Abstract
Stable transfection of a new, chimeric reporter in the human malaria parasite Plasmodium falciparum confers green fluorescence and methotrexate resistance that can be quantitated by Western blotting and flow cytometry. This provides a sensitive, live reporter for exploitation of genomic and high-throughput assays for the identification of new pathogenic determinants.
Malaria is a major health problem worldwide, and the spread of resistance to available drugs increases the need to develop new drugs and effective vaccines. Sequencing the genome of the human malarial parasite Plasmodium falciparum provides a major step forward, but there is a paucity of functional assays to exploit this genetic information. With the development of transformation techniques, reverse genetics provides potentially powerful tools to develop functional assays in Plasmodium (2–4, 8, 9–11, 14, 16, 17). Both rodent and human malaria can be transfected with exogenous DNA to stably express and disrupt plasmodial genes. However, there is no reporter expressed in live human malaria that can be detected by flow cytometry to allow the high-throughput analysis and automation required for rapid screens. This approach combined with genomics has enabled the identification of new determinants of virulence and pathogenesis in bacterial systems (13).
Green fluorescent protein (GFP) from Aequorea victoria is a valuable reporter whose relative stability and lack of toxicity have enabled studies of gene expression and protein localization in a large variety of cell types. Stable episomal or integrated expression of GFP has been reported in the rodent malaria parasite Plasmodium berghei (5, 12). However, P. falciparum does not produce stable infection in rodents. Furthermore, since control regions that regulate gene expression in the two plasmodial species are quite distinct, separate systems need to be developed for use with human malaria.
Construction of pHDGFP and its expression in P. falciparum.
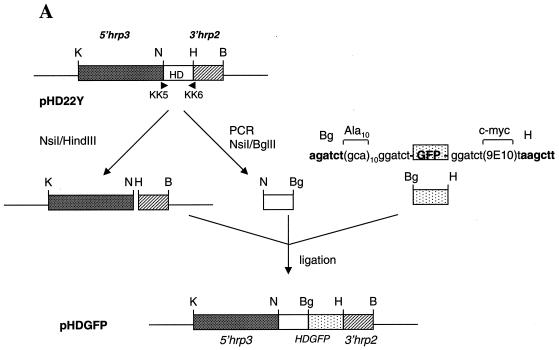
We had previously constructed a plasmid, pHRPGFPm2, in which gfp is flanked by 5′hrp3 and 3′hrp2 control regions. Transfection of this plasmid into P. falciparum-infected erythrocytes results in transient expression of green fluorescence in the parasite (15). To facilitate stable maintenance of the plasmid, we designed a chimeric molecule of human dihydrofolate reductase (DHFR) and GFP (called HDGFP) flanked by one set of gene control regions derived from 5′hrp3 and 3′hrp2 of P. falciparum (Fig. 1A). Human DHFR has been expressed in P. falciparum, and it confers resistance to WR99210 or methotrexate (6, 7). Cultures containing the pHDGFP construct showed viable methotrexate-resistant parasites 2 to 3 weeks after transfection (Fig. 1B). Thus, HDGFP appeared to provide a bonafide cytosolic marker in the parasite.
FIG. 1.
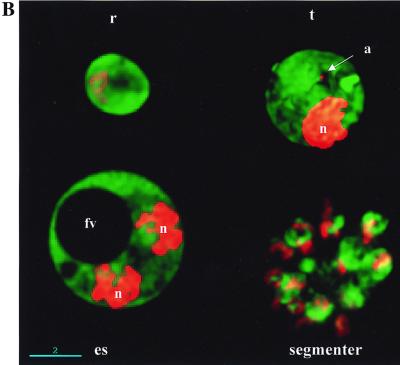
Construction of pHDGFP and its expression in P. falciparum. (A) Human DHFR (HD) sequences were amplified by PCR from the plasmid pHD22Y (6) by using primers 5′-GTATGCATGGTTCGCTAAACTGC (KK5) and 5′-TTAAGATCTATCATTCTTCTCATATACTTC (KK6), which conferred the flanking restriction sites NsiI (N) and BglII (Bg), indicated by boldface letters. The GFP mutant m2 (1) gene carrying an Ala(10) linker at the N terminus and a c-myc 9E10 epitope at the C terminus was derived by BglII-HindIII (Bg-H) digestion of plasmid pHRPExGFP (M. Kadekoppala and K. Haldar, unpublished data). The resulting HD and GFP fragments were ligated to the pHD22Y backbone to obtain pHDGFP. K, KpnI. (B) Cultures transfected with pHDGFP show green fluorescence detected at the ring (r), trophozoite (t), and late segmenter stages. Live cells were viewed by DeltaVision microscopy (Applied Precision) with filter settings for fluorescein isothiocyanate. Twenty optical sections of 0.3 μm were taken along the z axis, and the raw images were subjected to deconvolution and presented as 0° projections of three-dimensional reconstructions. Nucleic acid staining (Hoechst 33342) is shown in red in the nucleus (n) and the apicoplast (a). The food vacuole (fv) is indicated in a single optical section of an early schizont (es). Scale bar, 2 μm.
