative CC group was significantly lower than that in the HPV typing-positive group [P=0.002, χ2=9.752, 95% confidence interval (CI): 0.848–1.027]. However, this significant difference only existed in adenocarcinoma, and the incidence of positive RISH in HPV typing-negative SCC was the same as that in the HPV typing-positive group. No significant difference was seen in the expression of p16INK4a and Ki-67 (P=0.291, χ2=1.113, 95% CI: 0.863–1.047 and P=0.174, χ2=1.850, 95% CI: 0.905–1.033).
TABLE 2.
Descriptive statistics of the detection of RISH, p16INK4a, and Ki-67 in different groups
| Research group | Control group | ||||
|---|---|---|---|---|---|
| SCC | AC | SCC | AC | P | |
| RISH | |||||
| + | 22 | 6 | 232 | 46 | 0.002 |
| − | 0 | 2 | 0 | 0 | |
| p16INK4a | |||||
| + | 22 | 6 | 227 | 46 | 0.291 |
| − | 0 | 2 | 5 | 0 | |
| Ki-67 | |||||
| + | 22 | 7 | 232 | 46 | 0.174 |
| − | 0 | 1 | 0 | 0 | |
− indicates negative; +, positive; AC, adenocarcinoma; RISH, mRNA in situ hybridization; SCC, squamous cell carcinoma.
A total of 232/278 (83.4%) of the patients had a positive HPV typing of SCC, and the positive staining of RISH is presented in Figure 2. The malignant cells showed brown punctuate dot-like nuclear and/or cytoplasmic positivity. p16INK4a positivity showed diffuse and even staining in the nuclei and cytoplasm in 100% of the tumor cells. The representative positive staining of the RISH of adenocarcinoma patients who were positive for HPV typing presented with p16INK4a and Ki-67 strongly positive staining, as shown in Figure 2. In addition, the expression of RISH and Ki-67 was clearly positive, but p16INK4a was negative, and no block-like staining was observed in SCC with positive HPV typing, as shown in Figure 2.
FIG. 2.

Human papillomavirus typing–positive cervical cancer staining results. Squamous cell carcinoma specimens (A–C), A: mRNA in situ hybridization (+), B: p16INK4a (+), C: Ki-67 (+); adenocarcinoma specimens (D–F), D: mRNA in situ hybridization (+), E: p16INK4a (+), F: Ki-67 (+); squamous cell carcinoma specimens (G–I), G: mRNA in situ hybridization (+), H: p16INK4a (−), I: Ki-67 (+).
The characteristics of the RISH-positive staining of cervical SCC and adenocarcinoma patients who had negative HPV typing presented with positive p16INK4a and Ki-67 staining, as shown in Figure 3. Otherwise, the staining of adenocarcinoma was in conformity, with no positive RISH signals presented. Ki-67 was also negative, while p16INK4a expressed block-like staining, as shown in Figure 3.
FIG. 3.
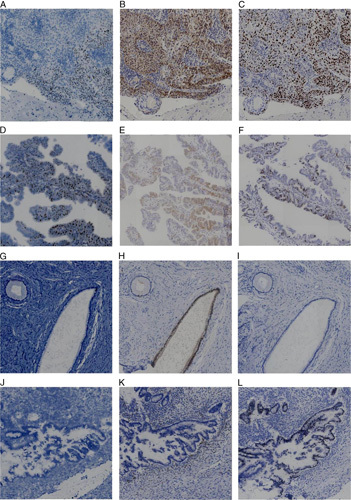
Human papillomavirus typing–negative cervical cancer staining results. Squamous cell carcinoma specimens (A–C), A: mRNA in situ hybridization (+), B: p16INK4a (+), C: Ki-67 (+); adenocarcinoma specimens (D–F), D: mRNA in situ hybridization (+), E: p16INK4a (+), F: Ki-67 (+); adenocarcinoma specimens (G–I), G: mRNA in situ hybridization (−), H: p16INK4a (+), I: Ki-67 (−); adenocarcinoma specimens (J–L), J: mRNA in situ hybridization (−), K: p16INK4a (−), L: Ki-67 (+).
