Abstract
In vivo expression technology was used for testing Pseudomonas aeruginosa in the rat lung model of chronic infection and in a mouse model of systemic infection. Three of the eight ivi proteins found showed sequence identity to known virulence factors involved in iron acquisition via an open reading frame (called pvdI) implicated in pyoverdine biosynthesis, membrane biogenesis (FtsY), and adhesion (Hag2).
Pseudomonas aeruginosa is an opportunistic pathogen important in cystic fibrosis patients, for whom chronic P. aeruginosa infections remain the major cause of acute pneumonia, leading to debilitating lung malfunction and premature death. Although several P. aeruginosa virulence factors have been extensively studied in vitro, less is known about virulence factors during infection. Several approaches have been reported to allow the recovery, identification, and characterization of genes that are expressed in the host (2–4). We have utilized the in vivo expression technology (IVET) purA promoter trap system (5) to identify P. aeruginosa genes that are specifically induced during mucosal and/or systemic infections. Here, we present evidence that the DNA fragments cloned in the promoter trap carry ivi genes in both animal models used.
Generation of chromosomal cointegrated P. aeruginosa PAO909 library.
A library of random genomic DNA fragments from P. aeruginosa were cloned to the promoterless purA-lacZY into pIVET1. Genomic DNA fragments from P. aeruginosa strain PAO1 from 1 to 4 kb were size selected, purified, ligated with pIVET1, and electroporated into Escherichia coli DH5αλpir (strains and plasmids are listed in Table 1). Analysis of 48 recombinant plasmids confirmed that 99% had different P. aeruginosa DNA fragments ranging between 1 and 4 kb (data not shown). This random pool of plasmids was transformed into E. coli SM10λpir and transferred by conjugation into the purA mutant P. aeruginosa strain PAO909. The resulting chromosomal cointegrated library was represented by at least 2 × 105 colonies of P. aeruginosa transformants.
TABLE 1.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5αλpir | F− φ80 ΔlacZ ΔM15 endA1 recA1 hsdR17(rK− mK+) supE44 thi-1 λ− gyrA96 relA1 Δ(lacZYA-argF)U169; recipient | J. J. Mekalanos, Harvard University |
| SM10λpir | F−araD Δ(lac pro) argE(Am) recA56 RifrnalA; recipient | J. J. Mekalanos, Harvard University |
| P. aeruginosa | ||
| PAO1 | Wild-type P. aeruginosa | B. W. Holloway, Monash University |
| PAO909 | purA pur-67 E79 tv-2; transduction mutant of PAO910 | Pseudomonas Genetic Stock Center |
| 100 | PAO909 [purA+lacZ+Y+(Am+)]; Lac− control strain | This study |
| 101 | PAO909 [purA+lacZ+Y+(Am+)]; Lac− control strain | This study |
| 102 | PAO909 [purA+lacZ+Y+(Am+)]; Lac+ control strain | This study |
| 131-8 | PAO909 [purA+lacZ+Y+(Am+) pIVI131-8] | This study |
| 131-14 | PAO909 [purA+lacZ+Y+(Am+) pIVI-131-14] | This study |
| 131-15 | PAO909 [purA+lacZ+Y+(Am+) pIVI-131-15] | This study |
| 131-17 | PAO909 [purA+lacZ+Y+(Am+) pIVI-131-17] | This study |
| 131-19 | PAO909 [purA+lacZ+Y+(Am+) pIVI-131-19] | This study |
| 134-21 | PAO909 [purA+lacZ+Y+(Am+) pIVI-134-21] | This study |
| 152-1 | PAO909 [purA+lacZ+Y+(Am+) pIVI-152-1] | This study |
| 153-1 | PAO909 [purA+lacZ+Y+(Am+) pIVI-153-1] | This study |
| Plasmids | ||
| pIVET1 | ′purA-lacZY; suicide plasmid pGP704 oriR6K Mob bla pir | 5 |
| pIVI-131-8 | DH5αλpir rescued from 131-8; PAO1 DNA-purA-lacZY fusion | This study |
| pIVI-131-14 | DH5αλpir rescued from 131-14; PAO1 DNA-purA-lacZY fusion | This study |
| pIVI-131-15 | DH5αλpir rescued from 131-15; PAO1 DNA-purA-lacZY fusion | This study |
| pIVI-131-17 | DH5αλpir rescued from 131-17; PAO1 DNA-purA-lacZY fusion | This study |
| pIVI-131-19 | DH5αλpir rescued from 131-19; PAO1 DNA-purA-lacZY fusion | This study |
| pIVI-134-21 | DH5αλpir rescued from 134-21; PAO1 DNA-purA-lacZY fusion | This study |
| pIVI-152-1 | DH5αλpir rescued from 152-1; PAO1 DNA-purA-lacZY fusion | This study |
| pIVI-153-1 | DH5αλpir rescued from 153-1; PAO1 DNA-purA-lacZY fusion | This study |
Selection of P. aeruginosa in vivo-induced genes.
