Abstract
The coronavirus (COVID-19) global pandemic has impacted the health of almost everyone, including changes in their salivary microbiota. Since 2019, there has been an increase in the number of new COVID-19 cases in Thailand. Therefore, COVID-19 active case finding is important for early detection and epidemic control. Moreover, the dynamic changes of salivary bacteriome in asymptomatic COVID-19 cases are largely unknown. This research aimed to investigate and compare the salivary bacteriome and the co-infectious bacterial pathogens in the asymptomatic COVID-19 positive group to the negative group, based on novel nanopore sequencing. This cohort was a cross-sectional study including saliva samples collected from 82 asymptomatic participants (39 COVID-19 positive and 43 COVID-19 negative cases). All samples were sequenced for the full-length bacterial 16S rDNA. The alpha and beta diversity analyses were not significantly different between groups. The three major species in salivary bacteriome including Veillonella parvula, Streptococcus mitis, and Prevotella melaninogenica were observed in both groups. Interestingly, Lautropia mirabilis was a significantly enriched species in the saliva of the asymptomatic COVID-19-positive cases based on linear discriminant analysis effect size (LEfSe) analysis. The results suggested that L. mirabilis was a co-infectious agent in the asymptomatic COVID-19 group. However, the potential role of L. mirabilis should be validated in further experimental studies.
Keywords: SARS-CoV-2, full-length, 16S rRNA, microbiota, oral bacteriome, MinION
Impact Statement
To date, there is little data on the association of salivary bacteriome in asymptomatic COVID-19 cases. In previous studies, the salivary bacteriome in COVID-19 patients was classified based on short-read sequencing which limits bacterial classification at the genus level. These problems can be improved within this study using long-read nanopore sequencing. The full-length 16S rDNA of salivary bacteriome in the asymptomatic COVID-19 groups were successfully sequenced and then classified into bacterial species. Lautropia mirabilis was significantly increased in the asymptomatic COVID-19-positive group. This species was a respiratory pathogen found in HIV and cystic fibrosis patients and COVID-19 cases. In contrast, Kingella oralis and Granulicatella adiacens were found only in the negative group. They were oral commensal and did not cause disease in a healthy host. Our results demonstrated the association between salivary bacteriome and asymptomatic COVID-19, which might be helpful for prognosticating health status.
Introduction
COVID-19 is an acute respiratory disease caused by a novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). In the winter of 2019, coronavirus disease (COVID-19) first emerged in Wuhan, China, and has rapidly spread to become a global health crisis resulting in a pandemic. 1 At the worldwide level, increases in the number of new cases and deaths were reported to the World Health Organization (WHO), with more than 546 million confirmed cases and over 6.3 million deaths recorded (https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19—6-july-2022). It generally manifests as an asymptomatic infection or mild respiratory symptoms. However, it can develop into severe pneumonia leading to death in those over 60 years old or those who have comorbidities. Currently, the treatments for COVID-19 are based on symptomatic and supportive treatments. 2
Recent studies have reported the detection of SARS-CoV-2 in saliva and the viral transmission via saliva droplets and potentially aerosols. 3 Therefore, the oral cavity might be an important reservoir of SARS-CoV-2 and infection site. Several microbial communities colonize the oral surfaces. The microbiota plays an important role in modulating human health through maintaining homeostasis and the immune system. 4 During respiratory viral infections, the host microbiome has been changed and associated with the viral infection or disease severity.5,6 As represented in a previous report, the bacterial ecosystem contributes to the onset of respiratory syncytial virus (RSV), avian influenza A virus (H7N9), and influenza A virus (IAV) infections.7 –9
Several microecological studies on COVID-19 have suggested that the microbial community plays an important role in COVID-19 and might serve as a new target of treatment.10,11 The intestinal and airway microbiomes in COVID-19 patients significantly changed in terms of richness and diversity. Recent reports showed that the proportion of beneficial bacteria tends to decrease, while pathogenic bacteria tend to increase in the saliva of COVID-19 patients.12 –14 The oral microbiome changes in patients with COVID-19 were recently studied in Chinese cases. The results revealed that the oral microbiome collected from tongue-coating samples was imbalanced in COVID-19 patients. Compared with the healthy group, the diversity of oral microbiome in COVID-19 patients was significantly lower than those found in the control group. In addition, butyric acid–producing bacteria decreased, and lipopolysaccharide-producing bacteria increased in the oral cavity of COVID-19 patients. 13 Other studies reported that Fusobacterium periodonticum might promote susceptibility to COVID-19 infection. 14 Similarly, the composition of oral microflora was changed in COVID-19 patients, as shown in other reports.15,16 Oral dysbiosis is involved in periodontitis and systemic diseases, including COVID-19. Moreover, oral dysbiosis potentially affects the severity of COVID-19.17,18 As the susceptibility to the SARS-CoV-2 viral infection differs from person to person, the characterization of the microbiome in COVID-19 patients will be useful for a better understanding of this and may also contribute to improving the prevention and treatment of COVID-19.
Currently, the Oxford nanopore third-generation sequencing represents a new and fast technology used to identify microbial profiling with high sensitivity and long-read data. For microbiome analysis, the Oxford nanopore platform can sequence the amplified full-length 16S rDNA gene with high accuracy and sensitivity. 19 In addition, the long-read 16S rDNA also allows bacterial classification at the species level, which is usually unachievable by short-read sequencing. 20 This study aims to investigate the salivary bacteriome and potential co-infected bacterial pathogens in asymptomatic COVID-19 cases compared with negative cases using novel nanopore sequencing technology.
Materials and methods
Sample collections
Eighty-two participants were enrolled in this study between July and August 2021 from the COVID-19 active screening program. 21 The Institutional Review Board of the Faculty of Medicine, Chulalongkorn University, approved this study (IRB No. 302/63). All participants complied with, and signed, the informed consent for the screening test. This study was conducted based on the practice of maintaining the security and confidentiality of participant records. The identity and information were concealed by sample ID. Participants with informed consent self-collected their saliva (approximately 0.5 mL) via saliva collection kits following the manufacturer’s instruction. The saliva collection tube contained 0.5 mL of 2× DNA/RNA shield (Zymo Research, USA) for viral inactivation and nucleic acid stabilization. All nucleic acids were extracted from 0.2 mL of saliva suspension within 24 h after saliva collection and stored at −80°C until use.
