Highlights
-
•
A novel gut bacterium Citrobacter amalonaticus CJ25 isolated from healthy human fecal sample.
-
•
CJ25 grows on choline as the sole carbon and energy source.
-
•
CJ25 does not produce the harmful proatherogenic metabolite trimethylamine.
Keywords: Choline, Quaternary amine, Anaerobic culturing, Trimethylamine, Gut microbiome, Pyruvate-formate lyase, Choline trimethylamine lyase, CutC, Glycyl radical enzymes
Abstract
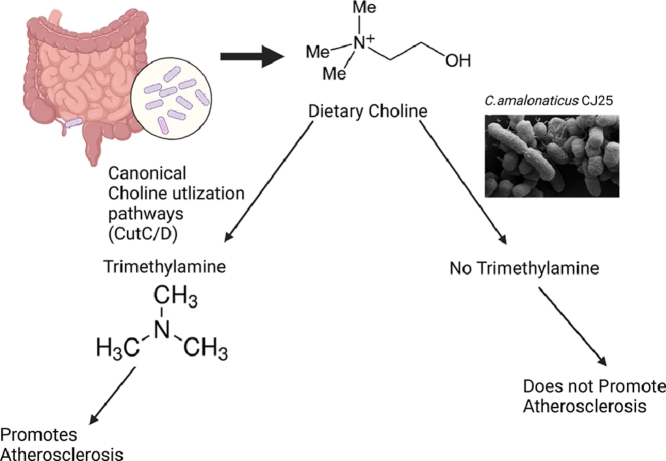
Gut microbiota metabolism can have profound effects on human health. Choline, a quaternary amine (QA) highly abundant in our diet, is canonically cleaved by a glycyl radical enzyme, choline trimethylamine lyase (CutC), and its SAM-dependent radical activator, CutD. CutC cleaves choline to form trimethylamine (TMA) and acetaldehyde. TMA is oxidized to TMAO by FMO3 in the liver, which plays a role in causing atherosclerosis. We hypothesized that alternative pathways for choline degradation occur within gut microbes and that certain gut microbiota can anaerobically respire or ferment QAs, such as choline. Based on this prediction we established QA-supplemented enrichment cultures using fecal material from healthy volunteers as the inocula. We have isolated, from a choline-supplemented enrichment of a human fecal sample, a strain of Citrobacter amalonaticus, that we have designated CJ25. This strain is capable of anaerobically utilizing choline as its sole carbon and energy source. Its genome does not contain the cutCD genes or genes encoding any COG5598 methyltransferases. We have confirmed the degradation of choline and production of acetate by the organism during growth of the strain. However, we used multiple analytical methods to confirm that no TMA accumulated in the medium during growth. Hence, strain CJ25 is a unique bacterium that degrades choline without the production of the proatherogenic metabolite TMA.
Graphical abstract
Introduction
The human gut includes trillions of microbes such as bacteria, archaea, viruses and eukaryotes. They play important, but generally poorly defined, roles in the physiology, defenses, and functioning of the human body. They impact the immune system, metabolic activities and protect us against pathogens (Li et al., 2015; Avula et al., 2018). Significant progress has been made to characterize the human gut microbiome and has provided great insights into the host-microbiome functional interactions. These studies have paved the way to better understand the role of the human gut microbiome in disease pathogenesis, thereby helping in better disease management (Durack and Lynch et al., 2019). In the recent years, the metabolism of the gut microbiome has been linked to the development of cardiovascular diseases (CVDs), by producing metabolites such as trimethylamine (TMA), trimethylamine-N-oxide (TMAO), bile acids (BA) and short chain fatty acids (SCFA) from the breakdown of diet rich in red meat (Hoyles et al., 2018; Ahmad et al., 2019). A number of microbes degrade the quaternary amines (QAs) like choline, carnitine, gamma-butyrobetaine and glycine betaine leading to production of TMA. These QAs are highly abundant in red meat, seafood, and poultry (Rath et al., 2017). There are four well-characterized pathways in gut bacteria for breakdown of quaternary amines and production of TMA: 1) choline TMA-lyase (CutC), a specific glycyl radical enzyme, which uses choline as a substrate to generate TMA (Rath et al., 2017); 2) a Rieske-type oxygenase/reductase (CntA/B) that utilizes carnitine as a substrate to generate TMA (Y. Zhu et al., 2014) 3); another two-component system termed YeaW/X that generates TMA from carnitine, gamma butyrobetaine, or choline (Y. Zhu et al., 2014); and 4) enzymes encoded by the bbu gene cluster that convert L-carnitine derived gamma-butyrobetaine to TMA (Rajakovich et al., 2021).
Quaternary amines can also be degraded by gut bacteria in a manner in which no TMA is generated. In the gut bacterium Eubacterium limosum, pyrrolysine-lacking enzymes of the COG5598 superfamily have been shown to demethylate the QAs carnitine, gamma-butyrobetaine, and proline betaine in corrinoid-dependent methyl transfer pathways (Kountz et al., 2020; Picking et al., 2019; Ellenbogen et al., 2021). The organism also encodes an MtgB for degradation of glycine betaine. These alternative methylotrophic pathways for QA degradation highlight the potential for other pathways utilized by gut microbes.
Choline is an essential component in the human diet because of its many roles, such as a methyl group donor, a precursor for the neurotransmitter acetylcholine, and as a component of cell membranes (Zeisel and Da Costaet al., 2009; Oliphant and Allen-Vercoe et al., 2019). The canonical route of choline degradation by gut bacteria is via the CutC/D enzymes where TMA and acetaldehyde are produced. In addition to the above stated enzymes, more cut genes in the cut cluster have been characterized that are involved in a bacterial microcompartment (BMC) system (Jameson et al., 2016; Herring et al., 2018). Choline diffuses into the BMC where it is broken down into TMA and acetaldehyde by CutC/D, TMA is then released from the BMC and acetaldehyde is converted to ethanol via CutO and acetate via CutH and CutF. Ethanol and acetate are released from the BMC along with generation of 1 ATP molecule for growth (Herring et al., 2018). These BMCs are encoded in large operons that encode shell proteins and the pathway proteins as well. The cutC gene is often considered a diagnostic indicator for choline degraders in the gut that promote TMA production, which is subsequently oxidized to TMAO in the liver via flavin monooxygenases (Anwar et al., 2018). These metabolites have been linked with cardiovascular diseases in the gut, although the complete cellular mechanisms are not fully characterized (Zhu et al., 2016).
