Abstract
The diversity of lignicolous freshwater fungi in northwestern Yunnan, China, has been studied for several years in the College of Agriculture and Biological Science, at Dali University. Over the last 5 years, we published two new genera and nine new species of Tubeufiaceae from northwestern Yunnan. This study focused on introducing tubeufia-like hyphomycetous fungi found in freshwater lakes in the northwestern Yunnan plateau. Eleven fresh collections of tubeufiaceous taxa were gathered and identified. Among them, a new genus, Neomanoharachariella, is introduced to accommodate Neomanoharachariella aquatica, which is characterized by a light brown to dark brown color, dictyoseptate, and broadly oval to ellipsoid and well-developed conidiophores. Two new species, viz., Neohelicosporium suae and Parahelicomyces suae, one new record, Helicoma rufum, and three new collections, namely, H. rugosum, P. hyalosporus, and Tubeufia cylindrothecia are introduced based on morphological evidence and molecular phylogenetic analysis of combined ITS, LSU, tef 1-α, and RPB2 sequence data. Detailed descriptions and illustrations of these species are provided, and a morphological comparison with similar taxa is discussed.
Keywords: Dothideomycetes, lignicolous freshwater fungi, helicosporous hyphomycetes, morphology, multigene phylogeny
Introduction
Lignicolous freshwater fungi are an important group of organisms, involved in nutrient cycling by decaying submerged wood (Hyde et al., 2016a; Shen et al., 2022). Yunnan Province is one of the richest biodiversity hotspots, containing abundant resources of lignicolous freshwater fungi, with more than 281 species reported since 1986 (Shen et al., 2022). Among lignicolous freshwater fungi, Tubeufiales is one of the most species-rich groups in Dothideomycetes. Tubeufiales was introduced by Boonmee et al. (2014) based on molecular phylogenetic analysis to accommodate Tubeufiaceae. Liu et al. (2017) treated Bezerromycetaceae and Wiesneriomycetaceae as accepted families in Tubeufiales based on divergence time estimates. To date, Tubeufiales contains three families, viz., Bezerromycetaceae, Tubeufiaceae, and Wiesneriomycetaceae. The majority of Tubeufiaceae comprised freshwater taxa (Doilom et al., 2017; Lu et al., 2018a,b; Dong et al., 2020; Hongsanan et al., 2020). The family was established by Barr (1979) based on the generic type Tubeufia (Penzig and Saccardo, 1897). In the last decade, several studies of Tubeufiaceae have been published, with many species reported in freshwater habitats; most of them were asexual morphs (Boonmee et al., 2011; Hyde et al., 2016b, 2017; Brahmanage et al., 2017; Luo et al., 2017; Liu et al., 2018; Lu et al., 2018a,b). Lu et al. (2018b) reappraised and provided an updated phylogenetic tree for Tubeufiales which included 13 new genera, and expanded the circumscription of the type family Tubeufiaceae. To date, Tubeufiaceae includes 47 genera. They are widely distributed in tropical, subtropical, and temperate regions (Boonmee et al., 2011, 2014; Luo et al., 2017; Lu et al., 2018b), and most taxa are saprobic on woody substrates in terrestrial and freshwater habitats (Cai et al., 2003; Zhao et al., 2007; Lu et al., 2018b).
Members of Tubeufiaceae are a group of microfungi that are morphologically fascinating (Zhao et al., 2007) and have helicosporous hyphomycetes. Tubeufiaceae has been reported as sexual and asexual morphs. Asexual morphologies are mostly found as helicosporous hyphomycetes, while some are phragmosporous and chlamydosporous conidia (Lu et al., 2018b; Dong et al., 2020). Helicosporous hyphomycetes make up a large part of the order Tubeufiales. They are known to be present in many genera, such as Acanthohelicosporium, Berkleasmium, Chlamydotubeufia, Dematiohelicosporum, Helicangiospora, Helicodochium, Helicohyalinum, Helicoma, Helicomyces, Helicosporium, Helicotubeufia, Neoacanthostigma, Neohelicomyces, Neohelicosporium, Parahelicomyces, and Tubeufia (Boonmee et al., 2011, 2014; Brahmanage et al., 2017; Lu et al., 2017a,b,c, 2018a,b; Luo et al., 2017; Liu et al., 2018). Chlamydosporous and phragmosporous hyphomycetes in Tubeufiaceae are reported in Aquaphila, Berkleasmium, Chlamydotubeufia, Dictyospora, Helicoma, Kamalomyces, Neochlamydotubeufia, Tamhinispora, and Tubeufia (Lu et al., 2018b). Their sexual morphs are characterized by superficial ascomata, bitunicate asci, and hyaline to pale brown, elongate, obovoid or oblong, and septate ascospores (Barr, 1980; Kodsueb et al., 2006; Boonmee et al., 2011, 2014; Brahmanage et al., 2017; Lu et al., 2018b).
Helicoma was introduced by Corda (1837) with the type species H. muelleri. It is one of the earliest described helicosporous genus (Morgan, 1892; Linder, 1929; Moore, 1955). Helicoma includes two asexual morphs, one is characterized by conidia pleurogenous, helicoid, becoming loosely coiled in water, conidiogenous cells with denticles, and tooth-like protrusions. Other conidia are acrogenous, helicoid, circinate, tapering toward the apex, truncating at the base, and not becoming loose in water (Lu et al., 2018b). Neohelicosporium was introduced by Lu et al. (2018a) based on phylogenetic and morphological evidence. Currently, 24 species are accepted in the genus, of which 11 species were reported in freshwater habitats. Pseudohelicomyces was established by Lu et al. (2018b) to accommodate five species, viz., Ps. aquaticus, Ps. hyalosporus, Ps. indicus, Ps. paludosus, and Ps. talbotii (type species) based on multi-gene phylogenetic analysis. However, following previous publications, this generic name has an older homonym: Pseudohelicomyces (Valenzuela and Garnica, 2000), and this rendered the Pseudohelicomyces described by Lu et al. illegitimate. Lu et al. (2020) provided a proposal to conserve Pseudohelicomyces (Tubeufiaceae) against Pseudohelicomyces (Hymenogastraceae). Hsieh et al. (2021) established Parahelicomyces to replace Pseudohelicomyces and transferred all species of Pseudohelicomyces to Parahelicomyces. Until recently, nine species are accepted in Parahelicomyces (Lu et al., 2018b; Li et al., 2022; Tian et al., 2022). Tubeufia is the largest genus in Tubeufiaceae and is commonly reported as saprobes on submerged decaying wood in freshwater habitats (Ho et al., 2001; Cai et al., 2002; Liu et al., 2018; Lu et al., 2018b; Jayasiri et al., 2019). Members of Tubeufiaceae are mostly saprobic and widely distributed and are often found on woody substrates in terrestrial and freshwater habitats (Lu et al., 2018b). The southern China areas of Guangdong, Guangxi, Guizhou, Hubei, Yunnan, and other subtropical or tropical regions are very suitable for the growth and distribution of Tubeufiaceae fungi (Cai et al., 2002; Liu et al., 2018; Lu et al., 2018a,b).
During our investigation of freshwater fungi on submerged decaying wood, more than 100 specimens of freshwater hyphomycetes were collected from the lakes in the northwestern Yunnan plateau. This article aims to introduce eleven helicosporous hyphomycetes which were collected from the Luguhu and Shuduhu lakes. Phylogenetic analyses of combined ITS, LSU, tef 1-α, and RPB2 sequence data place them in Helicoma, Neohelicosporium, Parahelicomyces, and Tubeufia. A new genus Neomanoharachariella and three new species, viz., Neomanoharachariella aquatica, Neohelicosporium suae, and Parahelicomyces suae are introduced with morphological and phylogenetic evidence. Helicoma rufum is newly recorded in freshwater habitats for the first time in China. In addition, we combine Helicoma sp. (HKUCC 9118) as H. rugosum (HKUCC 9118) according to multi-gene phylogeny analysis and morphological evidence. Three known species, namely, Helicoma rugosum, Parahelicomyces hyalosporus, and Tubeufia cylindrothecia, are also accounted. Full descriptions, color photo plates of the species, and an updated phylogenetic tree for Tubeufiaceae are provided. This study provides a case study for lakes as a worthwhile niche area for the further study of hyphomycetous associations and hints that these lakes in the Yunnan plateau may potentially host numerous unknown fungal species.
Materials and methods
Collection, isolation, and morphology
Specimens of submerged decaying wood were collected from the Luguhu and Shuduhu lakes in the northwestern Yunnan province of China and were taken to the laboratory in ziplock plastic bags. The specimens were incubated at room temperature for 1 week in plastic boxes lined with moistened tissue paper. Specimen observations and isolation were performed by following the protocols provided by Luo et al. (2018) and Senanayake et al. (2020). Macromorphological characteristics of samples were observed using an Optec SZ 760 compound stereomicroscope. Temporarily prepared microscope slides were placed under a Nikon ECLIPSE Ni-U compound stereomicroscope for observation and micro-morphological-photography. The morphologies of colonies on native substrates were photographed with a Nikon SMZ1000 stereo zoom microscope. Single spore isolation was performed according to the following steps: the conidia suspension from specimens was transported using a sterilized pipette, placed on potato dextrose agar (PDA), and incubated at room temperature overnight. Germinated conidia were transferred to new PDA/malt extract agar (MEA) (Beijing land bridge technology CO., LTD., China) plates and incubated at room temperature (25°C). The specimens were deposited in the Herbarium of Cryptogams Kunming Institute of Botany, Academia Sinica (KUN-HKAS), Kunming, China. Living cultures were deposited in the China General Microbiological Culture Collection Center (CGMCC), Beijing, China, and the Kunming Institute of Botany Culture Collection Center, Kunming, China (KUNCC). Mycobank numbers were registered (https://www.mycobank.org). New species were established following the recommendations outlined by Chethana et al. (2021).
DNA extraction, PCR amplification, and sequencing
Fungal mycelium was removed from the surfaces of colonies that were grown on PDA or MEA for 4–6 weeks and transferred to a 1.5 ml centrifuge tube. A Trelief TM Plant Genomic DNA Kit (TSP101-50) was used to extract DNA from the ground mycelium according to the manufacturer's instructions. Four gene regions; ITS, LSU, tef 1-α, and RPB2 were amplified using ITS5/ITS4, LR0R/LR5 (Vilgalys and Hester, 1990), 983F/2218R, and fRPB2-5F/fRPB2-7cR (Liu et al., 1999). The PCR mixture was prepared as follows: 12.5 μl of 2 × Taq Master Mix (Genes and Biotech Co., Ltd), 1 μl of each primer, 1 μl of genomic DNA extract, and 9.5 μl of deionized water. The PCRs of ITS, LSU, tef 1-α, and RPB2 genes were processed as described in Su et al. (2015). PCR amplification was confirmed on 1% agarose electrophoresis gels stained with ethidium bromide. Sequencing was carried out by Tsingke Biological Engineering Technology and Services Co., Ltd (Yunnan, P.R. China).
Sequence alignment
Sequences were assembled using BioEdit. A BLAST search was performed on sequences with high similarity indices to find the closest matches with taxa in Tubeufiaceae and in recently published data (Luo et al., 2017; Lu et al., 2018b; Dong et al., 2020). All consensus sequences and the reference sequences were automatically aligned with MAFFT version 7.0 (Kuraku et al., 2013; Katoh et al., 2019). Aligned sequences of each gene region (ITS, LSU, tef 1-α, and RPB2) were combined and manually improved using BioEdit v. 7.0 (Hall, 1999). Ambiguous regions were excluded from the analysis and gaps were treated as missing data.
