Abstract
In this perspective, we provide an overview of a recently established National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) initiative, the Human Pancreas Analysis Program - Type 2 Diabetes (HPAP-T2D). This program is designed to define the molecular pathogenesis of islet dysfunction by studying human pancreatic tissue samples from organ donors with T2D. HPAP-T2D generates detailed datasets of physiological, histological, transcriptomic, epigenomic, and genomic information. Importantly, all data collected, generated, and analyzed by HPAP-T2D are made immediately and freely available through a centralized database, PANC-DB, thus providing a comprehensive data resource for the diabetes research community.
Introduction
Type 2 diabetes (T2D) is a clinically heterogeneous disease that is increasing in frequency worldwide and is characterized by deficient β-cell insulin secretion, insulin resistance in peripheral tissues, and excessive glucagon secretion and action (Kahn, 2003). These physiological derangements lead to persistent hyperglycemia and a series of associated comorbidities (e.g., retinopathy, neuropathy, and vascular disease) that increase mortality in individuals with T2D. The exact etiology of T2D is unknown but there are strong environmental (e.g., diet, physical activity) and genetic contributors (Erion and Corkey, 2018; Halban et al., 2014). The correlation between excess adiposity/insulin resistance with T2D is clear, with the increasing prevalence of obesity accompanied by a concomitant surge in T2D (Schnurr et al., 2020). However, most individuals with obesity do not develop T2D, indicating that the islet’s compensatory response to insulin resistance is a critical determinant of whether hyperglycemia develops. While new therapeutic approaches exist for T2D, knowledge gaps regarding the molecular defects responsible for islet dysfunction and insulin resistance limit targeted interventions and precision medicine approaches.
In 2019, the NIDDK established a new initiative, the Human Pancreas Analysis Program - Type 2 Diabetes (HPAP-T2D). HPAP-T2D is designed to define the molecular pathogenesis of islet dysfunction by procuring human pancreas and curating de-identified clinical information from donors with T2D, generating detailed datasets of physiological, morphological, transcriptomic, epigenomic, and genomic information, and making the data immediately available to the scientific community. This perspective provides an overview of the rationale and endeavors of the HPAP-T2D program—we note that restrictions on length and scope of this manuscript prevents a comprehensive review of T2D.
Rationale for HPAP-T2D
T2D, a heterogeneous disease with a lack of clearly defined criteria, is often a diagnosis of exclusion. Adding to the challenge of understanding the mechanistic pathogenesis of T2D is the difficulty of molecularly profiling the cells and tissues of interest, especially the pancreatic islet. Studying pancreatic samples from living individuals is challenging and the propensity of the pancreas to auto-digest makes organs obtained at autopsy unsuitable for interrogation by many experimental techniques. In addition, studies with cell lines or animal models do not recapitulate all features of islet dysfunction in human T2D. Thus, the key motivations underlying the creation of HPAP-T2D were to (1) procure and comprehensively study the human pancreas and pancreatic islet in T2D, and to (2) integrate data with the clinical history from de-identified donors. A critical aspect of this effort involves the processing of pancreata from deceased organ donors in such a way that both viable and functional islets are isolated, while carefully fixed and frozen tissue samples are procured from the same pancreas. In the past, experimental approaches were directed at analysis of either isolated islets or pancreatic tissue, but not both simultaneously from the same organ. Our approach allows for studies of human islets such as hormone secretion and single-cell profiling, while also enabling the analysis of pancreatic tissue to define cellular neighborhoods (endocrine cells, immune cells, etc.), vascularization, innervation, and known T2D features such as amyloid deposits. The success of a related program, the Human Pancreas Analysis Program - Type 1 Diabetes (HPAP-T1D) (Kaestner et al., 2019), provided a valuable template for studying the human pancreas from donors with T2D.
Overview of HPAP-T2D
HPAP-T2D unites a complementary team of surgeons and clinicians with experimental and computational scientists into a multi-institutional effort led by investigators at the University of Pennsylvania (Penn), Vanderbilt University, the University of Florida, Stanford University, the University of Alberta, and the Icahn School of Medicine at Mt. Sinai. HPAP-T2D is part of the Human Pancreas Analysis Consortium (HPAC), one of five consortia in the Human Islet Research Network (https://www.hirnetwork.org). The core missions of HPAP-T2D are (1) to perform deep phenotyping of the human endocrine pancreas from donors without diabetes or donors with T2D, and (2) to generate, analyze, and deliver high-value datasets to the diabetes research community (Figure 1).
Figure 1.
HPAP Workflow
The foundation of this program is the identification of deceased organ donors with T2D, followed by skilled processing of the pancreas to isolate viable and functional islets, while carefully fixing tissue samples in ways that allow for subsequent multimodal analyses. An alternative approach to the HPAP-T2D efforts is the analysis of small pancreatic tissue fragments obtained from individuals undergoing surgical resection of the pancreas to remove pancreatic tumors (Wigger et al., 2021). This approach allows for assessment of glucose tolerance prior to surgery. Considerations related to this approach and the HPAP-T2D approach of studying both islets and tissue from the entire pancreas have been discussed recently (Gloyn et al., 2022; Gloyn and Powers, 2021).
