Abstract
We previously described three patients affected by metastatic colorectal cancer (mCRC) who experienced spontaneous tumour shrinkage during severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. Thereafter, the patients were closely monitored and no systemic treatments were applied. Here, we report follow-up clinical information about these patients as well as genetic characterization of their primary tumours through the TruSigt™Oncology 500 Next Generation Sequencing test targeting 523 cancer-relevant genes. An Illumina NovaSeq 6000 platform was used to perform sequencing. Time-to-progression was 23 and 2 months, respectively, in Patients 2 and 3 while it was not reached in Patient 1. Patients 1 and 2 had the greatest anti-SARS-CoV-2 IgG titres. Assessment of genetic landscapes evidenced common mutation in BARD1 gene (p.Val507Met) in Patients 1 and 2. Although our report is descriptive in its nature, we suggest that complex and unexplored interactions between genetic background and components of the immune response to SARS-CoV-2 infection could be responsible of unexpected rare mCRC shrinkage.
Keywords: colorectal cancer, COVID-19, genetics, next generation sequencing, SARS-CoV-2
Introduction
The impact of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection in cancer patients is relevant. In fact, cancer patients are more prone both to infection and development of severe symptoms compared to healthy population or non-cancer patients because of their immunosuppressive state (malignancy related and/or treatment related).1–3
Therefore, the scientific community, beside the urgent need for vaccine development and its administration among cancer patients, promoted institutional policies oriented to regulate the timing between SARS-CoV-2 infection course and chemotherapy, particularly in advanced patients.4 In our Institute, the patients positive for molecular pharyngo-nasal swab, even if asymptomatic, cannot receive chemotherapy. The treatment is started (or restored) in the absence of any coronavirus disease 19 (COVID-19)-related symptoms and after three consecutive negative molecular pharyngo-nasal swabs.
In this scenario, we previously reported histopathologic, immunologic and radiologic features of three patients affected by metastatic colorectal cancer (mCRC) who showed a spontaneous disease burden shrinkage during COVID-19.5
Here, we report follow-up clinical and radiological information as well as comprehensive genetic characterization of their primary tumours by Next Generation Sequencing (NGS).
Cases’ description
Cases’ follow-up and TIER 1–3 genetic landscape of tumours
After documenting tumour regressions, patients underwent to close surveillance. No systemic treatments were administered. They were monitored with standard total body computed tomography (CT) scan every 3 months. The final evaluation of disease status was done according to comparison with basal CT scans and Response Evaluation Criteria In Solid Tumours v1.1). In Table 1, we synthesized the clinicopathological characteristics of patients experiencing spontaneous tumour shrinkages.
Table 1.
Clinicopathological characteristics of patients.
| Pat ID | Age (years) | Sex | pT pN staging/side | MS status | RAS/BRAF mut* | Prev surgery/date | Metastatic sites | Prev CT | Disease status before COVID-19 | IgG anti-SARS-CoV-2 titre (U/ml) by ECLIA |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 68 | M | pT4a pN1b Right |
MSS | No | Only PT October 2019 |
Liver | Yes | SD | 1455.00 |
| 2 | 61 | M | pT3 pN0 Left |
MSS | Yes KRAS$ |
PT February 2019 Liver mets November 2019 |
Liver | Yes | PD | 188.40 |
| 3 | 63 | F | pT3 pN2a Left |
MSS | No | Only PT March 2019 |
Lung, peritoneum | Yes | SD | 1216.00 |
Both N- and K-RAS genes were assessed.
KRAS mutation (p.G12D) was revealed in the metastatic tissue by means of a tumour DNA sequencing restricted panel tests.
COVID-19, coronavirus disease 19; CT, chemotherapy; ECLIA, enhanced chemiluminescence immunoassay; mut, mutations; PT, primary tumour.
Here we describe the clinical follow-up description and the genetic landscape of tumours for each patient.
Patient #1 showed a CR during severe COVID-19 (November 2020) with regression of hepatic lesions. At last follow-up (June 2022), there was no progression of disease at any site; the liver was persistently free from disease (Figure 1). The patient presented two stop gain variations in APC gene (p.Ser1196Ter and p.Glu1374Ter), two missense variants in BARD1 (p.Val507Met) and NTRK3 (p.Arg46Pro), respectively, and a frameshift variant of p53 (p.Gly108ValfsTer15).
