Abstract
Purpose of Review
In this study, we planned and carried out a scoping review of the literature to learn how machine learning (ML) has been investigated in cardiovascular imaging (CVI).
Recent Findings
During our search, we found numerous studies that developed or utilized existing ML models for segmentation, classification, object detection, generation, and regression applications involving cardiovascular imaging data. We first quantitatively investigated the different aspects of study characteristics, data handling, model development, and performance evaluation in all studies that were included in our review. We then supplemented these findings with a qualitative synthesis to highlight the common themes in the studied literature and provided recommendations to pave the way for upcoming research.
Summary
ML is a subfield of artificial intelligence (AI) that enables computers to learn human-like decision-making from data. Due to its novel applications, ML is gaining more and more attention from researchers in the healthcare industry. Cardiovascular imaging is an active area of research in medical imaging with lots of room for incorporating new technologies, like ML.
Supplementary Information
The online version contains supplementary material available at 10.1007/s40134-022-00407-8.
Keywords: Scoping review, Cardiovascular imaging, Artificial intelligence, Machine learning, Deep learning, Radiology, Cardiology
Introduction
The use of artificial intelligence (AI) in healthcare has exploded in recent years. Since 1995, the output of AI publications on healthcare has increased by an average of 17.02% per year, and the growth rate of research papers in this field has significantly accelerated to 45.15% from 2014 to 2019 [1].
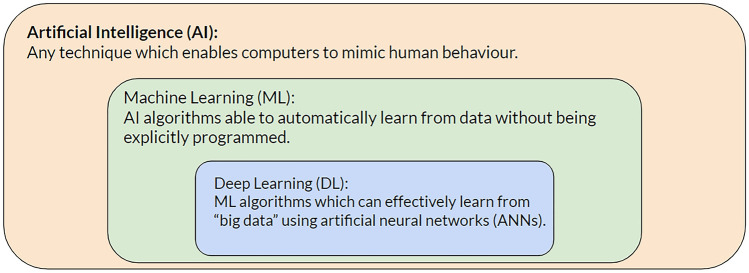
AI is a general term for any technique that enables computers to mimic human-like behavior [2]. Machine learning (ML) is a subset of AI that can learn human-like decision-making from data. Deep learning (DL) is a subset of ML that incorporates artificial neural network algorithms. Conventional ML, on the other hand, refers to the subfield of ML that does not involve neural networks (Fig. 1). Tree-based models, support vector machines, and K-nearest neighbors are famous examples of conventional ML algorithms.
Fig. 1.
An arbitrary framework to describe artificial intelligence, machine learning, deep learning, and conventional machine learning
Most ML algorithms can be thought of as parametric models that produce one or more quantities as their outputs while using data as their input variables [3]. During an iterative process known as model training, ML algorithms gradually encounter a carefully compiled set of data and discover the most optimal parameter values that can explain that dataset. ML algorithms can be distinguished from each other based on their mathematical expressions (a.k.a. architectures), input variables, and parameters. One can theoretically train many valid ML algorithms for the same task and on the same data, and this is what makes ML both an esthetic and scientific area. Aligned with the conventional routines, we hereafter use the word “model” to denote ML and DL algorithms in this report.
The major distinction between DL and conventional ML is their respective computational complexity [4]. In contrast to conventional ML models, which have a limited potential for data-driven learning, DL models are more complicated and can have millions of parameters. This increased capacity lets DL models learn more as they are exposed to additional data. However, the intricacies of DL models necessitate training them with larger datasets and more sophisticated hardware technology, such as graphics processing units.
The learning process of ML models is described as supervised when their training data are labeled. This strategy could be exemplified by a DL model that has been trained on chest X-ray (CXR) data from both normal and pneumonia patients and has similarly learned to label any CXR it encounters for the first time as normal or pneumonia indicating. While supervised learning is the most common strategy for training ML models, other strategies like unsupervised, semi-supervised, and self-supervised learning also exist and have diverse applications. Using only unlabeled training data, for instance, an unsupervised ML model can learn to cluster CXRs into arbitrary but still meaningful classes [5].
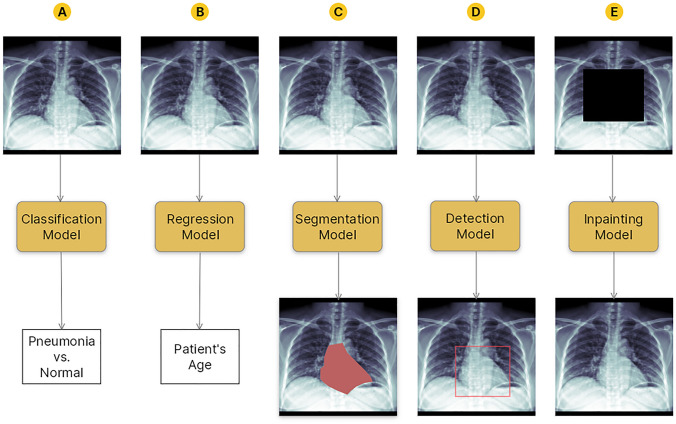
Another perspective for classifying ML models is based on their applications. Computer vision is a subset of ML that deals with imaging data. As illustrated in Fig. 2, computer vision models may be used for various tasks, the most common of which are as follows [6]:
Classification an input image is labeled with one or more categorical labels, e.g., to distinguish input CXRs based on whether they are presented with cardiomegaly or normal hearts.
Regression an input image is labeled with one or more quantitative labels, e.g., to predict the age of a patient by looking at input CXRs.
Semantic segmentation the entire surface areas or volumes of some objects of interest are delineated in an input image, e.g., to segment the entire heart area in input CXRs.
Object detection the locations of some objects of interest are approximated in an input image using key points or bounding boxes, e.g., to localize the heart in input CXRs.