Edwardsiella tarda ATCC 23,685 has recently been shown to produce TMA when grown on choline but the organism appears to lack the canonical cutCD cluster that is responsible for converting choline to TMA (Romano et al., 2015; Jameson et al., 2018; Herring et al., 2018). The organism encodes enzymes from the GRE family but the sequence alignment with the known CutCs showed only a 33% identity to the CutC from Proteus mirabilis and a poor consensus with the conserved residues (Jameson et al., 2018). Several of the glycyl radical enzymes (GREs) in the E. tarda genome were annotated as pyruvate formate lyases and are speculated to be novel GREs that convert choline to TMA, although this has not been experimentally demonstrated (Jameson et al., 2018).
In our study, we have isolated a novel gut strain of Citrobacter amalonaticus that is capable of degrading choline with no detectable production of TMA. The organism does not encode a canonical CutC or a COG5598 methyltransferase and therefore degrades choline via an unknown mechanism in which the risk of atherosclerosis in the host is likely not increased.
Materials and methods
Sample collection
A deidentified fecal sample (patient 25) was obtained from a healthy volunteer with no gastrointestinal disease from the University of Cincinnati and was stored at −80 °C until further use. The patient identity was kept confidential.
Anaerobic media components
One liter of media contained: 1.0 g NH4Cl2, 0.1 g NaCl, 0.1 g MgCl2·6H2O, 0.05 g CaCl2, 1X SL-10 trace elements solution (DSMZ medium 722), 1X selenite-tungstate solution (DSMZ medium 385), and resazurin (0.1% w/v). The media was sparged with ultra-high purity 100% N2 gas for 30 min. The following components were added to the base medium in the anaerobic chamber: 2 mM DTT; 3 mM Cysteine-HCl; 0.30 g Na2S·9H2O. Inside an anaerobic chamber, 10 mL of media were aliquoted in 27 mL glass Balch-type tubes, sealed with butyl rubber stoppers, and capped with aluminum crimp caps. The headspaces of the tubes were exchanged with 100% N2 for three 1 min. gas exchange cycles on a vacuum and gas manifold system. The tubes were autoclaved and stored at room temperature. All chemical reagents were purchased from Sigma-Aldrich or Fisher Scientific.
Growth and isolation of the culture
The fecal sample was moved to the anaerobic chamber, weighed and mixed with saline to make a fecal slurry. The slurry was then strained through SureStrain™ premium cell strainer (MTC Biotech). The anaerobic media tubes were inoculated, in triplicate, with 100 µL of fecal slurry of the concentration (0.03 g/ml). The media tubes were supplemented with 30 mM choline, 30 mM Na2SO3, 1X Vitamins (DSMZ medium 141), and 22 mM KH2PO4 buffer pH 7.2. For subculturing, 1% of the inoculum was taken from the previous enrichment tube. The cultures were incubated at 37 °C and subcultured three times before beginning to monitor absorbance at 600 nm by placing tubes directly into a Spectronic 20. The cultures were plated in anaerobic bottles with the media components stated above with addition of 1.5% agar. The medium was made in slants in 150 ml stoppered serum bottles to allow more surface area for bacterial growth. Isolated colonies were picked and cultured in liquid media to obtain a pure isolate.
Genomic DNA isolation and sequencing
The initial identity of the culture was obtained by carrying out 16S rRNA Sanger sequencing, using 27F and 1492R primers (Frank et al., 2008) and, for the V4 region, 515F (GTGCCAGCMGCCGCGGTAA) and 806R (GGACTACHVGGGTWTCTAAT) were used. The 16S rRNA amplicon was purified using a Promega clean up kit and the sequenced at the Center of Bioinformatic and Functional Genomics (CBFG) at Miami University. Genomic DNA was isolated using a protocol provided by the JGI for Bacterial genomic DNA isolation using CTAB (William et al., 2004). The gDNA was quantitated using Qubit 4.0 and the Qubit dsDNA high sensitivity quantitation kit from Thermo Fisher Scientific. The gDNA was then sent to the University of Delaware sequencing and genotyping center to sequence the whole genome using PacBio RS II technology. The library preparation, sequencing, assembly, and annotation was carried out by University of Delaware sequencing center. The whole genome of C. amalonaticus CJ25 was deposited to NCBI and JGI and is publicly available. NCBI (Assembly GCA_014859035.1 and WGS JADAQU000000000) and JGI/IMG (Study ID: Gs0145800 Project ID: Gp0489996 Analysis ID: Ga0439030 ).
High resolution melt curve analysis
HRM was performed on 72-well Rotor Gene Q (Qiagen). A culture of C. amalonaticus CJ25 was grown to stationary phase and the gDNA was isolated as described above. The 16S rRNA V4 region was amplified using the 515F and 806R primers. The HRM reaction was set up in 0.1 mL strip cap tubes from Qiagen. The components of the reaction were: 50 ng of amplified V4 segments, 1 µM 515F and 806F primers, 1X Evagreen dye, and nuclease-free H2O, making the total reaction volume 20 µL. The reaction conditions were: HRM cycle ramp temperature from 65 °C to 95 °C, rising for 0.1 °C degree each step, wait for 90 s of pre-melt conditioning on the first step and wait for 2 s for each step afterwards, cycling conditions were 95 °C for 20 s, 58 °C for 20 s and 72 °C for 20 s for 30 cycles. The total run time was 96 min.
Scanning electron microscopy examination
C. amalonaticus CJ25 was seeded on poly-l-lysine coated coverslips submerged in LB broth and incubated 37 °C in a humidified incubator. After 6 h, the growth medium was aspirated and the coverslips were gently washed with phosphate buffered saline and submerged in 0.2% glutaraldehyde, 2% paraformaldehyde fixative solution for 15 min at room temperature, followed by a 45 min incubation at 4 °C. The coverslips were subjected to four 10 min washes with 100 mM sodium cacodylate, followed by a series of 10 min ethanol dehydrations, increasing in concentration from 25%−100%. Once the samples were dehydrated, the samples were dried with CO2 at the critical point, followed by gold sputter-coating. Images of C. amalonaticus CJ25 were taken on a Zeiss Supra 35 VP FEG SEM at the centre for Advanced Microscopy and Imaging at Miami University.