For donor identification, HPAP-T2D leverages the longstanding expertise of the Network for Pancreatic Organ donors with Diabetes program (nPOD; https://www.jdrfnpod.org/), connecting HPAP-T2D with every organ procurement organization in the United States to identify donors with T2D, emphasizing diabetes of relatively short duration (< 10 years) and/or treatment with agents other than insulin. Currently, we exclude donors with T2D who are treated with insulin. Our rationale is two-fold: (1) Within the category of individuals receiving insulin treatment, the donor phenotype is quite broad: some individuals with obesity are very insulin resistant, but still produce substantial insulin, while other non-obese individuals are very insulin deficient. We have reasoned that including the range of donors treated with insulin will create unacceptable donor heterogeneity, and (2) in our experience thus far, we have found that if a patient is taking insulin, our procurement team is less likely to obtain a sufficient number of islets for the broad number of assays that HPAP performs (detailed below). Age (35–65 years) and body mass index (25–40) are also considered as inclusion criteria. To avoid potentially confounding co-morbidities that might influence the pancreatic islet, donors with a history of pancreatitis, cirrhosis, very severe fatty liver disease, excessive alcohol consumption, renal failure, very severe cardiovascular disease, or prolonged hospitalization are excluded (Table 1). Since the program was initiated in 2016, HPAP has processed pancreas samples from 121 organ donors: 68 donors with no history of diabetes (of which 18 were positive for islet autoantibodies), 26 donors with Type 1 diabetes, and 27 donors with Type 2 diabetes. HPAP aims to process pancreata from 32–40 donors per year, divided as evenly as possible between the T1D and T2D programs. Importantly, access to a nationwide network is enabling HPAP-T2D to collect data from diverse race and ethnicity groups. Of the donors processed by HPAP-T2D to date, 16% were reported as African American, 22% as Hispanic, and 62% as European ancestry, mirroring the current composition of the U.S. population.
Table 1.
HPAP Inclusion and Exclusion Criteria
| Donors without Diabetes | Age 35–65 years |
| BMI 25–40 | |
| A1C < 5.7 | |
| Donors with Type 2 Diabetes | Age 35–65 years |
| BMI 25–40 | |
| A1C ≥ 6.5 (could be < 6.5 with treatment) | |
| Specific Exclusion Criteria | Active COVID infection |
| Severe renal disease; on chronic dialysis prior to admission (hemo- or peritoneal) | |
| Heavy ETOH use (daily) – at discretion of HPAP team | |
| Visual inspection of pancreas (severely fatty, intraoperative damage) | |
| History of pancreatitis or cirrhosis | |
| Admission to cross clamp > 7 days | |
| Ventilator time > 7 days | |
| Hemodynamic instability treated with multiple pressors – at discretion of HPAP team | |
| Acute respiratory distress syndrome (ARDS) | |
| Positive serologies (HCV, HIV, hepatitis B) | |
| Amylase or lipase > 2X upper limits of normal | |
| DCD donor > 45 minutes | |
| Donor “down time” > 30 minutes | |
| Cold ischemia time > 20 hours |
HPAP-T2D has established a centralized pancreas processing workflow such that all donor pancreata are processed at Penn, ensuring consistency and reproducibility. Since the donors for the HPAP-T2D program are obtained from sites throughout the United States, this requires coordinated efforts for pancreas removal, usually by the same surgical team that is procuring other organs (e.g., heart, lung, liver, kidney) from the donor, and pancreas shipment to Penn. The time from pancreas removal to initiation of pancreas processing (cold ischemia time) is usually less than 20 hours to maximize the quality of material available for downstream analysis. In addition to studying islets and pancreatic tissue from the same donor, another distinctive feature of HPAP-T2D compared to other tissue collection programs is that tissue is not distributed outside HPAP-T2D, as the biological samples are completely used in the extensive studies performed by the HPAP-T2D team.
Mapping the biology of the pancreas through multidimensional phenotyping
Foundational to HPAP-T2D efforts is the integration of genomics, functional and secretory phenotyping, as well as tissue characterization to define the molecular signatures of T2D in islet cells, islets, and pancreas. Here, we briefly describe these experimental approaches; detailed standard protocols are available on the HPAP-T2D website (https://hpap.pmacs.upenn.edu; see Supplemental Table 1).
Genomics --
HPAP-T2D performs transcriptomic profiling and epigenomic analysis of the human endocrine pancreas in order to gain a deep understanding of the molecular changes that occur across the spectrum of T2D. For all donor pancreata, islets are sorted to purify α- and β-cells for bulk RNA-sequencing. By pooling cells, 15,000–20,000 distinct transcripts are detected per cell type, making this a sensitive approach to identify gene expression differences between donors and disease states. This method is complemented by single-cell RNA sequencing (scRNA-seq) to obtain transcriptional profiles of individual cells, an approach best suited to detect and profile rare populations within the heterogenous islet sample including delta, epsilon, and ductal cells (Fasolino et al., 2022; Segerstolpe et al., 2016). For example, scRNA-seq analysis previously revealed that α- and β-cells from donors with T2D have an immature transcriptional signature similar to that of donors from childhood, suggesting a dedifferentiation feature of the disease (Avrahami et al., 2020; Wang et al., 2016b).
