Abstract
Microplastics (MPs) have become emerging pollutants of public health concern, due to their impact on aqua-terrestrial ecosystems and integration into the food web, with evidence of human exposure and unrevealed health implications. There is a paucity of information regarding the effects of MPs exposure on the gut system using metagenomic and metabolomic approaches. In this study, Javanese medaka fish was exposed to 5 µm beads of polystyrene microplastics (PS-MPs) suspensions, at concentrations of 100 μg/L (MP-LOW), 500 μg/L (MP-MED), and 1000 μg/L (MP-HIGH), for a duration of 21 days, and evaluated for gut microbiome and metabolome responses. The results revealed a significant reduction (p < 0.05) in richness and diversity of the gut microbiome in the MP-HIGH group, and identification of 7 bacterial genera as differential features by the Linear discriminant analysis Effect Size (LEfSe). The gut metabolic profile revealed upregulation of 9 metabolites related to energy metabolism, via tricarboxylic acid cycle (TCA), creatine pathway, and urea cycle, as determined by the pathway analysis. Furthermore, positive correlation was found between the genus Aeromonas and glucose, lactate, and creatine metabolites. The study revealed that PS-MPs exposure resulted in altered bacterial microbiome and metabolic disorder related to energy metabolism. It further provided additional data on gut bacterial genera and metabolites associated with MPs toxicity in aquatic organism, which will inevitably enable its future health risks assessment in animals and possibly humans.
Keywords: Microplastics, Exposure, Gut, Microbiome, Metabolome, Javanese medaka
Graphical Abstract
Highlights
-
•
Polystyrene MPs induces a significant reduction in the richness and diversity of the gut bacterial microbiota.
-
•
The gut metabolic profile revealed upregulation of metabolites associated with energy metabolism and metabolic disorder.
-
•
Significant positive correlation was found between the genus Aeromonas and glucose, lactate, and creatine metabolites.
-
•
The gut microbiome and metabolomics biomarkers identified were related to hypoxia, oxidative stress, and inflammation.
1. Introduction
Plastics have become emerging pollutants of concern due to their adverse effect on the aquatic and terrestrial ecosystems, with global production estimated to reach 270 million metric tons by 2045, a threefold increase from its current status [33]. Larger plastics break down to generate microplastics [17], which are plastics of size ≤ 5 µm that are widely distributed on terrestrial and aquatic ecosystems, and eventually integral part of the food web [11], [53].
Studies have found MPs in seafood [2], [27], [7], fruits and vegetables [46], commercial salt [70], and drinking water [52], a probable reason for their presence in the human stool [40], [51] and colon specimens [20]. These have generated a lot of concern by various stakeholders across the globe, including the World Health Organisation (WHO) that called for the assessment of the impact of MPs in the environment, and on human health [67]. Additionally, it has culminated to abarrage of animal studies that explores the adverse effects of MPs on living organisms.
MPs have been shown to adversely affect growth and reproduction [10], [36], [79], induce oxidative stress and inflammatory changes in the gut [23], [24], [34], [73], [79]. Recently, it has been observed through histological and biochemical investigation to affect multiple organs, including the brain [62].
The metagenomics approach has recently been recognized as a method of understanding the toxicity of environmental pollutants through gut microbiome alterations [75], [78]. The host's diverse gastrointestinal microbial community is associated with its immune functions, metabolism, and supplementation of epithelial barrier function [30]. Additionally, the gut microbiota generates a huge amount of compounds that play a vital role in the microbial selection and metabolic signaling construct, with their activities affected by environmental stressors leading to the generation of a large number of compounds that influence host metabolome and health [63].
Studies that investigated the effects of MPs on organisms using metabolomic and metagenomic approaches are scarce [49], as a result, it provided a niche that requires exploration, so as to have a deeper understanding of how MPs exposure influence the host’s gut microbiome and metabolome. This will inevitably provide information on the usefulness of the approaches; enable biomarkers identification, reveal the relationship between microbiome and metabolome alterations, and possible future health implications. This study was aimed to decipher the exposure effect of PS-MPs on gut microbiome and metabolome changes using Next generation sequencing and proton Nuclear Magnetic resonance spectroscopy (1HNMR) methods respectively, to identify biomarkers, and the relationship between the identified gut microbiome and metabolite biomarkers in Javanese medaka fish (Oryzias javanicus Bleeker, 1854), a promising model fish [18], [31] with the ability to live in fresh and brackish/seawater [59] widely being used as a test organism [21] in ecotoxicological studies.
2. Methodology
2.1. Microplastics used in the study
Polystyrene microplastics beads 5 µm size obtained from Sigma Aldrich (Industriestrasse, Buchs (SG), Switzerland) were used in this study. Polystyrene is a commonly used plastic in our society, and has been used by several studies [12], [49], [62]. In addition, MPs of size 5 µm, are of public health concern. Their size made them readily accessible and ingestible by all aquatic organisms [15], and became transferred across the food web [11], [53]. More so, humans are exposed to MPs fibres, particles, and debris of size 1 µm–5 mm with unrevealed health implications [55]. The above reasons influence the choice of the specified PS-MPs in this study, as it will enable the exploration of its exposure effect in Javanese medaka fish.
The PS-MPs was validated in terms of size, morphology, and chemical composition as described in our previous study using scanning electron microscopy (SEM), (JOEL JSM 6400 SEM Japan), and Fourier transform infrared spectroscopy (FTIR) (Thermo Fisher Scientific, Nicolet 6700) [62]. UV-sterilized tap water was used to prepare PS-MPs suspensions, which were sonicated to ensure even dispersion of particles before use.
2.2. Medaka fish maintenance and exposure
Wild Javanese medaka fish of adult age (0.5–0.7 g in weight; 3–4 cm in length; both sexes), collected from Sepang estuary (2°37′15.38′ N, 101°42′38.33′ E) Malaysia were maintained in a semi-static circulating system at a temperature of 26 ± 1 °C, pH; 7.8–8.0, DO; 5.5–6.0 mg L−1 and photoperiod of 14:10 h light/dark cycle [3], [62]. Fish were acclimatized for two weeks, the same period of which the salinity was gradually reduced by 2ppt daily from Sepang value of 14.45 ± 0.5, till it reaches a zero value, using UV-sterilised well-aerated dechlorinated tap that was used throughout the experiment to enable total adaptability [43]. This is due to the Javanese medaka fish's inherent adaptability to live and procreate in brackish/Sea and freshwater [42], [54]. Fish were fed commercial feed (Aquadene) twice daily at 1.0% body weight.
Following acclimatization, fish were randomly assigned into the control group (PS-MPs free), and 3 PS-MPs exposed groups (4 fish in 1 L glass tank), for a duration of 21 days. The water and test suspensions were refreshed every 48 h. The concentrations of the PS-MPs exposed groups of 100 µg/L (1.46 × 103, MP-LOW), 500 µg/L (7.3 × 103, MP-MED), and 1000 µg/L (1.46 × 104, MP-HIGH), were formed by suspending the particles in UV-sterilised dechlorinated tap water, with the chosen concentrations guided by previous literature [34], [49], [62], and concentrations relevant to the environment (0.2–4137.3 item/L), as detected in surface water [76]. Tanks were continuously aerated to ensure optimal dissolved oxygen, and even dispersion of PS-MPs particles. Other parameters were maintained as they were during the acclimatization period. The research was strictly performed as per the Institutional Animal Care and Use Committee of Universiti Putra Malaysia guidelines, following their due approval (UPM/IACUC/AUP-R021/2020).