Nuclear magnetic resonance 400 MHz
C. amalonaticus CJ25 cultures were grown in the anaerobic medium described above supplemented with 0.1% yeast extract and 30 mM choline. The CJ25 cultures were grown at 37 °C and collected at 0, 4, 8, 12, 16, 20, and 24 h time points. A standard curve for choline was made with 0 mM, 10 mM, 15 mM, 20 mM, 25 mM and 30 mM choline. They were spun down at 12,000 rpm for 5 min. and the supernatant was transferred to another microcentrifuge tube. The NMR sample was prepared using 440 µl of HPLC grade water, 60 µL heavy water (D2O), and 100 µL of culture supernatant. The 1D proton spectra were measured using water suppression. NMR settings included: probe heater on, gas flow 400 L/h. CDCl3 shims were used and locked for samples in 90% H20 + 10% D2O. Sixteen scans were obtained for each sample. The spectra were analyzed and the choline concentrations were calculated over time.
Phylogenetic analyses
The 16S rRNA sequence for C. amalonaticus CJ25 was used as a query sequence to identify closely related C. amalonaticus strains. A total of 15 different strains were recorded and 16S rRNA gene sequences were obtained from JGI/IMG (Mavromatis et al., 2009). The genomes of all the strains were obtained from NCBI and the KBASE Fast Tree 2 suite (Price et al., 2010) was used to construct the phylogenetic tree using 49 core SNPs. A maximum likelihood tree was constructed in Mega 7 (Kumar et al., 2016) for CutCs and formate C-acetyltransferases from CJ25 and E. tarda sp . The bootstrap value was 1000 and with gamma distribution among sites Tamura-nei was used as the substitution model. The tree visualization was done with iTOL. The CutC from Proteus mirabilis (WP_012368484.1) and Desulfovibrio alaskensis (pdb|5FAU|D) were used as query sequences for a blastp searches specific for gamma and delta-proteobacteria and matches with 88% and above identities were selected. The sequences were aligned with clustalW (Larkin et al., 2007).
Gas chromatography and mass spectrometry
A Thermo Scientific TRACE 1300 gas chromatograph (GC) was used to assess TMA production in CJ25 culture supernatants. The column used was TG-5MS AMINE (length 30 m, I.D. 0.53 mm and film thickness 3.0 µm) and was purchased from Thermo Scientific. One microliter of the standard or culture supernatant was injected into the GC and the signal was measured using the flame ionization detector (FID). The oven temperature was 150 °C. The TMA standards used were 0 mM, 5 mM, 10 mM, 15 mM, 20 mM and 30 mM TMA. Additionally, a lower range of TMA standards from 100 µM – 1000 µM was also run to assess the lowest detection limit. The culture supernatants were collected every 24 h and spun down at 12,000 rpm for 5 min and stored at −80 °C until they were used for analysis. A more sensitive LC-MS/MS approach was performed to detect TMA concentrations from culture supernatants. CJ25 culture supernatants were harvested at 0 and 24 h. Additionally, media blank, 100 µg/mL TMA standard and a media blank with choline as a substrate were used as controls. The samples were spun twice at 20,000 RPM followed by filtering the supernatant through a 0.22-µm filter. The supernatants were assessed for TMA using a targeted high performance liquid chromatagraphy tandem mass spectrometry (HPLC-MS/MS) approach at the Mass Spectrometry and Proteomics Facility at The Ohio State University. For LC-MS/MS sample preparation, 100 µL of sample was added to 100 µL of spiking solution containing 2 µg/mL of 13C; D9 TMA (Cambridge Isotope Laboratories, Inc.). For the calibration solutions, different concentrations of TMA were prepared (10 µg/mL to 0.64 ng/mL) with 1.0 µg/mL of labeled TMA. The derived standard curve was used to calculate the TMA concentration in samples. All samples and calibration solutions were injected (5 µL) in triplicate for LC-MS/MS. The chromatographic separation was performed on a Thermo Ultimate 3000 HPLC system fitted with an Agilent Poroshell 120, HILIC-Z 2.7 µm column (2.1 mm x 100 mm). A gradient of 0–100% B was run over 20 min at 200 µl/min. Mobile phase A was 0.1% formic acid in water and mobile phase B was 0.1% formic acid in acetonitrile. Mass spectrometry was performed on a Thermo Scientific QE Plus instrument with ESI and operated in positive multiple reaction monitoring (MRM) mode. Thermo Scientific Xcalibur software was used for data acquisition and processing.
Results
Isolation of Citrobacter amalonaticus CJ25
Previous studies have shown that some gut microbes can degrade choline and form TMA as an end product. However, given the volume and diversity of gut microbes, we hypothesized that organisms with other pathways of choline degradation in the gut exist that remain undiscovered. Choline is known to be an important precursor to TMA and TMAO production, leading to atherosclerosis in humans (Rath et al., 2017). Therefore, it is critical that we understand all the potential routes of choline degradation in the gut and identify the organisms capable of carrying them out. This gap in knowledge encouraged us to isolate choline degraders from fecal samples, collected from healthy volunteers. We enriched the fecal sample from patient 25, which was chosen due to sample abundance. We chose to enrich the fecal sample on DSM 720 medium and supplemented with 30 mM choline as the sole energy source under strict anaerobic conditions under a N2 headspace. The initial 3 subcultures showed ample growth, but eventual transfers led to a decrease in biomass as the culture became more homogenous. After several subcultures, 500 µL aliquots of the culture were plated on anaerobic DSM 720 medium agar in sealed serum bottles and multiple isolated colonies were picked and transferred to liquid DSM 720 medium. Analysis of the 16S rRNA of this isolate was found to be that of C. amalonaticus CJ25, a gammaproteobacterium. The purity of the culture was tested using high resolution melt curve analysis using the V4 region, along with DNA from commercially bought E. coli EC100 as a positive control, a mixed culture as an experimental control and another impure fecal culture of E. fergusonii, isolated in our laboratory, was tested as a negative control. The results indicated only one V4 region belonging to C. amalonaticus CJ25 in the culture. Multiple peaks in a curve indicate that there could be other species present in the culture. This technique is not sensitive enough to differentiate between different strains of the same species (Fig. S2).