In addition, it is worth noting that adenocarcinoma with no staining of RISH and p16INK4a, in which p16INK4a staining was not expressed in every gland, had no patchy or block-like staining in the nucleus or cytoplasm of cancer cells, while Ki-67-positive staining is presented in Figure 3.
The positive percentages for RISH were 93.3% and 100% in the HPV typing-negative and HPV typing-positive groups, respectively (Table 3). The different markers in the same group had no statistically significant differences in RISH, p16INK4a, p16INK4a/Ki-67, and different combinations (all P>0.05) (Table 3).
TABLE 3.
Statistical descriptions of RISH, p16INK4a, and Ki-67
| Group | |||||
|---|---|---|---|---|---|
| HPV typing-negative | HPV typing-positive | Research (%) | Control (%) | P * | |
| RISH | |||||
| + | 28 | 278 | 93.3 | 100.0 | 1.0 |
| − | 2 | 0 | |||
| p16INK4a | |||||
| + | 28 | 273 | 93.3 | 98.2 | 1.0 |
| − | 2 | 5 | |||
| p16INK4a/Ki-67 | |||||
| + | 27 | 273 | 90 | 98.2 | 1.0 |
| − | 3 | 5 | |||
| RISH/p16INK4a | |||||
| + | 27 | 273 | 90 | 98.2 | 1.0 |
| − | 3 | 5 | |||
| RISH/p16INK4a/Ki-67 | |||||
| + | 27 | 273 | 90 | 98.2 | 1.0 |
| − | 3 | 5 | |||
P-value compared with different detection methods in the research group.
− indicates negative; +, positive; HPV, human papillomavirus; RISH, mRNA in situ hybridization.
DISCUSSION
In this study, we concluded 2 major findings: (1) HPV E6/E7 mRNA was expressed not only in CC with positive HPV typing but also in the majority of CC with negative HPV typing. HPV E6/E7 mRNA positive expression might reflect the phase of HPV infection, which “contributes to” the development of HPV-associated malignancy. (2) The negative expression of p16INK4a does not exclude HPV infection, and p16INK4a positivity does not necessarily indicate HPV infection.
It is well established that persistent infection with oncogenic HPV subtypes is necessary for the contribution to CC 7 and results in nearly all cases of CC 1. A multicentre, open-label, randomized clinical trial concluded that the primary screening of HR-HPV for CC was effective 15, and HPV typing detection is widely used in CC screening programs at present. However, a previous study in SCC using FDA-approved cervical screening technologies showed that HPV testing can be negative in patients with SCC diagnosed by liquid-based cytology 16. HPV-negative CC is ∼5% to 30% and may be related to differences in HPV testing methodology 6–8. In our study, HR-HPV–negative cases accounted for 9.74% of HPV typing CC, which was nearly consistent with previous studies 6–8. Another study demonstrated that HPV DNA, using different molecular methods in tissue samples, accounted for 45.9% of SCCs, which were previously diagnosed as HPV-negative SCCs on cytology material. Those authors had concluded that the higher percentage of HPV-negative results in some series was due to the high number of false-negative cases considering sampling variability, low virus load, rapidly progressive cancers developing between CC screening intervals, and the rarity of truly HR-HPV–negative SCC 17. Recently, a previous study of the integrated genomic and molecular characterization of CC showed that 95% of CCs are HPV-positive 18. These studies concluded that a subset of patients exists in which HR-HPV is not detectable by current laboratory methods used in the clinic.