The significant environmental contribution to T2D risk emphasizes the importance of investigating epigenetic mechanisms as a readout of diet, exercise, the microbiome, and other environmental exposures. Indeed, a number of diet-derived metabolites act as co-factors for epigenetic enzymes and thus influence downstream gene expression patterns, with evidence mounting that metabolism can instruct cell fate decisions (Dai et al., 2020). HPAP-T2D assays chromatin accessibility variation between non-diabetic pancreata and diseased organs using the Assay for Transposase-Accessible Chromatin using Sequencing (ATAC-Seq) (Ackermann et al., 2016).
This technique interrogates nucleosome positioning patterns to map accessible promoters and enhancers, as well as to identify putative transcription factor binding sites. Single-cell ATAC-seq (scATAC-seq) adapts this protocol to separate individual nuclei for analysis, allowing for the resolution of gene accessibility patterns within specific pancreatic cell types such as α-cells, β-cells, acinar, immune cells, and others. The integration of regulatory predictions based on scATAC-seq data and clustering analysis using scRNA-seq data in islets and immune cells creates a high-resolution map of the regulatory landscape that defines the inflammatory signature of T2D.
Of all epigenomic modifications, methylation of cytosine residues is particularly informative as DNA methylation is relatively stable in human α- and β-cells, but appears to be dysregulated in T2D (Avrahami and Kaestner, 2019). Multiple studies have addressed changes in DNA methylation in whole T2D islets (Ling et al., 2008; Volkmar et al., 2012). For example, previous work demonstrated that T2D donor islets feature increased methylation and decreased expression of the imprinted MEG3 microRNA cluster, correlating with β-cell dysfunction (Kameswaran et al., 2014), and comprehensive mapping by whole genome bisulfite sequencing (WGBS) of whole islets identified differentially methylated regions of genes with known islet function (Volkov et al., 2017). However, given that human islets vary greatly in their cellular composition from donor to donor (Brissova et al., 2005), the whole islet methylome approach is of limited analytical power. Therefore, HPAP-T2D performs WGBS on sorted α- and β-cell fractions from every donor, allowing us to connect distinct clinical phenotypes with DNA methylation patterns of the most relevant islet cell type such that small differences can be discerned.
Islet and Islet Cell Function --
Endocrine cells integrate fluctuations in energy balance for stimulus secretion coupling; thus, islet phenotyping requires a comprehensive examination of both the key regulatory steps that govern hormone secretion (glycolysis, mitochondrial respiration) and the hormonal output from α-, β-, and δ-cells. HPAP-T2D utilizes two complementary islet perifusion approaches to assess stimulated insulin and glucagon secretion in isolated islets. At Penn, islet perifusion analysis simultaneously measures fuel-stimulated insulin and glucagon secretion and oxygen consumption, with the response of intracellular islet calcium to fuel stimulation also measured by dual-wavelength fluorescence microscopy. Because the percentage of endocrine cell types varies between both individuals and within islets within a single donor (Brissova et al., 2005), HPAP-T2D uses cytometry by time-of-flight (CyTOF) to precisely define islet composition and enable accurate interpretation of hormone secretion data (Wang et al., 2016a). At Vanderbilt, HPAP-T2D islets are perifused and hormone secretion assessed with the protocol used by the Human Islet Phenotyping Program (HIPP) of the Integrated Islet Distribution Program (IIDP; https://iidp.coh.org/) which has evaluated more than 300 human islet preparations over the past five years. Thus, the islet hormone secretory pattern by islets in the HPAP-T2D program can be compared to the large number of islet preparations distributed by IIDP. This also provides an opportunity for the detailed molecular data of HPAP-T2D to be connected to HIPP-studied islets.
To link islet cell functional variation with transcriptomic heterogeneity, HPAP-T2D utilizes islet Patch-seq, a technique adapted from studies in neuronal cells that integrates single-cell RNA sequencing with the electrophysiological assessment of ion channel activities and single-cell exocytosis as proxy for hormone release. This allows for simultaneous measurement of cell membrane properties and transcriptome profiles in single islet cells, and has delineated transcriptional profiles linked to β-cell (Camunas-Soler et al., 2020) and α-cell (Dai et al., 2022) dysfunction in T2D. Using this approach, HPAP-T2D seeks to establish the relationships between electrical properties and hormone secretory profiles of endocrine cells, and to integrate this with single-cell epigenomic analyses. Taken together, these approaches will reveal compensatory mechanisms adopted by islet cells in response to insulin resistance, and pathological changes that precipitate islet failure in T2D.