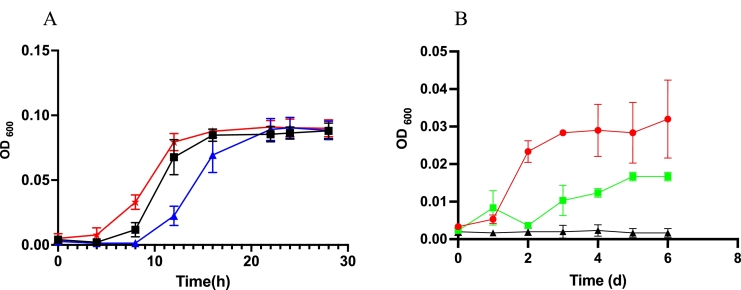
Pure colonies grown on anaerobic agar media bottles of this isolate were light yellow in color, smooth, and slightly raised. The organism stained negative in the gram stain. In addition to choline, this bacterium showed growth on other substrates such as carnitine and glucose. We cultured CJ25 initially without yeast extract supplementation to examine its ability to utilize choline as a sole carbon and energy source and later 0.1% yeast extract was supplemented to increase the biomass (Fig. 1). A draft of the whole genome of CJ25 was obtained at University of Delaware using PacBio sequencing technology and was assembled into two contiguous sequences. The genome was 5.05 megabases. Table 1 illustrates the similarities between the genome of CJ25 and other closely related C. amalonaticus strains. The draft genome was uploaded to NCBI (Assembly GCA_014859035.1 and WGS JADAQU000000000) and JGI/IMG (Study ID: Gs0145800 Project ID: Gp0489996 Analysis ID: Ga0439030).
Fig. 1.
A: Growth curve of CJ25 on 30 mM choline (black closed squares) and carnitine (red stars) with addition of 0.1% yeast extract, and with 0.1% yeast extract alone (blue closed triangles); B: growth of CJ25 on 30 mM choline (red closed circles) and carnitine (green closed squares) with no addition of yeast extract. The closed black triangles indicate the no substrate control.
Table 1.
Comparison of genomes of Citrobacter amalonaticus strains.
| Genome Name | IMG Genome ID | NCBI Assembly Accession | Genome Size | Gene Count |
|---|---|---|---|---|
| Citrobacter amalonaticus 3e8A | 2,690,316,252 | GCA_000936345.1 | 4,755,707 | 5006 |
| Citrobacter amalonaticus 3e8B | 2,660,238,606 | GCA_001373155.1 | 4,924,078 | 5484 |
| Citrobacter amalonaticus S1285 | 2,876,528,337 | GCA_002918935.1 | 5,536,114 | 5452 |
| Citrobacter amalonaticus GTA-817-RBA-P2 | 2,713,896,732 | GCA_000972645.1 | 4,999,216 | 4913 |
| Citrobacter amalonaticus YG8 | 2,648,501,729 | GCA_001276125.1 | 4,949,960 | 4847 |
| Citrobacter amalonaticus YG6 | 2,648,501,867 | GCA_001276105.1 | 4,947,518 | 4848 |
| Citrobacter amalonaticus FDAARGOS_490 | 2,871,664,767 | GCA_003938795.1 | 5,202,648 | 5242 |
| Citrobacter amalonaticus CJ25 | 2,893,839,411 | GCA_014859035.1 | 5,054,116 | 5234 |
| Citrobacter amalonaticus NcTC 10,805 | NA | GCA_900,460,855.1 | 5,093,340 | 5330 |
Identification and phylogenetic analysis
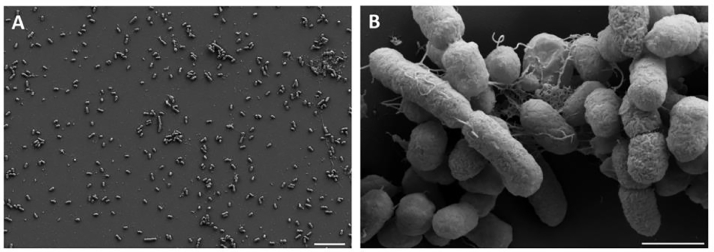
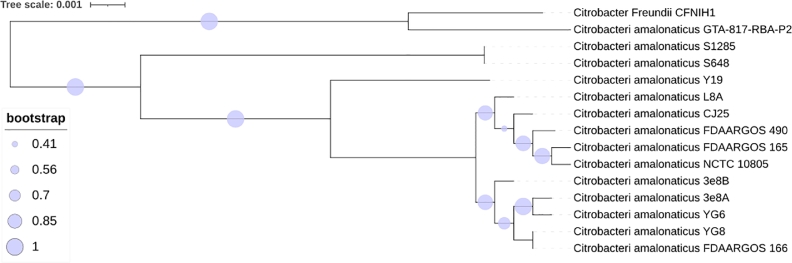
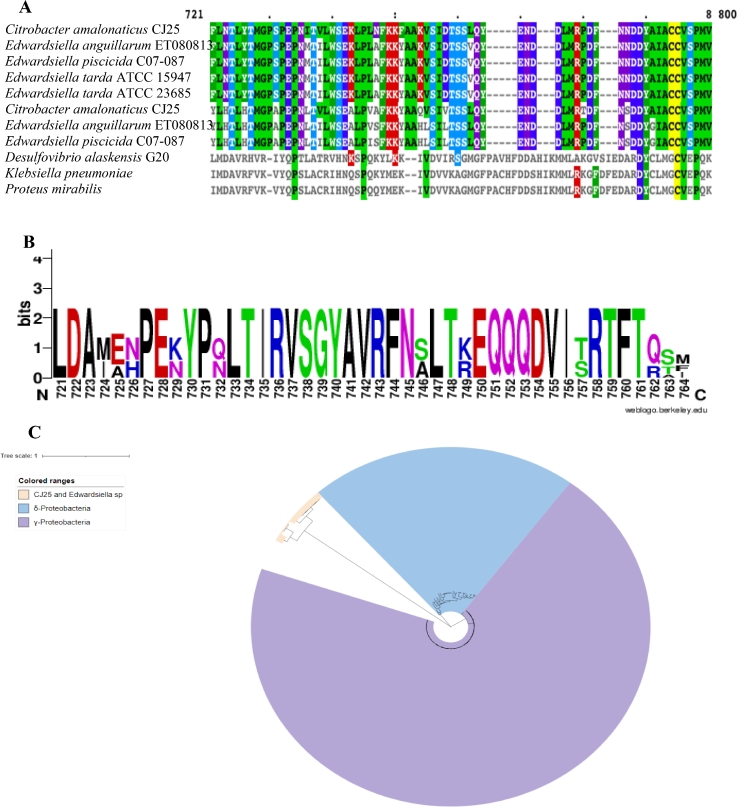
The identity of the fecal isolate was initially determined by 16S rRNA gene sequence analysis. The 16S rRNA was amplified using bacterial universal primers 1492F/27R (Lane et al., 1985) and the universal primers for the V4 region 515F/806R(Frank et al., 2008) and we obtained a single gene product from each primer pair. The amplicons were purified and sequenced using Sanger sequencing and the sequences obtained were checked for identity using NCBI Blastn (Altschul et al., 1990), the E-value was 0 with 99% identity to Citrobacter amalonaticus. The whole genome of this isolate was sequenced as the 16S rRNA region and did not show a 100% match with any of the known Citrobacter strains, as also evidenced by the pairwise ANI scores as well (Supplementary Table 2), hence we predict it to be a novel strain. We isolated the whole genome using the JGI Bacterial genomic DNA isolation using CTAB method (William et al., 2004). The average molecular weight of the whole genome fragments (Fig. S1) obtained was 20 kb detected via fragment analyzer and the whole genome was sequenced using PacBio RS II at University of Delaware. The core genome SNP tree from our sequenced whole genome was constructed using the KBase Fast Tree 2 suite (Price et al., 2010), the genomes of the 13 publicly available C. amalonaticus strains were obtained from NCBI (Fig. 2). The genome from Citrobacter freundii CFNIH1 was chosen as the outgroup for this tree. Strain CJ25 clusters closely with the strains FDAARGOS_490, FDAARGOS_165 S1285, and L8A but was unique and therefore the strain was named C. amalonaticus CJ25. Microscopic examination of CJ25 showed short rods. The bacteria were grown under aerobic and anaerobic conditions and no apparent changes in the morphology of the organism were observed. Upon close examination of the genome, we were unable to detect any cutCD gene clusters in its genome. CutC from