In the early stage of HPV infection, the virus is mostly present in transient or latent status, and the HPV genome is in a free state in the nucleus 11,19. HPV E6/E7 genes are regulated by E1/E2 genes, and their expression is inhibited during this stage 11,20,21. HPV E6/E7 mRNA is often undetectable in normal cervical tissues or low-grade squamous intraepithelial lesion 11,22. During the persistent infection stage, HR-HPV DNA integrates into the host genome with the disruption and loss of the E2 gene, which leads to HPV E6/E7 mRNA overexpression and L1 capsid protein loss 9,23. In some studies, an in situ hybridization test has been proven to have higher sensitivity and specificity in the detection of HPV infection, and the HPV E6/E7 mRNA test was considered a potential tool 23,24. In our study, the typical HPV E6/E7 mRNA RISH image of CC presented with weak-to-strong nuclear and cytoplasmic dot-like signals within malignant cells. Our study also showed that a subset of women may have a false-negative HPV typing result that may be associated with fluctuations in HR-HPV positivity and HPV L1 capsid protein loss. Moreover, HPV E6E7 mRNA RISH was negative in 2 (25%) HPV typing-negative cervical adenocarcinomas, which was in accordance with the previous study that CC cases were not all HPV-infected 6,23, and potential contributing factors included infection with an HPV genotype not currently contained in standard testing platforms, variations in sampling, low viral load or decreased transcriptional activity, excluding E6/E7 genes, host p53, and retinoblastoma protein (Rb protein) mechanisms, and metastatic neoplasms in vivo in early or truly HR-HPV–negative carcinoma 6,9,20,23. In contrast to a previous study, which showed that cervical SCC cases were not all HPV-infected 25, all SCCs in our study were positive for RISH, which could reveal that the HPV E6/E7 mRNA test had superior detection in HPV-associated SCC.
Histopathology is the gold standard for the diagnosis of CC. However, pathologists may use some markers to make a differential diagnosis in some cases with equivocal pathologic features, including p16INK4a and Ki-67; however, such markers are not specific. p16INK4a is regarded as a reliable surrogate method to detect HR-HPV with a sensitivity approaching 100% 26, and the use of p16INK4a has been reported in several studies on cytology and histologic samples as a biomarker assisting in the diagnosis of high-grade epithelial cervical lesions 27. In our study, most CCs with p16INK4a-positive staining were 93.3% and 98.2% in the 2 groups, respectively, which was concordant with previous studies reporting p16INK4a-positive staining from 80% to 100% in invasive carcinoma 28. The variation in expression rates may partly depend on the criteria defining positive expression. In this research, nuclear and continuous diffuse cytoplasmic staining of the cells was considered positive, while some studies required nuclear or cytoplasmic staining to be positive 28. However, negative cases were detected in adenocarcinoma. Many previous studies insisted that the overexpression of p16INK4a, as a multitumour suppressor gene, may be closely correlated with HPV oncoprotein E7 inactivating cell cycle regulation Rb protein 29 and was directly involved in the regulation of the cell cycle, inhibiting the activity of cyclin-dependent protein kinase CDK4/CDK6. In our study, we thought that p16INK4a negative staining might be evoked via pathways other than HPV infection through gene mutation or hypermethylation. Currently, the specific mechanism is not fully understood, and the correlation between p16INK4a and HPV infection history remains unclear. It is also controversial whether the p16INK4a expression level can accurately predict the potential risk of precancerous lesions or CC.
Ki-67 positivity is highly dependent on reactive changes and cell proliferation 30. As a nuclear proliferation antigen, Ki-67 is exclusively expressed in the proliferation phase of the cell cycle. Some previous data revealed that the Ki-67-positive staining percentage in normal cervical tissues and cervical intraepithelial neoplasia and CC tissues gradually increases with the aggravation of the disease 28. In our study, the levels of Ki-67-positive staining were 96.7% and 100% in the 2 groups, respectively. These results were better concordant with previous studies that found Ki-67 in 90% to 100% of invasive carcinomas 28. Ki-67 and p16INK4a/Ki-67 are the most widely used biomarkers that significantly improve the accuracy of the pathologic diagnosis of cervical lesions 28. The cotest of Ki-67-positive and p16INK4a-positive staining usually indicates that the cell cycle is out of control, which occurs in abnormal cell proliferation. However, our study showed that Ki-67 and p16INK4a/Ki-67 exhibited no significant improvement over p16INK4a alone in the diagnosis of CC. Hence, the routine addition of Ki-67 to p16INK4a was not recommended, which was consistent with the results of previous studies 11. In addition, some cases expressed positive staining of p16INK4a, while HPV E6/E7 mRNA was not expressed, a high correspondence between p16INK4a and the RISH test was shown in the literature; however, even in only a few cases, discordant results were found in our study and some previous studies 13,31–33, which may have resulted from HPV detection methods. In other words, the negative expression of p16INK4a does not exclude HPV infection, and p16INK4a positivity does not necessarily indicate HPV infection 34,35.