At the end of the exposure, fish were sampled and washed thoroughly with purified water (MilliQ), followed by euthanization in ice water at 4 °C. Subsequently, the guts were dissected for metabolomic analysis, and the faecal samples were collected for microbiome analysis. Samples were stored at − 80 °C until further investigation.
2.3. Microbiome analysis
The microbiome analysis included four biological replicates in each group, with each replicate composing of pooled faecal samples from 4 fish. Fast soil DNA soil kit (MP Biomedicals, USA) was used for total genomic DNA extraction following provided protocol. The V3-V4 region of 16rRNA of the DNA samples as the target was amplified, with loci-specific primers as described by previous study [57], with some modifications. Libraries were normalized and pooled per Illumina protocol, and sequenced on Illumina MiSeq platform 300PE (Illumina, USA). Adapters and low-quality reads were removed using The BBDuk of the BBTools package (https://sourceforge.net/projects/bbmap/), followed by subsequent merging of the paired-end reads using USEARCH v11.0.667 (https://www.drive5.com/usearch/). The bacterial 16S rRNA region was aligned using the SILVA release 132 database, with the chimeric error inspected using VSEARCH v2.6.2. The UPARSE v11.0.667 was used to process the reads through quality assessment steps, and thereafter clustered de novo at a similarity of 97% into the operational taxonomic unit (OTUs) [6], [9]. Finally, OTUs were assigned against the SILVA 16rRNA database (release 132) using QIIME V1.9.1 [50], [9]. The microbiomeanalyst, web-based software was used to conduct the gut microbiota analyses at the genus level in this study, this is to get higher confidence in the taxonomic assignment based on the 16SrRNA reads, a significant change in the gut microbiota at the genus level will signify a change in multiple gut bacteria at the species level [39]. Concomitantly, Alpha diversity (Simpson and Shannon), and LEfSe, were performed to determine the richness and diversity of the bacterial microbiome and potential biomarkers responsible for class separation at the genus level, respectively. Sequencing data were lodged at the Sequence Read Archive of the National Centre for Biotechnology Information (NCBI, Accession number PRJNA733698, and submission ID: SUB9745861).
2.4. Metabolomic analysis
The metabolomic analysis included 4 replicates in each group, with each replicate containing pooled intestinal tissues of 6 fish for metabolite extraction. The extraction process was as described previously [65], with some modifications. It entails the homogenization of freeze-dried gut samples of medaka fish in a mixture of 800 mL and 200 mL of cold water, and methanol, in a 1.5 mL centrifuge tube respectively. Subsequently, it was sonicated for 5 min, followed by removal of protein and tissue debris, after which it was kept on ice for 20 min. After that, it was centrifuged at 10,000 rpm for 10 min at 4 °C, and a clean centrifuge tube was used to collect the supernatant, with the same procedure repeated one more time. The obtained supernatant was dried, and reconstituted in 550 mL of 100 mM sodium phosphate buffer (pH 7.4), composing D2O (a lock signal), and 1.08 mM TSP-d4 as chemical shift reference. Thereafter, it was subjected to another centrifugation at 10,000 rpm at 4 °C for 10 min, and the supernatants was transferred into 5 mm NMR tubes.
The 500 MHz Varian INOVA NMR spectroscopy (Varian Inc., California, USA) was used to conduct 1 H NMR measurements, with the temperature maintained at 26 °C. Before NMR data acquisition, gradient shimming was performed using D2O as an internal lock. PRESAT pulse sequence, utilizing low power selective radiation at 4.85 ppm throughout the recycle delay; with chemical shift referenced to the internal standard of TSP= 0.0PPM, was further conducted on the 1NMR data acquisition to suppress large water resonance. Subsequently, the Carr-Purcell-Meiboom-Gill (CPMG) experiments, were used to acquire the relaxation measurements of T2 using the σ of 0.0004, and big σ of 0.8, as parameters, with 128 transient scans obtained for each sample [32].
Baseline and phase correction with a line broadening factor of 1.0 Hz of the spectra was conducted using Chenomx software (v. 8.1, Alberta, Canada). The region corresponding to the water peak residual of 4.7–5.0 ppm was removed, and all the spectra were subsequently binned, and exported as an Excel file. Multivariate data analysis (MVDA), using the Excel file binned integrals of the 1NMR was performed using SIMCA-P + version 13.0 (Umetrics AB, Umea, Sweden) Software. The principal component analysis (PCA), as an unsupervised pattern recognition method was conducted in order to obtain an overview of inherent clustering among different groups [65]. This is followed by Orthogonal partial least squares-discriminant analysis (OPLS-DA), used to strengthen the class separation, flatten the dataset, and ultimately determine potential biomarkers [35]. The quality of the model was validated using R2Ycum (goodness of fit), and cumulated Q2 (Q2cum, goodness of prediction), with a widely accepted threshold of 0.5 used in the model classification, with Q2cum of ≥ 0.5, and ≤ 0.5, been considered as having good and poor predictive capacity respectively [44], [60],72]. A permutation test (200 times), was conducted to minimize the risk of over-fitting, with a valid model having Q2 values of the permutated data set to the left lower than the Q2 value of the actual data set to the right, and a regression line with a negative value of intercept of the y-axis [41]. The supervised investigations were used to determine the variable importance in projection (VIP), as the output of contribution of each variable on the x-axis to the model, which is summed over all components, and weighted to the Y accounted for by every single component [5]. Metabolites with VIP> 1, were selected as major contributors and discriminants of the model, this is as described by previous studies [49,64,65,72]. Web-based software for metabolomics, the Metaboanalyst, was used for Heatmap and impact pathway analysis [69], with a cut-off value of > 0.1, considered significant for the pathway analysis [37]. The MestReNova software (v.6.0.2, Mestrelab Research, Santiago, Spain), was used for data evaluation [1]. Metabolites were identified through their corresponding chemical shifts from the online database, Human Metabolome Database (HMDB), Chenomx database (v. 8.1, Alberta, Canada), by routine peak fitting, and similitude with previous literature [1], [32], [49],65].
2.5. Statistical analysis
The effect of PS-MPs exposure on gut metabolite was tested for significance using One-way ANOVA with a p-value < 0.05 considered significant. Pearson's correlation between gut metabolites and bacterial composition was assessed using GRAPHPAD PRISM version 5.00 (GRAPHPAD Software, San Diego, CA, USA)[58].
