Abstract
Clostridioides difficile infection (CDI) is a life-threatening disease caused by the Gram-positive, opportunistic intestinal pathogen C. difficile. Despite the availability of antimicrobial drugs to treat CDI, such as vancomycin, metronidazole, and fidaxomicin, recurrence of infection remains a significant clinical challenge. The use of live commensal microorganisms, or probiotics, is one of the most investigated non-antibiotic therapeutic options to balance gastrointestinal (GI) microbiota and subsequently tackle dysbiosis. In this review, we will discuss major commensal probiotic strains that have the potential to prevent and/or treat CDI and its recurrence, reassess the efficacy of probiotics supplementation as a CDI intervention, delve into lessons learned from probiotic modulation of the immune system, explore avenues like genome-scale metabolic network reconstructions, genome sequencing, and multi-omics to identify novel strains and understand their functionality, and discuss the current regulatory framework, challenges, and future directions.
Keywords: Probiotics, C. difficile, inhibition, virulence, immunomodulatory, genomic exploration, discovery informatics, metabolomics, multi-omics, regulatory requirements
Introduction:
Clostridioides difficile (C. difficile), also known as Clostridium difficile, is the most common infectious agent that leads to healthcare-associated diarrhea [1, 2]. Although first identified in 1935 [3], it was not until recently that the bacterium was implicated as the causative agent of pseudomembranous colitis with the severity and rate of C. difficile-associated disease (CDAD) increasing since the emergence of hypervirulent strains [4, 5]. As per recent estimates provided by the U.S. Centers for Disease Control and Prevention (CDC), 223,900 people were hospitalized with C. difficile infection (CDI) in 2017, which resulted in 12,800 fatalities in the U.S. alone [6] .
CDI has been primarily associated with the use of antibiotics to treat other bacterial infections and is highly prevalent in healthcare settings [7, 8] . Broad-spectrum antibiotics modulate the gut microflora, which facilitates C. difficile colonization in the large intestine [9–11]. Indeed, antibiotic treatment induces class-specific dysbiosis in the microbiota and a recent study noted remarkably low abundance of Ruminococcus, Blautia, Porphyromonas, Bifidobacteria, Ezakiella and Ozoribacter spp. along with a higher abundance of Enterococcus in the baseline samples collected from CDI patients [12]. Furthermore, antibiotics might promote C. difficile growth by altering bile acid metabolism. Specific bile acid signals are the prerequisite for the germination of C. difficile spores into vegetative cells [13]. Antibiotic use and abuse induce intestinal microbiome dysbiosis which results in population decline for gut microorganisms that possess 7-α dehydroxylase activity, which is involved in bile metabolism. This, in turn, brings about a reduction in secondary bile acids like deoxycholate. The decrease in deoxycholate levels is accompanied by an increase in taurocholate, a primary bile acid. Taurocholate stimulates the germination of C. difficile spores to metabolically active and toxin-producing cells [10, 14, 15].
Despite the pitfalls associated with antibiotics, they remain the treatment of choice for CDI. Current therapeutic options for mild-to-moderate CDI cases include the glycopeptide vancomycin or the macrolide fidaxomicin [16, 17]. However, there is no effective countermeasure for CDI recurrence wherein 1 out of 5 patients experience recrudescence of infection [18]. Treatment of subsequent recurrent infections can be challenging as the patient fails to mount an effective immune response against the toxins secreted by vegetative C. difficile cells [19, 20].
One biotherapeutic approach that has garnered considerable attention in recent years is fecal microbiota transplantation (FMT) [21, 22]. FMT is based on infusion of liquid filtrate feces from a healthy donor into the gut of a recipient to treat/control disorders where there has been a disruption in the community structure of the gut microflora [23, 24]. FMT is effective in resolving ~80 % of recurrent infections [25, 26]. However, FMT administration can result in the transmission of enteropathogenic Escherichia coli (EPEC) and Shiga toxin-producing E. coli (STEC). In fact, investigational FMT administration led to the development of invasive antibiotic-resistant E. coli infections in two immunocompromised individuals, with one individual dying [27]. These fatalities have prompted the need for intensive clinical work to comply with all related regulatory agencies. For example, the U.S. Food and Drug Administration (FDA) has mandated additional requirements for FMT that include the screening of donors for risk and carriage of multidrug-resistant organisms [27]. A recent population evidence-based study noted that washed microbiota preparation via microfiltration significantly reduced FMT-related adverse events as compared to conventional FMT [28]. However, there is a lack of standardization of FMT protocols and different methods of delivery, such as colonoscopy, nasogastric tube, retention enema, or orally delivered encapsulated stool, have different levels of efficacy and associated risks [29, 30]. Thus, numerous questions regarding FMT remain to be addressed highlighting the need for therapeutics that can provide sustained resolution of clinical symptoms.
The dwindling number of antibiotics effective against CDI and CDI recurrence has led to an increase in probiotic testing and research. As defined by the Food and Agriculture Organization of the United Nations and the World Health Organization (FAO/WHO) in 2002, probiotics are “live microorganisms which, when administered in adequate amounts, confer a health benefit on the host” [31–33]. Currently, the commercialization of probiotics has led to the development of a multibillion-dollar industry, and probiotics now constitute one of the most consumed supplements [34–37].
Emerging insights from probiotic research interventions provide mechanistic aspects for their putative health benefits [38–42]. However, clinical trials that investigated the effect of supplementing probiotics to antibiotics therapy in treating CDI revealed mixed results; some trials validated the benefits of probiotics while other trials found negative results. In this perspective, we will delve into studies conducted on different probiotic strains to evaluate their ability to inhibit C. difficile in vitro and in vivo, highlight the efficacy of probiotics from a clinical context, and focus on the immunomodulatory properties of probiotics. We will also consider avenues like genome-scale metabolic network reconstructions that can identify novel strains of probiotics, discuss the role of genome sequencing in understanding the functionality of probiotics, highlight the importance of a multi-omics approach in understanding the C. difficile-lumen-intestine interaction as a measure to evaluate probiotic efficacy, and explore the current regulatory framework and possible future directions.
Probiotic efficacy in a preclinical context:
Corthier et al., in 1985, conducted one of the first studies that revealed the positive effect of probiotics in mice infected with C. difficile. In this study, gnotobiotic mice that were inoculated with Bifidobacterium bifidum or E. coli exhibited reduced cecal cytotoxin levels and survived C. difficile challenge without exhibiting diarrhea [43]. Since this study was published, numerous studies have been conducted to investigate the ability of different probiotic strains to attenuate virulence or directly inhibit growth of C. difficile. The following section highlights the notable probiotic strains that have been proposed as plausible interventions based on findings conducted mostly using in vitro cell culture studies and a few in vivo models.
Lactobacilli:
The intestinal commensal organisms Lacticaseibacillus paracasei and Lactiplantibacillus plantarum demonstrated antagonistic activity against C. difficile strains in vitro. The antagonistic activity of the Lactobacillus sp. was strain-dependent and correlated with their ability to produce lactic acid and hydrogen peroxide (H2O2) indicating the probable role of lactic acid and H2O2 behind the antagonistic interaction (Fig. 1) [44]. A different study concluded that the cell-free supernatant (CFS) of L. plantarum, Lacticaseibacillus rhamnosus, Lacticaseibacillus casei, and Ligilactobacillus salivarius exhibits a direct inhibitory effect on C. difficile in vitro in a pH-dependent manner hinting towards the production of organic acids[45].
Fig.1:
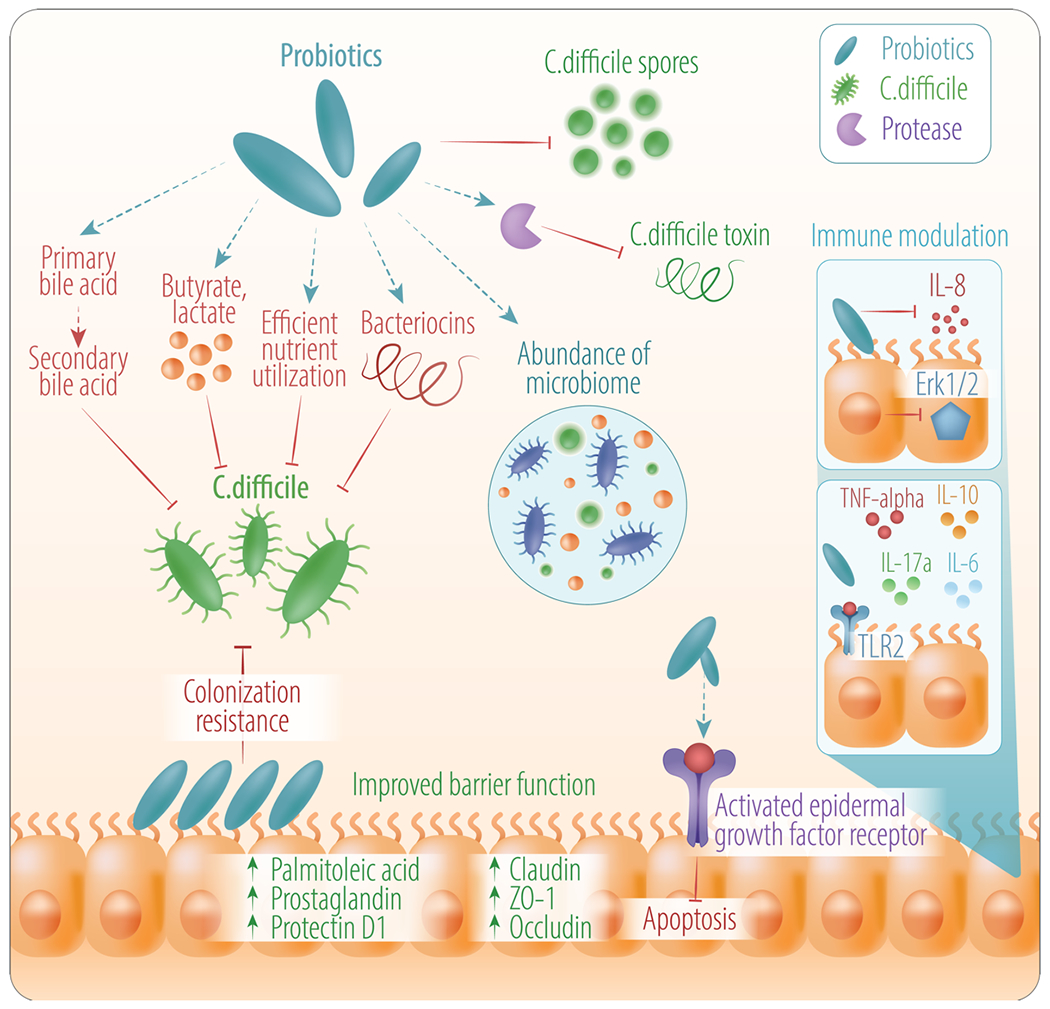
Schematic depiction of the effect of probiotics in the context of treating/preventing CDI. Probiotics can have several effects that can be useful for treating/preventing C. difficile including competition for nutrients, resulting in inhibition of pathogen outgrowth through conversion of bile acids, improved barrier function, abundance of microbiome, and modulation of the immune system. Probiotics an also play a potential role in inhibition of C. difficile virulence factors (toxins and spores).
Limosilactobacillus reuteri 17938 is one of the most studied probiotics in clinical trials for different applications including inhibiting growth of C. difficile. A study by Spinler et al., 2017 identified L. reuteri 17938 as a potential candidate for an adjunct probiotic therapy to be used alongside antibiotics to treat CDI [46]. L. reuteri converts glycerol into a potent broad-spectrum antimicrobial compound, reuterin, that inhibits the growth of both Gram-positive and Gram-negative bacteria. Using fecal mini-bioreactor arrays (MBRA), the investigators demonstrated a 5-log10 reduction in C. difficile count in glycerol-treated reactors containing L. reuteri. Ex vivo inhibition of C. difficile growth and germination was noted in the cecal contents procured from germ-free mice when treated with a combination of L. reuteri and glycerol. Moreover, supplementation of L. reuteri with glycerol did not show a drastic and statistically significant change in community structure in the MBRA samples [46]. A more recent study noted the ability of reuterin to inhibit the growth of metabolically active C. difficile by inducing reactive oxygen species (ROS) production resulting in a shift in carbon metabolism followed by reduced toxin synthesis . Reuterin could also inhibit C. difficile outgrowth from spores. Importantly, L. reuteri is resistant to the antibiotics used for CDI treatment, including vancomycin, fidaxomicin, and metronidazole; furthermore, the susceptibility of the vegetative pathogen to these antibiotics was enhanced by L. reuteri, which makes it a promising strain for adjunct therapy [46, 47].