Generation synthetic but realistic-looking imaging data is generated, e.g., to inpaint covered parts of input CXRs as if those parts came from real radiographs.
Fig. 2.
An illustration of different machine learning tasks on an arbitrary chest X-ray (CXR) example. A An example classifier model can learn to distinguish CXRs presenting with pneumonia from normal-appearing CXRs; B a regressor model can learn to predict a patient’s age by looking at their CXR; C an example segmentation model can learn to segment the heart on an input CXR; D an example object detector model can learn to localize the heart on an input CXR using a bounding box; E an example generator (inpainting) model can learn to inpaint a covered area of a CXR as if it came from a real radiograph
Cardiovascular imaging (CVI) is a rapidly expanding subspecialty of medical imaging that has made substantial contributions to translational research, risk assessment, diagnosis, prognosis, and therapeutic planning studies in structural and functional cardiovascular diseases [7]. In recent years, advanced medical imaging technologies have paved the way for improved phenotyping of cardiovascular pathologies. Given this context and the daily expansion of AI applications in healthcare, one can anticipate an increase in the incorporation of AI into CVI [8]. To comprehend the scope of AI applications in CVI, we conducted a scoping review of the available peer-reviewed literature. Our report attempts to provide an overview of ML research in CVI and to set the stage for future research in this field.
Methods
In accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) extension for scoping reviews [9], we designed a scoping review to answer two overarching research questions: (1) How are ML models used to analyze CVI? And (2) what is the quality of the research used to develop and report these models?
To answer the aforementioned questions, we combed through peer-reviewed original studies indexed in MEDLINE from January 1, 2012 to May 31, 2022, which examined the application of ML in CVI. Studies on non-human subjects, non-radiological data (e.g., histology data), fetal cardiovascular topics, non-peer-reviewed articles, non-English articles, review articles, and book chapters were excluded. The search was conducted using the PubMed search engine and a search term comprised MeSH terms and keywords related to cardiovascular organs, radiology imaging, and ML (Supplements 1).
Following an initial check for duplicate removal, six reviewers (PR, BK, SF, MM, SV, and EM) independently reviewed the titles and abstracts of the captured search results based on the inclusion and exclusion criteria stated previously. To make this process more reliable, 50 random studies were initially selected, and their data elements were charted by the authors and discussed in a focus group discussion to level their understanding of the required fields.
The full text of all eligible articles was then retrieved and revisited by reviewers for final evaluation and data extraction. Due to the lack of appropriate bias assessment tools for ML studies and the desire to maximize the inclusion of relevant articles, no assessment of the risk of bias was performed at this stage. A database of included studies was created, and several aspects of the study characteristics, data handling, model development, and performance evaluation of each article were extracted based on reviewer consensus. The detailed data elements which were charted in each of these categories are introduced in Table 1.
Table 1.
Detailed data elements extracted from the eligible studies during the data charting phase
| Category | Data elements |
|---|---|
| Study characteristics |
Publication year Country of the corresponding author Study design Clinical application Studied organs Studied pathologies |
| Data handling |
Studied imaging modalities The use of multi-modality models Dataset size |
| Model development |
Machine learning tasks The use of deep learning, conventional machine learning, or both The source of transfer learning (if any) The use of a standardized checklist for model development |
| Performance evaluation |
The use of cross-validation The use of external validation The provision of interpretation maps for deep learning models The provision of uncertainty measures for deep learning models |
Due to the heterogeneous scopes, applications, and methodologies of the included articles, no meta-analysis was conducted. Instead, the review results were reported using descriptive statistics. We also offered a narrative synthesis of the statistical findings to help non-technical readers better interpret our analyses.
Results
A systematic search of the scientific literature yielded 845 distinct papers (no duplicates), of which 215 were excluded after assessing their titles and abstracts. The full text of 41 studies was unavailable with our institutional access, yielding 589 eligible studies for final data extraction (Table 2). A database of all eligible articles and their extracted items is provided in Supplement 2.
Table 2.
Number of excluded articles in the current report based on their reason for exclusion
| Reasons for exclusion | Number (%) of excluded studies |
|---|---|
| Not radiology study | 113 (44.1%) |
| No full-text available | 41 (16.0%) |
| Not cardiovascular study | 34 (13.3%) |
| No human subject | 34 (13.3%) |
| Not machine learning study | 25 (9.8%) |
| Not original study | 9 (3.5%) |
Out of 845 identified studies, 256 were excluded, yielding a final pool of 589 eligible articles for data extraction
Study Characteristics
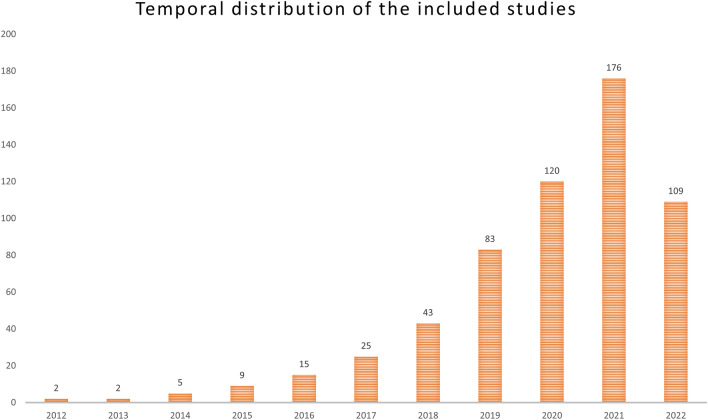
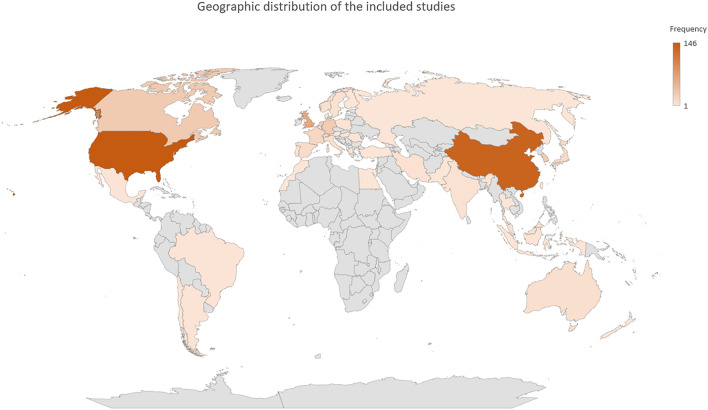
Figure 3 depicts the distribution of publication years for the collected articles. The number of published articles exhibits a consistent annual increase, with almost 69% of all articles published since 2020. The USA and China contributed the most publications among all countries. The geographic distribution of all published manuscripts is depicted in Fig. 4. The frequency of study designs, clinical applications, studied organs, and studied diseases is depicted in Table 3.