Citrobacter freundii (Anwar et al., 2018) was used to blast against the CJ25 genome but no CutC/D were found to be encoded. However, by performing a blastp search using a CutC from Klebsiella pneumonia (Kalnins et al., 2015), we identified GRE homologs annotated as pyruvate formate lyases, but the identities were low, ranging from 20% - 36%, when compared to CutC. CJ25 appeared to have three genes that were annotated to encode pyruvate formate lyases and Clustal Omega (McWilliam et al., 2013) was used to do a multiple sequence alignment of the 2 most probable pyruvate formate lyases with 3 known CutCs from K. pneumoniae, Proteus mirabilis and Desulfovibrio alaskenis G20 (Kalnins et al., 2015; Jameson et al., 2016; Craciun and Balskus, 2012). The conserved residues differed significantly from the CutCs that are well characterized. Interestingly, Edwardsiella tarda was recently shown to produce TMA when grown with choline via an unknown pathway but the organism encodes pyruvate formate lyases in its genome and no evident cut gene clusters. These pyruvate formate lyases showed a 33% identity with characterized CutC of P. mirabilis (Jameson et al., 2018). A multiple sequence alignment (MSA) of pyruvate formate lyases sequences obtained from accession IDs (AIJ06655.1, AGH74192.1, GAC65756.1, EFE22433.1, AIJ09213.1) provided in Jameson et al., 2018 and the pyruvate formate lyases from CJ25 showed a very high similarity and conserved regions, The 3 CutCs from Klebsiella pneumoniae, Proteus mirabilis and Desulfovibrio alaskenis G20 were also included in the MSA (Fig. 4), critical CutC residues were missing from the sequences in CJ25 and Edwardsiella species. Accession IDs of all sequences used are presented in Table S1. The WebLogo image shows the conserved amino acid residues throughout the sequences in CJ25 and Edwardsiella species. The phylogenetic tree (Fig. 4) shows the clustering of the potential enzymes from CJ25 and Edwardsiella species (orange) along with CutC sequences from gammaproteobacteria (purple) and deltaproteobacteria (blue).
Fig. 3.
Microscopic examination of C. amalonaticus CJ25 (A) SEM image of strain CJ25 at 1000x magnification. Scale bar indicates 10 μm. (B) SEM image of strain CJ25 at 20,000x magnification. Scale bar indicates 200 nm. .
Fig. 2.
Core genome SNP tree of publicly available strains of C. amalonaticus. The tree was constructed using FastTree 2 and the tree scale and bootstrap values are indicated.
Fig. 4.
A: Multiple Sequence alignment of pyruvate formate lyases from CJ25 and characterized CutCs from Klebsiella pneumoniae, Proteus mirabilis, and Desulfovibrio alaskenis G20. B: Multiple sequence alignment between E. tarda sp and the same dataset in 4a and a WebLogo showing conserved residues between pyruvate-formate lyases from CJ25 and pyruvate-formate lyases from E. tarda sp. C: Phylogenetic tree of pyruvate formate lyases and CutC enzymes from gammaproteobacteria (blue) and delta proteobacteria (purple).
Investigation of choline degradation by C. amalonaticus CJ25
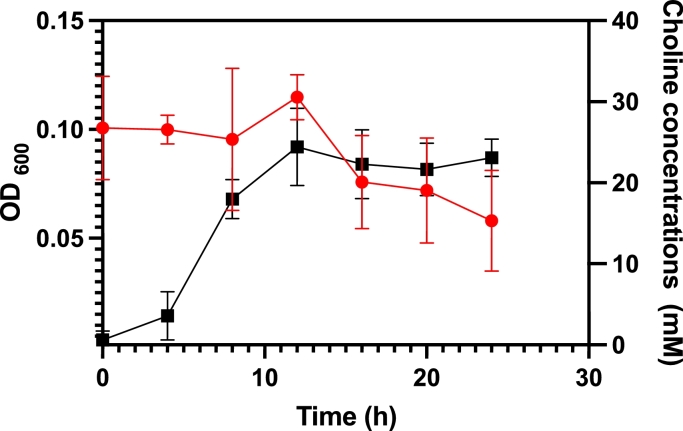
Although the genome of CJ25 does not encode any known CutC/D it can utilize choline as a sole carbon and energy source. This was confirmed using NMR 400 MHz to assess decreasing choline concentrations over time (Fig. 5). Choline depletion coincided with growth of the organism at the exponential phase which suggests that CJ25 utilizes choline while being metabolically active (Fig. S3). An abiotic control experiment with choline in the medium, but no culture, at 37 °C showed no loss of choline (data not shown). The NMR 400 MHz spectra also showed accumulation of acetate as choline is depleted, but the amount of acetate produced does not appear to be stoichiometric to choline utilization. GC analysis did not show any TMA production by CJ25 after 24 h. E. fergusonii was used as a positive control organism, which showed TMA production (Fig. S4). This supports our hypothesis that CJ25 is not generating TMA during choline degradation. The lower limit of detection of TMA by GC, using this method, was approximately 500 µM. To further investigate the potential TMA production, LC-MS/MS was also employed as a more sensitive technique. The results confirmed that no TMA accumulated during the growth of CJ25 on choline. The medium contained trace amounts of TMA but the concentration actually decreased slightly from 0 h to 24 h (Fig. S5).
