SUMMARY
Congenital Zika virus (ZIKV) infection results in neurodevelopmental deficits in up to 14% of infants born to ZIKV-infected mothers. Neutralizing antibodies are a critical component of protective immunity. Here, we demonstrate that plasma IgM contributes to ZIKV immunity in pregnancy, mediating neutralization up to three months post symptoms. From a ZIKV-infected pregnant woman, we isolated a pentameric ZIKV-specific IgM (DH1017.IgM) that exhibited ultrapotent ZIKV neutralization dependent on the IgM isotype. DH1017.IgM targets an envelope dimer epitope within Domain II. The epitope arrangement on the virion is compatible with concurrent engagement of all ten antigen-binding sites of DH1017.IgM, a solution not available to IgG. DH1017.IgM protected mice against viremia upon lethal ZIKV challenge more efficiently than when expressed as an IgG. Our findings identify a role for antibodies of the IgM isotype in protection against ZIKV and posit DH1017.IgM as a safe and effective candidate immunotherapeutic, particularly during pregnancy.
Graphical Abstract
eTOC
Singh et al. demonstrate that plasma IgM contributes to early Zika virus neutralization during pregnancy and isolate a pentameric IgM from a Zika virus-infected pregnant woman whose child was born without evidence of congenital Zika syndrome. This IgM’s ultrapotent neutralization in vitro and efficient protection from virus challenge in mice depends on its isotype, which enables modes of antigen recognition not available to IgG.
INTRODUCTION
Zika virus (ZIKV) emergence in the Americas revealed it to be congenitally transmitted, and the cause of microcephaly and other birth defects in up to 14% of infants born to ZIKV-infected pregnancies (Reynolds et al., 2017). Even infants born seemingly healthy following maternal ZIKV infection may develop neurodevelopmental defects years later (Nielsen-Saines et al., 2019). As ZIKV results mostly in mild febrile disease in healthy adults, its greatest disease burden arises through infections in pregnancy. In the 2015-2016 epidemic, ZIKV re-emergence in a susceptible population led to 11,000 cases of microcephaly in Brazil alone (Campos et al., 2018). There is no licensed vaccine for ZIKV, and the vaccine development and testing pipeline has stalled due to limited ZIKV circulation (Morabito and Graham, 2017).
Protective immunity in pregnancy could prevent ZIKV infection, congenital transmission, and lifelong congenital Zika syndrome sequelae. Neutralizing antibodies (nAbs) have been shown to be a critical aspect of protective immune responses against ZIKV and other flavivirus infections (Abbink et al., 2016; Hombach et al., 2005; Katzelnick et al., 2017; Kreil et al., 1997; Kwek et al., 2018; Larocca et al., 2016; Mason et al., 1973; Richner et al., 2017. NAb studies have largely focused on IgG antibodies. However, flavivirus infections are also characterized by virus-specific and prolonged IgM responses (Griffin et al., 2019a, 2019b; Murray et al., 2013; Stone et al., 2020). IgM are pentameric molecules with 5-times as many potential antigen binding sites compared to IgG. Conventionally, IgM are thought of as early, short-lived, and low-affinity antibodies that are not somatically matured and thus unable to confer potent viral neutralization. Yet, early neutralizing IgM responses have been identified for other flaviviruses such as West Nile virus and yellow fever virus (Diamond et al., 2003; Gasser et al., 2020; Monath, 1971; Wec et al., 2020). A common feature is that these viruses present dense and repetitive structures on their surface, which may favor avid multivalent interactions with B cell receptors and influence the functional antibody profile via B cell stimulation and clonal selection. Interestingly, long-lasting ZIKV-reactive IgM responses have been independently identified in two distinct populations (Griffin et al., 2019a; Stone et al., 2020). The presence of ZIKV-specific IgM lasting up to multiple years, when the typical half-life of IgM is 5 days, suggests that ZIKV-reactive IgM expressing B cells are specifically activated and expanded upon ZIKV infection (Lobo et al., 2004). While neutralizing activity is primarily attributed to IgG isotype antibodies, IgM may have an underappreciated role in ZIKV immunity.
The role of ZIKV IgM and IgM-producing B cells is especially understudied in pregnancy, a period of differential immunomodulation where ZIKV infections lead to their greatest disease burden. Early in pregnancy B cells are stimulated to produce IL-10, and subsequently B cell lymphopoiesis is suppressed, which promotes survival of mature B cells and decreases circulating naive B cell frequency (Christiansen et al., 1976; Lima et al., 2016; Nguyen et al., 2013). Retention of the IgM isotype diminishes as B cells differentiate from naive to memory and antibody-secreting cells, which shapes B cell clonal selection in response to infection (King et al., 2021), impacting the magnitude and quality of ZIKV-specific IgM and IgG immunity in pregnancy.
In this study, we evaluated the contribution of plasma IgM to ZIKV neutralization in pregnant women over time and demonstrated that, in pregnancy, plasma IgM contributes to ZIKV neutralization primarily within the first 3 months of infection, regardless of prior exposure to other flaviviruses. We probed the B cell repertoire from peripheral blood of mothers with primary and secondary ZIKV infections and established 9 ZIKV-binding B lymphoblastoid cell lines (B-LCL). One of them produced an IgM antibody, DH1017.IgM, in its native pentameric conformation. DH1017.IgM was somatically mutated, did not cross-react with other flaviviruses, displayed ultrapotent ZIKV neutralization that depended on its isotype and protected mice from viremia more efficiently than the respective IgG upon lethal ZIKV challenge. Structural studies identified a mode of antigen recognition on the ZIKV virion surface compatible with the concurrent engagement of all ten antigen-binding sites, a solution that is not available to antibodies of the IgG isotype. The ultrapotent activity and protection in mice pose DH1017.IgM as a candidate for anti-ZIKV immunotherapy.
RESULTS
IgM contributes to plasma ZIKV neutralization during early phases of infection in pregnant women.
We previously described a cohort of pregnant women from the 2015-2016 ZIKV outbreak in Brazil who presented with ZIKV-like symptoms in pregnancy and were serologically confirmed for ZIKV infection (Singh et al., 2019). Serologic evidence of prior exposure to DENV was demonstrated in most cases, and termed secondary ZIKV (Singh et al., 2019). Both primary (P24 and P54) and secondary (P14, P15, P17, P19, P23, P50, P56 and P73) ZIKV infection cases were included in this study. Eight women were diagnosed during acute infection by serum PCR and two were identified as ZIKV-exposed based on ZIKV and DENV 1-4 neutralization titers (Singh et al., 2019).
Plasma samples were collected between 8 and 406 days post-symptoms (DPS). For subject P73, multiple longitudinal samples were collected that revealed prolonged viremia (up to 42 DPS), while all other subjects were viremic only up to their first visit (Singh et al., 2019). All subjects had high levels of ZIKV-neutralizing plasma antibodies at delivery (median FRNT50 = 4566; range: 650–14,959) (Singh et al., 2019).
ZIKV-binding plasma IgM was present in all 10 subjects, and up to 406 days post-infection in subject P73 (Figure 1A). To evaluate the contribution of plasma IgM to ZIKV neutralization in this cohort, we measured total IgM and IgG concentrations, ZIKV-binding IgG, and ZIKV neutralization of paired samples after mock and IgM depletion (Table S1). Subject P73 displayed the highest percent IgM-mediated contribution to ZIKV plasma neutralization, which started modestly at 8 DPS (7%), then rapidly peaked to 78% at 14 DPS, ranged from 3% to 52% through 112 DPS, and waned thereafter to undetectable levels. (Figure 1B). An important contribution of IgM to plasma ZIKV neutralization was confirmed across all subjects up to 100 DPS regardless of serostatus (i.e., primary or secondary ZIKV infection) (Figure 1B,C). During late convalescence (≥100 DPS), plasma IgM contribution to ZIKV neutralization waned significantly (p<0.05). Late plasma IgM-mediated ZIKV neutralization was detected only in 4 of 7 subjects up to 209 DPS (P23, 28%) and all of them had a prior documented exposure to DENV (Figure 1C).
Figure 1. Contribution of plasma IgM to ZIKV neutralization from acute infection through late convalescence.
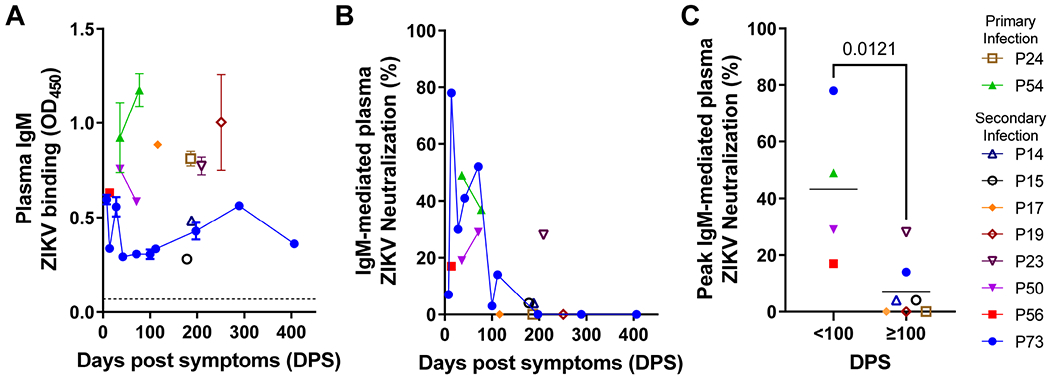
A. Plasma IgM ZIKV binding across ZIKV-infected mothers (n=10) from 8 to 406 days post symptoms (DPS). Plasma was tested at 1:10 dilution in two replicates. Error bars: standard deviation (SD). Dotted line: limit of detection. B. Percent ZIKV neutralization attributable to plasma IgM. Each sample was tested in three replicates. C. Comparison of early (<100 DPS) and late (≥100 DPS) plasma IgM-mediated ZIKV neutralization. When multiple samples from the same individual were collected within each time frame, only peak responses are shown. Statistical significance was measured using an unpaired Mann-Whitney test. P value is shown. See also Table S1 and Figure S1.
The magnitude of ZIKV-reactive plasma IgM and percent IgM contribution to ZIKV neutralization did not correlate (Figure S1), indicating that neutralizing IgM antibodies did not primarily drive the polyclonal ZIKV-reactive IgM plasma response.
Isolation of ZIKV-specific, ultrapotent neutralizing mAb DH1017.IgM.
We next probed the B cell compartment of subject P73 to identify B cells producing ZIKV-neutralizing IgM. In all, we selected four ZIKV-infected pregnant women from the same cohort (Singh et al., 2019), including one woman with primary ZIKV infection (P54: 29 and 77 DPS) and three women with secondary ZIKV infection (P34 at 162 DPS, P56 at 19 DPS, and P73 at 28 and 71 DPS) for a comparative analysis. We enriched for ZIKV-reactive B cells by sorting either unfractionated or memory B cells (MBCs) using a fluorescent ZIKV virion as bait and cultured cells at limiting dilution to derive human B-LCL through EBV transformation as described (Bonsignori et al., 2011, 2018).
Across subjects, ZIKV-reactive B cells constituted 0.6-1.7% of the circulating unfractionated B cell pool (Figure 2A). Regardless of serostatus and time of sample collection relative to symptoms onset, the frequency of ZIKV-reactive MBCs out of total MBCs was uniformly higher than the ZIKV-reactive B cells out of the total unfractionated B cell pool (0.7-2.6% vs 0.6-1.7%; p<0.03, paired t-test) (Figure 2A). On average, ZIKV-reactive memory B cells comprised 52% of total ZIKV-reactive B cells with higher frequencies observed in secondary infections (P73, P56, P34) than in the primary ZIKV infection case (P54) (46-69% vs 34%; Figure 2B), compatible with a re-engagement of pre-existing cross-reactive memory B cells in secondary infection.
Figure 2: Frequencies of ZIKV-reactive B cells.
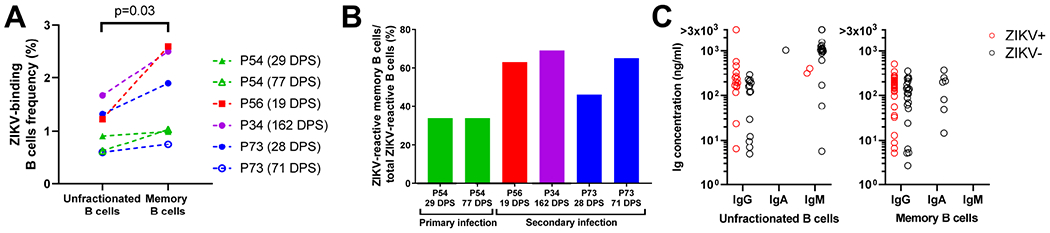
A. Frequency of peripheral blood ZIKV-binding unfractionated and memory B cells. Days-post symptoms (DPS) of PBMC collection are indicated for each pregnant woman. B. Proportion of ZIKV-reactive memory B cells (CD3−/CD14−/CD16−/CD19+/IgD−/ZIKV+) over total B cells (CD3−/CD14−/CD16−/CD19+/ZIKV+). C. Immunoglobulin (Ig) concentration in supernatants of cultured unfractionated (left) and memory (right) B cells, clustered by isotype. Red circles: cultures for which ZIKV specificity was confirmed (OD450 range = 0.44-2.5).
In vitro B cell stimulation across all 4 subjects yielded a total of 97 Ig-secreting culture supernatants, of which 37 were from unfractionated B cells and 60 from memory B cells. Among unfractionated cultures, 13 cultures contained both IgG and IgM, whereas all cultures from memory B cells were of a single isotype. Ig concentrations ranged from 3 to >3000 ng/ml with a geometric mean of 146 ng/ml. Overall, 49 of 97 (50.5%) Ig-secreting culture supernatants confirmed ZIKV reactivity: 17 (15 IgG and 2 IgM) from unfractionated B cells and 32 (all IgG) from memory B cell cultures (Figure 2C). From ZIKV-reactive Ig-secreting cultures, we established 9 B-LCL, 8 of which produced IgG monoclonal antibodies (mAbs) and one, termed DH1017.IgM, produced an IgM mAb.
None of the B-LCLs were clonally related (Table 1). Diverse heavy chain variable gene segments were used and paired with either κ or λ light chains (VL), without VH/VL pairing preferences. CDRH3 lengths ranged between 12 and 22 amino acids (median: 15 amino acids). Overall, VH and VL somatic hypermutation (SHM) frequency ranged from 3.1% to 10.8%, and 1.5% to 8.0%, respectively (Table 1). The VH SHM frequency of sequences obtained from unfractionated B cells and memory B cells was statistically similar (p>0.05, Kolmogorov-Smirnov test). DH1017.IgM used the VH4-31 gene segment paired with Vλ 1-51 and had a 15 amino acid-long CDRH3 (Table 1). While the B cell that expressed DH1017.IgM was isolated from unfractionated B cells and we could not determine its origin from the naïve or memory B cell compartment, the Ig V(D)J SHM frequencies (3.8% for the heavy chain and 3.4% for the light chain) were within the range of other IgG mAbs that we isolated, with 7 nucleotide mutations in the VH and 9 in the VH. All but two of these nucleotide changes were substitution mutations. Nine of the 16 nucleotide substitutions (56%) occurred in canonical activation-induced cytidine deaminase (AID) hotspot motifs with high mutability rates (Yaari et al., 2013). The remaining 7 substitutions occurred in neutral or cold-spot motifs (Table S2).
Table 1.
Immunogenetics of monoclonal antibodies isolated from B-LCLs. See also Table S2.
| Antibody ID | PTID | Source* | VH | D | JH | VH mutation frequency (%) | CDRH3 length (aa) | Isotype | k/l | VL | JL | VL mutation frequency (%) | CDRL3 length (aa) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DH1017.IgM | P73 | UBC | 4-31 | 7-27 | 3 | 3.8 | 15 | M | lambda | 1-51 | 2 | 3.37 | 11 |
| 123-3-G2.IgG | P73 | MBC | 3-30-3 | 3-3 | 4 | 3.1 | 12 | G1 | kappa | 2-30 | 4 | 3.6 | 9 |
| 119-4-D6.IgG | P34 | UBC | 3-64 | 2-2 | 6 | 9.4 | 13 | G1 | lambda | 3-21 | 1 | 7.97 | 11 |
| 119-1-D7.IgG | P34 | UBC | 3-21 | 2-15 | 3 | 8.4 | 17 | G1 | kappa | 3-20 | 2 | 6.74 | 10 |
| 119-5-C5.IgG | P34 | UBC | 3-21 | 6-13 | 6 | 3.1 | 22 | G1 | kappa | 3-11 | 4 | 1.52 | 9 |
| 124-4-C8.IgG | P56 | MBC | 3-30 | 3-16 | 6 | 9.3 | 22 | G1 | kappa | 2-28 | 4 | 3.0 | 10 |
| 124-1-C2.IgG | P56 | MBC | 4-39 | 1-26 | 4 | 7.2 | 15 | G1 | lambda | 2-14 | 3 | 5.2 | 10 |
| 124-2-G3.IgG | P56 | MBC | 3-23 | 3-10 | 4 | 10.8 | 12 | G1 | kappa | 2-30 | 1 | 5.4 | 8 |
| 126-1-D11.IgG | P54 | MBC | 3-48 | 4-17 | 6 | 4.5 | 16 | G1 | kappa | 1-39 | 2 | 3.8 | 9 |
UBC: unfractionated B cells. MBC: memory B cells
MAbs were produced and purified from the 9 B-LCLs for functional characterization. In a native gel, mAb DH1017.IgM yielded two bands at ~970 kDa and ~1048 KDa (Figure 3A), which is compatible with pentameric and hexameric IgM isoforms (Keyt et al., 2020; Wiersma et al., 1998). Negative stain electron microscopy class average analysis supported the presence of both pentameric and hexameric isoforms, with hexamers representing 18% and pentamers representing 73% of all images (Figure 3A).
Figure 3. Characterization of B-LCL-derived ZIKV-specific monoclonal antibodies.