Phylogenetic analyses
Phylogenetic analyses were performed using maximum likelihood (ML) and Bayesian tree building criteria. Maximum likelihood (ML) analysis was carried out using RAxML-HPC2 on XSEDE (8.2.12) (Stamatakis, 2006; Stamatakis et al., 2008) on the CIPRES Science Gateway website (Miller et al., 2010: http://www.phylo.org/portal2) and the estimated proportion of invariant sites was determined using the GTRGAMMA+I model. Bayesian analyses were performed using MrBayes v. 3.1.2. (Ronquist and Huelsenbeck, 2003). The model of each gene was estimated using MrModeltest 2.3, and the GTR + I + G model was the best-fit model for ITS, LSU, tef 1-α, and RPB2 Bayesian analyses. Posterior probabilities (PP) (Ranala and Yang, 1996) were performed by Markov chain Monte Carlo sampling (BMCMC) in MrBayes v.3.1.2 (Liu et al., 2012). Six simultaneous Markov chains were run for 10 million generations, and trees were sampled every 100th generation (resulting in 100,000 trees). The first 20,000 trees, representing the burn-in phase of the analyses, were discarded and the remaining 80,000 (post-burning) trees were used for calculating PP in the majority rule consensus tree (Cai et al., 2006; Liu et al., 2012). Phylogenetic trees were represented by FigTree v. 1.4.0 and edited in Microsoft Office PowerPoint 2016. Newly-generated sequences in this study were submitted to GenBank, and the strain information used in this paper is provided in Table 1.
Table 1.
GenBank numbers and culture collection accession numbers of species included in the phylogenetic study.
| Taxa | Strain | GenBank Accession No. | |||
|---|---|---|---|---|---|
| ITS | LSU | tef 1-α | RPB2 | ||
| Acanthohelicosporapinicola T | MFLUCC 10–0116 | KF301526 | KF301534 | KF301555 | – |
| Acanthohelicospora scopula | ANM 386 | GQ856141 | GQ850489 | – | – |
| Acanthostigmina multiseptatum | ANM 475 | GQ856145 | GQ850492 | – | – |
| Acanthostigmina multiseptatum | ANM 665 | GQ856144 | GQ850493 | – | – |
| Acanthotubeufiafiliforme T | ANM 101 | – | GQ850495 | – | – |
| Acanthotubeufia filiforme | ANM 514 | GQ856146 | GQ850494 | – | – |
| Acanthotubeufia albicans | BCC 3463 | DQ341097 | DQ341100 | – | – |
| Acanthotubeufia albicans | BCC 3520 | DQ341098 | DQ341102 | – | – |
| Acanthotubeufia albicans | BCC 3543 | DQ341096 | DQ341101 | – | – |
| Acanthotubeufia albicans | MFLUCC 16–0010 | KX454165 | KX454166 | KY117034 | MF535255 |
| Acanthotubeufia albicans | MFLUCC 16–0020 | KX454167 | KX454168 | – | MF535256 |
| Berkleasmiumaquaticum T | MFLUCC 17–0049 | KY790444 | KY790432 | KY792608 | MF535268 |
| Berkleasmiumfusiforme T | MFLUCC 17–1978 | MH558693 | MH558820 | MH550884 | MH551007 |
| Berkleasmiumguangxiense T | MFLUCC 17–0042 | KY790448 | KY790436 | KY792612 | MF535270 |
| Berkleasmiumlongisporum T | MFLUCC 17–1999 | MH558698 | MH558825 | MH550889 | MH551012 |
| Boerlagiomycesmacrospora T | MFLUCC 12–0388 | KU144927 | KU764712 | KU872750 | – |
| Botryosphaeria dothidea | CBS 115476 | KF766151 | DQ678051 | DQ767637 | DQ677944 |
| Chlamydotubeufiacylindrica T | MFLUCC 16–1130 | MH558702 | MH558830 | MH550893 | MH551018 |
| Chlamydotubeufiahuaikangplaensis T | MFLUCC 10–0926 | JN865210 | JN865198 | – | – |
| Chlamydotubeufiakrabiensis T | MFLUCC 16–1134 | KY678767 | KY678759 | KY792598 | MF535261 |
| Dematiohelicoma pulchrum | MUCL 39827 | AY916457 | AY856872 | – | – |
| Dematiohelicomyceshelicosporus T | MFLUCC 16–0213 | KX454169 | KX454170 | KY117035 | MF535258 |
| Dematiohelicomyces helicosporus | MFLUCC 16–0003 | MH558703 | MH558831 | MH550894 | MH551019 |
| Dematiohelicomyces helicosporus | MFLUCC 16–0007 | MH558704 | MH558832 | MH550895 | MH551020 |
| Dematiohelicosporumguttulatum T | MFLUCC 17–2011 | MH558705 | MH558833 | MH550896 | MH551021 |
| Dematiotubeufiachiangraiensis T | MFLUCC 10–0115 | JN865200 | JN865188 | KF301551 | – |
| Dictyosporathailandica T | MFLUCC 16–0001 | KY873627 | KY873622 | KY873286 | MH551023 |
| Dictyospora thailandica | MFLUCC 11–0512 | KF301528 | KF301536 | – | – |
| Dictyospora thailandica | MFLUCC 16–0215 | KY873628 | KY873623 | KY873287 | – |
| Helicangiosporalignicola T | MFLUCC 11–0378 | KF301523 | KF301531 | KF301552 | – |
| Helicoarctatusaquaticus T | MFLUCC 17–1996 | MH558707 | MH558835 | MH550898 | MH551024 |
| Helicoarctatusthailandicus T | MFLUCC 18–0332 | – | ON764311 | MK541685 | – |
| Helicodochium aquaticum | MFLUCC 16–0008 | MH558708 | MH558836 | MH550899 | MH551025 |
| Helicodochiumaquaticum T | MFLUCC 17–2016 | MH558709 | MH558837 | MH550900 | MH551026 |
| Helicohyalinum aquaticum | MFLUCC 16–1131 | KY873625 | KY873620 | KY873284 | MF535257 |
| Helicohyalinuminfundibulum T | MFLUCC 16–1133 | MH558712 | MH558840 | MH550903 | MH551029 |
| Helicoma ambiens | UAMH 10533 | AY916451 | AY856916 | – | – |
| Helicoma ambiens | UAMH 10534 | AY916450 | AY856869 | – | – |
| Helicomaaquaticum T | MFLUCC 17–2025 | MH558713 | MH558841 | MH550904 | MH551030 |
| Helicomabrunneisporum T | MFLUCC 17–1983 | MH558714 | MH558842 | MH550905 | MH551031 |
| Helicoma dennisii | NBRC 30667 | AY916455 | AY856897 | – | – |
| Helicoma fusiforme T | MFLUCC 17–1981 | MH558715 | – | MH550906 | – |
| Helicomaguttulatum T | MFLUCC 16–0022 | KX454171 | KX454172 | MF535254 | MH551032 |
| Helicoma hongkongense | MFLUCC 17–2005 | MH558716 | MH558843 | MH550907 | MH551033 |
| Helicomainthanonense T | MFLUCC 11–0003 | JN865211 | JN865199 | – | – |
| Helicomakhunkornensis T | MFLUCC 10–0119 | JN865203 | JN865191 | KF301559 | – |
| Helicoma linderi | NBRC 9207 | AY916454 | AY856895 | – | – |
| Helicoma longisporum | MFLUCC 16–0002 | MH558717 | MH558844 | MH550908 | MH551034 |
| Helicoma longisporum | MFLUCC 16–0005 | MH558718 | – | MH550909 | MH551035 |
| Helicoma longisporum | MFLUCC 16–0211 | MH558719 | MH558845 | MH550910 | MH551036 |
| Helicomalongisporum T | MFLUCC 17–1997 | MH558720 | MH558846 | MH550911 | MH551037 |
| Helicomamiscanthi T | MFLUCC 11–0375 | KF301525 | KF301533 | KF301554 | – |
| Helicoma muelleri | CBS 964.69 | AY916453 | AY856877 | – | – |
| Helicoma muelleri | UBC F13877 | AY916452 | AY856917 | – | – |
| Helicomamultiseptatum T | GZCC 16–0080 | MH558721 | MH558847 | MH550912 | MH551038 |
| Helicoma nematosporum | MFLUCC 16–0011 | MH558722 | MH558848 | MH550913 | MH551039 |
| Helicomarubriappendiculatum T | MFLUCC 18–0491 | MH558723 | MH558849 | MH550914 | MH551040 |
| Helicomarufum T | MFLUCC 17–1806 | MH558724 | MH558850 | MH550915 | – |
| Helicoma rufum | CGMCC 3.23543 | OP184080 | OP184069 | OP186053 | OP186061 |
| Helicoma rugosum | ANM 196 | GQ856138 | GQ850482 | – | – |
| Helicoma rugosum | ANM 953 | GQ856139 | GQ850483 | – | – |
| Helicoma rugosum | ANM 1169 | – | GQ850484 | – | – |
| Helicoma rugosum | JCM 2739 | – | AY856888 | – | – |
| Helicoma rugosum | KUNCC 22–12445 | OP184078 | OP184067 | OP186051 | – |
| Helicoma rugosum | HKUCC 9118 | – | AY849966 | – | – |
| Helicoma septoconstrictum | MFLUCC 17–1991 | MH558725 | MH558851 | MH550916 | MH551041 |
| Helicomaseptoconstrictum T | MFLUCC 17–2001 | MH558726 | MH558852 | MH550917 | MH551042 |
| Helicomasiamense T | MFLUCC 10–0120 | JN865204 | JN865192 | KF301558 | – |
| Helicomatectonae T | MFLUCC 12–0563 | KU144928 | KU764713 | KU872751 | – |
| Helicoma vaccinii | CBS 216.90 | AY916486 | AY856879 | – | – |
| Helicomyces hyalosporus | GZCC 16–0070 | MH558728 | MH558854 | MH550919 | MH551044 |
| Helicomyceshyalosporus T | MFLUCC 17–0051 | MH558731 | MH558857 | MH550922 | MH551047 |
| Helicomyces torquatus | MFLUCC 16–0217 | MH558732 | MH558858 | MH550923 | MH551048 |
| Helicomyceschiayiensis T | BCRC FU30842 | LC316604 | – | – | – |
| Helicomyces colligatus | MFLUCC 16–1132 | MH558727 | MH558853 | MH550918 | MH551043 |
