Abstract
The Staphylococcus aureus repeat (STAR) element is a sequence identified in two intergenic regions in S. aureus. The element is found in 13 to 21 copies in individual S. aureus strains, and elements in the homologous intergenic location are variable in length. The element sequence consists of several small and unusually GC-rich direct repeats with recurring intervening sequences. In addition, STAR-like elements may be present in related staphylococcal species.
Nosocomial Staphylococcus aureus infections require rapid identification and source recognition so that further spread, particularly of multiply resistant or unusually pathogenic strains, can be controlled. A number of methods to identify and distinguish bacterial strains have been developed. More-traditional phenotypic characterization has been gradually replaced by DNA sequence-based technology. PCR amplification has been particularly useful for strain typing in a clinical setting because it is rapid, relatively easy to automate, and less labor-intensive (14) than methods such as pulsed-field gel electrophoresis or Southern blotting (12). As DNA sequencing technology becomes increasingly more available in a clinical setting, nucleotide-level analysis is likely to be used more often, perhaps in combination with PCR fingerprinting, for strain identification.
A strategy currently used for strain typing of S. aureus relies on the amplification of defined tandemly repeated DNA sequences, often within surface-associated proteins, that are present in a variable number of copies in different strain isolates (for reviews, see references 7 and 13). This technique usually requires the analysis of several marker loci to differentiate strains that may by chance alone be identical at one locus.
Here, we describe a DNA sequence that is highly variable among S. aureus strains and that is found in many copies throughout the genome. An analysis of PCR products whose sizes may differ by only a few nucleotides and/or nucleotide sequence analysis at several loci would allow the rapid identification and detailed differentiation of clinical isolates.
Methods.
The primers listed in Fig. 1A lie within the uvrA and hprK coding regions and were used to amplify the uvrA-hprK intergenic sequence for sizing as well as subcloning and sequencing. The primers shown in Fig. 1B lie just downstream of the icaC and geh coding sequences and were used for fragment sizing. The following two primers within the coding region of each gene were used to amplify a slightly larger fragment from each strain for subcloning and sequencing: SA18, 5′-GTATAGGTGTCGGCATGATATTGCGTG; SA19, 5′-T GAAAACTGAAAAGCTGACTGATACTAAGC. Primers were obtained from MWG-Biotech (Ebersberg, Germany). Fragments were amplified using Taq (AGS, Heidelberg, Germany) or Vent (New England Biolabs GmbH, Schwalbach, Germany) polymerase and annealing temperatures of 62 (uvrA-hprK) or 52°C (icaC-geh). PCR products were blunt-end ligated into vector pUC18 using the SureClone ligation kit (Amersham Pharmacia Biotech, Braunschweig, Germany) and sequenced on both strands by MWG-Biotech, with the following exceptions. The uvrA-hprK intergenic sequence from strain 601055 (Zeneca Pharmaceuticals strain collection) was obtained using Pathoseq S. aureus contig sequence information available from Incyte Pharmaceuticals (Palo Alto, Calif.) and contained on contig Sau1c0627. The icaC-geh intergenic sequence from strain ATCC 35556 was previously published by our group and was not resequenced for this work (2). Computer sequence analysis was performed using Multalin, version 3.5.5 (1), with manual adjustments. Preliminary S. aureus sequence data were obtained from The Institute for Genomic Research at http://www.tigr.org and the S. aureus Genome Sequencing Project (University of Oklahoma) at http://www.genome.ou.edu. Southern blots were hybridized and washed (0.1× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]–0.1% sodium dodecyl sulfate) at 55°C and detected using the DIG DNA labeling and detection kit from Boehringer (Mannheim, Germany).
FIG. 1.