Lactobacilli-derived molecules can also attenuate virulence (Fig. 1). Lactobacilli have been found to inhibit the expression of virulence factors of C. difficile both in vitro and in vivo. Banerjee et al., in 2009, investigated the efficacy of six commercially available and conventional lactobacilli strains in mediating C. difficile-induced toxicity on a human enterocyte-like Caco-2 (human intestinal epithelial cell line) cell culture model. Indeed, CFS derived from Lactobacillus delbrueckii ssp. protected Caco-2 cells from the cytotoxic effects of C. difficile toxins [48]. When Caco-2 cells were exposed to the CFS from a co-culture of L. delbrueckii and C. difficile, similar results were generated, which hinted at the presence of one or more antitoxic components in L. delbrueckii-conditioned medium that possibly exerted a proteolytic effect on the C. difficile toxins. Furthermore, CFS from L. delbrueckii inhibited adhesion of C. difficile to the Caco-2 monolayer [49]. However, the mechanistic aspects that could decrease the adherence of C. difficile to Caco-2 cells and detoxification remain to be elucidated.
Another study investigated the ability of Lactobacillus acidophilus in modulating C. difficile virulence [50]. At a low pH, the probiotic inhibited growth of the pathogen, which was attributed to the production of lactic acid (Fig. 1). The cell extract of L. acidophilus GP1B had an inhibitory effect on the production of the quorum sensing regulator autoinducer-2 (AI-2) in C. difficile and resulted in downregulation of gene expression associated with the production of AI-2 and toxins A and B (tcdA, tcdB, and sigma factor txeR). L. acidophilus GP1B was further found to confer protection to C. difficile-infected mice, which was evident from the structural integrity of the intestinal epithelial cells in L. acidophilus-treated mice. [50]. Another recent study revealed the efficacy of CFS from L. acidophilus La-5 in reducing attachment of C. difficile to human epithelial cells and conferring protection to two human epithelial cell lines, HT-29 and Caco-2, from the cytopathic and cytotoxic effect of C. difficile CFS [51].
The surface layer proteins (S-layer) of Lactobacillus kefir, a member of the microbiota of kefir grains, antagonized the effect of both C. difficile-spent culture supernatant (SCS) and purified toxins on the African green monkey kidney epithelial (Vero) cells in an in vitro study. The inhibitory effect was higher for the S-layer proteins belonging to the aggregating L. kefir strains (strains harboring auto aggregation capacity) as compared to the non-aggregating strains, while whole cells of L. kefir failed to confer any protection [52, 53]. In a different study, L. lactis Centro de Investigación y Desarrollo en Criotecnología de Alimentos (CIDCA) 8221 and a mixture of kefir-isolated bacterial and yeast strains (L. kefir, L. plantarum, L. lactis ssp. lactis, Saccharomyces cerevisiae, and Kluveromyces marxianus) antagonized the cytotoxic effect of C. difficile toxins on Vero cells. The anti-toxin activity was attributed to the heat sensitive, protease-resistant components (greater than 10 kDa) present in the L. lactis CIDCA 8221 supernatant, which provided new insights into the anti-virulence activity of the kefir microbiome [54]. Additionally, the mixture of the kefir-isolated microorganisms conferred protection in vivo against CDI in a hamster model where hamsters treated with kefir-isolated microorganisms had a 100% survival rate as compared to the placebo group which had a survival rate of 28.5% [55].
Composed of three bacterial strains (L. acidophilus CL 1285, L. casei LBC80R, and L. rhamnosus CLR2), Bio-K+ is a commercial probiotic used as a supplement to prevent CDI. The probiotic formulation inhibited toxin A/B-producing strains of C. difficile. Furthermore, CFS from the three lactobacilli strains harbored anti-cytotoxic activity that protected Caco-2 cells from the cytotoxic effects of CFS obtained from C. difficile [56].
Saccharomyces boulardii:
S. boulardii, a non-pathogenic yeast, confers protection against C. difficile via indirect inhibitory mechanisms like attenuation of C. difficile virulence factors and modulation of the host immune response (Fig. 1). S. boulardii inhibited C. difficile toxin A-mediated receptor binding and the associated enterotoxic effects like intestinal secretion and mannitol permeability in a rat ileal loop model [57]. The significant reduction of the histologic damages caused by toxin A was attributed to a S. boulardii serine protease that hydrolyzed the toxin and thus prevented it from binding to glycoproteins present on the brush borders of the host epithelium [58]. S. boulardii-conditioned media also inhibited interleukin-8 (IL-8) production, induced by C. difficile toxin A in human mucosal epithelial colonocyte cells, in a dose-dependent manner. S. boulardii modulated the host signaling pathways pivotal to the intestinal inflammatory response and involved inhibition of ERK1/2 activation. S. boulardii supernatant also normalized toxin A-mediated fluid secretion and prevented the associated histopathologic changes in toxin A-treated ileal mucosa of mice [59]. Moreover, S. boulardii protease possessed enzymatic activity against C. difficile TcdB and inhibited the cytotoxic effects of both TcdA and TcdB on native human colonic mucosa and on colonic epithelial cells in vitro [60].
Bifidobacterium spp.:
Bifidobacteria have been evaluated for their ability to reduce the cytotoxic effect of clostridial toxins (Fig. 1). Bifidobacterium longum and Bifidobacterium breve reduced the toxic effect of C. difficile supernatant when exposed to a human intestinal epithelial cell line (HT-29) [61]. Of the strains that were investigated, B. longum IPLA20022 demonstrated the strongest ability to counteract the cytotoxic effect of clostridial toxins. The neutralized cell free supernatant (NCFS) obtained from the B. longum IPLA20022 strain permitted the least damage to the HT-29 cells, even after 22 hours of incubation with C. difficile supernatant; furthermore, the NCFS significantly reduced TcdA levels (12% remnant TcdA). Additionally, HT-29 cells incubated with the NCFS from B. longum IPLA20022 displayed an intact F-actin cytoskeleton and retained the integrity of the epithelial monolayer, which could be correlated to the reduced amount of C. difficile toxins [61]. B. bifidum CIDCA 5310 and L. plantarum CIDCA 83114 also antagonized the biological activity of C. difficile in a separate study [62]. Detection of C. difficile toxin concentration in spent co-cultures of the pathogen and the probiotics revealed that B. bifidum CIDCA 5310 significantly reduced the concentration of TcdA in the supernatant. The integrity of the F-actin network in Vero cells was maintained when the pathogen was co-cultured with the B. bifidum strain. Additionally, co-culturing with both B. bifidum CIDCA 5310 and L. plantarum CIDCA 83114 successfully inhibited cell rounding and detachment that occurred in the presence of the C. difficile supernatant [62]. Another study by Wei et al., 2018 demonstrated the efficacy of a commercial B. longum JDM301 strain in attenuating CDI in vivo. Mice infected with C. difficile and treated with B. longum had a survival rate of 75% compared to 43% for untreated mice [63].
Bifidobacterium-mediated direct inhibition of C. difficile growth was observed for Bifidobacterium animalis ssp. lactis [45]. Another study assessed the antagonistic activity of SCS from Bifidobacterium strains isolated from newborn babies [64]. All the Bifidobacterium strains used in this study showed strain-dependent growth inhibition of C. difficile. The supernatant from Bifidobacterium CIDCA 5323 exhibited the most potent inhibitory effect with a 4-log10 decrease in viable C. difficile colony forming units (CFU) observed within 1-hour post-incubation. Additionally, some of the probiotic strains significantly reduced the adherence of C. difficile to colonic epithelial cells (Caco-2) [64].
Pediococcus pentosaceus:
P. pentosaceus, a potential probiotic belonging to the family Lactobacillaceae, is a human gut-borne bacterium that is known to possess antimicrobial activity against enteropathogens [65]. The efficacy of P. pentosaceus LI05 in a mouse model of CDI was recently assessed [66]. Interestingly, a 100% survival rate and reduced histopathological symptoms were observed for mice who were challenged with C. difficile VPI10463 and were orally administered P. pentosaceus LI05. The P. pentosaceus LI05-treated mice showed substantial alleviation of inflammatory mediators in the colon, inflammatory cytokines and chemokines in the serum, and CDI-induced disruption of zonula ocludens-1 (ZO-1), occludin, and claudin-1, which led to reduced histopathological symptoms (Fig. 1). Additionally, treatment with P. pentosaceus LI05 resulted in an increase in the abundance of beneficial microbiota like Lactobacillus, Prevotella, and Paraprevotella and curbed the proliferation of opportunistic pathogens like Escherichia and Flavonifractor (Fig. 1) [66].
Streptococcus thermophilus:
S. thermophilus, a thermophilic lactic acid bacterium, is traditionally used in the food industry and has been clinically established as a probiotic formulation that intervenes with the progression of chronic kidney disease [67, 68]. S. thermophilus filtrates have been found to exhibit a dose-dependent growth inhibitory effect on C. difficile along with a significant decrease in tcdA expression (Fig. 1). In addition, C. difficile-infected mice treated with S. thermophilus exhibited significantly less diarrhea and attenuated pathological features in the colon and cecum when compared to the controls [69]. The high levels of lactate (used as a marker for lactic acid which exhibits a bactericidal effect on C. difficile) observed in the lumen of mice treated with S. thermophilus was attributed to the better experimental outcome observed in terms of significantly less weight loss (46%), diarrhea, toxin levels, and cecal pathology [69].
Non-pathogenic Clostridium spp.:
The endogenous intestinal microflora of humans is dominated by members of the Firmicutes and Bacteroidetes phyla. Of the inferred microorganisms in the Firmicutes phylum, most belong to known butyrate-producing bacteria and are members of the Clostridia class [70]. Clostridium butyricum is one such non-pathogenic butyric acid-producing Clostridium spp. found in the intestine of humans that has been used clinically to prevent human gastrointestinal diseases like antibiotic-associated diarrhea [71]. C. butyricum MIYAIRI 588 (CBM588), when co-cultured with C. difficile, resulted in a complete loss of C. difficile toxicity. Spores of CBM588 and C. difficile, when co-inoculated, exhibited a greater inhibitory effect which suggests that C. butyricum spores can probably inhibit C. difficile spore germination in the gut [72]. Moreover, CBM588 prevented CDI in gnotobiotic mice (80% survival in mice pre-infected with CBM588 vs. 14.2% survival in untreated) [73]; furthermore, administration of CBM588 reduced the incidence of watery diarrhea and the fecal-free water content in rats infected with C. difficile [74].
Colonization resistance using non-toxigenic strains of C. difficile successfully prevented colonization of toxigenic C. difficile in a hamster model of CDI (Fig. 1) [75]. The hamsters were colonized with non-toxigenic C. difficile strains M3, M23, and T7 which conferred protection ranging between 87-97% against the toxigenic strains.
Clostridium scindens is a bile acid 7α-dehydroxylating intestinal bacterium that possesses potent anticlostridial activity. C. scindens expresses enzymes that can convert the primary bile acids cholate and chenodeoxycholate to two secondary bile acids, deoxycholate and lithocholate. Albeit the fact that all cholate conjugates act as germinants for C. difficile spores, the secondary bile acid deoxycholate inhibited the growth of vegetative C. difficile cells [13]. C. scindens strains, because of their ability to efficiently convert cholate to deoxycholate, inhibited C. difficile growth and pathogenesis in vitro, ex vivo, in gnotobiotic mice, and in a C. difficile murine model (Fig. 1) [76–78]. Additionally, C. scindens and Clostridium sordelli secrete the tryptophan-derived antibiotics turbomycin A and B that mediate growth inhibition of C. difficile by interfering with the process of cell division. The activity of turbomycin A and B was augmented in the presence of deoxycholic acid and lithocholic acid, which indicates a second mechanism by which C. scindens prevents CDI [79].
Enterococcus spp.:
A component of the human and the animal gut microflora, Enterococcus spp., particularly Enterococcus faecium and Enterococcus faecalis, have been investigated for their ability to inhibit C. difficile growth [80, 81]. A screening assay identified the CFS of three isolates from infant fecal samples, E. faecalis NM815, E. faecalis NM915, and E. faecium NM1015, that exhibited more than 50% inhibition of C. difficile growth in vitro. The inhibitory action was presumed to be due to antibacterial substances and organic acids secreted by the enterococcal strains. A co-culture of the three probiotic enterococcal strains with C. difficile resulted in a 5-log10 reduction in the C. difficile CFU count. Administration of a mixture of the three enterococcal strains to mice protected the epithelial cells of mice from dense inflammation and mucin depletion when compared to the untreated group [81].