RT-qPCR
Using a quantitative reverse transcription polymerase chain reaction (RT-qPCR), the COVID-19-positive samples were identified by amplifying the SARS-CoV-2 nucleocapsid (N1) gene. The N1 primer-probe sets and thermal cycling were set up as in a previous report. 22 Briefly, the RT-qPCR reaction mixture consisted of 1× Luna® Universal Probe qPCR Master Mix (New England Biolabs, USA), 7.5 U WarmStart® RTx Reverse Transcriptase (New England Biolabs, USA), 0.5 µM N1F primer (5′-GACCCCAAAATCAGCGAAAT-3′), 0.5 µM N1-R primer (5′-TCTGGTTACTGCCAGTTGAATCTG-3′), 0.125 µM TaqMan probe (5′-FAM-ACCCCGCATTACGTTTGGTGGACC-BHQ1-3′), and 2.5 µL of extracted nucleic acid in the final volume of 10 µL. The reaction was run in the CFX96™ Real-Time PCR instrument (Bio-Rad, USA) using the thermal profile as follows: 55°C for 10 min, 95°C for 1 min, followed by 45 cycles of 95°C for 10 s, and 55°C for 1 min. The cut-off cycle thresholds for positive samples were lower than 36.
Amplicon sequencing based on the MinION nanopore
The full-length bacterial 16S rDNA was amplified using the tailed primers: 5′-TTTCTGTTGGTGCTGATATTGCAGRGTTYGATYMTGGCTCAG-3′and 5′-ACTTGCCTGTCGCTCTATCTTCCGGYTACCTTGTTACGACTT-3′ modified from a previous study. 20 The 20 µL PCR reaction consisted of 1× Phusion™ Plus buffer (Thermo Fisher Scientific, USA), 0.2 mM dNTPs (Promega, USA), 0.25 µM each of forward and reverse primers, 0.2 µL of Phusion™ Plus DNA Polymerase (Thermo Fisher Scientific, USA), and 1 µL of the nucleic acid template. The PCR temperature cycling conditions were as follows: 98°C for 30 s, followed by 25 cycles of 98°C for 10 s, 60°C for 10 s, 72°C for 45 s, and final extension at 72°C for 5 min. Then, the amplicons were barcoded by 5-cycle PCR using the barcode primers based on PCR Barcoding Expansion 1-96 (EXP-PBC096) kit (Oxford Nanopore Technologies, UK). The barcoded libraries were purified by QIAquick PCR Purification Kit (Qiagen, Germany). The purified libraries were measured using Quant-iT™ dsDNA HS Assay Kits (Invitrogen, USA) and then equimolarly pooled for multiplexing. The pooled library was purified by 0.5× Agencourt AMPure XP beads (Beckman Coulter, USA). After that, the library was subjected to end repair and adaptor ligation steps using Ligation Sequencing Kit (SQK-LSK109). Finally, the library was loaded onto the R10.3 flow cell and sequenced on the MinION Mk1C sequencer (Oxford Nanopore Technologies, UK).
Data analysis
The FAST5 data were base called by guppy basecaller v5.0.16 (Oxford Nanopore Technologies, UK) with a super-accuracy model to generate the FASTQ files. The quality of reads was examined by MinIONQC. 23 Then, FASTQ sequences were demultiplexed and adaptor-trimmed using porechop v0.2.4 (https://github.com/rrwick/Porechop). The filtered reads were then clustered, polished, and taxonomically classified by NanoCLUST 24 based on the reference sequences from SILVA database v138. The abundance and taxonomic assignment data were then converted into QIIME data format. The alpha and beta diversity were evaluated by the QIIME2 analysis plugin (QIIME2 v2021.2). The data were visualized and illustrated by MicrobiomeAnalyst. 25 The differential abundance analysis was performed using linear discriminant analysis effect size (LEfSe) with P < 0.05 and Linear discriminant analysis (LDA) score > 2. 26 Pairwise comparisons of eight species between groups were analyzed using the Mann–Whitney U test using GraphPad Prism 6 software.
Results
Study cohort
At the beginning of 2021, a coronavirus disease (COVID-19) outbreak occurred in Thailand, necessitating the COVID-19-active screening program for early detection and epidemic management. Eighty-two asymptomatic participants were included in this study from July to August 2021. Of all the saliva samples, 39 (47.6%) asymptomatic cases were detected with SARS-CoV-2, comprising the asymptomatic COVID-19-positive group. Meanwhile, 43 (52.4%) samples were found negative for this virus and represent the negative group. The average ages of the asymptomatic COVID-19 positive group and the negative group were 34.7 ± 12.4 and 36.2 ± 2.7 years, respectively (P = 0.450, t-test) as shown in Table 1. Moreover, the characteristics of all participants (N = 82) are presented in Supplementary Table 1.
Table 1.
Summary of study cohorts.