Pancreatic Tissue Analysis --
To understand the molecular, structural cellular, and histological changes in T2D at the tissue level, HPAP-T2D is profiling tissue samples from the head, body, and tail regions of the pancreas by conventional immunohistochemistry (IHC) and two high-resolution multiplexed imaging technologies. First, HPAP-T2D applies imaging mass cytometry (IMC), a highly multiplexed imaging method using antibodies conjugated to heavy metal isotopes combined with mass spectrometry. This technique significantly reduces background signals and spectral overlap seen with traditional fluorescence methods (Giesen et al., 2014). Tissue samples can be probed with up to forty antibodies at one time, permitting visualization of multiple cell types (immune, islet, endocrine, stromal, etc.) simultaneously in the native tissue microenvironment. Building on efforts of the HPAP-T1D program that had previously developed a panel of 33 disease-relevant antibodies to study interactions between endocrine cells and the immune system in pancreata from donors diagnosed with T1D (Wang et al., 2019), HPAP-T2D recently analyzed pancreata from age-matched donors without diabetes and donors with T2D, revealing marked differences in cell type composition and immune infiltration between the two groups (Wu et al., 2021). The list of optimized antibodies used in IMC experiments can be found in the “Workflow and Protocols” section of PANC-DB (Supplemental Table 1.)
HPAP-T2D is also employing Co-Detection by Indexing (CODEX), a multiplexed staining technique based on DNA-barcoded antibodies (Goltsev et al., 2018). CODEX can provide a comprehensive map of spatiotemporal changes that occur within pancreatic islets, exocrine tissue, nerves, and vasculature in T2D as evidenced by vascular changes in the microenvironment in short duration T2D (Walker et al., 2021).
Genetics --
30x whole-genome sequencing (WGS) with the NovaSeq 6000 platform and comprehensive array-genotyping are performed on every HPAP-T2D donor to characterize genetic variation (e.g., single nucleotide variants or short insertions and deletions) and calculate overall and partitioned genetic risk scores for T2D (Mahajan et al., 2018; Vujkovic et al., 2020). The availability of paired genetic and functional genomic datasets is valuable in a number of ways. When paired with the appropriate functional genomic readout generated by HPAP-T2D (e.g., RNA-seq, ATAC-seq, and WGBS), variant call sets enable quantitative trait locus (QTL) analysis to identify genetic variation associated with a change in gene expression (eQTLs), proportion of alternatively spliced isoforms (sQTLs), degree of DNA methylation (meQTLs), and accessibility of chromatin (acQTLs), facilitating a unique ability to map gene regulatory mechanisms. Moreover, the HPAP-T2D cohort collected is not only multi-ancestry, but also characterized for a collection of physiological trait and disease endpoints that can be further interrogated for QTL based discovery efforts. For example, by integrating genomic outputs, HPAP-T2D has the capability to (1) correlate marks of accessible chromatin and transcription to identify cis-regulatory elements, wherein changes in chromatin state may alter transcription of a cognate gene in a given cell population, (2) identify loci where differences in accessible chromatin correlate with changes in DNA methylation and downstream transcriptional state, and (3) map causal interaction between regions of accessible chromatin-mediated through acQTL genetic variation. Because HPAP performs many diverse and complementary assays on every pancreas, and because the number of donors analyzed by HPAP-T2D is relatively modest (e.g., ∼20 per year for T2D), we are more limited in sample size numbers than other large consortia. As a result, this approach is less powered for discovery of QTL effects for low-frequency or rare genetic variants. That said, the catalog of diverse types of QTLs in a common set of individuals is a key strength to the effort. Moreover, cell-type specificity to common variant QTLs will allow discoveries that will augment other programmatic QTL discovery efforts such as GTEx (https://gtexportal.org/).
Importantly, the assays performed by HPAP-T2D are complemented by related open-access human islet resources throughout the world (Supplemental Table 1). The Translational human pancreatic Islet Genotype tissue-Expression Resource (TIGER; http://tiger.bsc.es) provides over 500 comprehensive genomic datasets from donors without diabetes and donors with T2D, including eQTL meta-analysis and allele-specific expression data (Alonso et al., 2021). In addition, the Islet Regulome Browser (http://www.isletregulome.com) is a tool that integrates data from different labs to enable fast and user-friendly exploration of epigenomic and transcriptomic data from human islets (Mularoni et al., 2017). A more comprehensive discussion and summary of existing human islet analysis programs was recently published (Gloyn et al., 2022). Taken together, the rich availability of genomic, genetic, and functional data from various research initiatives will greatly advance efforts to understand the islet abnormalities in T2D and identify therapeutic targets for T2D. Future efforts that integrate data obtained from the different programs will increase the impact of these studies even further.