Bacillus spp.:
Bacillus spores may be a potential probiotic option to prevent CDI. Oral delivery of Bacillus subtilis PXN21 spores, a component of a commercial probiotic product, improved the survival rate for mice infected with C. difficile in a study conducted by Chen et al., 2008 [82]. Mice treated with B. subtilis spores, prior to being infected with C. difficile, exhibited a survival rate of 41.6%, whereas treatment of mice post-infection with B. subtilis spores resulted in a 66.6% survival rate. Furthermore, damage to cell structure and tissue integrity was reduced in mice treated with B. subtilis spores compared to the untreated group. Additionally, the live PXN21 spores upregulated the expression of the Toll-like receptor 2 (TLR2) gene, which led to the induction of two proinflammatory cytokines, IL-6 and TNF-α, thereby stimulating the innate immune response of the host [83].
Bacillus clausii, has also been investigated for both direct inhibition of C. difficile and mitigation of its virulence factors. B. clausii CFS previously was shown to exhibit an inhibitory effect on growth of Gram-positive pathogens, including C. difficile [84]. Further studies revealed the presence of serine protease (M protease) in the supernatant of B. clausii that prevented damage induced by C. difficile toxins on Vero and Caco-2 cells. [85].
A spore-forming novel probiotic species, Bacillus coagulans, has recently gained attention in probiotic-associated industries, including food and pharmaceuticals, due to the bacterium’s stability and resistance to stress. B. coagulans attenuated chemokine release and reduced neutrophil influx and COX-2 expression in C. difficile-induced colitis in mice [86, 87]. The strain B. coagulans BC30 improved C. difficile-induced colitis in mice, with 66.7% of treated mice demonstrating normal stools on day 12 compared to 13.0% in the control group [86]. B. coagulans GBI-30, 6086 (GanedenBC30), also enhanced indices of C. difficile-induced colitis in mice and prevented recurrence after vancomycin withdrawal [86, 87].
Lachnospiraceae D4:
A component of the murine gut microflora and a member of the Lachnospiraceae family, Lachnospiraceae D4 was found to confer protection to mice challenged with C. difficile. Mice that were treated with an antibiotic cocktail of kanamycin, gentamicin, colistin, metronidazole, and vancomycin in water for three days followed by a single intra-peritoneal dose of clindamycin and subsequent inoculation with C. difficile either developed lethal CDI or exhibited mild forms of the infection. The microbial flora of mice that exhibited mild symptoms were found to be colonized with Lachnospiraceae [88]. To further verify this, Lachnospiraceae and E. coli were isolated from the ceca of mice, and their ability to mediate colonization resistance against C. difficile was evaluated. Interestingly, 80% of mice pre-colonized with Lachnospiraceae, but not E. coli, had lower levels of C. difficile colonization, less severe forms of CDI, exhibited minimal weight loss, and were clinically healthy two days post-infection [89]. In contrast, mice pre-colonized with murine E. coli strains and then subsequently infected with C. difficile succumbed to the infection. Efficient nutrient utilization by Lachnospiraceae was proposed as a plausible reason for the lower levels of C. difficile colonization observed (Fig. 1). Alternatively, the production of metabolites or a different mechanism altogether may have provided colonization resistance to C. difficile [89]. Although there are limited studies regarding the role of human Lachnospiraceae isolates in preventing C. difficile colonization, it is known that a depletion of Lachnospiraceae, along with Ruminococcaceae and other butyrate-producing bacteria, is common for individuals with CDI and non-infectious diarrhea [90]. Hence, members of the human Lachnospiraceae family need to be investigated as this might provide a lead for a clinically efficacious probiotic to limit CDI.
The role of Synbiotics in preventing C. difficile infection:
While a prebiotic can be defined as a selectable, fermented, non-digestible food ingredient that triggers beneficial changes in the composition of the gut microflora, a synbiotic can be defined as a “synergistic combination of prebiotics and probiotics” [33, 91, 92]. Synbiotics have been the subject of extensive research because of their beneficial effect on the host by improving the survival of live commensal microorganisms administered as supplements [93].
Synbiotics, such as the prebiotic xylitol combined with the probiotic L. plantarum Inducia, have been found to inhibit the germination of C. difficile spores in vitro and to protect hamsters from CDI in vivo [94]. In another study, a synbiotic formulation consisting of the probiotics L. plantarum F44, L. paracasei F8, B. breve 46, B. lactis 8:8, and the prebiotics galacto-oligosaccharides, isomalto-oligosaccharides, and resistant starch conferred protection against C. difficile in a mouse model of CDI with significant toxin inhibition observed [95]. Additionally, B. longum and B. breve, in the presence of short chain fructo-oligosaccharides, inhibited the growth of toxigenic C. difficile [96]. Furthermore, Faecalibacterium prausnitzii along with potato starch protected against C. difficile colonization in an antibiotic-treated mouse model, likely through immunomodulation [97].
Breast milk supplies human milk oligosaccharides (HMOs), a family of structurally diverse glycans that support infant gut colonization and are active in innate immunity. HMOs are prebiotics, and they promote the growth of certain bifidobacteria, including Bifidobacterium infantis and B. bifidum [98]. Furthermore, the human breast milk microbiome is rich in Staphylococcus, Streptococcus, and Propionibacterium; beneficial lactobacilli and bifidobacterial species have also been identified [99]. Accordingly, human milk is a synbiotic. In fact, in neonates, the formation of the gut microbiota is supported by the mode of feeding, where breast-fed infants have a higher abundance of bifidobacteria than formula-fed infants. Additionally, formula-fed infants have higher levels of C. difficile [100–102].
Clinical efficacy:
Assessment of probiotics in clinical trials for preventing CDI:
Several meta-analyses have been conducted with the aim of deciphering the collective outcome of the use of probiotics in preventing CDI and its associated morbidity. A 2013 meta-analysis of 4,492 cases conducted by Goldenberg et al., concluded that probiotics were effective in preventing CDAD with moderate certainty evidence (RR 0.36; 95% CI: 0.26, 0.51) [103]. A follow-up analysis of 8,672 cases concluded with moderate certainty that probiotics were effective in preventing CDAD. However, a post hoc subgroup analyses revealed that probiotics had no significant effect on preventing CDAD in trials that had a low-to-moderate baseline risk [104]. A recent meta-regression analysis demonstrated that timely administration of probiotics can lower the risk of CDI by >50% in hospitalized patients that were administered antibiotics [105]. Another meta-analysis of six randomized control trials concluded that only S. boulardii is effective in preventing CDI recurrence when administered in combination with a standard antibiotic [106]. A later meta-analyses related to CDAD prevention found that S. boulardii significantly reduced the risk of CDAD only in children [107], with a low quality of evidence [108]. Another recent study noted that a 3-strain probiotic preparation of L. acidophilus CL1285, L. casei LBC8OR, and L. rhamnosus CLR2, commercially known as Bio-K+ 50 Billion (50 billion CFU per capsule), can lower the incidence of healthcare associated-CDI (HA-CDI) for at least half of the patients on multiple-antibiotic regimens. Similarly, patients on ≥1 high-risk antibiotic regimen had a lower incidence of infection when on Bio-K+ during the intervention [0.9%, odds ratio (OR), 0.49] [109]. Contrary to this is the finding from another study which do not support the use of the Bio-K+ probiotic formulation for prevention of primary CDI in hospitalized adults >50 years receiving antibiotics in a setting which has relatively low baseline CDI rates [110] The fact that the majority of these meta-analyses tested a variety of probiotic strains and the variable baseline risks of CDAD among cohorts may explain the difference in the outcomes between these studies.
A few companies have initiated clinical trials for microbiome-based therapeutics. VE303 is the most advanced drug candidate, composed of an eight cryopreserved bacterial strain mixture, which successfully met its primary efficacy endpoint of preventing CDI recurrence in a Phase II clinical trial [111]. Two newer candidates added to the field of clinical probiotics are the investigational oral microbiome therapeutics CP101 and SER109. CP101 restored colonization resistance in a randomized, multicenter, placebo-controlled trial and has been granted fast track designation and breakthrough therapy designation by the FDA for preventing recurrent CDI [112]. SER109, on the other hand, constitutes the first targeted microbiome drug candidate that successfully met its Phase III primary endpoint that resulted in a statistically significant decrease in the proportion of patients experiencing recurrence as compared to the placebo (Clinical Trials.gov identifier: NCT03183128). There are two additional microbiota-based therapies for recurrent CDI, RBX2660 and RBX7455. RBX2660 is a microbiota suspension delivered via enema that is currently in Phase III clinical trials (Clinical Trials.gov identifier: NCT03931941). RBX7455 is a room temperature stable, orally administered probiotic that was recently found to be safe and effective at preventing recurrent CDI in a Phase I clinical trial (Clinical Trials.gov identifier: NCT02981316).
Probiotics for preventing CDI recurrence in the pediatric population:
The clinical outcomes of CDI in children are not sufficiently explored. A common clinical assumption is that symptomatic CDI does not occur in young infants due to the hypothesis that they lack C. difficile toxin receptors. As a result, testing for C. difficile is not typically performed in infants up to 2 years of age who present with diarrhea. However, there have been multiple cases of young infants presenting with recurrent diarrhea who tested positive for C. difficile [157]. Additionally, several studies have shown a rise in CDI in pediatric populations, which warrants further investigation to understand the clinical significance and optimal treatment for CDI in children. Case reports have found that probiotics have been successfully used to prevent CDAD and treat recurrent CDI in children [113, 114]. One study showed effective symptom management and reduced CDI recurrence when the probiotic S. boulardii was administered to a child with surgically treated Hirschsprung disease [115]. Further research is necessary to compare the efficacy of various probiotic agents in the treatment and prevention of CDI and its recurrence.
Advancing the understanding of probiotic mechanisms in the context of treating CDI:
Probiotic modulation of the immune system:
An improvement in the application of probiotics as a medical intervention requires us to develop a comprehensive understanding of the probiotic molecules and the response they can elicit in the host at the molecular and physiological levels. Primary studies have identified different probiotic strains that functionally modulate the host cells’ response via strengthening the epithelial barrier, protecting against cytokine-chemical stress, and enteric pathogen induced disruption of the epithelial barrier (Fig. 1) [116–118].
L. plantarum strains produce plantaricin, which is a bacteriocin that modulates the level of the anti-inflammatory cytokine IL-10 [119, 120]. The secreted protein p40 of L. rhamnosus GG (LGG) activates the epidermal growth factor receptor (EGFR), which could reduce cytokine-induced epithelial cell apoptosis and thus protect against experimental colitis [121]. LGG can also adhere to macrophages; this interaction has been found to induce IL-10 mRNA and reduce IL-6 mRNA in murine macrophages, thereby promoting an anti-inflammatory effect [122]. LGG, in an ex vivo and in vivo trial with ulcerative colitis (UC) patients, demonstrated anti-inflammatory effects via a significant reduction of pro-inflammatory cytokines like TNF-α and IL-17 [123] The protease lactocepin secreted by L. paracasei degrades an array of pro-inflammatory chemokines and protected mice against colitis [124]. L. salivarius Ls33 has been found to harbor anti-inflammatory properties; it can confer protection to mice from chemically induced colitis via a nucleotide-binding oligomerization domain- containing protein 2 (NOD2)-IL-10-dependent response [125]. Interestingly, candidate probiotics (L. acidophilus, L. casei, L. reuteri, B. bifidum, and S. thermophilus) or a mixture of them can specifically upregulate CD4+Foxp3+ regulatory T cells (Tregs), thus harboring potent anti-inflammatory effects [126]. Similarly, L.casei DN-114 001 can alleviate disease severity in dinitrobenzene sulfonic acid (DNBS)-induced colitis through induction and expansion of colonic CD4+Foxp3+ Treg cells and reduce cytokine production and T cell apoptosis in vitro in intestinal tissue specimens from Crohn’s disease patients [127–129]. Additionally, both F. prausnitzii and Streptococcus salivarius JIM8772 has been found to elicit strong anti-inflammatory responses in murine colitis models [42, 130–132].
In addition to its anti-toxin and anti-spore activity against C. difficile, CBM588 harbors immunomodulatory properties like inducing IL-17A-producing γδ T cells and IL-17A-producing CD4 cells, which can enhance gut epithelial barrier function. CBM588 modulates the composition of the gut microbiome with an increased abundance of Bifidobacterium, Lactobacillus, and Lactococcus spp. CBM588 also upregulates anti-inflammatory lipid metabolites (palmitoleic acid, 15d prostaglandin J2, and protectin D1), enhances TGFβ1 expression, and reduces inflammatory cytokines [133].
Probiotics with anti-inflammatory properties might harbor therapeutic potential and should be investigated for their role in ameliorating diseases associated with inflammation, like CDI. The choice of which probiotic strains to use has mostly been arbitrary. It should be noted that probiotics can have pro-inflammatory or anti-inflammatory properties. In addition, the therapeutic effect of probiotics also depends on its ability to provide colonization resistance against the pathogen, produce organic acids and antimicrobial substances, restoration of microbial community structure, and regulation of enzyme activity related to bile salt metabolism (Fig 1.) [134] . Hence, the choice of strains to use in clinical trials plays a crucial role in coming up with a probiotic that may be clinically efficacious.