| COVID-19 positive | COVID-19 negative | |
|---|---|---|
| N | 39 | 43 |
| Age (years) | 34.7 ± 12.4 | 36.2 ± 2.7 |
| Gender | ||
| Female | 16 (41%) | 33 (77%) |
| Male | 23 (59%) | 10 (23%) |
Diversity of the salivary bacteriome
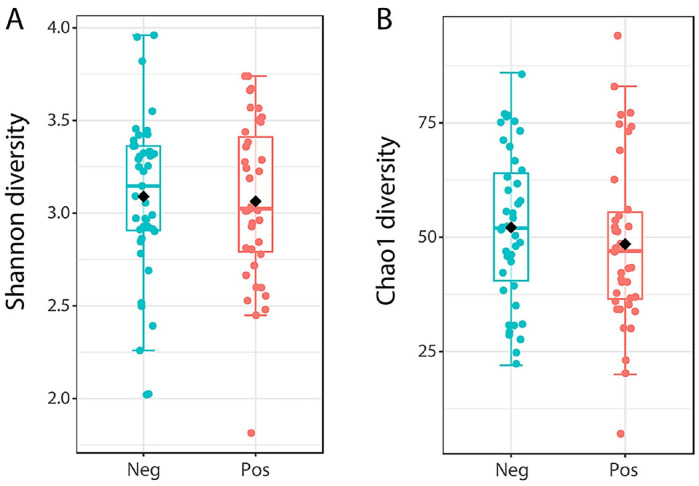
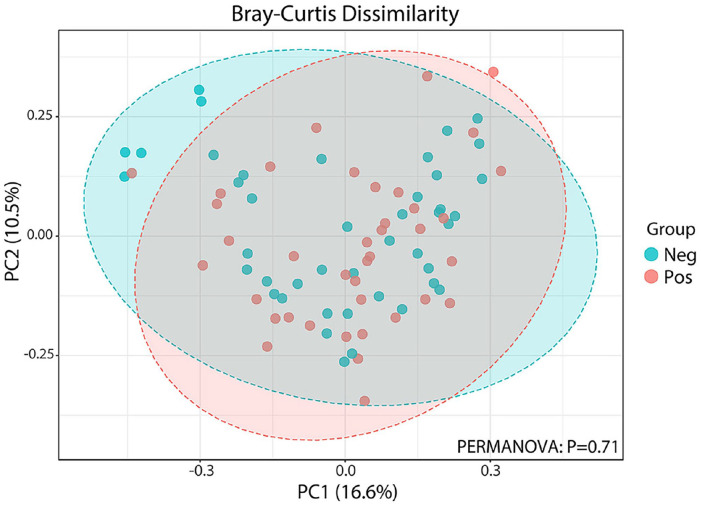
All samples were successfully sequenced with the full-length bacterial 16S rDNA with a high-throughput nanopore sequencing. In total, 853,898 reads were obtained with the average reads per sample of 10,413. The average classified reads were 9730 reads per sample (93.4%), and the percentage of classified reads ranged from 87% to 98%, as shown in Supplementary Table 1. The rarefaction analysis showed adequate sequencing depth for diversity for all the samples, as shown in Supplementary Figure 1. Alpha diversity comparisons between asymptomatic COVID-19 positive and negative groups, including richness and evenness analysis, showed no statistical differences (Mann–Whitney U test) in Shannon (P = 0.781) and Chao1 (P = 0.285) indexes as shown in Figure 1(A) and (B). When comparing the microbial community compositions between asymptomatic COVID-19 positive and negative groups, we performed beta diversity analyses using the Bray-Curtis cluster analysis index (Figure 2). The obtained result revealed that beta diversity analyses were not significantly distinguished between asymptomatic COVID-19 positive and negative groups (P = 0.71, permutational multivariate analysis of variance [PERMANOVA]).
Figure 1.
Alpha diversity of salivary bacteriome in asymptomatic cases with positive versus negative COVID-19. Boxplots present the Shannon (A) and Chao1 (B) diversities of microbial communities in asymptomatic COVID-19 positive (pink) and negative (blue) groups. Mann–Whitney U test was used to compare statistically alpha diversity. (A color version of this figure is available in the online journal.)
Figure 2.
UniFrac beta diversity in asymptomatic cases with positive versus negative COVID-19. Multidimensional scaling plot from Principal Coordinates Analysis (PCoA) based on Bray–Curtis distances, indicating a clustering pattern among samples obtained from asymptomatic COVID-19 positive (pink) and negative (blue) groups. Permutational ANOVA (PERMANOVA) was used to compare the beta diversity. (A color version of this figure is available in the online journal.)
Relative abundance of the salivary bacteriome
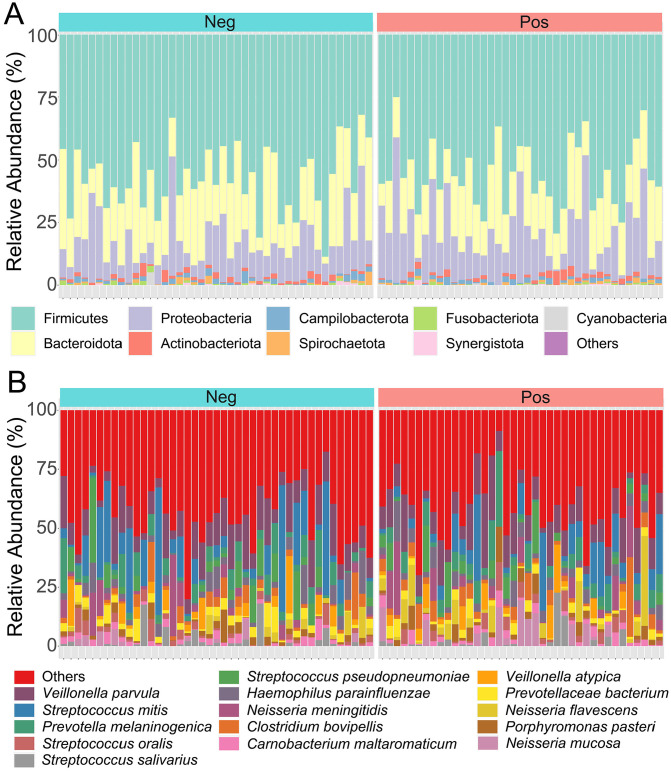
Microbial composition and relative abundance (%RA) of all samples were identified to characterize the oral bacterial profiles in asymptomatic COVID-19 positive and negative groups. At the phylum level, the top three dominant bacteria in both groups were Firmicutes, Bacteroidota, and Proteobacteria (Figure 3(A)). The relative species abundance of salivary bacteriome in both groups is illustrated in Figure 3(B). The result shows that Veillonella parvula, Streptococcus mitis, and Prevotella melaninogenica were the three most common species in the asymptomatic COVID-19 positive group (accounting for 23.8% of the community) and the negative group (accounting for 26.5% of the community).
Figure 3.