HPAP-T2D data analysis and access for the scientific community
Making comprehensive and high-quality datasets immediately accessible to the scientific community is central to the mission of HPAP-T2D. HPAP-T2D is also dedicated to open access sharing of all protocols and computational tools with the broader scientific community. To these ends, all data collected and analyzed by HPAP-T2D are made immediately and freely available through a centralized database, PANC-DB (https://hpap.pmacs.upenn.edu). The goal of PANC-DB is to provide a platform that is user-friendly and able to meet the diverse needs of its users. Clinical information, histology, genomic, and functional data are deposited into PANC-DB for web-based visualization, or files can be downloaded directly for processing with the user’s software of choice. PANC-DB provides both a “donor-centric” browsing mode, which allows users to select a specific donor to analyze, or an “assay-centric” browsing mode, which allows users to simultaneously download all data from one assay (e.g., all 27 scRNA-seq datasets from donors with T2D can be downloaded at once). In addition, all standard operating procedures, experimental protocols, and technical resources are available on PANC-DB and through the website protocols.io (Supplemental Table 1). PANC-DB also provides answers to frequently asked questions and a “Getting Started” tutorial with navigation tips and more.
HPAP-T2D’s goal is for the data and analysis tools distributed through PANC-DB to enable researchers to autonomously address outstanding questions from their own research and generate and test new hypotheses. To date, HPAP has distributed over 50,000 freely accessible files, comprising 6.5 terabytes of data, with PANC-DB hosting 530 registered users who have cumulatively performed over 2,000 downloads from the database. While not all assays can be performed on every donor due to biological and technical reasons, the “data status” page on PANC-DB indicates data that are available, in progress (in the sequencing queue, being analyzed, etc.) or data that will not be available (for example, if insufficient islets were obtained). This provides transparency for the user about the progress of HPAP-T2D. For example, if a user wonders how many scRNA-seq datasets are available from donors with T2D, he or she can filter on “T2D” and see a complete picture of the data status for all donors. Because of IRB and patient privacy considerations, the whole genome sequencing data and variant call sets are not available on PANC-DB, but are instead deposited in the Database of Genotypes and Phenotypes (dbGaP; https://www.ncbi.nlm.nih.gov/gap/).
We have recently expanded the role of PANC-DB by introducing the first interactive data analysis portal for pancreatic single-cell RNA-seq data. HPAP has already generated scRNA-seq data from over 270,000 cells from more than 70 donors. In order to enable researchers to easily visualize gene expression patterns, either for a selected subset of cells or the entire dataset, we have adopted CellxGene, a platform developed for this purpose by the Chan Zuckerberg initiative. CellxGene allows researchers to annotate cell types as they define them in the HPAP dataset and provides multiple analysis tools, including the filtering of cells by disease status or any other variable, ‘on the fly’ selection of cells for analysis, and differential gene expression analysis between any two clusters of cells selected by the user.
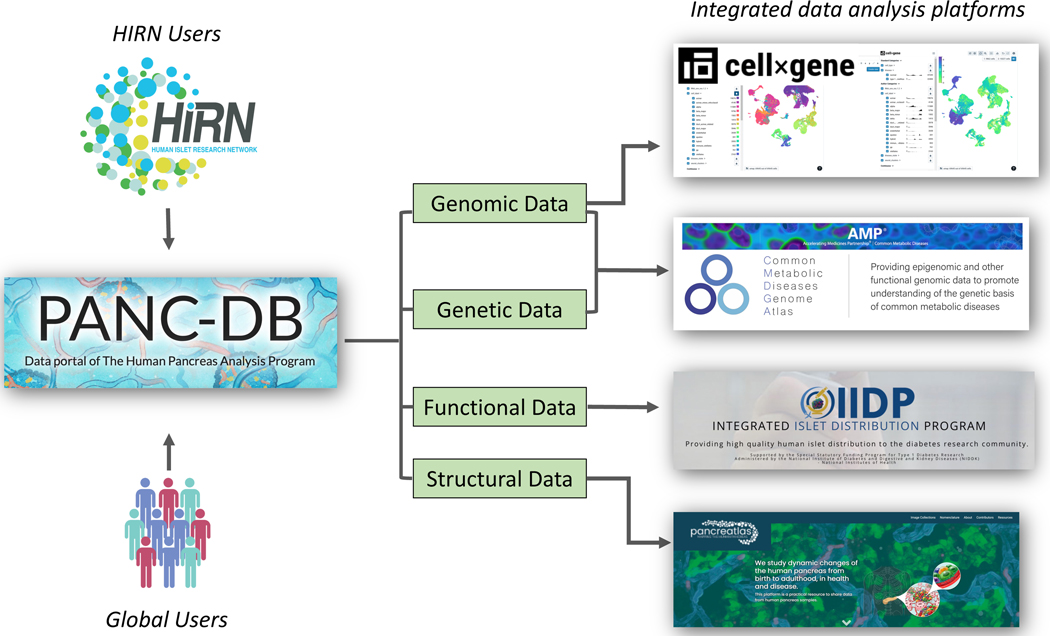
A major focus of PANC-DB is harmonizing and integrating with other related resources (Figure 2). For example, PANC-DB is linked to Pancreatlas™, a multifaceted image viewing platform that allows users to explore immunofluorescence, histology, imaging mass cytometry, and 3-D renderings from the human pancreas (https://www.pancreatlas.org) (Saunders et al., 2020). Pathviewer, the interactive image viewer on Pancreatlas, supports > 40 color channels. In addition, Pancreatlas can accommodate diverse image collections, and features have been developed to enhance data sharing via PANC-DB. Thus, the user can easily turn individual channels on or off, as well as select biologically relevant channel groupings. PANC-DB is also working to integrate islet perifusion results with the interactive HIPP/IIDP database. Transcriptomic data curated in PANC-DB are further linked to the Common Metabolic Diseases Genome Atlas (https://cmdga.org), which provides users with additional visualization and analysis modules. Taken together, PANC-DB serves as a wide-reaching, comprehensive resource for the diabetes research community and a model for open-access data sharing.