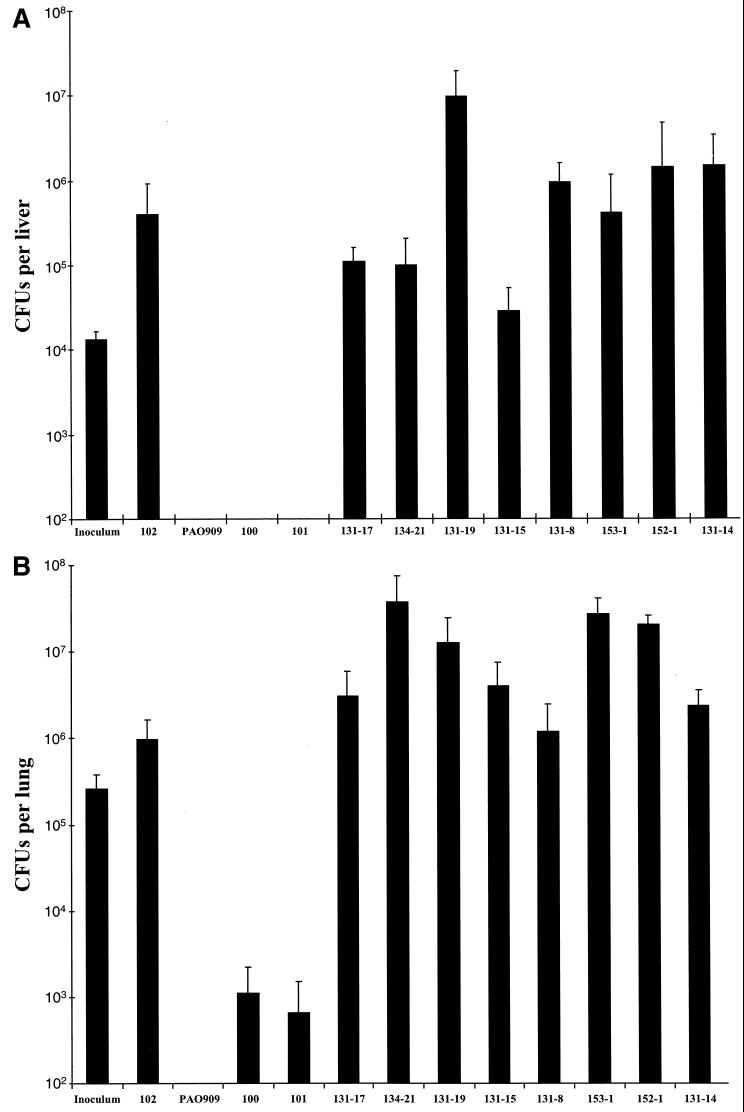
The cointegrated PAO909 library was used to infect BALB/c mice weighing 18 to 20 g (a septicemia model) intraperitoneally with 106 to 107 bacteria/mouse and to infect Sprague-Dawley rats intratracheally with 105 bacteria enmeshed into agar beads per lung (a chronic lung infection model [1]). After incubation, bacteria recovered from mouse livers and rat lungs were plated on rich selective medium containing the sensitive chromogenic substrate, 5-bromo-4-chloro-3-indoyl-β-d-galactopyranoside (X-Gal). A collection of 100 ivi fusions were recovered from infected mouse livers and infected rat lungs (Fig. 1).