The taxonomic composition of the salivary bacteriome. Bar charts of the relative abundance (%) of bacteria phyla (A) and species (B) were obtained from 82 saliva samples, including asymptomatic COVID-19 positive and negative subjects. Different colored bars represent the different salivary bacteriome taxa. (A color version of this figure is available in the online journal.)
Differential salivary bacteria between asymptomatic COVID-19 positive and negative groups
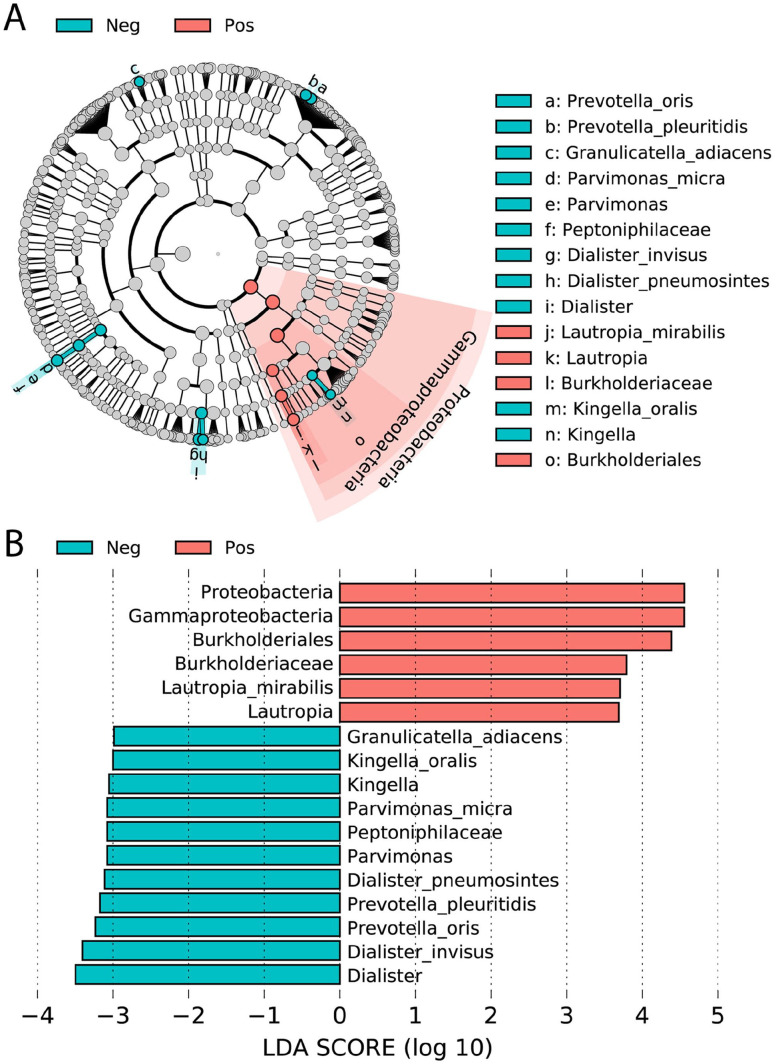
We accomplished the LEfSe analysis to distinguish the significant salivary bacteriome in the asymptomatic COVID-19 positive group in comparison with the negative group. The bar graph and cladogram of different taxonomic levels with LDA scores > 2 are presented in Figure 4(A) and (B), respectively. The results show that the phylum Proteobacteria, class Gammaproteobacteria, order Burkholderiales, family Burkholderiaceae, genus Lautropia, and species Lautropia mirabilis were significantly dominant in the asymptomatic COVID-19-positive group, whereas multiple taxa were significantly more abundant in the saliva of the COVID-19 negative group. The enriched species which were found in this group included Granulicatella adiacens, Kingella oralis, Parvimonas micra, Dialister pneumosintes, Prevotella pleuritidis, Prevotella oris, and Dialister invisus.
Figure 4.
Linear discriminant analysis Effect Size (LEfSe) of bacterial OTUs. The most differentially abundant taxa between asymptomatic COVID-19 positive (pink) and negative (blue) groups were investigated and represented in the bar graph (A). Cladogram (B) indicated the differentially abundant microbiota at the different taxonomic levels. The LDA score showed the effect size and ranking of each differentially abundant taxon (LDA score > 2). (A color version of this figure is available in the online journal.)
Pairwise comparisons of relative species abundances between groups
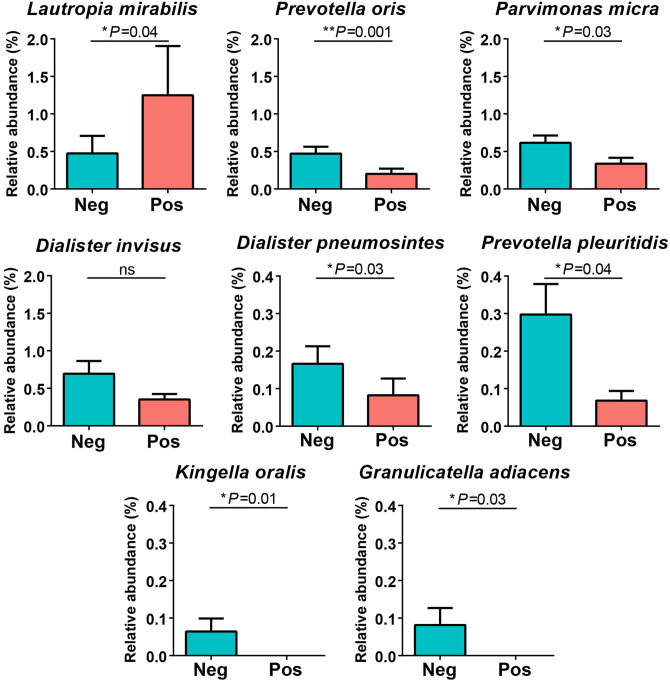
The differential relative abundances of bacterial species were statistically compared between groups based on the Mann–Whitney U test (Figure 5). The proportion of L. mirabilis in the asymptomatic COVID-19 positive group was significantly elevated compared to the negative group (P = 0.04). In contrast, the relative abundances of six species including P. oris, P. micra, D. pneumosintes, P. pleuritidis, K. oralis, and G. adiacens in the asymptomatic COVID-19 positive group were significantly decreased compared to the negative group (P = 0.001, 0.03, 0.03, 0.04, 0.01, and 0.03, respectively). Meanwhile, no statistical difference in Dialister invisus was observed between groups.
