Summary
Yamanaka factors are essential for establishing pluripotency in embryonic stem cells, but their function in multipotent stem cell populations is poorly understood. Here we show that OCT4 and SOX2 cooperate with tissue-specific transcription factors to promote neural crest formation. By assessing avian and human neural crest cells at distinct developmental stages, we characterized the epigenomic changes that occur during their specification, migration, and early differentiation. This analysis determined that the OCT4-SOX2 dimer is required to establish a neural crest epigenomic signature that is lost upon cell fate commitment. The OCT4-SOX2 genomic targets in the neural crest differ from those of embryonic stem cells, indicating the dimer displays context-specific functions. Binding of OCT4-SOX2 to neural crest enhancers requires pioneer factor TFAP2A, which physically interacts with the dimer to modify its genomic targets. Our results demonstrate how Yamanaka factors are repurposed in multipotent cells to control chromatin organization and define their developmental potential.
eTOC
Utilizing genomic analysis to survey neural crest development, Hovland and Bhattacharya et al. identify Yamanaka factors OCT4 and SOX2 as key regulators of multipotency. The authors show how components of the pluripotency network cooperate with tissue specific pioneer transcription factors to generate the neural crest stem cell state.
Introduction
In embryonic stem cells (ESCs), pluripotency is achieved by the action of the Yamanaka factors (OCT4, SOX2, KLF4, c-MYC), which form a core regulatory network that prevents cell fate commitment (Takahashi and Yamanaka, 2006). However, the mechanisms supporting the enhanced plasticity of multipotent stem cell populations are less understood. A multipotent stem cell population of remarkable developmental potential is the neural crest. These ectodermal progenitors can circumvent the boundaries between germ layers to give rise to cells and tissues that are generally formed by the mesoderm. Neural crest derivatives include the craniofacial skeleton, the peripheral nervous system, the pigmentation of the skin, and cardiac tissue (Le Douarin and Kalcheim, 1999). Classic experiments involving the transplantation of quail cells to chick embryos identified the extensive array of derivatives formed by the neural crest (Le Douarin, 1973). In vivo cell labeling by intracellular injection of vital dyes demonstrated that individual neural crest cells give rise to multiple cell types including neurons, glia, and pigmented cells (Bronner-Fraser and Fraser, 1988). More recently, genetic fate mapping of trunk neural crest cells with Confetti mice confirmed that both pre-migratory and migratory neural crest cells are multipotent, and that fate restriction takes place at later stages of development (Baggiolini et al., 2015).
The formation and differentiation of neural crest cells is orchestrated by a complex gene regulatory network (GRN), which has been substantially expanded in the last years with genomic analysis and functional studies (Martik and Bronner, 2017; Simoes-Costa and Bronner, 2015; Williams et al., 2019a). Yet, the features of this GRN that support multipotency remain unresolved. While some studies suggest that neural crest cells share part of the ESC pluripotency program, others propose that their developmental plasticity is controlled by a tissue-specific regulatory state that emerges later in development. Buitrago-Delgado and colleagues proposed that neural crest cells retain blastula-stage genes that are important for pluripotency (Buitrago-Delgado et al., 2015). Zalc and colleagues examined the expression and function of OCT4 and suggested that the ESC pluripotency program is reactivated in the cranial neural crest lineage (Zalc et al., 2021). Both studies provide support for the idea that neural crest multipotency is related to the ESC pluripotency network. This contrasts with the classical view that neural crest multipotency is controlled by a unique regulatory circuit that emerges during specification (Dupin et al., 2018). This view is supported by functional studies showing that disruption of known neural crest factors like PAX7, FOXD3, and SOX10 leads to an imbalance in multipotency, suggesting an independent mechanism from that of ESCs (Dottori et al., 2001; Honoré et al., 2003; Kondoh et al., 2012; Lukoseviciute et al., 2018). Furthermore, developmental trajectories from single-cell transcriptome analysis showed that neural crest cells diverge from the ESC-state during ectoderm specification (Briggs et al., 2018).
Here, we re-examine these models by surveying the transcriptional and epigenomic dynamics that accompany the transition of neural crest cells to a differentiated state. This allowed for the identification of pluripotency factors OCT4 and SOX2 (Soufi et al., 2012) as major regulators of the epigenomic landscape in naive neural crest cells. We found that the OCT4-SOX2 dimer plays an important role in neural crest formation, although its genomic targets in the neural crest are distinct from those in ESCs. Thus, while Yamanaka factors are important for neural crest developmental plasticity, they promote a unique chromatin landscape specific to this cell population. These results illustrate how components of the pluripotency network are co-opted to regulate the epigenome of multipotent stem cells.
Results
Transcriptomic and epigenomic dynamics of avian neural crest development
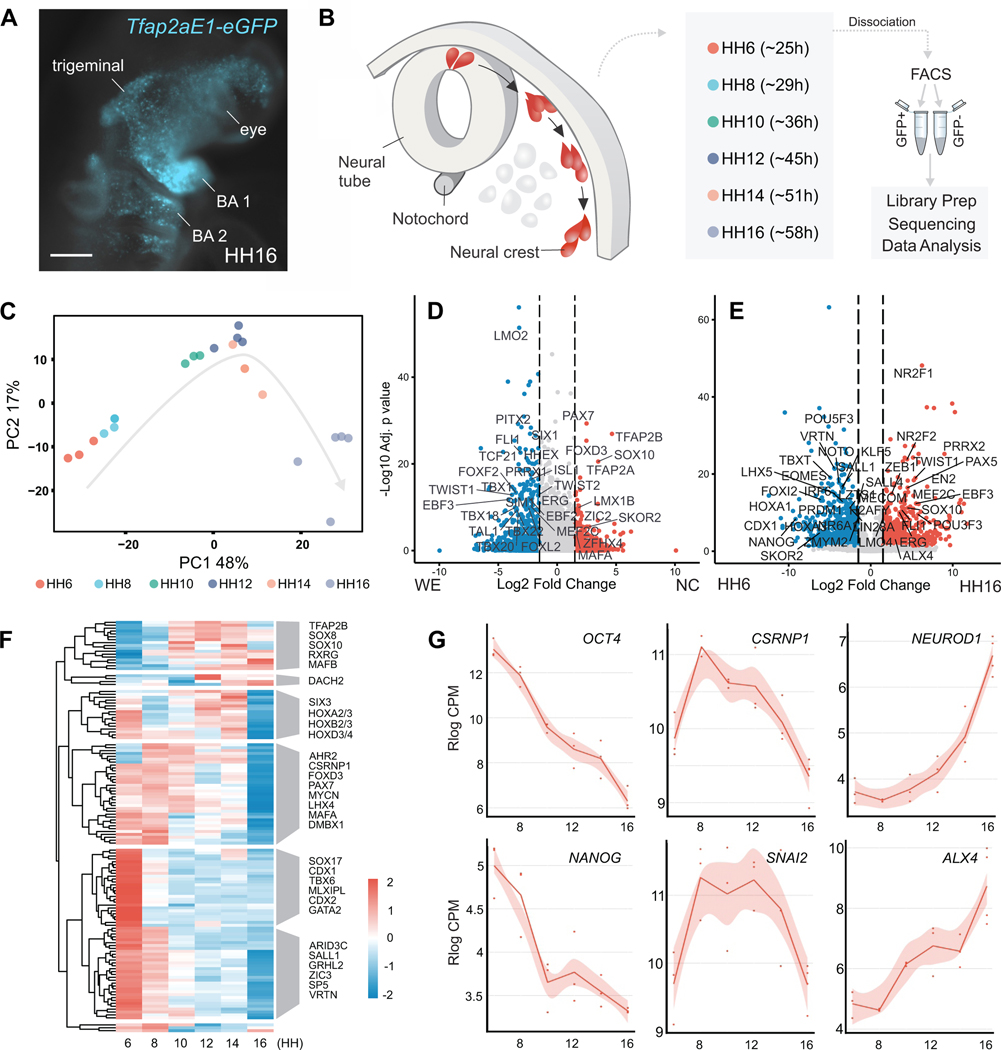
Genomic studies have identified additional players in neural crest development that allowed for substantial expansion of GRN circuitry (Hovland et al., 2020; Simoes-Costa and Bronner, 2016; Soldatov et al., 2019; Williams et al., 2019a). Yet, existing datasets lack the temporal resolution for examining the transition from multipotency to differentiation. To address this, we assembled an RNA-Seq time-course of avian neural crest cells at stages corresponding to induction, specification, migration, and early differentiation. We labeled neural crest cells from Hamburger and Hamilton (HH) stage 6 to HH16 using a neural crest-specific enhancer (Tfap2aE1) driving GFP expression (Attanasio et al., 2013; Hamburger and Hamilton, 1951; Rothstein and Simoes-Costa, 2020) (Figure 1A). Unlike other neural crest enhancers (Barembaum and Bronner, 2013; Simões-Costa et al., 2012), Tfap2aE1 is active throughout neural crest development (Figures S1A and S1B).
Figure 1 – Transcriptional dynamics of cranial neural crest development.
(A) Ex ovo electroporation of the Tfap2aE1 enhancer driving GFP faithfully labels neural crest derivatives in HH16 embryos. Expression is seen in the facial mesenchyme, the trigeminal ganglia, and branchial arches (BA1, BA2). Scale bar 200 μm.
(B) Schematic of the cross-section of the developing avian embryo head. Neural crest (red) and whole embryo cells (grey) were isolated at six developmental stages by FACS sorting for GFP+ neural crest or GFP− cells from the embryonic head.
(C) Principal Component Analysis (PCA) of GFP+ neural crest RNA-Seq libraries.
(D) Volcano plot of the likelihood ratio test (LRT) comparing neural crest (NC) and embryonic head (WE) RNA-Seq samples. Significantly (p-adj* < 0.05 & log2FC > 1.5) enriched (red) or depleted (blue) genes are colored, and most transcription factors with a p-adj* of 1e-8 or lower are labeled.
(E) Volcano plot of showing genes significantly (p-adj* < 0.05 & log2FC > 1.5) enriched (red) or depleted (blue) in differentiating neural crest. Most transcription factors with a p-adj* of 1e-8 or lower are labeled.
(F) Hierarchical clustering of significantly enriched (p-adj* < 0.05) transcription factors identified from ImpulseDE2.
(G) Visualization of pluripotency (OCT4, NANOG), neural crest (CSRNP1, SNAI2), and differentiation (NEUROD1, ALX4) markers’ expression from our RNA-Seq Shiny app.
HH, Hamburger and Hamilton stage; *, Bonferroni-corrected p-value; NC, neural crest; WE, whole embryo
We electroporated chicken embryos at HH4 with Tfap2aE1-GFP and dissected them at the desired stages (HH6, HH8, HH10, HH12, HH14, and HH16). GFP+ cranial neural crest cells and GFP- cells were isolated from embryonic heads with fluorescence-activated cell sorting (FACS). We performed bulk RNA-seq with at least 5,000 cells per sample and triplicates for each timepoint (Supplemental Table 1) (Figure 1B). Principal Component Analysis (PCA) of our RNA-seq datasets showed that GFP+ samples align chronologically along the first principal component (Figure 1C), while the GFP+ neural crest and GFP- embryonic head samples separate along the second principal component (Figure S1C). Next, we compared the transcriptomes of GFP+ and GFP- samples using a likelihood ratio test (LRT), to identify a neural crest-specific gene signature (Supplemental Table 2) (Love et al., 2014). The resulting gene set of 279 genes (padj <0.05 and Log2FC > 1) included bona fide markers such as SOX10, FOXD3, and TFAP2B as well as 38 other transcription factors (Figure 1D).
Next, to characterize the transcriptional changes associated with loss of neural crest multipotency, we performed pairwise differential gene expression analysis between neural plate border (HH6) and differentiating neural crest cells (HH16) using a negative binomial Wald test (Figure 1E). This analysis showed that neural plate border cells have high expression of several canonical pluripotency genes like OCT4 (POU5F3 in chick), SOX2, LIN28A, and KLF5, which are lost at later developmental stages. We also observed increased transcription of lineage-specific genes such as NEUROD1, ALX1, and PRRX2 in HH16 cells, corresponding to the early stages of neural crest differentiation (Soldatov et al., 2019) (Figures 1E and 1G).
We further examined these gene expression changes by performing hierarchical clustering analysis with neural crest-enriched transcription factors identified by the LRT test enriched compared to our whole-embryo samples (Figure 1F). This allowed us to group components of the GRN into distinct temporal modules. The largest module containing 59 genes, included general pluripotency factors that are downregulated upon differentiation. This analysis also confirmed the sequential activation of early (PAX7, CSRNP1, and TFAP2A) and late (TFAP2B, SOX10, and RXRG) specifier genes (Figures 1F and 1G). Consistent with previous reports (Bhattacharya et al., 2018), we also observed that both pluripotency and neural crest specification genes are downregulated in late neural crest cells (Figures 1F and 1G). In situ hybridization analysis confirmed that previously uncharacterized neural crest genes like SKOR2, MAFA, and KAT6B are expressed in this cell population (Figure S1D). Taken together, these analyses identified the transcriptional shifts that characterize the cell state changes observed during neural crest development. These datasets have been compiled into an easy-to-use public R Shiny app, which can be accessed at ash274.shinyapps.io/RNA-Seq_App/.