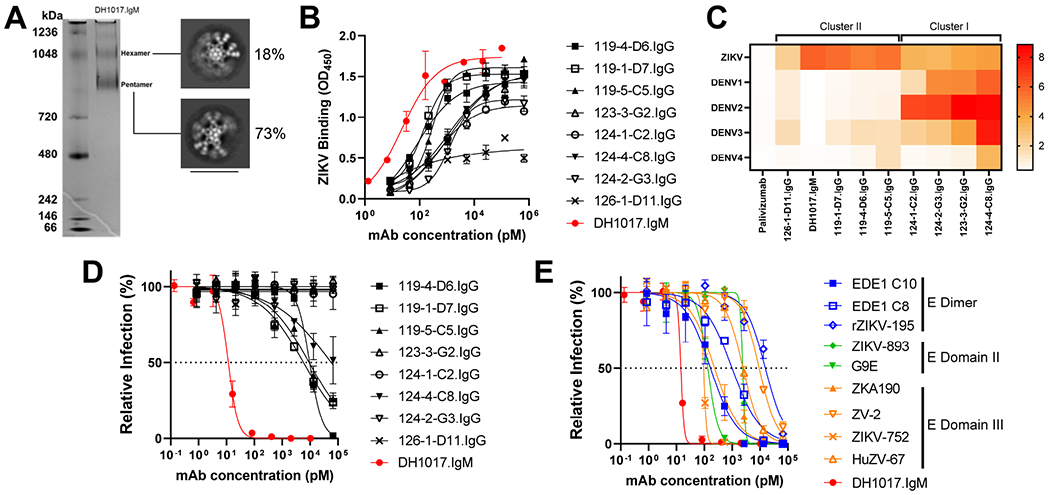
A. Left: DH1017.IgM native PAGE gel in non-reducing conditions. Ladder on the left. Right: Negative stain electron microscopy of purified DH1017.IgM showing representative class averages of hexameric and pentameric particles. Scale bar is 40nm. B. Binding to whole ZIKV PRVABC59 virions by B-LCL-derived mAbs (n=9). DH1017.IgM is shown in red. Error bars: SD of two replicates. Data representative of duplicate experiments. C. Heatmap showing binding to ZIKV and DENV serotypes 1-4 virions by the nine B-LCL-derived mAbs. Palivizumab: negative control. D. ZIKV PRVABC59 neutralization by the 9 B-LCL-derived mAbs expressed as percentage of the number of foci relative to the virus alone condition. Dotted line: 50% viral inhibition. Error bars: SD of three replicates. Data representative of duplicate experiments. E. ZIKV PRVABC59 strain neutralization curves of DH1017.IgM (red) and 9 previously described IgG ZIKV neutralizing mAbs. Error bars: SD of three replicates. Data representative of duplicate experiments. See also Figure S2.
All purified mAbs confirmed binding to ZIKV. While 126-1-D11.IgG bound weakly even at high concentrations (LogAUC = 2.0), all other mAbs bound with LogAUC ranging from 2.7 to 5.9, with the strongest ZIKV-binding mAb being DH1017.IgM (Figure 3B). Since plasma antibody displayed substantial cross-reactivity with DENV in all subjects except primary infection subject P54 (Figure S2A), we determined cross-reactivity of the nine mAbs with DENV 1-4 strains. Based on their binding profile, the mAbs segregated into two clusters (Figure 3C). Cluster I comprised mAbs 123-3-G2.IgG, 124-4-C8.IgG, 124-1-C2.IgG, and 124-2-G3.IgG that bound to one or more DENV 1-4 strains better than to ZIKV. These mAbs were isolated from memory B cells of mothers with secondary ZIKV-infection (P73 and P56), which parallels the plasma cross-reactivity profile and further implies re-engagement of pre-existing DENV-reactive memory B cells. Conversely, cluster II mAbs bound more strongly to ZIKV than to DENV, for which cross-reactivity was either at or below limit of detection, which suggests that these mAbs may constitute a de novo immune response to ZIKV. Notably, DH1017.IgM did not cross-react with or neutralize any of the four DENV serotypes (Figure 3C and Figure S2B).
Five of the nine ZIKV-binding mAbs mediated neutralization of the ZIKV PRVABC59 strain (Figure 3D). Mab 124-4-C8.IgG neutralized weakly with FRNT50 >10 μM. Mabs 119-1-D7.IgG, 119-5-C5.IgG and 119-4-D6.IgG neutralized with FRNT50 = 5.8 nM, 9.2 nM and 9.7 nM, respectively. Remarkably, DH1017.IgM neutralized ZIKV with ~500- to 1000-fold higher potency (FRNT50=12 pM) (Figure 3D). DH1017.IgM neutralization of the ZIKV PRVABC59 strain was repeated multiple times (n=10) and potent neutralization was independently confirmed by multiple operators (geometric mean [GM] FRNT50 = 12 pM; range: 4-31 pM). DH1017.IgM also neutralized the ZIKV H/PF/2013 strain with comparable potency (GM FRNT50 = 14 pM; range: 8-26 pM) (Figure S2C). We compared DH1017.IgM potency with a panel of well characterized ZIKV-neutralizing IgG mAbs: EDE1 C8, EDE1 C10, ZIKV-893, rZIKV-195, G9E, ZV-2, ZIKV-752, ZKA190, and HuZV-67 (Collins et al., 2019; Gilchuk et al., 2020; Hasan et al., 2017; Long et al., 2019; Rouvinski et al., 2015; Sapparapu et al., 2016; Wang et al., 2017; Zhao et al., 2016). In this side-by-side comparison, DH1017.IgM neutralized 8 to >10,000 fold more potently than mAbs in the reference panel (FRNT50 range: 95 pM–15.5 nM) (Figure 3E). Neutralizing antibodies with IC50 <10 ng/ml (i.e., 66.7pM) are defined as ultrapotent (Smith et al., 2015). Hence, DH1017.IgM meets the definition of ultrapotent ZIKV-neutralizing antibody.
Impact of isotype on DH1017.IgM functions.
We recombinantly produced the DH1017 Fab and IgG1 isoforms (DH1017.Fab and DH1017.IgG) to study the impact of antibody isotype on function. First, we assessed the ability to bind to whole Zika PRVABC59 virion. DH1017.Fab only interacted weakly with ZIKV (EC50 >2 mM) (Figure 4A). DH1017.IgG bound with EC50 = 112 pM (Figure 4B) whereas DH1017.IgM bound with EC50 = 22 pM (Figure 4C). Critically, DH1017.IgM neutralization potency (FRNT50 = 9 pM) was >40-fold higher than DH1017.IgG (FRNT50 = 366 pM), whereas DH1017.Fab yielded a shallow neutralization curve (FRNT50 = 93 nM) with 10,000-fold worse potency than DH1017.IgM (Figure 4D). These data demonstrate that DH1017.IgM in vitro ultrapotent neutralization depended on the IgM isotype.
Figure 4. Functional characterization of DH1017.IgM, DH1017.IgG, and DH1017.Fab.
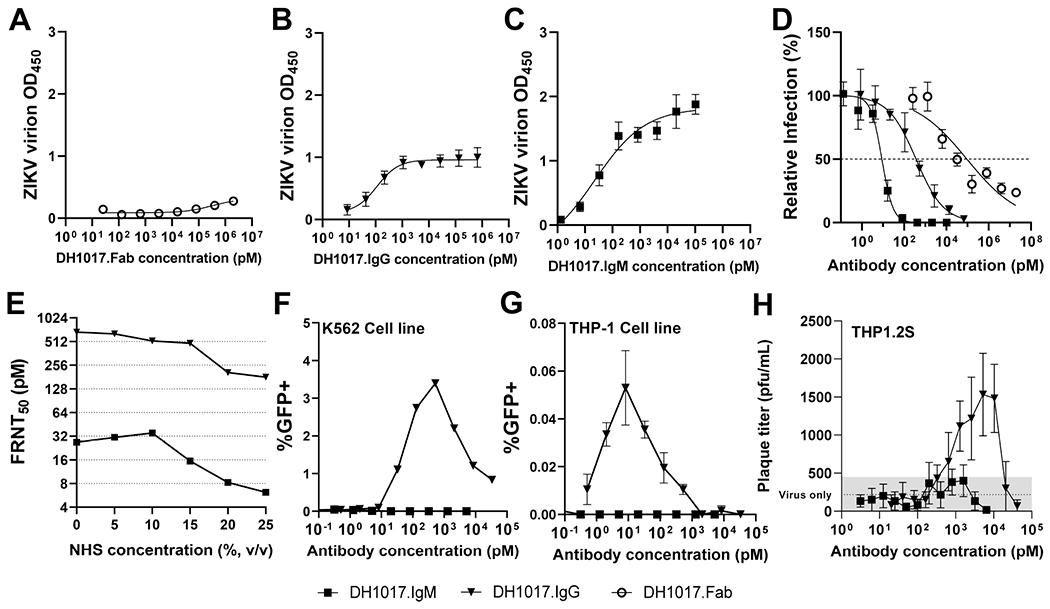
ZIKV virion binding of DH1017.Fab (A), DH1017.IgG (B) and DH1017.IgM (C). Error bars: SD of two experiments with 2 (DH1017.Fab) or 6 (DH1017.IgM and DH1017.IgG) replicates each. D. ZIKV neutralization of Fab, IgG and IgM versions of DH1017. Dotted line: 50% relative infection compared to virus alone (FRNT50). Error bars: SD from 3 replicates. Data representative of duplicate experiments. E. DH1017.IgM and DH1017.IgG-mediated FRNT50 (y-axis) over increasing complement concentrations supplemented through Normal Human Serum (NHS) (x-axis). MAbs were run in triplicate. F, G. Antibody-dependent enhancement (ADE) of infection using ZIKV H/PF/2013 reporter virus particles on K562 (F) and THP-1 (G) cells. Data representative of at least three experiments. H. ADE tested on THP1.2S cells measured by plaque assay. Error bars: SD from 6 replicates. Dotted line: virus-only control (grey area: ≤1 SD). See also Figure S3.
To test the contribution of the Fc region of DH1017.IgM to its binding, we compared the DH1017.IgM and DH1017.IgG binding kinetics to ZIKV E dimer using a BLI format with immobilized mAb. Under these conditions, DH1017.IgM and DH1017.IgG displayed similar dissociation kinetic (koff = 1.03 and 0.91 s−1x10−2, respectively) and affinity (Kd = 106 and 123 nM, respectively). We also confirmed that DH1017.IgG retained ZIKV-specificity and lacked cross-reactivity with DENV1-4, as observed for DH1017.IgM (Figure S3A). Thus, the Fc region of DH1017 does not impact affinity or alter cross-reactivity.
Since antibody-mediated complement deposition can reduce the amount of mAb required to neutralize virus, we tested the effect of complement from normal human serum (NHS) on DH1017.IgG and DH1017.IgM neutralizing activities (Mehlhop et al., 2009). Exogenous human complement from NHS enhanced ZIKV neutralization potency of both mAbs in a dose-dependent manner (Figure 4E). At all doses of NHS tested (5-25% v/v), DH1017.IgM retained more potent neutralization compared to DH1017.IgG. The largest improvement in neutralization potency was observed at 25% NHS with 4.3-fold and 3.7-fold increase in potency for DH1017.IgM and DH1017.IgG, respectively (Figure 4E). These data demonstrate that neutralization potency of the DH1017 mAb in its IgG and IgM isoforms is enhanced in presence of complement.
Sub-neutralizing concentrations of flavivirus-neutralizing IgG can mediate antibody-dependent enhancement (ADE) of viral infection through interactions of immune complexes with cell membrane-anchored FcγR, particularly on monocytes (Chan et al., 2015; Halstead, 1988; Halstead and O’Rourke, 1977). DH1017.IgG mediated ADE in both K562 and THP-1 cells whereas DH1017.IgM did not (Figures 4F,G). DH1017.IgM also did not mediate ADE in THP1.2S cells, a subclone of THP-1 cells with increased sensitivity to ADE (Chan et al., 2014) (Figure 4H). Similarly, DH1017.IgM did not mediate ADE in primary monocytes (Figure S3B). Finally, to further characterize the safety profile of DH1017.IgM, DH1017.IgG and DH1017.Fab, we measured binding to nine autoantigens associated with autoimmune diseases. All isotypes tested negative for reactivity to all autoantigens (Figure S3C). Further, they did not demonstrate intracellular immunofluorescent staining of Hep-2 cells (Figure S3D). Thus, DH1017.IgM does not mediate in vitro ADE like its IgG isoform, and the DH1017 clone does not react with human autoantigens.
DH1017.IgM controls Zika viremia in mice more efficiently than DH1017.IgG.
We sought to evaluate whether DH1017.IgM could protect against ZIKV infection in mice. We administered 103 pmol (100 μg) of DH1017.IgM or a non-ZIKV binding human IgM (DH1036.IgM) to Ifnar1−/− mice 1 day prior and 1 day following infection with 1000 focus forming units (FFU) of ZIKV H/PF/2013 and measured viremia and lethality. DH1017.IgM conferred protection from lethal challenge to all treated mice (n=7) and controlled viremia to the limit of detection (3.2-3.6 Log10 viral copies/mL). In contrast, all mice in the control IgM group (n=4) developed high viremia (6.3-6.9 Log10 viral copies/mL) and succumbed to infection (Figure 5A,B). Human IgM was maintained in vivo at detectable levels up to 4 days post challenge in both groups, or 3 days after the last administration (Figure 5C). Next, we dosed down DH1017. IgM to define its limit of in vivo protection. At half dosage (51.5 pmol, 50 μg), DH1017.IgM still protected from lethality but protection from viremia was incomplete (4.3-6.1 Log10 viral copies/ml in 3 of 5 mice) (Figure 5A,B). Thus, DH1017.IgM protects mice from lethal ZIKV challenge when administered at 103 or 51 pmol/dose prior to infection, but controls viremia more effectively at the higher dose.
Figure 5: DH1017.IgM protects mice more efficiently than DH1017.IgG against viremia upon lethal ZIKV challenge.
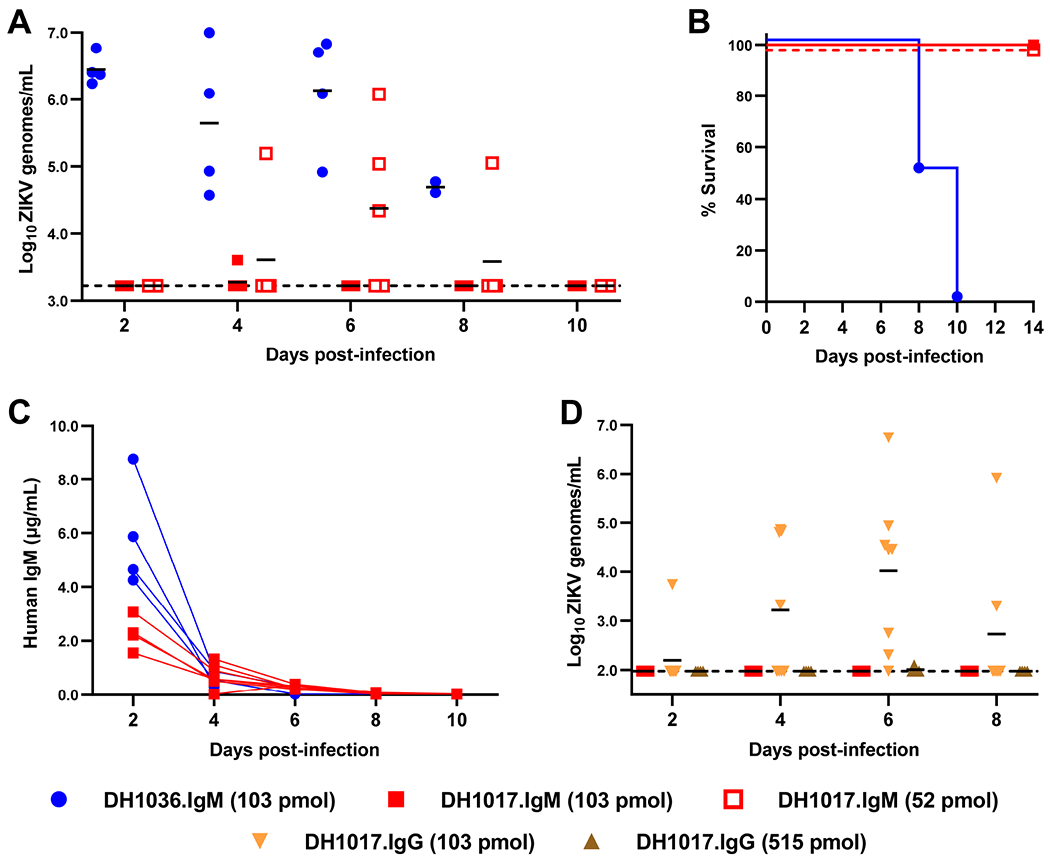
A. Serum viral load in ZIKV infected 5-week-old Ifnar1−/− mice treated with DH1017.IgM at 103 pmol/dose (n=7) or 52 pmol/dose (n=5), or non-ZIKV reactive mAb DH1036.IgM (103 pmol/dose, n=4). Line at mean. Dotted line at limit of detection. B. Survival curves for each IgM intervention group. C. Human IgM serum concentrations in mice treated with 103 pmol/dose of DH1017.IgM or DH1036.IgM measured up to 10 days post-infection. D. Serum viral load in mice receiving of DH1017.IgG at 103 pmol/dose (n=8) or 515 pmol/dose (n=4). Additional 4 mice treated with 103 pmol/dose DH1017.IgM are shown. Line at mean. Dotted line at limit of detection.
To evaluate the impact of isotype on protection and viral control, we tested DH1017.IgG at the equimolar amount of the DH1017.IgM optimal dose (103 pmol). While DH1017.IgG still protected from lethality, it failed to protect from viremia: all treated mice (n=8) developed high viral loads (3.7-6.7 Log10 virus copies/ml; Figure 5D). However, DH1017.IgG protected from viremia when administered at 515.4 pmol/dose (n=4), which is a 5-fold higher amount than the protective dose of DH1017.IgM and matches the number of antigen-binding sites in the administered antibody.
Thus, pentameric DH1017.IgM protects against ZIKV disease in mice and controls viremia more efficiently than the bivalent DH1017.IgG. These data demonstrate that superior viral control in vivo is conferred by the multivalent isotype of DH1017.IgM.
Structural characterization of the DH1017 epitope and mode of antigen recognition.
First, we characterized the DH1017 epitope using cryo-electron microscopy (cryo-EM) and single particle reconstruction. A set of 4104 ZIKV H/PF/2013 particles bound to DH1017.Fab were identified and used to generate a cryo-EM density map. A surface shaded view of the virus-Fab complex is shown in Figure 6A and demonstrates the sites of Fab binding to the surface of the particle. The cryo-EM density map resolution is 5.3 Å as determined from the Fourier Shell Correlation co-efficient 0.143 (Figure S4A). The Cα backbone of the E glycoprotein ectodomains from the ZIKV asymmetric unit (PDB 6CO8; Sevvana et al., 2018) is composed of three E monomers comprising chains A, C, and E (Figure 6B). Their position in the density map is shown in Figure S4B. In this asymmetric unit, Chain A lies alongside the antiparallel homodimer formed by chains C and E (Figure 6B). The fitted asymmetric unit is repeated 60 times within the density map demonstrating icosahedral symmetry like previous Zika virus structures (Sevvana et al., 2018; Sirohi et al., 2016).
Figure 6. DH1017 clone interacts with envelope dimer.