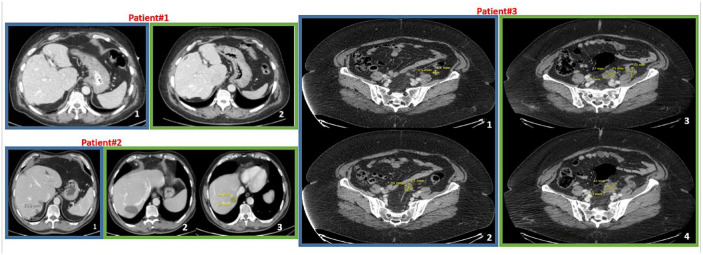
Figure 1.
Radiologic follow-up of patients who experienced disease burden reduction after COVID-19. In blue frames, representative basal CT scans after recovery from COVID-19; in green frames, CT scans at last follow-up or progression. Patient#1. CT scans show absence of liver metastases (section 2 versus section 1). Patient#2. CT scans in green frame show a residual cystic area (section 2) after surgical removal of the lesion at segment VII (section 1) but a new metastatic liver lesions at segment VI (section 3) with the relative measures (16 × 19 mm) (August 2020). Patient#3. Sections 3 and 4 evidence both new and increased size nodules on the peritoneum surface compared to basal sections (1 and 2) (relative measures are reported into the images).
COVID-19, coronavirus disease 19; CT, computed tomography.
Patient#2 had a reduction of liver disease during mild symptomatic COVID-19 (August 2020). He received metastases resection of segment VII in September 2020. At last follow-up (July 2022), there was disappearance of segment VII (where an area resembling a residual cyst was evidenced) but a new lesion (17 × 18 mm) at segment VI (Figure 1). Thus, the patient had a progressive disease. The time elapsed from the date of spontaneous regression documentation to progression was 23 months.
The genetic landscape of Patient 2 was characterized by missense variants of PIK3CA (p.Thr 1052Lys), BARD1 (p.Val507Met), SMO (p.Asn271His), ZFHX3 (p.His3611Asn), IDH1 (p.Arg132His), LRP1B (p.Val4461Leu), ROS1 (p.Pro1929Ala), SDHB (p.Ser193Thr), RASA1 (p.Gly413Arg), p53 (p.Cys135Tyr), NOTCH3 (p.Arg61Trp), stop gain in APC gene (p.Gln1338Ter), AKT3 (p.Glu17Ter) and inframe deletion of ZBTB7A (p.Glu543_Asp546del).
Patient#3 recovered from mild COVID-19 at the end of October 2020. She had spontaneous reduction of peritoneal and lung metastases. In December 2021, she presented progression of disease because of new lesions and increased size of pre-existing measurable nodules on the peritoneum surface (Figure 1). The time elapsed from the date of spontaneous regression documentation to progression was very short for this patient (2 months). She started a second-line chemotherapy with capecitabine and oxaliplatin (oral capecitabine 1000 mg/m2 twice daily on days 1–14 and oxaliplatin 130 mg/m2 on day 1 every 3 weeks) on January 2022 obtaining disease stabilization until June 2022 when CT scan showed PD. In July 2022, the patient started regorafenib per os at 120 mg/day in a 3 weeks on/1 week off schedule. The genetic landscape of the patient through NGS sequencing of the primary tumour revealed missense variants in AR (p.Leu548Val), FAT1 (p.Val1043Met), AKT3 (p.Ser403Asn), p53 (p.Gly245Ser) and a frameshift variant of ATRX (p.Lys1931ArgfsTer11).
Complete genetic sequences of patient (TIER 1–4 and non-coding variants) are available in Supplemental File S1.