| Helicosporiumflavum T | MFLUCC 16–1230 | KY873626 | KY873621 | KY873285 | – |
| Helicosporiumluteosporum T | MFLUCC 16–0226 | KY321324 | KY321327 | KY792601 | MH551056 |
| Helicosporiumvesicarium T | MFLUCC 17–1795 | MH558739 | MH558864 | MH550930 | MH551055 |
| Helicotruncatum palmigenum | NBRC 32663 | AY916480 | AY856898 | – | – |
| Helicotubeufiaguangxiensis T | MFLUCC 17–0040 | MH290018 | MH290023 | MH290028 | MH290033 |
| Helicotubeufiahydei T | MFLUCC 17–1980 | MH290021 | MH290026 | MH290031 | MH290036 |
| Helicotubeufiajonesii T | MFLUCC 17–0043 | MH290020 | MH290025 | MH290030 | MH290035 |
| Kamalomyces thailandicus | MFLUCC 11–0158 | MF506883 | MF506881 | MF506885 | – |
| Kamalomycesthailandicus T | MFLUCC 13–0233 | MF506884 | MF506882 | MF506886 | – |
| Manoharachariellatectonae T | MFLUCC 12–0170 | KU144935 | KU764705 | KU872762 | – |
| Muripulchra aquatica | DLUCC 0571 | KY320531 | KY320548 | – | – |
| Muripulchra aquatica | KUMCC 15–0245 | KY320533 | KY320550 | KY320563 | MH551057 |
| Muripulchra aquatica | KUMCC 15–0276 | KY320534 | KY320551 | KY320564 | MH551058 |
| Muripulchraaquatica T | MFLUCC 15–0249 | KY320532 | KY320549 | – | – |
| Neoacanthostigmafusiforme T | MFLUCC 11–0510 | KF301529 | KF301537 | – | – |
| N eochlamydotubeufiafusiformis T | MFLUCC 16–0016 | MH558740 | MH558865 | MH550931 | MH551059 |
| Neochlamydotubeufia fusiformis | MFLUCC 16–0214 | MH558741 | MH558866 | MH550932 | MH551060 |
| Neochlamydotubeufiakhunkornensis T | MFLUCC 10–0118 | JN865202 | JN865190 | KF301564 | – |
| Neochlamydotubeufia khunkornensis | MFLUCC 16–0025 | MH558742 | MH558867 | MH550933 | MH551061 |
| Neohelicoma fagacearum | MFLUCC 11–0379 | KF301524 | KF301532 | KF301553 | – |
| Neohelicomycesaquaticus T | MFLUCC 16–0993 | KY320528 | KY320545 | KY320561 | MH551066 |
| Neohelicomycesgrandisporus T | KUMCC 15–0470 | KX454173 | KX454174 | – | MH551067 |
| Neohelicomycessubmersus T | MFLUCC 16–1106 | KY320530 | KY320547 | – | MH551068 |
| Neohelicosporium abuense | CBS 101688 | AY916470 | AY916085 | – | – |
| Neohelicosporiumacrogenisporum T | MFLUCC 17–2019 | MH558746 | MH558871 | MH550937 | MH551069 |
| Neohelicosporiumaquaticum T | MFLUCC 17–1519 | MF467916 | MF467929 | MF535242 | MF535272 |
| Neohelicosporiumastrictum T | MFLUCC 17–2004 | NR_160377 | NG_068566 | MH550938 | MH551070 |
| Neohelicosporium aurantiellum | ANM 718 | GQ856140 | GQ850485 | – | – |
| Neohelicosporiumbambusicola T | MFLUCC 21–0156 | OL606157 | OL606146 | OL964517 | OL964523 |
| Neohelicosporiumellipsoideum T | MFLUCC 16–0229 | MH558748 | MH558873 | MH550939 | MH551071 |
| Neohelicosporiumfusisporum T | MFUCC 16–0642 | MG017612 | MG017613 | MG017614 | – |
| Neohelicosporium griseum | UAMH 1694 | AY916473 | AY856902 | – | – |
| Neohelicosporium guangxiense | GZCC 16–0068 | MH558749 | MH558874 | MH550940 | MH551072 |
| Neohelicosporiumguangxiense T | MFLUCC 17–1522 | MF467922 | MF467935 | MF535248 | MF535278 |
| Neohelicosporiumhyalosporum T | GZCC 16–0076 | MF467923 | MF467936 | MF535249 | MF535279 |
| Neohelicosporiumirregulare T | MFLUCC 17–1796 | MH55875 | MH558877 | MH550943 | MH551075 |
| Neohelicosporium krabiense | MFLUCC 16–0224 | MH558754 | MH558879 | MH550945 | MH551077 |
| Neohelicosporiumlaxisporum T | MFLUCC 17–2027 | MH558755 | MH558880 | MH550946 | MH551078 |
| Neohelicosporium morganii | CBS 281.54 | MH857331 | MH868874 | – | – |
| Neohelicosporium morganii | CBS 222.58 | AY916469 | AY856880 | – | – |
| Neohelicosporiumovoideum T | GZCC 16–0064 | MH558756 | MH558881 | MH550947 | MH551079 |
| Neohelicosporium panacheum | CBS 257.59 | MH857857 | – | – | – |
| Neohelicosporium parvisporum | GZCC 16–0078 | MF467924 | MF467937 | MF535250 | MF535280 |
| Neohelicosporium parvisporum | MFLUCC 17–1523 | MF467926 | MF467939 | MF535252 | MF535282 |
| Neohelicosporium sp. | HKUCC 10235 | – | AY849942 | – | – |
| Neohelicosporium sp. | CBS 189.95 | AY916472 | AY856882 | – | – |
| Neohelicosporium submersum | MFLUCC 17–2376 | NR_171979 | MN913738 | – | – |
| Neohelicosporiumsuae T | CGMCC 3.23541 | OP184079 | OP184068 | OP186052 | OP265702 |
| Neohelicosporiumtaiwanense T | BCRC FU30841 | LC316603 | – | – | – |
| Neohelicosporiumthailandicum T | MFLUCC 16–0221 | MF467928 | MF467941 | MF535253 | MF535283 |
| Neomanoharachariellaaquatica T | CGMCC 3.23539 | OP184074 | OP184063 | OP186047 | OP186058 |
| Neomanoharachariella aquatica | CGMCC 3.23540 | OP184075 | OP184064 | OP186048 | OP186059 |
| Neotubeufiakrabiensis T | MFLUCC 16–1125 | MG012031 | MG012024 | MG012010 | MG012017 |
| Parahelicomycesaquaticus T | MFLUCC 16–0234 | MH558766 | MH558891 | MH550958 | MH551092 |
| Parahelicomyceschiangmaiensis T | MFLUCC 21–0159 | OL697884 | OL606145 | OL964516 | OL964522 |
| Parahelicomyces hyalosporus | CBS 283.51 | AY916464 | AY856881 | DQ677928 | DQ677981 |
| Parahelicomyces hyalosporus | KUMCC 15–0281 | KY320526 | KY320543 | KY320559 | MH551089 |
| Parahelicomyces hyalosporus | KUMCC 15–0322 | KY320525 | KY320542 | KY320558 | – |
| Parahelicomyces hyalosporus | KUMCC 15–0411 | KY320527 | KY320544 | KY320560 | – |
| Parahelicomyces hyalosporus | KUMCC 15–0430 | KY320524 | KY320541 | KY320557 | MH551090 |
| Parahelicomyceshyalosporus T | MFLUCC 15–0343 | KY320523 | KY320540 | – | – |
| Parahelicomyces hyalosporus | CGMCC 3.23535 | OP184073 | OP184062 | OP186046 | OP186057 |
| Parahelicomyces hyalosporus | KUNCC 22–12443 | OP184076 | OP184065 | OP186049 | – |
| Parahelicomyces hyalosporus | KUNCC 22–12444 | OP184077 | OP184066 | OP186050 | OP186060 |
| Parahelicomyces indicus | CBS 374.93 | AY916477 | AY856885 | – | – |
| Parahelicomycesmenglunicus T | KUN HKAS 85795 | MK335914 | – | MK335916 | – |
| Parahelicomyces paludosus | CBS 120503 | DQ341095 | DQ341103 | – | – |
| Parahelicomyces quercus | MFUCC 17–0895 | MK347720 | MK347934 | MK360077 | MK434906 |
| Parahelicomycessuae T | CGMCC 3.23534 | OP184072 | OP184061 | OP186045 | OP186056 |
| Parahelicomyces suae | CGMCC 3.23538 | OP184081 | OP184070 | OP186054 | – |
| Parahelicomyces talbotii | MUCL 33010 | AY916465 | AY856874 | – | – |
| Parahelicomyces talbotii T | MFLUCC 17–2021 | MH558765 | MH558890 | MH550957 | MH551091 |
| Parahelicomycesyunnanensis T | CGMCC 3.20429 | MZ092717 | MZ841658 | – | OM022000 |
| Pleurohelicosporiumparvisporum T | MFLUCC 17–1982 | MH558764 | MH558889 | MH550956 | MH551088 |
| Pseudohelicoon gigantisporum | BCC 3550 | AY916467 | AY856904 | – | – |
| Pseudohelicoonsubglobosum T | BCRC FU30843 | LC316607 | LC316610 | – | – |
| Tamhinispora indica | NFCCI 2924 | KC469282 | KC469283 | – | – |
| Tamhinispora srinivasanii | NFCCI 4231 | MG763746 | MG763745 | – | – |
| Thaxteriellopsis lignicola | MFLUCC 10–0123 | JN865207 | JN865195 | KF301562 | – |
| Thaxteriellopsis lignicola | MFLUCC 10–0124 | JN865208 | JN865196 | KF301561 | – |
| Tubeufiaabundata T | MFLUCC 17–2024 | MH558769 | MH558894 | MH550961 | MH551095 |
| Tubeufia amazonensis | ATCC 42524 | AY916458 | AY856911 | – | – |
| Tubeufiaaquatica T | MFLUCC 16–1249 | KY320522 | KY320539 | KY320556 | MH551142 |
| Tubeufia aquatica | MFLUCC 17–1794 | MH558770 | MH558895 | MH550962 | MH551096 |
| Tubeufiabambusicola T | MFLUCC 17–1803 | MH558771 | MH558896 | MH550963 | MH551097 |
| Tubeufiabrevis T | MFLUCC 17–1799 | MH558772 | MH558897 | MH550964 | MH551098 |
| Tubeufiabrunnea T | MFLUCC 17–2022 | MH558773 | MH558898 | MH550965 | MH551099 |
| Tubeufiachiangmaiensis T | MFLUCC 11–0514 | KF301530 | KF301538 | KF301557 | – |
| Tubeufia chiangmaiensis | MFLUCC 17–1801 | MH558774 | MH558899 | MH550966 | MH551100 |
| Tubeufiachlamydospora T | MFLUCC 16–0223 | MH558775 | MH558900 | MH550967 | MH551101 |
| Tubeufiacocois T | MFLUCC 22–0001 | OM102541 | OL985957 | OM355486 | OM355491 |