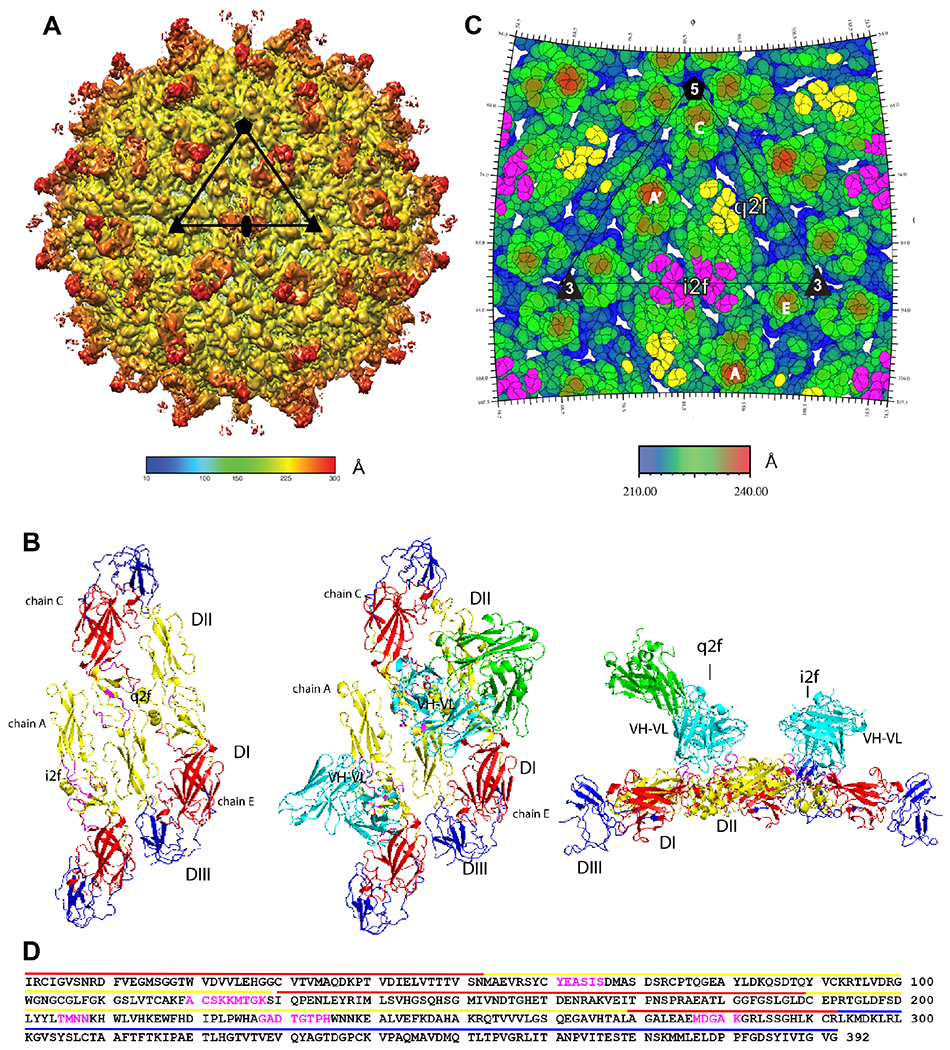
A. Surface-shaded view of the Zika virion bound with the DH1017 Fab. Shading represents the distance to the virion center (scale bar in Å). Black triangle: asymmetric unit; pentagon: five-fold axis; triangle: three-fold axis; oval: two-fold axis. B. Surface representation of the ZIKV E ectodomain asymmetric unit (PDB 6CO8) shown in top view unbound (left), bound with the DH1017.Fab variable (cyan) and constant (green) domains (middle), and in bound side view (right). E ectodomain DI: red, DII: yellow, and DIII: blue. DH1017.Fab footprint residues: magenta. C. Radially colored roadmap (scale bar in Å). The icosahedral and quasi two-fold axes are labelled i2f and q2f, respectively. The monomer chains of two E dimers are labeled A’ and A at the i2f, and C and E at the q2f. Residues on the surface of the virus within 6 Å of the variable domain structure fit to the density map are colored yellow on chain C at the q2f axis and magenta on chain A’ and A at the i2f axis. D. Fab DH1017 epitope shown on the primary sequence of E ectodomain. Epitope residues: magenta. Domains DI, DII, and DIII: red, yellow and blue lines, respectively. See also Figures S4, S5, S6 and Table S3.
The Cα backbone of the homology model of the DH1017.Fab was fit into the cryo-EM density map (Figure S4B). It shows that the DH1017 variable domain interacts primarily with DII of all three E monomers in the asymmetric unit, and at the interface of DII and DI on chains A and C (Figure 6B,S4B). The DH1017.Fab epitope footprint was defined by residues of the E ectodomain glycoprotein on the surface of the virus within 6 Å from the Cα backbone of the DH1017 variable domain structure (Table S3, Figure 6B,C,D). The DH1017.Fab paratope footprint was defined by Fab residues within 6 Å from the Cα backbone of the E ectodomain glycoprotein of the virus surface and included all three heavy chain CDRs and FRH3 (Table S3).
There are two 2-fold axes of symmetry on the virus surface: the icosahedral 2-fold (i2f) axis delineates symmetry at the juncture of two antiparallel asymmetric units on the repetitive virion surface between A and A’, whereas the quasi 2-fold (q2f) axis describes symmetry within the antiparallel E dimer between chains C and E (Figure 6C). Each asymmetric unit contains two epitope footprints, at the i2f and q2f symmetry axes, respectively (Figure 6C). At the i2f axis one half of the epitope footprint is located on chain A’ and the other half of on chain A (Figure 6C), whereas at the q2f axis, most epitope residues are on chain C, with some on chain E (Figure 6C, S4B). At the i2f axis, one bound Fab will exclude the other i2f site related by two-fold symmetry from being bound. At the q2f axis, the position on chain C and E is fully occupied. The result is an occupancy of 1.5 Fab per asymmetric unit.
Cryo-EM density data at the q2f axis identified as the Fab constant and variable domains occupied a single position (Figure 6A,S4B). However, two distinct positions of Fab constant domain densities could be identified at the i2f axis (Figure 6A,S4B). Hence, at the i2f axis the epitope can be approached from two angles but at the q2f it is only approached at one angle. At the i2f axis, only one apparent position for variable domain density was identified. It is most likely an average of two distinct variable domains bound to the virus surface at different constant domain angles (Figure S4B).
A top-down view of the virus particle shows that Fab densities at the q2f and i2f axes form a ring around the 5-fold axis of symmetry (i5f) with the constant domains pointing toward the i5f axis (Figure S4C). Computational modeling of the DH1017.IgM pentamer (PDB 2CRJ) above the surface of the virion suggests the Fc domain can be centered and positioned above the virion i5f axis with the Cμ3 ends above the constant domains of the Fabs (Figure S5A). The IgM Cμ2 domains are flexible and can bend up to 90° relative to the planar Cμ4-Cμ3-J portion, facilitating a bent umbrella-like conformation (Keyt et al., 2020; Murphy et al., 2012; Sharp et al., 2019). The flexibility at Cμ2 domain suggests that the arms of the DH1017.IgM pentamer can bend toward the surface of the virus and that each arm can contact the epitopes at q2f and i2f of neighboring asymmetric units (Figure S5A). Such arrangement of the epitopes on the virion surface may allow Fab pairs of a pentamer arm to contact epitope pairs on the virus surface either at an i2f and q2f axes between neighboring asymmetric units or at the i2f and q2f axes within the same asymmetric unit simultaneously, enabling a decavalent mode of epitope recognition. Hence, the DH1017 IgM pentamer can contact up to five epitope pairs compared to a single epitope pair for the bivalent DH1017.IgG.
We experimentally validated this multivalent mode of antigen recognition by applying subsaturating concentrations of DH1017.IgM to Zika virion and examining the electron micrographs (Figures S5B,C). We observed large and discrete density patches lying parallel to the virion surface (Figure S5C, red arrows). This configuration contrasts with the uniform decoration of the virion surface observed with DH1017.Fab (Figure 6A) and is compatible with the predicted multivalent mode of antigen recognition shown in Figure S5A. In this bent conformation, a theoretical maximum of three pentameric IgM can bind concurrently to the same virion, occupying 30 of the 90 epitopes present on the virion surface (Figure S5D). The simultaneous engagement of multiple E dimers at the 5-fold axis of viral symmetry represents a mechanism of IgM-mediated antigen recognition of ZIKV that is not available to an IgG molecule.
In addition, we observed a second distinct mode of antigen recognition. In a planar pentamer conformation, DH1017.IgM may also bind one or more pairs of epitopes on different virus particles, effectively cross-linking multiple virions into aggregates (Figure S6A). The electron micrographs revealed virus particle aggregates with interspersed densities outside the virion compatible with IgM mAbs in planar conformation (Figure S6B). In contrast to the multivalent mode of antigen recognition, these densities were oriented perpendicularly to the virion surface and bridged adjacent virions (Figure S6B, blue arrow). Further, we observed both the multivalent and aggregation modes of antigen recognition on the same virus particle (Figure S6B, red arrow) indicating that these two modes of antigen recognition are not mutually exclusive.
DISCUSSION
In this study, we evaluated the contribution of IgM responses to ZIKV neutralization in pregnant women, and described a ZIKV-neutralizing IgM monoclonal antibody, DH1017.IgM in its native multimeric form. Unlike previous investigations of plasma IgM, the isolation of a human IgM mAb enabled direct characterization of the impact of the IgM isotype on antibody function against ZIKV. DH1017.IgM ultrapotent ZIKV neutralization depended on its isotype. Importantly, DH1017 protected mice from lethal ZIKV challenge and the pentameric IgM mAb controlled viremia more efficiently than the monomeric IgG. We mapped the footprint of the DH1017 discontinuous epitope within the asymmetric unit and identified modes of antigen recognition not available to IgG. These findings identify a functional niche for IgM antibodies in protection against ZIKV.
The humoral response to ZIKV infection, like other flaviviruses, is characterized by the development of persistent IgM serum antibodies (Gibney et al., 2012; Griffin et al., 2019a; Monath, 1971; Stone et al., 2020). While persisting ZIKV-IgM antibodies can contribute to ZIKV neutralization (Calvert et al., 2021; Malafa et al., 2020, Stone et al., 2020), prior studies did not focus on pregnancy. ZIKV infections during pregnancy are of particular concern due to the risk of congenital transmission and subsequent burden of disease in children. In our cohort of pregnant women, circulating IgM antibodies were present up to 408 days post-symptoms, and contributed to ZIKV neutralization primarily within the first 3 months of infection. The temporal association between clearance of ZIKV infection (10-14 days post infection) in most individuals (Lessler et al., 2016; Paz-Bailey et al., 2018) and the early peak of serum IgM, which precedes peak IgG responses (Ravichandran et al., 2019; Tonnerre et al., 2020), are compatible with early IgM neutralizing responses playing a role in early control of infection. Our data extend such temporal association in the setting of pregnancy.
Subject P73, from whom DH1017.IgM was isolated, had prolonged viremia that outlasted the first peak of relative IgM contribution to neutralization at 14 DPS. A second peak of relative IgM contribution to neutralization ensued at 71 DPS, and this is the timepoint from which DH1017.IgM was isolated. It is possible that the DH1017 clone may have emerged after the first peak of relative IgM contribution to neutralization and aided in resolution of viremia. However, the relationship between quality of plasma IgM and control of prolonged viremia should be further investigated.
While serum IgM in flavivirus infection is being studied, ZIKV-specific IgM mAbs have not previously been characterized. Thus, characterization of DH1017.IgM addresses this gap in understanding of the mechanisms of action of this isotype against ZIKV. While subject P73 had a prior exposure to DENV, DH1017.IgM did not bind to DENV and was therefore not part of a recall response from pre-existing DENV immunity. The original DH1017 B cell was sorted from unfractionated B cells, which included naïve, and both class-switched and non-class-switched memory B cells. DH1017.IgM was somatically mutated and, while naive B cells can undergo SHM and class-switch in vitro in culturing conditions similar to our study (Heath et al., 2012), we did not identify additional nucleotide mutations or class-switched DH1017.IgG mAb from the DH1017.IgM LCL throughout multiple rounds of cell expansions. Hence, while we cannot exclude an origin from a naïve B cell, these observations suggest that DH1017.IgM LCL was derived from an IgM-expressing memory B cell. We speculate that the non-class switched IgM+ memory B cell and IgM+ plasma cell pools provide a source of neutralizing antibodies that interact with virions in an isotype-exclusive manner. Future studies should address if the IgM+ memory B cell compartment is a preferential source of ZIKV IgM neutralizing antibodies.
DH1017.IgM binds to a quaternary epitope in the E dimer that is primarily Domain II. While this epitope differs from that of known potently ZIKV-neutralizing mAbs, recognition of a quaternary epitope on ZIKV is the defining feature of a class of potently neutralizing IgG mAbs (Collins et al., 2019; Hasan et al., 2017; Long et al., 2019; Rogers et al., 2017; Sapparapu et al., 2016; Stettler et al., 2016). Upon viral entry into cellular endosomes, E dimers rearrange into fusogenic trimers to fuse with host membrane (Smit et al., 2011) and DH1017.IgM binding may cross-link ZIKV E monomers and restrict conformational changes to prevent infection (Zhang et al., 2015). Such mechanism has also been proposed for ZIKV-117 and ZIKV-195 IgG mAbs (Hasan et al., 2017; Long et al., 2019).
Importantly, the superior in vitro and in vivo activity of DH1017.IgM to DH1017.IgG demonstrated a functional advantage conferred by the IgM isotype, supported by its multimeric structure and ability to bind to virions with high orders of valency. Computational modelling and NSEM micrographs showed that five E dimers could be bound concurrently through an IgM-exclusive decavalent antigen recognition. While competition with other ZIKV-binding antibodies in vivo may interfere with decavalent binding of a single virion, this mode of antigen recognition may outcompete other less avid IgG mAbs. Intriguingly, the arrangement of the DH1017 epitope relative to the five-fold axis of symmetry on the virion surface is critical for multivalent antibody interactions. Whether neutralizing epitopes other than the DH1017 epitope are permissive to multivalent interactions needs to be determined. In addition to the effect of multivalent binding, the large molecular size of an IgM may interfere more efficiently than an IgG with virion attachment to the cell membrane and prevent cell entry by making the particle more rigid. This can sterically hinder interactions with the cell surface to prevent binding or fusion with the host cell, even with a less than decavalent mode of binding. Finally, viral aggregation is another potential mode of neutralization by which aggregated virions would not efficiently internalize into cells via endosomes. NSEM micrographs of a ZIKV aggregate in presence of DH1017.IgM bridging virions supports this mode of binding. That a 5-fold excess dose of DH1017.IgG achieved a biological effect comparable to DH1017.IgM further validates that the main determinant of DH1017.IgM ultrapotency is its multivalent isotype. Altogether, our studies suggest a functional niche that is exclusive to IgM in the context of pathogens with repetitive proximal structures.
Neutralizing antibodies mediate protection against ZIKV as demonstrated by passive transfer of immune plasma in the non-human primate model (Larocca et al., 2016; Richner et al., 2017). However, IgG neutralizing antibodies at sub-neutralizing concentrations can enhance flavivirus replication via ADE through FcγR mediated endocytosis of the immune complex. In vitro, DH1017.IgG mediated ADE, whereas DH1017.IgM did not. We note that these experimental results may be influenced by differences in the relative expression of FcγR and FcμR/Fcα/μR on cell surface. Nonetheless, DH1017.IgM in vitro neutralization potency is increased in the presence of complement in a dose-dependent manner. Since complement lowers the stoichiometry requirements for neutralization (Mehlhop et al., 2009), and DH1017.IgM is already potently neutralizing, the risk of DH1017.IgM-mediated ADE in vivo may be further reduced. Importantly, DH1017.IgM-mediated protection against lethal ZIKV challenge in mice recapitulates protection and viral control conferred by potent IgG neutralizing antibodies (Collins et al., 2019; Long et al., 2019; Robbiani et al., 2017; Sapparapu et al., 2016; Swanstrom et al., 2016).
Since efficacy trials for ZIKV vaccines are limited by currently low levels of endemic circulation, therapeutic interventions that can be safely deployed to pregnant women to mitigate the risks of congenital ZIKV transmission are urgently needed. We propose that DH1017.IgM may be suitable as a passive intervention for protection against ZIKV infection. In particular, since IgM isotype antibodies are not transferred across the placenta, the potential risks of fetal toxicity and ADE in infancy after transplacental IgG transfer are expected to be mitigated. Thus far, 20 IgM mAb interventions have been tested in clinical trials and demonstrate that infused IgM are safe and well tolerated (Keyt et al., 2020). The IgM half-life is considerably shorter than IgG. Consequently, antibody engineering to prolong the half-life would be likely required for effective prophylactic countermeasures. However, the potential issue of short half-life would be less relevant for a therapeutic intervention, administered at the time of diagnosis, aimed at rapid clearance of viremia and reduction in the time of fetal exposure to circulating virus. Further, our findings provide proof of concept for the development of engineered multimeric antibody formulations as potential prophylactic and therapeutic strategies.
In summary, our demonstration of the importance of IgM to early ZIKV neutralization during pregnancy and isolation of this ultrapotent ZIKV-neutralizing IgM mAb that protected mice from viremia upon lethal challenge revealed that the ultrapotent in vitro function and efficient in vivo viral control depended on the IgM isotype. We defined a conceptual framework by which the spatial arrangement of quaternary epitopes on the virion enables DH1017.IgM to access distinct modes of antigen recognition in an isotype-specific manner. As congenital transmission of ZIKV in pregnancy is the source of the greatest burden of ZIKV disease, further studies are warranted to assess whether DH1017.IgM can protect against fetal infection in pregnant animal models and mitigate fetal damage by rapidly reducing maternal viremia (Van Rompay et al., 2020). With the experience of delayed roll out of vaccines to pregnant women for emerging outbreaks such as Ebola and more recently the SARS-CoV-2 pandemic (Craig et al., 2021; Gomes et al., 2017), prophylactic and therapeutic interventions that are rapid, protective, and safe for use pregnancy will be particularly needed when ZIKV re-emerges.
Limitations of the study:
This study focused on prototypic ZIKV ultrapotent IgM monoclonal antibody DH1017.IgM isolated during pregnancy. Evaluating the prevalence of DH1017-like responses at population level, both in and outside pregnancy, was outside the scope of the current report and will require additional adequately powered studies.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact.
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Mattia Bonsignori (mattia.bonsignori@nih.gov).
Materials availability.
The research materials used in this study are available, contingent to product availability, upon written request and subsequent execution of an appropriate materials transfer agreement.
Data and code availability.