3. Results
3.1. Gut microbiome alterations induced by PS-MPs exposure
The relative abundance at the genera level provided visual information on the percentage of microbiome that is made up of specific organism, and the corresponding change in relation to different exposure concentrations of PS-MPs, as depicted in Fig. 1A. The top 10 genera in both the control and the PS-MPs exposed groups were dominated by the g_Aeromonas, g__Ralstonia, g_Mycobacterium, g_Gemmobacter, g_ Gordonia, g_Reyranella, and g_ Bosea. The g_Aeromonas showed increase abundance in the PS-MPs exposed groups compared to the control, with a much higher increase seen in the MP-HIGH group. LEfSe, as shown in Fig. 1B, revealed 7 bacterial genera to be responsible for variation between control and PS-MPs exposed groups, thus regarded as our biomarkers. The g_Aeromonas was the unique feature in the MP-HIGH group, whereas the g_Ralstonia, g_ Paraburkholderia, g_Pelmonas, g_Staphylococcus, g_Bradyrhizobium, and g_Pararhizobium were found as the unique features in the MP-LOW group. Simpson and Shannon indexes, as shown in Fig. 1C and D revealed the dynamics of α-diversity of gut bacterial microbiota at the genera level in Javanese medaka fish. The two indexes showed the PS-MPs exposed groups to have much lower values than the control group, with significant difference (p < 0.05) found between the control and MP-HIGH group of both the two indexes. This thus implies that, PS-MPs exposure altered gut microbial composition, differentially enriched microbiome, and decreases the richness and diversity of gut bacterial genera in Javanese medaka fish.
Fig. 1.
Bacterial Gut Microbiome Perturbation following PS-MPs Exposure. (A) Top 10 compositional profiles at genera level (B) Differential bacterial composition at genera level using LEfSe (C) Shannon diversity index (D) Simpson diversity index. Asterisk (*) signify significant difference (p < 0.05) between the control and PS-MPs exposed groups.
3.2. Metabolomic alterations induced by PS-MPs exposure
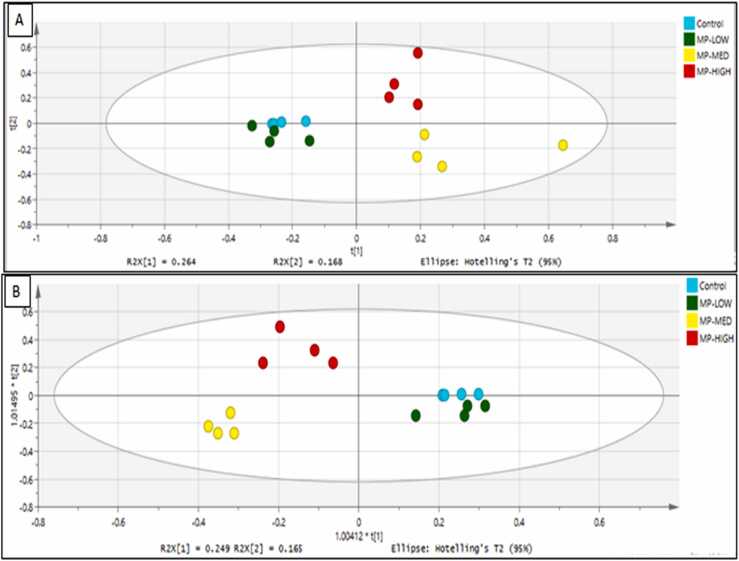
Multivariate Data Analysis (MVDA), using the unsupervised principal component analysis (PCA) revealed distinct clustering of the gut metabolites of Javanese medaka fish, as shown in Fig. 2A. The closeness of the biological replicates observed in the PCA score plot, signifies reproducibility of the biological replicates. The MP-MED exposed groups were seen on the right quadrant separated thorough PC1 from the control group, with the MP-LOW group on the left. The control group has its tightly clustered biological replicates near the MP-LOW group separated through PC2. The Orthogonal Partial Least Square Discriminant Analysis (OPLS-DA), as supervised method has further strengthened the MVDA, revealing a clear separation between the control, MP-MED and MP-HIGH groups by PC1 axis. The MP-LOW is separated by PC2 from the control, as depicted in Fig. 2B. The model parameters R2Y (cum) = 0.85, Q2 (cum) = 0.64, and the sum of predictive and orthogonal components R2X= 0.51, qualifies the model as statistically adequate. The permutation test further demonstrated a good validity level of the built model, as shown in Fig. 2C. The heat map of hierarchical clustering of metabolites changes within each group, except the MP-MED, showed clustering of the biological replicates. The control group is clustered in close proximity to the MP-LOW, and MP-LOW to the MP-HIGH. MP-MED medium has two distinct clustering close to the control and the MP-HIGH groups (Fig. 2D). Thus, it can summarily be deduced that PS-MPs exposure induces metabolic changes in Javanese medaka fish.
Fig. 2.
Principal Component Analysis (PCA), Orthogonal Partial Least Square Analysis (OPLS-DA) and heat Map Analysis. (A) PCA score plot (B) OPLS-DA score plot showing clear separation between control and PS-MPs exposed groups (C) OPLS-DA model validation with 200 permutations (D) Heatmap showing the concentration of VIP metabolites in the control and PS-MPs exposed groups. C, T1, T2 and T3 represent the replicates in control, MP-LOW, MP-MED and MP-HIGH respectively.
A total of 9 metabolites (anserine, glucose, creatine, glucuronate, glutamate, alanine, lactate, valine, and 2-hydroxyvalerate) were responsible for contributing to metabolomic alteration due to PS-MPs exposure as determined by VIP of OPLS-DA. The metabolites serving as major contributors from the VIP, and responsible for separation of the control and PS-MPs exposed groups were relatively quantified through normalization with the internal standard (TSP). The relative quantification of VIP metabolites changes were shown in Fig. 3. The box plots showed significant increase (p< 0.05) in glucose and lactate metabolites in all the PS-MPs exposed groups compared to the control. Furthermore, significant increase (p < 0.05) was found in alanine, glutamate, glucoronate and valine in MP-MED group, anserine in MP-HIGH group, and 2-hydoxyvalerate in MP-MED and MP-HIGH groups compared to the control.
Fig. 3.
Box plot showing relative quantification of VIP metabolites in the control and PS-MPs exposed Javanese medaka fish. Asterisk (*) signify significant difference (p < 0.05) between control and PS-MPs exposed groups.
The Log 2-fold change, as depicted in Table 1, showed an initial decrease in the level of the metabolites except glucose and valine in the MP-LOW group, followed by an increase in the level of all the metabolites with the exception of anserine in the MP-MED and MP-HIGH groups compared to the control. Among the metabolites, glucose and lactate showed a statistically significant increase (p< 0.05), with glucose having more than a fourfold increase (Log 2 fold change >2) in all the PS-MPs exposed groups, and lactate having more than 2-fold increase (Log 2 fold change >1) in MP-MED, and MP-HIGH exposed groups.
Table1.
Log 2 Fold of altered metabolites due to PS-MPs exposure in relation to control group.
|
Log2 Fold Change |
|||||
|---|---|---|---|---|---|
| Metabolite | MP-LOW/Control | MP-MED/Control | MP-HIGH/Control | ||
| Anserine | 4.47(s), 7.79(s) | -0.08 | + 0.16 | -0.04 * * | |
| Glucose | 4.63(d) | + 2.46 * * | + 2.62 * * | + 2.24 * * | |
| Creatine | 3.02(s) | -4.18 | + 0.35 | + 0.44 | |
| Glucoronate | 5.25(d) | -0.08 | + 1.01 * * | -0.07 | |
| Glutamate | 2.35(m) 2.01(m) | -0.45 | + 0.64 * * | + 0.17 | |
| Alanine | 1.46(d) | -1.23 | + 0.73 * * | + 0.24 | |
| Lactate | 1.31(d) | -0.46 * * | + 1.66 * * | + 1.16 * * | |
| Valine | 1.03(d) | + 0.13 | + 0.81 * * | + 0.28 | |
| 2-hydroxyvalerate | 0.95(d) | -0.28 | + 0.91 * * | + 0.29 * * | |
Note: (s) single; (d) doublet; (m) multiplet; (**) denotes significant (p<0.05) between groups.