Fig. 5.
Growth of CJ25 coincides with loss of choline in the culture over time. Choline was quantified using 400 MHz NMR.
Discussion
The isolation of C. amalonaticus CJ25 by enrichment of fecal samples from healthy patient 25 demonstrates the potential for undiscovered members of the gut microbiota, and gammaproteobacteria specifically, that can degrade choline. QAs such as choline, carnitine, and gamma-butyrobetaine are dietary components and have been directly linked with cardiovascular diseases in humans (Ahmad et al., 2019; Koeth et al., 2013) as these QAs are present in the highest concentrations in red meat and its consumption amounted to 111.4 pounds per capita in 2020 alone (per-Capita-Consumption-of-Red-Meat-in-the-Us-2017–2030.Pdf, n.d.). Choline, being a significant component of red meat, can lead to formation of TMA during its breakdown in the gut via bacterial choline trimethylamine lyase (CutC) enzymes and their cognate activator (CutD) enzymes (Rath et al., 2017). The genes of the cut cluster are found in a diverse range of anaerobic and facultative microorganisms in the gut that have been identified using bioinformatics tools (Martínez-del Campo et al., 2015). Additionally, there is growing evidence that, due to horizontal gene transfer between microbes in the gut, a number of genes encoding metabolic pathways are exchanged in the gut environment (Groussin et al., 2021). A majority of the bacterial species possessing the cut clusters have been reported to have mobile genetic elements encoded near these clusters which makes it plausible for bacteria to acquire cut genes even if they are not known to have any encoded in their genomes (Martínez-del Campo et al., 2015). To explore the potential choline degradation pathway in CJ25, we sequenced the genome of this organism and analyzed its genome on NCBI and JGI. A Blastp search against the well-characterized CutC enzymes from K. pneumoniae, P. mirabilis and D. alaskenis G20 showed the presence of genes encoding three GRE enzymes but did not reveal any genes encoding an apparent CutC, because all lacked the critical residues known to be involved in the CutC enzymatic mechanism (Craciun and Balskus, 2012). Furthermore, no putative COG5598 or other corrinoid dependent methyltransferases were found encoded in the genome when bona fide MttB, MtbB, or MtmB methyltransferases were used as query sequences (Ticak et al., 2014). However, we were able to identify genes encoding GRE homologs annotated as pyruvate formate lyases that show very high similarity with enzymes encoded in Edwardsiella tarda ATCC23685 and other strains reported by Jameson et al., 2018. Furthermore, genomic analysis indicated the presence of microcompartment genes in CJ25. However, only acetate production was observed by NMR 400 MHz and TMA production was not detected via either GC or NMR 400 or 500 MHz. Further analysis by LC-MS/MS showed that although minimal TMA could be detected, no TMA accumulated during the growth of the organism on choline. It is possible that CJ25 could be using a CutC-like GRE enzyme to degrade choline via an unknown mechanism. Furthermore, anaerobic bacteria in the gut use oxygen-tolerant or -intolerant GREs that play a critical role in metabolism and growth (Dragičević et al., 2015). In Firmicutes, fermentation of glutamate via 3-methylaspartate pathways or 2-hydroxyglutarate pathway yields ammonia, acetate, butyrate, and CO2 as the end products (Buckel, 2021). Additionally, many oxygen-tolerant and -intolerant GREs catalyze the same reactions or act as alternative pathways leading to formation of the same end products. A pyruvate formate lyase homolog from CJ25 may give the organism the potential to ferment choline anaerobically, yielding acetate as one of the end products. Genome analysis of CJ25 revealed genes encoding bacterial microcompartment proteins that were clustered separately from the pyruvate formate lyase genes in the genome. These microcompartment genes are analogous to the ethanolamine utilization cluster and 1,2- propanediol cluster (Dank et al., 2021; Kaval et al., 2019) which is also encoded in the genome. One possibility is that CJ25 is using these microcompartments to diffuse choline and using the pyruvate formate lyases to break it down into acetate and an unknown product(s). Since this organism does not accumulate TMA during growth on choline, it represents a paradigm shift in the current model for choline degradation and holds a potential for use as a microbe for probiotic therapy. The human gut microbiome is a vastly underexplored field and, by discovering more members of the human microbiota like CJ25, we can explore possibilities of potentially beneficial bacteria and implement them as part of therapeutic approaches for individuals suffering from or prone to CVD. There are a couple of possibilities of how choline could be utilized through other routes and there is evidence of alternate choline metabolism pathways where choline is oxidized to betaine via choline monooxygenase or demethylated to dimethylethanolamine via choline specific methyltransferases (Rajaie and Esmaillzadeh et al., 2011; K.A. Shipkowski et al., 2011). Future work, including proteomic analysis, will be crucial in providing insights into the proteins that are highly produced during the growth of CJ25 on choline and other quaternary amines like carnitine. Transmission electron microscopy can also be applied to examine if CJ25 forms microcompartments during growth on choline, as two different operons are found in its genome. This novel choline-degrading isolate C. amalonaticus CJ25 does not accumulate TMA and therefore utilizes an unknown non-atherogenic mechanism for choline utilization.
Funding
This research was supported by the National Institutes of Health (grant number: 1R01DK109345). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This material is based upon work supported by the National Science Foundation under Grant No. (Award Number: 1818178). Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation. This work was also supported by internal fudning from Miami University.