Sequences of Ig V(D)J rearrangements have been deposited in GenBank and are publicly available as of the date of publication. Accession numbers are listed in the key resource table. The Zika virus glycoprotein E and DH1017.Fab structures have been deposited in Protein Data Bank and are publicly available as of the date of publication. The accession number is listed in the key resource table. The density map of DH1017.Fab in complex with Zika virions has been deposited in EMDataResource and is publicly available as of the date of publication. The accession number is listed in the key resource table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| IgD-PE (clone IA6-2) | BD Biosciences | 555779 |
| CD10 PE-CF594 (clone HI10a) | BD Biosciences | 562396 |
| CD3 PE-Cy5 (clone HIT3a) | BD Biosciences | 555341 |
| CD14 BV605 (clone M5E2) | Biolegend | 301834 |
| CD16 BV570 (clone 3G8) | Biolegend | 302035 |
| CD27 PE-Cy7 (clone O323) | Thermo Fisher Scientific | 25-0279-42 |
| CD38 APC-AF700 (clone LS198-4-3) | Beckman Coulter | B23489 |
| CD19 APC-Cy7 (clone SJ25C1) | BD Biosciences | 557791 |
| (HRP)-conjugated goat anti-human IgG antibody | Jackson ImmunoResearch Laboratories | 109-035-008 |
| HRP-conjugated goat anti human IgM antibody | Jackson ImmunoResearch Laboratories | 109-035-129 |
| HRP-conjugated goat anti-human IgA antibody | Jackson ImmunoResearch Laboratories | 109-035-011 |
| C10 | Absolute Antibody | AB00677-10.0 |
| Anti NS1 IgM | myBiosource | MBS6120634 |
| Palivizumab | Duke Pharmacy, Durham NC | N/A |
| 1M7 | Smith et al., 2013; Produced from a hybridoma cell line at the UNC Protein Expression Core Facility | N/A |
| ZV-2 | Zhao et al., 2016; Produced from a hybridoma cell line at the UNC Protein Expression Core Facility | N/A |
| ZV-64 | Zhao et al., 2016; Produced from a hybridoma cell line at the UNC Protein Expression Core Facility | N/A |
| ZKA190 | Wang et al., 2017; Recombinantly produced by the UNC Protein Expression Core Facility | N/A |
| ZIKA-752 | Kindly provided by Dr. James Crowe, Vanderbilt University | N/A |
| ZIKA-893 | Kindly provided by Dr. James Crowe, Vanderbilt University | N/A |
| rZIKV-195 | Kindly provided by Dr. James Crowe, Vanderbilt University | N/A |
| DH1017.IgG | Recombinantly produced in this study | N/A |
| DH1017.Fab | Recombinantly produced in this study | N/A |
| 4G2 | ATCC | HB-112 |
| humanized IgG1 4G2 | Produced in-house | N/A |
| 3H5 | ATCC | HB-46 |
| ChromPure Human IgM (myeloma), whole molecule (2 mg) | Jackson ImmunoResearch Laboratories | 009-000-012 |
| goat anti-human Ig polyvalent antibody | Thermo Fisher Scientific | H17000 |
| goat anti-human H+L | Promega | W403B |
| IgG1 mAb 2F5 | Produced in-house | N/A |
| IgG1 mAb 17B | Produced in-house | N/A |
| Anti-Centromere B | Immunovision | HCT-0100 |
| Anti-single stranded DNA | Immunovision | HSS-0100 |
| Anti-Histone | Immunovision | HIS-0100 |
| Anti-Jo 1 | Immunovision | HJO-0100 |
| Anti-SRC | Immunovision | HRN-0100 |
| Anti-Scl 70 | Immunovision | HSC-0100 |
| Anti-Sm | Immunovision | HSM-0100 |
| Anti-SSA | Immunovision | HSA-0100 |
| Anti-SSB | Immunovision | HSB-0100 |
| Bacterial and virus strains | ||
| DENV1 (WestPac74) | Kindly provided by Dr. Aravinda de Silva, UNC | N/A |
| DENV2 (S-16603) | Kindly provided by Dr. Aravinda de Silva, UNC | N/A |
| DENV3 (CH54389) | Kindly provided by Dr. Aravinda de Silva, UNC | N/A |
| DENV4 (TVP-360) | Kindly provided by Dr. Aravinda de Silva, UNC | N/A |
| ZIKV (PRVABC59) | BEI Resources | NR-50240 |
| DENV1 (Hawaii NR-82) | BEI Resources | NR-82 |
| DENV2 (New Guinea C) | BEI Resources | NR-84 |
| DENV3 (Philippines) | BEI Resources | NR-80 |
| DENV4 (H241) | BEI Resources | NR-86 |
| ZIKV (PD13/251013-18) | Kindly provided by Dr. Didier Musso, IHU Mediterranee Infection Foundation, Tahiti, French Polynesia | N/A |
| ZIKV (H/PF/2013) | US Centers for Disease Control and Prevention; Baronti et al., 2014 | N/A |
| Biological samples | ||
| Human plasma and PBMCs from pregnant women with ZIKV infection | IRB-Approved prospective cohort led by Dr. S.R. Permar and Dr. R. Dietze | N/A |
| Normal human serum from human male AB plasma, USA origin, sterile-filtered | Sigma | H4522 |
| Chemicals, peptides, and recombinant proteins | ||
| ZIKV RVP | Dowd et al., 2016 | N/A |
| ZIKV envelope protein dimer | Premkumar L et al., 2018; recombinantly produced in-house | N/A |
| DNA | Worthington | LS002105 |
| Centromere B | Prospec | PRO-390 |
| Histone | Sigma | H9250 |
| Jo-1 | Immunovision | JO1-3000 |
| RnP/Sm | Immunovision | SRC-3000 |
| Scl-70 | Immunovision | SCL-3000 |
| Sm | Immunovision | SMA-3000 |
| SSA | Immunovision | SSA-3000 |
| SSB | Immunovision | SSB-3000 |
| Poly-L-Lysine | Sigma | P6282 |
| Critical commercial assays | ||
| ANA Hep-2 test system | Zeuss Scientific | FA2400 |
| Deposited data | ||
| Zika virus glycoprotein E and DH1017.Fab structures | Protein Data Bank | PDB: 7T17 |
| Density map of DH1017.Fab in complex with Zika virions | EMDataResource | EMD-25606 |
| 119-4-D6.IgG | GenBank | OP612786, OP612795 |
| 119-1-D7.IgG | GenBank | OP612787, OP612796 |
| 119-5-C5.IgG | GenBank | OP612788, OP612797 |
| DH1017.IgM | GenBank | OP612789, OP612798 |
| 123-3-G2.IgG | GenBank | OP612790, OP612799 |
| 124-4-C8.IgG | GenBank | OP612791, OP612800 |
| 124-1-C2.IgG | GenBank | OP612792, OP612801 |
| 124-2-G3.IgG | GenBank | OP612793, OP612802 |
| 126-1-D11.IgG | GenBank | OP612794, OP612803 |
| Experimental models: Cell lines | ||
| Vero African green monkey kidney epithelial cells | ATCC | CCL-81 |
| C6/36 Aedes albopictus insect cells | ATCC | CRL-1660 |
| THP-1 monocytic cells | ATCC | TIB-202 |
| THP1.2 monocytic cells subclone | Chan et al. 2014 | N/A |
| CD40 ligand-expressing MS40L cells | Luo et al., 2009 | N/A |
| K-562 cells | ATCC | CCL-243 |
| BHK-21 fibroblasts | ATCC | CCL-10 |
| 119-4-D6.IgG B-LCL | This manuscript | N/A |
| 119-1-D7.IgG B-LCL | This manuscript | N/A |
| 119-5-C5.IgG B-LCL | This manuscript | N/A |
| 124-4-C8.IgG B-LCL | This manuscript | N/A |
| 124-1-C2.IgG B-LCL | This manuscript | N/A |
| 124-2-G3.IgG B-LCL | This manuscript | N/A |
| 126-1-D11.IgG B-LCL | This manuscript | N/A |
| 123-3-G2.IgG B-LCL | This manuscript | N/A |
| DH1017.IgM B-LCL | This manuscript | N/A |
| 119-4-G11.IgM B-LCL | This manuscript | N/A |
| Vero-Furin cells | Mukherjee et al., 2016 | N/A |
| Expi 293i cells | Invitrogen | A14527 |
| Experimental models: Organisms/strains | ||
| Ifnar1−/− C57BL/6 mice (males) | Kindly provided by Dr. Jason Whitmire (UNC) and bred in-house. | N/A |
| Oligonucleotides | ||
| forward primer CCGCTGCCCAACACAAG | Integrated DNA Technologies; Carbaugh et al., 2019 | N/A |
| reverse primer CCACTAACGTTCTTTTGCAGACAT | Integrated DNA Technologies; Carbaugh et al., 2019 | N/A |
| probe AGCCTACCTTGACAAGCAATCAGACACTCAA | Integrated DNA Technologies; Carbaugh et al., 2019 | N/A |
| Software and algorithms | ||
| Prism version 9 | GraphPad Software | https://www.graphpad.com |
| BD FACSDiva | BD Biosciences | https://www.bdbiosciences.com/en-us/products/software/instrument-software/bd-facsdiva-software |
| FlowJo version 10 | FlowJo, LLC | https://flowjo.com |
| Cloanalyst | Kepler, 2013 | https://www.bu.edu/computationalimmunology/research/software/ |
| I-TASSER | Roy et al., 2010; Yang et al., 2014 and Zhang, 2008 | https://zhanggroup.org/I-TASSER/ |
| PYMOL version 2 | Schrodinger, LLC | https://pymol.org/2/ |
| RIVEM | Xiao and Rossmann, 2007 | N/A |
| BioRender | BioRender.com | http://biorender.com |
| Other | ||
| BioMek FX | Beckman Coulter | N/A |
| Cryoplunge 3 system | Gatan | N/A |
| Titan Krios | Thermofisher | N/A |
| Talos F200C | Thermofisher | N/A |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human samples.
All human serum and PBMC samples used in this study were derived from an IRB-approved protocol to investigate maternal ZIKV immunity during infections in pregnancy. We focused specifically on female and pregnant women in our sample collections as infections in this population lead to the greatest burden of disease in society, among newborns. Participants in this study were enrolled into our prospective cohort during the ZIKV epidemic from July 2016 to October 2017 in the city of Vitória, in Brazil. All participants were female and pregnant, and their ages ranged from 18 to 39 years. Pregnant women were enrolled based on symptoms suggestive of ZIKV infection, such as rash, arthralgia, and fever. Details on our sample collection, enrollment, and follow-up strategy are previously reported (Singh et al., 2019). Specimen volumes have been largely used for these studies and cannot be further shared.
In vivo mouse samples.
Five-week-old male Ifnar1−/− C57BL/6 mice were used in this study, and previously defined as a model of lethal ZIKV challenge (Lazear et al., 2016). We used this developmental stage because the phenotype of lethality has been well characterized and optimized previously. This study was carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health and under protocols approved by the Institutional Animal Care and Use Committee of the University of North Carolina at Chapel Hill.
Cell lines.
Vero African green monkey kidney epithelial cells (CCL-81; adult), C6/36 Aedes albopictus insect cells (CRL-1660), golden hamster BHK-21 kidney fibroblast cells (CCL-10), human K-562 lymphoblasts (CCL-243) and human THP-1 monocytic cells (TIB-202; male 1-year) were obtained from ATCC, and growth conditions are reported in our study methods. THP1.2 monocytic cells are a subclone of THP-1 cells (Chan et al., 2014).
B-LCL isolation and growth conditions are described in the methods details. All B-LCLs were isolated from pregnant women with confirmed ZIKV infection (age range: 18-39).
METHODS DETAILS
Donors and Sample Information.
Participants in this study were enrolled from July 2016 to October 2017 in the city of Vitória, State of Espírito Santo, in Brazil. In this prospective cohort, we enrolled pregnant women with Zika-like symptoms of rash and/or fever and collected blood samples through pregnancy, delivery, and postpartum. Cohort design, recruitment, enrollment and sample collection are detailed in our prior study (Singh et al., 2019). Plasma and peripheral blood mononuclear cells (PBMCs) from 11 mothers who were serologically confirmed for ZIKV infection previously (Singh et al., 2019) were included in this study. The infant born to subject P14 demonstrated microcephaly at birth, and the rest had no known signs of congenital Zika syndrome. DH1017.IgM was isolated from the PBMCs of P73, who presented with a symptomatic ZIKV infection at 22 weeks of gestation and had prolonged viral replication with vRNA detected in serum/urine up to 42 DPS. She delivered an apparently healthy baby at 38.5 weeks of gestation with normal head circumference. All other mothers had detectable ZIKV vRNA in plasma at the first visit only.
Ethics Statement.
The Institutional Review Boards of Hospital Cassiano Antonio Moraes (Brazilian National Research Ethics Committee (CEP/CONEP) Registration number: 52841716.0.0000.5071) and the Duke University Medical Center (Pro00100218) each approved this prospective cohort study and sampling. Pregnant women with rash or fever, who were a minimum of 18 years of age, and provided a willingness to participate through a written informed consent were approved for inclusion into this study. To protect the privacy of all subjects, publicly available identifiers are twice removed from the participant. All practices conducted as part of this study are compliant with ethical principles for medical research involving human subjects as outlined in the Declaration of Helsinki.
Cell culture and virus stocks.
Vero-81 cells were grown in Dulbecco’s Modified Eagle Media (Gibco, 11965092) supplemented with 10% heat-inactivated fetal bovine serum (Sigma, F4135), 1x penicillin-streptomycin (Gibco, 15140-122), and 1x MEM non-essential amino acids solution (Gibco, 11140-050). Viruses used for the focus reduction neutralization test were DENV1 (WestPac74), DENV2 (S-16803), DENV3 (CH54389), DENV4 (TVP-360), provided by Dr. Aravinda de Silva (University of North Carolina at Chapel Hill); ZIKV H/PF/2013 and PRVABC59 strains were obtained from the Biodefense and Emerging Infections Research Resources Repository (BEI) and from the US Centers for Disease Control and Prevention. Virion binding antibodies were detected using the following viruses from BEI: ZIKV (PRVABC59), DENV1 (Hawaii NR-82), DENV2 (New Guinea C), DENV3 (Philippines), and DENV4 (H241). ZIKV was grown in Vero-81 cells supplemented with 10% heat-inactivated fetal bovine serum (FBS) and 10 mM HEPES (Sigma, H0887). Dengue viruses were grown in C6/36 cells cultured in RPMI 1640 (Gibco, 11875-093), L-Glutamine (Gibco, 25030-081), 25 mM HEPES (Gibco, 22400-089), 1x penicillin-streptomycin (Gibco, 15140-122), and 20% FBS. During DENV infection, RPMI was supplemented with 25mM HEPES and 2% FBS. 0.5 mL of viruses were added to 80-90% confluent cells. Cells were infected with DENV2, DENV3, or DENV4 for 7-9 days and DENV1 for 11 days. ZIKV stock infections were stopped when cytopathic effect was observed (3-6 days post infection). Cell supernatant containing virus was harvested, centrifuged brought to a final concentration of 20% FBS, and filtered through 0.22 μm filter prior to storage at −80°C.
Fluorescent labelling of ZIKV for sorting B cells.
A previously developed approach to label DENV with Alexa Fluor 488 (AF488) dye was adapted to label ZIKV (Zhang et al., 2010). ZIKV (Strain: PF13/251013-18) was propagated in Vero cells and purified through 30% sucrose. Virus titer was determined using the BHK-21 cell plaque forming assay as previously described (Chan et al., 2011). Briefly, AF488 succinimidyl ester was reconstituted in 0.2M sodium bicarbonate buffer (pH 8.3) and added to 3 x 108 PFU of ZIKV at a final dye concentration of 100 μM. The mixture was incubated at RT for 1 h with gentle inversions every 15 min. Labelled ZIKV was purified by size exclusion chromatography using Sephadex G-25 columns (Amersham, GE Healthcare) to remove the excess dye and titrated on BHK-21 cells. Thereafter, ZIKV was UV inactivated (254 nm) for 1 min on ice. Inactivation of virus was verified by serial passaging of virus on C6/36 mosquito cell line (ATCC), without detection of infection on this susceptible cell line.
Staining and sorting ZIKV-reactive B cells.
Thawed PBMCs were stained with Aqua Vital Dye (Invitrogen), IgD-PE (clone IA6-2; BD Biosciences), CD10 PE-CF594 (clone HI10a; BD Biosciences; CD3 PE-Cy5 (clone HIT3a; BD Biosciences), CD14 BV605 (clone M5E2; BD Biolegend), CD16 BV570 (clone 3G8; Biolegend), CD27 PE-Cy7 (clone O323; Thermo Fisher Scientific), CD38 APC-AF700 (clone LS198-4-3; Beckman Coulter), CD19 APC-Cy7 (clone SJ25C1; BD Biosciences) and 1 x 106 PFU of freshly thawed UV-inactivated ZIKV labelled with AF488. Additionally, 5 μM of Chk2 kinase inhibitor II (Calbiochem/EMD Chemicals) was added to prevent cell death. Unfractionated B cells were defined as CD14−/CD16−/CD3−/CD19+. For memory B cells an additional IgD−/CD27all gate was applied. The ZIKV-reactive AF488 gate was set using a fluorescence minus one condition. Cells were sorted on a Beckton Dickinson FACS Aria II and analysis was performed in FlowJo.
Cell cultures.
ZIKV-reactive B cells were cultured as previously described (Bonsignori et al., 2011, 2016). Briefly, cells were sorted in wells pre-seeded with human CD40 ligand-expressing MS40L cells (5,000 cells/well) and containing 2.5 μg/ml CpG ODN2006 (InvivoGen, tlrl-2006), 5 μM Chk2 kinase inhibitor II and 50 ng/ml of recombinant human IL-21 (Peprotech, 200-21) in RPMI/15% FBS. EBV was added for the overnight incubation. After overnight incubation in bulk at 37°C, 5% CO2, B cells were distributed by limiting dilution at a calculated concentration of 1 cell/well into 96-well round-bottom tissue culture plates in the presence of MS40L cells and complete medium described above. Medium was refreshed after 7 days. Supernatants were collected after 14 days. Cell culture supernatants were assessed for IgG, IgM, and IgA levels in ELISA as previously described (Bonsignori et al., 2016) and Ig+ wells were further evaluated for ZIKV-reactivity with a virion binding ELISA. Culture supernatants were screened undiluted. The cut-off for ZIKV-positivity was the highest value of mean plate blanks optical density (OD) at 450 nm (OD450) + 2 SD and OD450 not less than 0.4. From each culture well, half of the cells were harvested and preserved in RNAlater (Qiagen) at day 14 and half were maintained in culture. Cell clones that immortalized were expanded and cloned by using a standard limiting dilution method. Reactivity and IgV(D)J sequences were checked periodically. We derived three IgG+ B-LCLs from P34 (termed 119-4-D6.IgG, 119-1-D7.IgG, and 119-5-C5.IgG), three IgG+ B-LCL from P56 (124-4-C8.IgG, 124-1-C2.IgG and 124-2-G3.IgG), and one IgG+ B-LCL from P54 (126-1-D11.IgG), From subject P73 we derived two B-LCLs: one expressing IgG from memory B cells collected 28 DPS (123-3-G2.IgG), and one IgM+ B-LCL (termed DH1017) from unfractionated B cells collected at 71 DPS.
Isolation of V(D)J immunoglobulin regions.
RNA from positive cultures was extracted by using standard procedures (RNeasy minikit; QIAGEN), and the genes encoding Ig VHDJH and VLJL rearrangements were amplified by RT and nested PCR as previously reported (Liao et al., 2009). Ig V(D)J sequences (Genewiz) were analyzed using the Cloanalyst software package (http://www.bu.edu/computationalimmunology/research/software/).
Monoclonal antibody production.