Genetic concordance
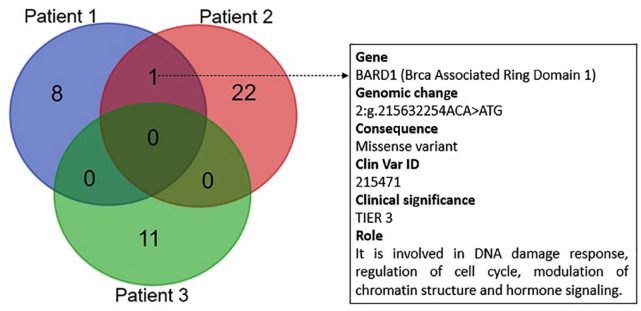
Since the genetic diversity of tumours can be involved in determining heterogeneity in their clinical course, the genetic sharing between the analysed tumours was studied. The aim was to identify any common genetic features putatively associated with the reported COVID-19-related spontaneous tumour shrinkages and to generate hypotheses. A descriptive analysis of the genetic concordance was done thorough the Venn diagram (Figure 2). The genetic sharing between primary tumours was low. The most common genetic variant (shared by Patients 1 and 2) was BARD1 p.Val507Met.
Figure 2.
Venn diagram reporting the number of shared genetic variant between the analysed patients. The most common genetic variant was BARD1 p.Val507Met. Gene nomenclature, genomic change, consequence, Clin Var ID, priority and biological role are reported to the right of the diagram. DNA was extracted form FFPE primary tissue specimens through the MGF03-Genomic DNA FFPE One-Step Kit (MagCoreDiatech). Libraries were prepared with TruSigtTMOncology 500 kit that analyses 523 cancer-relevant genes and sequenced on an Illumina NovaSeq 6000 (San Diego, USA) platform. A median of 90 million reads were generated for each sample and the coverage in the target region was above manufacturer’s suggested threshold of 150×. Sequences were aligned to the human reference genome GRCh37 (http://www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/human/index.shtml) using the Burrows–Wheeler Aligner tool. The prioritization of variants was done according to a four-tiered structure, adopting the joint consensus recommendation by AMP/ACMG (Association for Molecular Pathology and American College of Medical Genetics).
AMP, Association for Molecular Pathology; FFPE, formalin-fixed and paraffin-embedded.
Discussion
We previously reported unexpected spontaneous CRC regression of metastatic lesions during SARS-CoV-2 infection.5 Since angiotensin-converting enzyme 2 (ACE-2), used by SARS-CoV-2 to attach and entry into host cells,6 was expressed also by the analysed CRC cells, we hypothesized a direct cytotoxic effect against the infected tumour cells. We also argued that a cross-reactivity with viral antigens might be involved in inducing NK cell activation via antibody-dependent cellular cytotoxicity against opsonized CRC cells. The last hypothesis was reinforced by the observation that the humoral response of the selected patients was high (Patient 1: anti-SARS-CoV-2 IgG 1455.00 U/ml; Patient 2: IgG 188.40 U/ml; Patient 3: 1216.00 U/ml) as well as the degranulation activity of their lymphocytes towards ACE-2/NRP-1-positive CRC cell lines.5
At clinical follow-up, we observed that time-to-progression was not reached in Patient 1, 23 and 2 months, respectively, in Patients 2 and 3. Interestingly, anti-SARS-CoV-2 IgG evaluations at last follow-up visit or at progression evidenced that Patients 1 and 2 still retained high IgG titres (1002.50 U/ml and 45.22 U/ml) while Patient 3 had a decreased humoral response (7.20 U/ml). Notably, all patients received the COVID-19 mRNA BNT162b2-based vaccine (Patient 1: 3 doses, first dose after 2 months from COVID-19 recovery; Patients 2: 2 doses, first dose after 5 months; Patient 3: 2 doses, first dose after 6 months).
Genetic heterogeneity of tumours may account for inter-individual variability in the quality of cancer/host relationships, extent of the anti-cancer immune response, characteristics of tumour micro-environment, response to therapies, and, finally, clinical outcomes. For this reason, we believed it was useful for the scientific community to explore the genetic signatures of these interesting patients.