FIG. 1.
Induction of ivi genes is required for survival in the animal. The vertical axis represents the number of CFU recovered from the organ of interest after inoculation. The inoculum bar represents the number of CFU of P. aeruginosa injected into each animal. Results are from the BALB/c mouse model of septicemia induced by intraperitoneal injection (104 CFU/mouse; 3 days) (A) and the Sprague-Dawley rat model of chronic lung infection induced via intratracheal instillation of bacterial cells enmeshed in agar beads (5 × 105 CFU/rat; 5 days) (B). Cells were grown overnight at 37°C in rich (adenine-supplemented) laboratory medium. Strains 100 and 101 (Lac−) and strain 102 (Lac+) were preselected purA-lac fusion strains. PAO909 is a P. aeruginosa auxotroph for adenine. Data are presented as averages of two to five independent assays ± standard deviations.
Characterization of ivi genes.
Plasmid preparations from in vivo-selected PAO909 clones were electroporated into E. coli DH5αλpir to allow plasmid rescue. Next, rescued plasmids which had unique restriction patterns and which gave a Lac− phenotype in vitro were selected for further analysis by DNA sequencing. ivi junctions were sequenced using primers homologous to the 5′ region of the purA gene. Similarity searches with the P. aeruginosa genome were performed at the National Center for Biotechnology Information using the uncompleted P. aeruginosa sequence genome database (http://www.pseudomonas.com). Bioinformatics analysis was done using GeneMark and software in the University of Wisconsin Genetics Computer Group package (version 10.0). We identified three ivi genes with homology to known sequences: pvdD, ftsY, and hag2. The remaining six ivi genes were open reading frames (ORFs) found to have no DNA or protein similarity (Table 2).
TABLE 2.
P. aeruginosa in vivo-induced genes isolated in both animal modelsa
| ivi fusion strain | Homolog | % Identity | Possible function | Possible role |
|---|---|---|---|---|
| Recovered from both animal models | ||||
| 131-17 | P. aeruginosa (PudD) | 60 | Pyoverdine biosynthesis | Iron scavenging |
| 134-21 | H. influenzae (FtsY) | 66 | Docking protein | Transport-secretion |
| 131-19 | E. corrodens (Hag2) | 43 | Adhesion | Colonization |
| 131-15 | ORF | Unknown | Unknown | |
| 153-1 | ORF | Unknown | Unknown | |
| 131-8 | ORF | Unknown | Unknown | |
| Recovered exclusively from mouse model | ||||
| 152-1 | ORF | Unknown | Unknown | |
| 131-14 | ORF | Unknown | Unknown |
Strain 131-17, identified by IVET (henceforth IVET 131-17), contains an unidentified ORF of 15,450 nucleotides (named pvdI) coding for a 5,150-amino-acid synthetase having 43% identity with PvdD of P. aeruginosa. We refer to this synthetase gene located upstream and in the same orientation as pvdD. The PvdD pyoverdine synthetase is involved in the synthesis of the fluorescent siderophore pyoverdine that is essential for iron uptake (6, 7). The independent isolation of IVET 131-17 from both animal models reflects the relative importance of iron acquisition in the establishment and/or maintenance of P. aeruginosa mucosal and systemic infections. Large-scale isolation of candidate virulence genes of P. aeruginosa strain PAK identified the pyochelin receptor (fptA), known to be inducible under iron-deprived conditions, providing further evidence that animal host tissues are deficient in free iron due to the presence of high-affinity iron binding proteins like transferrin (12, 13).