Fig. 3.
The distribution of publication year across all eligible studies included in our review. The number of published articles exhibits a consistent annual increase, with almost 69% of all articles published since 2020
Fig. 4.
The geographic distribution of the corresponding countries for all studies included in the final review pool. The intensity of color for each country correlates with the number of publications from that country
Table 3.
The frequency of study designs, clinical applications, studied organs, and studied diseases across all eligible articles
| Category | Data elements | Number (%) of included studies |
|---|---|---|
| Study design | Observational | 510 (86.5%) |
| Retrospective | 50 (8.5%) | |
| cohort | 18 (3.0%) | |
| Prospective cohort | 7 (1.2%) | |
| Case–control Trial | 4 (0.7%) | |
| Clinical application | Informatics | 270 (45.8%) |
| Diagnosis | 223 (37.9%) | |
| Prognosis | 26 (4.4%) | |
| Primary prevention | 25 (4.2%) | |
| Treatment | 24 (4.1%) | |
| Education | 1 (0.2%) | |
| Combined | 16 (2.7%) | |
| Others | 4 (0.7%) | |
| Studied organ | Heart | 413 (70.1%) |
| Coronary vasculature | 120 (20.4%) | |
| Aorta | 24 (4.1%) | |
| Pulmonary vasculature | 9 (1.5%) | |
| Pericardial fat | 6 (1.0%) | |
| Conduction system | 1 (0.2%) | |
| Combined | 16 (2.7%) | |
| Studied pathology | No Pathology | 192 (33.2%) |
| Atherosclerosis | 109 (18.9%) | |
| Valvular disorders | 34 (5.9%) | |
| Heart failure | 28 (4.8%) | |
| Ischemic heart disease | 22 (3.8%) | |
| Arrhythmia | 17 (2.9%) | |
| Cardiomyopathy | 16 (2.9%) | |
| Cancer or mass | 12 (2.0%) | |
| Multiple pathologies | 103 (17.8%) | |
| Other pathologies | 45 (7.8%) |
While the majority of included studies (510 [86.5%]) were observational, trials were the least common study design (4 [0.7%]). Only 24 (4.1%) of the articles focused on treatment-related applications, while 223 (37.9%) developed ML models for diagnostic purposes. The uses of ML models with no direct clinical application (e.g., for segmenting organs to construct an atlas or for generating synthetic imaging data) were categorized as informatic in 270 (45.8%) of the papers. The heart and atherosclerosis were the most researched organs and pathologies, respectively.
Data Handling
Table 4 illustrates the distribution of included publications based on the researched imaging modality. In the majority of articles (244 [41.5%]), Magnetic Resonance Imaging (MRI) was the most studied modality, while Single-photon Emission Computed Tomography (SPECT) received the least amount of attention (8 [1.4%]). Only 79 (13.4%) of the included studies presented a multimodal ML method. Table 4 illustrates the distribution of dataset sizes for all articles. Most assessed publications developed their models using 100–1000 examinations.
Table 4.
The distribution of imaging modalities and dataset size (number of reported examinations) across all eligible studies
| Category | Data elements | Number of included studies |
|---|---|---|
| Imaging modality | MRI | 244 (41.4%) |
| Echocardiography/Ultrasound | 102 (17.3%) | |
| CT-angiography | 77 (13.1%) | |
| Chest CT | 41 (6.7%) | |
| Cardiac CT | 36 (6.2%) | |
| Coronary angiography | 15 (2.6%) | |
| Chest x-ray | 11 (1.9%) | |
| OCT | 9 (1.6%) | |
| SPECT | 8 (1.4%) | |
| Combined | 41 (7.0%) | |
| Others | 5 (0.8%) | |
| Dataset size | < 100 | 138 (23.4%) |
| 100–1000 | 219 (37.2%) | |
| 1000–10,000 | 122 (20.7%) | |
| 10,000–100,000 | 45 (7.6%) | |
| 100,000–1,000,000 | 9 (1.6%) | |
| > 1,000,000 | 2 (0.3%) | |
| Not Reported | 54 (9.2%) |
Model Development
A total of 60 (10.2%) articles did not develop an ML model. Of the remaining articles, the majority (393 [67.1%]) developed a DL model, whereas 102 (17.4%) only developed conventional ML techniques and 31 (5.3%) employed a combination of both approaches. Segmentation tasks were the most common across all studies (224 [42.6%]), followed by classification (115 [21.9%]), a combination of tasks (80 [15.2%]), generation (58 [11.0%]), regression (35 [6.6%]), and object detection (14 [2.7%]). Only 3 (0.5%) of the included studies reported adherence to a standard checklist or protocol when building and (or) evaluating their ML models. Sixty-three (11.9%) articles reported using transfer learning to train their ML models, 36 (57.1%) of which used models pretrained on clinical data.