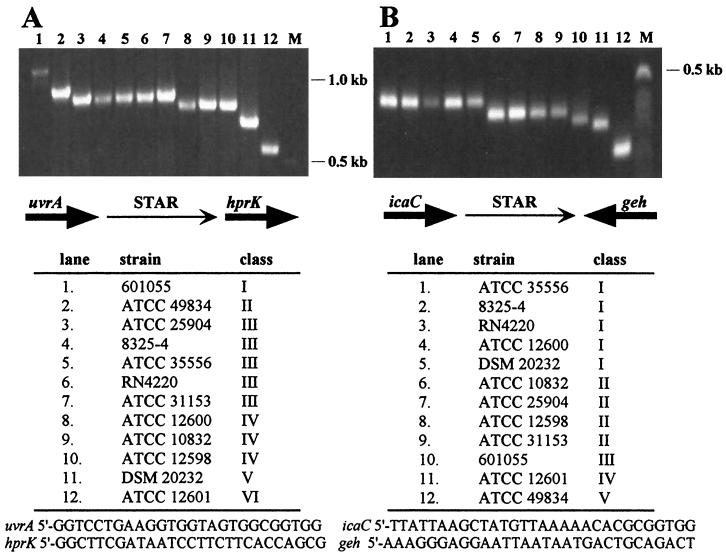
Intraspecies diversity in STAR element size. The intergenic sequences between uvrA-hprK (A) and icaC-geh (B) in 12 S. aureus strains, listed below each agarose gel, were amplified by PCR. The fragments were grouped into size classes (I to VI and I to V, respectively), and a representative from each class was sequenced. A schematic representation of the chromosomal organization in each region is shown beneath each agarose gel. The primers used for PCR amplification are listed at the bottom.
Identification of the STAR element.
A comparison of the hprK sequence from Staphylococcus xylosus (3a) with the S. aureus sequence available from a commercial provider (Incyte Pharmaceuticals) identified a noncoding intervening sequence in the region between uvrA and the hprK promoter that was not present in S. xylosus. The sequence contained two large and several smaller direct repeats that were unusually GC rich, including several copies of the sequence GGGGCCCC. The presence of multiple copies of this sequence, which contains the recognition site for restriction enzyme ApaI (GGGCC^C), in close proximity is highly improbable in these AT-rich organisms.
Public database searches identified a Tn557 attachment site (6) and significant similarity to a noncoding sequence included in a database entry for the S. aureus lipase (geh) (9). Other work with the neighboring S. aureus intercellular adhesion (ica) genes identified the same database entry and led to a closer examination of the intergenic sequences at the two loci (2). In the meantime, public databases show a third example of what we have called the S. aureus repeat (STAR) element near the trxB gene in S. aureus (accession no. AJ223781.1). From these dispersed representatives, we concluded that STAR is a repetitive element present in multiple copies in the S. aureus genome.
Size variability.
Initial sequence comparisons revealed that the intergenic regions between icaC and geh in strain ATCC 35556 and strain U500 in the database were not identical, the latter being smaller by a 58-bp directly repeated sequence. This led to a size analysis of the icaC-geh intergenic sequences by PCR amplification of a number of different S. aureus strains, which revealed that the sequence showed significant size variability. A similar PCR analysis, which also showed size variability among strains, was carried out at the uvrA-hprK locus. Figure 1A shows six different size classes ranging from 573 to 1,127 bp among representative strains in which the intergenic region between uvrA and hprK was examined. Five size classes (380 to 577 bp) were discernible among the same strains when the intergenic region between icaC and geh was investigated (Fig. 1B). In addition, a sixth size class (520 bp) is represented by the icaC-geh intergenic sequence of strain U500 (9), not shown here.
Sequence comparison.
The sizes of each sequenced fragment and the corresponding STAR element are listed in Table 1, along with the database nomenclature. A sequence comparison of the STAR elements, including part of each intergenic region, is shown in Fig. 2. The alignment is structured around the largest STAR element, star01, from the uvrA-hprK intergenic region of strain 601055. Figure 3 graphically depicts the structure of the star01 sequence, including the sequences flanking the STAR element.
TABLE 1.