Integration of pHDGFP and stable expression of HDGFP in P. falciparum.
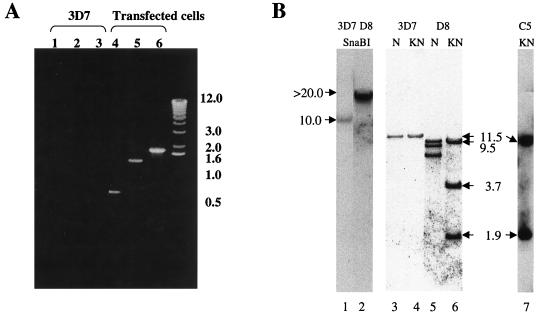
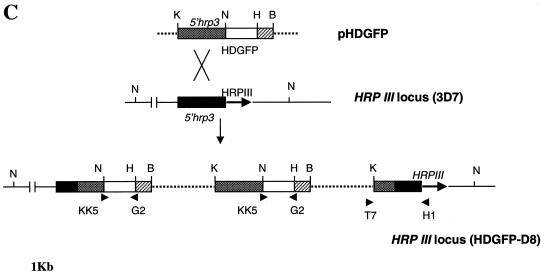
Previous studies suggested that integration in P. falciparum occurs by homologous recombination in both coding and noncoding regions (2, 16) and that the integrants can be selected ∼5 to 6 weeks after transfection. However, noncoding regions are not preferred, and there are no homologous coding regions available in pHDGFP. Thus, to increase the chance of chromosomal integration, we maintained transformed cultures continuously in a drug for 12 weeks. After this time, we investigated whether the plasmid integrated into the 5′hrp3 site. To do this, we designed a primer, H1, in the coding region of HRP III and used it in conjunction with a T7 promoter primer in PCRs. A fragment of the predicted size (1.9 kb) was amplified from the DNA of transfected (Fig. 2A, lane 6) but not nontransfected (lane 3) cells, suggesting that the plasmid did indeed integrate into the 5′hrp3 site.
FIG. 2.
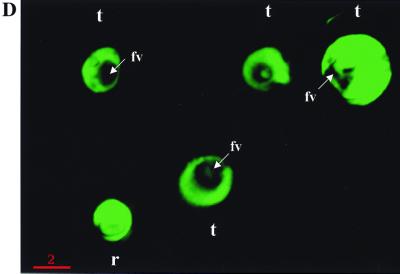
Integration of pHDGFP and its stable expression in P. falciparum. (A) PCR analysis of plasmid integration after transfection. Template genomic DNA from either the parent strain 3D7 (lanes 1 to 3) or transfected cells (lanes 4 to 6) was used to amplify (i) human DHFR by using primers KK5 and KK6 (lanes 1 and 4), (ii) HDGFP by using KK5 and G2 (5′-CTGCCTCGAGTTATAAATCTTCTTCAGATATTAATTTTTGTTC) (lanes 2 and 5), or (iii) the integrated fusion cassette by using the T7 promoter primer (5′-TAATACGACTCACTATAGGGAGA) and H1 (5′-GCGGATCCGTTATCTAACAAAAGTACGG; obtained from the coding region of HRP III) (lanes 3 and 6). Primer KK6 is indicated in Fig. 1A. The remaining primers are shown in panel C. (B) Southern analysis of parasite DNA from clones containing integrated or episomal pHDGFP. Genomic DNA from the parent strain 3D7 (lanes 1, 3, and 4) or a clone containing integrated pHDGFP (D8; lanes 2, 5, and 6) was digested with SnaBI (lanes 1 and 2) or NsiI (N; lanes 3 and 5) or double digested with KpnI-NsiI (KN; lanes 4 and 6) and probed with the full-length 5′hrp3 NsiI-KpnI fragment from pHDGFP (or a fragment of the gfp coding region; not shown). DNA from a clone containing episomal pHDGFP (C5) was double digested with KpnI-NsiI (lane 7) and probed as shown for lanes 1 to 6. Values to the left and right of panels A and B are in kilobases. (C) Physical map of the HRP III locus of clone P. falciparum D8. This was derived from Southern analysis of genomic DNA of clone D8 presented in panel B. (D) Green fluorescence detected in clone D8. Cells were imaged, and 0° projections of three-dimensional reconstructions of ring (r)- and trophozoite (t)-infected erythrocytes were obtained as described for Fig. 1B.
