In eukaryotes, gene expression is highly orchestrated not only by genomic promoters and enhancers but also by covalent modifications added to either chromatin or RNAs. Traditionally, “epigenetics” refers to the chemical modifications that govern heritable changes in gene expression independent of the DNA sequence; “epitranscriptomics” indicates the covalent decorations in RNA, which plays a central role in posttranscriptional gene regulation. To date, >170 RNA chemical modifications have been characterized. Most of these modifications were originally identified in highly abundant noncoding RNA species, such as ribosomal RNAs (rRNAs), transfer RNAs (tRNAs), and small nuclear RNA (snRNAs). Recently, the substantial advances in high-throughput sequencing and analytical chemistry have enabled the precise detection and characterization of chemical modifications in messenger RNA (mRNA). Indeed, a considerable number of mRNA decorations have been documented, including N6-methyladenosine (m6A); N1-methyladenosine (m1A); N6,2′-O-dimethyladenosine (m6Am); 3-methylcytidine (m3C); 5-methylcytidine (m5C); 5-hydroxymethylcytidine (hm5C); N4-acetylcytidine (ac4C); Adenosine-to-inosine (A-to-I) editing; pseudouridine (Ψ); N7-methylguanosine (m7G) and 2′-O-methylated nucleotides (Nm) (Fig. 1A). The studies from us and other researchers have unveiled that mRNA modifications play important roles in myeloid malignancies.1–11 Here, we highlight recent findings focusing on the functions and regulatory mechanisms of mRNA modifications (with an emphasis on m6A) and provide our insights to better elucidate epitranscriptomics during leukemogenesis.
Figure 1.
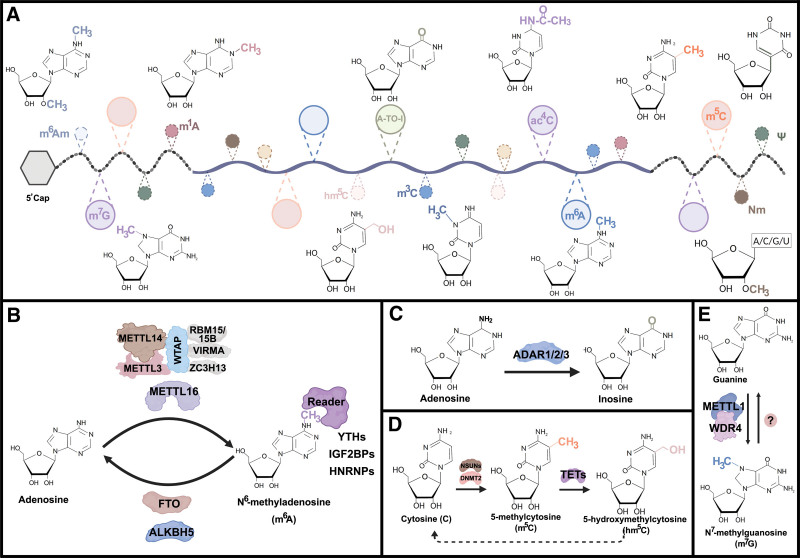
The covalent chemical modifications in mRNA. (A) A schematic view of chemical structures of RNA modifications (m6Am, m1A, A-to-I, ac4C, m5C, Ψ, m6A, hm5C, m3C, m7G and Nm) in mRNA. (B) The regulation of m6A by “writers,” “erasers,” and “readers.” The m6A modification is installed by writers, multicomponent m6A MTC (composed of METTL3, METTL14, WTAP, RBM15/15B, KIAA1429, and ZC3H13) or METTL16 alone. The two demethylases (eraser), FTO and ALKBH5, remove m6A modifications. The m6A residue is recognized by the three main classes, including the YTH domain family, IGF2BP family and HNRP family. (C) ADAR enzymes catalyze the A-to-I hydrolytic deamination reaction. (D) The NSUN methyltransferases and DNMT2 catalyze methylation of cytosine-5 and TET family function as dioxygenases catalyzing m5C to hm5C. (E) The formation of m7G is catalyzed by methyltransferases complex, which is composed of METTL1 and WDR4. ADAR = adenosine deaminases acting on RNA.