The present study has several limitations. As a retrospective study, specimens were taken from samples in the past that were not tested by HC-II, HPV DNA in situ hybridization, and HPV E6/E7 mRNA (APTIMA) to further confirm the presence or absence of HR-HPV. Another limitation was that our study had no detailed history information on HPV infection and had limited available data from Pap screening tests and colposcope examinations, which reflected both the small percentage of patients who were diagnosed with CC who were RISH-negative overall and the fact that the majority of patients who developed CC had inadequate screening. Previous data also revealed that 60% of women diagnosed with CC had never received Pap screenings or had done so at irregular intervals 36. These data would certainly assist further in the interpretation of the correlation of HR-HPV–associated CC (false-negative) and IHC findings. A multicentre, large-scale, and further prospective study with more appropriate and accurate data should be conducted.
CONCLUSIONS
HPV typing tests could cause misdiagnoses in ∼9.74% of CC patients, and HPV E6/E7 mRNA could detect all cervical SCC and most cervical adenocarcinoma with HPV-related status even when HPV typing is negative. This approach could provide a novel approach to accurately detect HR-HPVs in cervical tumors and help to eliminate the percentage of misdiagnosed HPV-related cases.
Footnotes
Supported by a grant from the National Natural Science Fund of China (No. 81572559) and the Scientific Research Foundation of Weifang Medical University (2017BSQD43).
The authors declare no conflict of interest.
Contributor Information
Yating Xu, Email: 645921656@qq.com.
Yonghong Sun, Email: syhdoctor2021@163.com.
Hui Chang, Email: lzchdoctor1987@163.com.
Jingjing Cai, Email: cjjdoctor1989@163.com.
Chengcheng Cao, Email: ccc2021fk@163.com.
Baogang Zhang, Email: zbg10438@163.com.
Youzhong Zhang, Email: zyzsduw@163.com.
Yuzhen Liu, Email: lyz0412@wfmc.edu.cn.
REFERENCES
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7–34. [DOI] [PubMed] [Google Scholar]
- 2. Stolnicu S, Hoang L, Hanko-Bauer O, et al. Cervical adenosquamous carcinoma: detailed analysis of morphology, immunohistochemical profile, and clinical outcomes in 59 cases. Mod Pathol 2019;32:269–79. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 3. Bulkmans NW, Berkhof J, Rozendaal L, et al. Human papillomavirus DNA testing for the detection of cervical intraepithelial neoplasia grade 3 and cancer: 5-year follow-up of a randomised controlled implementation trial. Lancet 2007;370:1764–72. [DOI] [PubMed] [Google Scholar]
- 4. Ogilvie GS, van Niekerk D, Krajden M, et al. Effect of screening with primary cervical HPV testing vs cytology testing on high-grade cervical intraepithelial neoplasia at 48 months: The HPV FOCAL Randomized Clinical Trial [published correction appears in JAMA. 2018;320:2273]. JAMA 2018;320:43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dijkstra MG, van Zummeren M, Rozendaal L, et al. Safety of extending screening intervals beyond five years in cervical screening programmes with testing for high risk human papillomavirus: 14 year follow-up of population based randomised cohort in the Netherlands [published correction appears in BMJ. 2016;355:i5782]. BMJ 2016;355:i4924. [DOI] [PubMed] [Google Scholar]
- 6. Alexander C, White M, Maleki Z, et al. HPV-ISH-negative invasive cervical squamous cell carcinoma: histologic and Pap test results. Acta Cytol 2019;63:417–23. [DOI] [PubMed] [Google Scholar]