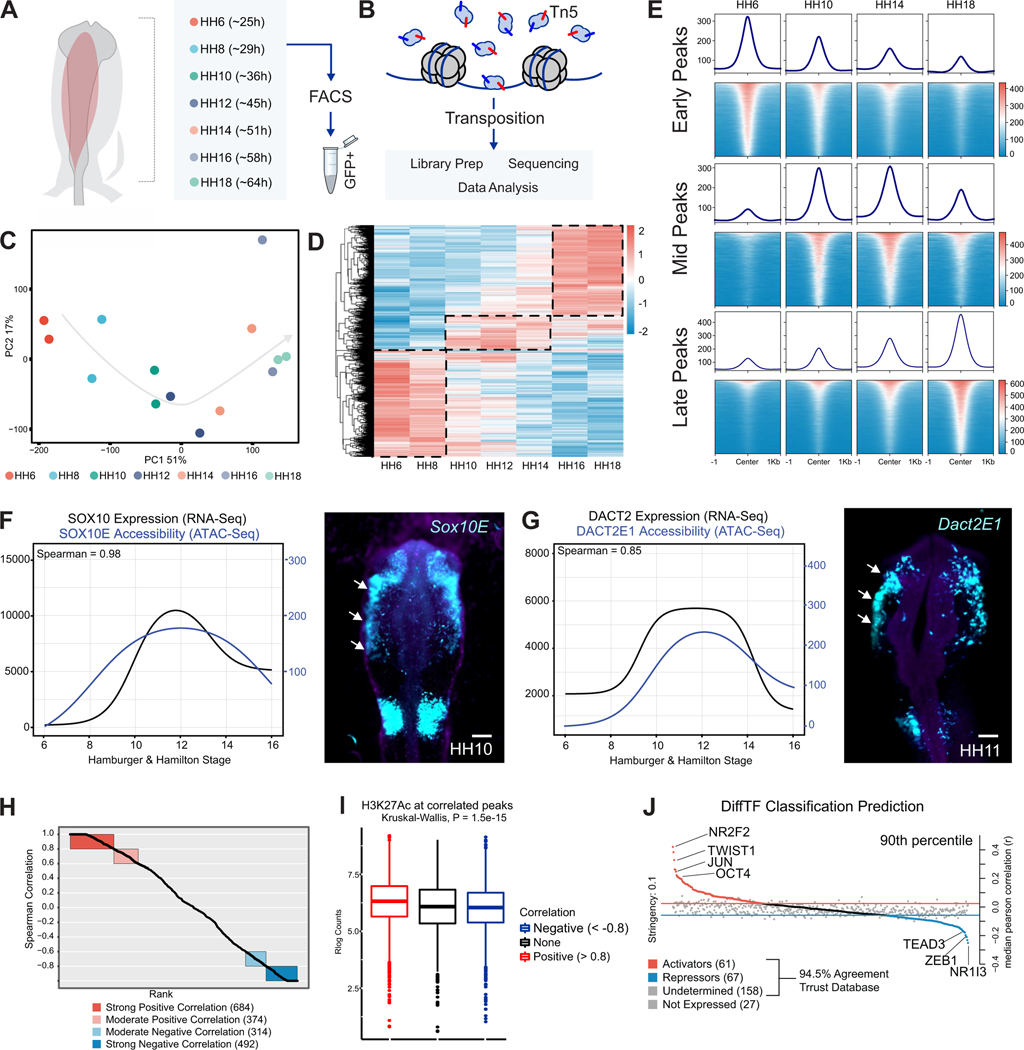
To understand how developmental transitions are regulated at the chromatin level, we assayed the epigenomic landscape of neural crest at distinct developmental stages. We employed the Omni-ATAC protocol (Corces et al., 2017) to map chromatin accessibility in Tfap2aE1-GFP labeled cranial neural crest cells isolated at seven embryonic timepoints (HH6-HH18) (Figures 2A and 2B; Figure S2A; Supplemental Table 3). PCA analysis of ATAC-seq datasets showed that the samples segregated according to developmental stage in the first principal component, representing 51% of the variance (Figure 2C). We combined all peaks across developmental stages, filtered out peaks present in whole embryo datasets, and performed differential peak enrichment analysis using DiffBind (Rory and Gord, 2011; Ross-Innes et al., 2012). LRT analysis was used to identify peaks enriched across all stages of neural crest development. Next, we performed hierarchical clustering of these peaks, which identified three main groups: early, mid, and late peaks (Figure 2D; Figure S2B). Consistent with our RNA-seq analysis (Figure 1F) many ‘early’ peaks displayed high accessibility in multipotent neural crest (HH6-HH8) but were closed upon differentiation (Figures 2D and 2E). Alternatively, ‘mid’ and ‘late’ peaks displayed increased accessibility at stages HH10-H12 and HH14-HH18, respectively (Figures 2D and 2E).
Figure 2 – Profiling the chromatin landscape of cranial neural crest cells with Omni ATAC-seq.
(A) Ex ovo electroporation of Tfap2aE1-GFP was used to label developing neural crest cells. GFP+ neural crest cells were isolated at seven developmental stages via FACS.
(B) FACS isolated nuclei from each stage were subjected to Tn5-mediated transposition with Omni-ATAC-Seq and were processed for short-read sequencing.
(C) Principal Component Analysis (PCA) of neural crest ATAC-Seq libraries. The first principal component, representing 51% of sample variation, separates samples according to developmental time.
(D) Averaged, Rlog normalized counts for each timepoint at differential peaks (FDR < 0.05) from an LRT test across all timepoints. Hierarchical clustering shows three large clusters of peaks (dashed lines) that were grouped together into early, mid, and late peak sets.
(E) Tornado plots showing the genome-wide accessibility profile of each peak set (from D) at four different stages of neural crest development.
(F) ImpulseDE2 models of the Sox10E2 enhancer and the SOX10 gene show a highly positive (0.98) Spearman correlation. Electroporation of this element driving a GFP reporter in vivo recapitulates SOX10 expression. Scale bar 100 μm.
(G) ImpulseDE2 models of the Dact2E1 enhancer and the DACT2 gene also share a highly positive (0.85) Spearman correlation and similar in vivo reporter activity compared to DACT2 expression. Scale bar 100 μm.
(H) ImpulseDE2 models for both expression (RNA-Seq) and accessibility (ATAC-Seq) of 146 neural crest genes and their nearby genomic elements identify 684 elements with a strong (>0.8) positive correlation.
(I) Boxplots showing that activating H3K27AC mark is enriched in positively correlated elements compared to negative correlations or a random background.
(J) Plot showing strongly activating (NR2F2, TWIST1, and OCT4) and repressing transcription factors (TEAD3, ZEB1, and NR1I3) in neural crest cells, as determined by DiffTF analysis.
***P≤0.001
To examine how these epigenomic changes are coupled with shifts in gene expression, we integrated our ATAC-seq and RNA-seq datasets using ImpulseDE2 (Fischer et al., 2018). By considering the five nearest genes to every ATAC peak, we correlated changes in chromatin accessibility to temporal patterns of gene expression (Supplemental Table 4). We observed that the accessibility of known neural crest enhancers, like Sox10E2 and Dact2E1, displayed a strong correlation with the expression of their cognate genes (Figures 2F and 2G) (Betancur et al., 2010; Rothstein and Simoes-Costa, 2020). This prompted us to expand our analysis to identify putative enhancers of significantly enriched neural crest genes identified by RNA-Seq (Supplemental Table 4). We found that 684 out of 3526 accessible peaks tested displayed a strong positive correlation (Spearman > 0.8) with expression levels of 146 neural crest genes (Figure 2H). These peaks included enhancers for neural crest genes like MSX1, FOXD3, SOX8, SOX9 (Azambuja and Simoes-Costa, 2021) (Figures S2D, S2E, S2F, S2G), and multiple enhancers from the SOX10 locus (Figure S2H) (Williams et al., 2019b). To test if cis-regulatory elements with strong correlation scores were more likely to act as enhancers, we measured levels of H3K27AC deposition in ATAC-seq peaks using a previously published dataset (Rothstein and Simoes-Costa, 2020). Indeed, we observed that these peaks displayed higher H3K2Ac levels than regions with weak or negative correlations (Figure 2I).
Finally, we employed DiffTF to identify the transcription factors with the strongest activating or repressing activities during neural crest development (Berest et al., 2019). This tool correlates the expression levels of transcription factors across time with the accessibility of peaks that contain their binding motifs. DiffTF identified OCT4, NR2F2, TWIST1, and JUN as activators in the neural crest GRN, whereas TEAD3, ZEB1, and NR1I3 were assigned as repressors (Figure 2J and Supplemental Table 5). These regulators are active at distinct developmental stages. For instance, the motif scores of DiffTF activators indicate that while some are active in the early neural crest, others seem to be involved in cell fate commitment (Figure S2C). Taken together, these results describe the epigenomic and transcriptomic changes inherent to neural crest development and identify genes that may play essential roles in the transition from multipotency to the differentiated state.
The OCT4-SOX2 dimer regulates the early neural crest epigenome
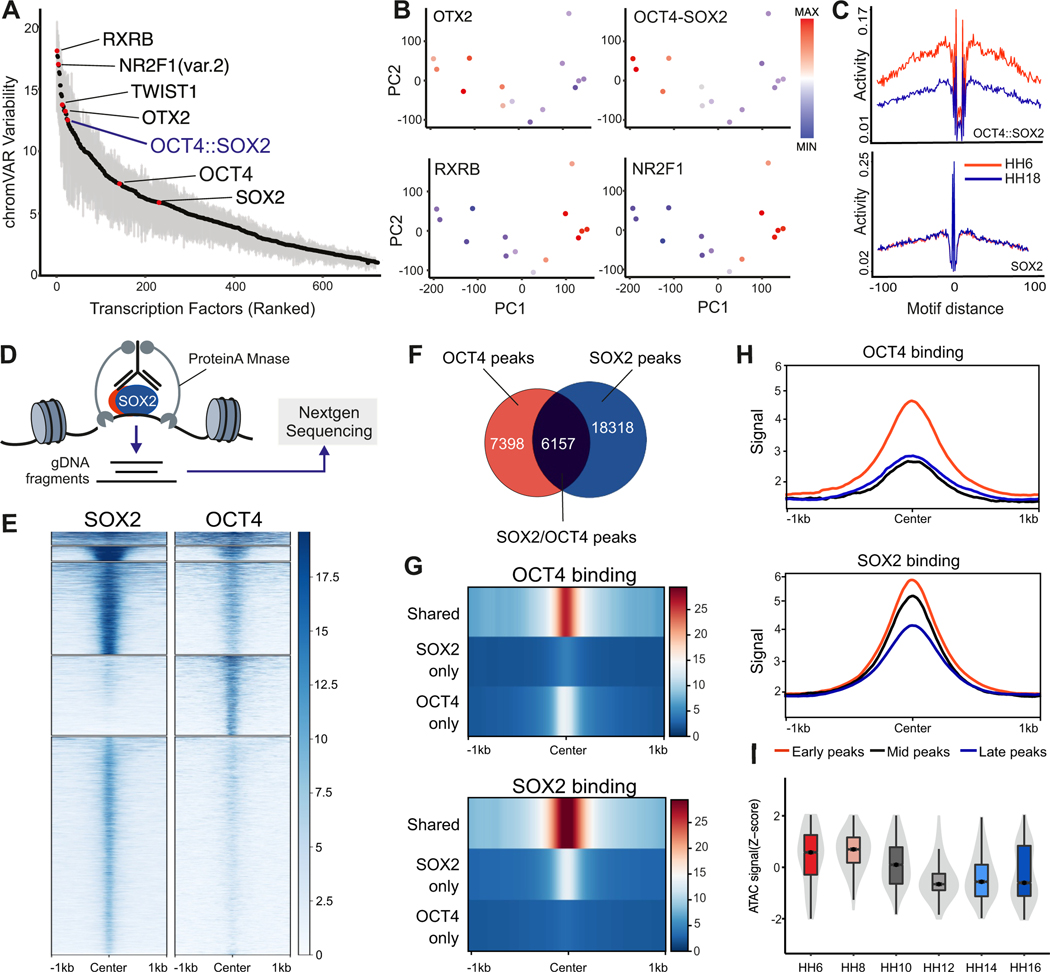
Next, we searched for the transcription factors that act as major regulators of neural crest identity. We utilized chromVAR to find transcription factor motifs that display high variability across developmental stages (Schep et al., 2017). The top-ranked motifs included RXRB, NR2F1, OTX2, and the motif for the OCT4-SOX2 dimer (Figure 3A). Consistent with their expression dynamics, the variability scores of OTX2 and the OCT4-SOX2 dimer motifs were highest in early neural crest cells, while the scores for the RXRB and NR2F1 motifs increased upon differentiation (Figure 3B). We next investigated the chromatin footprint for these motifs using HINT-ATAC (Li et al., 2019). An unbiased comparison between our ATAC-seq data from the earliest (HH6) and latest (HH18) stages confirmed that one of the highest enriched motif footprints in early neural crest cells was the OCT4-SOX2 dimer (Figure S3A), with SOX2 alone displaying a weaker footprint (Figure 3C). Corresponding footprints of OTX2 and NR2F1 were active in HH6 or HH18, respectively (Figures S3B and S3C). These results are consistent with our DiffTF results that place OCT4 as an important GRN activator and suggest that it works with SOX2 to promote neural crest identity.