Performance Evaluation
A total of 196 (32.7%) of the articles tested their ML models in a cross-validation scenario, and external validation of ML models was reported by 63 (10.6%) of all publications. Only 17 (17.7%) of the studies that utilized classification and regression DL models (96 articles) offered interpretation maps of their performance, and only 29 (4.9%) reported uncertainty measures for their algorithms.
Synthesis
Study Characteristics
CVI is a medical imaging domain with enormous potential for ML application. The yearly growth in the number of publications reviewed demonstrates an increasing interest in conducting such interdisciplinary research. Although the bulk of articles was published by scientific institutes in developed nations, the constraints of conducting ML research in underprivileged locations are quickly diminishing. On the one hand, ML requires less advanced on-site technology than it once did, and many ML operations can be conveniently executed via cloud services [10]. On the other hand, more public medical datasets are made available every day, which can enable ML research in locations that lack access to extensive institutional data [11].
Most ML studies with direct clinical applications had an observational setup with a diagnostic focus. This was not unexpected given that observational studies are the most viable study design, and classification is the most prevalent application of ML. However, we should stress the avenues of research that can leverage other study types or focus on outcomes other than diagnosis. For example, Commandeur et al. designed a prospective ML study to predict the long-term risk of myocardial infarction and cardiac death based on clinical risk, coronary calcium, and epicardial adipose tissue [12]. Their study is a good example of applying ML for primary prevention and also in a prospective setting. As another example, Lee et al. applied conventional ML to non-invasive measurements from Computed Tomography (CT) images and electrocardiograms (ECGs) to predict patients’ responses to cardiac resynchronization therapy. Their work exemplifies how ML models could help with different therapeutic planning scenarios [13].
Lastly, we observed a significant amount of research in areas like segmenting heart chambers, quantifying epicardial fat, and coronary artery calcium scoring by applying ML to various imaging modalities. Despite the undeniable significance of such topics, we strongly encourage ML researchers to investigate other study fields as well. In light of this proposal, we can provide an example of an innovative study by Pyrros et al., which applied ML to CXRs to analyze racial/ethnic and socioeconomic differences in the prevalence of atherosclerotic vascular disease [14]. Even among diagnostic studies, innovation and high-impact research are not rare. Recent research by Liebig et al. demonstrates that collaboration between ML models and radiologists improves the performance of mammography-based breast cancer screening compared to relying solely on radiologists’ decisions [15]. They endorsed a novel retrospective interventional design for their investigation, which could be replicated in CVI studies.
Data Handling
Most of the examined papers focused on MRIs and echocardiograms. AI can provide many solutions for image acquisition, reconstruction, and analysis of cardiac MRI studies [16]. Echocardiography, on the other hand, has various limitations (such as a longer process duration, high operator subjectivity, and vast observation ranges) that could be ameliorated by artificial intelligence [17]. Apart from these two modalities, chest CT scans are commonly ordered for a variety of thoracic diseases, including lung diseases. Automated ML models that can evaluate the heart and circulation on chest CT scans are therefore excellent candidates for opportunistic cardiovascular disease screening. Examples of such applications could be a study by Commandeur et al., which leveraged DL to quantify epicardial fat on non-contrast chest CT scans of asymptomatic individuals [18], and another study by Aquino et al., which measured the size of the left atrium on chest CT scans for prediction of cardiovascular outcomes [19].
We found a few publications that built and assessed multimodal ML models. Multi-modal (or fusion) models are a group of models that comprise data from multiple imaging modalities or any non-imaging data (e.g., clinical variables or textual data) in addition to imaging data [20]. ML models and, in particular, DL models can simultaneously analyze numerous kinds of data, much like human medical experts who frequently rely on multiple pieces of data from a single patient to reach a diagnostic or therapeutic conclusion. Input data to ML models may be distinct imaging modalities; for instance, Puyol-Anton et al. created a DL model to predict patients’ responses to cardiac resynchronization therapy using 2D echocardiography and cardiac MRI [21]. A blend of imaging and non-imaging data can also be used to train multimodal ML models. For instance, Huang et al. developed a DL model capable of detecting pulmonary embolisms in CT Pulmonary Angiography examinations while also leveraging information from the patient’s electronic health records [22]. They demonstrated that the performance of their multimodal model was superior to that of an identical model trained just on CT imaging.
The number of examinations utilized by the eligible articles for training or evaluating their ML models varied considerably. Aside from the differences in the actual size of the data researchers had access to, this variability could be attributed to two other factors: (1) the reviewed articles did not share their dataset size in a consistent manner. While some articles simply reported the number of examinations used, many others only reported the number of patients included in their study (without clarifying how many exams had been obtained from each patient). Furthermore, a few instances described the size of their datasets using inaccurate terms, such as subjects or scans. Such terms may refer to both the number of patients and the number of images, which might confuse the reader. (2) Terms like scan, image, and imaging are not used consistently in the medical imaging literature. In addition to referring to a single two-dimensional (2D) image, these words can also imply a three-dimensional (3D) volume. As a best practice, we encourage ML researchers to always supply separate patient and examination numbers for their research. The terminology used to describe 2D and 3D imaging data in research should also be clarified. These tips can considerably improve the reproducibility of ML research and lessen its susceptibility to bias [23•].
Finally, we would like to emphasize the significance of considering publicly accessible medical datasets when undertaking ML research in medical imaging. Multiple free public datasets are available to ML researchers for various medical imaging modalities [11]. Therefore, researchers who lack sufficient internal data to train their models may find comparable data from other institutions. Using data from such an external source will not only increase the training size of ML models but also make them more generalizable. The EchoNet-Dynamic dataset of more than 10,000 echocardiograms [24], the Lung Image Database Consortium image collection (LIDC-IDRI) dataset of more than 1000 chest CT scans [25], the ImageTBAD dataset of more than 100 CT angiographies with type-B aortic dissection [26], and the Cardiac Atlas Project (CAP) dataset of more than 80 cardiac MRIs [27] are among the public datasets that were introduced in different articles we reviewed. However, this is not an exhaustive list of available datasets, and we encourage researchers to seek appropriate public datasets before conducting any study. ML competition websites such as Kaggle are also an excellent place to hunt for public data, although using such datasets in ML research should be undertaken with extreme caution [23•].