To obtain stable clones, the transformed cells were subjected to limiting dilution in the absence of drug (16). PCR analysis was used to identify potential integrants, and Southern analysis of one representative clone (designated HDGFP-D8, or D8) is shown in Fig. 2B. The blots were probed with the 1.9-kb 5′hrp3 fragment released by digesting pHDGFP with KpnI and NsiI. Genomic DNA digested with SnaBI released two fragments of 10 and >20 kb from the parent 3D7 and clone D8 respectively (Fig. 2B, lanes 1 and 2). The difference in the size of these fragments suggests disruption of the hrp3 locus in D8. NsiI or KpnI-NsiI digestion of parent 3D7 (lanes 3 and 4) results in an 11.5-kb fragment. However parallel digestions of clone 8 (lanes 5 and 6) released three fragments, indicating the presence of two copies of 5′hrp3 in addition to the resident gene. Furthermore, the double digest of D8 DNA gave fragments with sizes of 9.5, 3.7, and 1.9 kb. The 1.9-kb fragment corresponds to the size of the full-length probe, indicating that one copy of 5′hrp3 was not disrupted. The 3.7-kb fragment suggests disruption of the second copy. On the other hand, a single cell fluorescent line (HDGFP-C5) carrying episomal pHDGFP showed no disruption of the resident HRP III locus and released only a 1.9-kb hybridizable fragment corresponding to the 5′hrp3 region of the plasmid (which is the same as the probe; Fig. 2B, lane 7). The gfp probe hybridized to a single 4.8-kb fragment in clone D8 double digested with KpnI and NsiI (data not shown). The results are summarized in the schematic in Fig. 2C and indicate tandem insertion of two copies of pHDGFP into the HRP III locus. Single-copy insertions were not detected. It should be noted that previous studies by Wu et al. (16) reporting integration into the 5′hrp3 site also detected insertion of two copies of a DHFR-TS cassette (pDT.Dd2) at this locus. Although the precise reason for insertion of two copies is not clear, it might be linked to specific properties of this locus. The pHDGFP insertions were stably maintained in the absence of drug, all detected parasites contain green fluorescence, and there was uniform intensity of cytosolic expression during asexual development (Fig. 2D).
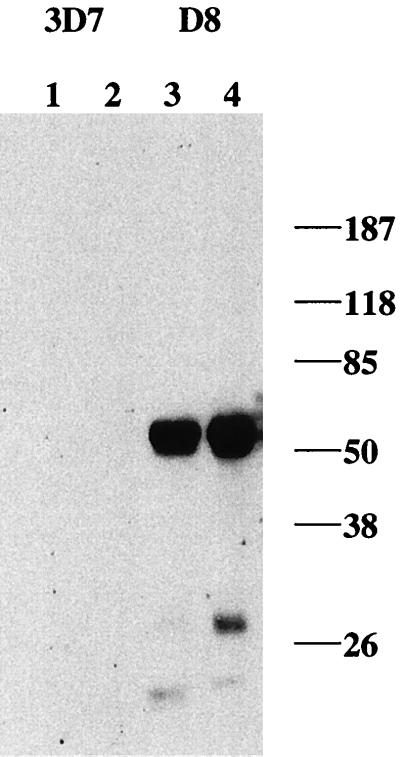
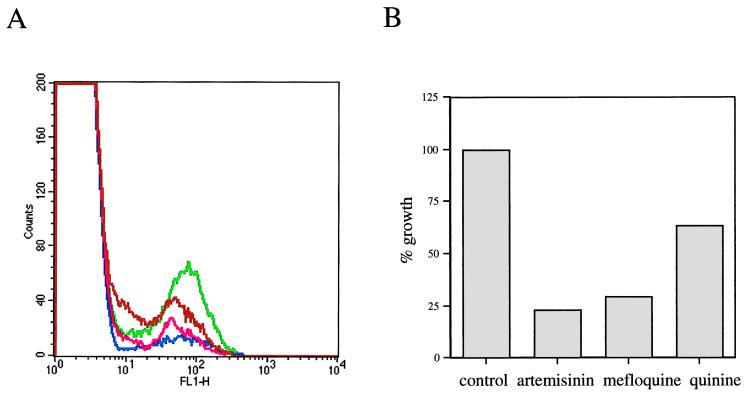
Stable, quantitative expression of HDGFP during asexual development was further analyzed by Western blotting and flow cytometry. Immunoblots probed with antibody to c-myc detected a predicted chimeric myc-tagged HDGFP protein (∼52 kDa) in saponin (0.01%)-lysed ring- and trophozoite-stage parasites isolated from the transformed D8 clone, but not the parent 3D7 parasites: larger amounts were seen at the trophozoite stage (Fig. 3). In addition, fluorescence-activated cell sorter (FACS) analysis indicated that fluorescence associated with D8 parasites was shifted by at least 1 log relative to that of nontransformed cells and could easily be detected by flow cytometry (Fig. 4). There was a clear decrease in the population of fluorescent cells in cultures treated with inhibitors, indicating that accumulation of fluorescence is a good index of parasite proliferation. The relative effects on growth in the FACS assay are the same as those found by measuring radioactive hypoxanthine (a conventional method of determining parasite growth).