The enriched metabolites (VIP>1) were subjected to impact pathway analysis. The result is presented in the metabolome analysis view (Fig. 4A). Most of the significantly perturbed pathways (pathway impact>0.1), were related to amino acid metabolism. This includes alanine, aspartate, and glutamate metabolism; arginine and proline metabolism; D-glutamine and D-glutamate metabolism; ascorbate and aldarate metabolism; arginine biosynthesis; and pentose and glucuronate interconversions. The impacted pathways were found to be associated with energy metabolism, through their intermediary metabolites, specifically by the tricarboxylic acid cycle (TCA), creatine pathway, and urea cycle, with the generation of lactate, possibly through aerobic and anaerobic glycolysis. This is schematically represented in Fig. 4B.
Fig. 4.
(A) Summary of pathway analysis (B) Altered pathways with differential metabolites following PS-MPs exposure. The altered metabolites were shown in italics, with red arrows signifying increase in the metabolite.
The Pearson's correlation coefficient, showed significant (p< 0.05) positive correlation between the enriched g_ Aeromonas in the PS-MPs exposed groups and the glucose, creatine, and lactate metabolites. In contrast, the enriched genera in the control group (g_Leucobacter and g_Obscurbacteraceae) revealed a significant (p< 0.05) negative correlation with the glucose metabolite. This is illustrated in Fig. 5.
Fig. 5.
The correlation between differential gut bacterial genera and metabolites. Asterisk (*) denotes significant difference (p-value of <0.05).
4. Discussion
Studies on the exposure effects of microplastics concerning gut microbiome and metabolome responses are generally scanty. This study revealed a reduction in the richness and diversity of bacterial gut microbiome exposed to PS-MPs at the genera level in Javanese medaka fish. Studies on PS-MPs exposure effect on adult zebra fish [25], [49], and closely related Marine medaka (Oryzias malastigma) exposed to polyethylene microplastics [74] also showed a reduction in richness and diversity of the bacterial gut microbiome. The distortion in richness and diversity observed in this study are as demonstrated in previous studies [49], [62], and similar to the reports of distorted gut microbiome in inflammatory bowel diseases (IBDs) [28], [47]. However, whether inflammation lead to alteration of gut microbiome or reversely, still remains a subject of debate that requires further exploration.
The enriched genera in this study were found to be linked to inflammation and metabolic disorders, and this shows the possibility of MPs to exert and manifest their toxicity in the short and long-term. Zebra fish were shown to have increase in the bacterial genera Pseudomonas, Aeromonas, Ralstonia and Burkholderia [26], however, in addition to aforementioned, this study have discovered more unique features in the PS-MPs exposed groups that included, g_Pelmonas, g_Staphylococcus, g_Bradyrhizobium, and g_Pararhizobium. The genera Streptococcaceae and Burkholderiaceae enriched in PS-MPs exposed groups in this study were found to be abundantly increased in Crohn's disease and ulcerative colitis, respectively [28]. The Streptococcus species were also found as abundant gut microbiome that drives local inflammation in joint disease mediated through their metabolites [8]. The genus Aeromonas have been an opportunist pathogen in the elderly and a public health problem, due to the pathogenic nature of its species having several virulence factors, making them capable to override host immunity [4]. Ralstonia pickettii, an organism under the enriched genera Ralstonia, was found to be increase in abundance in the faeces of obese, pre-diabetics, and type 2 diabetes mellitus (T2DM) patients, and has been causally linked to obesity and T2DM. Ralstonia has also been demonstrated to reduce glucose tolerance in mice [61], reduce renal function in patients with ulcerative colitis [29], and linked to nosocomial infections in immunocompromised individuals [14]. Ironically, the enriched genus Leucobacter found in our control group was found to be among the most abundant genera in women's breast milk with a healthy body mass index, without subclinical mastitis, and with a more diverse microbiota, in contrast to their counterparts [38].
Similar to our study, glucose creatine and alanine were found to significantly increase in the gut of zebra fish exposed to PS-MPs, the study further showcases the altered metabolites to be drastically increase in medium exposure group compared to the highest [49], this could possibly be due to MPs exhibiting their toxicity through a negative feedback effect on gut metabolome, or in a non-dose dependant manner. The study further revealed lactate, anserine, valine, glutamate, glucoronate and 2-hydroxyvalerate as new set of metabolites that increase. The metabolites alteration revealed MPs as possible agents of causing metabolic disorder, inflammation and oxidative damage. MPs fibres were found to up-regulate glycerophospholipids metabolism, which are known to aggravate inflammation and oxidative damage [77]. Further in support of this, PS-MPs exposure in Litopenaeus vannamei have been found to change the metabolism of amino acids valine, and D-glutamine and D-glutamate metabolism, and the depletion of ornithine, a similar pattern observed in our study, which might possibly disrupt cellular proliferation and lead to metabolic diseases [13]. Glucose enrichment has been reported in proinflammatory states [19,71], to meet the requirements and increase uptake by the activated immune cells that largely depend on it. The immune cells utilize glucose for glycolysis as a major source of energy, and in the process pyruvate is converted to lactate for rapid generation of ATP aerobically (Warburg effect) [66], and anaerobically, the lactate of which has been shown to have downstream effect in inducing inflammatory response [48]. In addition to inflammatory factors, microbial products were also found to induce glycolysis [45]. Similarly, activation of inflammation and hypoxia in disease states such as, chronic inflammation, and tissue injury were found to cause dramatic increase in aerobic and anaerobic glycolysis, respectively, both of which increases the production and release of lactate, and acid generation in extracellular sites [22]. Lactate has been recognized as metabolic end product that accumulates in tissues with on-going inflammation; it increases the production of IL-17 as proinflammatory cytokines by T-cells, which are responsible for the features seen in chronic inflammatory infiltrates [16]. Creatine enrichment in this study may be as a result of its role in providing energy and intermediaries of energy metabolism in states of increasing demand, this is particularly evident in pathological states such as hypoxia, tissue ischemia, and increase oxidative stress where it serves a conserved role to replenish the depleted intracellular ATP, reverse internal acidification, and enhance protein synthesis and free radical scavenging that will stabilise cellular membranes after ischemic injury [30]. It also increases the generation of sarcosine and glycine in several organisms [68]. The activation and enhancement of the urea cycle, alanine, and valine in this study were reportedly found to be related to increased oxidative stress in perinatal asphyxia [56].
In a nutshell, the originality and significance of the current work explore the methodology of new generation sequencing (NGS) and NMR spectroscopy for the interpretation of gut microbiome and metabolome responses, enabling the identification and characterization of bacterial genera and metabolites that can be used as biomarkers in relation to microplastics exposure with potential implications for diseases risk in the future.