Figure 2.
Programs connected to HPAP/PANC-DB
Concluding remarks regarding HPAP-T2D
The availability of transplant-quality human pancreata, made possible through the gift of organ donation, is allowing HPAP-T2D to greatly expand the knowledge about the pancreatic islet in T2D. As discussed below, HPAP-T2D is evolving to improve and have an even greater impact. As new metabolic phenotyping and genomic approaches emerge, HPAP-T2D will continue to incorporate new analytical approaches. For example, HPAP-T2D is working to (1) optimize new base-resolution sequencing methods for 5-hydroxymethylcytosine to complement DNA methylation analyses and identify regulatory elements, and (2) apply seqFISH, a platform that will enable single-cell resolution gene expression analyses of human tissue to analyze a large number of genes simultaneously while retaining location information. In support of these experimental advances, PANC-DB is building state-of-the-art bioinformatic tools for data analysis and integration to further enhance capabilities for data exploration, and visual analytics to facilitate the identification of data patterns. Our goal is to generate and share a compendium of high-quality datasets to enable molecular phenotyping of the human pancreas at greater and greater resolution, thus facilitating discoveries that lead to new ways to prevent and/or treat T2D.
Future efforts and unresolved questions related to the islet dysfunction in T2D
By applying new technologies to study human tissue and cells, HPAP-T2D and other initiatives are poised to dramatically improve our understanding of islet physiology and pathophysiology. For example, one outstanding question is whether T2D results in fewer numbers of β-cells, impaired function of existing β-cells, or a combination of both. Through our analyses, HPAP is poised to shed light on this issue, which could inform clinical efforts to either increase β-cell numbers and/or function. Looking to the future, several important conceptual, experimental, clinical, and organizational questions should be considered.
Supplementary Material
Conceptual:
The recognized clinical heterogeneity of T2D has likely correlates on the molecular level (Udler et al., 2018). Currently, we have little understanding of the molecular underpinnings of islet dysfunction in the (at least 5) clinical subgroups of T2D (Dennis et al., 2019; Philipson, 2020).
Patients progress from pre-diabetes to short- and then long-duration T2D, which are responsive to dietary interventions, oral medications or only insulin therapy, respectively. What is the parallel progression of islet dysfunction on the molecular and epigenetic levels?
How do therapies that are successful in reversing T2D (weight loss, low carbohydrate diet, bariatric surgery) impact islet physiology and pathophysiology?
Experimental and Clinical:
What is the role of islet architecture and the microenvironment in islet dysfunction in T2D?
How do the epigenetic and transcriptomic profiles impact islet biology in T2D?
Can advances in proteomics, lipidomics, and metabolomics increase our understanding of islet failure in T2D?
How do we leverage epidemiological studies and GWAS results to precisely define molecular abnormalities in islet dysfunction?
How can the hypotheses generated through HPAP data be tested in model systems (in vitro and in vivo) or in human patients?
How can results uncovered by HPAP be translated to inform clinical assessments and/or interventions?
Organizational:
How do we integrate information from studies of isolated islets with those from pancreatic fragments and pancreatic slices?
How is data from multiple experimental modalities co-registered, integrated, and shared?
How are the multi-disciplinary teams needed to conduct these types of studies supported and organized and individual as well as team efforts recognized?
Acknowledgments:
We thank the donors and their families for their gift to the research community. We are grateful to our colleagues at nPOD and the University of Florida for their role in organ procurement. We note that due to space limitations, we were unable to cite many important contributions to the field by many outstanding investigators. This manuscript describes the work of our colleagues in the HPAP-T2D program, and we thank them for their efforts. The members of the HPAP-T2D team are listed in Supplemental Table 2. This work was supported by the Human Islet Research Network (RRID:SCR_014393), the Human Pancreas Analysis Program (RRID:SCR_016202), DK112217, DK123716, DK112232, DK123594, DK20593 (Vanderbilt Diabetes Research and Training Center), and DK019525 (PENN Diabetes Research Center).