B-LCLs were expanded in CELLine bioreactors (Wheaton) following the manufacturer’s recommendations. Monoclonal antibodies were purified using protein A resin columns for IgG and CaptureSelect beads (Thermo Scientific) for DH1017.IgM following manufacturer’s recommendations. DH1017.IgG and DH1017.Fab heavy and light chain plasmids (GenScript) were co-transfected at an equal ratio in suspension Expi 293i cells (Invitrogen) using ExpiFectamine 293 transfection reagents (Life Technologies) according to the manufacturer’s protocols. Transfected cells were gently shaken overnight for 16-18 h and incubated at 37°C, 8% CO2. After adding the enhancer provided in the kit, cells were incubated for additional 4-6 days. Supernatants were harvested and filtered, and co-incubated with a Protein A affinity resin (Thermo Fisher Scientific) for IgG antibody or LambdaFabSelect Agarose Beads (GE Healthcare Life Sciences) for Fab at 4°C on a rotating shaker overnight. The bead and supernatant mixture was then loaded onto a column for purification. Following a Tris/NaCl Buffer (pH 7.0) wash, mAb was eluted from beads using Trizma HCl (pH 8.0; VWR) and concentrated in the Vivaspin Turbo 15 Concentrator (Thermo Fisher Scientific) with a pH neutralizing buffer exchange using Citrate Buffer (pH 6.0). Purified antibody concentration was determined by Nanodrop and product was evaluated by reducing and nonreducing SDS-polyacrylamide gel electrophoresis and Coomassie Blue staining for appropriate size.
Native PAGE and Coomassie.
DH1017.IgM was run under non-reducing conditions using a NuPAGE 3-8% Tris-Acetate Gel (Invitrogen) with 1x Tris-Glycine Native Running Buffer (Novex) at 130V for 2.5 h. DH1017.IgM (5μg/lane) was prepared with Native Tris-Glycine Sample Buffer (Novex) and NativeMark Protein Standard (Invitrogen) was used as the ladder. Gel was subsequently stained with Coomassie and imaged using ChemiDoc (Bio-Rad).
Negative stain electron microscopy.
DH1017.IgM was equilibrated to RT and diluted to 20-40 μg/ml with buffer containing 10 mM HEPES, pH 7.4, 150 mM NaCl, and 0.02% ruthenium red. A 5 μl drop of diluted sample was applied to a glow-discharged carbon-coated grid for 8-10 s, blotted, then rinsed with two drops of buffer containing 1 mM HEPES, pH 7.4 and 7.5 mM NaCl, and finally stained with one drop of 2% uranyl formate for 60 s, then blotted and air dried. Images were obtained with a Philips EM420 electron microscope at 120 kV, 82,000x magnification, and captured with a 2k x 2k CCD camera at 4.02 Å/pixel. The RELION program (Scheres, 2016) was used for particle picking and 2D class averaging.
ZIKV and DENV virion capture ELISA.
The virion capture ELISA methods were previously described (Singh et al., 2019). Briefly, high-binding 96-well plates (Greiner) were coated with 40 ng/well of 4G2 antibody (clone D1-4G2-4-15) in 0.1 M carbonate buffer, pH 9.6 overnight at 4°C. Plates were blocked in Tris-buffered saline containing 0.05% Tween-20 and 5% normal goat serum for 1 h at 37°C, followed by an incubation with optimal dilutions of either ZIKV (PRVABC59), DENV1 (Hawaii), DENV2 (New Guinea C), DENV3 (Philippines), and DENV4 (H241) from BEI Resources for 1 h at 37°C. 50 μL/well of samples were added and incubated for 1 h at 37°C. Plasma and purified mAbs were tested in duplicate using eight-point serial dilutions starting at 1:25 with 3-, or 5-fold dilutions for plasma and at 100 μg/mL with 5-fold serial dilutions for mAbs. Horseradish peroxidase (HRP)-conjugated goat anti-human IgG antibody (Jackson ImmunoResearch Laboratories, 109-035-008), HRP-conjugated goat anti-human IgM antibody (Jackson ImmunoResearch Laboratories, 109-035-129), or HRP-conjugated goat anti-human IgA antibody (Jackson ImmunoResearch Laboratories, 109-035-011) were all used at a 1:5,000 dilution followed by the addition of SureBlue Reserve TMB substrate (KPL). Reactions were stopped using Stop Solution (KPL) after 5 min and OD was detected at 450 nm (Perkin Elmer, Victor). ZIKV-specific mAb C10 (Absolute Antibody, AB00677-10.0) was used as positive control. Negative controls included diluent alone and respiratory syncytial virus-specific IgG Palivizumab. EC50 was calculated with a sigmoidal dose-response (variable slope) curve in Prism 7 (GraphPad) using a least squares fit.
Reactivity to autoantigens.
ELISA was performed in 384-well plates (Corning) as previously described (Bonsignori et al., 2011). Briefly, plates were coated overnight at 4°C with 15 μl purified proteins at optimized concentrations in 0.1M Sodium Bicarbonate: DNA (Worthington, LS002105) at 5 μg/mL, Centromere B (Prospec, PRO-390) at 0.15 μg/mL, Histone (Sigma, H9250) at 0.2 μg/mL, Jo-1 (Immunovision, JO1-3000) at 0.05 units/well, RnP/Sm (Immunovision, SRC-3000) at 0.2 units/well, Scl-70 (Immunovision, SCL-3000) at 0.4 units/well, Sm (Immunovision, SMA-3000) at 0.1 units/well, SSA (Immunovision, SSA-3000) at 0.2 units/well, and SSB (Immunovision, SSB-3000) at 0.1 units/well. Subsequently, plates were blocked (50 μL/well) with assay diluent (PBS containing 4% [w/v] whey protein, 15% normal goat serum, 0.5% Tween 20) for 2 h at RT. For DNA, plates were pre-coated with Poly-L-Lysine (Sigma, P6282) at 2 μg/mL in PBS overnight at 4°C, and the assay diluent was PBS containing 2% [w/v] BSA and 0.05% Tween-20. Monoclonal antibodies were tested using 3-fold serial dilutions starting at 10 μg/ml. 10 μl of primary antibodies were added to each well and incubated for 1 h at RT. The following positive control antibodies from Immunovision were all tested at a 1:25 starting dilution with 3-fold serial dilutions: Anti-Centromere B (HCT-0100), Anti-single stranded DNA (HSS-0100), Anti-Histone (HIS-0100), Anti-Jo 1 (HJO-0100), Anti-SRC (HRN-0100), Anti-Scl 70 (HSC-0100), Anti-Sm (HSM-0100), Anti-SSA (HSA-0100), and Anti-SSB (HSB-0100). The negative control was assay diluent alone. Plates were developed using 15 μl/well of combination of HRP–conjugated antibodies in assay diluent comprising goat anti-human IgG (Jackson ImmunoResearch Laboratories, 109-035-098) at 1:10,000 dilution, goat anti-human IgM (Jackson ImmunoResearch Laboratories, 109-035-129) at 1:10,000 dilution, and goat anti-human H+L (Promega, W403B) at 1:3,000 dilution. After a 1 h incubation, plates were developed with 20 μl/well of SureBlue Reserve TMB substrate (KPL) and stopped by 20 μl/well Stop Solution (KPL) after 5 min. OD was detected at 450 nm (Perkin Elmer, Victor).
HEp-2 Cell Staining.
Indirect immunofluorescence (Zeuss Scientific) binding of DH1017 mAbs to HEp-2 cells was performed as previously described (Bonsignori et al., 2014). IgG1 mAbs 2F5 and 17B were used as positive and negative controls, respectively. All mAbs were tested at 25 μg/ml and 50 μg/mL. Images were acquired for 8 s with a 40x objective.
Focus reduction neutralization test.
We used previously described methods for FRNT50 in a 96-well plate (Singh et al., 2019). Briefly, serial dilutions of heat-inactivated plasma or purified mAbs were added to 30-60 focus forming units of either DENV serotypes 1-4 or ZIKV (H/PF/2013). Plasma was used at a starting dilution of 1:25 with subsequent 5-fold or 7-fold dilutions. MAbs were tested at 5 μg/mL or 10 μg/mL with 5-fold dilution series. DH1017.Fab was tested at 1 mg/mL with a 5-fold dilution series. Negative control was media alone, and positive controls were known ZIKV-neutralizing mAbs and plasma from ZIKV-infected subjects. Virus and plasma or mAb samples were co-incubated for 1 h at 37°C, then transferred to a 96-well plate (Greiner Bio One) with confluent Vero-81 cells and incubated for 1 h at 37°C. Plates were overlayed with 1% methylcellulose and incubated at 37°C for 40-42 h (ZIKV and DENV4), 51-53 h (DENV1), or 48 h (DENV2 and DENV3). Cells were fixed with 2% paraformaldehyde for 30 min and stained with 0.5 μg/mL of 4G2 or E60 mouse monoclonal antibodies. Foci were detected with an anti-mouse IgG conjugated to HRP at a 1:5000 dilution (Sigma), followed by True Blue substrate (KPL). Foci were counted using an ImmunoSpot plate reader (Cellular Technology Limited). FRNT50 values were calculated with the sigmoidal dose-response (variable slope) curve in Prism (GraphPad), constraining values between 0 and 100% relative infection. Percent relative infection curves were considered to pass quality control criteria for FRNT50 determination if R2 >0.65, absolute value of hill slope >0.5, and curve crossed 50% relative infection within the range of the plasma dilutions in the assay. Samples were repeated up to 3 times to quantify a valid FRNT50.
Reporter Virus Particle (RVP) production.
ZIKV RVPs incorporating the structural proteins (C-prM-E) of ZIKV strain H/PF/2013 were produced by genetic complementation of a GFP-expressing WNV lineage II sub-genomic replicon with the virus structural gene plasmids as previously described (Dowd et al., 2016). Briefly, the WNV replicon and C-prM-E plasmids were co-transfected into HEK-293T cells using Lipofectamine 3000TM transfection reagent (Invitrogen). Transfected cells were incubated at 30°C and RVP-containing supernatants were harvested 3-6 days post-transfection and pooled. RVP-containing supernatants were passed through a 0.2 μm filter (Millipore) and stored at −80°C.
RVP-based Antibody-Dependent Enhancement (ADE) assay.
RVP-based ADE assays were performed by incubating green fluorescent protein (GFP)-encoding RVPs with serial dilutions of mAbs for 1 h at 37°C. FcγR+ K562 or THP-1 cells were infected with RVP immune complexes and incubated at 37°C for 36-48 h. Cells were fixed with paraformaldehyde, and GFP expression was detected by flow cytometry. Antibody enhanced infection was scored as detectable if the number of GFP positive cells was ≥3-fold above background (defined as the average percent GFP positive cells in the absence of antibody).
Plaque assay-based ADE assay.
DH1017.IgM and DH1017.IgG were serially diluted 2-fold over 6.25 – 0.003 μg/mL and 10 μL of diluted mAb were co-incubated in duplicate with 2 x 105 PFU of ZIKV (Strain: PD13/251013-18) in a round bottom 96 well-plate for 1 h at 37°C to form immune complexes. Thereafter, 2 x 104 cells of THP1.2S or primary monocytes were added to the ZIKV and mAb immune complexes for a 72-h infection at 37°C. THP1.2S cells are a subclone of the THP1 monocyte cell line (ATCC) more susceptible to flavivirus infection (Chan et al., 2014). Collection and processing of primary monocytes has been previously described and was conducted with approval of the NUS Institutional Review Board under reference code B-15-227 (Chan et al., 2019). Supernatants were collected, and infectious virus was titrated via plaque forming assays on BHK-1 cells (ATCC) in 24 well plates in quadruplicate, as previously described (Chan et al., 2011). For the positive control we used humanized IgG1 4G2 mAb, and for the negative control we used 3H5 (DENV-specific mAb) as well as the virus only condition, without any mAb present.
Antibody-dependent complement antiviral activity.
Focus reduction neutralization test (FRNT) in the presence of increasing concentrations [volume/volume] of complement from normal human serum (Sigma), including 0, 5, 10, 15, 20 and 25% final concentration were tested. Percent infection for each antibody treatment was calculated relative to virus alone using matched concentrations of NHS. Samples were run in triplicate and experiments were independently repeated for each concentration of complement. Positive control was DH1017.IgM and DH1017.IgG run in the absence of complement, and negative control was the virus and complement only conditions. The rest of the analysis and approach are identical to the FRNT approach described above.
Determining the contribution of plasma IgM to ZIKV neutralization.
Each plasma sample was split into 2 aliquots, with one portion depleted of IgM isotype antibodies and another portion mock depleted. First, each sample was heat inactivated for 30 min at 56°C, diluted 1:1 with sterile PBS, and centrifuged at 10,000 x g for 10 mins to remove debris. Depletion beads were packed into sterile 0.5 mL centrifugal filter devices with a 0.22 μm pore PVDF membrane (Millipore) and equilibrated with 3x sterile PBS washes (pH 7.2). 200 mg of CaptureSelect IgM affinity beads (ThermoFisher Scientific) were used for IgM depletion, and 66 mg of corresponding beads of the same size (200-400 mesh) and material (polystyrene divinylbenzene 1% cross linked beads; Alfa Aesar) were used for mock depletion. Samples were co-incubated with beads for 10 min at RT with gentle inversions, and then the depleted fraction was centrifuged at 10,000 x g for 10 min.
Depletion of IgM was confirmed by IgM ELISA, and non-specific losses to ZIKV-binding IgG were quantified through virion binding ELISA. The limit of detection of 0.12 μg/mL was based on the linear range of a sigmoidal standard curve of human IgM (Jackson ImmunoResearch Laboratories). Magnitude of ZIKV-binding IgG was assessed by virion-binding ELISA and neutralization potency was assessed using the Focus Reduction Neutralization Test. Due to slight differences in ZIKV-binding IgG across IgM and mock depleted fractions, each fraction was adjusted to the magnitude of ZIKV-binding IgG in the same sample such that differences in neutralization activity could be attributed to differences in IgM isotype antibodies. Thus, the percent neutralization attributable to IgM that is reported in this study was calculated as follows:
Detection of antibody isotype from sera.
Human IgM antibodies were detected in sera from passively transferred mice using high-binding 384-well plates (VWR) coated with 0.5 μg/well of goat anti-human Ig polyvalent antibody (ThermoFisher, H17000) in 0.1 M carbonate buffer, pH 9.6 overnight at 4°C. Plates were blocked the next day (2 h at RT) with 10X PBS containing 4% whey protein, 15% goat serum, and 0.5% Tween-20. Mouse serum samples were diluted 1:30, serially diluted three-fold, then added to the plate (10 μL/well) and incubated for 1 h at 37°C. HRP-conjugated goat anti-human IgM antibody (Jackson ImmunoResearch Laboratories, 109-035-129) was used at a 1:10,000 dilution (10 μL/well) followed by the addition of SureBlue Reserve TMB substrate (KPL). Reactions were stopped by Stop Solution (KPL) after 5 min and OD was detected at 450 nm. DH1017.IgM was used as a positive control and standard. Blank wells were used as a negative control. Antibody concentrations were interpolated from the standard curve, which was fit using a 5-parameter fit sigmoidal curve. Concentrations were interpolated from OD450 values within the linear range of the sigmoidal curve at dilution 90, or alternatively 30, for samples at 2-6 DPI, and at dilution 10 for samples at 8+ DPI. A passing quality control criterion of replicate OD450 values less than or equal to 25% variance was applied. Limit of detection for this assay was 0.08 μg/mL across assays, based on the linear range of the standard curve.
To measure concentration of IgM from human plasma, the same assay was applied with the following modifications. All original and IgM/mock depleted samples were diluted 1:30 relative to original plasma with 3-fold serial dilutions. Human IgM (Jackson ImmunoResearch Laboratories, 009-000-012) was used as positive control and standard. IgM concentration in sample was inferred based upon the 1:2430, or alternatively the 1:270 and 1:810 serum dilutions, which were all in the linear range of the standard curve.
Biolayer interferometry.
BLI assays were performed on the ForteBio Octet Red96 instrument at 25°C with a shake speed of 1,000 RPM. Goat Anti-Human Ig polyvalent (20 μg/mL; Thermo Fisher) was amine coupled to AR2G sensor tips as follows: sensor tips were activated with s-NHS/EDC (300 s), incubated in Anti-Human Ig (600 s), and quenched with ethanolamine (300 s). DH1017.IgM, DH1017.IgG, and a negative control IgG (palivizumab) were diluted to 50 μg/mL in PBS and captured on the biosensors (300 s), followed by a PBS wash (60 s). A baseline signal was recorded for 2 min in PBS (pH 7.4). Biosensors were then immersed into a two-fold dilution series of ZIKV E protein dimer in PBS (50-0.78μg/mL) to measure association (400 s), followed by immersion into PBS to measure dissociation (600 s). ZIKV E protein dimer purity and size were verified by size exclusion chromatography prior to the assay. All binding profiles were corrected by double reference subtraction using the signal obtained in a PBS control (without E protein) and the signal obtained from the palivizumab control sensor. All affinities were calculated using the fast kinetic components of the heterogeneous ligand model fit. Kinetics and affinity data are the result of a single measurement.
Size exclusion chromatography.
ZIKV E dimer was screened for size on Superdex 200 Inc 10/300 GL column: 100 μL of E dimer was loaded onto the column at 0.13 mg/mL and was run at a flow rate of 0.5 mL/min in PBS.
Assessing protection in ZIKV mouse model.
Animal husbandry and experiments were performed under the approval of the University of North Carolina at Chapel Hill Institutional Animal Care and Use Committee. Five-week-old male Ifnar1−/− mice were inoculated with a lethal dose of ZIKV strain H/PF/2013 (1 x 103 FFU) subcutaneously in the footpad on day 0 (Lazear et al., 2016). On days −1 and +1, antibody was delivered intravenously via the retro-orbital route. Serum was collected every two days for up to 11 days to monitor viremia by qRT-PCR and survival was monitored for 15 days. Animals that lost ≥30% of their starting weight or that exhibited severe disease signs were euthanized.
Measuring viral burden in mice.
RNA from the serum of ZIKV-infected mice was extracted with the QIAGEN viral RNA minikit. ZIKV RNA was quantified by TaqMan one-step qRT-PCR using a CFX96 Touch real-time PCR detection system (Bio-Rad). Genome copies per ml of serum on a log10 scale was determined by comparison with a standard curve generated by using serial 10-fold dilutions of a ZIKV plasmid and previously reported primers: forward primer CCGCTGCCCAACACAAG, reverse primer CCACTAACGTTCTTTTGCAGACAT, and probe AGCCTACCTTGACAAGCAATCAGACACTCAA (Integrated DNA Technologies) (Carbaugh et al., 2019).
Virus propagation and purification for cryo electron microscopy.
ZIKV strain H/PF/2013 was propagated in Vero-Furin cells (Mukherjee et al., 2016). Approximately 109 Vero-Furin cells, grown in 10% FBS (Sigma, F0926), Dulbecco’s modified Eagle medium (DMEM) media (Thermofisher) and 50 μg/mL blasticidin (Invivogen, anti-bl-1) at 37°C 5% CO2, were infected with a MOI of 0.1. Virus particles in 2% FBS/DMEM/1mM Pen/Strep (Thermofisher, 15140122) were incubated with cells for 2 h at RT. After 2 h, infected cells were incubated at 37°C, 5% CO2 for 36 h. At 36 h post infection (hpi), infectious media was collected and replaced with fresh media every 12 h up to 84 hpi. Virus particles were purified from media collected at 60 and 72 hpi.