Figure 3 – OCT4 and SOX2 regulate the epigenetic landscape of early neural crest cells.
(A) Variability ranked-motif plot of TF motifs enriched in neural crest cells. Some highly variable motifs across neural crest development include: RXRB, OTX2, and OCT4-SOX2.
(B) Motif Z-scores for OTX2, OCT4-SOX2, RXRB, and NR2F1 projected onto the ATAC-Seq PCA of neural crest samples.
(C) Individual footprinting plots for the OCT4-SOX2 and SOX2 motifs at the earliest (HH6) and latest (HH18) developmental timepoints. These plots display a specific enrichment for the OCT4-SOX2 motif at HH6.
(D) Diagram of CUT&RUN experiments to map genome occupancy of transcription factors of interest.
(E) Tornado plots depicting the genomic occupancy of SOX2 and OCT4 in HH8 neural crest cells.
(F) Venn diagram showing the overlap in SOX2 and OCT4 CUT&RUN peaks in HH8 neural fold samples.
(G) Compressed genomic occupancy profiles show that the strongest binding events occur at shared OCT4 and SOX2 peaks.
(H) CUT&RUN signal enrichment centered at the previously identified early, mid, and late ATAC-seq peak sets show that both OCT4 and SOX2 have highest binding at early ATAC peaks.
(I) Normalized ATAC-Seq accessibility of shared OCT4 and SOX2 peaks over the neural crest developmental time course.
*P≤0.05, **P≤0.01, ***P≤0.001
To test if OCT4 and SOX2 directly modulate the neural crest epigenome, we mapped their genomic occupancy in HH8 neural crest cells with Cleavage Under Targets & Release Using Nuclease (CUT&RUN) (Skene and Henikoff, 2017) (Figure 3D; Figures S3D and S3E). Peak calling identified ~24,000 and ~13,000 peaks for SOX2 and OCT4 respectively, of which 6,157 were shared between the two factors (Figures 3E and 3F). Consistent with our previous analysis indicating the importance of OCT4-SOX2 dimerization, binding of both OCT4 and SOX2 was significantly higher at co-occupied regions (Figure 3G). Integration of our neural crest-specific early, mid, and late ATAC peaks with OCT4 and SOX2 CUT&RUN showed that these factors had the highest binding in regions accessible early during neural crest development (Figure 3H). The shared OCT4-SOX2 CUT&RUN peaks were specifically open in multipotent cells and lost accessibility over neural crest differentiation (Figure 3I). These results support the cooperative action of the two factors in early neural crest cells.
To confirm that SOX2 and OCT4 are indeed functioning as heterodimers in neural crest cells, we performed Enhanced Chromatin Occupancy (EChO) with our CUT&RUN datasets to determine transcription factor binding events at a base-pair resolution (Meers et al., 2019). This analysis identified over 12,000 foci within the shared CUT&RUN peaks that were simultaneously bound by OCT4 and SOX2 in neural crest cells (Supplemental Table 6). We observed co-binding events of these factors around neural plate border genes such as PAX7, SP5, and ZIC1, consistent with the role of the heterodimer as a direct regulator of the early neural crest GRN (Figures S3F and S3G). Lastly, we performed Proximity Ligation Assays (PLAs) to identify putative molecular interactions between endogenous OCT4 and SOX2 proteins (Alam, 2018). A comparison of HH8 neural folds and surrounding mesoderm showed increased proximity between OCT4 and SOX2 in early neural crest cells (Figures S3H and S3I). Taken together, these observations indicate that the OCT4-SOX2 heterodimer regulates the epigenomic signature of multipotent neural crest cells.
OCT4-SOX2 cooperates with TFAP2A to activate the neural crest GRN
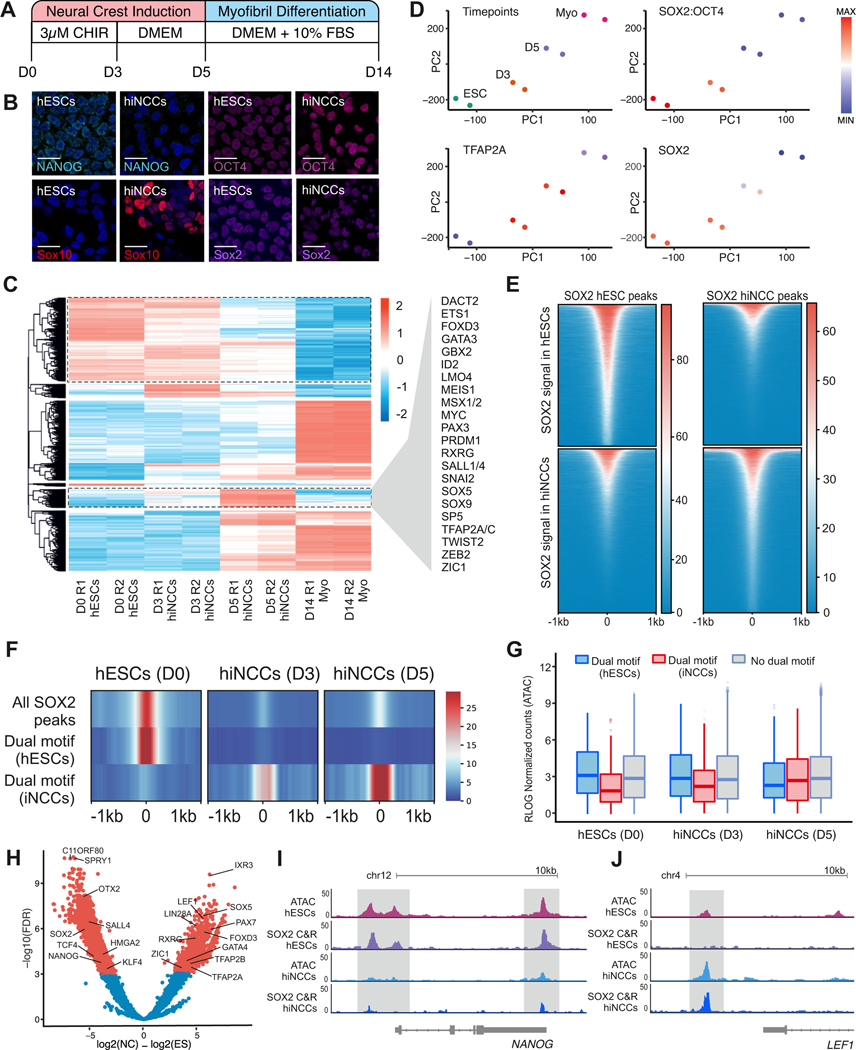
We next examined if the OCT4-SOX2 dimer displayed conserved or divergent functions in neural crest and ESCs. To define whether neural crest formation involves (i) the retention of a subset of the ESC pluripotency program or (ii) the establishment of a new, tissue-specific epigenomic signature, we employed an in vitro model of neural crest formation from hESCs using the WNT agonist CHIR99021 (Gomez et al., 2019) (Figure 4A). Human induced neural crest cells (hiNCCs) displayed loss of pluripotency factor NANOG but showed expression of SOX10, OCT4, and SOX2 by Day 5 of induction (Figure 4B; Figure S4A). To examine accessibility dynamics along in vitro neural crest induction, we employed ATAC-seq to profile chromatin accessibility in cells from D0 (hESCs), D3 (early hiNCCs), D5 (hiNCCs), and D14 (differentiated myoblasts) of the induction protocol (Figure 4A; Figures S4B and S4C). We performed pairwise comparisons using DiffBind for each stage, identifying over 42,900 unique dynamic loci (adj-pval < 1e-6). This analysis demonstrated extensive remodeling of the epigenomic landscape during neural crest induction (Figure 4C).
Figure 4 – SOX2 and OCT4 control distinct sets of regulatory regions in pluripotent ES cells and multipotent neural crest cells.
(A) Schematic of the 5-day protocol used for obtaining induced human neural crest cells (hiNCCs) from human embryonic stem cells (hESCs).
(B) Immunohistochemistry for NANOG, OCT4, SOX10, and SOX2 in hESCs (D0) and hiNCCs (D5) show a loss in NANOG expression and gain in SOX10 upon neural crest induction. OCT4 and SOX2 levels were comparable between D0 and D5 of induction. Scale bar 20
(C) Heatmap showing the differentially accessible ATAC-seq peaks between D0, D3, D5, and D14 hiNCCs. Neural crest genes associated with D5 specific peaks are highlighted.
(D) PCA plot depicting the progression of hESC to fully differentiated Myofibrils after fourteen days. ChromVAR analysis shows enrichment of OCT4-SOX2, TFAP2A, and SOX2 motifs in the different ATAC samples.
(E) Tornado plot showing CUT&RUN signal of SOX2 at genomic peaks bound by the factor at D0 hESCs and D5 iNCCs.
(F) Compressed genomic occupancy plot of SOX2 CUT&RUN signal at all SOX2 peaks, SOX2 peaks that are enriched in hESCs with an OCT4-SOX2 dual motif, and SOX2 peaks that are enriched in hiNCCs with an OCT4-SOX2 dual motif.
(G) Normalized ATAC-Seq signal at sites containing hESC-specific dual motifs, hiNCC-specific dual motifs, and peaks without a dual motif at D0, D3, and D5.
(H) Volcano plot showing the distinct targets of SOX2 at D0 and D5 of neural crest induction.
(I) Genome browser view of ATAC-Seq and SOX2 CUT&RUN at the Nanog locus.
(J) Genome browser view of ATAC-Seq and SOX2 CUT&RUN at the Lef1 locus.
*P≤0.05, **P≤0.01, ***P≤0.001
We next examined how OCT4-SOX2 regulates the neural crest epigenome during induction. Analyzing motif enrichment via chromVAR, we found high scores for the OCT4-SOX2 motif in both hESCs and early hiNCCs (Figure 4D). The motif score for the neural crest-specific pioneer factor TFAP2A increased as early as D3 during hiNCC induction. We next wanted to determine whether OCT4-SOX2 (i) promotes maintenance of the ESC signature, or (ii) promotes accessibility of neural crest-specific regulatory regions. We thus examined how the occupancy of OCT4-SOX2 changes as cells transition from hESC to hiNCCs during the induction process. To this end, we first performed CUT&RUN for SOX2 at D5 hiNCCs and compared it to previously published SOX2 CUT&RUN in hESCs (Meers et al., 2019) (Figures S4D and S4E). The results show a shift in the patterns of SOX2 occupancy in hESCs and hiNCCs. The genomic regions bound by SOX2 in hESCs lost transcription factor binding during induction, while a distinct set of genomic regions gained SOX2 binding in Day5 hiNCCs (Figure 4E).
Next, we used FIMO to scan for occurrences of the OCT4-SOX2 dual motif within all SOX2 peaks, SOX2 peaks enriched in hESCs, and SOX2 peaks enriched in hiNCCs (Grant et al., 2011). When we quantified the SOX2 CUT&RUN signal at these locations, we observed this factor relocates to unique dual-motif locations in D5 hiNCCs (Figure 4F). Consistent with the well-documented role of SOX2 as a driver of chromatin accessibility (Blassberg et al., 2020; Bunina et al., 2020; Zhang et al., 2019), we also found that ATAC signal at dimer-associated regions changed according to SOX2 occupancy (Figure 4G). These data support the hypothesis that OCT4-SOX2 dimers bind to a new set of neural crest cis-regulatory regions upon specification. We next used DiffBind to quantify SOX2 CUT&RUN signal in hESCs and D5 hiNCCs and identify differentially bound regions (Figure 4H). This analysis demonstrated enrichment of SOX2 binding near many neural crest genes (LEF1, FOXD3, and TFAP2A) and loss of SOX2 binding near canonical pluripotency targets (NANOG, SOX2, and KLF4). Inspection of ATAC-Seq and SOX2 binding at the NANOG and LEF1 loci demonstrated individual regulatory elements nearby with a loss or gain of signal across both assays, respectively (Figures 4I and 4J). These results indicate that OCT4-SOX2 dimer promotes distinct epigenomic states in ESCs vs. neural crest cells.