FIG. 3.
Western blots of protein expression in clone D8. Cell lysates (30 μg of protein) of infected erythrocytes at the ring (lanes 1 and 3) and trophozoite (lanes 2 and 4) stages were probed with a commercially available mouse monoclonal antibody to c-myc (9E10) to detect the tagged chimeric HDGFP. Values to the right are in kilodaltons.
FIG. 4.
Fluorescence-based assay for malarial growth. (A) Cultures of P. falciparum clone D8 were subjected to FACS on a FACScalibur, and data were processed with CellQuest version 3.1 (Becton Dickinson). For growth assays, ring-infected cells were mock treated (green) or incubated with known inhibitors, such as 1 μM quinine (brown), 50 nM artemisinin (blue), and 50 nM mefloquine (pink), for 48 h and then subjected to FACS analysis. Fluorescence associated with the parent strain 3D7 was 1 log lower than that seen with D8 (data not shown). (B) Histogram showing the effect of inhibitors on growth of P. falciparum D8. Untreated or treated fluorescent parasites were FACS counted, and the numbers of fluorescent parasites of untreated cultures were taken to reflect 100% growth.
Concluding remarks.
Expression of a chimeric protein, HDGFP, either by stable integration into the chromosome or episomal maintenance, provides methotrexate resistance and green fluorescence in P. falciparum. This chimeric reporter is driven by the 5′hrp3 regulatory region, and it could be used as a promoter trap for blood-stage genes in P. falciparum. Given the sensitivity of flow cytometry, multiple rounds of negative and positive selection should be possible. Thus, analogous to bacterial systems (13), the reporter may enable the detection of virulence determinants that are expressed in vivo (in primates) but not in vitro. It may also be used as a trap for promoters expressed in the sexual stage, but not in asexual stages that are important in gametocyte differentiation. The minimal levels of detection are at ∼0.05% parasitemia, assuming 105 cells are scanned at a rate of 5,000 per s. This provides a live cell reporter with capacity for both high throughput and sensitivity. Additionally, stably transformed clones can be used in combinatorial assays to screen for new antimalarial agents as well as to define mutation frequencies and mechanisms of drug resistance.
Acknowledgments
We thank David Fidock and Thomas Wellems for the pHD22Y plasmid and members of the Haldar laboratory for careful reading of the text.
This work was supported by grants from the NIH (AI 26670, 39073) and a Burroughs Wellcome New Initiatives in Malaria Award to K.H.