Footnotes
Using a collaborative, multi-institutional approach, the Human Pancreas Analysis Program for Type 2 Diabetes (HPAP-T2D) studies the molecular pathogenesis of islet dysfunction in type 2 diabetes. All data generated by the program are made immediately and freely accessible, thus providing a valuable resource for the diabetes research community.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ackermann AM, Wang Z, Schug J, Naji A, and Kaestner KH. (2016). Integration of ATAC-seq and RNA-seq identifies human alpha cell and beta cell signature genes. Mol Metab 5, 233–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso L, Piron A, Moran I, Guindo-Martinez M, Bonas-Guarch S, Atla G, Miguel-Escalada I, Royo R, Puiggros M, Garcia-Hurtado X, et al. (2021). TIGER: The gene expression regulatory variation landscape of human pancreatic islets. Cell Rep 37, 109807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avrahami D, and Kaestner KH. (2019). The dynamic methylome of islets in health and disease. Mol Metab 27S, S25–S32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avrahami D, Wang YJ, Schug J, Feleke E, Gao L, Liu C, Consortium H, Naji A, Glaser B, and Kaestner KH. (2020). Single-cell transcriptomics of human islet ontogeny defines the molecular basis of beta-cell dedifferentiation in T2D. Mol Metab 42, 101057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brissova M, Fowler MJ, Nicholson WE, Chu A, Hirshberg B, Harlan DM, and Powers AC. (2005). Assessment of human pancreatic islet architecture and composition by laser scanning confocal microscopy. J Histochem Cytochem 53, 1087–1097. [DOI] [PubMed] [Google Scholar]
- Camunas-Soler J, Dai XQ, Hang Y, Bautista A, Lyon J, Suzuki K, Kim SK, Quake SR, and MacDonald PE. (2020). Patch-Seq Links Single-Cell Transcriptomes to Human Islet Dysfunction in Diabetes. Cell Metab 31, 1017–1031 e1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai XQ., Camunas-Soler J., Briant LJB., Dos Santos T., Spigelman AF., Walker EM., Arrojo EDR., Bautista A., Jones RC., Avrahami D., et al. (2022). Heterogenous impairment of alpha cell function in type 2 diabetes is linked to cell maturation state. Cell Metab 34, 256–268 e255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Z, Ramesh V, and Locasale JW. (2020). The evolving metabolic landscape of chromatin biology and epigenetics. Nat Rev Genet 21, 737–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis JM, Shields BM, Henley WE, Jones AG, and Hattersley AT. (2019). Disease progression and treatment response in data-driven subgroups of type 2 diabetes compared with models based on simple clinical features: an analysis using clinical trial data. The Lancet Diabetes & Endocrinology 7, 442–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erion K, and Corkey BE. (2018). beta-Cell Failure or beta-Cell Abuse? Front Endocrinol (Lausanne) 9, 532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasolino M, Schwartz GW, Patil AR, Mongia A, Golson ML, Wang YJ, Morgan A, Liu C, Schug J, Liu J, et al. (2022). Single-cell multi-omics analysis of human pancreatic islets reveals novel cellular states in type 1 diabetes. Nat Metab 4, 284–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giesen C, Wang HAO, Schapiro D, Zivanovic N, Jacobs A, Hattendorf B, Schüffler PJ, Grolimund D, Buhmann JM, Brandt S, et al. (2014). Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nature Methods 11, 417–422. [DOI] [PubMed] [Google Scholar]
- Gloyn AL, Mark I, Marchetti P, Powers AC, Rorsman P, Sander M, and Solimena M. (2022). Every Islet Matters: Improving the Impact of Human Islet Research. Nature Metabolism In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloyn AL, and Powers AC. (2021). There is more than one way to reach type 2 diabetes. Nat Metab 3, 894–895. [DOI] [PubMed] [Google Scholar]
- Goltsev Y, Samusik N, Kennedy-Darling J, Bhate S, Hale M, Vazquez G, Black S, and Nolan GP. (2018). Deep Profiling of Mouse Splenic Architecture with CODEX Multiplexed Imaging. Cell 174, 968–981 e915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halban PA, Polonsky KS, Bowden DW, Hawkins MA, Ling C, Mather KJ, Powers AC, Rhodes CJ, Sussel L, and Weir GC. (2014). beta-cell failure in type 2 diabetes: postulated mechanisms and prospects for prevention and treatment. Diabetes Care 37, 1751–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaestner KH, Powers AC, Naji A, Consortium H, and Atkinson MA. (2019). NIH Initiative to Improve Understanding of the Pancreas, Islet, and Autoimmunity in Type 1 Diabetes: The Human Pancreas Analysis Program (HPAP). Diabetes 68, 1394–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn SE. (2003). The relative contributions of insulin resistance and beta-cell dysfunction to the pathophysiology of Type 2 diabetes. Diabetologia 46, 3–19. [DOI] [PubMed] [Google Scholar]