m6A, the most abundant and best-characterized mRNA modification, was initially discovered in 1974.12 Owing to the lack of effective and sensitive technologies, recognition of functional significance of m6A has been largely absent over the past decades. Until 2011, the identification of fat mass- and obesity-associated protein (FTO) as the first m6A demethylase13 has profoundly revived the interest in the biological relevance of m6A and RNA chemical modifications. Both FTO and AlkB homolog 5 (ALKBH5) can catalyze the demethylation of m6A, demonstrating the reversible and dynamic posttranscriptional modification in RNA (Fig. 1B). Methyltransferase-like 3 (METTL3) forms a heterodimer with METTL14 to deposit m6A residues in mRNAs. METTL3 acts as the catalytically active methyltransferase with METTL14 serving as an allosteric activator to enhance catalysis.14 The large methyltransferase complex (MTC) contains additional subunits, including Wilms tumor 1-associated protein (WTAP), zinc finger CCCH-type contain 13 (ZC33H13), vir like m6A methyltransferase associated (VIRMA) and RNA-binding motif protein 15/15B (RBM15/15B) (Fig. 1B). More recently, METTL16 has been identified as a new m6A writer, which could exert its enzymatic activity independently and catalyze m6A formation on limited mRNAs and noncoding RNAs15,16 (Fig. 1B). Characterization of m6A “reader” proteins, including YT521-B homology (YTH) domain, insulin-like growth factor 2 mRNA-binding protein (IGF2BP) family, and heterogeneous nuclear ribonucleoprotein (HNRP) family, has provided valuable insights into understanding the underlying mechanism of m6A-mediated posttranscriptional gene regulation.
The dysregulation of m6A regulators has been reported to be extensively involved in the pathogenesis of myeloid malignancies. We first reported that FTO is highly expressed in acute myeloid leukemia (AML) with t(11q23)/MLL-rearrangements, t(15;17)/PML-RARA, FLT3-ITD and/or NPM1 mutations, and enhances leukemic malignant transformation and leukemogenesis as an m6A demethylase.3 Subsequently, we showed that R-2-hydroxyglutarate (R-2HG), an oncometabolite of mutant isocitrate dehydrogenase 1/2 (IDH1/2), exerts a broad and intrinsic antileukemic activity through competitively inhibiting the demethylase activity of FTO.1 Such identification indicates that FTO might act as a druggable target for leukemia therapy. Moreover, FTO drives and sustains tyrosine kinase inhibitor (TKI) tolerance in leukemia cells by enhancing mRNA stability or translation efficiency of antiapoptotic genes via m6A decoration.17 Analogous to FTO, the other m6A “eraser” ALKBH5 also exerts a tumor-promoting role in leukemia.4,18 ALKBH5 is highly expressed in AML, especially in leukemia stem/initiating cells (LSCs/LICs), and maintains LSC/LIC frequency by posttranscriptional regulation of its critical targets, such as TACC3 and AXL.4,18 Two independent studies have reported that METTL3 exerts an essential oncogenic role in AML.6,7 Mechanistically, METTL3 could deposit m6A modification on its mRNA targets, including MYC and BCL2, which, in turn, leads to their translational activation to block myeloid differentiation and promote leukemogenesis.7 Another recent study in AML showed that a portion of METTL3 is recruited to the promoter regions of ~80 genes by CAATT enhancers binding protein zeta (CEBPZ) to activate MYC signaling.6 METTL14, another essential component of m6A MTC, is highly expressed in AML and also plays a critical oncogenic role during leukemogenesis.5 Genetic depletion of METTL14 inhibits cell proliferation, induces myeloid differentiation and cell apoptosis, suppresses self-renewal of LSCs/LICs and delays AML progression In vivo via an m6A-deppendent manner.5 Several other studies have revealed the oncogenic role of WTAP and RBM15, another 2 components of m6A MTC, in leukemia pathogenesis.19,20 However, whether such oncogenic roles are attributed to m6A decoration remains to be elucidated. Via genome-wide CRISPR-Cas9 screen, METTL16 has been identified as one of the most essential genes for the survival of leukemia cells.6 Yet further studies are required to address its role in hematological malignancies. In addition, YTHDF2 is highly expressed in a broad spectrum of human AMLs and required for LSC/LIC self-renewal and AML initiation and propagation via shortening the half-life of m6A-modified transcripts.8 Another reader protein YTHDC1 is also required for the survival of AML cells.21,22 YTHDC1 could undergo liquid–liquid phase separation and form nuclear YTHDC1-m6A condensates to protect MYC from the PAXT complex and exosome-associated RNA degradation.21 Moreover, YTHDC1 drives leukemogenesis through controlling stability of MCM4.22 Taken together with these reported studies on m6A regulators, both m6A writers and erasers function as oncoproteins in leukemia, despite their opposite role in mRNA methylation. It might be highly possible that different m6A writers, especially erasers and readers regulate different groups of target genes, and different m6A regulators preferentially bind to distinct regions of the same transcript and result in divergent fates. Actually, IGF2BP preferentially binds to the 3′ UTR region of MYC and increases its stability,23 whereas YTHDF2 prefers to bind to the 5′ UTR and middle exons of MYC and leads to its decay.1 Thus, it will be very important and interesting to systemically characterize the specific targets of each m6A regulator and clarify how a particular m6A site in a given transcript fine-tunes its metabolism and determines its fate.