5. Conclusion
This study has reported the first application of NGS and NMR spectroscopy to investigate gut microbiome and metabolome responses, and biomarkers identification in Javanese medaka fish exposed to PS-MPs. The exposed fish showed gut microbiome perturbations as reduction in richness and diversity of the microbiota. Furthermore, discriminant analyses have revealed 7 bacterial genera, and 9 metabolites, that change significantly in the gut due to PS-MPs exposure. The biomarkers of bacterial genera and metabolites identified were related to hypoxia, oxidative stress, inflammation, and deranged energy metabolism. This thus, highlighted the potential disease risks that microplastics exposure may incur in the future, with the need to explore further.
Funding
The research was funded by the Malaysian Ministry of Higher Education under the Fundamental Research Grant Scheme (grant no: FRGS/1/2019/SKK06/UPM/02/21) on the project entitled Deciphering exposure effect of polystyrene microplastics (PS-MPs) towards Javanese medaka fish (Oryzias javanicus) using combinations of metabolomic and metagenomic approaches.
CRediT authorship contribution statement
Conceptualisation, S.U. and A.F.A.R.; Data curation, A.F.A.R., K.S., M.N.A.A., M.Z.S., N.M.I. and M.F.N.; Formal analysis, S.U., A.F.A.R., and K.S.; Funding acquisition, A.F.A.R.; Investigation, S.U.; Methodology, S.U., A.F.A.R., K.S., M.N.A.A. and M.Z.S.; Supervision, A.F.A.R., K.S., M.N.A.A., M.Z.S. and N.M.I.; Visualisation, A.F.A.R., K.S. and M.N.A.A.; Writing – original draft, S.U. and A.F.A.R.; Writing – review & editing, S.U., A.F.A.R., K.S., M.N.A.A., M.Z.S., N.M.I. and M.F.N.; All authors have read and agreed to the published version of the manuscript.
Institutional review board statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Institutional Animal Care and Use Committee (IACUC) of Universiti Putra Malaysia (UPM), with the approval number: UPM/IACUC/AUP-R021/2020 dated 17 May 2020.
Informed consent statement
Not applicable.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Contributor Information
Sunusi Usman, Email: usunusi.bch@buk.edu.ng.
Ahmad Faizal Abdull Razis, Email: madfaizal@upm.edu.my.
Khozirah Shaari, Email: khozirah@upm.edu.my.
Mohammad Noor Amal Azmai, Email: mnamal@upm.edu.my.
Mohd Zamri Saad, Email: mzamri@upm.edu.my.
Nurulfiza M. Isa, Email: nurulfiza@upm.edu.my.
Muhammad Farhan Nazarudin, Email: m_farhannaza@upm.edu.my.
References
- 1.Abdul-Hamid N.A., Abas F., Ismail I.S., Tham C.L., Maulidiani M., Mediani A., Swarup S., Umashankar S. 1H NMR-based metabolomics to investigate the effects of Phoenix dactylifera seed extracts in LPS-IFN-γ-induced RAW 264.7 cells. Food Res. Int. 2019;125(February) doi: 10.1016/j.foodres.2019.108565. [DOI] [PubMed] [Google Scholar]
- 2.Abidli S., Lahbib Y., Trigui El Menif N. Microplastics in commercial molluscs from the lagoon of Bizerte (Northern Tunisia) Mar. Pollut. Bull. 2019;142(March):243–252. doi: 10.1016/j.marpolbul.2019.03.048. [DOI] [PubMed] [Google Scholar]
- 3.Adamu M., Zahmir S., Noor M., Azmai A., Mohamat-yusuff F., Ismail A. Embryonic toxicity of 3, 4-dichloroaniline ( 3, 4-DCA) on Javanese medaka. Toxicol. Rep. 2020;7:1039–1045. doi: 10.1016/j.toxrep.2020.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baldelli V., Scaldaferri F., Putignani L., Del Chierico F. The role of enterobacteriaceae in gut microbiota dysbiosis in inflammatory bowel diseases. Microorganisms. 2021;9(4):1–15. doi: 10.3390/microorganisms9040697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barnes, S., Benton, H. P., Casazza, K., Cooper, S. J., Cui, X., Du, X., Engler, J., Kabarowski, J. H., Li, S., Pathmasiri, W., Prasain, J. K., Renfrow, M. B., & Tiwari, H. K. (2016). Training in metabolomics research. II. Processing and statistical analysis of metabolomics data, metabolite identification, pathway analysis, applications of metabolomics and its future. Journal of Mass Spectrometry: JMS, 51(8), 535–548. https://doi.org/10.1002/jms.3780. [DOI] [PMC free article] [PubMed]
- 6.Behnke A., Engel M., Christen R., Nebel M., Klein R.R., Stoeck T. Depicting more accurate pictures of protistan community complexity using pyrosequencing of hypervariable SSU rRNA gene regions. Environ. Microbiol. 2011;13(2):340–349. doi: 10.1111/j.1462-2920.2010.02332.x. [DOI] [PubMed] [Google Scholar]
- 7.Bessa F., Barría P., Neto J.M., Frias J.P.G.L., Otero V., Sobral P., Marques J.C. Occurrence of microplastics in commercial fish from a natural estuarine environment. Mar. Pollut. Bull. 2018;128(January):575–584. doi: 10.1016/j.marpolbul.2018.01.044. [DOI] [PubMed] [Google Scholar]
- 8.Boer C.G., Radjabzadeh D., Medina-Gomez C., Garmaeva S., Schiphof D., Arp P., Koet T., Kurilshikov A., Fu J., Ikram M.A., Bierma-Zeinstra S., Uitterlinden A.G., Kraaij R., Zhernakova A., van Meurs J.B.J. Intestinal microbiome composition and its relation to joint pain and inflammation. Nat. Commun. 2019;10(1):1–9. doi: 10.1038/s41467-019-12873-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bokulich N.A., Subramanian S., Faith J.J., Gevers D., Gordon J.I., Knight R., Mills D.A., Caporaso J.G. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods. 2013;10(1):57–59. doi: 10.1038/nmeth.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cong Y., Jin F., Tian M., Wang J., Shi H., Wang Y., Mu J. Ingestion, egestion and post-exposure effects of polystyrene microspheres on marine medaka (Oryzias melastigma) Chemosphere. 2019;228:93–100. doi: 10.1016/j.chemosphere.2019.04.098. [DOI] [PubMed] [Google Scholar]
- 11.Cverenkárová K., Valachovičová M., Mackul’ak T., Žemlička L., Bírošová L. Microplastics in the food chain. Life. 2021;11(12):1–18. doi: 10.3390/life11121349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng Y., Zhang Y., Lemos B., Ren H. Tissue accumulation of microplastics in mice and biomarker responses suggest widespread health risks of exposure. Sci. Rep. 2017;7(October 2016):1–10. doi: 10.1038/srep46687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duan Y., Xiong D., Wang Y., Zhang Z., Li H., Dong H., Zhang J. Toxicological effects of microplastics in Litopenaeus vannamei as indicated by an integrated microbiome, proteomic and metabolomic approach. Sci. Total Environ. 2021;761 doi: 10.1016/j.scitotenv.2020.143311. [DOI] [PubMed] [Google Scholar]
- 14.Fang Q., Feng Y., Feng P., Wang X., Zong Z. Nosocomial bloodstream infection and the emerging carbapenem-resistant pathogen Ralstonia insidiosa. BMC Infect. Dis. 2019;19(1):1–9. doi: 10.1186/s12879-019-3985-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.GESAMP Joint Group of Experts on the Scientific Aspects of Marine Environmental Protection. (2016). Sources, fate and effects of microplastics in the marine environment: part 2 of a global assessment. (IMO, FAO/UNESCO-IOC/UNIDO/WMO/IAEA/UN/UNEP/UNDP). In: Kershaw, P.J. (Ed.), Rep. Stud. GESAMP No. 90 (96 pp). Reports and Studies GESAMP, No. 93, 96 P., 93.