It is also challenging to predict the effect of probiotics in humans based on in vitro and in vivo immunomodulation measurement designs. To determine the potential clinical impact of the probiotic strains, human mucosal responses should be investigated. Investigating the effect of probiotic strains in human volunteers at the transcriptome and proteomic levels can advance the knowledge base required to decipher a strain that might be clinically relevant [135, 136].
Genomic exploration of probiotic mechanisms:
In recent years, genome sequencing has been used to shed light on the presence of numerous genetic markers attributed to genuine probiotic features, such as bile and acid tolerance, epithelial cell adhesion, and the production of bacteriocins [137–139]. Defining core and pan-genomes in probiotic strains will undoubtedly provide a new baseline to examine the evolution and functionality of probiotics. Sun et al., in 2015, completed a comparative analysis of bifidobacterial genomes (45 strains) and identified 402 core-genes and > 20,000 pan-genes that could be essential for colonization and survival in the human gastrointestinal tract [140]. Further functional analysis of these genes will provide insights into how probiotics exert a protective effect against C. difficile. Mining of the B. breve UCC2003.31 probiotic genome sequences supports the notion that genetic adaptation to the colonization and persistence of B. breve in the human gut is mediated by fimbria-like structures encoded by the tad gene cluster [141]. Genome mining of B. animalis ssp. lactis AD011, a probiotic strain derived from an infant fecal sample, revealed the presence of several glycosylases and the fos gene cluster which are involved in processing of fructo-oligosaccharides [142, 143]. In another study, prolonged culture of B. longum DJO10A, an intestinal isolate, loses its functionality by gene loss identified by genome sequencing. The mutant strain showed a significantly decreased competitive ability against C. difficile and E. coli; this suggests that genomic deletions in probiotics, which is often mediated by genetic mobile elements, confers a loss of competitive abilities in the human gut [144, 145].
Comparative genome analysis of probiotic genomes would provide insights into unique probiotic attributes that are representative of the organisms’ niche. For example, in silico analysis revealed that intestinal lactobacilli showed enrichment of genes that encoded for mucus-binding proteins, bile salt hydrolase, and other enzymes that are involved in bile acid deconjugation and breakdown of carbohydrates; in contrast, milk-adapted lactobacilli showed enrichment of genes specific to metabolism of milk-derived sugars and other carbohydrates [146–149] . Huang et al., in 2021, used metagenome shotgun sequencing and whole genome sequencing approaches and identified that L. plantarum can utilize a highly convergent adaptive evolution strategy in a diverse array of host environments (human, mouse, and zebra fish); this confers fitness advantages to the strain and permits positive interactions between gut microbes and host cells [150]
In silico analysis of probiotic genomes could reveal a genetic repertoire that is essential for probiotic survival during transit through the harsh environment in the gastrointestinal tract. For example, analysis of the B. coagulans HS243 genome identified genes in the arginine deiminase (ADI) pathway, bacteriocin-producing genes, multi-subunit ATPases, adhesion proteins, and chologlycine hydrolase, which are required for survival and colonization of B. coagulans HS243 in the gastrointestinal tract [151]. Undoubtedly, analysis of the probiotic genome is the most valuable tool for safety assessments of probiotic products, which could alleviate the rising level of concerns over the risks of using probiotic products. Recently, Wang et al., in 2021, conducted a whole-genome analysis of commercially available probiotics and identified genes related to virulence factors, toxins, and resistance to antibiotics, which poses a potentially serious health issue [152].
Genomic exploration and genetic manipulation of probiotic microorganisms are crucial for defining their functional roles and also facilitates the rapid discovery of essential genes or genetic loci that can be exploited for bio-therapeutics and precision medicine strategy [153]. In the field of probiogenomics, it is noteworthy that CRISPR-Cas tools can be used to precisely tweak the probiotic genomes based on their known genetic sequences to generate next-generation probiotic strains that are more effective and capable of achieving more desirable traits including their improved adherence to host GIT, bile salt tolerance, enhanced antimicrobial activities, fine-tuning immunomodulatory properties through surface protein expression/suppression, and increased shelf life. [154–156]. CRISPR-engineered probiotics for therapeutic benefits are still in infancy stage, however, the massive expansion of probiotic genome datasets and advancement in genome-editing tools gives us advantages to create next-generation probiotics. This can eliminate specific pathogenic bacterial strains through targeted delivery of CRISPR-Cas system or lytic bacteriophages, which will open new avenues for the development of novel therapies against CDI. SNIPR Biome, a preclinical start-up based out of Denmark, focuses on targeting endogenous microbiome using CRISPR based vectors to selectively kill pathogenic strains without targeting other species in the microbiome. In addition, Van Pijkeren Laboratory at the University of Wisconsin-Madison has been developing a probiotic bacterium containing bacteriophage, which can carry a customized CRISPR message (knock down genetic message) to tackle antibiotic resistance in pathogens. When the engineered probiotic bacterium (with bacteriophage) reaches the intestinal tract, the bacteriophage within the probiotic bacterium would burst and infect any nearby C. difficile, causing them to degrade their own genetic material (commit suicide) [157]. Besides these, Novome Biotechnologies has been developing Genetically Engineered Microbial Medicines (GEMMs) that uses genetically engineered Bacteroides (native gut bacteria) carrying a gene cassette to metabolize porphyran and create new ecological niche. It has been shown that Bacteroides GEMMs can compete with the resident microbes to durably colonize the GIT and has potential to deliver therapies in wide range of disease [158]. Neil et al., in 2021 generated CRISPR-cas9 delivery vehicle that eliminated > 99.9% of targeted antibiotic-resistant E. coli in the mouse gut microbiota. The same research group has also applied this strategy to a Citrobacter rodentium infection model and achieved complete elimination of C. rodentium within 4 days of treatment [159]. Similar approach can also be used to eliminate C. difficile from the microbiota by targeting its essential genes or genetic loci of defined functional roles using CRISPR-Cas conjugative delivery vehicle, which could aid in development of preventive strategies for CDI. The development of genetic disruption techniques such as CRISPR-interference (CRISPRi) and CRISPR-activation (CRISPRa) to repress or activate genes has immense potential to quickly screen probiotics for phenotypes of both essential and non-essential genes. Applications of CRISPRi and CRISPRa have been successfully demonstrated in diverse probiotic bacteria, which provides deep insight into biology and the physiology of the probiotic species necessary for further strain improvement and exploitation [160]. Thus, the combination of probiogenomics and CRISPR-Cas tools enables selection of potential probiotic candidates and further enhancing their functional properties creating engineered probiotic strains with improved applicability in preventing CDI and its recurrence.
Emerging approaches to advance knowledge on probiotic use for the management of CDI:
Discovery informatics to identify novel probiotic strains:
Recently, several groups have deployed informatics-driven probiotic interventions to suppress C. difficile infection. Steinway et al. constructed a Boolean dynamic model utilizing time-series metagenomic data to capture therapeutic targets of probiotics. Genome-scale metabolic network reconstructions predicted the interaction between C. difficile and Barnesiella; this approach can be used to identify new probiotic strains to use as therapeutics [161]. A network meta-analysis was used to compare and rank the relative efficacy and tolerability of probiotic agents in antibiotic-associated diarrhea. L. casei exhibited better efficacy and medium tolerance in reducing C. difficile infection [162, 163]. Johnson et al., performed a meta-analysis of randomized controlled trials that supports the efficacy of L. casei, L. rhamnosus and S. boulardii formulations in demonstrating protective effects. In one study, 30,000 isolates from a human fecal sample were screened for anti-clostridial activity and one isolate was identified that produced a strong narrow-spectrum bacteriocin that can be used for treatment of CDI [164]. Koenigsknecht et al., performed time-course metagenomic sequencing and found that Lactobacillaceae and C. difficile dominated the small intestine and the large intestine, respectively. Furthermore, metabolomic analysis revealed antibiotic-induced alteration of the secondary bile acid compositions that inhibit vegetative C. difficile cells [165].
Metabolomics to identify colonization resistance metabolites:
Metabolomics offers an assessment of microbial activity through analysis of microbial metabolites. Furthermore, microbial impact on host metabolism can be investigated through host primary and secondary metabolites. The use of metabolomics in probiotic discovery identified a different response in fecal bile acids and SCFA upon probiotic administration that decreased colonization of C. difficile [166]. Additionally, metabolomics could be used to predict metabolic derivatives from diet or prebiotics, including bioactive microbial metabolites that affect microbiota composition and function [167].
Exploring the tripartite C. difficile-lumen-intestine interaction holistically via multi-omics to evaluate clinical efficacy of probiotics:
C. difficile is a pathobiont that lives primarily in peace with its host. Given an advantage, however, strains of C. difficile can exploit this advantage to potentially thrive and damage the host. The lack of microbial competitors, modifications in the chemical environment, and impedance of the intestinal host defenses can tip the balance towards C. difficile pathogenicity similar to other opportunistic pathogens; for example, Pseudomonas aeruginosa pathogenicity is dependent on the specific strain, chemical environment, and host defenses [168, 169]. A holistic personalized assessment of the intestinal environment should support evaluating the effectiveness of probiotics in a clinical setting. In the personalized assessment, the following questions could be asked to better understand person-to-person and patient-to-patient variation: What is the source of C. difficile strain-to-strain variation in virulence beyond the mere presence or absence of a virulence effector? For example, the metabolic capacity of the strain might be important to its virulence [168]. Also, what is the role of diet and the chemical environment before and after antibiotic treatment and concomitant CDI? Measuring the chemical output of a specific diet in the intestinal lumen may provide critical information that is relevant to treatment [169].
The effect of secreted factors from C. difficile on the host epithelium, muscles, and immune cells can potentially be simulated in model organisms. For example, the expression of the catalytic subunit of the C. difficile toxin, CDTa, in the Drosophila midgut reduces body weight, fecal output, and overall survival. Mechanistically, the Drosophila enterocytes are informative to illustrate that CDTa induces F-actin network collapse, eliminates the intestinal brush border, and disrupts intercellular junctions by re-distributing Rab11 to the enterocytes’ apical surface and the activation of the calmodulin/calcineurin pathway [170].
Thus, non-vertebrate or vertebrate model hosts can be used complementary to clinical development. Drosophila or mice exposed to with C. difficile secreted factors in multidimensional screens involving microbiota, prebiotics, and probiotics and the study of host epithelium response can teach us about the plasticity of C. difficile pathogenicity. Modulating host epithelium genetics or metabolism to alter the impact of CDI or virulence factors on the host is also a viable option, since genetic inhibition of Rab11 or chemical inhibition of the calmodulin/calcineurin pathway by cyclosporin A or FK506 reduces CDTa phenotypes [170].
Regulatory requirements and oversight for the approval of probiotics for C. difficile-associated diarrhea:
Most probiotic products are regulated as dietary supplements or foods for use by the general healthy population to improve their immunity or gastrointestinal flora. The increase in antibiotic-resistant infections, including the higher incidence of CDI, has resulted in probiotics being prescribed for the prevention and treatment of recurrent or refractory diseases in hospitalized patients [171]. The regulation of probiotics depends on multiple factors including the intended use, degree of health claims, warning statements on the label, dosage regimen, and geographical region. Based on the intended use, probiotics can be subclassified into probiotic drugs (therapeutic use), probiotic foods (use as foods, food ingredients, and dietary supplements), probiotic feeds (for animal use), and designed probiotics (genetically modified probiotics) [172].
In Australia and New Zealand, probiotics are regulated as foods and covered by the appropriate Food Standards Code when used as foods with or without associated general level claims (enhance immunity and/or regulate gastrointestinal flora). Probiotic foods associated with a high level of health claims require approval from the Food Standards Australia New Zealand (FSANZ). Furthermore, probiotic drugs with a health claim, statements or instructions for use, and the dosage regimen are considered as therapeutic and hence listed on the Australian Register of Therapeutic Goods (ARTG) and are regulated as complementary medicine by the Therapeutic Goods Administration [173].
In the United States, probiotics that are intended for use as dietary supplements are regulated by the FDA’s Center for Food Safety and Applied Nutrition. Unlike therapeutic drugs, the FDA does not need to approve dietary supplements; however, the regulatory body must be notified before a supplement can be marketed in the U.S., and the manufacturer should ensure product quality, efficiency, and safety [171].