| Tubeufia cylindrothecia | MFLUCC 16–1253 | KY320519 | KY320536 | KY320553 | – |
| Tubeufia cylindrothecia | MFLUCC 16–1283 | KY320518 | KY320535 | KY320552 | MH551143 |
| Tubeufia cylindrothecia | MFLUCC 17–1792 | MH558776 | MH558901 | MH550968 | MH551102 |
| Tubeufia cylindrothecia | MFLUCC 11–0076 | MT627709 | MN913702 | – | – |
| Tubeufia cylindrothecia | MFLUCC 10–0919 | MT627710 | MN913701 | – | – |
| Tubeufia cylindrothecia | CGMCC 3.23552 | OP184071 | OP184060 | OP186044 | OP186055 |
| Tubeufiadictyospora T | MFLUCC 17–1805 | MH558778 | MH558903 | MH550970 | MH551104 |
| Tubeufia eccentrica | GZCC 16–0084 | MH558781 | MH558906 | MH550973 | MH551107 |
| Tubeufiaeccentrica T | MFLUCC 17–1524 | MH558782 | MH558907 | MH550974 | MH551108 |
| Tubeufia entadae | MFLU 18–2102 | NR163323 | – | – | – |
| Tubeufiafangchengensis T | MFLUCC 17–0047 | MH558783 | MH558908 | MH550975 | MH551109 |
| Tubeufiafiliformis T | MFLUCC 16–1128 | – | KY092407 | KY117028 | MF535284 |
| Tubeufia filiformis | MFLUCC 16–1135 | KY092416 | KY092411 | KY117032 | MF535285 |
| Tubeufiageniculata T | BCRC FU30849 | LC335817 | – | – | – |
| Tubeufia geniculata | NCYU U2–1B | LC335816 | – | – | – |
| Tubeufia guangxiensis | MFLUCC 17–0045 | MG012025 | MG012018 | – | – |
| Tubeufiahechiensis T | MFLUCC 17–0052 | MH558785 | MH558910 | MH550978 | MH551112 |
| Tubeufiahyalospora T | MFLUCC 15–1250 | MH558786 | MH558911 | MH550979 | – |
| Tubeufia inaequalis | MFLUCC 17–0053 | MH558789 | MH558914 | MH550982 | MH551115 |
| Tubeufia inaequalis | MFLUCC 17–1998 | MH558791 | MH558916 | MH550984 | MH551117 |
| Tubeufia javanica | MFLUCC 12–0545 | KJ880034 | KJ880036 | – | – |
| Tubeufiakrabiensis T | MFLUCC 16–0228 | MH558792 | MH558917 | MH550985 | MH551118 |
| Tubeufialatispora T | MFLUCC 16–0027 | KY092417 | KY092412 | KY117033 | MH551119 |
| Tubeufia laxispora | MFLUCC 16–0219 | KY092414 | KY092409 | KY117030 | MF535286 |
| Tubeufialaxispora T | MFLUCC 16–0232 | KY092413 | KY092408 | KY117029 | MF535287 |
| Tubeufia laxispora | MFLUCC 17–2023 | MH558794 | MH558919 | MH550987 | MH551121 |
| Tubeufia lilliputea | NBRC 32664 | AY916483 | AY856899 | – | – |
| Tubeufialongihelicospora T | MFLUCC 16–0753 | MZ538531 | MZ538565 | MZ567106 | – |
| Tubeufia longihelicospora | MFLUCC 21–0151 | OL606156 | OL606149 | OL964520 | OL964526 |
| Tubeufialongiseta T | MFLUCC 15–0188 | KU940133 | – | – | – |
| Tubeufia machaerinae | MFLUCC 17–0055 | MH558795 | MH558920 | MH550988 | MH551122 |
| Tubeufiamackenziei T | MFLUCC 16–0222 | KY092415 | KY092410 | KY117031 | MF535288 |
| Tubeufianigroseptum T | CGMCC 3.20430 | MZ092716 | MZ853187 | OM022002 | OM022001 |
| Tubeufia parvispora | MFLUCC 17–1992 | MH558796 | MH558921 | MH550989 | MH551123 |
| Tubeufia parvispora | MFLUCC 17–2009 | MH558798 | MH558923 | MH550991 | MH551125 |
| Tubeufiaroseohelicospora T | MFLUCC 15–1247 | KX454177 | KX454178 | – | MH551144 |
| Tubeufiarubra T | GZCC 16–0081 | MH558801 | MH558926 | MH550994 | MH551128 |
| Tubeufiasahyadriensis T | NFCCI 4252 | MH033849 | MH033850 | MH033851 | – |
| Tubeufia sahyadriensis | RAJ 99.2 | MN393081 | MN393082 | MN393083 | – |
| Tubeufia sessilis | MFLUCC 16–0021 | MH558803 | – | MH550996 | MH551130 |
| Tubeufia sympodihylospora | GZCC 16–0049 | MH558804 | MH558928 | MH550997 | MH551131 |
| Tubeufia sympodihylospora | GZCC 16–0051 | MH558805 | MH558929 | MH550998 | MH551132 |
| Tubeufia sympodihylospora | MFLUCC 17–0044 | MH558806 | MH558930 | MH550999 | MH551133 |
| Tubeufiasympodilaxispora T | MFLUCC 17–0048 | MH558808 | MH558932 | MH551001 | MH551135 |
| Tubeufia taiwanensis | BCRC FU30844 | LC316605 | – | – | – |
| Tubeufiatectonae T | MFLUCC 12–0392 | KU144923 | KU764706 | KU872763 | – |
| Tubeufia tectonae | MFLUCC 16–0235 | MH558809 | MH558933 | MH551002 | MH551136 |
| Tubeufia tectonae | MFLUCC 15–0974 | – | MN913688 | MT954376 | – |
| Tubeufiatratensis T | MFLUCC 17–1993 | MH558811 | MH558935 | MH551004 | MH551138 |
| Tubeufia xylophila | GZCC 16–0038 | MH558812 | MH558936 | MH551005 | MH551139 |
| Tubeufia xylophila | MFLUCC 17–1520 | MH558813 | MH558937 | MH551006 | MH551140 |
Ex-type strains are indicated by T after the species name. Newly generated sequences are indicated in bold. The symbol “–” indicates information unavailable.
Results
Phylogenetic analyses
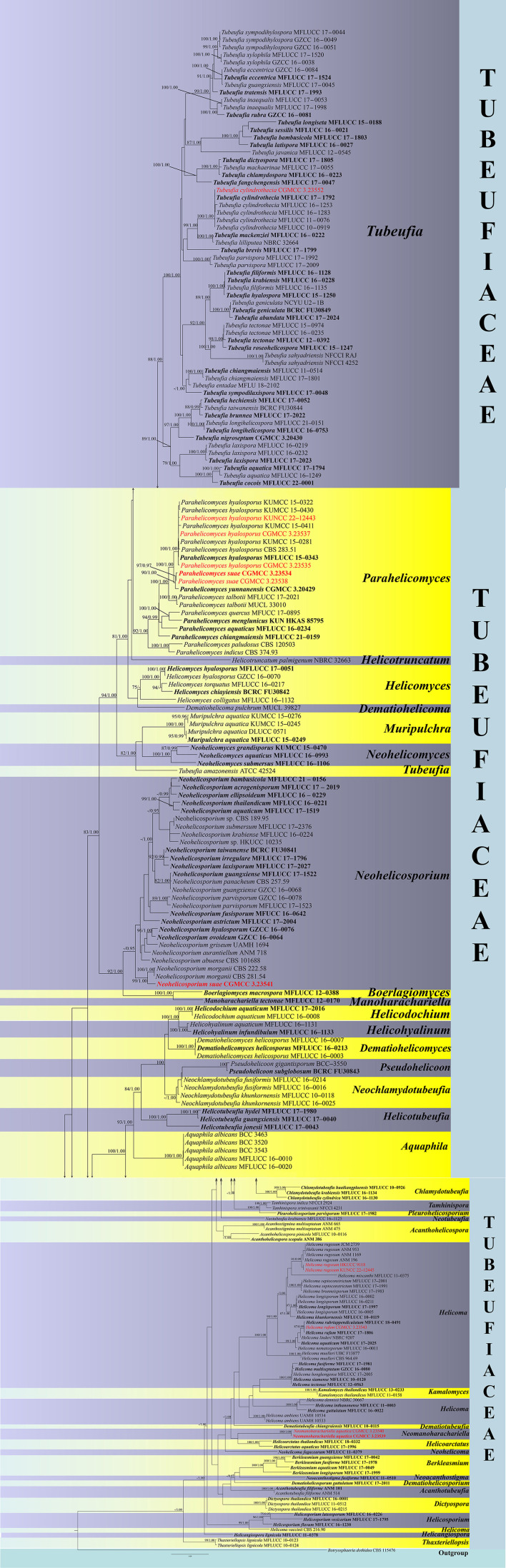
Phylogenetic analyses of combined ITS, LSU, tef 1-α, and RPB2 sequences comprised a total of 3,316 characters including gaps, ITS (1–534 bp), LSU (535–1,362 bp), tef 1-α (1,363–2,273 bp), and RPB2 (2,274–3,316 bp) including 217 strains, with Botryosphaeria dothidea (CBS 115476) as the outgroup taxon. RAxML and Bayesian analyses of the combined dataset resulted in phylogenetic reconstructions with largely similar topologies. The result of ML analyses with a final likelihood value of −53,732.520635 is shown in Figure 1. Alignment exhibits 1,618 distinct alignment patterns; the proportion of gaps and completely undetermined characters in this alignment is 27.38%. Gamma distribution shape parameter: α = 0.226507; tree-length: 6.955943; estimated base frequencies: A = 0.242825, C = 0.253033, G = 0.260763, and T = 0.243379; substitution rates: AC = 1.238257, AG = 6.612700, AT = 2.116761, CG = 0.859127, CT = 10.120846, and GT = 1.000000. Bootstrap support values for RAxML >75% and Bayesian PP >0.95 are given at each node (Figure 1).
Figure 1.
Phylogram generated from maximum likelihood analysis (RAxML) of Tubeufiaceae based on ITS, LSU, tef 1-α, and RPB2 sequence data. Maximum likelihood bootstrap values equal to or above 75% and Bayesian posterior probabilities (PP) equal to or above 0.95 are given above the nodes. The tree is rooted at Botryosphaeria dothidea CBS 115476. Newly-generated sequences are indicated in red. Ex-type strains are indicated in black/red bold.
Phylogenetic analyses showed that the new isolates were nested in Tubeufiaceae with close affinities to four exciting genera, viz., Helicoma, Neohelicosporium, Parahelicomyces, Tubeufia, and the new genus Neomanoharachariella, forming a distinct clade among the genera of Tubeufiaceae. KUNCC 22–12445 and CGMCC 3.23543 clustered within Helicoma, sister to Helicom rugosum (ANM 196, ANM 953, ANM 1169, and JCM 2739) with 97% ML and 0.99 PP support values. Another strain, CGMCC 3.23543 nested in H. rubriappendiculatum (MFLUCC 18–0491) and H. rufum (MFLUCC 17–1806) with 87% ML and 0.99 PP support values. CGMC3.23541 nested in N. morganii (CBS 281.54) with strong bootstrap support (100% ML/1.00 PP). CGMC3.23539 and CGMCC 3.23540 clustered as a monophyletic clade sister to Helicoarctatus aquaticus (MFLUCC 17–1996) and H. tailandicus (MFLUCC 18–0332). Three new collections (CGMCC 3.23535, KUNCC 22–12443, and KUNCC 22–12444) clustered with Parahelicomyces hyalosporus (CBS 283.51, MFLUCC 15–0343, KUMCC 15–0430, KUMCC 15–0411, KUMCC 15–0322, and KUMCC 15–0281) with 100% ML and 1.00 PP support. CGMCC 3.23534 and CGMCC 3.23538 formed a sister lineage to Parahelicomyces yunnanensis (CGMCC 3.20429) with 90% ML and 1.00 PP support. CGMCC 3.23552 clustered with five strains of Tubeufia cylindrothecia (MFLUCC 10–0919, MFLUCC 11–0076, MFLUCC 16–1253, MFLUCC 16–1283, and MFLUCC 17–1792) with 100% ML and 1.00 PP support.
Taxonomy
Helicoma rugosum (C. Booth) Boonmee and K.D. Hyde [as 'rugosa'], Fungal Divers. 68: 266 (2014), Figure 2
Figure 2.
Helicoma rugosum (KUN-HKAS 124608). (a,b) Colony on decaying wood. (c–f) Conidiophores with attached conidia. (g,h) Conidiogenous cells. (i–m) Conidia. (n) Germinating conidium. (o,p) Colony on PDA observed from above and below. Scale bars: (c,d) 30 μm, (e) 50 μm, (f) 30 μm, (g,h) 10 μm, and (i–n) 20 μm.