CRediT authorship contribution statement
Jyoti Kashyap: Investigation, Methodology, Writing – review & editing. Jeffery R. Ringiesn: Investigation, Methodology. Nathan Schwab: Methodology. Donald J. Ferguson: Supervision, Conceptualization, Funding acquisition, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We acknowledge and thank the staff (Dr. Andor Kiss & Ms. Xiaoyun Deng) of the Center for Bioinformatics & Functional Genomics (CBFG) at Miami University for instrumentation and computational support. We thank Dr. Olga Shevchenko and Dr. Brewster Kingham in the University of Delaware DNA Sequencing and Genotyping Center for assistance with sequencing the whole genome of C. amalonaticus CJ25. We thank Mr. Roshan Timsina for his preparation of samples for HPLC-MS/MS analysis of TMA levels. We thank Dr. Gong Wu and colleagues at The Ohio State University Campus Chemical Instrument Center Mass Spectrometry and Proteomics Facility (CCIC MSP) for assistance with the HPLC-MS/MS measurements. The CCIC MSP is supported by NIH Award Number Grant P30 CA016058.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.crmicr.2022.100157.
Appendix. Supplementary materials
References
- Ahmad A.F., Dwivedi G., O'Gara F., Caparros-Martin J., Ward N.C. The gut microbiome and cardiovascular disease: current knowledge and clinical potential. Am. J. Physiol. - Heart Circulat. Physiol. 2019;317(5):H923–H938. doi: 10.1152/AJPHEART.00376.2019. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Anwar S., Bhandari U., Panda B.P., Dubey K., Khan W., Ahmad S. Trigonelline inhibits intestinal microbial metabolism of choline and its associated cardiovascular risk. J. Pharm. Biomed. Anal. 2018;159:100–112. doi: 10.1016/j.jpba.2018.06.027. [DOI] [PubMed] [Google Scholar]
- Avula, Ziegler C., Bremer E., Krämer R., Zeisel S.H., Da Costa K.A., Heijthuijsen J.H.F.G., Hansen T.A., Knauf G.A., Cunningham A.L., Kazi M.I., Riddington I.M., Crofts A.A., Cattoir V., Trent M.S., Davies B.W., Rajakovich L.J., Fu B., Bollenbach M., …, Arias J.L. The diet – microbe morbid union Industry-compatible graphene transistors. Appl. Environ. Microbiol. 2018;109(1):1–29. doi: 10.1186/s40168-019-0704-8. [DOI] [Google Scholar]
- Buckel W. Energy conservation in fermentations of anaerobic bacteria. Front. Microbiol. 2021;12(September):1–16. doi: 10.3389/fmicb.2021.703525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craciun S., Balskus E.P. Microbial conversion of choline to trimethylamine requires a glycyl radical enzyme. Proc. Natl. Acad. Sci. U.S.A. 2012;109(52):21307–21312. doi: 10.1073/pnas.1215689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dank A., Zeng Z., Boeren S., Notebaart R.A., Smid E.J., Abee T. Bacterial microcompartment-dependent 1,2-propanediol utilization of propionibacterium freudenreichii. Front. Microbiol. 2021;12(May):1–10. doi: 10.3389/fmicb.2021.679827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragičević I., Barić D., Kovačević B., Golding B.T., Smith D.M. Non-enzymatic ribonucleotide reduction in the prebiotic context. Chem. Eur. J. 2015;21(16):6132–6143. doi: 10.1002/chem.201405741. [DOI] [PubMed] [Google Scholar]
- Durack J., Lynch S.V. The gut microbiome: relationships with disease and opportunities for therapy. J. Exp. Med. 2019;216(1):20–40. doi: 10.1084/jem.20180448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenbogen J.B., Jiang R., Kountz D.J., Zhang L., Krzycki J.A. The MttB superfamily member MtyB from the human gut symbiont Eubacterium limosum is a cobalamin-dependent γ-butyrobetaine methyltransferase. J. Biol. Chem. 2021;101327 doi: 10.1016/j.jbc.2021.101327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J.A., Reich C.I., Sharma S., Weisbaum J.S., Wilson B.A., Olsen G.J. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl. Environ. Microbiol. 2008;74(8):2461–2470. doi: 10.1128/AEM.02272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groussin M., Poyet M., Sistiaga A., Kearney S.M., Moniz K., Noel M., Hooker J., Gibbons S.M., Segurel L., Froment A., Mohamed R.S., Fezeu A., Juimo V.A., Lafosse S., Tabe F.E., Girard C., Iqaluk D., Nguyen L.T.T., Shapiro B.J., Alm E.J. Elevated rates of horizontal gene transfer in the industrialized human microbiome. Cell. 2021;184(8):2053–2067. doi: 10.1016/j.cell.2021.02.052. e18. [DOI] [PubMed] [Google Scholar]
- Herring T.I., Harris T.N., Chowdhury C., Mohanty S.K. Crossm A bacterial microcompartment is used for choline. J. Bacteriol. 2018;200(10):1–13. doi: 10.1128/JB.00764-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyles L., Jiménez-Pranteda M.L., Chilloux J., Brial F., Myridakis A., Aranias T., Magnan C., Gibson G.R., Sanderson J.D., Nicholson J.K., Gauguier D., McCartney A.L., Dumas M.E. Metabolic retroconversion of trimethylamine N-oxide and the gut microbiota. Microbiome. 2018;6(1):73. doi: 10.1186/s40168-018-0461-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson E., Fu T., Brown I.R., Paszkiewicz K., Purdy K.J., Frank S., Chen Y. Anaerobic choline metabolism in microcompartments promotes growth and swarming of Proteus mirabilis. Environ. Microbiol. 2016;18(9):2886–2898. doi: 10.1111/1462-2920.13059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson E., Quareshy M., Chen Y. Methodological considerations for the identification of choline and carnitine-degrading bacteria in the gut. Methods. 2018;149:42–48. doi: 10.1016/j.ymeth.2018.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipkowski1 K.A., Sanders1 M., McDonald2 J.D., Garner2 C.E., Doyle-Eisele2 M., Wegerski2 S.W. Comparative disposition of dimethylaminoethanol and choline in rats and mice following oral or intravenous administration. Physiol. Behav. 2011;176(10):139–148. doi: 10.1016/j.taap.2019.05.011.Comparative. C. J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalnins G., Kuka J., Grinberga S., Makrecka-Kuka M., Liepinsh E., Dambrova M., Tars K. Structure and function of CutC choline lyase from human microbiota bacterium Klebsiella pneumoniae. J. Biol. Chem. 2015;290(35):21732–21740. doi: 10.1074/jbc.M115.670471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaval, K.G., Gebbie, M., Goodson, J.R., Cruz, M.R., Winkler, W.C., & Garsin, A. (2019). Ethanolamine utilization and bacterial microcompartment. 201(10), 1–13. [DOI] [PMC free article] [PubMed]