Model Development
As previously noted, DL models are much more complicated than conventional ML models and have a higher learning capacity. This explains why DL models have been more popular than conventional ML methods in the reviewed articles. However, DL is not always the go to option in ML research and selecting the appropriate ML model is more task specific. Although DL models are widely regarded as state-of-the-art computer solutions for automated decision-making in many different fields, there are some circumstances where conventional ML models or a combination of DL and conventional ML methods offer higher performance or are more affordable than DL methods [4, 28]. In a study by Gao et al., for instance, tree-based traditional ML techniques such as gradient boosting machines outperformed neural networks for vessel segmentation on X-ray coronary angiography [29]. It is also commonly believed that, when applied to tabular data, the same tree-based models are frequently superior to or on par with DL approaches [30].
Segmentation and classification were the most researched ML applications across all studies. Nonetheless, ML has additional intriguing uses in CVI. Among the least investigated ML tasks were object detection methods. Similar to segmentation models, these models pinpoint items of interest in input images while requiring less annotation effort (one draws rectangles around target objects rather than paints over all their pixels). For instance, Nizar et al. introduced an object detection technique for real-time aortic valve detection on echocardiography [31]. Another interesting area of research in medical imaging is image generation. Although current research efforts have utilized generative DL for objectives, such as removing artifacts from imaging data [32], raising the resolution of imaging [33], producing synthetic imaging datasets [34, 35], and improving segmentation outcomes [36], and the capabilities of generative models are far broader. It has been demonstrated that generative models can convert two biplanar CXRs to a natural-looking chest CT scan and even incorporate synthetic tumoral lesions into normal imaging data [37, 38].
Transfer learning is the technical term for when a DL model is first trained (pretrained) on a different dataset and its parameters are then fine-tuned on the dataset of interest [39]. This technique enables DL models to be trained more quickly (assuming the time to train the pretrained model is not included), with less training data, and with greater generalizability to unseen data [40]. The quality and similarity of the initial dataset that the model was pretrained on are a significant factor in determining the efficiency of transfer learning. Transfer learning is more beneficial for DL models when the original dataset is large and has imaging features similar to those of the dataset of interest.
Surprisingly, several of the examined articles did not disclose whether or not they used transfer learning. A few of those who reported employing transfer learning had pretrained their models using non-medical imaging datasets, such as ImageNet. Medical images, however, are fundamentally different from natural photographs, and pretraining DL models on natural photographs may not be the best transfer learning choice for medical images. Pretraining DL models using public medical image datasets is a more effective technique. Ankenbrand et al. illustrate this strategy by transferring weights from a DL model pretrained on a public medical image dataset to train their DL model for segmenting the heart on cardiac MRI data [41].
Almost none of the examined research acknowledged developing their ML models using a systematic checklist like the Checklist for Artificial Intelligence in Medical Imaging (CLAIM) [42•]. While many aspects of ML studies, including but not limited to model development, are susceptible to systematic biases [23•, 43•, 44•], documenting study adherence to a set of predefined standards helps reassure both researchers and their audience about the validity and reliability of a model's performance. In addition, publicly sharing the code, datasets, and weights of the trained ML models can further improve the repeatability of the researchers’ work, but we recognize that such public disclosures may not always be possible due to institutional policies that hold sway.
Performance Evaluation
It is typical in ML to sequester a random subset of data as the test set and train the ML models on the remaining data with or without validation sets). This allows the trained model to be evaluated against an untouched test set. When datasets are small or very imbalanced, this typical method of data partitioning is not the most effective. Instead, cross-validation approaches can provide a more accurate evaluation of a model’s performance under such conditions [23•]. Even though many medical datasets are either small or highly imbalanced, few of the examined articles utilized cross-validation strategies. This is a crucial shortcoming that diminishes confidence in their reported performance. We should note, though, that we also identified a few studies that employed more sophisticated and reliable kinds of cross-validation, such as nested cross-validation [45, 46].
Similar to cross-validation, few studies documented external validation of their ML models. External validation is a valuable performance measure as it helps to demonstrate a model’s generalizability to unseen data [47]. For example, assume that a DL model performs well on internal test data, but its performance declines significantly when applied to data from other institutions. A likely explanation for this observation could be the discrepancy in medical imaging devices and vendors across different institutions. A well-trained ML model should have minimal (or acceptable) sensitivity to the vendor-specific characteristics of the input imaging data and be able to extract relevant signals from that data regardless of its acquisition properties.
Even if the internal and external performance of a DL model is exceptional, there is no assurance that these models have learned to pick relevant and meaningful imaging signals. For instance, the apparent superior performance of a DL model in identifying pneumonia may be attributable to its attention to radiology markers present in CXR imaging [48]. Even though DL models are commonly referred to as black boxes, there are ways to visualize the regions of the imaging data to which they contribute the most when making predictions. This is called DL interpretation (or explanation) mapping [49]. Despite its uttermost importance, the majority of research employing DL classifier or regressor models did not report interpretation mapping for their models. Although interpretation maps have limitations [50], they can often shed more light on what a DL model has learned and whether or not it appears valid to human experts.
Finally, DL models could have uncertain performance due to their inherent properties or when they encounter data points that were not adequately represented in their training data [51•]. For example, a classifier designed to distinguish between non-COVID-19 and COVID-19 viral pneumonia on the CXR may be unreliable when used on a CXR presenting with bacterial pneumonia. Even though neither label could be accurate, this classifier will still predict a label for this CXR and without adequate uncertainty quantification, a naive user may accept that prediction. Although several techniques exist for uncertainty quantification of DL models, they have not been thoroughly studied for medical purposes and therefore, it is not surprising that only a small number of studies in our pool have employed such techniques (52). The need to quantify the uncertainty of DL models, however, will likely become more and more important to healthcare researchers.