Index Fungorum: IF 340543; Facesoffungi number: FoF 02650
Saprobic on submerged decaying wood in the lake. Asexual morph: Hyphomycetous, helicosporous. Colonies on natural substrate superficial, effuse, discrete, dilute, and light brown to brown. Mycelium composed of partly immersed, partly superficial, septate, pale brown to brown, branched hyphae, with masses of crowded, glistening conidia. Conidiophores 95–151 μm long, 5.4–6.8 μm wide ( = 122.6 × 6 μm, n = 20), macronematous, mononematous, straight to slightly bent, unbranched, septate, cylindrical, erect, pale brown to brown, and smooth-walled. Conidiogenous cells 9–12 μm long, 5–6 μm wide, holoblastic, mono- to polyblastic, integrated, intercalary, cylindrical, with denticles, tiny tooth-like protrusions (0.9–2.6 μm long, 0.5–1.7 μm wide), brown, and smooth-walled. Conidia 60.7–85.5 μm diameter, conidial filament 4–4.8 μm wide ( = 73 × 4.4 μm, n = 20), 216–290 μm long, slightly coiled 1.0–2.5 times, pleurogenous, helicoid, rounded at tip, septate, becoming loosely coiled in water, guttulate, pale brown, and smooth-walled. Sexual morph: not observed.
Culture characteristics: Conidia germinating on PDA and germ tubes produced from conidia within 12 h. Colonies growing on PDA, irregular, center umbonate, with a rough surface, wrinkle, edge undulate, reaching 10–15 mm in 2 weeks at 26°C, and pale brown to brown in the PDA medium. Mycelium superficial and partially immersed, branched, septate, hyaline to pale brown, and smooth-walled.
Material examined: China, Yunnan Province, Luguhu lake, on submerged decaying wood, 22 October 2021 (Altitude: 2,625 m, 27°42'41“N, 100°46'48“E), Long-Li Li, L-1013 (KUN-HKAS 124608), living culture, KUNCC 22–12445.
Notes: Helicoma rugosum was reported by Boonmee et al. (2014) to combine Sphaeria helicoma, Thaxteriella helicoma, and Tubeufia rugosa based on phylogenetic and morphological evidence. H. rugosum (KUNCC 22–12445) resembles H. rufum, presenting macronematous, mononematous, unbranched or branched, septate conidiophores, holoblastic, mono- to ployblastic conidiogenous cells, helicoid, and septate conidia. However, H. rugosum (KUNCC 22–12445) is distinct from H. rufum as it has shorter and narrower conidiophores (95–151 × 5.4–6.8 vs. 110–210 × 7–8.5 μm), longer and wider conidia (60.7–85.5 × 4–4.8 vs. 35–45 × 4–5.5 μm), and shorter conidial filaments (216–290 × 4–5 vs. 240–410 × 4–5.5 μm). Furthermore, H. rufum produces a reddish brown pigment in the PDA medium in 7 days but H. rugosum lacks this characteristic. In the phylogenetic analyses, H. rugosum (KUNCC 22–12445) cluster together with H. rugosum (ANM 196, ANM 1169, ANM 953, and JCM 2739) and Helicoma sp. (HKUCC 9118) with strong support (91% ML and 0.99 PP). In this study, we introduce our new collection with Helicoma sp. (HKUCC 9118) as H. rugosum because of identical LSU nucleotide sequences and morphological characteristics. Our fresh collection is morphologically similar to Helicoma sp. (HKUCC 9118) (Kodsueb et al., 2004) in terms of conidiogenous cells with tiny tooth-like protrusions, dentical, conidiophores brownish-gray, upright, and the same conidia size (61–86 × 4–5 vs. 37–86.4 × 4.6–5.4 μm). Furthermore, both of their morphologies fit into the generic group Helicoma, and the analyses show that they should be the same species.
Helicoma rufum Y.Z. Lu, J.C. Kang, and K.D. Hyde, Fungal Divers. 92: 183 (2018), Figure 3
Figure 3.
Helicoma rufum (KUN-HKAS 124609). (a,b) Colony rises from mycelium on natural wood substrate. (c–f) Conidiophores with attached conidia. (g,h) Conidiogenous cells. (i–l) Conidia. (m) Germinating conidium. (n,o) Culture on PDA. Scale bars: (c–f) 60 μm, (g,h) 10 μm, and (i–m) 20 μm.
Index Fungorum: IF 554843; Facesoffungi number: FoF 04718
Saprobic on submerged decaying wood in the lake. Asexual morph: Hyphomycetous, helicosporous. Colonies superficial, effuse, gregarious, and brown. Mycelium composed of immersed, partly superficial, hyaline to pale brown, septate, branched hyphae, with masses of crowded, glistening conidia. Conidiophores 136–209 μm long, 6–7 μm wide ( = 173 × 6.5 μm, n = 30), macronematous, mononematous, cylindrical, erect, straight to slightly bent, mostly unbranched, septate, the lower part brown and the upper part pale yellow, and smooth-walled. Conidiogenous cells 12–14 μm long, 5–7 μm wide, holoblastic, mono- to polyblastic, integrated, intercalary, cylindrical, with denticles, rising laterally from the lower portion of conidiophores as tiny tooth-like protrusions (2.7–3.9 μm long, 1.5–2.3 μm wide), brown, and smooth-walled. Conidia 57–104 μm diameter, conidial filament 3.4–5.2 μm wide ( = 80.6 × 4.3 μm, n = 20), 248–327 μm long, solitary, pleurogenous, helicoid, rounded at tip, septate, slightly constricted at septa, loosely coiled 1.5–3.5 times, becoming loosely coiled in water, guttulate, hyaline to pale brown, and smooth-walled. Sexual morph: not observed.
Culture characteristics: Conidia germinating on PDA within 12 h and many germ tubes produced from conidium cells. Colonies growing on PDA, reaching 25 mm, and started producing reddish brown pigment in 3 weeks at 26°C, brown to reddish brown in the PDA medium, irregular, with a flat surface, edge slightly undulate. Mycelium superficial and partially immersed, branched, septate, hyaline to pale brown, and smooth-walled.
Material examined: China, Yunnan Province, Luguhu lake, on submerged decaying wood (Altitude: 2,717 m, 27°42'41“N, 100°46'48“E), 21 October 2021, Long-Li Li, L-1032 (KUN-HKAS 124609), living cultures, CGMCC 3.23543 = KUNCC 22–12439.
Notes: Helicoma rufum was introduced by Lu et al. (2018b) on decaying wood in a mountain in Thailand. The new isolate L-1032 collected from freshwater habitats was identified as H. rufum based on the phylogenetic analyses and the morphological features. Our new collection CGMCC 3.23543 clusters in the same clade with H. rufum (MFLUCC 17–1806) and H. rubriappendiculatum (MFLUCC 18–0491) with bootstrap support (87% ML and 0.99 PP). Morphologically, our new collection is almost identical to H. rufum (MFLUCC 17–1806) except for the conidia diameter (57–104 vs. 35–45 μm long). The nucleotide comparisons show 4 bp, 1 bp, and 2 bp of ITS, LSU, and tef 1-α differences between the new isolate CGMCC 3.23543 and H. rufum (MFLUCC 17–1806). Between H. rubriappendiculatum (MFLUCC 18–0491) and H. rufum (CGMCC 3.23543), there are 4, 2, and 6 bp of ITS, LSU, and tef 1-α differences; compared with H. rubriappendiculatum, H. rufum (CGMCC 3.23543) produces a reddish brown pigment in the PDA medium and presents a longer conidia diameter (57–104 vs. 25–35 μm), lacking the characteristic red appendant near the apex in conidiophores. Thus, we identify the new isolate as H. rufum based on both phylogenetic analyses and morphological characteristics. This is the first report of H. rufum in freshwater habitats and its occurrence in China.
Neohelicosporium suae L.L. Li, H.W. Shen and Z.L. Luo, sp. nov.
MycoBank number: MB 845321, Figure 4
Figure 4.
Neohelicosporium suae (KUN-HKAS 124610, holotype). (a) Colony on decaying wood. (b,c,e) Conidiophores with attached conidia. (d) Conidiophores. (f–h) Conidiogenous cells. (i–l) Conidia. (m) Germinating conidium. (n,o) Colony on PDA observed from above and below. Scale bars: (b,c) 30 μm, (d,e) 20 μm, and (f–m) 10 μm.
Holotype—KUN-HKAS 124610
Etymology—“suae” (Lat.) in memory of the Chinese mycologist Prof. Hong-Yan Su (4 April 1967–3 May 2022).
Saprobic on submerged decaying wood in the lake. Asexual morph: Hyphomycetous, helicosporous. Colonies on substratum superficial, effuse, and white. Mycelium composed of superficial, partly immersed, brown, septate, branched hyphae, with crowded by conidial masses. Conidiophores 52–97 μm long, 4.2–5.1 μm wide ( = 75 × 4.7 μm, n = 20), macronematous, mononematous, erect, cylindrical, unbranched or less branched, 3–6-septate, hyaline to pale brown, and smooth-walled. Conidiogenous cells 15–27 μm long, 3.5–5 μm wide ( = 21 × 4.2 μm, n = 20), holoblastic, mono- to polyblastic, cylindrical, truncate at apex after conidial secession, integrated, sympodial, terminal, cylindrical, with denticles 2–3 × 1.5–2.4 μm, hyaline to pale brown, and smooth-walled. Conidia 45–55 μm diameter, conidial filaments 5–7 μm wide ( = 50 × 6 μm, n = 20), 212–268 μm long, tightly coiled 2–2.5 times, helicoid, rounded at tip, multi-septate, slightly constricted at septa, guttulate, hyaline, not becoming loose in water, and smooth-walled. Sexual morph: not observed.
Culture characteristics: Conidia germinating on PDA within 8 h. Colonies growing on PDA, circular, with a flat surface, edge entire, reaching 28 mm in 3 weeks at room temperature, pale brown to brown in the MEA medium. Mycelium superficial and partially immersed, branched, septate, hyaline to pale brown, and smooth-walled.
Material examined: China, Yunnan Province, Luguhu lake, on submerged decaying wood in the lake (Altitude: 2,242 m, 26°48'29“N, 100°43'4.8“E), 21 October 2021, Long-Li Li, L-1030 (KUN-HKAS 124610, holotype), ex-type cultures, CGMCC 3.23541 = KUNCC 22–12438.
Notes: Neohelicosporium suae is introduced as a new species based on morphological and phylogenetic evidence. In phylogeny, N. suae (CGMCC 3.23541) is a sister to N. morganii with strong bootstrap support (100% ML and 1.00 PP). Based on pairwise nucleotide comparisons, the new strain N. suiae (CGMCC3.23541) is different from N. morganii (CBS 281.54) in 9/532 bp (1.69%) of the ITS and 3/804 bp (0.37%) of the LSU. Morphologically, N. suae can be distinguished from N. morganii; the conidiophores of N. suae are unbranched or less branched, the latter are branched and shorter (52–97 μm long, 4.2–5.1 μm wide vs. up to 145 μm long, 5–7 μm wide) (Zhao et al., 2007), and the number of septa is more than 6. The conidiogenous cells of N. suiae are 15–27 μm long, swollen, with longer and wider denticles (2–3 × 1.5–2.4 vs. 1–2.5 × 0.5–1.5 μm), terminal, whereas N. morganii displays no swelling. Furthermore, N. suiae is distinct from N. morganii, presenting distinguished conidia characteristics in terms of a larger diameter (45–55 × 5–7 vs. 17–23 × 3–4 μm).
Neomanoharachariella L.L. Li, H.W. Shen, and Z.L. Luo, gen. nov.
Mycobank number: MB 845535
Etymology—The generic epithet, neo (Lat., new), refers to the similarity to Manoharachariella.
Saprobic on decaying wood in the lake. Asexual morph: Hyphomycetous, dictyosporous. Colonies on the substratum superficial, effuse, and dark brown. Conidiophores macronematous, mononematous, erect, cylindrical, unbranched, straight or flexuous, paler, and smooth-walled. Conidiogenous cells monoblastic, integrated, terminal, cylindrical, subhyaline to pale brown, and smooth-walled. Conidia holoblastic smooth, shiny, simple, broadly oval to ellipsoid, muriform, tuberculous at the top, white and pale brown when immature, becoming dark to black when mature, and pale yellow at the basal cell and brown at other parts. Sexual morph: not observed.