The European Union (EU) possesses one of the strictest, if not the strictest, health claim regulations globally, has only authorized a small fraction of the health claims out of several claims submitted, and possesses no EU-wide legal framework that defines probiotic bacteria or probiotic foods. However, a list of bacteria at the species level, considered safe in foods and feeds, is kept and updated annually by the EU [174]. Health claim legislation in the EU revolves around consumer safety with an emphasis on controlled human intervention research and identification of the probiotic strains to the strain level by genotypic and phenotypic analyses, before approval [175].
According to the Joint Food and Agriculture Organization of the United Nations/World Health Organization Expert Consultation on Evaluation of Health and Nutritional Properties of Probiotics, the summary of the guidelines for evaluating probiotics that could lead to substantial health claims should include probiotic strain identification at the genus, species, and strain levels, in vitro testing to understand the mechanisms of action, clinical health benefits determined by human trials, and probiotic safety with respect to patterns of antimicrobial drug resistance, metabolic activities, side effects during clinical and post-clinical trials, toxicology studies, and investigation to determine a lack of infectivity in animal trials [171].
Challenges and future directions:
Several bacterial species confer protection against C. difficile and symptoms associated with CDI. Nevertheless, human studies using probiotics have generated contradictory results. Till date, no conventional probiotics have been clearly proven to be an effective prophylactic option for CDI prevention [176]. This may be in part due to disparity in study design, but it is also because of our limited understanding of the specificity and the mechanisms by which probiotics exert their action. Thus, a better understanding is needed regarding the gut microflora secreted effector molecules, their bactericidal/bacteriostatic effect, involvement in the modulation of the host immune response, and modulation of the receptor signaling cascades. What should be noted is that most of the current mechanistic studies are based on in vitro cell culture models and will most certainly require obtaining better mechanistic insights into their effects in humans.
The probiotics currently on the market cater to healthy individuals and claim to reduce risk of diseases prophylactically without any validation of its prophylactic or therapeutic effects. What is currently lacking in the field is biologically coherent data on the ability of probiotics to prevent diseases and/or modulate immune responses in the host and thereby provide therapeutic benefits. Clinical studies to obtain such data must follow defined study parameters like population size, volume and duration of dose administration, control groups, and disease outcome. Well designed, scientifically sound clinical trials are necessary to minimize bias in assessing the effects of probiotic interventions [177].
Additionally, specific studies are required to validate probiotics as an adjunctive therapy to the existing treatment repertoire. This will require assessing the combination of probiotics with standard-of-care antibiotics used to treat CDI. Genetic manipulation of current probiotic strains (designer probiotics) can also offer intriguing prospects for circumventing the side effects associated with antibiotic therapy and preventing CDAD. Comparative analysis of probiotic genomes will provide insights into unique probiotic attributes and the genetic repertoire that is essential for probiotic survival in the intestine and confer a protective effect against C. difficile. Another limitation in the use of probiotics is the broad-spectrum effect of their secreted metabolites on gut microflora strains. This has been observed with reuterin, produced by L. reuteri, which exhibits broad-spectrum antimicrobial activity against intestinal flora [178]. Several antimicrobial peptide (AMP)-producing microflora strains are effective against C. difficile but addressing off-target effects associated with these strains remains a challenge. This issue might be addressed by genetically engineering the AMP to specifically target C. difficile without harming the beneficial microbiota [179]. Novel members of the host microbiota identified from recent advancements in microbiome research can also be investigated to determine their inhibitory effect on C. difficile.
One notable limitation of different probiotic formulations is the successful delivery of viable cells at a high enough burden to the gut. Thus, novel delivery systems that have been used to treat other enteric diseases should be investigated for probiotics [180].
Another important question regarding probiotic use is whether to use a single strain or multi-strain formulations to treat CDI. It is postulated that because of the complexity of the intestinal microbiota, the use of multi-strain probiotic formulations might confer an advantage over the use of single strains in restoring the gut microbiome [181]. Clinical studies of probiotic use for CDI prevention involve the use of both single and multi-strain probiotics. In fact, a systematic review and meta-analysis study by Johnston et al., in 2012, concluded that clinical trials that used multiple species were more effective than clinical trials that used a single species [114]. However, as noted by the authors, the meta-analysis conducted was based on between-studies rather than within-studies comparison. Hence, there is no conclusive evidence regarding the benefit of multi-strain species over a single strain. Therefore, affirmative clinical studies comparing the same strains administered alone versus in a mixture need to be conducted.
Overall, probiotics might harbor potential to be utilized for the prevention of CDAD and treating recurrent CDI alongside antibiotics. However, not all probiotics in the current repertoire can be expected to deliver promising results in human clinical trials. Probiotic is an umbrella term, and a careful selection of specific organisms using a mechanistic-based approach coupled with multicenter, randomized, controlled trials can address the potential of probiotics’ supplementation as an option for treating or preventing CDI and its recurrence. In conclusion, a comprehensive understanding of the dynamic microbial community, deciphering the mechanistic relationship between the host microflora and C. difficile, and more evidence-based studies can help develop preemptive measures to better thwart the threat of CDI.
Table 1:
Summary of probiotic strains used in human clinical trials to treat CDAD.
| Strain(s) | Clinical Trial Application | Outcome/Results | Reference(s) |
|---|---|---|---|
| Lactobacillus plantarum 299v | Reducing colonization of C. difficile in critically ill patients treated with antibiotics. | None of the patients who received the probiotic were colonized with C. difficile compared to 19% colonization was present in the control group. | [182] |
| Cocktail of 4 strains L. acidophilus NCFM, L. paracasei Lpc-37, B. lactis Bi-07 and B. lactis BI-04 | Probiotic supplement for C. difficile infection in adults. | Probiotic adjunct therapy was associated with a significant improvement in diarrhea (rate and duration). | [183] |
| Cocktail of two strains: L. acidophilus CL1285 and L. casei LBC80R | Proprietary probiotic formulation for reducing CDAD in hospitalized patients on antibiotics. | Probiotic blend effective in lowering CDAD incidence in patients treated with probiotics vs. placebo. | [184, 185] |
| Probiotic formulation composed of L. acidophilus CL1285, L. casei LBC80R, and L. rhamnosus CLR2 | Adult hospitalized patients were treated with the probiotic formulation from the initiation of antibiotic use to prevent nosocomial CDI. | Highly effective infection prevention strategy with the lowest rate of nosocomial CDI on average over the past 15 years. | [186] |
| Cocktail of L. casei DN114-001 (L. casei immunitass), L. bulgaricus, and S. thermophilus | Consumption of the probiotic cocktail by hospitalized patients taking antibiotics. | Patients on probiotics did not develop CDAD as compared to placebo (17%). | [187] |
| Lactobacillus GG | Patients with recurrent C. difficile were treated with Lactobacillus GG. | Treatment showed a high cure rate for recurrent CDI. | [113, 188] |
| Saccharomyces boulardii | Supplement to prevent antibiotic associated diarrhea including CDAD in children. | Patients had lower incidence of CDAD (3.4%) as compared to the placebo (17.3%). | [189, 190] |
| Saccharomyces boulardii | Probiotic used with antibiotics (vancomycin or metronidazole) in adult patients with active C. difficile disease (CDD) or one prior CDD episode. | Significant improvement in recurrence rate in patients treated with probiotic (34.6%) vs. placebo (64.7%); however, no significant outcome in adults with initial CDD. | [191] |
| Saccharomyces boulardii | Hospitalized patients exposed to antibiotics were administered this probiotic. | No significant evidence was observed in prevention of CDAD in a population of hospitalized patients on antibiotics. | [192] |
| Spores of non-toxigenic C. difficile strain M3 (NTCD M3) | Patients diagnosed with CDI were administered three different doses of NTCD M3 spores. | Significant reduction in CDI recurrence in the combined NTCD M3 groups (11%) vs placebo (30%). Recurrence was also significantly lower in patients who developed colonization to NTCD (31% vs. 2%). | [193] |
| Multi-strain preparation of Lactobacilli and Bifidobacteria (L. acidophilus CUL60, B. bifidum CUL20 and B. lactis CUL34) | Pragmatic efficacy trial of the probiotic preparation in elderly hospitalized patients (≥65 years) exposed to oral or parenteral antibiotics. | No significant evidence suggesting the efficacy of the multi-strain probiotic in preventing CDAD. | [194] |
Acknowledgements:
Funding was provided by National Institute of Allergy and Infectious Diseases (Grant No. R01AI130186) for MNS.
Footnotes
Disclosure statement: The authors report no competing interest.
References:
- 1.Bartlett JG, et al. , Role of Clostridium difficile in antibiotic-associated pseudomembranous colitis. Gastroenterology, 1978. 75(5): p. 778–82. [PubMed] [Google Scholar]
- 2.Bartlett JG, et al. , Antibiotic-associated pseudomembranous colitis due to toxin-producing clostridia. N Engl J Med, 1978. 298(10): p. 531–4. [DOI] [PubMed] [Google Scholar]
- 3.Hall IC and O’TOOLE E, Intestinal flora in new-born infants: with a description of a new pathogenic anaerobe, Bacillus difficilis. American journal of diseases of children, 1935. 49(2): p. 390–402. [Google Scholar]
- 4.Kuijper EJ, et al. , Emergence of Clostridium difficile-associated disease in North America and Europe. Clin Microbiol Infect, 2006. 12 Suppl 6: p. 2–18. [DOI] [PubMed] [Google Scholar]
- 5.He M, et al. , Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nat Genet, 2013. 45(1): p. 109–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.CDC. Antibiotic Resistance Threats in the United States, 2019. Atlanta, GA: U.S. Department of Health and Human Services, CDC. 2019. [Google Scholar]
- 7.McDonald LC, Owings M, and Jernigan DB, Clostridium difficile infection in patients discharged from US short-stay hospitals, 1996–2003. Emerg Infect Dis, 2006. 12(3): p. 409–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldstein EJ, et al. , Pathway to Prevention of Nosocomial Clostridium difficile Infection. Clin Infect Dis, 2015. 60 Suppl 2: p. S148–58. [DOI] [PubMed] [Google Scholar]
- 9.Theriot CM, et al. , Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat Commun, 2014. 5: p. 3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Theriot CM, Bowman AA, and Young VB, Antibiotic-Induced Alterations of the Gut Microbiota Alter Secondary Bile Acid Production and Allow for Clostridium difficile Spore Germination and Outgrowth in the Large Intestine. mSphere, 2016. 1(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mullish BH and Williams HR, Clostridium difficile infection and antibiotic-associated diarrhoea. Clin Med (Lond), 2018. 18(3): p. 237–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berkell M, et al. , Microbiota-based markers predictive of development of Clostridioides difficile infection. Nat Commun, 2021. 12(1): p. 2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sorg JA and Sonenshein AL, Bile salts and glycine as cogerminants for Clostridium difficile spores. J Bacteriol, 2008. 190(7): p. 2505–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thanissery R, Winston JA, and Theriot CM, Inhibition of spore germination, growth, and toxin activity of clinically relevant C. difficile strains by gut microbiota derived secondary bile acids. Anaerobe, 2017. 45: p. 86–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilson KH, Efficiency of various bile salt preparations for stimulation of Clostridium difficile spore germination. Journal of clinical microbiology, 1983. 18(4): p. 1017–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McDonald LC, et al. , Clinical practice guidelines for Clostridium difficile infection in adults and children: 2017 update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA). Clinical Infectious Diseases, 2018. 66(7): p. e1–e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Prehn J, et al. , European Society of Clinical Microbiology and Infectious Diseases: 2021 update on the treatment guidance document for Clostridioides difficile infection in adults. Clin Microbiol Infect, 2021. 27 Suppl 2: p. S1–S21. [DOI] [PubMed] [Google Scholar]
- 18.CDC, Nearly half a million Americans suffered from Clostridium difficile infections in a single year. 2015, Feb 25.