- 16.Haas R., Smith J., Rocher-Ros V., Nadkarni S., Montero-Melendez T., D’Acquisto F., Bland E.J., Bombardieri M., Pitzalis C., Perretti M., Marelli-Berg F.M., Mauro C. Lactate regulates metabolic and proinflammatory circuits in control of T cell migration and effector functions. PLoS Biol. 2015;13(7):1–24. doi: 10.1371/journal.pbio.1002202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hartmann N.B., Hüffer T., Thompson R.C., Hassellöv M., Verschoor A., Daugaard A.E., Rist S., Karlsson T., Brennholt N., Cole M., Herrling M.P., Hess M.C., Ivleva N.P., Lusher A.L., Wagner M. Are we speaking the same language? recommendations for a definition and categorization framework for plastic debris. Environ. Sci. Technol. 2019;53(3):1039–1047. doi: 10.1021/acs.est.8b05297. [DOI] [PubMed] [Google Scholar]
- 18.Horie Y., Kanazawa N., Yamagishi T., Yonekura K., Tatarazako N. Ecotoxicological test assay using OECD TG 212 in marine Java Medaka (Oryzias javanicus) and freshwater Japanese medaka (Oryzias latipes) Bull. Environ. Contam. Toxicol. 2018;101(3):344–348. doi: 10.1007/s00128-018-2398-1. [DOI] [PubMed] [Google Scholar]
- 19.Hotamisligil G.S. Inflammation and metabolic disorders. Nature. 2006;444(7121):860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- 20.Ibrahim Y.S., Tuan Anuar S., Azmi A.A., Wan Mohd Khalik W.M.A., Lehata S., Hamzah S.R., Ismail D., Ma Z.F., Dzulkarnaen A., Zakaria Z., Mustaffa N., Tuan Sharif S.E., Lee Y.Y. Detection of microplastics in human colectomy specimens. JGH Open. 2021;5(1):116–121. doi: 10.1002/jgh3.12457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ismail A., Yusof S. Effect of mercury and cadmium on early life stages of Java medaka (Oryzias javanicus): a potential tropical test fish. Mar. Pollut. Bull. 2011;63(5–12):347–349. doi: 10.1016/j.marpolbul.2011.02.014. [DOI] [PubMed] [Google Scholar]
- 22.Ivashkiv L.B. The hypoxia–lactate axis tempers inflammation Lionel. Physiol. Behav. 2017;176(3):139–148. doi: 10.1038/s41577-019-0259-8.The. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jabeen K., Li B., Chen Q., Su L., Wu C., Hollert H., Shi H. Effects of virgin microplastics on goldfish (Carassius auratus) Chemosphere. 2018;213:323–332. doi: 10.1016/j.chemosphere.2018.09.031. [DOI] [PubMed] [Google Scholar]
- 24.Jin Y., Lu L., Tu W., Luo T., Fu Z. Impacts of polystyrene microplastic on the gut barrier, microbiota and metabolism of mice. Sci. Total Environ. 2019;649:308–317. doi: 10.1016/j.scitotenv.2018.08.353. [DOI] [PubMed] [Google Scholar]
- 25.Jin Y., Xia J., Pan Z., Yang J., Wang W., Fu Z. Polystyrene microplastics induce microbiota dysbiosis and in fl ammation in the gut of adult zebra fi sh *. Environ. Pollut. 2018;235:322–329. doi: 10.1016/j.envpol.2017.12.088. [DOI] [PubMed] [Google Scholar]
- 26.Jin Y., Xia J., Pan Z., Yang J., Wang W., Fu Z. Polystyrene microplastics induce microbiota dysbiosis and inflammation in the gut of adult zebrafish. Environ. Pollut. 2018;235:322–329. doi: 10.1016/j.envpol.2017.12.088. [DOI] [PubMed] [Google Scholar]
- 27.Karbalaei S., Golieskardi A., Watt D.U., Boiret M., Hanachi P., Walker T.R., Karami A. Analysis and inorganic composition of microplastics in commercial Malaysian fish meals. Mar. Pollut. Bull. 2020;150(October) doi: 10.1016/j.marpolbul.2019.110687. [DOI] [PubMed] [Google Scholar]
- 28.Kho Z.Y., Lal S.K. The human gut microbiome - a potential controller of wellness and disease. Front. Microbiol. 2018;9(AUG):1–23. doi: 10.3389/fmicb.2018.01835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim J.M., Rim J.H., Kim D.H., Kim H.Y., Choi S.K., Kim D.Y., Choi Y.J., Yu S., Gee H.Y., Cheon J.H. P157 microbiome analysis reveals that ralstonia is responsible for decreased renal function in patients with ulcerative colitis. J. Crohn’s Colitis. 2021;15(Supplement_1):S236–S238. doi: 10.1093/ecco-jcc/jjab076.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kitzenberg D., Colgan S.P., Glover L.E. Creatine kinase in ischemic and inflammatory disorders. Clin. Translational Med. 2016;5(1) doi: 10.1186/s40169-016-0114-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koyama J., Kawamata M., Imai S., Fukunaga M., Uno S., Kakuno A. Java medaka: a proposed new marine test fish for ecotoxicology. Environ. Toxicol. 2008;23:487–491. doi: 10.1002/tox.2036. [DOI] [PubMed] [Google Scholar]
- 32.Lauri I., Savorani F., Iaccarino N., Zizza P., Pavone L.M., Novellino E., Engelsen S.B., Randazzo A. Development of an optimized protocol for NMR metabolomics studies of human colon cancer cell lines and first insight from testing of the protocol using DNA G-quadruplex ligands as novel anti-cancer drugs. Metabolites. 2016;6(1):1–14. doi: 10.3390/metabo6010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lebreton L., Andrady A. Future scenarios of global plastic waste generation and disposal. Palgrave Commun. 2019;5(1):1–11. doi: 10.1057/s41599-018-0212-7. [DOI] [Google Scholar]
- 34.Lei L., Wu S., Lu S., Liu M., Song Y., Fu Z., Shi H., Raley-Susman K.M., He D. Microplastic particles cause intestinal damage and other adverse effects in zebrafish Danio rerio and nematode Caenorhabditis elegans. Sci. Total Environ. 2018;619–620:1–8. doi: 10.1016/j.scitotenv.2017.11.103. [DOI] [PubMed] [Google Scholar]
- 35.Li F., Gonzalez F.J., Ma X. LC–MS-based metabolomics in profiling of drug metabolism and bioactivation. Acta Pharmaceutica Sinica B. 2012;2(2):118–125. doi: 10.1016/j.apsb.2012.02.010. [DOI] [Google Scholar]
- 36.Li Y., Wang J., Yang G., Lu L., Zheng Y., Zhang Q., Zhang X., Tian H., Wang W., Ru S. Low level of polystyrene microplastics decreases early developmental toxicity of phenanthrene on marine medaka (Oryzias melastigma) J. Hazard. Mater. 2020;385 doi: 10.1016/j.jhazmat.2019.121586. [DOI] [PubMed] [Google Scholar]
- 37.Liu G., Lee D.P., Schmidt E., Prasad G.L. Pathway analysis of global metabolomic profiles identified enrichment of caffeine, energy, and arginine metabolism in smokers but not moist snuff consumers. Bioinform. Biol. Insights. 2019:13. doi: 10.1177/1177932219882961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lopez Leyva L., Gonzalez E., Li C., Ajeeb T., Solomons N.W., Agellon L.B., Scott M.E., Koski K.G. Human milk microbiota in an indigenous population is associated with maternal factors, stage of lactation, and breastfeeding practices. Curr. Dev. Nutr. 2021;5(4):1–15. doi: 10.1093/cdn/nzab013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu K., Abo R.P., Schlieper K.A., Graffam M.E., Levine S., Wishnok J.S., Swenberg J.A., Tannenbaum S.R., Fox J.G. Arsenic exposure perturbs the gut microbiome and its metabolic profile in mice: an integrated metagenomics and metabolomics analysis. Environ. Health Perspect. 2014;122(3):284–291. doi: 10.1289/ehp.1307429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luqman, A., Nugrahapraja, H., Wahyuono, R.A., Islami, I., Haekal, H., Fardiansyah, Y., Putri, B.Q., Amalludin, F.I., Rofiqa, E.A., Götz, F., & Wibowo, A.T. (2021). Microplastic Contamination in Human Stools, Foods, and Drinking Water Associated with Indonesian Coastal Population. 1–9.