Surprisingly, we found a BARD1 gene variant (p.Val507Met) shared between Patients 1 and 2. BARD1 encodes a protein binding to BRCA1 at its N-terminal and is involved in DNA damage response, regulation of cell cycle, modulation of chromatin structure and hormone signalling working both as tumour suppressor gene and oncogene depending from the biochemical and cellular context.7 Interestingly, BARD1 is altered in about 1% of all cancers with the greatest prevalence in lung adenocarcinoma (https://www.mycancergenome.org, accessed on July 2022). It has to be noticed that a variant of BARD1 (p.Arg378Ser) was associated with the overall survival of patients affected by chronic lymphocytic leukaemia after the first treatment.8 This finding suggests a correlation between BARD1 mutational status and the propensity or resistance of the transformed lymphocytes to undergo to apoptosis. The latter event is crucial in the modulation of the response of cancer/transformed cells to cytotoxic agents and can also account for a changed sensitivity to the cell death induced by other cell components of the tumour micro-environment (i.e. immunological infiltrates). Further studies are needed to explore any putative associations between this genetic variant and the possible unexpected interrelations between the composite immune response to SARS-CoV-2 infection and CRC shrinkage.
This report has a predominant descriptive aim. A major limitation to be evidenced is the size of our series, which allow us to exclusively describe and generate hypotheses. In fact, mutations express their force along with genetic polymorphisms and epigenetic factors, the final effect is difficult to predict with a ‘gradient’ of biological and clinical heterogeneity. To this regard, Patient 3 is obese. This is sufficient to introduce a burden of genetic/epigenetic factors potentially confounding any putative relevant mutational trajectory. The only way to overcome this problem is to characterize larger series.
We suggest that unexplored and profound connections between genetic background and components of the immune response to SARS-CoV-2 infection could be responsible of these extremely unexpected rare mCRC tumour shrinkages.
Supplemental Material
Supplemental material, sj-xlsx-1-tam-10.1177_17588359221138388 for Genetic landscape of colorectal cancer patients manifesting tumor shrinkage during SARS-Cov-2 infection by Alessandro Ottaiano, Mariachiara Santorsola, Luisa Circelli, Marco Cascella, Nadia Petrillo, Francesco Perri, Marika Casillo, Vincenza Granata, Monica Ianniello, Francesco Izzo, Carmine Picone, Marco Correra, Antonella Petrillo, Roberto Sirica, Gabriella Misso, Paolo Delrio, Guglielmo Nasti, Giovanni Savarese and Michele Caraglia in Therapeutic Advances in Medical Oncology
Supplemental material, sj-xlsx-2-tam-10.1177_17588359221138388 for Genetic landscape of colorectal cancer patients manifesting tumor shrinkage during SARS-Cov-2 infection by Alessandro Ottaiano, Mariachiara Santorsola, Luisa Circelli, Marco Cascella, Nadia Petrillo, Francesco Perri, Marika Casillo, Vincenza Granata, Monica Ianniello, Francesco Izzo, Carmine Picone, Marco Correra, Antonella Petrillo, Roberto Sirica, Gabriella Misso, Paolo Delrio, Guglielmo Nasti, Giovanni Savarese and Michele Caraglia in Therapeutic Advances in Medical Oncology
Supplemental material, sj-xlsx-3-tam-10.1177_17588359221138388 for Genetic landscape of colorectal cancer patients manifesting tumor shrinkage during SARS-Cov-2 infection by Alessandro Ottaiano, Mariachiara Santorsola, Luisa Circelli, Marco Cascella, Nadia Petrillo, Francesco Perri, Marika Casillo, Vincenza Granata, Monica Ianniello, Francesco Izzo, Carmine Picone, Marco Correra, Antonella Petrillo, Roberto Sirica, Gabriella Misso, Paolo Delrio, Guglielmo Nasti, Giovanni Savarese and Michele Caraglia in Therapeutic Advances in Medical Oncology
Acknowledgments
We thank Alessandra Trocino, librarian at the Istituto Nazionale Tumori di Napoli, IRCCS ‘G. Pascale’, Italy, for bibliographic assistance. We thank Daniela Capobianco for technical editing and writing assistance. We also acknowledge the ‘Lega Italiana per la Lotta contro i Tumori (LILT)-sezione di Napoli’ for their precious and unconditional collaboration in this work.