- 19.Leffler DA and Lamont JT, Clostridium difficile infection. N Engl J Med, 2015. 372(16): p. 1539–48. [DOI] [PubMed] [Google Scholar]
- 20.Walters BA, et al. , Relapse of antibiotic associated colitis: endogenous persistence of Clostridium difficile during vancomycin therapy. Gut, 1983. 24(3): p. 206–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kociolek LK and Gerding DN, Breakthroughs in the treatment and prevention of Clostridium difficile infection. Nat Rev Gastroenterol Hepatol, 2016. 13(3): p. 150–60. [DOI] [PubMed] [Google Scholar]
- 22.Schwan A, et al. , Relapsing clostridium difficile enterocolitis cured by rectal infusion of homologous faeces. Lancet, 1983. 2(8354): p. 845. [DOI] [PubMed] [Google Scholar]
- 23.Cammarota G, et al. , European consensus conference on faecal microbiota transplantation in clinical practice. Gut, 2017. 66(4): p. 569–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Nood E, et al. , Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med, 2013. 368(5): p. 407–15. [DOI] [PubMed] [Google Scholar]
- 25.Kassam Z, et al. , Fecal microbiota transplantation for Clostridium difficile infection: systematic review and meta-analysis. Am J Gastroenterol, 2013. 108(4): p. 500–8. [DOI] [PubMed] [Google Scholar]
- 26.Xiao Y, et al. , An ecological framework to understand the efficacy of fecal microbiota transplantation. Nat Commun, 2020. 11(1): p. 3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carlson PE Jr, Regulatory considerations for fecal microbiota transplantation products. Cell Host & Microbe, 2020. 27(2): p. 173–175. [DOI] [PubMed] [Google Scholar]
- 28.Zhang T, et al. , Washed microbiota transplantation vs. manual fecal microbiota transplantation: clinical findings, animal studies and in vitro screening. Protein Cell, 2020. 11(4): p. 251–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Borody TJ and Khoruts A, Fecal microbiota transplantation and emerging applications. Nat Rev Gastroenterol Hepatol, 2011. 9(2): p. 88–96. [DOI] [PubMed] [Google Scholar]
- 30.Gulati M, et al. , Delivery routes for faecal microbiota transplants: Available, anticipated and aspired. Pharmacol Res, 2020. 159: p. 104954. [DOI] [PubMed] [Google Scholar]
- 31.Hill C, et al. , Expert consensus document: The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nature Reviews Gastroenterology and Hepatology, 2014. 11(8): p. 506. [DOI] [PubMed] [Google Scholar]
- 32.Mills JP, Rao K, and Young VB, Probiotics for prevention of Clostridium difficile infection. Curr Opin Gastroenterol, 2018. 34(1): p. 3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Vrese M and Schrezenmeir J, Probiotics, prebiotics, and synbiotics. Adv Biochem Eng Biotechnol, 2008. 111: p. 1–66. [DOI] [PubMed] [Google Scholar]
- 34.Markets M.a. Probiotics Market worth $69.3 billion by 2023. Jan, 2019; Available from: https://www.marketsandmarkets.com/PressReleases/probiotics.asp.
- 35.Clarke TC, et al. , Trends in the use of complementary health approaches among adults: United States, 2002–2012. Natl Health Stat Report, 2015(79): p. 1–16. [PMC free article] [PubMed] [Google Scholar]
- 36.Zmora N, et al. , Personalized Gut Mucosal Colonization Resistance to Empiric Probiotics Is Associated with Unique Host and Microbiome Features. Cell, 2018. 174(6): p. 1388–1405 e21. [DOI] [PubMed] [Google Scholar]
- 37.Draper K, Ley C, and Parsonnet J, Probiotic guidelines and physician practice: a cross-sectional survey and overview of the literature. Benef Microbes, 2017. 8(4): p. 507–519. [DOI] [PubMed] [Google Scholar]
- 38.Zhao Y, et al. , GPR43 mediates microbiota metabolite SCFA regulation of antimicrobial peptide expression in intestinal epithelial cells via activation of mTOR and STAT3. Mucosal Immunol, 2018. 11(3): p. 752–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jacobson A, et al. , A Gut Commensal-Produced Metabolite Mediates Colonization Resistance to Salmonella Infection. Cell Host Microbe, 2018. 24(2): p. 296–307 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pagnini C, et al. , Probiotics promote gut health through stimulation of epithelial innate immunity. Proc Natl Acad Sci U S A, 2010. 107(1): p. 454–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Segawa S, et al. , Probiotic-derived polyphosphate enhances the epithelial barrier function and maintains intestinal homeostasis through integrin-p38 MAPK pathway. PLoS One, 2011. 6(8): p. e23278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sokol H, et al. , Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A, 2008. 105(43): p. 16731–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Corthier G, Dubos F, and Raibaud P, Modulation of cytotoxin production by Clostridium difficile in the intestinal tracts of gnotobiotic mice inoculated with various human intestinal bacteria. Appl Environ Microbiol, 1985. 49(1): p. 250–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Naaber P, et al. , Inhibition of Clostridium difficile strains by intestinal Lactobacillus species. J Med Microbiol, 2004. 53(Pt 6): p. 551–4. [DOI] [PubMed] [Google Scholar]
- 45.Schoster A, et al. , In vitro inhibition of Clostridium difficile and Clostridium perfringens by commercial probiotic strains. Anaerobe, 2013. 20: p. 36–41. [DOI] [PubMed] [Google Scholar]
- 46.Spinler JK, et al. , Next-Generation Probiotics Targeting Clostridium difficile through Precursor-Directed Antimicrobial Biosynthesis. Infect Immun, 2017. 85(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Engevik MA, et al. , Reuterin disrupts Clostridioides difficile metabolism and pathogenicity through reactive oxygen species generation. Gut Microbes, 2020. 12(1): p. 1788898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Banerjee P, Merkel GJ, and Bhunia AK, Lactobacillus delbrueckii ssp. bulgaricus B-30892 can inhibit cytotoxic effects and adhesion of pathogenic Clostridium difficile to Caco-2 cells. Gut Pathog, 2009. 1(1): p. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Banerjee P, Merkel GJ, and Bhunia AK, Lactobacillus delbrueckii ssp. bulgaricus B-30892 can inhibit cytotoxic effects and adhesion of pathogenic Clostridium difficile to Caco-2 cells. Gut pathogens, 2009. 1(1): p. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yun B, Oh S, and Griffiths MW, Lactobacillus acidophilus modulates the virulence of Clostridium difficile. J Dairy Sci, 2014. 97(8): p. 4745–58. [DOI] [PubMed] [Google Scholar]
- 51.Najarian A, Sharif S, and Griffiths MW, Evaluation of protective effect of Lactobacillus acidophilus La-5 on toxicity and colonization of Clostridium difficile in human epithelial cells in vitro. Anaerobe, 2019. 55: p. 142–151. [DOI] [PubMed] [Google Scholar]
- 52.Carasi P, et al. , Surface proteins from Lactobacillus kefir antagonize in vitro cytotoxic effect of Clostridium difficile toxins. Anaerobe, 2012. 18(1): p. 135–42. [DOI] [PubMed] [Google Scholar]
- 53.Mobili P, et al. , Heterogeneity of S-layer proteins from aggregating and non-aggregating Lactobacillus kefir strains. Antonie Van Leeuwenhoek, 2009. 95(4): p. 363–72. [DOI] [PubMed] [Google Scholar]
- 54.Bolla PA, et al. , Kefir-isolated Lactococcus lactis subsp. lactis inhibits the cytotoxic effect of Clostridium difficile in vitro. J Dairy Res, 2013. 80(1): p. 96–102. [DOI] [PubMed] [Google Scholar]
- 55.Bolla PA, et al. , Protective effect of a mixture of kefir-isolated lactic acid bacteria and yeasts in a hamster model of Clostridium difficile infection. Anaerobe, 2013. 21: p. 28–33. [DOI] [PubMed] [Google Scholar]
- 56.Auclair J, Frappier M, and Millette M, Lactobacillus acidophilus CL1285, Lactobacillus casei LBC80R, and Lactobacillus rhamnosus CLR2 (Bio-K+): Characterization, Manufacture, Mechanisms of Action, and Quality Control of a Specific Probiotic Combination for Primary Prevention of Clostridium difficile Infection. Clin Infect Dis, 2015. 60 Suppl 2: p. S135–43. [DOI] [PubMed] [Google Scholar]
- 57.Pothoulakis C, et al. , Saccharomyces boulardii inhibits Clostridium difficile toxin A binding and enterotoxicity in rat ileum. Gastroenterology, 1993. 104(4): p. 1108–15. [DOI] [PubMed] [Google Scholar]
- 58.Castagliuolo I, et al. , Saccharomyces boulardii protease inhibits Clostridium difficile toxin A effects in the rat ileum. Infect Immun, 1996. 64(12): p. 5225–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen X, et al. , Saccharomyces boulardii inhibits ERK1/2 mitogen-activated protein kinase activation both in vitro and in vivo and protects against Clostridium difficile toxin A-induced enteritis. J Biol Chem, 2006. 281(34): p. 24449–54. [DOI] [PubMed] [Google Scholar]
- 60.Castagliuolo I, et al. , Saccharomyces boulardii protease inhibits the effects of Clostridium difficile toxins A and B in human colonic mucosa. Infect Immun, 1999. 67(1): p. 302–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Valdes-Varela L, et al. , Screening of Bifidobacteria and Lactobacilli Able to Antagonize the Cytotoxic Effect of Clostridium difficile upon Intestinal Epithelial HT29 Monolayer. Front Microbiol, 2016. 7: p. 577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Trejo FM, Perez PF, and De Antoni GL, Co-culture with potentially probiotic microorganisms antagonises virulence factors of Clostridium difficile in vitro. Antonie Van Leeuwenhoek, 2010. 98(1): p. 19–29. [DOI] [PubMed] [Google Scholar]
- 63.Wei Y, et al. , Protective Effects of Bifidobacterial Strains Against Toxigenic Clostridium difficile. Front Microbiol, 2018. 9: p. 888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Trejo FM, et al. , Inhibition of Clostridium difficile growth and adhesion to enterocytes by Bifidobacterium supernatants. Anaerobe, 2006. 12(4): p. 186–93. [DOI] [PubMed] [Google Scholar]
- 65.Lv LX, et al. , Whole-genome sequence assembly of Pediococcus pentosaceus LI05 (CGMCC 7049) from the human gastrointestinal tract and comparative analysis with representative sequences from three food-borne strains. Gut Pathog, 2014. 6: p. 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu Q, et al. , Protective Effect of Pediococcus pentosaceus LI05 Against Clostridium difficile Infection in a Mouse Model. Front Microbiol, 2018. 9: p. 2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vitetta L, Llewellyn H, and Oldfield D, Gut Dysbiosis and the Intestinal Microbiome: Streptococcus thermophilus a Key Probiotic for Reducing Uremia. Microorganisms, 2019. 7(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rul F, et al. , Impact of the metabolic activity of Streptococcus thermophilus on the colon epithelium of gnotobiotic rats. J Biol Chem, 2011. 286(12): p. 10288–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kolling GL, et al. , Lactic acid production by Streptococcus thermophilus alters Clostridium difficile infection and in vitro Toxin A production. Gut Microbes, 2012. 3(6): p. 523–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Eckburg PB, et al. , Diversity of the human intestinal microbial flora. Science, 2005. 308(5728): p. 1635–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Seki H, et al. , Prevention of antibiotic-associated diarrhea in children by Clostridium butyricum MIYAIRI. Pediatr Int, 2003. 45(1): p. 86–90. [DOI] [PubMed] [Google Scholar]
- 72.Woo TD, et al. , Inhibition of the cytotoxic effect of Clostridium difficile in vitro by Clostridium butyricum MIYAIRI 588 strain. J Med Microbiol, 2011. 60(Pt 11): p. 1617–25. [DOI] [PubMed] [Google Scholar]
- 73.Kamiya S, Taguchi H, Yamaguchi H,Osaki T, Takahashi M, Nakamura S, Bacterioprophylaxis using Clostridum butyricum for lethal caecitis by Clostridium difficile in gnotobiotic mice. Reviews in Medical Microbiology, 1997: p. S57–S59. [Google Scholar]
- 74.Oka K, et al. , Establishment of an Endogenous Clostridium difficile Rat Infection Model and Evaluation of the Effects of Clostridium butyricum MIYAIRI 588 Probiotic Strain. Front Microbiol, 2018. 9: p. 1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sambol SP, et al. , Colonization for the prevention of Clostridium difficile disease in hamsters. J Infect Dis, 2002. 186(12): p. 1781–9. [DOI] [PubMed] [Google Scholar]