Briefly, cell debris were pelleted by centrifugation at 2,744 x g for 30 min. Infectious media was filtered with through a Steritop-GP 0.2 μm filter (Millipore, SCGPT05RE) and virus particles were precipitated overnight at 4°C with PEG 4000, 8% final concentration. PEG precipitated particles were concentrated at 8,891 x g for 50 min. Particles were concentrated through a 24% sucrose cushion by ultra-centrifugation at 126,144 x g for 2 h at 4°C. Virus particles were isolated with a discontinuous potassium-tartrate/glycerol/NTE buffer (120mM NaCl, 20mM Tris pH 8.0, 1mM EDTA) gradient in 5% increments between 35% and 10% potassium-tartrate and ultra-centrifugation at 126,144 x g for 2 h at 4°C. The particles were extracted from the 20% fraction of the discontinuous gradient and buffer exchanged and concentrated in NTE buffer with a 100 kDa MWCO centrifugal filter.
Single particle cryo-electron microscopy.
Prior to freezing, purified virus and DH1017.Fab fragments were incubated on ice for 2 h at a molar ratio of 0.2 mM Fab to 1 mM E protein. Samples were then plunge frozen in liquid ethane using a Cryoplunge 3 System (GATAN). Briefly, liquid nitrogen was used to liquefy ethane at −190°C. A 2.5 μL volume of sample was spotted on lacey carbon grids (Ultrathin C on Lacey Carbon Support film, 400 mesh, Cu. Ted Pella, Inc. Product number 01824) and blotted with Whatman grade 1, 200 mm circle filter paper (GE Healthcare Life Sciences, 1001-020) for 2.5 s. Blotting air pressure setting used was 100 psi.
Data were collected on a Titan Krios (Thermofisher) microscope equipped with a Gatan K3 detector using and Leginon software package (Suloway et al., 2005). A total of 1,929 cryo EM micrographs were collected with a nominal magnification of 64000x, 0.66 Å/pixel size, and an electron dose equivalent of 35 e-/Å2. Motion correction and CTF calculations estimations were performed using MotionCorr2 (Zheng et al., 2017) and CTFFIND4 (Rohou and Grigorieff, 2015) respectively.
Automated particle selection picking (Sigworth, 2004) performed with cisTEM (Grant et al., 2018) selected 34,474 particles. A maximum-likelihood algorithm based 2D classification (Scheres et al., 2005; Sigworth, 1998) was performed with using cisTEM. From a subset of 2D classes, 4104 particles were selected for further processing.
Single particle reconstruction was performed according to the ‘gold standard’ method using jspr (Guo and Jiang, 2014). Briefly, the particles were divided equally into two randomly selected independent particle sets. Twenty ab-initio models were generated from a random set of 700 particles selected from the set of 4,104 particles. Two ab-initio models were selected, and one ab-initio model was assigned to one independent particle set and the other ab-initio model was assigned to the other independent data set. Each dataset was refined iteratively assuming icosahedral symmetry. Refinement resulted in two independent models that converged on the same structure. Following corrections for astigmatism, elliptical distortion, defocus, and the masking of the disordered nucleocapsid core, the final models of each independent data set were combined into a single final model. The resolution of the map was calculated at 0.143 from the FSC curve (Rosenthal and Henderson, 2003).
Model fitting, refinement, and structure analysis.
Models used in fitting viral structural proteins in the cryo EM density map are the E glycoprotein ectodomain of the ZIKV asymmetric unit (PDB 6CO18) (Sevvana et al., 2018) and a homology model of DH1017.Fab fragment. The Fab fragment model is composed of individual domains variable heavy, variable light, C, and lambda domains. The homology models of each Fab fragment domain were generated with I-TASSER (Roy et al., 2010; Yang et al., 2014; Zhang, 2008) and aligned with PYMOL (version 2.0, Schrodinger LLC) to an IgM cryoglobulin Fab crystal structure (PDB 2AGJ) (Ramsland et al., 2006) to form the complete DH1017.Fab homology model. The Cα backbones of all structures were fit in the cryo-EM map using Chimera (Pettersen et al., 2004) fit-to-map function. The position of the Fab domains was further refined with Coot (Emsley, P 2010). Residues of the E ectodomain Cα backbone within 6 Å of the Fab fragment Cα backbone were identified with PYMOL. These residues are defined as the Fab footprint and mapped to the surface of the virus particle with RIVEM (Xiao and Rossmann, 2007). The binding of a human IgM pentamer to the virus surface at the five-fold icosahedral axis was modeled. The solution structure of an IgM pentamer (PDB 2CRJ) (Perkins et al., 1991) and the homology model of DH1017.Fab from this paper were used to build the model. The Fc domains Cμ4, Cμ3, and Cμ2 of PDB 2CRJ were modeled to similar positions of Cμ4 (PDB 4JVW), Cμ3 (PDB 4BA8), and Cμ2 (4JVU) domains fit to the 25A tomogram of an IgM pentamer bound to the surface of an artificial liposome (Müller et al., 2013; Sharp et al., 2019). One Fab domain from each monomer of the pentamer structure PDB 2CRJ were fit to the density map after being aligned manually to the fitted DH1017.Fab structure.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis was performed using GraphPad Prism using tests as reported in the manuscript. Hierarchical clustering was performed on Los Alamos Database heatmap tool: https://www.hiv.lanl.gov/content/sequence/HEATMAP/heatmap.html.
Supplementary Material
Figure S1. Lack of linear relationship between plasma ZIKV binding IgM levels and IgM-mediated neutralization. Related to Figure 1. ZIKV binding IgM levels were measured at 1:10 via ELISA and IgM mediated neutralization was measured by focus reduction neutralization tested on paired IgM- and mock-depleted samples. A. Spearman Rank correlation with all samples included in this study (n=21). B. Correlation with the subset of samples in which IgM contributed to ZIKV neutralization (n=15).
Figure S2. Cross-reactivity with DENV 1-4 serotypes. Related to Figure 3. A. ZIKV and DENV1-4 virion binding plasma IgG measured by ELISA and expressed as Log10 ED50 from all collections from which B cell responses were analyzed (n=6; DPS: days post symptoms). Maternal flavivirus serostatus as previously defined (Singh et al., 2019) is shown for each subject. Plasma samples were run in duplicate. B. DH1017.IgM neutralization of DENV 1-4 strains. Error bars: SD of technical replicates run in duplicate. C. DH1017.IgM repeated neutralization assays against the ZIKV PRVABC59 (n=10) and H/PF/2013 (n=2) strains. Line: geometric mean. Replicate experiments were independently performed by multiple operators and at different institutions for ZIKV H/PF/2013.
Figure S3. Functional in vitro characterization of DH1017.IgM and DH1017.IgG. Related to Figure 4. A. DH1017.IgM (left) and DH1017.IgG (right) binding to Zika and Dengue 1-4 virions at 100 μg/mL, measured by ELISA and expressed as optical density at 450nm (OD450). Mean of technical replicates is shown. Negative control is the mean of the diluent alone. B. Antibody-dependent enhancement of DH1017.IgM and DH1017.IgG on primary monocytes measured in a plaque assay. Dotted line: mean of six virus-only control replicates. Grey area: ≤1 SD. Error bars: SD from 6 replicates. C. DH1017.IgM, DH1017.IgG and DH1017.Fab binding to a panel of autoantigens (Sjogren’s syndrome antigens A and B [SSA and SSB, respectively], Smith antigen [Sm], ribonucleoprotein [RNP], centromere B, histone, scleroderma 70 [SCL-70], Jo-1 proteins, and DNA) measured in ELISA. Respective positive controls are shown in the right panel. Error bars: SD of duplicate observations. D. Hep2 cells were stained with DH1017.IgG and DH1017.IgM, 2F5 mAb (positive control), and 17b mAb (negative control) at a concentration of 50 μg/mL. Images shown were taken with a 40X objective with an 8s exposure and are representative of duplicate experiments.
Figure S4. DH1017.Fab interacts with ZIKV surface at multiple angles. Related to Figure 6. A. FCS curve calculated from two half maps of the final model at 1.32 Å/pixel with the RNA core masked out. B. Section of electron density map fit with the asymmetric ectodomain, and a model of the DH1017 variable domain structure at the i2f axis and the Fab model at the q2f axis. DI: red, DII: yellow, and DIII: blue. The DH1017 Fab fragment variable domain (VH, VL) is colored cyan, with the constant domain (c-domain) in green. The two different positions of constant domain density at the i2f axis are labeled c-a and c-b. The variable domain density at both axes is labeled VH-VL. The constant domain density at the q2f axis is labeled c-domain. The i2f and q2f axes are shown. C. Top-down view from the i5f axis of the density map with Fab DH1017 structures fit at the i2f axes (cyan) and q2f axes (green). Two neighboring asymmetric units are shown with open black triangles and the i5f and i3f axes are labeled as a pentagon and triangle, respectively.
Figure S5. DH1017.IgM pentamer multivalent binding to Zika virus particles. Related to Figure 6. A. A top down (left) and side view (right) of a model of the pentamer binding the ZIKV particle in an umbrella like conformation. The density map is shown in gray. The Fc domain is shown in yellow, domains Cμ3 and Cμ4 are labeled. The Cμ2 domain is colored magenta. The DH1017 Fab structure fit to the density map is shown in cyan at the i2f and in green at the q2f. B Electron micrographs of purified ZIKV particles incubated with IgM pentameric DH1017.IgM mAb. Numbered red boxes indicate selected fields showing potential multivalent pentamer binding. Micrographs were collected at 200kV accelerating voltage, magnification 47,000x with a Talos F200C (Thermofisher). C. Expanded view of micrographs of purified Zika particles incubated with IgM pentameric DH1017.IgM mAb. Numbering corresponds to the red boxes in Panel B. Red arrows point to densities predicted to be multivalent binding of pentameric DH1017.IgM. Scale bar is 200 nm. D. The density map of the Fab bound virus particle in an icosahedral cage with top-down views at each icosahedral axis, i2f (left), i5f (middle), and i3f (right). The green asterisk indicates a position where the pentamer is bound as shown in panel A. The red X indicates the position of five-fold axes related to the green asterisk by two-fold symmetry that do not have the i2f position available for Fab binding to form the umbrella like conformation. The i3f view shows positions of complete occupancy for the particle bound as shown in panel A.
Figure S6. DH1017.IgM pentamer cross-linking of Zika virus particles. Related to Figure 6. A. Schematic representation of virus particles cross-linked by pentamers. The Fc portion of the molecule is shown in red cross-linking virus. B. Aggregation of Zika particles possibly facilitated by pentamer crosslinking of particles. Blue arrow: density predicted to be IgM binding two virus particles. Red arrow: density suggesting multivalent binding to a single virion, as also shown in Figure S5C. Scale bar is 200 nm.
Highlights.
Plasma IgM contributes to early ZIKV neutralization during pregnancy.
Ultrapotent neutralization by pentameric DH1017.IgM mAb depends on its isotype.
DH1017.IgM can engage the ZIKV virion with all its binding sites concurrently.
DH1017.IgM protects mice more efficiently from ZIKV viremia than DH1017.IgG.
Acknowledgments:
This work was supported by a Duke Global Health Institute Award to S.R.P., a Duke Incubation Fund Award to M.B. and by the Division of Intramural Research, NIAID, NIH (M.B.); by NIH grants R21-AI132677 (S.R.P.), R21-AI144631 (H.M.L.), F31-AI143237 (C.A.L.), K12-GM000678 (S.J.D.) and R01-AI073755 (R.J.K., PI: M.S. Diamond), by NIH/NIAID contract HHSN272201700060C (R.J.K; PI: K. Satchell), by the Fundação de Amparo à Pesquisa do Espírito Santo (306/2016 to RD; Protocol Financial Support Number: 74910132/16), and by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, Brazil (C.G.). Part of this work was done while M.B. and S.R.P were employed by the Duke Human Vaccine Institute (DHVI). We thank Drs. Aravinda de Silva (UNC Chapel Hill) and James Crowe (Vanderbilt University) for sharing reagents; Dr. M. Anthony Moody (DHVI), the DHVI Research Flow Cytometry Facility, and Brian Watts of the DHVI BIA Core Facility for helpful scientific discussions and/or logistical support; Dr. Geeta K. Swamy, Kristin E. Weaver and Carolyn A. Weinbaum (Duke University) for clinical study and regulatory support; Juliana Carnielli, Solange Alves Vinhas and Keyla Fonseca for establishing cohort enrollment and sample collections at Federal University of Espírito Santo; the Cassiano Antonio de Moraes Hospital maternity ward staff for supporting the participants’ deliveries and sample collections, and the mothers who participated in this study and made efforts to provide samples for research during pregnancy.
Declaration of interests:
M.B., S.R.P., T.S. and K.K.H. have filed a patent application directed to antibodies that are related to this work. S.R.P. serves as a consultant to Moderna, Merck, Pfizer, GSK, Hoopika, and Dynavax CMV vaccine programs and leads a sponsored research program with Moderna and Merck. All other authors declare no conflict of interest.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Inclusion and Diversity: We support inclusive, diverse, and equitable conduct of research.
REFERENCES
- Abbink P, Larocca RA, De La Barrera RA, Bricault CA, Moseley ET, Boyd M, Kirilova M, Li Z, Ng’ang’a D, Nanayakkara O, et al. (2016). Protective efficacy of multiple vaccine platforms against Zika virus challenge in rhesus monkeys. Science. 353, 1129–1132. 10.1126/science.aah6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baronti C, Piorkowski G, Charrel RN, Boubis L, Leparc-Goffart I, and de Lamballerie X (2014). Complete coding sequence of Zika virus from a French Polynesia outbreak in 2013. Genome Announc. 2, 500–514. 10.1128/genomeA.00500-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonsignori M, Hwang K-K, Chen X, Tsao C-Y, Morris L, Gray E, Marshall DJ, Crump JA, Kapiga SH, Sam NE, et al. (2011). Analysis of a Clonal Lineage of HIV-1 Envelope V2/V3 Conformational Epitope-Specific Broadly Neutralizing Antibodies and Their Inferred Unmutated Common Ancestors. J. Virol 85, 9998–10009. 10.1128/JVI.05045-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonsignori M, Wiehe K, Grimm SK, Lynch R, Yang G, Kozink DM, Perrin F, Cooper AJ, Hwang KK, Chen X, et al. (2014). An autoreactive antibody from an SLE/HIV-1 individual broadly neutralizes HIV-1. J. Clin. Invest 124, 1835–1843. 10.1172/JCI73441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonsignori M, Zhou T, Sheng Z, Chen L, Gao F, Joyce MG, Ozorowski G, Chuang GY, Schramm CA, Wiehe K, et al. (2016). Maturation Pathway from Germline to Broad HIV-1 Neutralizer of a CD4-Mimic Antibody. Cell 165, 449–463. 10.1016/j.cell.2016.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonsignori M, Scott E, Wiehe K, Easterhoff D, Alam SM, Hwang KK, Cooper M, Xia SM, Zhang R, Montefiori DC, et al. (2018). Inference of the HIV-1 VRC01 Antibody Lineage Unmutated Common Ancestor Reveals Alternative Pathways to Overcome a Key Glycan Barrier. Immunity 49, 1162–1174.e8. 10.1016/j.immuni.2018.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvert AE, Horiuchi K, Boroughs KL, Ong YT, Anderson KM, Biggerstaff BJ, Stone M, Simmons G, Busch MP, and Huang CYH (2021). The Specificity of the Persistent IgM Neutralizing Antibody Response in Zika Virus Infections among Individuals with Prior Dengue Virus Exposure. J. Clin. Microbiol 59, e0040021. 10.1128/JCM.00400-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos MC, Dombrowski JG, Phelan J, Marinho CRF, Hibberd M, Clark TG, and Campino S (2018). Zika might not be acting alone: Using an ecological study approach to investigate potential co-acting risk factors for an unusual pattern of microcephaly in Brazil. PLoS One 13, e0201452. 10.1371/journal.pone.0201452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbaugh DL, Baric RS, and Lazear HM (2019). Envelope Protein Glycosylation Mediates Zika Virus Pathogenesis. J. Virol 93. 10.1128/jvi.00113-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CYY, Low JZH, Gan ES, Ong EZ, Zhang SL-X, Tan HC, Chai X, Ghosh S, Ooi EE, and Chan KR (2019). Antibody-Dependent Dengue Virus Entry Modulates Cell Intrinsic Responses for Enhanced Infection. MSphere 4, 528–547. 10.1128/msphere.00528-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KR, Zhang SL-X, Tan HC, Chan YK, Chow A, Lim APC, Vasudevan SG, Hanson BJ, and Ooi EE (2011). Ligation of Fc gamma receptor MB inhibits antibody-dependent enhancement of dengue virus infection. Proc. Natl. Acad. Sci 108, 12479–12484. 10.1073/pnas.1106568108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KR, Ong EZ, Tan HC, Zhang SL-X, Zhang Q, Tang KF, Kaliaperumal N, Lim APC, Hibberd ML, Chan SH, et al. (2014). Leukocyte immunoglobulin-like receptor B1 is critical for antibody-dependent dengue. Proc. Natl. Acad. Sci. U. S. A 111, 2722–2727. 10.1073/pnas.1317454111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KR, Ong EZ, Mok DZL, and Ooi EE (2015). Fc receptors and their influence on efficacy of therapeutic antibodies for treatment of viral diseases. Expert Rev. Anti. Infect. Ther 13, 1351–1360. 10.1586/14787210.2015.1079127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen JS, Andersen AR, Osther K, Peitersen B, Bach-Mortensen N, and Lebech PE (1976). The Relationship Between Pregnancy, HCS and B Lymphocytes. Acta Pathol. Microbiol. Scand. Sect. C Immunol. 84 C, 313–318. 10.1111/j.1699-0463.1976.tb00035.x. [DOI] [PubMed] [Google Scholar]
- Collins MH, Tu HA, Gimblet-Ochieng C, Liou GJA, Jadi RS, Metz SW, Thomas A, McElvany BD, Davidson E, Doranz BJ, et al. (2019). Human antibody response to Zika targets type-specific quaternary structure epitopes. JCI Insight 4. 10.1172/jci.insight.124588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig AM, Hughes BL, and Swamy GK (2021). Coronavirus disease 2019 vaccines in pregnancy. Am. J. Obstet. Gynecol. MFM 3, 100295. 10.1016/j.ajogmf.2020.100295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond MS, Sitati EM, Friend LD, Higgs S, Shrestha B, and Engle M (2003). A Critical Role for Induced IgM in the Protection against West Nile Virus Infection. J. Exp. Med 198, 1853–1862. 10.1084/jem.20031223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd KA, Demaso CR, Pelc RS, Speer SD, Smith ARY, Goo L, Platt DJ, Mascola JR, Graham BS, Mulligan MJ, et al. (2016). Broadly Neutralizing Activity of Zika Virus-Immune Sera Identifies a Single Viral Serotype HHS Public Access. Cell Rep 16, 1485–1491. 10.1016/j.celrep.2016.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr 66, 486–501. 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser R, Cloutier M, Prévost J, Fink C, Ducas É, Dussault N, Landry P, Tremblay T, Laforce-Lavoie A, Lewin A, et al. (2020). Major role of IgM in the neutralizing activity of convalescent plasma against SARS-CoV-2. Preprint on BioRxiv 2020.10.09.333278. 10.1101/2020.10.09.333278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibney KB, Edupuganti S, Panella AJ, Kosoy OI, Delorey MJ, Lanciotti RS, Mulligan MJ, Fischer M, and Staples JE (2012). Detection of anti-yellow fever virus immunoglobulin M antibodies at 3-4 years following yellow fever vaccination. Am. J. Trap. Med. Hyg 87, 1112–1115. 10.4269/ajtmh.2012.12-0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchuk P, Bombardi RG, Erasmus JH, Tan Q, Nargi R, Soto C, Abbink P, Suscovich TJ, Durnell LA, Khandhar A, et al. (2020). Integrated pipeline for the accelerated discovery of antiviral antibody therapeutics. Nat. Biomed. Eng 4, 1030–1043. 10.1038/S41551-020-0594-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes MF, De La Fuente-Núñez V, Saxena A, and Kuesel AC (2017). Protected to death: Systematic exclusion of pregnant women from Ebola virus disease trials. Reprod. Health 14. 10.1186/s12978-017-0430-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant T, Rohou A, and Grigorieff N (2018). CisTEM, user-friendly software for single-particle image processing. Elife 7. 10.7554/eLife.35383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin I, Martin SW., Fischer M, Chambers TV., Kosoy OL., Goldberg C, Falise A, Villamil V, Ponomareva O, Gillis LD., et al. (2019a). Zika virus IGM 25 months after symptom onset, Miami-Dade County, Florida, USA. Emerg. Infect. Dis 25, 2264–2265. 10.3201/eid2512.191022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin I, Martin SW., Fischer M, Chambers TV., Kosoy O, Falise A, Ponomareva O, Gillis LD., Blackmore C, and Jean R (2019b). Zika virus IgM detection and neutralizing antibody profiles 12–19 months after illness onset. Emerg. Infect. Dis 25, 299–303. 10.3201/eid2502.181286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, and Jiang W (2014). Single particle cryo-electron microscopy and 3-D reconstruction of viruses. Methods Mol. Biol 1117, 401–443. 10.1007/978-1-62703-776-1_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halstead SB (1988). Pathogenesis of dengue: Challenges to molecular biology. Science. 239, 476–481. 10.1126/science.3277268. [DOI] [PubMed] [Google Scholar]