- 7. Tao X, Griffith CC, Zhou X, et al. History of high-risk HPV and Pap test results in a large cohort of patients with invasive cervical carcinoma: experience from the largest women's hospital in China. Cancer Cytopathol 2015;123:421–7. [DOI] [PubMed] [Google Scholar]
- 8. Samimi SA, Mody RR, Goodman S, et al. Do infection patterns of human papillomavirus affect the cytologic detection of high-grade cervical lesions on Papanicolaou tests? Arch Pathol Lab Med 2018;142:347–52. [DOI] [PubMed] [Google Scholar]
- 9. Hoppe-Seyler K, Bossler F, Braun JA, et al. The HPV E6/E7 oncogenes: key factors for viral carcinogenesis and therapeutic targets. Trends Microbiol 2018;26:158–68. [DOI] [PubMed] [Google Scholar]
- 10. Randén-Brady R, Carpén T, Jouhi L, et al. In situ hybridization for high-risk HPV E6/E7 mRNA is a superior method for detecting transcriptionally active HPV in oropharyngeal cancer. Hum Pathol 2019;90:97–105. [DOI] [PubMed] [Google Scholar]
- 11. Hui C, Bai H, Liu J, et al. Accuracy of HPV E6/E7 mRNA examination using in situ hybridization in diagnosing cervical intraepithelial lesions. Diagn Pathol 2021;16:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Darragh TM, Colgan TJ, Thomas Cox J, et al. The Lower Anogenital Squamous Terminology Standardization project for HPV-associated lesions: background and consensus recommendations from the College of American Pathologists and the American Society for Colposcopy and Cervical Pathology [published correction appears in Int J Gynecol Pathol. 2013;32:432] [published correction appears in Int J Gynecol Pathol. 2013;32:241]. Int J Gynecol Pathol 2013;32:76–115. [DOI] [PubMed] [Google Scholar]
- 13. Mirghani H, Casiraghi O, Amen F, et al. Diagnosis of HPV-driven head and neck cancer with a single test in routine clinical practice. Mod Pathol 2015;28:1518–27. [DOI] [PubMed] [Google Scholar]
- 14. Zito Marino F, Ronchi A, Stilo M, et al. Multiplex HPV RNA in situ hybridization/p16 immunohistochemistry: a novel approach to detect papillomavirus in HPV-related cancers. A novel multiplex ISH/IHC assay to detect HPV. Infect Agent Cancer 2020;15:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang J, Zhao Y, Dai Y, et al. Effectiveness of high-risk human papillomavirus testing for cervical cancer screening in China: a multicenter, open-label, randomized clinical trial. JAMA Oncol 2021;7:263–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Austin RM, Onisko A, Zhao C. Enhanced detection of cervical cancer and precancer through use of imaged liquid-based cytology in routine cytology and HPV cotesting. Am J Clin Pathol 2018;150:385–92. [DOI] [PubMed] [Google Scholar]
- 17. Tao X, Zheng B, Yin F, et al. Polymerase chain reaction human papillomavirus (HPV) detection and hpv genotyping in invasive cervical cancers with prior negative HC2 test results. Am J Clin Pathol 2017;147:477–83. [DOI] [PubMed] [Google Scholar]
- 18. Cancer Genome Atlas Research Network; Albert Einstein College of Medicine; Analytical Biological Services. Integrated genomic and molecular characterization of cervical cancer. Nature 2017;543:378–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Woodman CB, Collins SI, Young LS. The natural history of cervical HPV infection: unresolved issues. Nat Rev Cancer 2007;7:11–22. [DOI] [PubMed] [Google Scholar]
- 20. Münger K, Werness BA, Dyson N, et al. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. EMBO J 1989;8:4099–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yeo-Teh NSL, Ito Y, Jha S. High-risk human papillomaviral oncogenes E6 and E7 target key cellular pathways to achieve oncogenesis. Int J Mol Sci 2018;19:1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Graham SV. The human papillomavirus replication cycle, and its links to cancer progression: a comprehensive review. Clin Sci (Lond) 2017;131:2201–21. [DOI] [PubMed] [Google Scholar]