Discussion
CVI is a rapidly growing area of medical imaging with ample opportunities for ML study. In this report, we presented the findings of a recent scoping review describing the applications of ML in CVI.
Our findings must be evaluated in light of two significant limitations. First, due to feasibility concerns, we limited our search to the MEDLINE database and English peer-reviewed manuscripts. Searching other databases, adding non-English and gray literature, and applying broader search terms would increase the number of articles eligible for inclusion. Despite this limitation, we believe that our combined pool of articles were sufficient to describe the broad trends in ML research for CVI. Second, we did not conduct a bias assessment of the individual articles included in our review. We considered this limitation acceptable because our objective was not to perform any meta-analyses on the results of the review but rather to assemble a more thorough list of factors that might make included studies more susceptible to bias.
In conclusion, we used quantitative statistics and qualitative synthesis to summarize four major aspects of ML research in CVI (study characteristics, data handling, model development, and performance evaluation) and attempted to sketch the big picture of current research gaps and future directions for similar studies.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Dr. Hani Abujudeh for reviewing the manuscript.
Abbreviations
- CVI
Cardiovascular imaging
- AI
Artificial intelligence
- ML
Machine learning
- DL
Deep learning
- CXR
Chest X-ray
- SPECT
Single-photon Emission Computed Tomography
- CT
Computed Tomography
- ECG
Electrocardiograms
- MRI
Magnetic Resonance Imaging
- 2D
Two-dimensional
- 3D
Three-dimensional
- COVID-19
Coronavirus Disease of 2019
- CLAIM
Checklist for Artificial Intelligence in Medical Imaging
- PRISMA
Preferred Reporting Items for Systematic Reviews and Meta-Analyses
Funding
N.A.
Data Availability
All data analyzed during this study are provided in the supplementary materials.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Research Involving Human and Animals Rights
This article does not contain any studies with human or animal subjects performed by any of the authors.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Pouria Rouzrokh and Bardia Khosravi have contributed equally to this work.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
- 1.Guo Y, Hao Z, Zhao S, Gong J, Yang F. Artificial intelligence in health care: bibliometric analysis. J Med Internet Res. 2020;22(7):e18228. doi: 10.2196/18228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Secinaro S, Calandra D, Secinaro A, Muthurangu V, Biancone P. The role of artificial intelligence in healthcare: a structured literature review. BMC Med Inform Decis Mak. 2021;21(1):125. doi: 10.1186/s12911-021-01488-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.•Weinan E. Machine Learning and Computational Mathematics. arXiv [math.NA]. 2020. http://arxiv.org/abs/2009.14596. This reference is a nice introduction to machine learning and their mathematical background.
- 4.•Chauhan NK, Singh K. A Review on Conventional Machine Learning vs Deep Learning. 2018 International Conference on Computing, Power and Communication Technologies (GUCON). 2018. p. 347–352. This reference is a nice introduction to deep learning and how it stands differently from conventional machine learning methods.
- 5.•King B, Barve S, Ford A, Jha R. Unsupervised Clustering of COVID-19 Chest X-Ray Images with a Self-Organizing Feature Map. 2020 IEEE 63rd International Midwest Symposium on Circuits and Systems (MWSCAS). 2020. p. 395–398. This reference provides an interesting example of unsupervised learning in medical imaging.
- 6.•Elyan, Vuttipittayamongkol. Computer vision and machine learning for medical image analysis: recent advances, challenges, and way forward. Artif Cells Blood Substit Immobil Biotechnol. https://rgu-repository.worktribe.com/output/1631673/computer-vision-and-machine-learning-for-medical-image-analysis-recent-advances-challenges-and-way-forward. This reference highlights the most recent applications of machine learning in medical image analysis.
- 7.Di Carli MF, Geva T, Davidoff R. The future of cardiovascular imaging. Circulation. 2016;133(25):2640–2661. doi: 10.1161/CIRCULATIONAHA.116.023511. [DOI] [PubMed] [Google Scholar]
- 8.Esteva A, Chou K, Yeung S, et al. Deep learning-enabled medical computer vision. NPJ Digit Med. 2021;4(1):5. doi: 10.1038/s41746-020-00376-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Page MJ, McKenzie JE, Bossuyt PM, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. Rev Esp Cardiol. 2021;74(9):790–799. doi: 10.1016/j.recesp.2021.06.016. [DOI] [PubMed] [Google Scholar]
- 10.Agavanakis KN, Karpetas GE, Taylor M, et al. Practical machine learning based on cloud computing resources. AIP Conf Proc. 2019;2123(1):020096. doi: 10.1063/1.5117023. [DOI] [Google Scholar]
- 11.••Li J, Zhu G, Hua C, et al. A Systematic Collection of Medical Image Datasets for Deep Learning. arXiv [eess.IV]. 2021. http://arxiv.org/abs/2106.12864. It introduces multiple public datasets for training deep learning models on medical imaging data.