Database sequence entries
| STAR element | Strain | Length (bp) of entire database entry | Accession no. | Length (bp) of STAR element | No. of signature sequences |
|---|---|---|---|---|---|
| uvrA-hprK | |||||
| star01 | 601055 | 1,127 | AF195957 | 434 | 5 |
| star02 | 8325-4 | 876 | AF195958 | 279 | 3 |
| star03 | ATCC 10832 | 825 | AF195959 | 226 | 2 |
| star04 | DSM 20232 | 720 | AF195960 | 109 | 1 |
| star05 | ATCC 49834 | 940 | AF195961 | 329 | 3 |
| star06 | ATCC 12601 | 573 | AF195962 | 0 | 0 |
| icaC-geh | |||||
| star07 | ATCC 10832 | 521 | AF195963 | 168 | 2 |
| star08 | 601055 | 494 | AF195964 | 157 | 3 |
| star09 | ATCC 49834 | 380 | AF195965 | 43 | 1 |
| star10 | ATCC 12601 | 464 | AF195966 | 108 | 1 |
| star11 | ATCC 35556 | 577 | AF195967 | 220 | 3 |
FIG. 2.
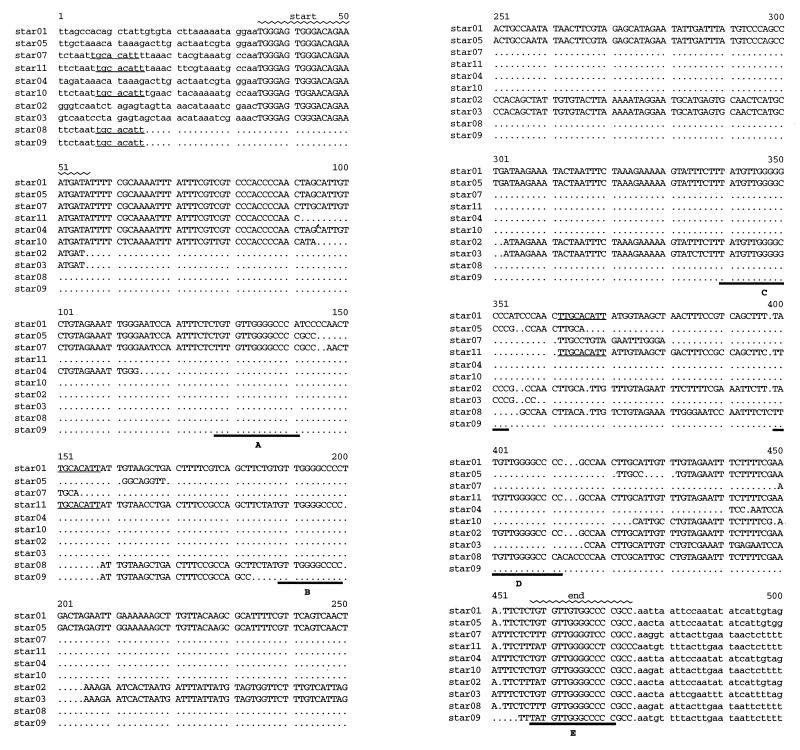
Comparison of sequenced STAR elements. Presented is an alignment of STAR sequences, showing the core element sequence (capital letters) and a short flanking sequence (lowercase letters) for each sequence, with the exception of the hprK region of strain 12601, which contains no detectable STAR element. The numbering refers to star01 (601055) and corresponds to bases 345 to 838 of the entire sequenced region included in the database entry. The beginning (start) and end of the element are indicated. Five (A to E) 14-bp signature sequence direct repeats are found in star01, and subsets are found in the other sequences (boldface underlining). A possible 8-bp recombination site that could account for the deletion of the “beginning” of the element in star08 and star09 is indicated for the strains in which it is present (lightface underlining).
FIG. 3.