Misregulation of OCT4 and SOX2 results in defects in neural crest fate commitment
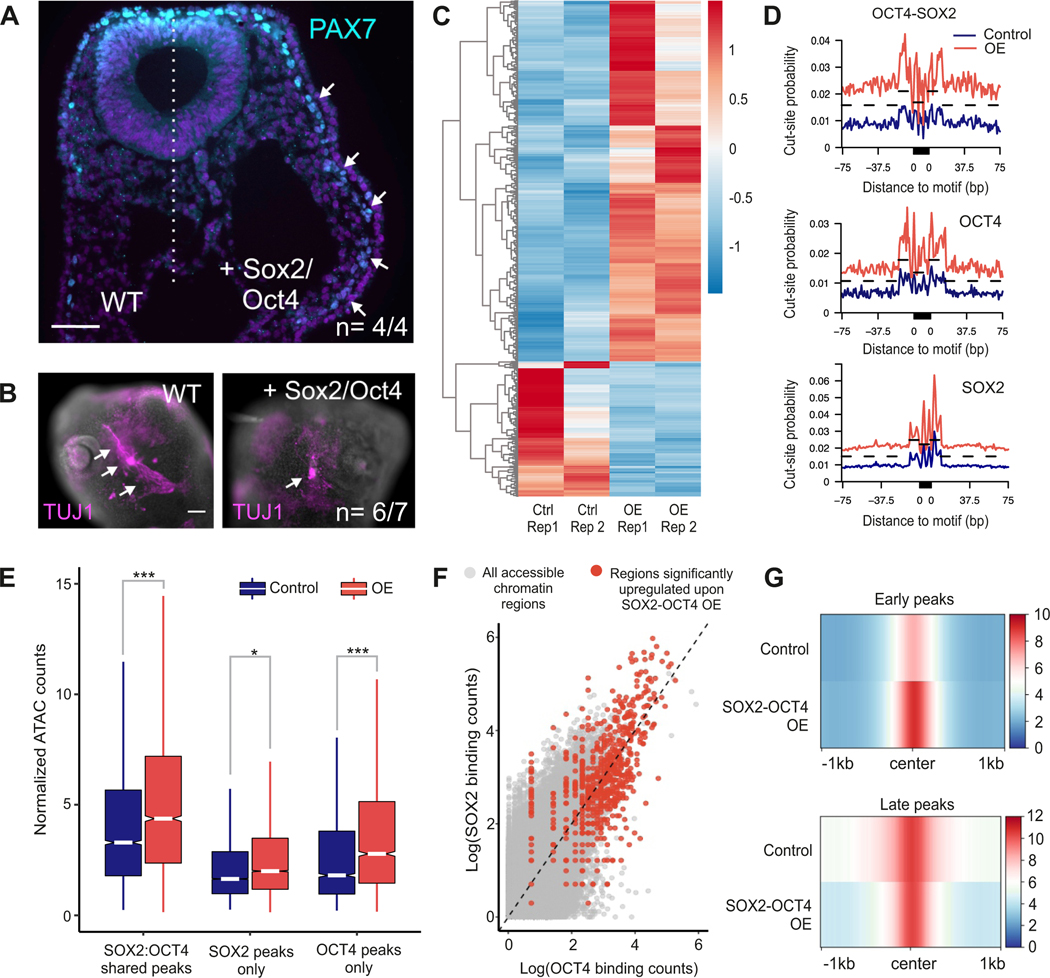
To establish the relevance of OCT4 and SOX2 in the regulation of the neural crest multipotent cell state, we manipulated their expression dynamics. We bilaterally electroporated gastrula-stage chicken embryos (HH4) with an expression vector encoding both SOX2 and OCT4 (pCI-OCT4-P2A-SOX2-H2B-RFP) to drive sustained expression of these genes in late neural crest cells (Figure S5A). Prolonged expression of SOX2 and OCT4 resulted in the maintenance of high levels of the early neural crest gene PAX7 in the late migratory neural crest (HH13), at stages when it would normally be downregulated (Figure 5A). This prevented proper neural crest differentiation as evidenced by gross morphological defects in the trigeminal ganglia in HH16 embryos. TUJ1 staining to label the cranial ganglia showed a reduced number of sensory neurons on the right (overexpression) side of the embryo (Figure 5B) compared to the control side (n=6/7). This defect in sensory neuron differentiation was not mediated by increased cell death or decreased proliferation (Figure S5B). These experiments indicate that OCT4 and SOX2 promote the expression of early GRN components and that their downregulation is required for the proper differentiation of neuronal derivatives. We thus postulated that the dimer helps to initiate and maintain chromatin accessibility that is characteristic of the early neural crest, which is lost as these cells become fate restricted.
Figure 5 – Overexpression of SOX2 and OCT4 sustains multipotency in differentiating neural crest cells.
(A) Chick embryos bilaterally electroporated with control (left side) and OCT4-SOX2 overexpression constructs (right side) show sustained expression of multipotent neural crest marker PAX7, on the overexpression side of the embryo. Scale bar 50 μm.
(B) Immunostaining for TUJ1, a marker of neural-crest derived trigeminal ganglia, shows improper differentiation of sensory neurons upon OCT4-SOX2 overexpression. Scale bar 100 μm.
(C) Heatmap showing ATAC signal at genomic regions whose accessibility was significantly altered (> 2-fold) upon OCT4-SOX2 overexpression.
(D) Differential motif footprinting analysis of OCT4-SOX2, OCT4, and SOX2 in control versus OE samples.
(E) Boxplot showing change in accessibility of different CUT&RUN peaks upon overexpression.
(F) Scatter plot depicting OCT4 and SOX2 binding at all ATAC peaks, with red dots being peaks that are significantly altered upon OE.
(G) Compressed genome plot showing accessibility of early and late ATAC peaks in control and OCT4-SOX2 overexpression samples respectively.
*P≤0.05, **P≤0.01, ***P≤0.001
To test this hypothesis, we next assessed whether sustained expression of OCT4 and SOX2 is sufficient to maintain the epigenomic landscape of multipotent cells in late migratory neural crest. We electroporated HH4 chick embryos with control and OCT4-SOX2 overexpression vectors (RFP) alongside the neural crest-specific Tfap2aE1-GFP reporter construct. We FACS sorted double-positive GFP+/RFP+ cells from each side of the embryonic head at HH13 to perform ATAC-Seq. Differential peak analysis determined many peaks (~700) whose accessibility significantly increased (> 2-fold) upon sustained expression of OCT4-SOX2 (Figure 5C). As expected, overexpression of OCT4 and SOX2 augmented chromatin footprinting scores for the OCT4-SOX2, OCT4, and SOX2 motifs, with the strongest effect seen for the dimer motif (Figure 5D). Assessing chromatin accessibility changes at the OCT4/SOX2 CUT&RUN peaks confirmed that shared peaks co-occupied by the two factors at HH8 maintained high ATAC signal even at HH13 in the overexpression samples (Figure 5E). The largest change in accessibility was observed for peaks that had the highest occupancy of OCT4 and SOX2 in early neural crest cells (Figure 5F). Sustained expression of OCT4-SOX2 specifically increased chromatin accessibility at early neural crest peaks without significantly altering that of the late peaks (Figure 5G). The data from these functional experiments support that the OCT4-SOX2 dimer is an important regulator of the epigenomic signature of early neural crest cells.
TFAP2A interacts with SOX2 and drives the reorganization of the neural crest epigenome.
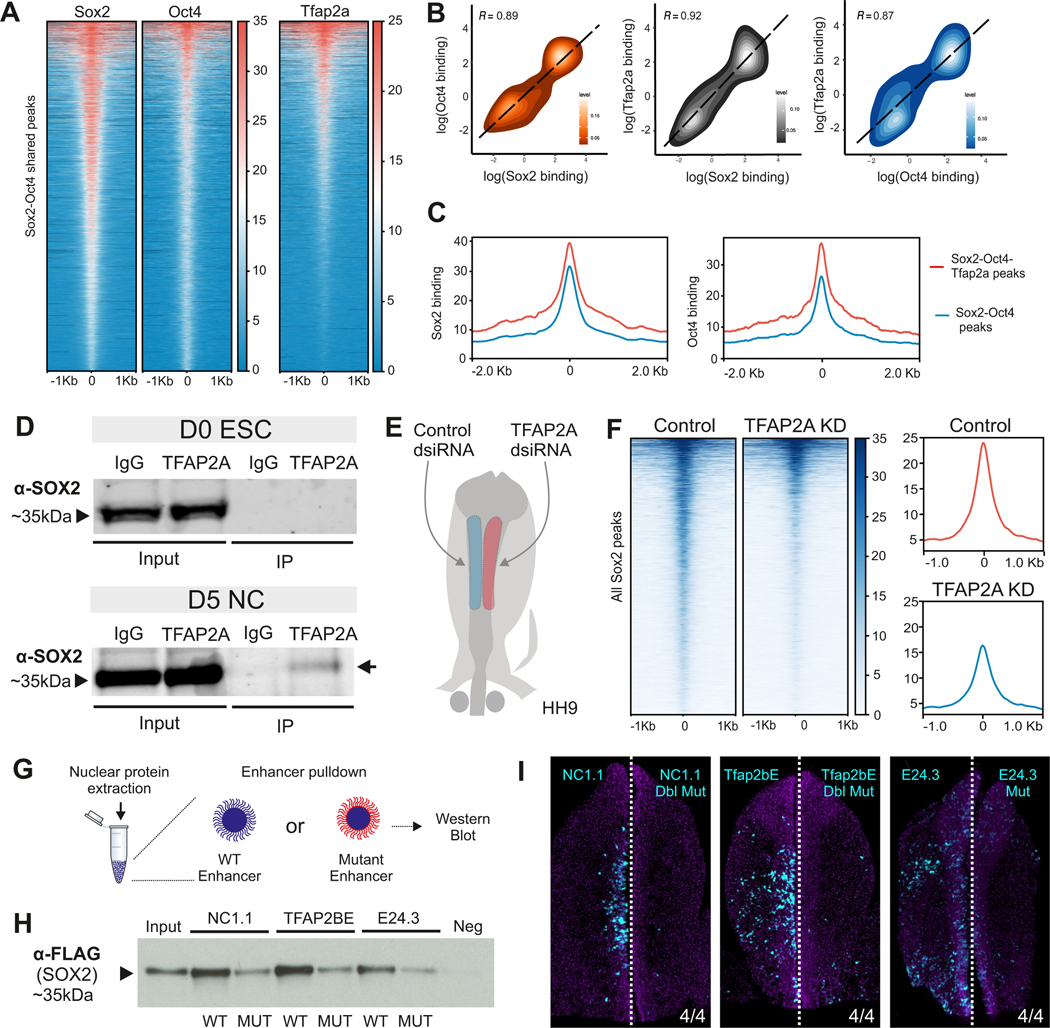
We next sought to identify the molecular players that allow for the translocation of the dimer to a new set of genomic regions during neural crest formation. A pioneer transcription factor that plays a central role in the formation of neural crest cells is TFAP2A (Li and Cornell, 2007; Mullins et al., 2019; Pla and Monsoro-Burq, 2018; Rothstein and Simoes-Costa, 2020). TFAP2A dimerizes with its paralogs TFAP2C and TFAP2B to regulate neural plate border and neural crest genes, respectively. Analysis of previously reported TFAP2A CUT&RUN in HH8 neural crest cells (Rothstein and Simoes-Costa, 2020) showed pioneer factor association at genomic loci co-occupied by OCT4-SOX2 (Figure 6A). We also observed a strong correlation in the binding of TFAP2A with that of OCT4 and SOX2 at shared regulatory regions (Figure 6B). SOX2 and OCT4 binding was higher at peaks co-occupied by the three factors (Figure 6C). Additionally, peaks where all three factors were bound displayed higher accessibility and BRD4 binding, indicating their potential as strong regulatory regions (Figure S6A). This suggested that TFAP2A cooperated with OCT4-SOX2 during neural crest formation.
Figure 6 – The pioneer factor TFAP2A regulates tissue-specific function of SOX2 and OCT4 in neural crest cells.
(A) Tornado plots showing the occupancy of SOX2, OCT4 and TFAP2A at shared OCT4-SOX2 peaks in neural crest cells.
(B) Density plots of pairwise correlations between all profiled transcription factors at shared peaks.
(C) Genome pile-up plot showing peaks that are bound by all three factors have the highest levels of SOX2 and OCT4 binding.
(D) Western Blot for SOX2 after an immunoprecipitation for TFAP2A in D0 ESCs and D5 hiNCCs.
(E) Electroporation scheme for a HH9 embryo in which control dsiRNA and TFAP2A targeting dsiRNA are electroporated on the left and right side of the embryo, respectively.
(F) Tornado plots comparing SOX2 signal at all SOX2 bound peaks in control vs TFAP2A dsiRNA treated NC cells. Summary plots (to the right) show a global decrease in SOX2 binding upon TFAP2A knockdown.
(G) Schematic of the enhancer pulldown experiment.
(H) Western blot for Flag-SOX2 following enhancer pulldown experiment with three different OCT4-SOX2 bound enhancers and their TFAP2A binding site mutated versions.