IVET 134-21 carries an ORF that encodes a protein sharing 65% identity with E. coli FtsY, a docking protein that interacts with the prokaryotic signal recognition particle-like complex involved in protein targeting and membrane biogenesis (10). Several ivi genes are involved in bacterial membrane modifications, presumably in response to overcoming environmental stresses imposed on the pathogen during infection (4).
IVET 131-19 carries a predicted peptide which has 43% identity with hemagglutinin Hag2 of Eikenella corrodens, an oral bacterium found in dental plaque (9). Similarly, the adherence of P. aeruginosa to the mucosa of the oropharynx is believed to be the initial step in colonization of the lower respiratory tract (14). The ivi of IVET 131-19 in both infection models suggests a mucosal and systemic requirement for P. aeruginosa adhesins, as is the case for other mucosal and systemic infection models (4). Cross talk of virulence factors between different in vivo pathogenesis models has been described previously using plants as hosts to identify P. aeruginosa virulence factors (8). The remaining six ivi genes code for proteins having no significant homology to reported proteins found in databases.
Induction of fusions is required for in vivo survival.
All eight ivi clones showed no or weak β-galactosidase activity when in vitro promoter activity was tested as described by Slauch et al. (11) (data not shown). Results shown in Fig. 1 indicate that the mutant P. aeruginosa purA strain PAO909 could not be recovered from mouse liver and rat lung tissues, confirming the efficacy of the selection in both animal models. Moreover, the eight ivi fusions showed a 103- to 105-fold growth advantage in both infection models. Thus, induction of all eight ivi fusions is required for survival in both animal models under conditions of the IVET selection.
These eight ivi genes were shown to be required for survival under the conditions of IVET selection in both animal models, suggesting that at least some host signals present during mouse systemic infection are also present in the rat respiratory mucosa. The propensity to isolate ivi genes coding for proteins related to the expression of surface proteins such as FtsY, PvdI, and Hag2 may suggest that they play a role in virulence by some unknown mechanisms. IVET selects bacterial ivi genes that presumably contribute to the in vivo fitness of the pathogen host tissues. Many of the ivi genes that have been recovered from several pathogens infecting a wide variety of animal models are unknown (4). The high possibility of recovering ivi genes of unknown function may reflect our limited knowledge of the bacterial functions required to survive during infection. Many of these presumably reflect the unique lifestyle of each individual pathogen during growth in the host and may not be shared by other pathogens. Thus, further studies on both known and unknown P. aeruginosa ivi gene products will contribute to a better understanding of the pathobiology of P. aeruginosa as an opportunistic pathogen.
Acknowledgments
We express our gratitude to Bruce Holloway, Monash University, for strain PAO1; John J. Mekalanos, Harvard University, for plasmid pIVET1 and E. coli strains DH5αλpir and SM10λpir; Paul Phibbs, Pseudomonas Genetic Stock Center, for strain PAO909; and J. Renaud and G. Cardinal, University of Laval, for excellent assistance in DNA sequencing.
This work was supported by the Medical Research Council of Canada. Work in R.C.L. lab is also funded by the Canadian Cystic Fibrosis Foundation and the Canadian Bacterial Diseases Network via the Canadian Centers of Excellence (R.C.L.) and by NIH grant AI36373, ACS Junior Faculty Research Award 554, and a Beckman Young Investigator Award (M.J.M.). R. C. Levesque is a Scholar of Exceptional Merit from Le Fonds de la Recherche en Santé du Québec, and M. Handfield obtained a studentship from the Canadian Cystic Fibrosis Foundation.
The first two authors (M.H. and D.E.L.) contributed equally to this work and are listed alphabetically.