Figure 5.
Pairwise comparison of eight differential bacterial species (one for asymptomatic COVID-19 positive (pink) and seven for negative (blue) groups). The significant differences in each species were analyzed and reported in the P-value after the Mann–Whitney U test. (A color version of this figure is available in the online journal.)
Discussion
The oral microbiota is the second most diverse microbial community in humans after the gut microbiota. 27 Bacterial composition exists within oral cavities and has been linked to host genetics, diet, lifestyle, immune system, antibiotic exposure, hormones, and other factors.27,28 Meanwhile, the changes in the oral microbiota are implicated in the severity and complications of COVID-19. 13 Recently, the oral microbiota was investigated from tongue-coating, nasal and throat swabs, and saliva samples of ongoing symptomatic, severe and post COVID-19 patients compared to healthy controls.12 –18 However, there are only limited data on the dynamic changes of salivary bacteriome in asymptomatic COVID-19 cases. All studies on the salivary bacteriome were performed on short-read sequencing using the Illumina MiSeq technology.29,30 Notably, there are limitations of the Illumina NGS in obtaining the short-read sequences and identifying bacterial classification at the genus level, which can be improved by long-read nanopore sequencing. Moreover, the species identification of salivary bacteriome using Oxford nanopore technology has not been reported previously. Therefore, this study focuses on the salivary bacteriome in asymptomatic COVID-19 cases using long-read nanopore sequencing. Almost half of the saliva samples from asymptomatic cases were detected with SARS-CoV-2. After demographic analysis, there is no significant difference in age between the asymptomatic and negative cohorts. Notably, we are the first to propose and successfully characterize the bacterial species microbiota in asymptomatic COVID-19 saliva using long-read nanopore sequencing.
In a previous report, oral bacterial compositions of newly hospitalized COVID-19 patients in New York City during the outbreak’s peak were unaltered compared to control patients. Moreover, there were no significant differences in alpha and beta diversities between groups. 29 However, alterations in the oral microbiome were associated with COVID-19, as previously reported in China, where the result showed that bacterial alpha diversity was significantly decreased in tongue-coating samples of active COVID-19 patients compared to healthy controls. In addition, the beta diversity of the oral microbiome showed a significant difference between both groups. 13 In our cohort, alpha and beta diversity analysis did not differ significantly in the salivary bacteriome between asymptomatic COVID-19 positive and negative groups. Our findings are consistent with a previous study of newly admitted COVID-19 patients in New York City. 29
Other viral infection reports also presented their effect on the microbiome. The upper respiratory tract bacteria changed dramatically in Influenza A and Influenza B virus-infected patients. 31 The microbial composition associated with the RSV group differed from the negative group. The result revealed that the bacterial ecosystem affected the severity of bronchiolitis. Because RSV colonization and infection lead to bacterial dysbiosis and favorable growth condition for some microbes (e.g. Streptococcus pneumoniae), increasing the risk of respiratory complications. 7 These suggested that respiratory viral infections might be associated with the change of respiratory bacterial community and susceptibility to secondary bacterial infections.
Characterizing the salivary bacterial profiles of asymptomatic COVID-19 positive and negative groups, it was found that V. parvula, S. mitis, and P. melaninogenica were core species common to both groups. Generally, Streptococcus, Veillonella, and Prevotella were previously reported as major genera in oral cavities of a healthy individual. 32 The differential species abundance analysis by LEfSe indicated the differences in major populations of the salivary bacteriome between asymptomatic COVID-19 positive and negative groups. This result suggested the salivary bacteriome dysbiosis during SARS-CoV-2 infection despite lacking symptoms. The phylum Proteobacteria, class Gammaproteobacteria, order Burkholderiales, family Burkholderiaceae, genus Lautropia, and species L. mirabilis appeared with higher abundances in the asymptomatic COVID-19 positive group. Members of these taxa have been found in many organs, such as skin, oral cavity, tongue, vaginal tract, and gut. 33 Interestingly, several important pathogens in these taxa are involved with lung diseases, such as asthma, chronic obstructive pulmonary disease (COPD), and cystic fibrosis.34 –36 Specifically, L. mirabilis was significantly enriched in saliva samples from the asymptomatic COVID-19 positive group, suggesting it has an important role and is a co-infectious agent in asymptomatic COVID-19 cases. In a previous study, L. mirabilis has evidently been found in the oral cavities of HIV patients and sputa of cystic fibrosis patients.37,38 Therefore, recent studies suggest a role in respiratory pathogens, possibly leading to lung infection among COVID-19 cases. On the other hand, many members of the genera Prevotella and Veillonella in oral microbiota were associated with ongoing symptomatic COVID-19 and long COVID-19. This evidence suggests that oral microbiota are correlated with the clinical severity of COVID-19.39,40 There are some differences in the core species in the COVID-19 group between our and previous studies. Because our cohort focuses on the asymptomatic COVID-19 group compared to the negative group, other studies consider symptomatic COVID-19 and long COVID-19 groups.