Footnotes
ORCID iDs: Alessandro Ottaiano  https://orcid.org/0000-0002-2901-3855
https://orcid.org/0000-0002-2901-3855
Marco Cascella  https://orcid.org/0000-0002-5236-3132
https://orcid.org/0000-0002-5236-3132
Vincenza Granata  https://orcid.org/0000-0002-6601-3221
https://orcid.org/0000-0002-6601-3221
Michele Caraglia  https://orcid.org/0000-0003-2408-6091
https://orcid.org/0000-0003-2408-6091
Supplemental material: Supplemental material for this article is available online.
Contributor Information
Alessandro Ottaiano, SSD-Innovative Therapies for Abdominal Metastases, Department of Abdominal Oncology, Istituto Nazionale Tumori di Napoli, IRCCS “G. Pascale”, via M. Semmola, Naples, 80131, Italy.
Mariachiara Santorsola, SSD-Innovative Therapies for Abdominal Metastases, Department of Abdominal Oncology, Istituto Nazionale Tumori di Napoli, IRCCS “G. Pascale”, Naples, Italy.
Luisa Circelli, AMES, Centro Polidiagnostico Strumentale, Casalnuovo Di Napoli, Naples, Italy.
Marco Cascella, Unit of Anestesiology and Pain Therapy, Istituto Nazionale Tumori di Napoli, IRCCS “G. Pascale”, Naples, Italy.
Nadia Petrillo, AMES, Centro Polidiagnostico Strumentale, Casalnuovo Di Napoli, Naples, Italy.
Francesco Perri, Medical and Experimental Head and Neck Oncology Unit, Istituto Nazionale Tumori di Napoli, IRCCS “G. Pascale”, Naples, Italy.
Marika Casillo, AMES, Centro Polidiagnostico Strumentale, Casalnuovo Di Napoli, Naples, Italy.
Vincenza Granata, Department of Radiology, Istituto Nazionale Tumori di Napoli, IRCCS “G. Pascale”, Naples, Italy.
Monica Ianniello, AMES, Centro Polidiagnostico Strumentale, Casalnuovo Di Napoli, Naples, Italy.
Francesco Izzo, Hepato-Biliary Surgey Unit, Istituto Nazionale Tumori di Napoli, IRCCS “G. Pascale”, Naples, Italy.
Carmine Picone, Department of Radiology, Istituto Nazionale Tumori di Napoli, IRCCS “G. Pascale”, Naples, Italy.
Marco Correra, Department of Radiology, Istituto Nazionale Tumori di Napoli, IRCCS “G. Pascale”, Naples, Italy.
Antonella Petrillo, Department of Radiology, Istituto Nazionale Tumori di Napoli, IRCCS “G. Pascale”, Naples, Italy.
Roberto Sirica, AMES, Centro Polidiagnostico Strumentale, Casalnuovo Di Napoli, Naples, Italy.
Gabriella Misso, Department of Precision Medicine, University of Campania “L. Vanvitelli”, Naples, Italy.
Paolo Delrio, Colorectal Surgical Oncology, Istituto Nazionale Tumori di Napoli, IRCCS “G. Pascale”, Naples, Italy.
Guglielmo Nasti, SSD-Innovative Therapies for Abdominal Metastases, Department of Abdominal Oncology, Istituto Nazionale Tumori di Napoli, IRCCS “G. Pascale”, Naples, Italy.
Giovanni Savarese, AMES, Centro Polidiagnostico Strumentale, Casalnuovo Di Napoli, Naples, Italy.
Michele Caraglia, Department of Precision Medicine, University of Campania “L. Vanvitelli”, Naples, Italy.
Declarations
Ethics approval and consent to participate: Ethics approval is not required for the present study according to internal institutional policies. All patients signed an informed consent before any diagnostic or interventional procedures.
Consent for publication: All patients gave consent for publication.
Author contribution(s): Alessandro Ottaiano: Conceptualization, Data curation, Methodology, Writing – original draft.
Mariachiara Santorsola: Conceptualization, Data curation, Methodology, Writing – original draft.
Luisa Circelli: Conceptualization, Data curation, Methodology, Writing – original draft.
Marco Cascella: Writing – review & editing.
Nadia Petrillo: Formal analysis, Investigation.
Francesco Perri: Writing – review & editing.
Marika Casillo: Formal analysis, Investigation.
Vincenza Granata: Investigation.
Monica Ianniello: Investigation.