- 12.Commandeur F, Slomka PJ, Goeller M, et al. Machine learning to predict the long-term risk of myocardial infarction and cardiac death based on clinical risk, coronary calcium, and epicardial adipose tissue: a prospective study. Cardiovasc Res. 2020 doi: 10.1093/cvr/cvz321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee AWC, Razeghi O, Solis-Lemus JA, et al. Non-invasive simulated electrical and measured mechanical indices predict response to cardiac resynchronization therapy. Comput Biol Med. 2021;138:104872. doi: 10.1016/j.compbiomed.2021.104872. [DOI] [PubMed] [Google Scholar]
- 14.Pyrros A, Rodríguez-Fernández JM, Borstelmann SM, et al. Detecting racial/ethnic health disparities using deep learning from frontal chest radiography. J Am Coll Radiol. 2022;19(1 Pt B):184–191. doi: 10.1016/j.jacr.2021.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leibig C, Brehmer M, Bunk S, Byng D, Pinker K, Umutlu L. Combining the strengths of radiologists and AI for breast cancer screening: a retrospective analysis. Lancet Digit Health. 2022;4(7):e507–e519. doi: 10.1016/S2589-7500(22)00070-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fotaki A, Puyol-Antón E, Chiribiri A, Botnar R, Pushparajah K, Prieto C. Artificial intelligence in cardiac MRI: is clinical adoption forthcoming? Front Cardiovasc Med. 2021;8:818765. doi: 10.3389/fcvm.2021.818765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou J, Du M, Chang S, Chen Z. Artificial intelligence in echocardiography: detection, functional evaluation, and disease diagnosis. Cardiovasc Ultrasound. 2021;19(1):29. doi: 10.1186/s12947-021-00261-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Commandeur F, Goeller M, Betancur J, et al. Deep learning for quantification of epicardial and thoracic adipose tissue from non-contrast CT. IEEE Trans Med Imaging. 2018;37(8):1835–1846. doi: 10.1109/TMI.2018.2804799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aquino GJ, Chamberlin J, Mercer M, et al. Deep learning model to quantify left atrium volume on routine non-contrast chest CT and predict adverse outcomes. J Cardiovasc Comput Tomogr. 2022;16(3):245–253. doi: 10.1016/j.jcct.2021.12.005. [DOI] [PubMed] [Google Scholar]
- 20.Huang S-C, Pareek A, Seyyedi S, Banerjee I, Lungren MP. Fusion of medical imaging and electronic health records using deep learning: a systematic review and implementation guidelines. NPJ Digit Med. 2020;3:136. doi: 10.1038/s41746-020-00341-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Puyol-Antón E, Sidhu BS, Gould J, et al. A multimodal deep learning model for cardiac resynchronisation therapy response prediction. Med Image Anal. 2022;79:102465. doi: 10.1016/j.media.2022.102465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang S-C, Pareek A, Zamanian R, Banerjee I, Lungren MP. Multimodal fusion with deep neural networks for leveraging CT imaging and electronic health record: a case-study in pulmonary embolism detection. Sci Rep. 2020;10(1):22147. doi: 10.1038/s41598-020-78888-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.•Rouzrokh P, Khosravi B, Faghani S, et al. Mitigating Bias in Radiology Machine Learning: 1. Data Handling. Radiology: Artificial Intelligence. Radiological Society of North America; 2022;4(5):e210290. This paper discusses how bias can happen and be mitigated during model development of machine learning models in medicine. [DOI] [PMC free article] [PubMed]
- 24.Ouyang D, He B, Ghorbani A, et al. Video-based AI for beat-to-beat assessment of cardiac function. Nature. 2020 doi: 10.1038/s41586-020-2145-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Armato SG, 3rd, McLennan G, Bidaut L, et al. The Lung Image Database Consortium (LIDC) and Image Database Resource Initiative (IDRI): a completed reference database of lung nodules on CT scans. Med Phys. 2011;38(2):915–931. doi: 10.1118/1.3528204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yao Z, Xie W, Zhang J, et al. ImageTBAD: A 3D computed tomography angiography image dataset for automatic segmentation of type-B aortic dissection. Front Physiol. 2021;12:732711. doi: 10.3389/fphys.2021.732711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fonseca CG, Backhaus M, Bluemke DA, et al. The Cardiac Atlas Project—an imaging database for computational modeling and statistical atlases of the heart. Bioinformatics. Oxford Academic. 2011;27(16):2288–2295. doi: 10.1093/bioinformatics/btr360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lai Y. A comparison of traditional machine learning and deep learning in image recognition. J Phys Conf Ser. IOP Publishing. 2019;1314(1):012148. doi: 10.1088/1742-6596/1314/1/012148. [DOI] [Google Scholar]
- 29.Gao Z, Wang L, Soroushmehr R, et al. Vessel segmentation for X-ray coronary angiography using ensemble methods with deep learning and filter-based features. BMC Med Imaging. 2022;22(1):10. doi: 10.1186/s12880-022-00734-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.•Grinsztajn L, Oyallon E, Varoquaux G. Why do tree-based models still outperform deep learning on tabular data? arXiv [cs.LG]. 2022. http://arxiv.org/abs/2207.08815. This reference lists several reasons for why conventional machine learning approaches like tree-based models are sometimes better in interpreting tabular data than deep learning models.