REFERENCES
- 1.Cash H A, Woods D E, McCullough B, Johanson W G, Jr, Bass J A. A rat model of chronic respiratory infection with Pseudomonas aeruginosa. Am Rev Respir Dis. 1979;119:453–459. doi: 10.1164/arrd.1979.119.3.453. [DOI] [PubMed] [Google Scholar]
- 2.Conner C P, Heithoff D M, Mahan M J. Bacterial infection: close encounters at the host-pathogen interface, vol. 225. In vivo gene expression: contributions to infection, virulence and pathogenesis. New York, N.Y: Springer-Verlag; 1997. pp. 1–12. [DOI] [PubMed] [Google Scholar]
- 3.Handfield M, Levesque R C. Strategies for isolation of in vivo expressed genes from bacteria. FEMS Microbiol Rev. 1999;23:69–91. doi: 10.1111/j.1574-6976.1999.tb00392.x. [DOI] [PubMed] [Google Scholar]
- 4.Heithoff D M, Conner C P, Mahan M J. Dissecting the biology of a pathogen during infection. Trends Microbiol. 1997;5:509–513. doi: 10.1016/S0966-842X(97)01153-0. [DOI] [PubMed] [Google Scholar]
- 5.Mahan M J, Slauch J M, Mekalanos J J. Selection of bacterial virulence genes that are specifically induced in host tissues. Science. 1993;259:686. doi: 10.1126/science.8430319. [DOI] [PubMed] [Google Scholar]
- 6.Merriman T R, Lamont I L. Construction and use of a self-cloning promoter probe vector for gram-negative bacteria. Gene. 1993;126:17–23. doi: 10.1016/0378-1119(93)90585-q. [DOI] [PubMed] [Google Scholar]
- 7.Meyer J M, Neely A, Stintzi A, Georges C, Holder I A. Pyoverdin is essential for virulence of Pseudomonas aeruginosa. Infect Immun. 1996;64:518–523. doi: 10.1128/iai.64.2.518-523.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rahme L G, Tan M W, Le L, Wong S M, Tompkins R G, Calderwood S B, Ausubel F M. Use of model plant hosts to identify Pseudomonas aeruginosa virulence factors. Proc Natl Acad Sci USA. 1997;94:13245–13250. doi: 10.1073/pnas.94.24.13245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rao V K, Whitlock J A, Proguske-Fox A. Cloning, characterization and sequencing of two haemagglutinin genes from Eikenella corrodens. J Gen Microbiol. 1993;139:639–650. doi: 10.1099/00221287-139-3-639. [DOI] [PubMed] [Google Scholar]
- 10.Seluanov A, Bibi E. FtsY, the prokaryotic signal recognition particle receptor homologue, is essential for biogenesis of membrane proteins. J Biol Chem. 1997;272:2053–2055. doi: 10.1074/jbc.272.4.2053. [DOI] [PubMed] [Google Scholar]
- 11.Slauch J M, Mahan M J, Mekalanos J J. Measurement of transcriptional activity in pathogenic bacteria recovered directly from infected host tissue. BioTechniques. 1994;16:641–644. [PubMed] [Google Scholar]
- 12.Wang J, Lory S, Ramphal R, Jin S. Isolation and characterization of Pseudomonas aeruginosa genes inducible by respiratory mucus derived from cystic fibrosis patients. Mol Microbiol. 1996;22:1005–1012. doi: 10.1046/j.1365-2958.1996.01533.x. [DOI] [PubMed] [Google Scholar]
- 13.Wang J Y, Mushegian A, Lory S, Jin S G. Large-scale isolation of candidate virulence genes of Pseudomonas aeruginosa by in vivo selection. Proc Natl Acad Sci USA. 1996;93:10434–10439. doi: 10.1073/pnas.93.19.10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woods D E, Bass J A, Johanson W G, Jr, Straus D C. Role of adherence in the pathogenesis of Pseudomonas aeruginosa lung infection in cystic fibrosis patients. Infect Immun. 1980;30:694–699. doi: 10.1128/iai.30.3.694-699.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]