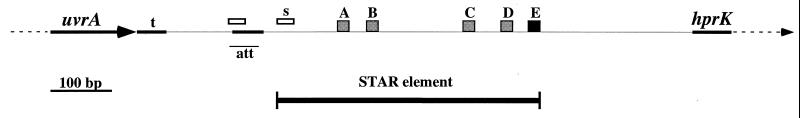
Graphic representation of the star01 sequence structure. The end and beginning of the uvrA and hprK coding regions are indicated with an arrow and an arrowhead, respectively. The portion of the sequence containing the STAR element is marked. Grey boxes, directly repeated 14-bp signature sequence; black box, extended signature/end sequence; white rectangles, two copies of a 22-bp sequence, the second of which is the beginning of the element(s). The Tn557 attachment site (att) is marked. Black bars, inverted repeats, including the uvrA transcription terminator (t).
The hallmark of the STAR element is the directly repeated signature sequence T(G/A/T)TGTTG(G/T)GGCCC(C/A), which is present in as many as five copies at one locus. The single exception found was strain 12601, which does not contain a signature sequence and, therefore, has no STAR element in the uvrA-hprK intergenic region.
We defined a beginning (TGGG[A/C]GTGGGACAGAAATGAT) for the STAR element starting at nucleotide 35, which is present in 8 of the 10 sequences, and an end (T[G/A/T]TGTTGGGGCCCCGCC), which is an extended signature sequence and present in all of the sequences, finishing at nucleotide 474. These borders are based on sequence similarity between the two loci and similarity in the flanking sequence only among sequences from the same locus. Signature sequences A/B and C/D in star01 may be part of larger 75-bp imperfect direct repeats. Longer repeats are also found in star02, star03, star05, star07, star08, and star11.
The intergenic region that contains the largest STAR element (star01) harbors an additional copy of the beginning sequence upstream of the defined element. An inverted repeat and a Tn557 attachment site overlap this sequence. Deletion of the DNA sequence between repeated beginning sequences in star01 would result in a STAR element that resembles the others examined. The sequences between direct repeats in the STAR elements show similarity in some but not all comparisons, as if the interrepeat sequences have been randomly deleted. For example, the interrepeat sequences of star01 and star05 are nearly identical between direct repeats B and C (Fig. 2), as are the sequences of star02 and star03, but the star01/star05 sequence shows no similarity to the star02/star03 sequence. We speculate that a large hypothetical “master element” that has not only distributed itself throughout the genome but also deleted interrepeat sequences via recombination at the signature direct repeats may exist (or may have existed in the past). In addition, a smaller sequence (TGCACATT) found in the upstream flanking sequences of icaC-geh elements and again elsewhere within the STAR sequences of star01 and star11 could explain, again via recombination/deletion, the absence of the initial sequences of star08 and star09 (Fig. 2).
Abundance of the STAR element in S. aureus.
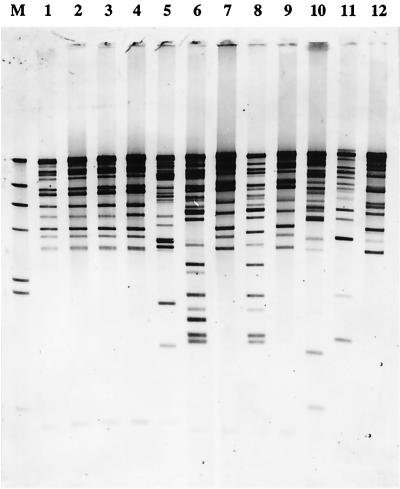
Due to the presence of two STAR elements in different and apparently unrelated chromosomal locations, it seemed reasonable to assume that there were more STAR elements elsewhere in the S. aureus genome. Chromosomal DNA from the strains shown in Fig. 1 was digested with restriction enzyme HindIII, blotted, and hybridized with the uvrA-hprK intergenic region from strain 601005, the largest STAR sequence from the two loci we investigated in detail. We expected that a single hybridizing band would be specific for the fragment containing the uvrA-hprK region used as the probe and that any other bands would represent other STAR elements. The Southern blot shown in Fig. 4 reveals 13 to 21 cross-hybridizing bands, depending on the strain. The number of cross-hybridizing bands may be an underestimate, however, because each band could contain more than one STAR element, but it is clear that the STAR element is present in tens but not thousands of copies in the S. aureus genome.