(I) Images of embryos bilaterally electroporated with WT (left) and TFAP2A-site mutated versions (left) of the enhancer reporters used in (H). Mutation of the TFAP2A site within these enhancers leads to loss of enhancer activity. Scale bar 100 μm.
*P≤0.05, **P≤0.01, ***P≤0.001
These results led us to postulate that during neural crest induction, the dimer switches from interacting with its pluripotency partners (Tapia et al., 2015) to cooperating with TFAP2A to promote a cell type-specific epigenomic signature. To test this, we performed PLA for OCT4/SOX2 and either NANOG or TFAP2A in hESCs (D0) and hiNCCs (D5) (Figures S6B and S6C). We observed a decrease in interactions of both SOX2 and OCT4 with NANOG, indicating the dissolution of the pluripotency protein complex. Concomitantly, we found increased interaction frequency between TFAP2A and both OCT4 and SOX2 in hiNCCs at D5 of induction (Figure S6D). Indeed, analysis of the predicted protein structure of these three factors with AlphaFold and 3DBioNotes (Jumper et al., 2021; Segura et al., 2019) identified a putative interaction domain between SOX2 (ASP107/ARG114/SER290) and TFAP2A (ARG128/TYR306/GLU311) (Supplemental Table 7). To confirm that these factors interact, we performed co-immunoprecipitation (co-IP) in hiNCCs. Consistent with our previous observations, IP of endogenous TFAP2A resulted in the pulldown of SOX2 in hiNCCs, but not in hESCs (Figure 6D; Figures S6E and S6F).
This evidence of interaction between TFAP2A and SOX2 led us to further hypothesize that TFAP2A could be required for the translocation of the dimer. To test this, we examined how loss-of-function (LOF) of TFAP2A affected the genome-wide occupancy of SOX2 in neural crest cells. We transfected the right side of avian embryos with a dsiRNA targeting TFAP2A (Figure S6G) and performed CUT&RUN for SOX2 in control and TFAP2A-depleted neural folds (Figure 6E). We observed that loss of TFAP2A resulted in a reduction of SOX2 binding in neural crest cells (Figure 6F). As an orthogonal approach, we performed enhancer pulldown experiments (Azambuja and Simoes-Costa, 2021) to test if TFAP2A is required for the interaction of the dimer with the neural crest epigenome (Figure 6G). Disruption of TFAP2A binding sites in three neural crest enhancers containing OCT4-SOX2 motifs (NC1.1, Tfap2bE1, and E24.3) resulted in a loss of SOX2 binding (Figure 6H) and affected enhancer activity (Figure 6I). These experiments indicate that TFAP2A is required for OCT4-SOX2 to interact with the cis-regulatory regions that comprise the neural crest epigenomic signature.
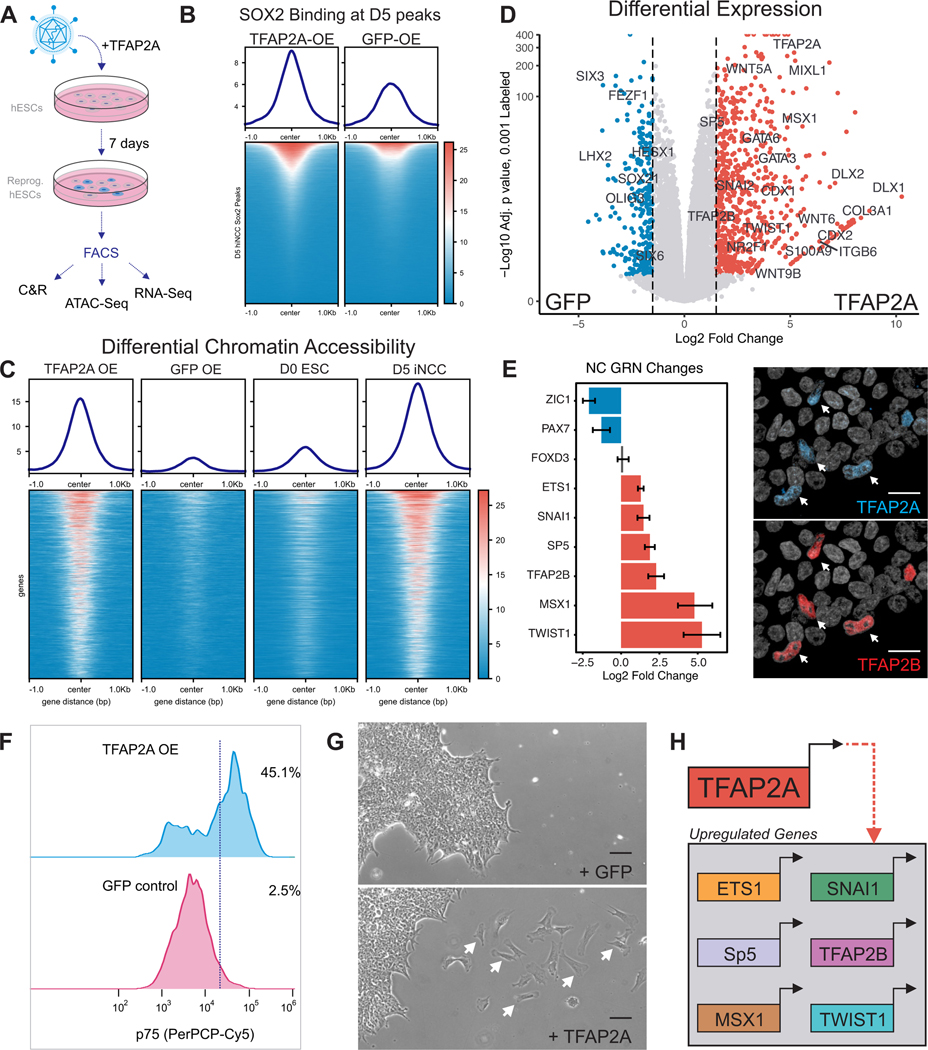
Finally, we performed reprogramming experiments to evaluate whether TFAP2A expression was sufficient to mediate dimer translocation and activation of the neural crest GRN in pluripotent ESCs. Human ESCs were transduced with a lentiviral vector driving the expression of TFAP2A. Seven days post-infection control and TFAP2A-expressing hESCs were subjected to genomic analysis (CUT&RUN for SOX2, ATAC-seq, and RNA-seq) (Figure 7A; Figure S7). Consistent with our hypothesis, we observed that introduction of TFAP2A in hESCs resulted in increased SOX2 occupancy at hiNCC-specific D5 peaks (Figure 7B). Furthermore, ATAC-seq analysis showed that TFAP2A-expressing hESCs displayed increased chromatin accessibility at many genomic regions (Figure 7C). These regions were also accessible in D5 hiNCCs but not in D0 hESCs, indicating that TFAP2A is sufficient to promote at least part of the neural crest epigenomic signature (Figure 7C). Indeed, transcriptomic analysis showed a partial deployment of the neural crest GRN, with genes like MSX1 and TFAP2B being activated by TFAP2A in hESCs (Figures 7D and 7E). The transcriptomic changes induced by TFAP2A were also evident at the protein level, indicated by positive staining for neural crest markers TFAP2B and p75 (NGFR) (Figures 7E and 7F). Strikingly, upon TFAP2A expression, reprogrammed cells delaminated from hESCs colonies and displayed migratory behaviors in culture (Figure 7G). Thus, TFAP2A promotes changes in genomic occupancy of SOX2 in hESCs and is sufficient to elicit partial activation of the neural crest GRN (Figure 7H).
Figure 7 – TFAP2A Overexpression Partially Reprograms hESCs to a Neural Crest-like State.
(A) Design of hESC reprogramming experiment by overexpression of TFAP2A.
(B) Tornado plot of SOX2 CUT&RUN signal at all SOX2 peaks identified from untreated conditions.
(C) Tornado plots of ATAC-Seq signal in overexpression and wild-type conditions at peaks enriched within untreated D5 neural crest.
(D) Volcano plot for RNA-Seq analysis between GFP and TFAP2A overexpression samples shows enrichment of the TFAP2A gene and several other members of the neural crest GRN.
(E) Bar chart of selected neural crest GRN components and their Log2 fold change in the TFAP2A overexpression condition. Immunohistochemistry staining of TFAP2A and TFAP2B in reprogrammed hESCs showing nuclear co-staining. Scale bar 20 μm.
(F) FACS analysis of neural crest marker p75 in control and TFAP2A overexpressing hESCs.
(G) Bright field image of GFP or TFAP2A overexpressing cells. In the TFAP2A condition, a large portion of cells undergo EMT and migrate across the substrate. Scale bar 20 μm.
(H) Graphical representation of neural crest GRN components found to be significantly upregulated upon TFAP2A overexpression in hESCs.
Discussion
Here we conducted time-resolved genomic analysis to delineate temporal modules within the neural crest GRN and to characterize the epigenomic changes that occur during cell state transitions. By integrating epigenomic and transcriptomic datasets, we identified a large number of cis and trans components of the network. The generation of impulse models (Fischer et al., 2018) of both gene expression and chromatin accessibility allowed us to curate a collection of active (H3K27AC+) enhancers that likely play a role in the temporal regulation of cognate genes. This strategy may be useful for enhancer-gene assignment from time-resolved datasets and can be used in the absence of physical linkage (Lieberman-Aiden et al., 2009) or single-cell accessibility data (Pliner et al., 2018). Finally, we took advantage of both accessibility and transcriptomic data to assign an activating or repressing role for transcription factors expressed during neural crest development (Berest et al., 2019).
This transcriptomic and epigenomic characterization of neural crest development resulted in mechanistic insights into the molecular control of multipotency. First, we found that the regulatory state of neural crest cells is distinct from that of ESCs, suggesting divergent mechanisms for developmental plasticity. Second, chromatin footprinting analysis and mapping of protein-DNA interactions demonstrated that the neural crest epigenome is regulated by the OCT4-SOX2 heterodimer. Finally, we identified a mechanism whereby the neural crest pioneer factor TFAP2A modulates the target specificity of the dimer. Thus, we propose a model for the control of neural crest plasticity where components of the ESC pluripotency machinery cooperate with tissue-specific factors to create a multipotent stem cell state.
Our study identifies an important role for OCT4 and SOX2 in neural crest stem cell identity. We observed that these pluripotency factors are expressed in early neural crest cells and that they exhibit context-specific functions. In the ectoderm, SOX2 has traditionally been described as a pro-neural factor, which promotes neural plate identity via a mutual cross-repression circuit involving neural crest genes (Hovland et al., 2020; Wakamatsu et al., 2004). Yet, recent studies have demonstrated that neural crest cells express SOX2 (Roellig et al., 2017), albeit at lower levels than neural plate progenitors. Here we show that, despite relatively low levels of expression, SOX2 is still critical for neural crest multipotency. We propose that different functions of this pioneer factor observed in neural vs. neural crest progenitors is due to its dimerization to OCT4. Consistent with this, OCT4 is required for proper development of the cranial neural crest (Zalc et al., 2021). While previous studies have proposed that the pluripotency program is either retained (Buitrago-Delgado et al., 2015) or regained (Zalc et al., 2021) during neural crest development, our results indicate that Yamanaka factors perform specialized functions in the neural crest that are distinct from their roles in ESCs.
Our model illustrates how pluripotency factors are repurposed to generate distinct chromatin landscapes in multipotent stem cells. In the neural crest trans environment, the OCT4-SOX2 dimer translocates to a distinct set of cis-regulatory regions, promoting epigenomic remodeling. This process is mediated by TFAP2 pioneer transcription factors, which physically interact with the dimer and are necessary and sufficient to induce changes in their genomic occupancy. The cooption of OCT4-SOX2 from the pluripotency circuit by neural crest cells may be due to their strong pioneering activity (Michael et al., 2020; Whyte et al., 2013), which would facilitate overarching changes in the chromatin landscape. Translocation of the OCT4-SOX2 dimer, along with loss of other canonical pluripotency factors (e.g., NANOG) may underlie the key differences between the neural crest multipotency GRN compared to the stem cell pluripotency circuit (Akberdin et al., 2018; Niwa et al., 2000; Thomson et al., 2011). Taken together, our results demonstrate how modifications in the ESC program by tissue-specific partners can generate distinct cellular states with variable degrees of developmental plasticity.
Limitations of the study:
Our results demonstrate that TFAP2A recruits the OCT4-SOX2 heterodimer to neural crest-specific enhancers. However, our study did not address if OCT4 and SOX2 expression is maintained from epiblast stem cells or reactivated in multipotent neural crest cells or how these pluripotency genes are silenced during cell fate commitment. Previous studies have invoked the role of Wnt signaling in the establishment of neural crest stem cells and let-7 miRNAs in silencing the multipotency program. The mechanisms through which these pathways intersect with the OCT4-SOX2-TFAP2A axis in early NC cells remain to be determined and could be the premise of future studies.
STAR METHODS
RESOURCE AVAILIBILITY
LEAD CONTACT
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Marcos Simoes-Costa (marcos@hms.harvard.edu).
MATERIAL AVAILIBILITY
This study did not generate any new reagents.