Type species: Neomanoharachariella aquatica L.L. Li, H.W. Shen, and Z.L. Luo.
Notes: Neomanoharachariella is morphologically similar to Chlamydotubeufia, Dictyospora, and Neochlamydotubeufia, presenting dictyoseptate, broadly oval to ellipsoid, and darkened to black when matured conidia. However, Neomanoharachariella can be distinguished from other chlamydosporous genera by well-developed conidiophores. The morphological characteristics allow the assignment of Neomanoharachariella to Tubeufiaceae. In phylogeny, it formed a well-separated clade from all other genera of Tubeufiaceae (Figure 5). The molecular phylogenetic studies indicate its placement in Tubeufiaceae as a genus that is phylogenetically close to the genera, Berkleasium, Dictyospora, Helicoarctatus, Helicoma, and Helicosporium.
Figure 5.
Neomanoharachariella aquatica (KUN-HKAS 124611, holotype). (a,b) Colony erect on decaying wood. (c–e) Conidiophores with attached conidia. (f,g) Conidiogenous cells. (h–m) Conidia. (n) Germinating conidium. (o,p) Culture on PDA. Scale bars: (c,e) 25 μm, (f,g) 5 μm, (h–j) 15 μm, and (d,k–n) 20 μm.
Neomanoharachariella aquatica L.L. Li, H.W. Shen, and Z.L. Luo, sp. nov.
Mycobank number: MB 845536, Figure 5
Holotype—KUN-HKAS 124611
Etymology—“aquatica” referring to the aquatic habitat of this fungus.
Saprobic on decaying woods in the lake. Asexual morph: hyphomycetous, dictyosporous. Colonies on the substratum superficial, effuse, and dark brown. Conidiophores 20–31 μm long, 3.5–4.2 μm wide ( = 25 × 4 μm, n = 20), macronematous, mononematous, erect, cylindrical, unbranched, straight or flexuous, paler, and smooth-walled. Conidiogenous cells monoblastic, integrated, terminal, cylindrical, subhyaline to pale brown, and smooth-walled. Conidia 37–61 μm long, 17–32 μm wide ( = 49 × 24 μm, n = 20), muriform 8–10-transversely septate, with 1–4-longitudinal septa, smooth, shiny, simple, broadly oval to ellipsoid, tuberculous at the top, hyaline to pale brown when immature, becoming dark to black when mature, and pale yellow at the basal cells and brown at other parts. Sexual morph: not observed.
Culture characteristics: Conidia germinating on PDA within 12 h. Colonies growing on PDA, circular, with a flat surface, edge entire, reaching 15 mm in 3 weeks at 26°C, and brown to dark brown in the PDA medium. Mycelium superficial and partially immersed, branched, septate, hyaline to pale brown, and smooth-walled.
Material examined: China, Yunnan Province, Shuduhu lake, on submerged decaying wood (Altitude: 3,578 m, 27°54'24“N, 99°57'15“E), 25 August 2020, Zheng-Quan Zhang, L-190 (KUN-HKAS 124611, holotype), ex-type cultures, CGMCC 3.23539 = KUNCC 22–12437; China, Yunnan Province, Shuduhu lake, on submerged decaying wood (Altitude: 3,578 m, 27°54'24“N, 99°57'15“E), 25 August 2020, Zheng-Quan Zhang, L-281 (KUN-HKAS 124612), living cultures, CGMCC 3.23540 = KUNCC 22–12442.
Notes: The new collection can be easily distinguished from other Tubeufiaceae genera by the long oval and dictyosporous conidia with well-developed conidiophores. In the phylogenetic analyses, Neomanoharachariella aquatica shares a sister relationship to Helicoarctatus aquaticus (MFLUCC 17–1996) and H. thailandicus (MFLUCC 18–0332). However, there are great differences in morphology; the asexual morph of H. aquaticus and H. thailandicus are helicosporous, and our new collection is dictyosporous. H. aquaticus and H. thailandicus are characterized by setiform, unbranched, septate conidiophores, holoblastic, mono- to poly-blastic, denticulate conidiogenous cells, pleurogenous, helicoid, multi-septate, guttulate, and hyaline conidia. Based on pairwise nucleotide comparisons, the new strain CGMCC 3.23540 is different from the type species Helicoarctatus aquaticus (MFLUCC 17–1996) in 30/541 bp (5.54%) of the ITS, 24/805 bp (2.98%) of the LSU, 74/875 bp (8.46%) of the tef 1-α, and 154/1045 bp (14.74%) of the RPB2. In addition, Neomanoharachariella aquatica is most similar to the asexual state of Chlamydotubeufia huaikangplaensis, but the conidia of N. aquatica are shorter (37–61 × 17–32 vs. 50–77 × 39–42) and presenting erect, unbranched, and smooth-walled conidiophores; the phylogenetic analyses also clearly segregate it from C. huaikangplaensis. We therefore identify the newly obtained taxon as Neomanoharachariella aquatica sp. nov.
Parahelicomyces hyalosporus (Y.Z. Lu, J.K. Liu, and K.D. Hyde) S. Y. Hsieh, Goh, and C. H. Kuo, Mycol. Prog. 20(2): 182 (2021) Figure 6
Figure 6.
Parahelicomyces hyalosporus (KUN-HKAS 124603). (a) Colony on decaying wood. (b–d) Conidiophores with attached conidia and lateral minute polyblastic denticles. (e,f,i,j) Conidiogenous cells. (g,h,k–p) Conidia. (p,q) Colony on PDA observed from above and below. Scale bars: (b) 50 μm, (c,d) 40 μm, and (e–p) 10 μm.
Index Fungorum: IF 554888; Facesoffungi number: FoF 04812
Saprobic on submerged decaying woods in the lake. Asexual morph: Hyphomycetous, helicosporous. Colonies on wood substrate superficial, effuse, gregarious, and hyaline to white. Mycelium composed of partly immersed, partly superficial, pale brown, septate, anastomosing, reapent, with masses of crowded conidia. Conidiophores 60–142 μm long, 4–5.2 μm wide ( = 101 × 4.6 μm, n = 10), macronematous, mononematous, cylindrical, branched, septate, hyaline to pale brown, and smooth-walled. Conidiogenous cells 5–10 μm long, 3–4 μm wide, holoblastic, mono-to polyblastic, integrated, terminal or intercalary, cylindrical, truncate at apex after conidial secession, hyaline to pale brown, and smooth-walled. Conidia 40–56.7 μm diameter, and conidial filaments 3.5–4.5 μm wide ( = 48 × 4 μm, n = 20), 145–180 μm long, loosely coiled 1–2.5 times, solitary, pleurogenous or acropleurogenous, helicoid, rounded at tip, multi-septate, becoming loosely coiled in water, guttulate, hyaline, and smooth-walled. Sexual morph: not observed.
Culture characteristics: Conidia germinating on PDA within 12 h; many germ tubes produced from conidium cells. Colonies growing on PDA, circular, with umbonate surface, edge dulate, and brown to dark brown in PDA medium, reaching 20 mm in 3 weeks at 26°C, and brown to dark brown in the PDA medium. Mycelium superficial and partially immersed, branched, septate, hyaline to pale brown, and smooth-walled.
Material examined: China, Yunnan Province, Luguhu lake, on submerged decaying wood (Altitude: 2,698 m, 27°41'11“N, 100°48'18“E), 5 March 2021, Zheng-Quan Zhang, L-159 (KUN-HKAS 124603), living cultures, CGMCC 3.23535 = KUNCC 22–12436; China, Yunnan Province, Luguhu lake, on submerged decaying wood (Altitude: 2734 m, 27°45'18“N, 100°46'42“E), 5 March 2021, Zheng-Quan Zhang, L-315 (KUN-HKAS 124606), living culture, KUNCC 22–12443; China, Yunnan Province, Luguhu lake, on submerged decaying wood (Altitude: 2,794 m, 27°45'02“N, 100°51'02“E), 5 March 2021, Zheng-Quan Zhang, L-326 (KUN-HKAS 124605), living cultures, CGMCC 3.23537 = KUNCC 22–12444.
Notes: Parahelicomyces hyalosporus was first introduced as Pseudohelicomyces hyalosporus by Lu et al. (2018b) based on morphological and phylogenetic evidence. Hsieh et al. (2021) transferred it to Parahelicomyces as the genus Pseudohelicomyces was an older homonym and illegitimate. In this paper, three newly-obtained isolates were identified as Parahelicomyces hyalosporus, and the morphology characteristics fit well with Parahelicomyces hyalosporus; the conidiophores macronematous, mononematous, branched, septate, conidiogenous cells with denticles, holoblastic, mono- to polyblastic, intercalary or terminal, determinate or sympodial and pleurogenous or acropleurogenous, conidia helicoid, multi-septate, and hyaline to pale brown. Species of the P. hyalosporus are widely found in lakes and streams of freshwater habitats in China and Thailand (Luo et al., 2017; Lu et al., 2018b; Li et al., 2022). Based on pairwise nucleotide comparisons, ITS and LSU are identical between the type species (MFLUCC 15–0343) and P. hyalosporus (CGMCC 3.23535).
Parahelicomyces suae L.L. Li, H.W. Shen, and Z.L. Luo, sp. nov.
Mycobank number: MB 845534, Figure 7
Figure 7.
Parahelicomyces suae (KUN-HKAS 124604, holotype). (a) Colony on decaying wood. (b–d) Conidiophores with attached conidia. (e–h) Conidiogenous cells. (i–m) Conidia. (n) Germinating conidium. (o,p) Colony on MEA observed from above and below. Scale bars: (b) 70 μm, (c) 60 μm, (d) 30 μm, (e–h,j–n) 10 μm, and (i) 15 μm.
Holotype—KUN-HKAS 124604
Etymology—“suae” (Lat.) in memory of the Chinese mycologist Prof. Hong-Yan Su (4 April 1967–3 May 2022).
Saprobic on submerged decaying woods in the lake. Asexual morph: Hyphomycetous, helicosporous. Colonies on the wood substratum superficial, effuse, gregarious, and white. Mycelium composed of partly immersed, partly superficial, hyaline to pale brown, septate, abundantly branched hyphae, with masses of crowded, glistening conidia. Conidiophores 114.8–173.5 μm long, 3–4 μm wide ( = 144 × 3.5 μm, n = 20), macronematous, mononematous, cylindrical, branched or unbranched, erect, septate, dark brown at base, becoming hyaline toward apex, and smooth-walled. Conidiogenous cells 12–18 μm long, 3–4 μm wide, sympodial, holoblastic, monoblastic, integrated, terminal, cylindrical, truncate at apex after conidial secession, denticles or bladder-like cells, hyaline to pale brown, and smooth-walled. Conidia 29–36 μm diameter, conidial filament 1.8–2.2 μm wide ( = 32.5 × 2 μm, n = 20), 103–121 μm long, coiled 1–3.5 times, solitary, helicoid, rounded at tip, young conidia have indistinct septate, not easily loosely coiled in water, guttulate, hyaline, and smooth-walled. Sexual morph: not observed.
Culture characteristics: Conidia germinating on PDA within 12 h and many germ tubes produced from conidium cells. Colonies growing on MEA, reaching 14 mm diameter in 2 weeks at 26°C, circular, with a flat surface, edge entire, and pale brown to brown in the MEA medium. Mycelium superficial and partially immersed, branched, septate, hyaline to pale brown, and smooth.