- Halstead SB, and O’Rourke EJ (1977). Antibody-enhanced dengue virus infection in primate leukocytes. Nature 265, 739–741. 10.1038/265739a0. [DOI] [PubMed] [Google Scholar]
- Hasan SS, Miller A, Sapparapu G, Fernandez E, Klose T, Long F, Fokine A, Porta JC, Jiang W, Diamond MS, et al. (2017a). A human antibody against Zika virus crosslinks the E protein to prevent infection. Nat. Commun 8, 14722. 10.1038/ncomms14722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath E, Begue-Pastor N, Chaganti S, Croom-Carter D, Shannon-Lowe C, Kube D, Feederle R, Delecluse H-J, Rickinson AB, and Bell AI (2012). Epstein-Barr Virus Infection of Naïve B Cells In Vitro Frequently Selects Clones with Mutated Immunoglobulin Genotypes: Implications for Virus Biology. PLOS Pathog. 8, e1002697. 10.1371/journal.ppat.1002697.t002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hombach J, Solomon T, Kurane I, Jacobson J, and Wood D (2005). Report on a WHO consultation on immunological endpoints for evaluation of new Japanese encephalitis vaccines, WHO, Geneva, 2-3 September, 2004. Vaccine 23, 5205–5211. 10.1016/j.vaccine.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Katzelnick LC, Harris E, Baric R, Coller BA, Coloma J, Crowe JE, Cummings DAT, Dean H, De Silva A, Diamond MS, et al. (2017). Immune correlates of protection for dengue: State of the art and research Agenda. In Vaccine, (Elsevier Ltd; ), pp. 4659–4669. 10.1016/j.vaccine.2017.07.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepler TB (2013). Reconstructing a B-cell clonal lineage. I. Statistical inference of unobserved ancestors. F1000Research 2, 103. 10.12688/f1000research.2-103.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyt BA, Baliga R, Sinclair AM, Carroll SF, and Peterson MS (2020). Structure, Function, and Therapeutic Use of IgM Antibodies. Antibodies 9, 53. 10.3390/antib9040053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King HW, Orban N, Riches JC, Clear AJ, Warnes G, Teichmann SA, and James LK (2021). Single-cell analysis of human B cell maturation predicts how antibody class switching shapes selection dynamics. Sci. Immunol 6. 10.1126/sciimmunol.abe6291. [DOI] [PubMed] [Google Scholar]
- Kreil TR, Burger I, Bachmann M, Fraiss S, and Eibl MM (1997). Antibodies protect mice against challenge with tick-borne encephalitis virus (TBEV)-infected macrophages. Clin. Exp. Immunol 110, 358–361. 10.1046/j.1365-2249.1997.4311446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwek S. Sen, Watanabe S, Chan KR, Ong EZ, Tan HC, Ng WC, Nguyen MTX, Gan ES, Zhang SL, Chan KWK, et al. (2018). A systematic approach to the development of a safe live attenuated Zika vaccine. Nat. Commun 9, 1031. 10.1038/s41467-018-03337-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larocca RA, Abbink P, Peron JPS, Zanotto PMDA, Iampietro MJ, Badamchi-Zadeh A, Boyd M, Ng’ang’a D, Kirilova M, Nityanandam R, et al. (2016). Vaccine protection against Zika virus from Brazil. Nature 536, 474–478. 10.1038/nature18952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazear HM, Govero J, Smith AM, Platt DJ, Fernandez E, Miner JJ, and Diamond MS (2016). A Mouse Model of Zika Virus Pathogenesis. Cell Host Microbe 19, 720–730. 10.1016/j.chom.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessler J, Ott CT, Carcelen AC, Konikoff JM, Williamson J, Bi Q, Kucirka LM, Cummings DAT, Reich NG, and Chaisson LH (2016). Times to key events in Zika virus infection and implications for blood donation: A systematic review. Bull. World Health Organ 94, 841–849. 10.2471/BLT.16.174540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H-X, Levesque MC, Nagel A, Dixon A, Zhang R, Walter E, Parks R, Whitesides J, Marshall DJ, Hwang K-K, et al. (2009). High-throughput isolation of immunoglobulin genes from single human B cells and expression as monoclonal antibodies. J Virol Methods 158, 171–179. 10.1016/jjviromet.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima J, Martins C, Leandro MJ, Nunes G, Sousa MJ, Branco JC, and Borrego LM (2016). Characterization of B cells in healthy pregnant women from late pregnancy to postpartum: A prospective observational study. BMC Pregnancy Childbirth 16, 139. 10.1186/s12884-016-0927-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo ED, Hansen RJ, and Balthasar JP (2004). Antibody pharmacokinetics and pharmacodynamics. J. Pharm. Sci 93, 2645–2668. 10.1002/jps.20178. [DOI] [PubMed] [Google Scholar]
- Long F, Doyle M, Fernandez E, Miller AS, Klose T, Sevvana M, Bryan A, Davidson E, Doranz BJ, Kuhn RJ, et al. (2019). Structural basis of a potent human monoclonal antibody against Zika virus targeting a quaternary epitope. Proc. Natl. Acad. Sci. U. S. A 116, 1591–1596. 10.1073/pnas.1815432116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo XM, Maarschalk E, O ’connell RM, Wang P, Yang L, Baltimore D, Connell RMO, Wang P, Yang L, and Baltimore D (2009). Engineering human hematopoietic stem / progenitor cells to produce a broadly neutralizing anti-HIV antibody after in vitro maturation to human B lymphocytes. Vaccine 113, 1422–1431. 10.1182/blood-2008-09-177139. [DOI] [PubMed] [Google Scholar]
- Malafa S, Medits I, Aberle JH, Aberle SW, Haslwanter D, Tsouchnikas G, Wölfel S, Huber KL, Percivalle E, Cherpillod P, et al. (2020). Impact of flavivirus vaccine-induced immunity on primary zika virus antibody response in humans. PLoS Negl. Trap. Dis 14, e0008034. 10.1371/journal.pntd.0008034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason RA, Tauraso NM, Spertzel RO, and Ginn RK (1973). Yellow fever vaccine: direct challenge of monkeys given graded doses of 17D vaccine. Appl. Microbiol 25, 539–544. 10.1128/am.25.4.539-544.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlhop E, Nelson S, Jost CA, Gorlatov S, Johnson S, Fremont DH, Diamond MS, and Pierson TC (2009). Complement Protein C1q Reduces the Stoichiometric Threshold for Antibody-Mediated Neutralization of West Nile Virus. Cell Host Microbe 6, 381–391. 10.1016/j.chom.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monath T (1971). Neutralizing antibody responses in the major immunoglobulin classes to Yellow Fever 17D vaccination of humans. Am. J. Epidemiol 93, 122–129. 10.1093/oxfordjournals.aje.a121232. [DOI] [PubMed] [Google Scholar]
- Morabito KM, and Graham BS (2017). Zika Virus Vaccine Development. J. Infect. Dis 216, S957–S963. 10.1093/infdis/jix464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Sirohi D, Dowd KA, Chen Z, Diamond MS, Kuhn RJ, and Pierson TC (2016). Enhancing dengue virus maturation using a stable furin over-expressing cell line. Virology 497, 33–40. 10.1016/j.virol.2016.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller R, Gräwert MA, Kern T, Madl T, Peschek J, Sattler M, Groll M, and Buchner J (2013). High-resolution structures of the IgM Fc domains reveal principles of its hexamer formation. Proc. Natl. Acad. Sci. U. S. A 110, 10183–10188. 10.1073/pnas.1300547110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K, Travers P, Walport M, and Janeway C (2012). Janeway’s Immunobiology (New York: Garland Science; ). [Google Scholar]
- Murray KO, Garcia MN, Yan C, and Gorchakov R (2013). Persistence of Detectable Immunoglobulin M Antibodies Up to 8 Years After Infection with West Nile Virus. Am. J. Trop. Med. Hyg 89, 996–1000. 10.4269/AJTMH.13-0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TG, Ward CM, and Morris JM (2013). To B or not to B cells-mediate a healthy start to life. Clin. Exp. Immunol 171, 124–134. 10.1111/cei.12001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen-Saines K, Brasil P, Kerin T, Vasconcelos Z, Gabaglia CR, Damasceno L, Pone M, Abreu de Carvalho LM, Pone SM, Zin AA, et al. (2019). Delayed childhood neurodevelopment and neurosensory alterations in the second year of life in a prospective cohort of ZIKV-exposed children. Nat. Med 25, 1213–1217. 10.1038/s41591-019-0496-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz-Bailey G, Rosenberg ES, Doyle K, Munoz-Jordan J, Santiago GA, Klein L, Perez-Padilla J, Medina FA, Waterman SH, Adams LE, et al. (2018). Persistence of Zika Virus in Body Fluids — Final Report. N. Engl. J. Med 379, 1234–1243. 10.1056/NEJMoal613108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins SJ, Nealis AS, Sutton BJ, and Feinstein A (1991). Solution structure of human and mouse immunoglobulin M by synchrotron X-ray scattering and molecular graphics modelling. A possible mechanism for complement activation. J. Mol. Biol 221, 1345–1366. 10.1016/0022-2836(91)90937-2. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera - A visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–1612. 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Premkumar L, Collins M, Graham S, Liou GJA, Lopez CA, Jadi R, Balmaseda A, Brackbill JA, Dietze R, Camacho E, et al. (2018). Development of envelope protein antigens to serologically differentiate zika virus infection from dengue virus infection. J. Clin. Microbiol 56, JCM.01504-17. 10.1128/JCM.01504-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsland PA, Terzyan SS, Cloud G, Bourne CR, Farrugia W, Tribbick G, Geysen HM, Moomaw CR, Slaughter CA, and Edmundson AB (2006). Crystal structure of a glycosylated Fab from an IgM cryoglobulin with properties of a natural proteolytic antibody. Biochem. J 395, 473–481. 10.1042/BJ20051739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravichandran S, Hahn M, Belaunzarán-Zamudio PF, Ramos-Castañeda J, Nájera-Cancino G, Caballero-Sosa S, Navarro-Fuentes KR, Ruiz-Palacios G, Golding H, Beigel JH, et al. (2019). Differential human antibody repertoires following Zika infection and the implications for serodiagnostics and disease outcome. Nat. Commun 10, 1–14. 10.1038/S41467-019-09914-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds MR, Jones AM, Petersen EE, Lee EH, Rice ME, Bingham A, Ellington SR, Evert N, Reagan-Steiner S, Oduyebo T, et al. (2017). Vital Signs: Update on Zika Virus–Associated Birth Defects and Evaluation of All U.S. Infants with Congenital Zika Virus Exposure — U.S. Zika Pregnancy Registry, 2016. MMWR. Morb. Mortal. Wkly. Rep 66, 366–373. 10.15585/mmwr.mm6613e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richner JM, Jagger BW, Shan C, Fontes CR, Dowd KA, Cao B, Himansu S, Caine EA, Nunes BTD, Medeiros DBA, et al. (2017). Vaccine Mediated Protection Against Zika Virus-Induced Congenital Disease. Cell 170, 273–283.e12. 10.1016/j.cell.2017.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani DF, Bozzacco L, Keeffe JR, Khouri R, Olsen PC, Gazumyan A, Schaefer-Babajew D, Avila-Rios S, Nogueira L, Patel R, et al. (2017). Recurrent Potent Human Neutralizing Antibodies to Zika Virus in Brazil and Mexico. Cell 169, 597–609.e11. 10.1016/j.cell.2017.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers TF, Goodwin EC, Briney B, Sok D, Beutler N, Strubel A, Nedellec R, Le K, Brown ME, Burton DR, et al. (2017). Zika virus activates de novo and cross-reactive memory B cell responses in dengue-experienced donors. Sci. Immunol 2. 10.1126/sciimmunol.aan6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohou A, and Grigorieff N (2015). CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol 192, 216–221. 10.1016/jjsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal PB, and Henderson R (2003). Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol 333, 721–745. 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- Rouvinski A, Guardado-Calvo P, Barba-Spaeth G, Duquerroy S, Vaney MC, Kikuti CM, Navarro Sanchez ME, Dejnirattisai W, Wongwiwat W, Haouz A, et al. (2015). Recognition determinants of broadly neutralizing human antibodies against dengue viruses. Nature 520, 109–113. 10.1038/nature14130. [DOI] [PubMed] [Google Scholar]
- Roy A, Kucukural A, and Zhang Y (2010). I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc 5, 725–738. 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapparapu G, Fernandez E, Kose N, Cao Bin, Fox JM, Bombardi RG, Zhao H, Nelson CA, Bryan AL, Barnes T, et al. (2016). Neutralizing human antibodies prevent Zika virus replication and fetal disease in mice. Nature 540, 443–447. 10.1038/nature20564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH. (2016). Methods in Enzymology, Vol. 579 (Cambridge, MA: Academic Press; ). [Google Scholar]
- Scheres SHW, Valle M, and Carazo JM (2005). Fast maximum-likelihood refinement of electron microscopy images. Bioinformatics 21. 10.1093/bioinformatics/bti1140. [DOI] [PubMed] [Google Scholar]
- Sevvana M, Long F, Miller AS, Klose T, Buda G, Sun L, Kuhn RJ, and Rossmann MG (2018). Refinement and Analysis of the Mature Zika Virus Cryo-EM Structure at 3.1 Å Resolution. Structure 26, 1169–1177.e3. 10.1016/j.str.2018.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp TH, Boyle AL, Diebolder CA, Kros A, Koster AJ, and Gros P (2019). Insights into IgM-mediated complement activation based on in situ structures of IgM-C1-C4b. Proc. Natl. Acad. Sci. U. S. A 116, 11900–11905. 10.1073/pnas.1901841116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigworth FJ (1998). A maximum-likelihood approach to single-particle image refinement. J. Struct. Biol 122, 328–339. 10.1006/jsbi.1998.4014. [DOI] [PubMed] [Google Scholar]
- Sigworth FJ (2004). Classical detection theory and the cryo-EM particle selection problem. In Journal of Structural Biology, (J Struct Biol), pp. 111–122. [DOI] [PubMed] [Google Scholar]
- Singh T, Lopez CA, Giuberti C, Dennis ML, Itell HL, Heimsath HJ, Webster HS, Roark HK, Merçon de Vargas PR, Hall A, et al. (2019). Efficient transplacental IgG transfer in women infected with Zika virus during pregnancy. PLoS Negl. Trop. Dis 13, e0007648. 10.1371/journal.pntd.0007648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirohi D, Chen Z, Sun L, Klose T, Pierson TC, Rossmann MG, and Kuhn RJ (2016). The 3.8Å resolution cryo-EM structure of Zika Virus. 352, 467–470. 10.1126/science.aaf5316.The.The. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit JM, Moesker B, Rodenhuis-Zybert I, and Wilschut J (2011). Flavivirus cell entry and membrane fusion. Viruses 3, 160–171. 10.3390/v3020160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SA, de Alwis AR, Kose N, Harris E, Ibarra KD, Kahle KM, Pfaf JM, Xiang X, Doranz BJ, de Silva AM, et al. (2013). The potent and broadly neutralizing human dengue virus-specific monoclonal antibody 1C19 reveals a unique cross-reactive epitope on the bc loop of domain II of the envelope protein. MBio 4. 10.1128/mBio.00873-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SA, Silva LA, Fox JM, Flyak AI, Kose N, Sapparapu G, Khomandiak S, Ashbrook AW, Kahle KM, Fong RH, et al. (2015). Isolation and characterization of broad and ultrapotent human monoclonal antibodies with therapeutic activity against chikungunya virus. Cell Host Microbe 18, 86–95. 10.1016/j.chom.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stettler K, Beltramello M, Espinosa DA, Graham V, Cassotta A, Bianchi S, Vanzetta F, Minola A, Jaconi S, Mele F, et al. (2016). Specificity, cross-reactivity, and function of antibodies elicited by Zika virus infection. Science. 353, 823–826. 10.1126/science.aaf8505. [DOI] [PubMed] [Google Scholar]
- Stone M, Bakkour S, Lanteri MC, Brambilla D, Simmons G, Bruhn R, Kaidarova Z, Lee TH, Orlando Alsina J, Williamson PC, et al. (2020). Zika virus RNA and IgM persistence in blood compartments and body fluids: a prospective observational study. Lancet Infect. Dis 20, 1446–1456. 10.1016/S1473-3099(19)30708-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, and Carragher B (2005). Automated molecular microscopy: The new Leginon system. J. Struct. Biol 151, 41–60. 10.1016/jjsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Swanstrom JA, Plante JA, Plante KS, Young EF, McGowan E, Gallichotte EN, Widman DG, Heise MT, de Silva AM, and Baric RS (2016). Dengue virus envelope dimer epitope monoclonal antibodies isolated from dengue patients are protective against zika virus. MBio 7. 10.1128/mBio.01123-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonnerre P, Melgaço JG, Torres-Cornejo A, Pinto MA, Yue C, Blümel J, de Sousa PSF, de Mello V. da M., Moran J, de Filippis AMB, et al. (2020). Evolution of the innate and adaptive immune response in women with acute Zika virus infection. Nat. Microbiol 5, 76–83. 10.1038/s41564-019-0618-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Rompay KKA, Coffey LL, Kapoor T, Gazumyan A, Keesler RI, Jurado A, Peace A, Agudelo M, Watanabe J, Usachenko J, et al. (2020). A combination of two human monoclonal antibodies limits fetal damage by Zika virus in macaques. Proc. Natl. Acad. Sci 117, 202000414. 10.1073/pnas.2000414117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Bardelli M, Espinosa DA, Pedotti M, Ng TS, Bianchi S, Simonelli L, Lim EXY, Foglierini M, Zatta F, et al. (2017). A Human Bi-specific Antibody against Zika Virus with High Therapeutic Potential. Cell 171, 229–241.e15. 10.1016/j.cell.2017.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wec AZ, Haslwanter D, Abdiche YN, Shehata L, Pedreño-Lopez N, Moyer CL, Bornholdt ZA, Lilov A, Nett JH, Jangra RK, et al. (2020). Longitudinal dynamics of the human B cell response to the yellow fever 17D vaccine. Proc. Natl. Acad. Sci. U. S. A 117, 6675–6685. 10.1073/pnas.1921388117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiersma EJ, Collins C, Fazel S, and Shulman MJ (1998). Structural and functional analysis of J chain-deficient IgM. J. Immunol 160, 5979–5989. . [PubMed] [Google Scholar]
- Xiao C, and Rossmann MG (2007). Interpretation of electron density with stereographic roadmap projections. J. Struct. Biol 158, 182–187. 10.1016/jjsb.2006.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaari G, Vander Heiden J, Uduman M, Gadala-Maria D, Gupta N, Stern JNH, O’Connor K, Hafler D, Laserson U, Vigneault F, et al. (2013). Models of Somatic Hypermutation Targeting and Substitution Based on Synonymous Mutations from High-Throughput Immunoglobulin Sequencing Data. Front. Immunol 0, 358. 10.3389/FIMMU.2013.00358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Yan R, Roy A, Xu D, Poisson J, and Zhang Y (2014). The I-TASSER suite: Protein structure and function prediction. Nat. Methods 12, 7–8. 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. (2008). I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9. 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SLX, Tan HC, Hanson BJ, and Ooi EE (2010). A simple method for Alexa Fluor dye labelling of dengue virus. J. Virol. Methods 167, 172–177. 10.1016/j.jviromet.2010.04.001. [DOI] [PubMed] [Google Scholar]
- Zhang X, Sheng J, Austin SK, Hoornweg TE, Smit JM, Kuhn RJ, Diamond MS, and Rossmann MG (2015). Structure of Acidic pH Dengue Virus Showing the Fusogenic Glycoprotein Trimers. J. Virol 89, 743–750. 10.1128/jvi.02411-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Fernandez E, Dowd KA, Speer SD, Platt DJ, Gorman MJ, Govero J, Nelson CA, Pierson TC, Diamond MS, et al. (2016). Structural Basis of Zika Virus-Specific Antibody Protection. Cell 166, 1016–1027. 10.1016/j.cell.2016.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, and Agard DA (2017). MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332. 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Lack of linear relationship between plasma ZIKV binding IgM levels and IgM-mediated neutralization. Related to Figure 1. ZIKV binding IgM levels were measured at 1:10 via ELISA and IgM mediated neutralization was measured by focus reduction neutralization tested on paired IgM- and mock-depleted samples. A. Spearman Rank correlation with all samples included in this study (n=21). B. Correlation with the subset of samples in which IgM contributed to ZIKV neutralization (n=15).