- 41.Mahadevan S., Shah S.L., Marrie T.J., Slupsky C.M. Analysis of metabolomic data using support vector machines. Anal. Chem. 2008;80(19):7562–7570. doi: 10.1021/ac800954c. [DOI] [PubMed] [Google Scholar]
- 42.Masato, K., Kenji, M., Kiyoshi, N., & Minoru, T. (Ed.). (2009). Medaka, Biology, Management and Experimental Protocol (1st Editio). Wiley Black-well.
- 43.Mohamat-Yusuff F., Sarah-Nabila A.G., Zulkifli S.Z., Azmai M.N.A., Ibrahim W.N.W., Yusof S., Ismail A. Acute toxicity test of copper pyrithione on Javanese medaka and the behavioural stress symptoms. Mar. Pollut. Bull. 2018;127(May 2017):150–153. doi: 10.1016/j.marpolbul.2017.11.046. [DOI] [PubMed] [Google Scholar]
- 44.Moltu S.J., Sachse D., Blakstad E.W., Strømmen K., Nakstad B., Almaas A.N., Westerberg A.C., Rønnestad A., Brække K., Veierød M.B., Iversen P.O., Rise F., Berg J.P., Drevon C.A. Urinary metabolite profiles in premature infants show early postnatal metabolic adaptation and maturation. Nutrients. 2014;6(5):1913–1930. doi: 10.3390/nu6051913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O’Neill L.A.J., Kishton R.J., Rathmell J. A guide to immunometabolism for immunologists. Nat. Rev. Immunol. 2016;16(9):553–565. doi: 10.1038/nri.2016.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oliveri Conti G., Ferrante M., Banni M., Favara C., Nicolosi I., Cristaldi A., Fiore M., Zuccarello P. Micro- and nano-plastics in edible fruit and vegetables. The first diet risks assessment for the general population. Environ. Res. 2020;187(April) doi: 10.1016/j.envres.2020.109677. [DOI] [PubMed] [Google Scholar]
- 47.Ott S.J., Musfeldt M., Wenderoth D.F., Hampe J., Brant O., Fölsch U.R., Timmis K.N., Schreiber S. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut. 2004;53(5):685–693. doi: 10.1136/gut.2003.025403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pucino V., Certo M., Bulusu V., Cucchi D., Goldmann K., Pontarini E., Haas R., Smith J., Headland S.E., Blighe K., Ruscica M., Humby F., Lewis M.J., Kamphorst J.J., Bombardieri M., Pitzalis C., Mauro C. Lactate buildup at the site of chronic inflammation promotes disease by inducing CD4+ T cell metabolic rewiring. Cell Metab. 2019;30(6):1055–1074. doi: 10.1016/j.cmet.2019.10.004. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qiao R., Sheng C., Lu Y., Zhang Y., Ren H., Lemos B. Microplastics induce intestinal inflammation, oxidative stress, and disorders of metabolome and microbiome in zebrafish. Sci. Total Environ. 2019;662:246–253. doi: 10.1016/j.scitotenv.2019.01.245. [DOI] [PubMed] [Google Scholar]
- 50.Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., Glöckner F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013;41(D1):590–596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schwabl P., Koppel S., Konigshofer P., Bucsics T., Trauner M., Reiberger T., Liebmann B. Detection of various microplastics in human stool: a prospective case series. Ann. Internal Med. 2019;171(7):453–457. doi: 10.7326/M19-0618. [DOI] [PubMed] [Google Scholar]
- 52.Schymanski D., Goldbeck C., Humpf H.U., Fürst P. Analysis of microplastics in water by micro-Raman spectroscopy: Release of plastic particles from different packaging into mineral water. Water Res. 2018:129. doi: 10.1016/j.watres.2017.11.011. [DOI] [PubMed] [Google Scholar]
- 53.Setälä O., Fleming-Lehtinen V., Lehtiniemi M. Ingestion and transfer of microplastics in the planktonic food web. Environ. Pollut. 2014;185:77–83. doi: 10.1016/j.envpol.2013.10.013. [DOI] [PubMed] [Google Scholar]
- 54.Y. Sidorova Detecting Oxidative Stress Biomarkers in Neurodegenerative Disease Models and Patients 2020 1 14. [DOI] [PMC free article] [PubMed]
- 55.Sobhani Z., Lei Y., Tang Y., Wu L., Zhang X., Naidu R. Microplastics generated when opening plastic packaging. Sci. Rep. 2020;10(4841):1–7. doi: 10.1038/s41598-020-61146-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Solevåg A.L., Zykova S.N., Thorsby P.M., Schmölzer G.M. Metabolomics to diagnose oxidative stress in perinatal asphyxia: Towards a non-invasive approach. Antioxidants. 2021;10(11):1–11. doi: 10.3390/antiox10111753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stephens W.Z., Burns A.R., Stagaman K., Wong S., Rawls J.F., Guillemin K., Bohannan B.J.M. The composition of the zebrafish intestinal microbial community varies across development. ISME J. 2016;10(3):644–654. doi: 10.1038/ismej.2015.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sun Y., Su Y., Zhu W. Microbiome-metabolome responses in the cecum and colon of pig to a high resistant starch diet. Front. Microbiol. 2016;7 doi: 10.3389/fmicb.2016.00779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takehana, Y., Zahm, M., Cabau, C., Klopp, C., Roques, C., Bouchez, O., Donnadieu, C., Barrachina, C., Journot, L., Kawaguchi, M., Yasumasu, S., Ansai, S., Naruse, K., Inoue, K., Shinzato, C., Schartl, M., Guiguen, Y., & Herpin, A. (2020). Genome Sequence of the Euryhaline Javafish Medaka, Oryzias javanicus: A Small Aquarium Fish Model for Studies on Adaptation To Salinity. G3: Genes=Genomes|Genetics, 10(March), g3.400725.2019. https://doi.org/10.1534/g3.119.400725. [DOI] [PMC free article] [PubMed]
- 60.Triba M.N., Le Moyec L., Amathieu R., Goossens C., Bouchemal N., Nahon P., Rutledge D.N., Savarin P. PLS/OPLS models in metabolomics: the impact of permutation of dataset rows on the K-fold cross-validation quality parameters. Mol. BioSyst. 2015;11(1):13–19. doi: 10.1039/c4mb00414k. [DOI] [PubMed] [Google Scholar]