Material examined: China, Yunnan Province, Luguhu lake, on submerged decaying wood in the lake (Altitude: 2,698 m, 27°41'11“N, 100°48'18“E), 3 March 2021, Sha Luan, L-158 (KUN-HKAS 124604, holotype), ex-type cultures, CGMCC 3.23534 = KUNCC 22–12435; China, Yunnan Province, Luguhu lake, on submerged decaying wood in the lake (Altitude: 2698 m, 27°42'43“N, 100°44'56“E), 3 March 2021, Long-Li Li, L-1038, (KUN-HKAS 124607), living cultures, CGMCC 3.23538 = KUNCC 22–12440.
Notes: Parahelicomyces suae is introduced as a new species from Luguhu lake in Yunnan, China. In phylogeny, P. suae constitutes a strongly supported independent lineage basal to P. yunnanensis. Compared with CGMCC 3.20429, there are 5/563 (0.89%), 11/1048 bp (1.05%) base pair differences in the ITS and RPB2 regions between these two species. Morphologically, compared with P. yunnanensis, the conidia of P. suae are shorter (103–121 vs. 104–156 μm). In addition, our isolate conidia are not easily loosely coiled in water, conidiogenous cells with denticulate, and hyaline. Therefore, we identify the isolate as a new species of P. suae.
Tubeufia cylindrothecia (Seaver) Höhn Sber. Akad. Wiss. Wien, Math.-naturw. Kl., Abt. 1 128: 562 (1919), Figure 8
Figure 8.
Tubeufia cylindrothecia (KUN-HKAS 124602). (a,b) Colony on decaying wood. (c) Conidiophores with attached conidia. (d) Conidiophores. (e–h) Conidiogenous cells. (i–m) Conidia. (n) Germinating conidium. (o,p) Colony on CMA observed from above and below. Scale bars: (c) 70 μm, (d,e) 20 μm, and (f–n) 10 μm.
Index Fungorum: IF 340543; Facesoffungi number: FoF 02650
Saprobic on decaying wood in the lake. Asexual morph: Hyphomycetous, helicosporous. Colonies on the substratum superficial, effuse, gregarious, and white to pale brown. Mycelium composed of partly immersed, partly superficial, hyaline to pale brown, septate, abundantly branched hyphae, with masses of crowded, glistening conidia. Conidiophores 97–200 μm long, 5–6 μm wide ( = 148 × 5.5 μm, n = 30), macronematous, mononematous, cylindrical, branched or unbranched, erect, flexuous, pale brown to brown, and smooth-walled. Conidiogenous cells 10.4–17 × 4–6 μm ( = 13.7 × 5 μm, n = 30), holoblastic, mono- to polyblastic, integrated, intercalary or terminal, cylindrical, repeatedly geniculate, truncate at the apex after conidial secession, each with single or several conidia hyaline to pale brown, and smooth-walled. Conidia 41.6–57.8 μm diameter and conidial filament 3.7–4.9 μm wide ( = 50 × 4.3 μm, n = 30), 105–206 μm long, coiled 1.5–3.5 times, solitary, acrogenous or acropleurogenous, helicoid, rounded at tip, becoming loosely coiled in water, guttulate, young Conidia hyaline and pale brown when edged, and smooth-walled. Sexual morph: not observed.
Culture characteristics: Conidia germinating on PDA within 12 h. Colonies growing slowly on CMA, reaching 15 mm diameter after 2 weeks at 26°C, effuse, the middle is dark, velvety to hairy, edge undulate, brown to dark brown in the CMA medium, mycelium superficial, effuse, with irregular edge, and hyphae pale yellow to brown.
Material examined: China, Yunnan Province, Luguhu lake, on submerged decaying wood (Altitude: 2,734 m, 27°45'18“N, 100°46'42“E), 5 March 2021, Zheng-Quan Zhang, L-157 (KUN-HKAS 124602), living cultures, CGMCC 3.23552 = KUNCC 22–12434.
Notes: The asexual morph of Tubeufa cylindrothecia was first reported by Luo et al. (2017) and later encountered by Lu et al. (2018b) in freshwater habitats. In this study, the newly obtained collection has longer conidiophores (97–200 vs. 50–81 μm) and shorter conidia (105–206 vs. 256–314 μm) compared with the holotype (Luo et al., 2017). However, their ITS, LSU, tef 1-α, and RPB2 sequence data are identical; we therefore identify it as Tubeufia cylindrothecia.
Discussion
The modern classification of Tubeufiaceae was established by Boonmee et al. (2014), based on phylogenetic analyses and morphology. However, there are still taxonomic confusions in this group, especially in those types with helicosporous asexual morphs; their morphologically-based intergeneric classifications are controversial. Some species have been transferred or are synonymous to other genera of Tubeufiaceae, for example, Helicosporium pannosum, Neohelicosporium griseum, and N. morganii have been transferred several times. The asexual state of Neomanoharachariella is dictyosporous conidia. It is a unique tubeufiaceous fungus with broadly oblong, elongate, multiseptate, muriform conidia, at first pale brown, becoming dark brown, with well-developed conidiophores, and basal cells are hyaline and bulging. These characteristics make it distinct from all related Tubeufiacceae genera and is hence proposed as a new genus. Phylogenetic analyses based on ITS, LSU, tef 1-α, and RPB2 sequence (Figure 1) also distinguish N. aquatica from other dictyosporous members of Tubeufiaceae. The new genus is related to Helicoarctatus aquaticus (MFLUCC 17–1996) and Helicoarctatus thailandicus (MFLUCC 18–0332) which formed a distinct clade. The phylogenetic analyses also clearly segregated other dictyosporous genera of Tubeufiaceae such as Chlamydotubeufia, Dictyospora, Manoharachariella, and Tamhinispora in well-differentiated monophyletic lineages.
An abundance of lakes is a major feature of the Yunnan plateau. In recent years, lignicolous freshwater fungi were investigated in Yunnan, in nine freshwater lakes on the plateau. These lakes are distributed in high-altitude areas and most of them are depression pools formed by the subsidence of faults, with no water channels connected (Yang et al., 2004; Shen et al., 2022). Because of their unique development, formation, and relativele isolation, each lake possesses its own unique species. In this study, we have also examined seven tubuefiaceous species collected from these plateau lakes. Of which, three were introduced as new species and a new genus Neomanoharachariella, while four were identified as existing species based on phylogenetic analyses and morphological characteristics. The nine species were placed in Helicoma, Neohelicosporium, Parahelicomyces, and Tubeufia. This study provides a case study for lakes as a worthwhile niche area of hyphomycetous associations. Parahelicomyces is well studied, and eight species in this genus have sequence data in the GenBank. For the common and confusing genera Helicoma, Neohelicosporium, and Tubeufia, morphological characteristics (conidiophores, conidiogenous cells, and conidia including size and color) and phylogenetic analyses are essential to distinguish them.
In conclusion, some tubeufiaceous species have the potential to produce new structural and active secondary metabolites (Mao et al., 2014; Lu et al., 2018a). Fang et al. (2019) tested and reported that most Tubeufiaceae species have certain antibacterial and anti-tumor activities in vitro. At present, few studies have reported secondary degradation products of Helicoma, Helicomyces, and Helicosporium species. In view of the potential to produce active compounds, and the reports on secondary metabolites of Tubeufiaceae, the prospect of active research is broad, and it is very possible to obtain new compounds with various biological activities from Tubeufiaceae.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author contributions
L-LL conducted the experiments, analyzed the data, and wrote the manuscript. D-FB, DW, and Y-ZL revised the manuscript. H-WS planned the experiments and analyzed the data. Z-LL planned and funded the experiments. YF conducted the experiments. All authors contributed to the article and approved the submitted version.
Funding
This work was mainly supported by the National Natural Science Foundation of China (Project ID: 32060005 and 31900020) and the Yunnan Fundamental Research Project (Grant Nos. 202101AU070137 and 202201AW070001).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
L-LL is grateful to Xi Fu and Jun He for sharing their knowledge of morphology and phylogeny. Sha Luan and Zheng-Quan Zhang are thanked for their help with sample collection. DW thanks the CAS President's International Fellowship Initiative (PIFI) for funding his postdoctoral research (number 2021FYB0005), the National Science Foundation of China (NSFC) under the project code 32150410362, and the Postdoctoral Fund from the Human Resources and Social Security Bureau of Yunnan Province. Xin-Wei Wan, Ming-Hui Chen, and Yuan-Yue Zhang are acknowledged for their help with DNA extraction and PCR amplification. The author also thank Shaun Pennycook for checking species names.