Figure S2. Cross-reactivity with DENV 1-4 serotypes. Related to Figure 3. A. ZIKV and DENV1-4 virion binding plasma IgG measured by ELISA and expressed as Log10 ED50 from all collections from which B cell responses were analyzed (n=6; DPS: days post symptoms). Maternal flavivirus serostatus as previously defined (Singh et al., 2019) is shown for each subject. Plasma samples were run in duplicate. B. DH1017.IgM neutralization of DENV 1-4 strains. Error bars: SD of technical replicates run in duplicate. C. DH1017.IgM repeated neutralization assays against the ZIKV PRVABC59 (n=10) and H/PF/2013 (n=2) strains. Line: geometric mean. Replicate experiments were independently performed by multiple operators and at different institutions for ZIKV H/PF/2013.
Figure S3. Functional in vitro characterization of DH1017.IgM and DH1017.IgG. Related to Figure 4. A. DH1017.IgM (left) and DH1017.IgG (right) binding to Zika and Dengue 1-4 virions at 100 μg/mL, measured by ELISA and expressed as optical density at 450nm (OD450). Mean of technical replicates is shown. Negative control is the mean of the diluent alone. B. Antibody-dependent enhancement of DH1017.IgM and DH1017.IgG on primary monocytes measured in a plaque assay. Dotted line: mean of six virus-only control replicates. Grey area: ≤1 SD. Error bars: SD from 6 replicates. C. DH1017.IgM, DH1017.IgG and DH1017.Fab binding to a panel of autoantigens (Sjogren’s syndrome antigens A and B [SSA and SSB, respectively], Smith antigen [Sm], ribonucleoprotein [RNP], centromere B, histone, scleroderma 70 [SCL-70], Jo-1 proteins, and DNA) measured in ELISA. Respective positive controls are shown in the right panel. Error bars: SD of duplicate observations. D. Hep2 cells were stained with DH1017.IgG and DH1017.IgM, 2F5 mAb (positive control), and 17b mAb (negative control) at a concentration of 50 μg/mL. Images shown were taken with a 40X objective with an 8s exposure and are representative of duplicate experiments.
Figure S4. DH1017.Fab interacts with ZIKV surface at multiple angles. Related to Figure 6. A. FCS curve calculated from two half maps of the final model at 1.32 Å/pixel with the RNA core masked out. B. Section of electron density map fit with the asymmetric ectodomain, and a model of the DH1017 variable domain structure at the i2f axis and the Fab model at the q2f axis. DI: red, DII: yellow, and DIII: blue. The DH1017 Fab fragment variable domain (VH, VL) is colored cyan, with the constant domain (c-domain) in green. The two different positions of constant domain density at the i2f axis are labeled c-a and c-b. The variable domain density at both axes is labeled VH-VL. The constant domain density at the q2f axis is labeled c-domain. The i2f and q2f axes are shown. C. Top-down view from the i5f axis of the density map with Fab DH1017 structures fit at the i2f axes (cyan) and q2f axes (green). Two neighboring asymmetric units are shown with open black triangles and the i5f and i3f axes are labeled as a pentagon and triangle, respectively.
Figure S5. DH1017.IgM pentamer multivalent binding to Zika virus particles. Related to Figure 6. A. A top down (left) and side view (right) of a model of the pentamer binding the ZIKV particle in an umbrella like conformation. The density map is shown in gray. The Fc domain is shown in yellow, domains Cμ3 and Cμ4 are labeled. The Cμ2 domain is colored magenta. The DH1017 Fab structure fit to the density map is shown in cyan at the i2f and in green at the q2f. B Electron micrographs of purified ZIKV particles incubated with IgM pentameric DH1017.IgM mAb. Numbered red boxes indicate selected fields showing potential multivalent pentamer binding. Micrographs were collected at 200kV accelerating voltage, magnification 47,000x with a Talos F200C (Thermofisher). C. Expanded view of micrographs of purified Zika particles incubated with IgM pentameric DH1017.IgM mAb. Numbering corresponds to the red boxes in Panel B. Red arrows point to densities predicted to be multivalent binding of pentameric DH1017.IgM. Scale bar is 200 nm. D. The density map of the Fab bound virus particle in an icosahedral cage with top-down views at each icosahedral axis, i2f (left), i5f (middle), and i3f (right). The green asterisk indicates a position where the pentamer is bound as shown in panel A. The red X indicates the position of five-fold axes related to the green asterisk by two-fold symmetry that do not have the i2f position available for Fab binding to form the umbrella like conformation. The i3f view shows positions of complete occupancy for the particle bound as shown in panel A.
Figure S6. DH1017.IgM pentamer cross-linking of Zika virus particles. Related to Figure 6. A. Schematic representation of virus particles cross-linked by pentamers. The Fc portion of the molecule is shown in red cross-linking virus. B. Aggregation of Zika particles possibly facilitated by pentamer crosslinking of particles. Blue arrow: density predicted to be IgM binding two virus particles. Red arrow: density suggesting multivalent binding to a single virion, as also shown in Figure S5C. Scale bar is 200 nm.
Data Availability Statement
Sequences of Ig V(D)J rearrangements have been deposited in GenBank and are publicly available as of the date of publication. Accession numbers are listed in the key resource table. The Zika virus glycoprotein E and DH1017.Fab structures have been deposited in Protein Data Bank and are publicly available as of the date of publication. The accession number is listed in the key resource table. The density map of DH1017.Fab in complex with Zika virions has been deposited in EMDataResource and is publicly available as of the date of publication. The accession number is listed in the key resource table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| IgD-PE (clone IA6-2) | BD Biosciences | 555779 |
| CD10 PE-CF594 (clone HI10a) | BD Biosciences | 562396 |
| CD3 PE-Cy5 (clone HIT3a) | BD Biosciences | 555341 |
| CD14 BV605 (clone M5E2) | Biolegend | 301834 |
| CD16 BV570 (clone 3G8) | Biolegend | 302035 |
| CD27 PE-Cy7 (clone O323) | Thermo Fisher Scientific | 25-0279-42 |
| CD38 APC-AF700 (clone LS198-4-3) | Beckman Coulter | B23489 |
| CD19 APC-Cy7 (clone SJ25C1) | BD Biosciences | 557791 |
| (HRP)-conjugated goat anti-human IgG antibody | Jackson ImmunoResearch Laboratories | 109-035-008 |
| HRP-conjugated goat anti human IgM antibody | Jackson ImmunoResearch Laboratories | 109-035-129 |
| HRP-conjugated goat anti-human IgA antibody | Jackson ImmunoResearch Laboratories | 109-035-011 |
| C10 | Absolute Antibody | AB00677-10.0 |
| Anti NS1 IgM | myBiosource | MBS6120634 |
| Palivizumab | Duke Pharmacy, Durham NC | N/A |
| 1M7 | Smith et al., 2013; Produced from a hybridoma cell line at the UNC Protein Expression Core Facility | N/A |
| ZV-2 | Zhao et al., 2016; Produced from a hybridoma cell line at the UNC Protein Expression Core Facility | N/A |
| ZV-64 | Zhao et al., 2016; Produced from a hybridoma cell line at the UNC Protein Expression Core Facility | N/A |
| ZKA190 | Wang et al., 2017; Recombinantly produced by the UNC Protein Expression Core Facility | N/A |
| ZIKA-752 | Kindly provided by Dr. James Crowe, Vanderbilt University | N/A |
| ZIKA-893 | Kindly provided by Dr. James Crowe, Vanderbilt University | N/A |
| rZIKV-195 | Kindly provided by Dr. James Crowe, Vanderbilt University | N/A |
| DH1017.IgG | Recombinantly produced in this study | N/A |
| DH1017.Fab | Recombinantly produced in this study | N/A |
| 4G2 | ATCC | HB-112 |
| humanized IgG1 4G2 | Produced in-house | N/A |
| 3H5 | ATCC | HB-46 |
| ChromPure Human IgM (myeloma), whole molecule (2 mg) | Jackson ImmunoResearch Laboratories | 009-000-012 |
| goat anti-human Ig polyvalent antibody | Thermo Fisher Scientific | H17000 |
| goat anti-human H+L | Promega | W403B |
| IgG1 mAb 2F5 | Produced in-house | N/A |
| IgG1 mAb 17B | Produced in-house | N/A |
| Anti-Centromere B | Immunovision | HCT-0100 |
| Anti-single stranded DNA | Immunovision | HSS-0100 |
| Anti-Histone | Immunovision | HIS-0100 |
| Anti-Jo 1 | Immunovision | HJO-0100 |
| Anti-SRC | Immunovision | HRN-0100 |
| Anti-Scl 70 | Immunovision | HSC-0100 |
| Anti-Sm | Immunovision | HSM-0100 |
| Anti-SSA | Immunovision | HSA-0100 |
| Anti-SSB | Immunovision | HSB-0100 |
| Bacterial and virus strains | ||
| DENV1 (WestPac74) | Kindly provided by Dr. Aravinda de Silva, UNC | N/A |
| DENV2 (S-16603) | Kindly provided by Dr. Aravinda de Silva, UNC | N/A |
| DENV3 (CH54389) | Kindly provided by Dr. Aravinda de Silva, UNC | N/A |
| DENV4 (TVP-360) | Kindly provided by Dr. Aravinda de Silva, UNC | N/A |
| ZIKV (PRVABC59) | BEI Resources | NR-50240 |
| DENV1 (Hawaii NR-82) | BEI Resources | NR-82 |
| DENV2 (New Guinea C) | BEI Resources | NR-84 |
| DENV3 (Philippines) | BEI Resources | NR-80 |
| DENV4 (H241) | BEI Resources | NR-86 |
| ZIKV (PD13/251013-18) | Kindly provided by Dr. Didier Musso, IHU Mediterranee Infection Foundation, Tahiti, French Polynesia | N/A |
| ZIKV (H/PF/2013) | US Centers for Disease Control and Prevention; Baronti et al., 2014 | N/A |
| Biological samples | ||
| Human plasma and PBMCs from pregnant women with ZIKV infection | IRB-Approved prospective cohort led by Dr. S.R. Permar and Dr. R. Dietze | N/A |
| Normal human serum from human male AB plasma, USA origin, sterile-filtered | Sigma | H4522 |
| Chemicals, peptides, and recombinant proteins | ||
| ZIKV RVP | Dowd et al., 2016 | N/A |
| ZIKV envelope protein dimer | Premkumar L et al., 2018; recombinantly produced in-house | N/A |
| DNA | Worthington | LS002105 |
| Centromere B | Prospec | PRO-390 |
| Histone | Sigma | H9250 |
| Jo-1 | Immunovision | JO1-3000 |
| RnP/Sm | Immunovision | SRC-3000 |
| Scl-70 | Immunovision | SCL-3000 |
| Sm | Immunovision | SMA-3000 |
| SSA | Immunovision | SSA-3000 |
| SSB | Immunovision | SSB-3000 |
| Poly-L-Lysine | Sigma | P6282 |
| Critical commercial assays | ||
| ANA Hep-2 test system | Zeuss Scientific | FA2400 |
| Deposited data | ||
| Zika virus glycoprotein E and DH1017.Fab structures | Protein Data Bank | PDB: 7T17 |
| Density map of DH1017.Fab in complex with Zika virions | EMDataResource | EMD-25606 |
| 119-4-D6.IgG | GenBank | OP612786, OP612795 |
| 119-1-D7.IgG | GenBank | OP612787, OP612796 |
| 119-5-C5.IgG | GenBank | OP612788, OP612797 |
| DH1017.IgM | GenBank | OP612789, OP612798 |
| 123-3-G2.IgG | GenBank | OP612790, OP612799 |
| 124-4-C8.IgG | GenBank | OP612791, OP612800 |
| 124-1-C2.IgG | GenBank | OP612792, OP612801 |
| 124-2-G3.IgG | GenBank | OP612793, OP612802 |
| 126-1-D11.IgG | GenBank | OP612794, OP612803 |
| Experimental models: Cell lines | ||
| Vero African green monkey kidney epithelial cells | ATCC | CCL-81 |
| C6/36 Aedes albopictus insect cells | ATCC | CRL-1660 |
| THP-1 monocytic cells | ATCC | TIB-202 |
| THP1.2 monocytic cells subclone | Chan et al. 2014 | N/A |
| CD40 ligand-expressing MS40L cells | Luo et al., 2009 | N/A |
| K-562 cells | ATCC | CCL-243 |
| BHK-21 fibroblasts | ATCC | CCL-10 |
| 119-4-D6.IgG B-LCL | This manuscript | N/A |
| 119-1-D7.IgG B-LCL | This manuscript | N/A |
| 119-5-C5.IgG B-LCL | This manuscript | N/A |
| 124-4-C8.IgG B-LCL | This manuscript | N/A |
| 124-1-C2.IgG B-LCL | This manuscript | N/A |
| 124-2-G3.IgG B-LCL | This manuscript | N/A |
| 126-1-D11.IgG B-LCL | This manuscript | N/A |
| 123-3-G2.IgG B-LCL | This manuscript | N/A |
| DH1017.IgM B-LCL | This manuscript | N/A |
| 119-4-G11.IgM B-LCL | This manuscript | N/A |
| Vero-Furin cells | Mukherjee et al., 2016 | N/A |
| Expi 293i cells | Invitrogen | A14527 |
| Experimental models: Organisms/strains | ||
| Ifnar1−/− C57BL/6 mice (males) | Kindly provided by Dr. Jason Whitmire (UNC) and bred in-house. | N/A |
| Oligonucleotides | ||
| forward primer CCGCTGCCCAACACAAG | Integrated DNA Technologies; Carbaugh et al., 2019 | N/A |
| reverse primer CCACTAACGTTCTTTTGCAGACAT | Integrated DNA Technologies; Carbaugh et al., 2019 | N/A |
| probe AGCCTACCTTGACAAGCAATCAGACACTCAA | Integrated DNA Technologies; Carbaugh et al., 2019 | N/A |
| Software and algorithms | ||
| Prism version 9 | GraphPad Software | https://www.graphpad.com |
| BD FACSDiva | BD Biosciences | https://www.bdbiosciences.com/en-us/products/software/instrument-software/bd-facsdiva-software |
| FlowJo version 10 | FlowJo, LLC | https://flowjo.com |
| Cloanalyst | Kepler, 2013 | https://www.bu.edu/computationalimmunology/research/software/ |
| I-TASSER | Roy et al., 2010; Yang et al., 2014 and Zhang, 2008 | https://zhanggroup.org/I-TASSER/ |
| PYMOL version 2 | Schrodinger, LLC | https://pymol.org/2/ |
| RIVEM | Xiao and Rossmann, 2007 | N/A |
| BioRender | BioRender.com | http://biorender.com |
| Other | ||
| BioMek FX | Beckman Coulter | N/A |
| Cryoplunge 3 system | Gatan | N/A |
| Titan Krios | Thermofisher | N/A |
| Talos F200C | Thermofisher | N/A |