Francesco Izzo: Investigation.
Carmine Picone: Investigation.
Marco Correra: Data curation, Software.
Antonella Petrillo: Investigation.
Roberto Sirica: Formal analysis.
Gabriella Misso: Investigation.
Paolo Delrio: Investigation.
Guglielmo Nasti: Supervision.
Giovanni Savarese: Data curation, Supervision, Writing – review & editing.
Michele Caraglia: Data curation, Supervision, Writing – review & editing.
Funding: The authors received no financial support for the research, authorship and/or publication of this article.
The authors declare that there is no conflict of interest.
Availability of data and materials: Complete genetic assessments of patients are available in the Supplemental File of this article.
References
- 1. Challen R, Brooks-Pollock E, Read JM, et al. Risk of mortality in patients infected with SARS-CoV-2 variant of concern 202012/1: matched cohort study. BMJ 2021; 372: 579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wiersinga WJ, Rhodes A, Cheng AC, et al. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): a review. JAMA 2020; 324: 782–793. [DOI] [PubMed] [Google Scholar]
- 3. Desai A, Gupta R, Advani S, et al. Mortality in hospitalized patients with cancer and coronavirus disease 2019: a systematic review and meta-analysis of cohort studies. Cancer 2021; 127: 1459–1468. [DOI] [PubMed] [Google Scholar]
- 4. Rajan S, Akhtar N, Sharma S, et al. COVID-19 vaccination for cancer patients: evidence, priority, and practice. Vaccine 2021; 39: 5075–5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ottaiano A, Scala S, D’Alterio C, et al. Unexpected tumor reduction in metastatic colorectal cancer patients during SARS-Cov-2 infection. Ther Adv Med Oncol 2021; 13: 17588359211011455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shang J, Wan Y, Luo C, et al. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci U S A 2020; 117: 11727–11734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hawsawi YM, Shams A, Theyab A, et al. BARD1 mystery: tumor suppressors are cancer susceptibility genes. BMC Cancer 2022; 22: 599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rasi S, Forconi F, Bruscaggin A, et al. Impact of the host genetic background on prognosis of chronic lymphocytic leukemia. Blood 2010; 115: 1106–1107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-xlsx-1-tam-10.1177_17588359221138388 for Genetic landscape of colorectal cancer patients manifesting tumor shrinkage during SARS-Cov-2 infection by Alessandro Ottaiano, Mariachiara Santorsola, Luisa Circelli, Marco Cascella, Nadia Petrillo, Francesco Perri, Marika Casillo, Vincenza Granata, Monica Ianniello, Francesco Izzo, Carmine Picone, Marco Correra, Antonella Petrillo, Roberto Sirica, Gabriella Misso, Paolo Delrio, Guglielmo Nasti, Giovanni Savarese and Michele Caraglia in Therapeutic Advances in Medical Oncology
Supplemental material, sj-xlsx-2-tam-10.1177_17588359221138388 for Genetic landscape of colorectal cancer patients manifesting tumor shrinkage during SARS-Cov-2 infection by Alessandro Ottaiano, Mariachiara Santorsola, Luisa Circelli, Marco Cascella, Nadia Petrillo, Francesco Perri, Marika Casillo, Vincenza Granata, Monica Ianniello, Francesco Izzo, Carmine Picone, Marco Correra, Antonella Petrillo, Roberto Sirica, Gabriella Misso, Paolo Delrio, Guglielmo Nasti, Giovanni Savarese and Michele Caraglia in Therapeutic Advances in Medical Oncology
Supplemental material, sj-xlsx-3-tam-10.1177_17588359221138388 for Genetic landscape of colorectal cancer patients manifesting tumor shrinkage during SARS-Cov-2 infection by Alessandro Ottaiano, Mariachiara Santorsola, Luisa Circelli, Marco Cascella, Nadia Petrillo, Francesco Perri, Marika Casillo, Vincenza Granata, Monica Ianniello, Francesco Izzo, Carmine Picone, Marco Correra, Antonella Petrillo, Roberto Sirica, Gabriella Misso, Paolo Delrio, Guglielmo Nasti, Giovanni Savarese and Michele Caraglia in Therapeutic Advances in Medical Oncology