- 23. Chen T, Li J, Wang S, et al. High-risk HPV E6/E7 mRNA in situ hybridization in endocervical glandular neoplasia: performance compared with p16INK4a and Ki67 immunochemistry. Am J Transl Res 2019;11:6498–506. [PMC free article] [PubMed] [Google Scholar]
- 24. Derbie A, Mekonnen D, Woldeamanuel Y, et al. HPV E6/E7 mRNA test for the detection of high grade cervical intraepithelial neoplasia (CIN2+): a systematic review. Infect Agent Cancer 2020;15:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lewis JS, Jr. Human papillomavirus testing in head and neck squamous cell carcinoma in 2020: where are we now and where are we going? Head Neck Pathol 2020;14:321–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Madrigal E, Bishop JA, Faquin WC. Head and neck cytopathology: human papillomavirus-positive carcinomas, including diagnostic updates, testing modalities, and recommendations. Surg Pathol Clin 2018;11:501–14. [DOI] [PubMed] [Google Scholar]
- 27. Hodgson A, Park KJ, Djordjevic B, et al. International endocervical adenocarcinoma criteria and classification: validation and interobserver reproducibility. Am J Surg Pathol 2019;43:75–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kanthiya K, Khunnarong J, Tangjitgamol S, et al. Expression of the p16 and Ki67 in cervical squamous intraepithelial lesions and cancer. Asian Pac J Cancer Prev 2016;17:3201–6. [PubMed] [Google Scholar]
- 29. Martin CM, O'Leary JJ. Histology of cervical intraepithelial neoplasia and the role of biomarkers. Best Pract Res Clin Obstet Gynaecol 2011;25:605–15. [DOI] [PubMed] [Google Scholar]
- 30. Goel MM, Mehrotra A. Immunohistochemical expression of MIB-1 and PCNA in precancerous and cancerous lesions of uterine cervix. Indian J Cancer 2013;50:200–5. [DOI] [PubMed] [Google Scholar]
- 31. Lewis JS, Jr, Ukpo OC, Ma XJ, et al. Transcriptionally-active high-risk human papillomavirus is rare in oral cavity and laryngeal/hypopharyngeal squamous cell carcinomas—a tissue microarray study utilizing E6/E7 mRNA in situ hybridization. Histopathology 2012;60:982–91. [DOI] [PubMed] [Google Scholar]
- 32. Bishop JA, Ma XJ, Wang H, et al. Detection of transcriptionally active high-risk HPV in patients with head and neck squamous cell carcinoma as visualized by a novel E6/E7 mRNA in situ hybridization method. Am J Surg Pathol 2012;36:1874–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ukpo OC, Flanagan JJ, Ma XJ, et al. High-risk human papillomavirus E6/E7 mRNA detection by a novel in situ hybridization assay strongly correlates with p16 expression and patient outcomes in oropharyngeal squamous cell carcinoma. Am J Surg Pathol 2011;35:1343–50. [DOI] [PubMed] [Google Scholar]
- 34. Castle PE, Adcock R, Cuzick J, et al. Relationships of p16 immunohistochemistry and other biomarkers with diagnoses of cervical abnormalities: implications for LAST terminology. Arch Pathol Lab Med 2020;144:725–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zafereo ME, Xu L, Dahlstrom KR, et al. Squamous cell carcinoma of the oral cavity often overexpresses p16 but is rarely driven by human papillomavirus. Oral Oncol 2016;56:47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wheeler CM, Hunt WC, Joste NE, et al. Human papillomavirus genotype distributions: implications for vaccination and cancer screening in the United States. J Natl Cancer Inst 2009;101:475–87. [DOI] [PMC free article] [PubMed] [Google Scholar]