DATA AND CODE AVAILIBILITY
Genomic data for this work is publicly available at the NCBI Gene Expression Omnibus (GEO) under the accession number GSE163961.
Data processing and analysis scripts can be found at https://github.com/Austin-s-h/NC_Timecourse (Hovland and Bhattacharya, 2022).
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODELS AND SUBJECT DETAILS
Chick embryo collection and electroporation
Fertilized chicken embryos (Leghorn White) were obtained from the Department of Animal Science at the University of Connecticut. Eggs were incubated at 37°C until they reached the desired Hamburger and Hamilton (HH) (Hamburger and Hamilton, 1951) developmental stage. Embryos were collected and cultured ex-ovo following the EC method (Chapman et al., 2001) for HH6–16 and a Cornish Pastry (MC) method for HH16+ (Nagai et al., 2011). Enhancer, overexpression, and morpholino constructs were electroporated into HH4 embryos as previously described (Bhattacharya et al., 2018). Briefly, these constructs were injected between the epiblast and vitelline membrane of HH4 chicken embryos and electroporated using platinum electrodes (five 50ms pulses at 5.1V, 100ms resting interval).
Generation of induced neural crest (hiNCC)
The induction of human neural crest cells from WA01 (H1) embryonic stem cells was performed as previously described (Gomez et al., 2019). Briefly, H1 cells cultured in mTESR were dissociated with Accumax, and plated on Matrigel coated dishes at a density of 20K cells/cm2 in DMEM/F12 media supplemented with 1% B27, 1% N2,0.5%BSA and 3uM CHIR (induction media). For the first 24h, 10uM ROCK inhibitor (Y-27632) was added to the induction media to improve cell survival. After 72 hours (Day 3), induction media was substituted for basal media constituting of DMEM/F12, 1% B27 and 0.5%BSA. The cells were grown in basal media for an additional 48 hours (Day 5), after which they were processed for downstream analysis. For Myoblast differentiation, induced neural crest cells (hiNCCs) were dissociated and plated at a density of 50K/cm2 in alpha-MEM and 10% FBS for an additional 14 days.
METHOD DETAILS
Cloning and expression constructs
To label neural crest cells, we electroporated an enhancer of the TFAP2A gene (TFAP2a-E1) driving GFP (Rothstein and Simoes-Costa, 2020) and allowed embryos to develop ex ovo until the desired stage. Selected enhancers were amplified from HH10 chicken genomic DNA and cloned in pTK-EGFP (Uchikawa et al., 2003). Mutant constructs were also cloned into pTKEGFP, and the mutated enhancer activity was compared to wild-type constructs co-transfected in the same embryo. Mutated binding sites were defined using the JASPAR database of transcription factor binding profiles (Fornes et al., 2019). The details of these enhancers can be found in Supplemental Table of Enhancer Names and Locations. To create an OCT4-SOX2 expression vector, we amplified the msSox2-P2A-msOct4 fragment from the FUW-SOKM plasmid (Addgene #20325) with Gibson assembly overhangs to a H2B-RFP expression vector. We assembled the vector, leading to a CMV-driven Sox2-P2A-Oct4-IRES-H2B-RFP ubiquitous expression vector with a fluorescent readout of electroporation efficiency.
Embryo dissociation and cell sorting
To isolate broad regions containing neural crest cells, we surgically dissected cranial regions. At HH6, we dissected tissue anterior to the node. For HH8–10 we dissected pre-somitic tissue. From HH12 onward, we dissected pre-otic tissue only, containing the first and second branchial arches. Neural crest cells were dissociated as previously described (Bhattacharya et al., 2018). Briefly, robustly GFP or RFP expressing embryos were dissected in Ringer’s solution, dissociated using Accumax (Accutase SCR006), filtered, and resuspended in Hank’s solution. Single-cell suspensions were then sorted using a Sony MA900. Forward and side scatter were used to exclude debris and doublets, and another gate of either GFP (TFAP2aE1-GFP) or RFP (OCT4-P2A-SOX2-H2B-RFP) was used to isolate cells of interest.
RNA extraction, library preparation, and sequencing
For chicken neural crest RNA-Sequencing, we FACS isolated at least 5,000 cranial neural crest cells into the lysis buffer from the RNAqueous micro kit (ThermoFisher #AM1931). RNA was extracted according to the manufacturer’s protocol, quantified using a Qubit RNA HS Assay (ThermoFisher #Q32852), and analyzed for quality on an ABI 3730×l DNA Analyzer. RNA-Sequencing libraries were prepared using the NEBNext® Ultra™ II Directional RNA Library Prep Kit (NEB #E7765) according to the manufacturer’s protocol. Depending on the input RNA amount, libraries were PCR amplified 13–16 cycles. Libraries were quantified using a Qubit DNA HS Assay (ThermoFisher #Q33230) and checked for fragment size distribution and quality on an ABI 3730×l DNA Analyzer. Individual samples were pooled at an equimolar ratio calculated using the KAPA Library Quantification Kit (Roche #07960336001) and sequenced in a single-end configuration on an Illumina NextSeq500 using the High Output 75bp kit. Sequencing was performed by the Biotechnology Resource Center (BRC) Genomics Facility (RRID:SCR_021727) at the Cornell Institute of Biotechnology.
ATAC-Seq library preparation and sequencing
For chicken neural crest ATAC-Seq, at least 5,000 cells were isolated via FACS, gently centrifuged at 500rcf, resuspended in CryoStor freezing media (Stem Cell Technologies #CS10), and frozen at −80°C until library preparation. For human ESC and iNCC ATAC-Seq, cells were dissociated using Accutase and then counted. One hundred thousand cells were resuspended in CryoStor media and frozen at −80°C until library preparation. Cells were processed using the OMNI-ATAC-Seq protocol (Corces et al., 2017). Frozen cells were quickly thawed at 37°C and ATAC-RSB buffer was added to a total volume of 1.5mL. Samples were centrifuged at 500rcf for 5 minutes at 4°C and the supernatant was removed. Next, cells were resuspended in 100μl of ATAC-RSB-LYSIS and kept on ice for 3–5 minutes, depending on input cell type. To stop lysis, 1mL of ATAC-RSB-WASH was added to each sample and they were again centrifuged for 5 minutes at 500rcf. The supernatant was removed, and cells were resuspended in 50μl of OMNI-ATAC Mix (~100nM concentration of Illumina TDE1 enzyme). Cells were then tagmented on a mixing (500rpm) thermoblock at 37°C for one hour. Tagmented DNA was recovered using a Qiagen MinElute Kit (#28204), with 21μl of elution buffer warmed to 55°C. Library amplification PCR was performed with the NEBNext Ultra II Q5 2X Master Mix (NEB #M0544S) using Nextera primers for 13–15 cycles. DNA concentration was measured using a Qubit DNA HS Assay (ThermoFisher #Q33230) and stereotypical nucleosomal banding was observed using an ABI 3730×l DNA Analyzer. Individual samples were pooled at an equimolar ratio calculated using the KAPA Library Quantification Kit (Roche #07960336001) and sequenced on an Illumina NextSeq500 at the Cornell Institute of Biotechnology using the 75bp kit in a paired end configuration (32×32).
CUT&RUN library preparation and sequencing
CUT&RUN was performed as previously described (Rothstein and Simoes-Costa, 2020). For chicken neural crest samples, 20 neural folds were dissected per sample. For human hESC and hiNCC samples, cultures were briefly dissociated using Accumax, counted, and 500,000 cells were used for each sample.
Immunohistochemistry
Whole-mount chicken embryos were isolated and fixed in PFA-PB for 10–20 minutes at room temperature. After dissection, embryos were washed in TBS containing 0.1X Triton X-100 and 1% DMSO (TBTD). Primary antibodies, diluted in 10% donkey serum blocking solution, were incubated at 4°C overnight, and secondary antibodies were incubated for 2 hours at room temperature.
Enhancer Pulldown
To detect the ability of proteins to bind to DNA sequence features, we performed enhancer pulldown experiments using biotinylated enhancer sequences (Azambuja and Simoes-Costa, 2021). First, known neural crest enhancer sequences (300–500bp) and their mutant versions were amplified in the presence of biotinylated nucleotides. Dynabeads MyOne Streptavidin T1 beads (Invitrogen, #65602) were washed and incubated with this biotinylated DNA for 30 minutes at room temperature. HH4 chicken embryos were electroporated with a FLAG-OCT4 and FLAG-SOX2 expression construct. Nuclear extracts were generated from these embryos at HH9 (Azambuja and Simoes-Costa, 2021). These nuclear extracts were incubated with bead-bound DNA for 1.5 hours at 4°C, washed three times, and eluted. Samples were size-separated via gel-electrophoresis and then subjected to a Western Blot for FLAG (the presence of SOX2/OCT4).
Co-immunoprecipitation (Co-IP)
To capture TFAP2A and SOX2 interactions, we utilized the protocol from Mohammed et. al. with minor modifications (Mohammed et al., 2016). For each IP, we started with 6×107 cells that were dissociated, counted, and then crosslinked with 1% formaldehyde. Before proceeding with the IP reactions, a 50uL aliquot of each lysate was reserved to serve as input. 7μg of TFAP2A mouse monoclonal antibody (sc-12726– Santa Cruz Biotechnology) and normal mouse IgG (12–371 – Millipore Sigma) antibodies were used in each IP reaction. After performing the IP reactions and wash steps, samples were eluted on Laemmli buffer and heated at 95°C for 10 minutes. All samples were resolved on a SDS-PAGE, and after the run proteins were transferred to an immobilon-FL PVDF membrane (Millipore Sigma - MA – USA). Sox2 Goat polyclonal antibody (AF2018 – R&D Systems - 1:1000 dilution) was used for protein detection. LICOR IRDye secondary antibodies (1:15000 dilution) were used to detect Sox2 primary antibody, according to manufacturer instructions. Membranes were scanned on the Odyssey DLx system (LI-COR Biosciences, NE-USA), and images were quantified on the Image Studio Lite software.
Immunocytochemistry
For NGFR (p75) cell surface marker staining across the neural crest induction time course, adhered cells were washed once with 1X PBS and dissociated with Accumax Cell dissociation solution (Accutase, SCR006) for 5 minutes at 37°C. The subsequent cell suspension was washed once with 0.1% BSA in PBS solution and spun down for 5 minutes at 400rcf. Cells were next incubated with appropriate dilutions of directly conjugated primary antibody in 0.1% BSA in PBS solution for 30 minutes at RT. The anti-p75 NGF receptor antibody (Abcam, ab234270) was used at a dilution of 1:100. Following primary antibody staining, the cells were washed once with 0.1% BSA in PBS solution and the antibody staining intensity was measured by FACS analysis using the Attune Nxt flow cytometer.
Proximity Ligation Assays (PLA)
To identify proximity between various combinations of transcription factors, we performed proximity ligation assays (PLAs) as previously described (Alam, 2018) using the DuoLink Fluorescence approach (Sigma-Aldrich, DUO92101) with minor modifications. For PLA in chick embryo sections, embryos were fixed with phosphate buffer (PB) containing 4% PFA for 20 minutes and cryosectioned in OCT compound. Primary antibody pairs used are as follows: goat anti-SOX2 (R&D Systems AF2018), rabbit anti-OCT4 (Invitrogen 701756). For PLA in human cells, we fixed cells at day 0 and day 5 of neural crest induction in 4% PFA for 10 minutes at room temperature. We then permeabilized the cells with 0.1% NP-40 in PBS for 30 minutes and 37°C, followed by blocking and primary antibody incubation in 1% BSA in PBS overnight at 4°C. Primary antibody pairs used are as follows: rabbit anti-SOX2 (Abcam ab97959), mouse anti-NANOG (Santa Cruz Biotechnology sc-293121); rabbit anti-OCT4 (Abcam ab109183), mouse anti-Nanog (Santa Cruz Biotechnology sc-293121); rabbit anti-SOX2 (Abcam ab97959), mouse anti-Tfap2a (DSHB 3B5); rabbit anti-Oct4 (Abcam ab109183), mouse anti-TFAP2A (DSHB 3B5)
QUANTIFICATION AND STATISTICAL ANALYSIS
RNA-Seq Analysis
Raw sequencing reads were demultiplexed by the Cornell Institute of Biotechnology and trimmed using CutAdapt (v2.10) (Martin, 2011) with the TruSeq sequencing adapter (AGATCGGAAGAGCACACGTCTGAACTCCAGTCAC), and a minimum read length of at least 25. Reads were then aligned to the ENSEMBL galGal6 genome using HiSat2 (Kim et al., 2019) using reverse strandedness and discarding unaligned reads. Counts were assigned to genes using featureCounts (Liao et al., 2014) at ENSEMBL gene annotation (v93). Downstream differential expression analysis, PCA dimensionality reduction, clustering, and GO term enrichment analysis were performed in R v4.2.0 (https://www.R-project.org/). Details can be found in the scripts available under Data Availability and Code.