FIG. 4.
Abundance of STAR element sequences in S. aureus. Chromosomal DNA from 12 S. aureus strains was digested with restriction enzyme HindIII and hybridized with a labeled probe corresponding to the sequence between the uvrA and hprK primers listed in Fig. 1A in strain 601055. Lanes 1 to 12 correspond, respectively, to the strains listed in Fig. 1B. Size marker (M), lambda/HindIII DNA.
A recent (December 1999) search of the two publicly accessible unfinished S. aureus genome projects revealed approximately 20 copies of the beginning sequence and up to 20 additional sequences where one or more copies of the signature sequence were present. This apparent frequency is in agreement with our results. The presence of hprK and trxB on the same sequenced contig (The Institute for Genomic Research unfinished microbial genomes), flanked by two STAR elements in the same orientation, STAR-hprK-lgt-yvoF-yvcD-trxB-STAR, suggests a potential mobility of large pieces of chromosome mediated by STAR sequences.
The first three strains shown in Fig. 4 are all derivatives of strain NCTC 8325 (4, 5, 10, 11) and show identical hybridization patterns. This indicates that the numbers and positions of STAR elements in these related strains have remained stable over many years of laboratory culture.
Presence and abundance of the STAR element in other staphylococci.
Cross-species staphylococcal DNA hybridization was also performed, using the same S. aureus uvrA-hprK STAR element as a probe. No signal was detected with DNA from S. carnosus, S. hyicus, S. saprophyticus, S. simulans, or S. xylosus; however, cross-hybridization with the STAR probe was observed with DNA from S. epidermidis (6 bands), S. haemolyticus (16 bands), S. hominis (1 band), S. intermedius (1 band), S. lentus (18 bands), S. sciuri (10 bands), and S. warneri (15 bands) (data not shown).
Hybridization to S. aureus chromosomal DNA was at least 10 times stronger than hybridization to that of the other species, although approximately one-third as much S. aureus DNA was loaded on the gel. The single band in S. hominis and S. intermedius may represent cross-hybridization with uvrA-hprK homologues. The probe contained the Tn557 attachment site; however these data suggest that portions of the STAR element may also be present in other members of this genus. Database searches of all the publicly available finished and unfinished genome projects failed to reveal evidence of this element in any organism other than S. aureus.
Concluding remarks.
With the ever-expanding number of microbial genome sequences available, it is likely that more small multicopy sequences will be identified. The Bacillus subtilis genome project has produced one such 190-bp sequence, present in 10 nonidentical copies in the strain sequenced (8). The genome of Mycobacterium leprae also contains 28 copies of a sequence that shows strain-to-strain variability (3, 15).
We have no evidence for the function of the GC-rich STAR element in S. aureus; however, its ubiquity suggests that it has (or had) an important function in microbial life. The element does not have a defined short sequence that is tandemly repeated a variable number of times, as is the case with many repeating-unit sequences that are currently used for strain identification, nor does it show similarity to known insertion elements. The diversity of sequences at the same locus among S. aureus strains suggests that the element changes quite rapidly in evolutionary terms and might thus be useful as a typing marker for the identification and pedigree analysis of clinical isolates. The presence of multiple STAR loci would greatly expand the specificity and accuracy of any strain analysis.
Nucleotide sequence accession numbers.
The sequences determined in this study have been submitted to the EMBL, GenBank, and DDBJ nucleotide sequence data libraries under the accession numbers listed in Table 1.
Acknowledgments
We thank Phuong Lan Huynh and Ulrike Pfitzner for technical assistance.
S.E.C. was supported by NRSA Postdoctoral Fellowship AI09626 from the National Institute of Allergy and Infectious Diseases. This project was supported by the German Bundesministerium für Bildung, Wissenschaft, Forschung und Technologie (DLR: 01KI9751/1) and by the Deutsche Forschungsgemeinschaft (BR 947/3-1).