- 31.Nizar MHA, Chan CK, Khalil A, Yusof AKM, Lai KW. Real-time detection of aortic valve in echocardiography using convolutional neural networks. Curr Med Imaging Rev. 2020;16(5):584–591. doi: 10.2174/1573405615666190114151255. [DOI] [PubMed] [Google Scholar]
- 32.Nezafat M, El-Rewaidy H, Kucukseymen S, Hauser TH, Fahmy AS. Deep convolution neural networks based artifact suppression in under-sampled radial acquisitions of myocardial T 1 mapping images. Phys Med Biol. 2020;65(22):225024. doi: 10.1088/1361-6560/abc04f. [DOI] [PubMed] [Google Scholar]
- 33.Mahapatra D, Bozorgtabar B, Garnavi R. Image super-resolution using progressive generative adversarial networks for medical image analysis. Comput Med Imaging Graph. 2019;71:30–39. doi: 10.1016/j.compmedimag.2018.10.005. [DOI] [PubMed] [Google Scholar]
- 34.Sun Y, Vixege F, Faraz K, et al. A pipeline for the generation of synthetic cardiac color doppler. IEEE Trans Ultrason Ferroelectr Freq Control. 2022;69(3):932–941. doi: 10.1109/TUFFC.2021.3136620. [DOI] [PubMed] [Google Scholar]
- 35.Prakosa A, Sermesant M, Delingette H, et al. Generation of synthetic but visually realistic time series of cardiac images combining a biophysical model and clinical images. IEEE Trans Med Imaging. 2013;32(1):99–109. doi: 10.1109/TMI.2012.2220375. [DOI] [PubMed] [Google Scholar]
- 36.Xu C, Xu L, Ohorodnyk P, Roth M, Chen B, Li S. Contrast agent-free synthesis and segmentation of ischemic heart disease images using progressive sequential causal GANs. Med Image Anal. 2020;62:101668. doi: 10.1016/j.media.2020.101668. [DOI] [PubMed] [Google Scholar]
- 37.•Ying, Guo, Ma, Wu, Weng. X2CT-GAN: reconstructing CT from biplanar X-rays with generative adversarial networks. Proc Estonian Acad Sci Biol Ecol. http://openaccess.thecvf.com/content_CVPR_2019/html/Ying_X2CT-GAN_Reconstructing_CT_From_Biplanar_X-Rays_With_Generative_Adversarial_Networks_CVPR_2019_paper.html. This reference introduces an interesting deep learning model that can build a three-dimensional CT scan from a two-dimensional Chest X-ray.
- 38.•Wolleb J, Sandkühler R, Bieder F, Cattin PC. The Swiss Army Knife for Image-to-Image Translation: Multi-Task Diffusion Models. arXiv [cs.CV]. 2022. http://arxiv.org/abs/2204.02641. This paper introduces different applications of diffusion models, including a couple of interesting applications in medicine.
- 39.Kim HE, Cosa-Linan A, Santhanam N, Jannesari M, Maros ME, Ganslandt T. Transfer learning for medical image classification: a literature review. BMC Med Imaging. 2022;22(1):69. doi: 10.1186/s12880-022-00793-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.•Iman M, Rasheed K, Arabnia HR. A Review of Deep Transfer Learning and Recent Advancements. arXiv [cs.LG]. 2022. http://arxiv.org/abs/2201.09679. This paper introduces transfer learning, a deep learning technique that enables new models leverage the knowledge of the already trained models.
- 41.Ankenbrand MJ, Lohr D, Schlötelburg W, Reiter T, Wech T, Schreiber LM. Deep learning-based cardiac cine segmentation: transfer learning application to 7T ultrahigh-field MRI. Magn Reson Med. 2021;86(4):2179–2191. doi: 10.1002/mrm.28822. [DOI] [PubMed] [Google Scholar]
- 42.•Mongan J, Moy L, Kahn CE Jr. Checklist for Artificial Intelligence in Medical Imaging (CLAIM): A Guide for Authors and Reviewers. Radiol Artif Intell. 2020;2(2):e200029. This paper introduces a checklist for standard development of machine learning models in medicine. [DOI] [PMC free article] [PubMed]
- 43.•Zhang K, Khosravi B, Vahdati S, et al. Mitigating Bias in Radiology Machine Learning: 2. Model Development. Radiology: Artificial Intelligence. Radiological Society of North America; 2022;e220010. This paper discusses how bias can happen and be mitigated during model development of machine learning models in medicine. [DOI] [PMC free article] [PubMed]
- 44.•Faghani S, Khosravi B, Zhang K, et al. Mitigating Bias in Radiology Machine Learning: 3. Performance Metrics. Radiology: Artificial Intelligence. Radiological Society of North America; 2022;e220061. This paper discusses how bias can happen and be mitigated in during the performance evaluation of machine learning models in medicine. [DOI] [PMC free article] [PubMed]
- 45.Larroza A, Materka A, López-Lereu MP, Monmeneu JV, Bodí V, Moratal D. Differentiation between acute and chronic myocardial infarction by means of texture analysis of late gadolinium enhancement and cine cardiac magnetic resonance imaging. Eur J Radiol. 2017 doi: 10.1016/j.ejrad.2017.04.024. [DOI] [PubMed] [Google Scholar]
- 46.Kotu LP, Engan K, Borhani R, et al. Cardiac magnetic resonance image-based classification of the risk of arrhythmias in post-myocardial infarction patients. Artif Intell Med. 2015;64(3):205–215. doi: 10.1016/j.artmed.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 47.Yu AC, Mohajer B, Eng J. External validation of deep learning algorithms for radiologic diagnosis: a systematic review. Radiol Artif Intell. 2022;4(3):e210064. doi: 10.1148/ryai.210064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zech JR, Badgeley MA, Liu M, Costa AB, Titano JJ, Oermann EK. Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: a cross-sectional study. PLoS Med. 2018;15(11):e1002683. doi: 10.1371/journal.pmed.1002683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ayyar MP, Benois-Pineau J, Zemmari A. Review of white box methods for explanations of convolutional neural networks in image classification tasks. JEI. 2021;30(5):050901. [Google Scholar]
- 50.•Saporta, Gui, Agrawal, Pareek, Truong. Deep learning saliency maps do not accurately highlight diagnostically relevant regions for medical image interpretation. MedRxiv. This reference explains how saliency maps, that are often used for explaining how deep learning models work, could be biased and how they should be interpreted with caution.
- 51.•Abdar M, Pourpanah F, Hussain S, et al. A review of uncertainty quantification in deep learning: Techniques, applications and challenges. Inf Fusion. 2021;76:243–297. This article discusses different applications of uncertainty quantification in deep learning.
- 52.Loftus TJ, Shickel B, Ruppert MM, et al. Uncertainty-aware deep learning in healthcare: a scoping review. PLOS Digit Health. 2022;1(8):0000085. doi: 10.1371/journal.pdig.0000085. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data analyzed during this study are provided in the supplementary materials.