- 76.Reed AD, et al. , Strain-Dependent Inhibition of Clostridioides difficile by Commensal Clostridia Carrying the Bile Acid-Inducible (bai) Operon. J Bacteriol, 2020. 202(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Studer N, et al. , Functional Intestinal Bile Acid 7alpha-Dehydroxylation by Clostridium scindens Associated with Protection from Clostridium difficile Infection in a Gnotobiotic Mouse Model. Front Cell Infect Microbiol, 2016. 6: p. 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Buffie CG, et al. , Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature, 2015. 517(7533): p. 205–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kang JD, et al. , Bile Acid 7alpha-Dehydroxylating Gut Bacteria Secrete Antibiotics that Inhibit Clostridium difficile: Role of Secondary Bile Acids. Cell Chem Biol, 2019. 26(1): p. 27–34 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Baccouri O, et al. , Probiotic Potential and Safety Evaluation of Enterococcus faecalis OB14 and OB15, Isolated From Traditional Tunisian Testouri Cheese and Rigouta, Using Physiological and Genomic Analysis. Front Microbiol, 2019. 10: p. 881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mansour NM, et al. , Inhibition of Clostridium difficile in Mice Using a Mixture of Potential Probiotic Strains Enterococcus faecalis NM815, E. faecalis NM915, and E. faecium NM1015: Novel Candidates to Control C. difficile Infection (CDI). Probiotics Antimicrob Proteins, 2018. 10(3): p. 511–522. [DOI] [PubMed] [Google Scholar]
- 82.Chen X, et al. , A mouse model of Clostridium difficile-associated disease. Gastroenterology, 2008. 135(6): p. 1984–92. [DOI] [PubMed] [Google Scholar]
- 83.Colenutt C and Cutting SM, Use of Bacillus subtilis PXN21 spores for suppression of Clostridium difficile infection symptoms in a murine model. FEMS Microbiol Lett, 2014. 358(2): p. 154–61. [DOI] [PubMed] [Google Scholar]
- 84.Urdaci MC, Bressollier P, and Pinchuk I, Bacillus clausii probiotic strains: antimicrobial and immunomodulatory activities. J Clin Gastroenterol, 2004. 38(6 Suppl): p. S86–90. [DOI] [PubMed] [Google Scholar]
- 85.Ripert G, et al. , Secreted Compounds of the Probiotic Bacillus clausii Strain O/C Inhibit the Cytotoxic Effects Induced by Clostridium difficile and Bacillus cereus Toxins. Antimicrob Agents Chemother, 2016. 60(6): p. 3445–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fitzpatrick LR, et al. , Bacillus coagulans GBI-30 (BC30) improves indices of Clostridium difficile-induced colitis in mice. Gut pathogens, 2011. 3(1): p. 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fitzpatrick LR, et al. , Bacillus coagulans GBI-30, 6086 limits the recurrence of Clostridium difficile-Induced colitis following vancomycin withdrawal in mice. Gut pathogens, 2012. 4(1): p. 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Reeves AE, et al. , The interplay between microbiome dynamics and pathogen dynamics in a murine model of Clostridium difficile Infection. Gut Microbes, 2011. 2(3): p. 145–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Reeves AE, et al. , Suppression of Clostridium difficile in the gastrointestinal tracts of germfree mice inoculated with a murine isolate from the family Lachnospiraceae. Infect Immun, 2012. 80(11): p. 3786–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Antharam VC, et al. , Intestinal dysbiosis and depletion of butyrogenic bacteria in Clostridium difficile infection and nosocomial diarrhea. J Clin Microbiol, 2013. 51(9): p. 2884–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gibson GR and Roberfroid MB, Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J Nutr, 1995. 125(6): p. 1401–12. [DOI] [PubMed] [Google Scholar]
- 92.Schrezenmeir J and de Vrese M, Probiotics, prebiotics, and synbiotics--approaching a definition. Am J Clin Nutr, 2001. 73(2 Suppl): p. 361S–364S. [DOI] [PubMed] [Google Scholar]
- 93.Pandey KR, Naik SR, and Vakil BV, Probiotics, prebiotics and synbiotics- a review. J Food Sci Technol, 2015. 52(12): p. 7577–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ratsep M, et al. , A combination of the probiotic and prebiotic product can prevent the germination of Clostridium difficile spores and infection. Anaerobe, 2017. 47: p. 94–103. [DOI] [PubMed] [Google Scholar]
- 95.Kondepudi KK, et al. , A novel multi-strain probiotic and synbiotic supplement for prevention of Clostridium difficile infection in a murine model. Microbiol Immunol, 2014. 58(10): p. 552–8. [DOI] [PubMed] [Google Scholar]
- 96.Valdes-Varela L, et al. , Effect of Bifidobacterium upon Clostridium difficile Growth and Toxicity When Co-cultured in Different Prebiotic Substrates. Front Microbiol, 2016. 7: p. 738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Roychowdhury S, et al. , Faecalibacterium prausnitzii and a Prebiotic Protect Intestinal Health in a Mouse Model of Antibiotic and Clostridium difficile Exposure. JPEN J Parenter Enteral Nutr, 2018. 42(7): p. 1156–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bode L, Human milk oligosaccharides: every baby needs a sugar mama. Glycobiology, 2012. 22(9): p. 1147–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ruiz L, Garcia-Carral C, and Rodriguez JM, Unfolding the Human Milk Microbiome Landscape in the Omics Era. Front Microbiol, 2019. 10: p. 1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bezirtzoglou E, Tsiotsias A, and Welling GW, Microbiota profile in feces of breast- and formula-fed newborns by using fluorescence in situ hybridization (FISH). Anaerobe, 2011. 17(6): p. 478–82. [DOI] [PubMed] [Google Scholar]
- 101.Penders J, et al. , Quantification of Bifidobacterium spp., Escherichia coli and Clostridium difficile in faecal samples of breast-fed and formula-fed infants by real-time PCR. FEMS Microbiol Lett, 2005. 243(1): p. 141–7. [DOI] [PubMed] [Google Scholar]
- 102.Backhed F, et al. , Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host Microbe, 2015. 17(5): p. 690–703. [DOI] [PubMed] [Google Scholar]
- 103.Goldenberg JZ, et al. , Probiotics for the prevention of Clostridium difficile-associated diarrhea in adults and children. Cochrane Database Syst Rev, 2013(5): p. CD006095. [DOI] [PubMed] [Google Scholar]
- 104.Goldenberg JZ, et al. , Probiotics for the prevention of Clostridium difficile-associated diarrhea in adults and children. Cochrane Database Syst Rev, 2017. 12: p. CD006095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shen NT, et al. , Timely Use of Probiotics in Hospitalized Adults Prevents Clostridium difficile Infection: A Systematic Review With Meta-Regression Analysis. Gastroenterology, 2017. 152(8): p. 1889–1900 e9. [DOI] [PubMed] [Google Scholar]
- 106.McFarland LV, Meta-analysis of probiotics for the prevention of antibiotic associated diarrhea and the treatment of Clostridium difficile disease. Am J Gastroenterol, 2006. 101(4): p. 812–22. [DOI] [PubMed] [Google Scholar]
- 107.Szajewska H and Kolodziej M, Systematic review with meta-analysis: Saccharomyces boulardii in the prevention of antibiotic-associated diarrhoea. Aliment Pharmacol Ther, 2015. 42(7): p. 793–801. [DOI] [PubMed] [Google Scholar]
- 108.Szajewska H, et al. , Probiotics for the Prevention of Antibiotic-Associated Diarrhea in Children. J Pediatr Gastroenterol Nutr, 2016. 62(3): p. 495–506. [DOI] [PubMed] [Google Scholar]
- 109.Maziade PJ, et al. , Enhanced Clostridioides difficile Infection Prevention With a Pharmacy-Controlled Policy That Adds a 3-Strain Lactobacillus Probiotic Concomitantly to Antibiotic Therapy. Clin Infect Dis, 2021. 73(8): p. 1524–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Heil EL, et al. , A Multicenter Evaluation of Probiotic Use for the Primary Prevention of Clostridioides difficile Infection. Clin Infect Dis, 2021. 73(8): p. 1330–1337. [DOI] [PubMed] [Google Scholar]
- 111.Chakradhar S, Positive selection. Nat Med, 2019. 25(2): p. 192–194. [DOI] [PubMed] [Google Scholar]
- 112.CP101 for recurrent C. difficile. 2020; Available from: https://finchtherapeutics.com/cp101.
- 113.Biller JA, et al. , Treatment of recurrent Clostridium difficile colitis with Lactobacillus GG. J Pediatr Gastroenterol Nutr, 1995. 21(2): p. 224–6. [DOI] [PubMed] [Google Scholar]
- 114.Johnston BC, et al. , Probiotics for the prevention of Clostridium difficile-associated diarrhea: a systematic review and meta-analysis. Ann Intern Med, 2012. 157(12): p. 878–88. [DOI] [PubMed] [Google Scholar]
- 115.Ooi CY, Dilley AV, and Day AS, Saccharomyces boulardii in a child with recurrent Clostridium difficile. Pediatr Int, 2009. 51(1): p. 156–8. [DOI] [PubMed] [Google Scholar]
- 116.Wang J, et al. , Probiotic Lactobacillus plantarum Promotes Intestinal Barrier Function by Strengthening the Epithelium and Modulating Gut Microbiota. Front Microbiol, 2018. 9: p. 1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.O’Hara AM, et al. , Functional modulation of human intestinal epithelial cell responses by Bifidobacterium infantis and Lactobacillus salivarius. Immunology, 2006. 118(2): p. 202–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Resta-Lenert S and Barrett KE, Probiotics and commensals reverse TNF-alpha- and IFN-gamma-induced dysfunction in human intestinal epithelial cells. Gastroenterology, 2006. 130(3): p. 731–46. [DOI] [PubMed] [Google Scholar]
- 119.Takano T, et al. , Lactobacillus plantarum OLL2712 induces IL-10 production by intestinal dendritic cells. Biosci Microbiota Food Health, 2020. 39(2): p. 39–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.van Hemert S, et al. , Identification of Lactobacillus plantarum genes modulating the cytokine response of human peripheral blood mononuclear cells. BMC Microbiol, 2010. 10: p. 293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yan F, et al. , Colon-specific delivery of a probiotic-derived soluble protein ameliorates intestinal inflammation in mice through an EGFR-dependent mechanism. J Clin Invest, 2011. 121(6): p. 2242–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Vargas Garcia CE, et al. , Piliation of Lactobacillus rhamnosus GG promotes adhesion, phagocytosis, and cytokine modulation in macrophages. Appl Environ Microbiol, 2015. 81(6): p. 2050–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pagnini C, et al. , Mucosal adhesion and anti-inflammatory effects of Lactobacillus rhamnosus GG in the human colonic mucosa: A proof-of-concept study. World J Gastroenterol, 2018. 24(41): p. 4652–4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.von Schillde MA, et al. , Lactocepin secreted by Lactobacillus exerts anti-inflammatory effects by selectively degrading proinflammatory chemokines. Cell Host Microbe, 2012. 11(4): p. 387–96. [DOI] [PubMed] [Google Scholar]
- 125.Macho Fernandez E, et al. , Anti-inflammatory capacity of selected lactobacilli in experimental colitis is driven by NOD2-mediated recognition of a specific peptidoglycan-derived muropeptide. Gut, 2011. 60(8): p. 1050–9. [DOI] [PubMed] [Google Scholar]
- 126.Kwon HK, et al. , Generation of regulatory dendritic cells and CD4+Foxp3+ T cells by probiotics administration suppresses immune disorders. Proc Natl Acad Sci U S A, 2010. 107(5): p. 2159–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hacini-Rachinel F, et al. , CD4+ T cells and Lactobacillus casei control relapsing colitis mediated by CD8+ T cells. J Immunol, 2009. 183(9): p. 5477–86. [DOI] [PubMed] [Google Scholar]
- 128.Carol M, et al. , Modulation of apoptosis in intestinal lymphocytes by a probiotic bacteria in Crohn’s disease. J Leukoc Biol, 2006. 79(5): p. 917–22. [DOI] [PubMed] [Google Scholar]
- 129.Llopis M, et al. , Lactobacillus casei downregulates commensals’ inflammatory signals in Crohn’s disease mucosa. Inflamm Bowel Dis, 2009. 15(2): p. 275–83. [DOI] [PubMed] [Google Scholar]
- 130.Kaci G, et al. , Anti-inflammatory properties of Streptococcus salivarius, a commensal bacterium of the oral cavity and digestive tract. Appl Environ Microbiol, 2014. 80(3): p. 928–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Rossi O, et al. , Faecalibacterium prausnitzii A2-165 has a high capacity to induce IL-10 in human and murine dendritic cells and modulates T cell responses. Sci Rep, 2016. 6: p. 18507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhang M, et al. , Faecalibacterium prausnitzii inhibits interleukin-17 to ameliorate colorectal colitis in rats. PLoS One, 2014. 9(10): p. e109146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hagihara M, et al. , Clostridium butyricum Modulates the Microbiome to Protect Intestinal Barrier Function in Mice with Antibiotic-Induced Dysbiosis. iScience, 2020. 23(1): p. 100772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yang J and Yang H, Non-antibiotic therapy for Clostridioides difficile infection: a review. Crit Rev Clin Lab Sci, 2019. 56(7): p. 493–509. [DOI] [PubMed] [Google Scholar]