- Kameswaran V, Bramswig NC, McKenna LB, Penn M, Schug J, Hand NJ, Chen Y, Choi I, Vourekas A, Won KJ, et al. (2014). Epigenetic regulation of the DLK1-MEG3 microRNA cluster in human type 2 diabetic islets. Cell Metab 19, 135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling C., Del Guerra S., Lupi R., Ronn T., Granhall C., Luthman H., Masiello P., Marchetti P., Groop L., and Del Prato S. (2008). Epigenetic regulation of PPARGC1A in human type 2 diabetic islets and effect on insulin secretion. Diabetologia 51, 615–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan A, Wessel J, Willems SM, Zhao W, Robertson NR, Chu AY, Gan W, Kitajima H, Taliun D, Rayner NW, et al. (2018). Refining the accuracy of validated target identification through coding variant fine-mapping in type 2 diabetes. Nat Genet 50, 559–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mularoni L, Ramos-Rodriguez M, and Pasquali L. (2017). The Pancreatic Islet Regulome Browser. Front Genet 8, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipson LH. (2020). Harnessing heterogeneity in type 2 diabetes mellitus. Nat Rev Endocrinol 16, 79–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders DC, Messmer J, Kusmartseva I, Beery ML, Yang M, Atkinson MA, Powers AC, Cartailler JP, and Brissova M. (2020). Pancreatlas: Applying an Adaptable Framework to Map the Human Pancreas in Health and Disease. Patterns (N Y) 1, 100120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnurr TM, Jakupovic H, Carrasquilla GD, Angquist L, Grarup N, Sorensen TIA, Tjonneland A, Overvad K, Pedersen O, Hansen T, et al. (2020). Obesity, unfavourable lifestyle and genetic risk of type 2 diabetes: a case-cohort study. Diabetologia 63, 1324–1332. [DOI] [PubMed] [Google Scholar]
- Segerstolpe A, Palasantza A, Eliasson P, Andersson EM, Andreasson AC, Sun X, Picelli S, Sabirsh A, Clausen M, Bjursell MK, et al. (2016). Single-Cell Transcriptome Profiling of Human Pancreatic Islets in Health and Type 2 Diabetes. Cell Metab 24, 593–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udler MS, Kim J, von Grotthuss M, Bonas-Guarch S, Cole JB, Chiou J, Christopher D.A.o.b.o.M.a.t.I., Boehnke M, Laakso M, Atzmon G, et al. (2018). Type 2 diabetes genetic loci informed by multi-trait associations point to disease mechanisms and subtypes: A soft clustering analysis. PLoS Med 15, e1002654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkmar M, Dedeurwaerder S, Cunha DA, Ndlovu MN, Defrance M, Deplus R, Calonne E, Volkmar U, Igoillo-Esteve M, Naamane N, et al. (2012). DNA methylation profiling identifies epigenetic dysregulation in pancreatic islets from type 2 diabetic patients. EMBO J 31, 1405–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkov P, Bacos K, Ofori JK, Esguerra JL, Eliasson L, Ronn T, and Ling C. (2017). Whole-Genome Bisulfite Sequencing of Human Pancreatic Islets Reveals Novel Differentially Methylated Regions in Type 2 Diabetes Pathogenesis. Diabetes 66, 1074–1085. [DOI] [PubMed] [Google Scholar]
- Vujkovic M., Keaton JM., Lynch JA., Miller DR., Zhou J., Tcheandjieu C., Huffman JE., Assimes TL., Lorenz K., Zhu X., et al. (2020). Discovery of 318 new risk loci for type 2 diabetes and related vascular outcomes among 1.4 million participants in a multi-ancestry meta-analysis. Nat Genet 52, 680–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JT, Saunders DC, Rai V, Dai C, Orchard P, Hopkirk AL, Reihsmann CV, Tao Y, Fan S, Shrestha S, et al. (2021). RFX6-mediated dysregulation defines human β cell dysfunction in early type 2 diabetes. bioRxiv. [Google Scholar]
- Wang YJ, Golson ML, Schug J, Traum D, Liu C, Vivek K, Dorrell C, Naji A, Powers AC, Chang KM, et al. (2016a). Single-Cell Mass Cytometry Analysis of the Human Endocrine Pancreas. Cell Metab 24, 616–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YJ, Schug J, Won KJ, Liu C, Naji A, Avrahami D, Golson ML, and Kaestner KH. (2016b). Single-Cell Transcriptomics of the Human Endocrine Pancreas. Diabetes 65, 3028–3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YJ, Traum D, Schug J, Gao L, Liu C, Consortium H, Atkinson MA, Powers AC, Feldman MD, Naji A, et al. (2019). Multiplexed In Situ Imaging Mass Cytometry Analysis of the Human Endocrine Pancreas and Immune System in Type 1 Diabetes. Cell Metab 29, 769–783 e764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigger L, Barovic M, Brunner AD, Marzetta F, Schoniger E, Mehl F, Kipke N, Friedland D, Burdet F, Kessler C, et al. (2021). Multi-omics profiling of living human pancreatic islet donors reveals heterogeneous beta cell trajectories towards type 2 diabetes. Nat Metab 3, 1017–1031. [DOI] [PubMed] [Google Scholar]
- Wu M, Lee MYY, Bahl V, Traum D, Schug J, Kusmartseva I, Atkinson MA, Fan G, Consortium H, and Kaestner KH. (2021). Single-cell analysis of the human pancreas in type 2 diabetes using multi-spectral imaging mass cytometry. Cell Rep 37, 109919. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.