In this study, K. oralis and G. adiacens were completely missing bacteria in the asymptomatic COVID-19 positive group but found in only the negative group. The salivary bacteriome showed a significant decrease of P. oris, P. micra, D. pneumosintes, and P. pleuritidis in the asymptomatic COVID-19 positive group when compared to the negative group. This result agrees with a previous report, which showed that P. oris was underrepresented in the saliva of hospitalized COVID-19 patients with detectable SARS-CoV-2. 29 P. oris is a gram-negative anaerobic, rod-shaped bacterium. It is commonly detected and localized in oral lesions and can cause periodontal disease in humans. 41
Interestingly, K. oralis is oral commensal and more frequently found in the healthy site of the child’s oral cavity than the diseased site. There is evidence of the protective potential of K. oralis in periodontal health, suggesting its role as a health-associated species. 42 In addition, G. adiacens has been previously found in the tongue microbiomes of adults. 43 It is a gram-positive coccus and normal flora in the human mouth, gastrointestinal tract, and genitourinary tract. Normally, G. adiacens does not cause disease in a healthy host, but it can infect an immunocompromised host. 44 The most common infections were sepsis and endocarditis as shown in prior reports.45,46 P. micra was a part of normal commensals in the oral cavity, gastrointestinal tract, genitourinary tract, and skin. Many reports presented P. micra to be associated with several diseases including periodontitis, 47 abdominal abscesses, 48 arthritis, 49 intracranial infection, 50 and bloodstream infection. 51 D. pneumosintes was reported to be frequently associated with gingivitis, periodontitis, and progression of periodontitis.52,53 So far, it has been reported to cause systemic infection. 54 Finally, P. pleuritidis can be commonly detected in the pleural fluid. 55 It has been rarely reported to be particularly important in the pathogenesis of lung abscesses. 56 Moreover, P. pleuritidis and others were significantly identified in smokers’ oral microbiota compared to non-smokers. 57 Concerning limitations of the study, the oral health, preexisting periodontal disease, and smoking data were unavailable, so these confounding factors might affect the obtained result. This study did not examine the association between oral health, smoking, and the salivary bacteriome in asymptomatic COVID-19. In addition, this work needs further confirmation that the salivary bacteriome plays a role in asymptomatic COVID-19 cases using an experimental study.
In summary, these findings provide a better understanding of the association between salivary bacteriome and asymptomatic COVID-19, which might help predict health conditions in the future.
Supplemental Material
Supplemental material, sj-pdf-1-ebm-10.1177_15353702221118091 for Classification of salivary bacteriome in asymptomatic COVID-19 cases based on long-read nanopore sequencing by Rungrat Jitvaropas, Oraphan Mayuramart, Vorthon Sawaswong, Pornchai Kaewsapsak and Sunchai Payungporn in Experimental Biology and Medicine
Acknowledgments
We would like to thank Dr Naphat Chantaravisoot, Dr Piriya Wongkongkathep, Miss Pattaraporn Nimsamer, Miss Suwanan Mankhong, Miss Nantinee Chomta, Miss Rungnapa Bootsri, and Mr Isara Alee from the Faculty of Medicine, Chulalongkorn University, for participating in the saliva sample collections.
Footnotes
Authors’ Contributions: SP conceived the study design. OM and PK participated in the sample collection and the experiments. RJ and VS carried out data analysis and illustrations. RJ and SP conducted the interpretation and discussion. RJ, OM, and VS drafted the manuscript. SP provided suggestions on manuscript preparation and coordinated the project. RJ, PK, and SP revised the article. All authors reviewed and approved the final version of this article.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethical Approval: This study was approved by the Institutional Review Board of the Faculty of Medicine, Chulalongkorn University (IRB No. 302/63).
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This research was supported by funding from the Health & Aging Platform (764002-HE08); Ratchadapisek Sompoch Endowment Fund, Chulalongkorn University; Research Grants for Talented Mid-Career Researchers, The National Research Council of Thailand (NRCT; N41A640077); and Administration and Capital Management Unit for Enhancing the Competitiveness of The Country (AEC) (C17F640216).
ORCID iDs: Rungrat Jitvaropas  https://orcid.org/0000-0001-7555-0048
https://orcid.org/0000-0001-7555-0048
Sunchai Payungporn  https://orcid.org/0000-0003-2668-110X
https://orcid.org/0000-0003-2668-110X
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol 2020;5:536–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, Liu L, Shan H, Lei CL, Hui DSC, Du B, Li LJ, Zeng G, Yuen KY, Chen RC, Tang CL, Wang T, Chen PY, Xiang J, Li SY, Wang JL, Liang ZJ, Peng YX, Wei L, Liu Y, Hu YH, Peng P, Wang JM, Liu JY, Chen Z, Li G, Zheng ZJ, Qiu SQ, Luo J, Ye CJ, Zhu SY, Zhong NS. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med 2020;382:1708–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Han P, Ivanovski S. Saliva-Friend and Foe in the COVID-19 Outbreak. Diagnostics 2020;10:290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Honda K, Littman DR. The microbiota in adaptive immune homeostasis and disease. Nature 2016;535:75–84 [DOI] [PubMed] [Google Scholar]
- 5. Dubourg G, Edouard S, Raoult D. Relationship between nasopharyngeal microbiota and patient’s susceptibility to viral infection. Expert Rev Anti Infect Ther 2019;17:437–47 [DOI] [PubMed] [Google Scholar]