REFERENCES
- 1.Cormack B P, Valdivia R H, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 2.Crabb B S, Cooke B M, Reeder J C, Waller R F, Caruana S R, Davern K M, Wickham M E, Brown G V, Coppel R L, Cowman A F. Targeted gene disruption shows that knobs enable malaria-infected red cells to cytoadhere under physiological shear stress. Cell. 1997;89:287–296. doi: 10.1016/s0092-8674(00)80207-x. [DOI] [PubMed] [Google Scholar]
- 3.Crabb B S, Cowman A F. Characterization of promoters and stable transfection by homologous and nonhomologous recombination in Plasmodium falciparum. Proc Natl Acad Sci USA. 1996;93:7289–7294. doi: 10.1073/pnas.93.14.7289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crabb B S, Triglia T, Waterkeyn J G, Cowman A F. Stable transgene expression in Plasmodium falciparum. Mol Biochem Parasitol. 1997;90:131–144. doi: 10.1016/s0166-6851(97)00143-6. [DOI] [PubMed] [Google Scholar]
- 5.de Koning-Ward T F, Thomas A W, Waters A P, Janse C J. Stable expression of green fluorescent protein in blood and mosquito stages of Plasmodium berghei. Mol Biochem Parasitol. 1998;97:247–252. doi: 10.1016/s0166-6851(98)00134-0. [DOI] [PubMed] [Google Scholar]
- 6.Fidock D A, Nomura T, Wellems T E. Cycloguanil and its parent compound proguanil demonstrate distinct activities against Plasmodium falciparum malaria parasites transformed with human dihydrofolate reductase. Mol Pharmacol. 1998;54:1140–1147. doi: 10.1124/mol.54.6.1140. [DOI] [PubMed] [Google Scholar]
- 7.Fidock D A, Wellems T E. Transformation with human dihydrofolate reductase renders malaria parasites insensitive to WR99210 but does not affect the intrinsic activity of proguanil. Proc Natl Acad Sci USA. 1997;94:10931–10936. doi: 10.1073/pnas.94.20.10931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goonewardene R, Daily J, Kaslow D, Sullivan T J, Duffy P, Carter R, Mendis K, Wirth D. Transfection of the malaria parasite and expression of firefly luciferase. Proc Natl Acad Sci USA. 1993;90:5234–5236. doi: 10.1073/pnas.90.11.5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mamoun C B, Gluzman I Y, Goyard S, Beverley S M, Goldberg D E. A set of independent selectable markers for transfection of the human malaria parasite Plasmodium falciparum. Proc Natl Acad Sci USA. 1999;96:8716–8720. doi: 10.1073/pnas.96.15.8716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Menard R, Sultan A A, Cortes C, Altszuler R, van Dijk M R, Janse C J, Waters A P, Nussenzweig R S, Nussenzweig V. Circumsporozoite protein is required for development of malaria sporozoites in mosquitoes. Nature (London) 1997;385:336–340. doi: 10.1038/385336a0. [DOI] [PubMed] [Google Scholar]
- 11.Sultan A A, Thathy V, Frevert U, Robson K J, Crisanti A, Nussenzweig V, Nussenzweig R S, Menard R. TRAP is necessary for gliding motility and infectivity of plasmodium sporozoites. Cell. 1997;90:511–522. doi: 10.1016/s0092-8674(00)80511-5. [DOI] [PubMed] [Google Scholar]
- 12.Sultan A A, Thathy V, Nussenzweig V, Menard R. Green fluorescent protein as a marker in Plasmodium berghei transformation. Infect Immun. 1999;67:2602–2606. doi: 10.1128/iai.67.5.2602-2606.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valdivia R H, Falkow S. Bacterial genetics by flow cytometry: rapid isolation of Salmonella typhimurium acid-inducible promoters by differential fluorescence induction. Mol Microbiol. 1996;22:367–378. doi: 10.1046/j.1365-2958.1996.00120.x. [DOI] [PubMed] [Google Scholar]
- 14.van Dijk M R, Waters A P, Janse C J. Stable transfection of malaria parasite blood stages. Science. 1995;268:1358–1362. doi: 10.1126/science.7761856. [DOI] [PubMed] [Google Scholar]
- 15.VanWye J D, Haldar K. Expression of green fluorescent protein in Plasmodium falciparum. Mol Biochem Parasitol. 1997;87:225–229. doi: 10.1016/s0166-6851(97)00059-5. [DOI] [PubMed] [Google Scholar]
- 16.Wu Y, Kirkman L A, Wellems T E. Transformation of Plasmodium falciparum malaria parasites by homologous integration of plasmids that confer resistance to pyrimethamine. Proc Natl Acad Sci USA. 1996;93:1130–1134. doi: 10.1073/pnas.93.3.1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu Y, Sifri C D, Lei H H, Su X Z, Wellems T E. Transfection of Plasmodium falciparum within human red blood cells. Proc Natl Acad Sci USA. 1995;92:973–977. doi: 10.1073/pnas.92.4.973. [DOI] [PMC free article] [PubMed] [Google Scholar]