- 135.van Baarlen P, et al. , Human mucosal in vivo transcriptome responses to three lactobacilli indicate how probiotics may modulate human cellular pathways. Proc Natl Acad Sci U S A, 2011. 108 Suppl 1: p. 4562–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.van Baarlen P, et al. , Differential NF-kappaB pathways induction by Lactobacillus plantarum in the duodenum of healthy humans correlating with immune tolerance. Proc Natl Acad Sci U S A, 2009. 106(7): p. 2371–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lebeer S, Vanderleyden J, and De Keersmaecker SC, Genes and molecules of lactobacilli supporting probiotic action. Microbiol Mol Biol Rev, 2008. 72(4): p. 728–64, Table of Contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Papadimitriou K, et al. , Discovering probiotic microorganisms: in vitro, in vivo, genetic and omics approaches. Front Microbiol, 2015. 6: p. 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bermudez-Brito M, et al. , Probiotic mechanisms of action. Ann Nutr Metab, 2012. 61(2): p. 160–74. [DOI] [PubMed] [Google Scholar]
- 140.Sun Z, et al. , Comparative genomic analysis of 45 type strains of the genus Bifidobacterium: a snapshot of its genetic diversity and evolution. PLoS One, 2015. 10(2): p. e0117912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.O’Connell Motherway M, et al. , Functional genome analysis of Bifidobacterium breve UCC2003 reveals type IVb tight adherence (Tad) pili as an essential and conserved host-colonization factor. Proc Natl Acad Sci U S A, 2011. 108(27): p. 11217–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kim JY, Choi YO, and Ji GE, Effect of oral probiotics (Bifidobacterium lactis AD011 and Lactobacillus acidophilus AD031) administration on ovalbumin-induced food allergy mouse model. J Microbiol Biotechnol, 2008. 18(8): p. 1393–400. [PubMed] [Google Scholar]
- 143.Kim JF, et al. , Genome sequence of the probiotic bacterium Bifidobacterium animalis subsp. lactis AD011. J Bacteriol, 2009. 191(2): p. 678–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lee JH, et al. , Comparative genomic analysis of the gut bacterium Bifidobacterium longum reveals loci susceptible to deletion during pure culture growth. BMC Genomics, 2008. 9: p. 247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.O’Sullivan DJ, Genomics can advance the potential for probiotic cultures to improve liver and overall health. Curr Pharm Des, 2008. 14(14): p. 1376–81. [DOI] [PubMed] [Google Scholar]
- 146.Boekhorst J, et al. , Comparative analysis of proteins with a mucus-binding domain found exclusively in lactic acid bacteria. Microbiology (Reading), 2006. 152(Pt 1): p. 273–280. [DOI] [PubMed] [Google Scholar]
- 147.Siezen R, et al. , Lactobacillus plantarum gene clusters encoding putative cell-surface protein complexes for carbohydrate utilization are conserved in specific gram-positive bacteria. BMC Genomics, 2006. 7: p. 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lambert JM, et al. , Improved annotation of conjugated bile acid hydrolase superfamily members in Gram-positive bacteria. Microbiology (Reading), 2008. 154(Pt 8): p. 2492–2500. [DOI] [PubMed] [Google Scholar]
- 149.Ventura M, et al. , Genome-scale analyses of health-promoting bacteria: probiogenomics. Nat Rev Microbiol, 2009. 7(1): p. 61–71. [DOI] [PubMed] [Google Scholar]
- 150.Huang S, et al. , Candidate probiotic Lactiplantibacillus plantarum HNU082 rapidly and convergently evolves within human, mice, and zebrafish gut but differentially influences the resident microbiome. Microbiome, 2021. 9(1): p. 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kapse NG, et al. , Functional annotation of the genome unravels probiotic potential of Bacillus coagulans HS243. Genomics, 2019. 111(4): p. 921–929. [DOI] [PubMed] [Google Scholar]
- 152.Wang Y, et al. , Whole-genome analysis of probiotic product isolates reveals the presence of genes related to antimicrobial resistance, virulence factors, and toxic metabolites, posing potential health risks. BMC Genomics, 2021. 22(1): p. 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Yadav R, et al. , Gene editing and genetic engineering approaches for advanced probiotics: A review. Crit Rev Food Sci Nutr, 2018. 58(10): p. 1735–1746. [DOI] [PubMed] [Google Scholar]
- 154.Wang P, Yi Y, and Lu X, CRISPR/Cas9-Based Genome Editing Platform for Companilactobacillus crustorum to Reveal the Molecular Mechanism of Its Probiotic Properties. J Agric Food Chem, 2021. 69(50): p. 15279–15289. [DOI] [PubMed] [Google Scholar]
- 155.Rottinghaus AG, et al. , Genetically stable CRISPR-based kill switches for engineered microbes. Nat Commun, 2022. 13(1): p. 672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Goh YJ and Barrangou R, Harnessing CRISPR-Cas systems for precision engineering of designer probiotic lactobacilli. Curr Opin Biotechnol, 2019. 56: p. 163–171. [DOI] [PubMed] [Google Scholar]
- 157.Mullin E Edible CRISPR could replace antibiotics. April 17, 2017; Available from: https://www.technologyreview.com/2017/04/17/106060/edible-crispr-could-replace-antibiotics/.
- 158.Creating GEMMs for chronic disease. December 2020.
- 159.Neil K, et al. , High-efficiency delivery of CRISPR-Cas9 by engineered probiotics enables precise microbiome editing. Mol Syst Biol, 2021. 17(10): p. e10335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ramachandran G and Bikard D, Editing the microbiome the CRISPR way. Philos Trans R Soc Lond B Biol Sci, 2019. 374(1772): p. 20180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Steinway SN, et al. , Inference of Network Dynamics and Metabolic Interactions in the Gut Microbiome. PLoS Comput Biol, 2015. 11(5): p. e1004338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Cai J, et al. , Comparative efficacy and tolerability of probiotics for antibiotic-associated diarrhea: Systematic review with network meta-analysis. United European Gastroenterol J, 2018. 6(2): p. 169–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Johnson S, et al. , Is primary prevention of Clostridium difficile infection possible with specific probiotics? Int J Infect Dis, 2012. 16(11): p. e786–92. [DOI] [PubMed] [Google Scholar]
- 164.Murphy K, et al. , Genome mining for radical SAM protein determinants reveals multiple sactibiotic-like gene clusters. PLoS One, 2011. 6(7): p. e20852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Koenigsknecht MJ, et al. , Dynamics and establishment of Clostridium difficile infection in the murine gastrointestinal tract. Infect Immun, 2015. 83(3): p. 934–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Li X, et al. , Consortium of Probiotics Attenuates Colonization of Clostridioides difficile. Front Microbiol, 2019. 10: p. 2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Klemashevich C, et al. , Rational identification of diet-derived postbiotics for improving intestinal microbiota function. Curr Opin Biotechnol, 2014. 26: p. 85–90. [DOI] [PubMed] [Google Scholar]
- 168.Panayidou S, et al. , Pseudomonas aeruginosa core metabolism exerts a widespread growth-independent control on virulence. Sci Rep, 2020. 10(1): p. 9505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Christofi T, et al. , Metabolic output defines Escherichia coli as a health-promoting microbe against intestinal Pseudomonas aeruginosa. Sci Rep, 2019. 9(1): p. 14463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Schwartz R, et al. , A Drosophila Model for Clostridium difficile Toxin CDT Reveals Interactions with Multiple Effector Pathways. iScience, 2020. 23(2): p. 100865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Venugopalan V, Shriner KA, and Wong-Beringer A, Regulatory oversight and safety of probiotic use. Emerging infectious diseases, 2010. 16(11): p. 1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Sanders ME, How do we know when something called “probiotic” is really a probiotic? A guideline for consumers and health care professionals. Functional Food Reviews, 2009. 1(1): p. 3–12. [Google Scholar]
- 173.Au C, et al. , Probiotics Regulation in Asian and Australasian Countries, in Lactic Acid Bacteria. 2019, CRC Press. p. 631–682. [Google Scholar]
- 174.Hazards E.P.o.B., Scientific Opinion on the maintenance of the list of QPS biological agents intentionally added to food and feed (2013 update). EFSA Journal, 2013. 11(11): p. 3449. [Google Scholar]
- 175.van Loveren H, Sanz Y, and Salminen S, Health claims in Europe: probiotics and prebiotics as case examples. Annual review of food science and technology, 2012. 3: p. 247–261. [DOI] [PubMed] [Google Scholar]
- 176.Reigadas E, et al. , How to: prophylactic interventions for prevention of Clostridioides difficile infection. Clin Microbiol Infect, 2021. 27(12): p. 1777–1783. [DOI] [PubMed] [Google Scholar]
- 177.Higgins JP, et al. , Cochrane handbook for systematic reviews of interventions. 2019: John Wiley & Sons. [Google Scholar]
- 178.Cleusix V, et al. , Inhibitory activity spectrum of reuterin produced by Lactobacillus reuteri against intestinal bacteria. BMC Microbiol, 2007. 7: p. 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Gebhart D, et al. , A modified R-type bacteriocin specifically targeting Clostridium difficile prevents colonization of mice without affecting gut microbiota diversity. MBio, 2015. 6(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Olson JK, et al. , An enhanced Lactobacillus reuteri biofilm formulation that increases protection against experimental necrotizing enterocolitis. Am J Physiol Gastrointest Liver Physiol, 2018. 315(3): p. G408–G419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.McFarland LV, Efficacy of Single-Strain Probiotics Versus Multi-Strain Mixtures: Systematic Review of Strain and Disease Specificity. Dig Dis Sci, 2020. [DOI] [PubMed] [Google Scholar]
- 182.Klarin B, et al. , Lactobacillus plantarum 299v reduces colonisation of Clostridium difficile in critically ill patients treated with antibiotics. Acta Anaesthesiol Scand, 2008. 52(8): p. 1096–102. [DOI] [PubMed] [Google Scholar]
- 183.Barker AK, et al. , A randomized controlled trial of probiotics for Clostridium difficile infection in adults (PICO). J Antimicrob Chemother, 2017. 72(11): p. 3177–3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Gao XW, et al. , Dose-response efficacy of a proprietary probiotic formula of Lactobacillus acidophilus CL1285 and Lactobacillus casei LBC80R for antibiotic-associated diarrhea and Clostridium difficile-associated diarrhea prophylaxis in adult patients. Am J Gastroenterol, 2010. 105(7): p. 1636–41. [DOI] [PubMed] [Google Scholar]
- 185.Beausoleil M, et al. , Effect of a fermented milk combining Lactobacillus acidophilus Cl1285 and Lactobacillus casei in the prevention of antibiotic-associated diarrhea: a randomized, double-blind, placebo-controlled trial. Can J Gastroenterol, 2007. 21(11): p. 732–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Maziade P-J, Lussier D, and Dubé F. 2417. Feasibility and Safety of Using a Probiotic Comprised of Lactobacillus acidophilus CL1285, L. casei LBC80R and L. rhamnosus CLR2 for C. difficile Infection Prevention Among Antibiotic Users: 15-Years of Prospective Results from a Single-Center. in Open Forum Infectious Diseases. 2019.
- 187.Hickson M, et al. , Use of probiotic Lactobacillus preparation to prevent diarrhoea associated with antibiotics: randomised double blind placebo controlled trial. BMJ, 2007. 335(7610): p. 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Gorbach SL, Chang TW, and Goldin B, Successful treatment of relapsing Clostridium difficile colitis with Lactobacillus GG. Lancet, 1987. 2(8574): p. 1519. [DOI] [PubMed] [Google Scholar]
- 189.Kotowska M, Albrecht P, and Szajewska H, Saccharomyces boulardii in the prevention of antibiotic-associated diarrhoea in children: a randomized double-blind placebo-controlled trial. Aliment Pharmacol Ther, 2005. 21(5): p. 583–90. [DOI] [PubMed] [Google Scholar]
- 190.McFarland LV, Can Saccharomyces boulardii prevent antibiotic-associated diarrhea in children? Nat Clin Pract Gastroenterol Hepatol, 2005. 2(6): p. 262–3. [DOI] [PubMed] [Google Scholar]
- 191.McFarland LV, et al. , A randomized placebo-controlled trial of Saccharomyces boulardii in combination with standard antibiotics for Clostridium difficile disease. JAMA, 1994. 271(24): p. 1913–8. [PubMed] [Google Scholar]
- 192.Ehrhardt S, et al. , Saccharomyces boulardii to Prevent Antibiotic-Associated Diarrhea: A Randomized, Double-Masked, Placebo-Controlled Trial. Open Forum Infect Dis, 2016. 3(1): p. ofw011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Gerding DN, et al. , Administration of spores of nontoxigenic Clostridium difficile strain M3 for prevention of recurrent C. difficile infection: a randomized clinical trial. JAMA, 2015. 313(17): p. 1719–27. [DOI] [PubMed] [Google Scholar]
- 194.Allen SJ, et al. , Lactobacilli and bifidobacteria in the prevention of antibiotic-associated diarrhoea and Clostridium difficile diarrhoea in older inpatients (PLACIDE): a randomised, double-blind, placebo-controlled, multicentre trial. Lancet, 2013. 382(9900): p. 1249–57. [DOI] [PubMed] [Google Scholar]