REFERENCES
- 1.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cramton S E, Gerke C, Schnell N F, Nichols W W, Götz F. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect Immun. 1999;67:5427–5433. doi: 10.1128/iai.67.10.5427-5433.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fsihi H, Cole S T. The Mycobacterium leprae genome: systematic sequence analysis identifies key catabolic enzymes, ATP-dependent transport systems and a novel polA locus associated with genomic variability. Mol Microbiol. 1995;16:909–919. doi: 10.1111/j.1365-2958.1995.tb02317.x. [DOI] [PubMed] [Google Scholar]
- 3a.Huynh, P. L., I. Jankovic, N. F. Schnell, and R. Brückner. Characterization of an HPr Kinase mutant of Staphylococcus xylosus. J. Bacteriol., in press. [DOI] [PMC free article] [PubMed]
- 4.Iordanescu S, Surdeanu M. Two restriction and modification systems in Staphylococcus aureus NCTC 8325. J Gen Microbiol. 1976;96:277–281. doi: 10.1099/00221287-96-2-277. [DOI] [PubMed] [Google Scholar]
- 5.Kreiswirth B N, Lofdahl S, Betley M J, O'Reilly M, Schlievert P M, Bergdoll M S, Novick R P. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature. 1983;305:709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- 6.Lindsay J A, Kreiswirth B N, Novick R P. The gene for toxic shock toxin is carried by a family of mobile pathogenicity islands in Staphylococcus aureus. Mol Microbiol. 1998;29:527–543. doi: 10.1046/j.1365-2958.1998.00947.x. [DOI] [PubMed] [Google Scholar]
- 7.Lupski J R, Weinstock G M. Short, interspersed repetitive DNA sequences in prokaryotic genomes. J Bacteriol. 1992;174:4525–4529. doi: 10.1128/jb.174.14.4525-4529.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moszer I. The complete genome of Bacillus subtilis: from sequence annotation to data management analysis. FEBS Lett. 1998;430:28–36. doi: 10.1016/s0014-5793(98)00620-6. [DOI] [PubMed] [Google Scholar]
- 9.Nikoleit K, Rosenstein R, Jerheij H M, Götz F. Comparative biochemical and molecular analysis of the Staphylococcus hyicus, Staphylococcus aureus and a hybrid lipase. Indication for a C-terminal phospholipase domain. Eur J Biochem. 1995;228:732–738. [PubMed] [Google Scholar]
- 10.Novick R P. Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology. 1967;33:155–166. doi: 10.1016/0042-6822(67)90105-5. [DOI] [PubMed] [Google Scholar]
- 11.Novick R P, Richmond M H. Nature and interactions of the genetic elements governing penicillinase synthesis in Staphylococcus aureus. J Bacteriol. 1965;90:467–480. doi: 10.1128/jb.90.2.467-480.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Belkum A. DNA fingerprinting of medically important microorganisms by use of PCR. Clin Microbiol Rev. 1994;7:174–184. doi: 10.1128/cmr.7.2.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Belkum A, Scherer S, van Alphen L, Verbrugh H. Short-sequence DNA repeats in prokaryotic genomes. Microbiol Mol Biol Rev. 1998;62:275–293. doi: 10.1128/mmbr.62.2.275-293.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van der Zee A, Verbakel H, van Zon J-C, Frenay I, van Belkum A, Peeters M, Buiting A, Bergmans A. Molecular genotyping of Staphylococcus aureus strains: comparison of repetitive element sequence-based PCR with various typing methods and isolation of a novel epidemicity marker. J Clin Microbiol. 1999;37:342–349. doi: 10.1128/jcm.37.2.342-349.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woods S A, Cole S T. A family of dispersed repeats in Mycobacterium leprae. Mol Microbiol. 1990;4:1745–1751. doi: 10.1111/j.1365-2958.1990.tb00552.x. [DOI] [PubMed] [Google Scholar]