- 6. Man WH, de Steenhuijsen Piters WA, Bogaert D. The microbiota of the respiratory tract: gatekeeper to respiratory health. Nat Rev Microbiol 2017;15:259–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schippa S, Frassanito A, Marazzato M, Nenna R, Petrarca L, Neroni B, Bonfiglio G, Guerrieri F, Frasca F, Oliveto G, Pierangeli A, Midulla F. Nasal microbiota in RSV bronchiolitis. Microorganisms 2020;8:731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lu HF, Li A, Zhang T, Ren ZG, He KX, Zhang H, Yang JZ, Luo QX, Zhou K, Chen CL, Chen XL, Wu ZW, Li LJ. Disordered oropharyngeal microbial communities in H7N9 patients with or without secondary bacterial lung infection. Emerg Microbes Infect 2017;6:e112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wen Z, Xie G, Zhou Q, Qiu C, Li J, Hu Q, Dai W, Li D, Zheng Y, Wen F. Distinct nasopharyngeal and oropharyngeal microbiota of children with influenza A virus compared with healthy children. Biomed Res Int 2018;2018:6362716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML, Zhang YL, Dai FH, Liu Y, Wang QM, Zheng JJ, Xu L, Holmes EC, Zhang YZ. A new coronavirus associated with human respiratory disease in China. Nature 2020;579:265–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McIlroy JR, Mullish BH, Goldenberg SD, Ianiro G, Marchesi JR. Intestinal microbiome transfer, a novel therapeutic strategy for COVID-19 induced hyperinflammation? In reply to, “COVID-19: immunology and treatment options,” Felsenstein, Herbert McNamara et al. Clin Immunol 2020;218:108542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xu R, Lu R, Zhang T, Wu Q, Cai W, Han X, Wan Z, Jin X, Zhang Z, Zhang C. Temporal association between human upper respiratory and gut bacterial microbiomes during the course of COVID-19 in adults. Commun Biol 2021;4:240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ren Z, Wang H, Cui G, Lu H, Wang L, Luo H, Chen X, Ren H, Sun R, Liu W, Liu X, Liu C, Li A, Wang X, Rao B, Yuan C, Zhang H, Sun J, Chen X, Li B, Hu C, Wu Z, Yu Z, Kan Q, Li L. Alterations in the human oral and gut microbiomes and lipidomics in COVID-19. Gut 2021;70:1253–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang H, Ai JW, Yang W, Zhou X, He F, Xie S, Zeng W, Li Y, Yu Y, Gou X, Li Y, Wang X, Su H, Zhu Z, Xu T, Zhang W. Metatranscriptomic characterization of coronavirus disease 2019 identified a host transcriptional classifier associated with immune signaling. Clin Infect Dis 2021;73:376–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nardelli C, Gentile I, Setaro M, Di Domenico C, Pinchera B, Buonomo AR, Zappulo E, Scotto R, Scaglione GL, Castaldo G, Capoluongo E. Nasopharyngeal microbiome signature in COVID-19 positive patients: can we definitively get a role to Fusobacterium periodonticum? Front Cell Infect Microbiol 2021;11:625581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Miao Q, Ma Y, Ling Y, Jin W, Su Y, Wang Q, Pan J, Zhang Y, Chen H, Yuan J, Wu H, Hu B. Evaluation of superinfection, antimicrobial usage, and airway microbiome with metagenomic sequencing in COVID-19 patients: a cohort study in Shanghai. J Microbiol Immunol Infect 2021;54:808–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kleinstein SE, Nelson KE, Freire M. Inflammatory networks linking oral microbiome with systemic health and disease. J Dent Res 2020;99: 1131–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xiang Z, Koo H, Chen Q, Zhou X, Liu Y, Simon-Soro A. Potential implications of SARS-CoV-2 oral infection in the host microbiota. J Oral Microbiol 2020;13:1853451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Benítez-Páez A, Sanz Y. Multi-locus and long amplicon sequencing approach to study microbial diversity at species level using the MinION™ portable nanopore sequencer. Gigascience 2017;6:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Matsuo Y, Komiya S, Yasumizu Y, Yasuoka Y, Mizushima K, Takagi T, Kryukov K, Fukuda A, Morimoto Y, Naito Y, Okada H, Bono H, Nakagawa S, Hirota K. Full-length 16S rRNA gene amplicon analysis of human gut microbiota using MinION™ nanopore sequencing confers species-level resolution. BMC Microbiol 2021;21:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chantaravisoot N, Kaewsapsak P, Mayuramart O, Nimsamer P, Mankhong S, Chomta N, Bootsri R, Alee I, Wongkongkathep P, Treeprasertsuk S, Payungporn S. COVID-19 active case findings based on self-collected saliva samples with CRISPR-Cas12a detection. Exp Biol Med (Maywood) 2022:15353702221090181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lu X, Wang L, Sakthivel SK, Whitaker B, Murray J, Kamili S, Lynch B, Malapati L, Burke SA, Harcourt J, Tamin A, Thornburg NJ, Villanueva JM, Lindstrom S. US CDC real-time reverse transcription PCR panel for detection of severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis 2020;26:1654–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lanfear R, Schalamun M, Kainer D, Wang W, Schwessinger B. MinIONQC: fast and simple quality control for MinION sequencing data. Bioinformatics 2018;35:523–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rodríguez-Pérez H, Ciuffreda L, Flores C. NanoCLUST: a species-level analysis of 16S rRNA nanopore sequencing data. Bioinformatics 2021;37:1600. [DOI] [PubMed] [Google Scholar]
- 25. Chong J, Liu P, Zhou G, Xia J. Using MicrobiomeAnalyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat Protoc 2020;15:799–821 [DOI] [PubMed] [Google Scholar]
- 26. Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol 2011;12:R60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Verma D, Garg PK, Dubey AK. Insights into the human oral microbiome. Arch Microbiol 2018;200:525–40 [DOI] [PubMed] [Google Scholar]
- 28. Wade WG. The oral microbiome in health and disease. Pharmacol Res 2013;69:137–43 [DOI] [PubMed] [Google Scholar]
- 29. Miller EH, Annavajhala MK, Chong AM, Park H, Nobel YR, Soroush A, Blackett JW, Krigel A, Phipps MM, Freedberg DE, Zucker J, Sano ED, Uhlemann AC, Abrams JA. Oral microbiome alterations and SARS-CoV-2 saliva viral load in patients with COVID-19. Microbiol Spectr 2021;9:e0005521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schult D, Reitmeier S, Koyumdzhieva P, Lahmer T, Middelhoff M, Erber J, Schneider J, Kager J, Frolova M, Horstmann J, Fricke L, Steiger K, Jesinghaus M, Janssen KP, Protzer U, Neuhaus K, Schmid RM, Haller D, Quante M. Gut bacterial dysbiosis and instability is associated with the onset of complications and mortality in COVID-19. Gut Microbes 2022;14:2031840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rattanaburi S, Sawaswong V, Chitcharoen S, Sivapornnukul P, Nimsamer P, Suntronwong N, Puenpa J, Poovorawan Y, Payungporn S. Bacterial microbiota in upper respiratory tract of COVID-19 and influenza patients. Exp Biol Med (Maywood) 2022;247:409–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zarco MF, Vess TJ, Ginsburg GS. The oral microbiome in health and disease and the potential impact on personalized dental medicine. Oral Dis 2012;18:109–20 [DOI] [PubMed] [Google Scholar]