Considering the critical roles of m6A modification and its machinery in hematological malignancies, targeting the dysregulated m6A regulator(s) may represent an attractive strategy for leukemia treatment. Indeed, several small molecular compounds targeting m6A machinery have been discovered. STM2457, a potent and selective inhibitor of METTL3 leads to reduced AML growth and an increase in differentiation and apoptosis.24 We have discovered several specific and highly efficient inhibitors targeting FTO to treat leukemia.25 More promisingly, m6A modification also modulates drug resistance and reprograms immune response.26 Therefore, combining such inhibitors targeting m6A machinery with chemotherapy and/or immunotherapy may lead to the development of more effective therapies to overcome therapy resistance and cure leukemia.
Aside from m6A, other RNA epigenetic marks, such as A-to-I editing, m5C and m7G, are also implicated in the pathogenesis of hematological malignancies. The adenosine deaminases acting on RNA 1 (ADAR1)-mediated A-to-I editing is correlated with a poor prognosis of chronic myeloid leukemia (CML) patients,9 and promotes LSC self-renewal and CML progression (Fig. 1C).10 The m5C modification in RNA is deposited by m5C methyltransferases (RCMTs), including NOL1/NOP2/SUN domain (NSUN) family and DNA methyltransferase homologue (DNMT2), oxidized by TET proteins,27 and recognized by Y-box binding protein 1 (YBX1)28 (Fig. 1D). NSUN3 and DNMT2 directly interact with hnRNPK to form a functional complex which is important for the survival and drug response of leukemia cells.11 Although extensive studies have reported the critical roles of TET protein in blood malignancies, it is totally unknown whether TET-induced demethylation of mRNA m5C is involved in leukemogenesis and if so, what the biological function would be. METTL1/WDR4-mediated m7G decoration in tRNA potentiates oncogenic transformation and tumorigenesis.29 METTL1 also acts as the methyltransferase for internal m7G modification in mRNA (Fig. 1E); while it is still underdetermined whether METTL1-induced m7G modification in mRNA is involved in leukemogenesis.
Within the past few years, tremendous efforts have been devoted to deciphering the role of RNA modifications in the pathogenesis, leading to rapid expansion of epitranscriptomics. Unlike the well-established methods to investigate m6A decoration, studies on other mRNA chemical modifications, such as m5C, m7G, m1A, and m6Am, are still at their infant stages due to the lack the transcriptome-wide sequencing approaches as well as the characterization of the modification machinery, especially the “eraser” and “reader” proteins. The development of novel tools that can precisely determine the landscape of RNA modifications at single-nucleotide resolution will greatly push the field forward. In addition, further unraveling the fundamental mechanisms of RNA epigenetic modifications and the related machinery may reveal the promising novel therapeutic strategies to treat leukemia and other life-threatening diseases.
ACKNOWLEDGMENTS
We apologize to colleagues whose work could not be cited due to space limitations.
Footnotes
J.C. received support from NIH (R01 CA243386, R01 CA214965, R01 CA236399, R01 CA211614, and R01 DK124116). R.S. received support from The Margaret E. Early Medical Research Trust.
Conflicts of interest: The authors declare that they have no conflict of interest.