ATAC-Seq sequence processing
Raw sequencing reads were demultiplexed by the Cornell Institute of Biotechnology and trimmed using CutAdapt (v2.10) in paired-end mode with the forward and reverse Nextera sequencing adapters (Fwd – CTGTCTCTTATACACATCT, Rev- AGATGTGTATAAGAGACAG) and a minimum read length of at least 25. Next, we used Bowtie2 (Langmead and Salzberg, 2012) to align paired-end reads to the ENSEMBL galGal6 or UCSC hg38 genomes for chicken and human samples, respectively. We used the options “--local --very-sensitive-local --no-unal --no-mixed --no-discordant” and excluded alignments > 850bp. PicardTools (https://github.com/broadinstitute/picard) was used to mark duplicates, which were then filtered from the BAM files using samtools (Li et al., 2009). MACS2 was used to call peaks genome-wide, with a q value of 0.05 and the arguments “-f BAMPE -g $GENOME_SIZE --nomodel --shift 37 --extsize 73” (Zhang et al., 2008). All downstream analysis including peak set generation, PCA, DiffBind, HOMER, chromVAR, and HINT-ATAC are available as scripts under Data Availability and Code (Heinz et al., 2010).
Peak to Gene Correlation Analysis (RNA-Seq and ATAC-Seq integration)
To generate peak to gene links, we utilized ImpulseDE2 generated continuous models of gene expression and peak accessibility across our time course. Briefly, we generated impulse expression models of all expressed genes across our time course and generated gene expression values for normalized time series (1–100). Next, we generated peak accessibility models and scaled them to a similar normalized time series (1–100). Next, we used Bedtools to generate a list of the nearest 5 genes for each peak to test for a strong correlation (Quinlan and Hall, 2010). We then compared the impulse models for each peak to each of the nearest five genes and reported the Spearman correlation. Correlations > 0.8 are considered strongly positive and in our testing is a reasonably accurate predictor of in-vivo specificity and activity. The exact code for this analysis can be found within our Data Availability and Code. For determining peak to gene linkage in human ATAC and CUT&RUN data, we annotated genomic regions using the GREAT algorithm (McLean et al., 2010).
CUT&RUN sequence processing
Raw sequencing reads were demultiplexed by the Cornell Institute of Biotechnology and trimmed using CutAdapt (v2.10) in paired-end mode with the forward and reverse TruSeq sequencing adapters (Fwd – AGATCGGAAGAGCACACGTCTGAACTCCAGTCA, Rev-AGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGT) and a minimum read length of at least 25. Next, chicken and human samples were aligned to ENSEMBL galGal6 or UCSC hg38, respectively, using Bowtie2. We used the following options “--local --very-sensitive-local –no-unal --no-mixed --no-discordant” and excluded alignments > 1000bp. PicardTools (https://github.com/broadinstitute/picard) was used to mark duplicates, which were then filtered from the BAM files using samtools. MACS2 was used to call peaks genome-wide, with a q value of 0.05 and the arguments “-f BAMPE -g $GENOME_SIZE -q 0.05 --call-summits.” Subsequent analysis including peak filtering, motif enrichment, clustering, and comparison to ATAC-Seq can be found as R scripts within Data Availability and Code.
Experimental Quantification
Details on statistical tests for each experiment can be found within the figure legends. Comparison of grouped samples were analyzed with a one-sided student’s t-test. Multiple hypothesis test correction was performed with Bonferroni multiple test correction. For RNA-Seq, at least 3 biological replicates were used for each time point and condition. For ATAC-Seq, two biological replicates were used for each time point. For CUT&RUN samples, at least 2 biological replicates were used for each condition. For hiNCC and TFAP2A-overexpression experiments, two independent inductions were performed.
Additional Resources
Gene Expression Shiny Application: https://ash274.shinyapps.io/RNA-Seq_App/
Supplementary Material
Supplemental Table 1 (Related to Figure 1) Raw counts for RNA-Seq of chicken neural crest time course.
Supplemental Table 2 (Related to Figure 1) Likelihood Ratio Test (LRT) results comparing GFP+ and GFP− samples.
Supplemental Table 3 (Related to Figure 2) Raw counts for ATAC-Seq of chicken neural crest time course.
Supplemental Table 4 (Related to Figure 2) Peak to gene correlation analysis for all genes and neural crest GRN genes (galGal6).
Supplemental Table 5 (Related to Figure 2) DiffTF results classifying transcription factors as activators or repressors during neural crest development.
Supplemental Table 6 (Related to Figure 3) Shared (~50 bp) foci of SOX2 and OCT4 binding from EcHO analysis (galGal6).
Supplemental Table 7 (Related to Figure 6) AlphaFold modeling of human TFAP2A and SOX2 in pdb format.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat Polyclonal anti-SOX2 (1:50 CUT&RUN, 1:200 PLA) | R&D Systems | Cat#AF2018 |
| Goat Polyclonal anti-OCT4 (1:50 CUT&RUN) | R&D Systems | Cat#AF1759 |
| Rabbit Monoclonal anti-H3K27Ac (1:50 CUT&RUN) | Abcam | Cat#ab177178 |
| Mouse Monoclonal anti-NANOG (1:50 PLA) | Santa Cruz Biotechnology | Cat#sc293121 |
| Mouse Monoclonal anti-TFAP2A (1:50 PLA) | DSHB | Cat#3B5c |
| Rabbit Polyclonal anti-SOX2 (1:200 PLA) | Abcam | Cat#ab97959 |
| Rabbit Monoclonal anti-OCT4 (1:200 PLA) | Invitrogen | Cat#701756 |
| Mouse Monoclonal anti-p75 (1:100 FACS) | Abcam | Cat#ab234270 |
| Biological samples | ||
| Chicken Eggs (Leghorn White) | University of Connecticut Department of Animal Science | N/A |
| Critical commercial assays | ||
| RNA Aqueous Micro Kit | ThermoFisher | Cat#AM1931 |
| NEBNext® Ultra™ II Directional RNA Library Prep Kit | New England Biolabs | Cat#E7765 |
| Illumina Tagment DNA Enzyme and Buffer Kit | Illumina | Cat# 20034197 |
| Deposited data | ||
| Raw and analyzed data | This paper | GEO: GSE163961 |
| Experimental models: Cell lines | ||
| Human embryonic stem cells – WA01 (H1) | WiCell | WAe001-A |
| Oligonucleotides | ||
| Primer: SOX2-OCT4-OH1-F GCGCGCCTTAATTAACGTTTATGTATAACATGATGGAGACGGAG | This paper | N/A |
| Primer: SOX2-OCT4-OH2-R AGCCATTTGCATGCATGTTTGCCGGAGCCGCATGCGTT | This paper | N/A |
| Recombinant DNA | ||
| TFAP2AE1 chr2: 63870453– 63871022 (galGal6) | Rothstein and Simoes-Costa, 2020 | N/A |
| SOX10E1+2 chr1:51065744–51067043 | Betancur et al., 2010 | N/A |
| DACT2E1 chr3:41956178–41958139 | Rothstein and Simoes-Costa, 2020 | N/A |
| E4.4 chr4:78729975–78730374 (MSX1) | Azambuja and Simoes-Costa, 2021 | N/A |
| E8.5 chr8:28018194–28018593 (FOXD3) | Azambuja and Simoes-Costa, 2021 | N/A |
| E14.1 chr14:6236095–6236494 (SOX8) | Azambuja and Simoes-Costa, 2021 | N/A |
| E18.1 chr18:8487307–8487706 (SOX9) | Azambuja and Simoes-Costa, 2021 | N/A |
| NC1.1 chr8:28111172–28111727 (FOXD3) | Simões-Costa et al., 2012 | N/A |
| NC1.1 (double mut) TFAP2AMut1-chr8:28111473–28111486 TFAP2AMut2-chr8:28111493–28111518 | This paper | N/A |
| TFAP2BE chr3:108021169–108021490 | Azambuja and Simoes-Costa, 2021 | N/A |
| TFAP2BE (double mut) TFAP2AMut1-chr3:108021273–108021300 TFAP2AMut2-chr3:108021375–108021400 | This paper | N/A |
| E24.3 chr24:397366–399331 (ETS1) | Azambuja and Simoes-Costa, 2021 | N/A |
| E24.3 (mut) TFAP2AMut1-chr24:397876–397890 | This paper | N/A |
| Software and algorithms | ||
| R (v 4.2.0) | R Foundation for Statistical Computing | https://www.R-project.org/ |
| Bioconductor | Huber et al. 2015 | https://www.bioconductor.org/ |
| Other | ||
| Data processing scripts and analysis notebooks | This paper | https://zenodo.org/record/7044924 |
Highlights.
The OCT4-SOX2 dimer regulates the epigenomic landscape of neural crest cells
Dimer targets in the neural crest differ from those of embryonic stem cells
Pioneer factor TFAP2A interacts with OCT4-SOX2 to modify its genomic occupancy
Acknowledgement:
We thank P. Schweitzer and the Biotechnology Resource Center (BRC) Genomics Facility (RRID:SCR_021727) at the Cornell institute of Biotechnology for NGS sequencing. We also thank members of the BRC Flow Cytometry facility (RRID:SCR_021740) for assistance with cell sorting. This work was supported by NIH grant R01DE028576 to M.S.-C. and a Cornell CVG training grant to A.H (T32HD057854).