- 33. Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012;486:207–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rizzatti G, Lopetuso LR, Gibiino G, Binda C, Gasbarrini A. Proteobacteria: a Common Factor in Human Diseases. Biomed Res Int 2017; 2017:9351507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Voronina OL, Kunda MS, Ryzhova NN, Aksenova EI, Sharapova NE, Semenov AN, Amelina EL, Chuchalin AG, Gintsburg AL. On Burkholderiales order microorganisms and cystic fibrosis in Russia. BMC Genomics 2018;19:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Eberl L, Vandamme P. Members of the genus Burkholderia: good and bad guys. F1000Res 2016;5:F1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rossmann SN, Wilson PH, Hicks J, Carter B, Cron SG, Simon C, Flaitz CM, Demmler GJ, Shearer WT, Kline MW. Isolation of Lautropia mirabilis from oral cavities of human immunodeficiency virus-infected children. J Clin Microbiol 1998;36:1756–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ben Dekhil SM, Peel MM, Lennox VA, Stackebrandt E, Sly LI. Isolation of Lautropia mirabilis from sputa of a cystic fibrosis patient. J Clin Microbiol 1997;35:1024–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Haran JP, Bradley E, Zeamer AL, Cincotta L, Salive MC, Dutta P, Mutaawe S, Anya O, Meza-Segura M, Moormann AM, Ward DV, McCormick BA, Bucci V. Inflammation-type dysbiosis of the oral microbiome associates with the duration of COVID-19 symptoms and long COVID. JCI Insight 2021;6:e152346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Khan AA, Khan Z. COVID-2019-associated overexpressed Prevotella proteins mediated host-pathogen interactions and their role in coronavirus outbreak. Bioinformatics 2020;36:4065–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yousefi-Mashouf R, Duerden BI, Eley A, Rawlinson A, Goodwin L. Incidence and distribution of non-pigmented Prevotella species in periodontal pockets before and after periodontal therapy. Microb Ecol Health Dis 1993;6:35–42 [Google Scholar]
- 42. Shaddox LM, Huang H, Lin T, Hou W, Harrison PL, Aukhil I, Walker CB, Klepac-Ceraj V, Paster BJ. Microbiological characterization in children with aggressive periodontitis. J Dent Res 2012;91:927–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hall MW, Singh N, Ng KF, Lam DK, Goldberg MB, Tenenbaum HC, Neufeld JDG, Beiko R, Senadheera DB. Inter-personal diversity and temporal dynamics of dental, tongue, and salivary microbiota in the healthy oral cavity. NPJ Biofilms Microbiomes 2017;3:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gardenier JC, Hranjec T, Sawyer RG, Bonatti H. Granulicatella adiacens bacteremia in an elderly trauma patient. Surg Infect (Larchmt) 2011;12:251–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Christensen JJ, Facklam RR. Granulicatella and Abiotrophia species from human clinical specimens. J Clin Microbiol 2001;39:3520–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Senn L, Entenza JM, Greub G, Jaton K, Wenger A, Bille J, Calandra T, Prod’hom G. Bloodstream and endovascular infections due to Abiotrophia defectiva and Granulicatella species. BMC Infect Dis 2006;6:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shinha T, Caine V. Pylephlebitis due to Parvimonas micra. Infect Dis Clin Pract 2016;24:54–6 [Google Scholar]
- 48. Ang MY, Dymock D, Tan JL, Thong MH, Tan QK, Wong GJ, Paterson IC, Choo SW. Genome sequence of Parvimonas micra strain A293, isolated from an abdominal abscess from a patient in the United Kingdom. Genome Announc 2013;1:e01025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Baghban A, Gupta S. Parvimonas micra: a rare cause of native joint septic arthritis. Anaerobe 2016;39:26–7 [DOI] [PubMed] [Google Scholar]
- 50. Prieto R, Callejas-Díaz A, Hassan R, de Vargas AP, López-Pájaro LF. Parvimonas micra: a potential causative pathogen to consider when diagnosing odontogenic brain abscesses. Surg Neurol Int 2020;11:140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. García Carretero R, Luna-Heredia E, Olid-Velilla M, Vazquez-Gomez O. Bacteraemia due to Parvimonas micra, a commensal pathogen, in a patient with an oesophageal tumour. BMJ Case Rep 2016;2016:bcr2016217740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Contreras A, Doan N, Chen C, Rusitanonta T, Flynn MJ, Slots J. Importance of Dialister pneumosintes in human periodontitis. Oral Microbiol Immunol 2000;15:269–72 [DOI] [PubMed] [Google Scholar]
- 53. Doan N, Contreras A, Flynn J, Slots J, Chen C. Molecular identification of Dialister pneumosintes in subgingival plaque of humans. J Clin Microbiol 2000;38:3043–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Park JE, Huh HJ, Ha YE, Kim WS, Ki CS, Lee NY. A case of bacteremia caused by Dialister pneumosintes with Streptococcus anginosus. Ann Clin Microbiol 2015;18:60–3 [Google Scholar]
- 55. Sakamoto M, Ohkusu K, Masaki T, Kako H, Ezaki T, Benno Y. Prevotella pleuritidis sp. nov., isolated from pleural fluid. Int J Syst Evol Microbiol 2007;57:1725–8 [DOI] [PubMed] [Google Scholar]
- 56. Asif AA, Roy M, Ahmad S. Rare case of Prevotella pleuritidis lung abscess. BMJ Case Rep 2020;13:e235960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Al Bataineh MT, Dash NR, Elkhazendar M, Alnusairat DMH, Darwish IMI, Al-Hajjaj MS, Hamid Q. Revealing oral microbiota composition and functionality associated with heavy cigarette smoking. J Transl Med 2020;18:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-ebm-10.1177_15353702221118091 for Classification of salivary bacteriome in asymptomatic COVID-19 cases based on long-read nanopore sequencing by Rungrat Jitvaropas, Oraphan Mayuramart, Vorthon Sawaswong, Pornchai Kaewsapsak and Sunchai Payungporn in Experimental Biology and Medicine