REFERENCES
- [1].Su R, Dong L, Li C, et al. R-2HG exhibits anti-tumor activity by targeting FTO/m(6)A/MYC/CEBPA signaling. Cell 2018;172:90–105.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Qing Y, Dong L, Gao L, et al. R-2-hydroxyglutarate attenuates aerobic glycolysis in leukemia by targeting the FTO/m(6)A/PFKP/LDHB axis. Mol Cell 2021;81:922–939.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Li Z, Weng H, Su R, et al. FTO plays an oncogenic role in acute myeloid leukemia as a N(6)-methyladenosine RNA demethylase. Cancer Cell 2017;31:127–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Shen C, Sheng Y, Zhu AC, et al. RNA demethylase ALKBH5 selectively promotes tumorigenesis and cancer stem cell self-renewal in acute myeloid leukemia. Cell Stem Cell 2020;27:64–80.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Weng H, Huang H, Wu H, et al. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m(6)A modification. Cell Stem Cell 2018;22:191–205.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Barbieri I, Tzelepis K, Pandolfini L, et al. Promoter-bound METTL3 maintains myeloid leukaemia by m(6)A-dependent translation control. Nature 2017;552:126–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Vu LP, Pickering BF, Cheng Y, et al. The N(6)-methyladenosine (m(6)A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med 2017;23:1369–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Paris J, Morgan M, Campos J, et al. Targeting the RNA m(6)A reader YTHDF2 selectively compromises cancer stem cells in acute myeloid leukemia. JCell Stem Cell 2019;25:137–148 e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Jiang Q, Crews LA, Barrett CL, et al. ADAR1 promotes malignant progenitor reprogramming in chronic myeloid leukemia. Proc Natl Acad Sci USA 2013;110:1041–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zipeto MA, Court AC, Sadarangani A, et al. ADAR1 activation drives leukemia stem cell self-renewal by impairing Let-7 biogenesis. Cell Stem Cell 2016;19:177–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Cheng JX, Chen L, Li Y, et al. RNA cytosine methylation and methyltransferases mediate chromatin organization and 5-azacytidine response and resistance in leukaemia. Nat Commun 2018;9:1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci USA 1974;71:3971–3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Jia G, Fu Y, Zhao X, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol 2011;7:885–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Liu J, Yue Y, Han D, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol 2014;10:93–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Pendleton KE, Chen B, Liu K, et al. The U6 snRNA m(6)A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell 2017;169:824–835 e814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Su R, Dong L, Li Y, et al. METTL16 exerts an m(6)A-independent function to facilitate translation and tumorigenesis. Nat Cell Biol 2022;24:205–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Yan F, Al-Kali A, Zhang Z, et al. A dynamic N(6)-methyladenosine methylome regulates intrinsic and acquired resistance to tyrosine kinase inhibitors. Cell Res 2018;28:1062–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Wang J, Li Y, Wang P, et al. Leukemogenic chromatin alterations promote AML leukemia stem cells via a KDM4C-ALKBH5-AXL signaling axis. Cell Stem Cell 2020;27:81–97 e88. [DOI] [PubMed] [Google Scholar]
- [19].Bansal H, Yihua Q, Iyer SP, et al. WTAP is a novel oncogenic protein in acute myeloid leukemia. Leukemia 2014;28:1171–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hsiao HH, Yang MY, Liu YC, et al. RBM15-MKL1 (OTT-MAL) fusion transcript in an adult acute myeloid leukemia patient. Am J Hematol 2005;79:43–45. [DOI] [PubMed] [Google Scholar]
- [21].Cheng Y, Xie W, Pickering BF, et al. N(6)-Methyladenosine on mRNA facilitates a phase-separated nuclear body that suppresses myeloid leukemic differentiation. Cancer Cell 2021;39:958–972 e958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Sheng Y, Wei J, Yu F, et al. A critical role of nuclear m6A reader YTHDC1 in leukemogenesis by regulating MCM complex-mediated DNA replication. Blood 2021;138:2838–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Huang H, Weng H, Sun W, et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol 2018;20:285–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Yankova E, Blackaby W, Albertella M, et al. Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature 2021;593:597–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Su R, Dong L, Li Y, et al. Targeting FTO suppresses cancer stem cell maintenance and immune evasion. Cancer Cell 2020;38:79–96 e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Li Y, Su R, Deng X, Chen Y, Chen J. FTO in cancer: functions, molecular mechanisms, and therapeutic implications. Trends Cancer 2022. In press. [DOI] [PubMed] [Google Scholar]
- [27].Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol 2017;18:31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Yang Y, Wang L, Han X, et al. RNA 5-methylcytosine facilitates the maternal-to-zygotic transition by preventing maternal mRNA decay. Mol Cell 2019;75:1188–1202 e1111. [DOI] [PubMed] [Google Scholar]
- [29].Orellana EA, Liu Q, Yankova E, et al. METTL1-mediated m(7)G modification of Arg-TCT tRNA drives oncogenic transformation. Mol Cell 2021;81:3323–3338 e3314. [DOI] [PMC free article] [PubMed] [Google Scholar]