References
- Barr M. E. (1979). Classification of Loculoascomycetes. Mycologia 71, 935–957. 10.1080/00275514.1979.12021099 [DOI] [Google Scholar]
- Barr M. E. (1980). On the family Tubeufiaceae (Pleosporales). Mycotaxon 12, 137–167. [Google Scholar]
- Boonmee S., Rossman A. Y., Liu J. K., Li W. J., Dai D. Q., Bhat D. J., et al. (2014). Tubeufiales, ord. nov., integrating sexual and asexual generic names. Fungal Divers. 68, 239–298. 10.1007/s13225-014-0304-7 [DOI] [Google Scholar]
- Boonmee S., Zhang Y., Chomnunti P., Chukeatirote E., Tsui C. K. M., Bahkali A. H., et al. (2011). Revision of lignicolous Tubeufiaceae based on morphological reexamination and phylogenetic analysis. Fungal Divers. 51, 63–102. 10.1007/s13225-011-0147-4 [DOI] [Google Scholar]
- Brahmanage R. S., Lu Y. Z., Bhat D. J., Wanasinghe D. N., Yan J. Y., Hyde K. D., et al. (2017). Phylogenetic investigations on freshwater fungi in Tubeufiaceae (Tubeufiales) reveals the new genus Dictyospora and new species Chlamydotubeufia aquatica and Helicosporium flavum. Mycosphere 8, 917–933. 10.5943/mycosphere/8/7/8 [DOI] [Google Scholar]
- Cai L., Jeewon R., Hyde K. D. (2006). Phylogenetic investigations of Sordariaceae based on multiple gene sequences and morphology. Mycol. Res. 110, 137–150. 10.1016/j.mycres.2005.09.014 [DOI] [PubMed] [Google Scholar]
- Cai L., Tsui C. K. M., Zhang K. Q., Hyde K. D. (2002). Aquatic fungi from Lake Fuxian, Yunnan, China. Fungal Divers. 9, 57–70. 10.1017/S095375620200565835597355 [DOI] [Google Scholar]
- Cai L., Zhang K., McKenzie E. H. C., Hyde K. D. (2003). Freshwater fungi from bamboo and wood submerged in the Liput River in the Philippines. Fungal Divers. 13, 1–12. 10.1016/S1567-1356(03)00107-7 [DOI] [Google Scholar]
- Chethana K. W. T., Manawasinghe I. S., Hurdeal V. G., Bhunjun C. S., Appadoo M. A., Gentekaki E., et al. (2021). What are fungal species and how to delineate them? Fungal Divers. 109, 1–25. 10.1007/s13225-021-00483-9 [DOI] [Google Scholar]
- Corda A. K. J. (1837). Incones Fungorum Hucusque Cognitorum, Vol. 1. Prague: J.G. Calve, 1–32. [Google Scholar]
- Doilom M., Dissanayake A. J., Wanasinghe D. N., Boonmee S., Liu J. K., Bhat D. J., et al. (2017). Microfungi on Tectona grandis (teak) in Northern Thailand. Fungal Divers. 82, 107–182. 10.1007/s13225-016-0368-7 [DOI] [Google Scholar]
- Dong W., Bin W., Hyde K. D., McKenzie E. H. C., Raja H. A., Tanaka K., et al. (2020). Freshwater Dothideomycetes. Fungal Divers. 105, 319–575. 10.1007/s13225-020-00463-5 [DOI] [Google Scholar]
- Fang C., Lu Y. Z., Kang J. C., Wang L., Lei B. X., Chen L. Z. (2019). Evaluation of bioactivity of secondary metabolites from fungi of Tubeufiaceae. Mycosystema 38, 560–574. 10.1007/s13205-016-0518-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 41, 95–98. 10.12691/ajmr-6-4-3 [DOI] [Google Scholar]
- Ho W. H., Hyde K. D., Hodgkiss I. J. (2001). Fungal communities on submerged wood from streams in Brunei, Hong Kong, and Malaysia. Mycol. Res. 105, 1492–1501. 10.1017/S095375620100507X [DOI] [Google Scholar]
- Hongsanan S., Hyde K. D., Phookamsak R., Wanasinghe D. N., McKenzie E. H. C., Sarma V. V., et al. (2020). Refined families of Dothideomycetes: orders and families incertae sedis in Dothideomycetes. Fungal Divers. 105, 17–318. 10.1007/s13225-020-00462-6 [DOI] [Google Scholar]
- Hsieh S. Y., Goh T. K., Kuo C. H. (2021). New species and records of Helicosporium sensu lato from Taiwan, with a reflection on current generic circumscription. Mycol. Prog. 20, 169–190. 10.1007/s11557-020-01663-8 [DOI] [Google Scholar]
- Hyde K. D., Fryar S., Tian Q., Bahkali A. H., Xu J. C. (2016a). Lignicolous freshwater fungi along a northsouth latitudinal gradient in the Asian/Australian region; can we predict the impact of global warming on biodiversity and function? Fungal Ecol. 19, 190–200. 10.1016/j.funeco.2015.07.002 [DOI] [Google Scholar]
- Hyde K. D., Hongsanan S., Jeewon R., Bhat D. J., McKenzie E. H. C., Jones E. B. G., et al. (2016b). Fungal diversity notes 367–490: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 80, 1–270. 10.1007/s13225-016-0373-x34899100 [DOI] [Google Scholar]
- Hyde K. D., Norphanphoun C., Abreu V. P., Bazzicalupo A., Chethana K. W. T., Clericuzio M., et al. (2017). Fungal diversity notes 603–708: taxonomic and phylogenetic notes on genera and species. Fungal Divers. 87, 1–235. 10.1007/s13225-017-0391-334899100 [DOI] [Google Scholar]
- Jayasiri S. C., Hyde K. D., Jones E. B. G., McKenzie E. H. C., Jeewon R., Phillips A. J. L., et al. (2019). Diversity, morphology and molecular phylogeny of Dothideomycetes on decaying wild seed pods and fruits. Mycosphere 10, 1–186. 10.5943/mycosphere/10/1/1 [DOI] [Google Scholar]
- Katoh K., Rozewicki J., Yamada K. D. (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 20, 1160–1166. 10.1093/bib/bbx108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodsueb R., Jeewon R., Vijaykrishna D., McKenzie E. H. C., Lumyong P., Lumyong S., et al. (2006). Systematic revision of Tubeufiaceae based on morphological and molecular data. Fungal Divers. 21, 105–130. [Google Scholar]
- Kodsueb R., Lumyong S., Lumyong P., Mckenzie E., Ho W. W. H., Hyde K. D. (2004). Acanthostigma and Tubeufia species, including T. claspisphaeria sp. nov., from submerged wood in Hong Kong. Mycologia 96, 667–674. 10.1080/15572536.2005.11832963 [DOI] [PubMed] [Google Scholar]
- Kuraku S., Zmasek C. M., Nishimura O., Katoh K. (2013). A leaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucl. Acids Res. 41, W22–W28. 10.1093/nar/gkt389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L. L., Shen H. W., Bao D. F., Lu Y. Z., Su H. Y., Luo Z. L. (2022). New species, Parahelicomyces yunnanensis sp. nov. and Tubeufia nigroseptum sp. nov. from freshwater habitats in Yunnan, China. Phytotaxa 530, 021–037. 10.11646/phytotaxa.530.1.2 [DOI] [Google Scholar]
- Linder D. H. (1929). A monograph of the helicosporous fungi imperfecti. Ann. Missouri Bot. Gard. 16, 227–388. [Google Scholar]
- Liu J. K., Hyde K. D., Jeewon R., Phillips A. J., Maharachchikumbura S. S., Ryberg M., et al. (2017). Ranking higher taxa using divergence times: a case study in Dothideomycetes. Fungal Divers. 84, 75–99. 10.1007/s13225-017-0385-1 [DOI] [Google Scholar]
- Liu J. K., Lu Y. Z., Cheewangkoon R., To-Anun C. (2018). Phylogeny and morphology of Helicotubeufia gen. nov, with three new species in Tubeufiaceae from aquatic habitats. Mycosphere 9, 495–509. 10.5943/mycosphere/9/3/4 [DOI] [Google Scholar]
- Liu J. K., Phookamsak R., Doilom M., Wikee S., Li Y. M., Ariyawansha H., et al. (2012). Towards a natural classification of Botryosphaeriales. Fungal Divers. 57, 149–210. 10.1007/s13225-012-0207-4 [DOI] [Google Scholar]
- Liu Y. J., Whelen S., Hall B. D. (1999). Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. Mol. Biol. Evol. 16, 1799–1808. [DOI] [PubMed] [Google Scholar]
- Lu Y. Z., Boonmee S., Bhat D. J., Hyde K. D., Kang J. C. (2017a). Helicosporium luteosporum sp. nov. and Acanthohelicospora aurea (Tubeufiaceae, Tubeufiales) from terrestrial habitats. Phytotaxa 319, 241–253. 10.11646/phytotaxa.319.3.3 [DOI] [Google Scholar]
- Lu Y. Z., Boonmee S., Dai D. Q., Liu J. K., Hyde K. D., Bhat D. J., et al. (2017b). Four new species of Tubeufia (Tubeufiaceae, Tubeufiales) from Thailand. Mycol. Prog. 16, 403–417. 10.1007/s11557-017-1280-6 [DOI] [Google Scholar]
- Lu Y. Z., Boonmee S., Liu J. K., Hyde K. D., Bhat D. J., Eungwanichayapant P. D., et al. (2017c). Novel Neoacanthostigma species from aquatic habitats. Crypt. Mycol. 38, 169–190. 10.7872/crym/v38.iss2.2017.169 [DOI] [Google Scholar]
- Lu Y. Z., Boonmee S., Liu J. K., Hyde K. D., McKenzie E. H., Eungwanichayapant P. D., et al. (2018a). Multi-gene phylogenetic analyses reveals Neohelicosporium gen. nov. and five new species of helicosporous hyphomycetes from aquatic habitats. Mycol. Prog. 17, 631–646. 10.1007/s11557-017-1366-1 [DOI] [Google Scholar]
- Lu Y. Z., Liu J. K., Hyde K. D. (2020). (2744) Proposal to conserve Pseudohelicomyces Y. Z. Lu; al. (Tubeufiaceae) against Pseudohelicomyces Garnica; E. Valenz. (Hymenogastraceae). Laxon 69, 615–616. 10.1002/tax.12268 [DOI] [Google Scholar]
- Lu Y. Z., Liu J. K., Hyde K. D., Jeewon R., Kang J. C., Fan C., et al. (2018b). A taxonomic reassessment of Tubeufiales based on multi-locus phylogeny and morphology. Fungal Divers. 92, 131–344. 10.1007/s13225-018-0411-y [DOI] [Google Scholar]
- Luo Z. L., Bhat D. J., Jeewon R., Boonmee S., Bao D. F., Zhao Y. C., et al. (2017). Molecular phylogeny and morphological characterization of asexual fungi (Tubeufiaceae) from freshwater habitats in Yunnan, China. Crypt. Mycol. 38, 27–53. 10.7872/crym/v38.iss1.2017.27 [DOI] [Google Scholar]
- Luo Z. L., Hyde K. D., Liu J. K., Bhat D. J., Bao D. F., Li W. L., et al. (2018). Lignicolous freshwater fungi from China II: novel Distoseptispora (Distoseptisporaceae) species from northwestern Yunnan Province and a suggested unified method for studying lignicolous freshwater fungi. Mycosphere 9, 444–461. 10.5943/mycosphere/9/3/2 [DOI] [Google Scholar]
- Mao Z. L., Sun W. B., Fu L. Y., Luo H. Y., Lai D. W., Zhou L. G. (2014). Natural dibenzo-α-pyrones and their bioactivities. Molecules 19, 5088–5108. 10.3390/molecules19045088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M. A., Pfeiffer W., Schwartz T. (2010). “Creating the CIPRES Science Gateway for inference of large phylogenetic trees,” in Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), 1–8. 10.1109/GCE.2010.5676129 [DOI] [Google Scholar]
- Moore R. T. (1955). Index to the helicosporae. Mycologia 47, 90–103. [Google Scholar]
- Morgan A. P. (1892). North American helicosporae. J. Cincinnati. Soc. Nat. Hist. 15, 39–52. [Google Scholar]
- Penzig O. A. J., Saccardo P. A. (1897). Diagnoses fungorum novorum in Insula Java collectorum. Series secunda. Malpighia 11, 491–530. [Google Scholar]
- Ranala B., Yang Z. (1996). Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. J. Mol. Evolut. 43, 304–311. 10.1007/BF02338839 [DOI] [PubMed] [Google Scholar]
- Ronquist F., Huelsenbeck J. P. (2003). MrBayes3: bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Senanayake I. C., Rathnayaka A. R., Marasinghe D. S., Calabon M. S., Gentekaki E., Wanasinghe D. N., et al. (2020). Morphological approaches in studying fungi: collection, examination, isolation, sporulation and preservation. Mycosphere 11, 2678–2754. 10.5943/mycosphere/11/1/20 [DOI] [Google Scholar]
- Shen H. W., Bao D. F., Bhat D. J., Su H. Y., Luo Z. L. (2022). Lignicolous freshwater fungi in Yunnan Province, China: an overview. Mycology 13, 119–132. 10.1080/21501203.2022.2058638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690. 10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- Stamatakis A., Hoover P., Rougemont J. (2008). A rapid bootstrap algorithm for the RAxML web-servers. Syst. Biol. 75, 758–771. 10.1080/10635150802429642 [DOI] [PubMed] [Google Scholar]
- Su H. Y., Udayanga D., Luo Z. L., Manamgoda D. S., Zhao Y. C., Yang J., et al. (2015). Hyphomycetes from aquatic habitats in Southern China: species of Curvularia (Pleosporaceae) and Phragmocephala (Melannomataceae). Phytotaxa 226, 201–216. 10.11646/phytotaxa.226.3.1 [DOI] [Google Scholar]
- Tian X. G., Karunarathna S. C., Xu R. J., Lu Y. Z., Suwannarach N., Mapook A., et al. (2022). Three new species, two new records and four new collections of Tubeufiaceae from Thailand and China. JOF 8, 206. 10.3390/jof8020206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela E., Garnica S. (2000). Pseudohelicomyces, a new anamorph of Psilocybe. Mycol. Res. 104, 738–741. 10.1017/S0953756299002117 [DOI] [Google Scholar]
- Vilgalys R., Hester M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 172, 4238–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Tian K., Hao J., Pei S., Yang Y. (2004). Biodiversity and biodiversity conservation in Yunnan, China. Biodivers. Conserv. 13, 813–826. 10.1023/B:BIOC.0000011728.46362.3c [DOI] [Google Scholar]
- Zhao G. Z., Liu X., Wu W. (2007). Helicosporous hyphomycetes from China. Fungal Divers. 26, 313–524.35205960 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.