- Koeth R.A., Wang Z., Levison B.S., Buffa J.A., Org E., Sheehy B.T., Britt E.B., Fu X., Wu Y., Li L., Smith J.D., Didonato J.A., Chen J., Li H., Wu G.D., Lewis J.D., Warrier M., Brown J.M., Krauss R.M., Hazen S.L. Intestinal microbiota metabolism of l-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 2013;19(5):576–585. doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kountz D.J., Behrman E.J., Zhang L., Krzycki J.A. MtcB, a member of the MttB superfamily from the human gut acetogen Eubacterium limosum, is a cobalamindependent carnitine demethylase. J. Biol. Chem. 2020;295(34):11971–11981. doi: 10.1074/jbc.RA120.012934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane D.J., Pace B., Olsen G.J., Stahlt D.A., Sogint M.L., Pace N.R. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses (reverse transcriptase/dideoxynudeotide) Evolution (N Y) 1985;82(October):6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M.A., Blackshields G., Brown N.P., Chenna R., Mcgettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., Thompson J.D., Gibson T.J., Higgins D.G. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Li E., Bestor T.H., Jaenisch R., Shreiner A.B., Kao J.Y., Young V.B., Turnbaugh P.J., Ley R.E., Mahowald M.A., Magrini V., Mardis E.R., Gordon J.I., Wilson R.C., Doudna J.A. Dicer (E-7): sc-393328. Cell. 2015;69(1) doi: 10.1097/MOG.0000000000000139.The. [DOI] [Google Scholar]
- Martínez-del Campo A., Bodea S., Hamer H.A., Marks J.A., Haiser H.J., Turnbaugh P.J., Balskusa E.P. Characterization and detection of a widely distributed gene cluster that predicts anaerobic choline utilization by human gut bacteria. MBio. 2015;6(2) doi: 10.1128/mBio.00042-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavromatis K., Ivanova N.N., Chen I.min A., Szeto E., Markowitz V.M., Kyrpides N.C. The DOE-JGI standard operating procedure for the annotations of microbial genomes. Stand. Genomic. Sci. 2009;1(1):63–67. doi: 10.4056/sigs.632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWilliam H., Li W., Uludag M., Squizzato S., Park Y.M., Buso N., Cowley A.P., Lopez R. Analysis tool web services from the EMBL-EBI. Nucleic Acids Res. 2013;41(Web Server issue):597–600. doi: 10.1093/nar/gkt376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliphant K., Allen-Vercoe E. Macronutrient metabolism by the human gut microbiome: major fermentation by-products and their impact on host health. Microbiome. 2019;7(1):1–15. doi: 10.1186/s40168-019-0704-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- per-capita-consumption-of-red-meat-in-the-us-2017-2030. (n.d.).
- Picking J.W., Behrman E.J., Zhang L., Krzycki J.A. MtpB, a member of the MttB superfamily from the human intestinal acetogen Eubacterium limosum, catalyzes proline betaine demethylation. J. Biol. Chem. 2019;294(37):13697–13707. doi: 10.1074/jbc.ra119.009886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price M.N., Dehal P.S., Arkin A.P. FastTree 2 - Approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;5(3) doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajaie S., Esmaillzadeh A. Dietary choline and betaine intakes and risk of cardiovascular diseases: review of epidemiological evidence. ARYA Atheroscler. 2011;7(2):78–86. [PMC free article] [PubMed] [Google Scholar]
- Rajakovich L.J., Fu B., Bollenbach M., Balskus E.P. Proceedings of the National Academy of Sciences of the United States of America (Vol. 118, Issue 32) 2021. Elucidation of an anaerobic pathway for metabolism of L-carnitine-derived γ-butyrobetaine to trimethylamine in human gut bacteria. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath S., Heidrich B., Pieper D.H., Vital M. Uncovering the trimethylamine-producing bacteria of the human gut microbiota. Microbiome. 2017;5(1):1–14. doi: 10.1186/S40168-017-0271-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano K.A., Vivas E.I., Amador-noguez D., Rey F.E. From diet and accumulation of the proatherogenic metabolite. Intest. Microb. Choline Metab. 2015;6(2):1–8. doi: 10.1128/mBio.02481-14.Romano. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ticak T., Kountz D.J., Girosky K.E., Krzycki J.A., Ferguson D.J. A nonpyrrolysine member of the widely distributed trimethylamine methyltransferase family is a glycine betaine methyltransferase. Proc. Natl. Acad. Sci. 2014;111(43):E4668–E4676. doi: 10.1073/pnas.1409642111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- William S., Feil H., Copeland A. Bacterial DNA isolation CTAB protocol bacterial genomic DNA isolation using CTAB materials & reagents. Doe Joint Genome Inst. 2004;(4) http://my.jgi.doe.gov/general/protocols/JGI-Bacterial-DNA-isolation-CTAB-Protocol-2012.pdf [Google Scholar]
- Zeisel S.H., Da Costa K.A. Choline: an essential nutrient for public health. Nutr. Rev. 2009;67(11):615–623. doi: 10.1111/j.1753-4887.2009.00246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W., Gregory J.C., Org E., Buffa J.A., Gupta N., Wang Z., Li L., Fu X., Wu Y., Mehrabian M., Sartor R.B., McIntyre T.M., Silverstein R.L., Tang W.H.W., Didonato J.A., Brown J.M., Lusis A.J., Hazen S.L. Gut microbial metabolite TMAO enhances platelet hyperreactivity and thrombosis risk. Cell. 2016;165(1):111–124. doi: 10.1016/j.cell.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Jameson E., Crosatti M., Schäfer H., Rajakumar K., Bugg T.D.H., Chen Y. Carnitine metabolism to trimethylamine by an unusual Rieske-type oxygenase from human microbiota. Proc. Natl. Acad. Sci. U.S.A. 2014;111(11):4268–4273. doi: 10.1073/pnas.1316569111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.