- 61.Udayappan S.D., Kovatcheva-Datchary P., Bakker G.J., Havik S.R., Herrema H., Cani P.D., Bouter K.E., Belzer C., Witjes J.J., Vrieze A., De Sonnaville N., Chaplin A., Van Raalte D.H., Aalvink S., Dallinga-Thie G.M., Heilig H.G.H.J., Bergström G., Van Der Meij S., Van Wagensveld B.A., Nieuwdorp M. Intestinal Ralstonia pickettii augments glucose intolerance in obesity. PLoS ONE. 2017;12(11) doi: 10.1371/journal.pone.0181693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Usman S., Faizal A., Razis A., Shaari K., Noor M., Amal A., Saad M.Z., Isa N.M., Nazarudin M.F. Polystyrene microplastics exposure: an insight into multiple organ histological alterations, oxidative stress and neurotoxicity in Javanese medaka fish ( Oryzias javanicus. Int. J. Environ. Res. Public Health Article. 2021;18:9449. doi: 10.3390/ijerph18189449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vernocchi P., Del Chierico F., Putignani L. Gut microbiota profiling: metabolomics based approach to unravel compounds affecting human health. Front. Microbiol. 2016;7(JUL) doi: 10.3389/fmicb.2016.01144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang H., Zhang H., Deng P., Liu C., Li D., Jie H., Zhang H., Zhou Z., Zhao Y.L. Tissue metabolic profiling of human gastric cancer assessed by 1H NMR. BMC Cancer. 2016;16(1):1–12. doi: 10.1186/s12885-016-2356-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang Y., Zhu W., Wang D., Teng M., Yan J., Miao J., Zhou Z. 1H NMR-based metabolomics analysis of adult zebrafish (Danio rerio) after exposure to diniconazole as well as its bioaccumulation behavior. Chemosphere. 2017;168:1571–1577. doi: 10.1016/j.chemosphere.2016.11.157. [DOI] [PubMed] [Google Scholar]
- 66.Warburg O. The metabolism of carcinoma cells 1. J. Cancer Res. 1925;9(1):148–163. doi: 10.1158/jcr.1925.148. [DOI] [Google Scholar]
- 67.WHO. (2019). WHO calls for more research into microplastics and a crackdown on plastic pollution. NEWS RELEASE. https://www.who.int/news/item/22–08-2019-who-calls-for-more-research-into-microplastics-and-a-crackdown-on-plastic-pollution.
- 68.Wyss M., Kaddurah-Daouk R. Creatine and creatinine metabolism. Physiol. Rev. 2000;80(3):1107–1213. doi: 10.1152/physrev.2000.80.3.1107. [DOI] [PubMed] [Google Scholar]
- 69.Xia, J., & Wishart, D. S. (2016). Using metaboanalyst 3.0 for comprehensive metabolomics data analysis. Current Protocols in Bioinformatics, 2016(September), 14.10.1–14.10.91. https://doi.org/10.1002/cpbi.11. [DOI] [PubMed]
- 70.Yang D., Shi H., Li L., Li J., Jabeen K., Kolandhasamy P. Microplastic pollution in table salts from China. Environ. Sci. Technol. 2015;49(22):13622–13627. doi: 10.1021/acs.est.5b03163. [DOI] [PubMed] [Google Scholar]
- 71.Yang J., Chi Y., Burkhardt B.R., Guan Y., Wolf B.A. Leucine metabolism in regulation of insulin secretion from pancreatic beta cells. Nutr. Rev. 2010;68(5):270–279. doi: 10.1111/j.1753-4887.2010.00282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang Y., Wu Z., Li S., Yang M., Xiao X., Lian C., Wen W., He H., Zeng J., Wang J., Zhang G. Targeted blood metabolomic study on retinopathy of prematurity. Investig. Ophthalmol. Visual Sci. 2020;61(2) doi: 10.1167/iovs.61.2.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang F., Wang X., Xu J., Zhu L., Peng G., Xu P., Li D. Food-web transfer of microplastics between wild caught fish and crustaceans in East China Sea. Mar. Pollut. Bull. 2019;146(May):173–182. doi: 10.1016/j.marpolbul.2019.05.061. [DOI] [PubMed] [Google Scholar]
- 74.Zhang X., Wen K., Ding D., Liu J., Lei Z., Chen X., Ye G., Zhang J., Shen H., Yan C., Dong S., Huang Q., Yi L. Size-dependent adverse effects of microplastics on intestinal microbiota and metabolic homeostasis in the marine medaka ( Oryzias melastigma) Environ. Int. 2021;151 doi: 10.1016/j.envint.2021.106452. [DOI] [PubMed] [Google Scholar]
- 75.Zhang Y., Zhao F., Deng Y., Zhao Y., Ren H. Metagenomic and metabolomic analysis of the toxic effects of trichloroacetamide-induced gut microbiome and urine metabolome perturbations in mice. J. Proteome Res. 2015;14(4):1752–1761. doi: 10.1021/pr5011263. [DOI] [PubMed] [Google Scholar]
- 76.Zhao S., Zhu L., Wang T., Li D. Suspended microplastics in the surface water of the Yangtze Estuary System, China: First observations on occurrence, distribution. Mar. Pollut. Bull. 2014;86(1–2):562–568. doi: 10.1016/j.marpolbul.2014.06.032. [DOI] [PubMed] [Google Scholar]
- 77.Zhao Y., Qiao R., Zhang S., Wang G. Metabolomic profiling reveals the intestinal toxicity of different length of microplastic fibers on zebrafish (Danio rerio) J. Hazard. Mater. 2021;403(August 2020) doi: 10.1016/j.jhazmat.2020.123663. [DOI] [PubMed] [Google Scholar]
- 78.Zhao Y., Zhang Y., Wang G., Han R., Xie X. Effects of chlorpyrifos on the gut microbiome and urine metabolome in mouse (Mus musculus) Chemosphere. 2016;153:287–293. doi: 10.1016/j.chemosphere.2016.03.055. [DOI] [PubMed] [Google Scholar]
- 79.Zhu M., Chernick M., Rittschof D., Hinton D.E. Chronic dietary exposure to polystyrene microplastics in maturing Japanese medaka (Oryzias latipes) Aquat. Toxicol. 2020;220(October 2019) doi: 10.1016/j.aquatox.2019.105396. [DOI] [PubMed] [Google Scholar]