Footnotes
Declaration of interests:
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akberdin IR, Omelyanchuk NA, Fadeev SI, Leskova NE, Oschepkova EA, Kazantsev FV, Matushkin YG, Afonnikov DA, and Kolchanov NA (2018). Pluripotency gene network dynamics: System views from parametric analysis. PLOS ONE 13, e0194464. 10.1371/journal.pone.0194464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam MS (2018). Proximity Ligation Assay (PLA). Current Protocols in Immunology 123, e58. 10.1002/cpim.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attanasio C, Nord AS, Zhu Y, Blow MJ, Li Z, Liberton DK, Morrison H, Plajzer-Frick I, Holt A, Hosseini R, et al. (2013). Fine Tuning of Craniofacial Morphology by Distant-Acting Enhancers. Science 342, 1241006–1241006. 10.1126/science.1241006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azambuja AP, and Simoes-Costa M.(2021). The connectome of neural crest enhancers reveals regulatory features of signaling systems. Developmental Cell 56, 1268–1282.e1266. 10.1016/j.devcel.2021.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggiolini A., Varum S., Mateos José M., Bettosini D, John N, Bonalli M, Ziegler U, Dimou L, Clevers H, Furrer R, and Sommer L. (2015). Premigratory and Migratory Neural Crest Cells Are Multipotent In Vivo. Cell Stem Cell 16, 314–322. 10.1016/j.stem.2015.02.017. [DOI] [PubMed] [Google Scholar]
- Barembaum M, and Bronner ME (2013). Identification and dissection of a key enhancer mediating cranial neural crest specific expression of transcription factor, Ets-1. Developmental Biology 382, 567–575. 10.1016/j.ydbio.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berest I, Arnold C, Reyes-Palomares A, Palla G, Rasmussen KD, Giles H, Bruch P-M, Huber W, Dietrich S, Helin K, and Zaugg JB (2019). Quantification of Differential Transcription Factor Activity and Multiomics-Based Classification into Activators and Repressors: diffTF. Cell Reports 29, 3147–3159.e3112. 10.1016/j.celrep.2019.10.106. [DOI] [PubMed] [Google Scholar]
- Betancur P, Bronner-Fraser M, and Sauka-Spengler T.(2010). Genomic code for Sox10 activation reveals a key regulatory enhancer for cranial neural crest. Proc Natl Acad Sci U S A 107, 3570–3575. 10.1073/pnas.0906596107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya D, Rothstein M, Azambuja AP, and Simoes-Costa M.(2018). Control of neural crest multipotency by Wnt signaling and the Lin28/let-7 axis. Elife 7. 10.7554/eLife.40556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blassberg R, Patel H, Watson T, Gouti M, Metzis V, Delás MJ, and Briscoe J.(2020). Sox2 levels configure the WNT response of epiblast progenitors responsible for vertebrate body formation. Cold Spring Harbor Laboratory. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs JA, Weinreb C, Wagner DE, Megason S, Peshkin L, Kirschner MW, and Klein AM (2018). The dynamics of gene expression in vertebrate embryogenesis at single-cell resolution. Science 360. 10.1126/science.aar5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronner-Fraser M, and Fraser SE (1988). Cell lineage analysis reveals multipotency of some avian neural crest cells. Nature 335, 161–164. 10.1038/335161a0. [DOI] [PubMed] [Google Scholar]
- Buitrago-Delgado E, Nordin K, Rao A, Geary L, and LaBonne C.(2015). Shared regulatory programs suggest retention of blastula-stage potential in neural crest cells. Science 348, 1332–1335. 10.1126/science.aaa3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunina D, Abazova N, Diaz N, Noh KM, Krijgsveld J, and Zaugg JB (2020). Genomic Rewiring of SOX2 Chromatin Interaction Network during Differentiation of ESCs to Postmitotic Neurons. Cell Syst 10, 480–494 e488. 10.1016/j.cels.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman SC, Collignon J.r.m., Schoenwolf GC, and Lumsden A.(2001). Improved method for chick whole-embryo culture using a filter paper carrier. Developmental Dynamics 220, 284–289. . [DOI] [PubMed] [Google Scholar]
- Corces MR, Trevino AE, Hamilton EG, Greenside PG, Sinnott-Armstrong NA, Vesuna S, Satpathy AT, Rubin AJ, Montine KS, Wu B, et al. (2017). An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat Methods 14, 959–962. 10.1038/nmeth.4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dottori M, Gross MK, Labosky P, and Goulding M.(2001). The winged-helix transcription factor Foxd3 suppresses interneuron differentiation and promotes neural crest cell fate. Development 128, 4127–4138. 11684651. [DOI] [PubMed] [Google Scholar]
- Dupin E, Calloni GW, Coelho-Aguiar JM, and Le Douarin NM (2018). The issue of the multipotency of the neural crest cells. Dev Biol 444 Suppl 1, S47–S59. 10.1016/j.ydbio.2018.03.024. [DOI] [PubMed] [Google Scholar]
- Fischer DS, Theis FJ, and Yosef N.(2018). Impulse model-based differential expression analysis of time course sequencing data. Nucleic Acids Research. 10.1093/nar/gky675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornes O, Castro-Mondragon JA, Khan A, van der Lee R, Zhang X, Richmond PA, Modi BP, Correard S, Gheorghe M, Baranašić D, et al. (2019). JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Research 48, D87–D92. 10.1093/nar/gkz1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez GA., Prasad MS., Wong M., Charney RM., Shelar PB., Sandhu N., Hackland JOS., Hernandez JC., Leung AW., and García-Castro MI. (2019). WNT/β-catenin modulates the axial identity of embryonic stem cell-derived human neural crest. Development 146, dev175604. 10.1242/dev.175604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, and Noble WS (2011). FIMO: scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018. 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger V, and Hamilton HL (1951). A series of normal stages in the development of the chick embryo. J Morphol 88, 49–92. [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, and Glass CK (2010). Simple Combinations of Lineage-Determining Transcription Factors Prime cis-Regulatory Elements Required for Macrophage and B Cell Identities. Molecular Cell 38, 576–589. 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honoré SM, Aybar MJ, and Mayor R.(2003). Sox10 is required for the early development of the prospective neural crest in Xenopus embryos. Developmental Biology 260, 79–96. 10.1016/s0012-1606(03)00247-1. [DOI] [PubMed] [Google Scholar]
- Hovland AS, Rothstein M, and Simoes-Costa M.(2020). Network architecture and regulatory logic in neural crest development. Wiley Interdiscip Rev Syst Biol Med 12, e1468. 10.1002/wsbm.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovland Austin, & Bhattacharya Debadrita. (2022). Austin-s-h/NC_Timecourse: Publication Release (v1.0.0). Zenodo. 10.5281/zenodo.7044924 [DOI] [Google Scholar]
- Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke T, et al. (2015). Orchestrating high-throughput genomic analysis with Bioconductor. Nature Methods 12, 115–121. 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, and Salzberg SL (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 37, 907–915. 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondoh H, Simões-Costa MS, McKeown SJ, Tan-Cabugao J, Sauka-Spengler T, and Bronner ME (2012). Dynamic and Differential Regulation of Stem Cell Factor FoxD3 in the Neural Crest Is Encrypted in the Genome. PLoS Genetics 8, e1003142. 10.1371/journal.pgen.1003142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359. 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Douarin N.(1973). A biological cell labeling technique and its use in experimental embryology. Developmental Biology 30, 217–222. 10.1016/0012-1606(73)90061-4. [DOI] [PubMed] [Google Scholar]
- Le Douarin NM, and Kalcheim C.(1999). The Neural Crest, 2nd Edition (Cambridge University Press; ). [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, and Genome Project Data Processing, S. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, and Cornell RA (2007). Redundant activities of Tfap2a and Tfap2c are required for neural crest induction and development of other non-neural ectoderm derivatives in zebrafish embryos. Dev Biol 304, 338–354. 10.1016/j.ydbio.2006.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Schulz MH, Look T, Begemann M, Zenke M, and Costa IG (2019). Identification of transcription factor binding sites using ATAC-seq. Genome Biology 20. 10.1186/s13059-0191642-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, and Shi W.(2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Lieberman-Aiden E, Van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. (2009). Comprehensive Mapping of Long-Range Interactions Reveals Folding Principles of the Human Genome. Science 326, 289–293. 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukoseviciute M, Gavriouchkina D, Williams RM, Hochgreb-Hagele T, Senanayake U, Chong-Morrison V., Thongjuea S., Repapi E., Mead A., and Sauka-Spengler T. (2018). From Pioneer to Repressor: Bimodal foxd3 Activity Dynamically Remodels Neural Crest Regulatory Landscape In Vivo. Developmental Cell 47, 608–628.e606. 10.1016/j.devcel.2018.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martik ML, and Bronner ME (2017). Regulatory Logic Underlying Diversification of the Neural Crest. Trends in Genetics 33, 715–727. 10.1016/j.tig.2017.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M.(2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10. 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, and Bejerano G.(2010). GREAT improves functional interpretation of cis-regulatory regions. Nature Biotechnology 28, 495–501. 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meers MP, Janssens DH, and Henikoff S.(2019). Pioneer Factor-Nucleosome Binding Events during Differentiation Are Motif Encoded. Mol Cell 75, 562–575 e565. 10.1016/j.molcel.2019.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael AK, Grand RS, Isbel L, Cavadini S, Kozicka Z, Kempf G, Bunker RD, Schenk AD, Graff-Meyer A, Pathare GR, et al. (2020). Mechanisms of OCT4-SOX2 motif readout on nucleosomes. Science 368, 1460–1465. 10.1126/science.abb0074. [DOI] [PubMed] [Google Scholar]
- Mohammed H, Taylor C, Brown GD, Papachristou EK, Carroll JS, and D’Santos CS (2016). Rapid immunoprecipitation mass spectrometry of endogenous proteins (RIME) for analysis of chromatin complexes. Nature Protocols 11, 316–326. 10.1038/nprot.2016.020. [DOI] [PubMed] [Google Scholar]
- Mullins MC, Dooley CM, Wali N, Sealy IM, White RJ, Stemple DL, Collins JE, and Busch-Nentwich EM (2019). The gene regulatory basis of genetic compensation during neural crest induction. PLOS Genetics 15, e1008213. 10.1371/journal.pgen.1008213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai H, Lin MC, and Sheng G.(2011). A modified cornish pasty method for ex ovo culture of the chick embryo. Genesis 49, 46–52. 10.1002/dvg.20690. [DOI] [PubMed] [Google Scholar]
- Niwa H, Miyazaki J-I, and Smith AG (2000). Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nature Genetics 24, 372–376. 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- Pla P, and Monsoro-Burq AH (2018). The neural border: Induction, specification and maturation of the territory that generates neural crest cells. Dev Biol 444 Suppl 1, S36–S46. 10.1016/j.ydbio.2018.05.018. [DOI] [PubMed] [Google Scholar]
- Pliner HA, Packer JS, McFaline-Figueroa JL, Cusanovich DA, Daza RM, Aghamirzaie D, Srivatsan S, Qiu X, Jackson D, Minkina A, et al. (2018). Cicero Predicts cis-Regulatory DNA Interactions from Single-Cell Chromatin Accessibility Data. Mol Cell 71, 858–871 e858. 10.1016/j.molcel.2018.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roellig D, Tan-Cabugao J, Esaian S, and Bronner ME (2017). Dynamic transcriptional signature and cell fate analysis reveals plasticity of individual neural plate border cells. Elife 6. 10.7554/eLife.21620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rory S, and Gord B.(2011). DiffBind: differential binding analysis of ChIP-Seq peak data. http://bioconductor.org/packages/release/bioc/html/DiffBind.html.
- Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, Brown GD, Gojis O, Ellis IO, Green AR, et al. (2012). Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 481, 389–393. 10.1038/nature10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein M, and Simoes-Costa M.(2020). Heterodimerization of TFAP2 pioneer factors drives epigenomic remodeling during neural crest specification. Genome Res 30, 35–48. 10.1101/gr.249680.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schep AN, Wu B, Buenrostro JD, and Greenleaf WJ (2017). chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat Methods 14, 975–978. 10.1038/nmeth.4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura J, Sanchez-Garcia R, Sorzano COS, and Carazo JM (2019). 3DBIONOTES v3.0: crossing molecular and structural biology data with genomic variations. Bioinformatics 35, 35123513. 10.1093/bioinformatics/btz118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoes-Costa M, and Bronner ME (2015). Establishing neural crest identity: a gene regulatory recipe. Development 142, 242–257. 10.1242/dev.105445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoes-Costa M, and Bronner ME (2016). Reprogramming of avian neural crest axial identity and cell fate. Science 352, 1570–1573. 10.1126/science.aaf2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simões-Costa MS., McKeown SJ., Tan-Cabugao J., Sauka-Spengler T., and Bronner ME. (2012). Dynamic and Differential Regulation of Stem Cell Factor FoxD3 in the Neural Crest Is Encrypted in the Genome. PLoS Genetics 8, e1003142. 10.1371/journal.pgen.1003142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene PJ, and Henikoff S.(2017). An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. eLife 6, e21856. 10.7554/eLife.21856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldatov R, Kaucka M, Kastriti ME, Petersen J, Chontorotzea T, Englmaier L, Akkuratova N, Yang Y, Haring M, Dyachuk V, et al. (2019). Spatiotemporal structure of cell fate decisions in murine neural crest. Science 364, 1–67. 10.1126/science.aas9536. [DOI] [PubMed] [Google Scholar]
- Soufi A, Donahue G, and Kenneth (2012). Facilitators and Impediments of the Pluripotency Reprogramming Factors’ Initial Engagement with the Genome. Cell 151, 994–1004. 10.1016/j.cell.2012.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, and Yamanaka S.(2006). Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 126, 663–676. 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tapia N, MacCarthy C, Esch D, Gabriele Marthaler A, Tiemann U, Arauzo-Bravo MJ, Jauch R, Cojocaru V, and Scholer HR (2015). Dissecting the role of distinct OCT4-SOX2 heterodimer configurations in pluripotency. Sci Rep 5, 13533. 10.1038/srep13533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson M, Siyuan L, Zou N, Smith Z, Meissner A, and Ramanathan S.(2011). Pluripotency Factors in Embryonic Stem Cells Regulate Differentiation into Germ Layers. Cell 145, 875–889. 10.1016/j.cell.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchikawa M, Ishida Y, Takemoto T, Kamachi Y, and Kondoh H.(2003). Functional analysis of chicken Sox2 enhancers highlights an array of diverse regulatory elements that are conserved in mammals. Dev Cell 4, 509–519. 10.1016/s1534-5807(03)00088-1. [DOI] [PubMed] [Google Scholar]
- Wakamatsu Y, Endo Y, Osumi N, and Weston JA (2004). Multiple roles of Sox2, an HMG-box transcription factor in avian neural crest development. Dev Dyn 229, 74–86. 10.1002/dvdy.10498. [DOI] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, and Young RA (2013). Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319. 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RM, Candido-Ferreira I, Repapi E, Gavriouchkina D, Senanayake U, Ling ITC, Telenius J, Taylor S, Hughes J, and Sauka-Spengler T.(2019a). Reconstruction of the Global Neural Crest Gene Regulatory Network In Vivo. Dev Cell 51, 255–276 e257. 10.1016/j.devcel.2019.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RM, Candido-Ferreira I, Repapi E, Gavriouchkina D, Senanayake U, Ling ITC, Telenius J, Taylor S, Hughes J, and Sauka-Spengler T.(2019b). Reconstruction of the Global Neural Crest Gene Regulatory Network In Vivo. Developmental Cell 51, 255–276.e257. 10.1016/j.devcel.2019.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalc A, Sinha R, Gulati GS, Wesche DJ, Daszczuk P, Swigut T, Weissman IL, and Wysocka J.(2021). Reactivation of the pluripotency program precedes formation of the cranial neural crest. Science 371, eabb4776. 10.1126/science.abb4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Bell E, Zhi H, Brown S, Imran SAM, Azuara V, and Cui W.(2019). OCT4 and PAX6 determine the dual function of SOX2 in human ESCs as a key pluripotent or neural factor. Stem Cell Research & Therapy 10. 10.1186/s13287-019-1228-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer CA., Eeckhoute J., Johnson DS., Bernstein BE., Nussbaum C., Myers RM., Brown M., Li W., and Liu XS. (2008). Model-based Analysis of ChIP-Seq (MACS). Genome Biology 9, R137. 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1 (Related to Figure 1) Raw counts for RNA-Seq of chicken neural crest time course.
Supplemental Table 2 (Related to Figure 1) Likelihood Ratio Test (LRT) results comparing GFP+ and GFP− samples.
Supplemental Table 3 (Related to Figure 2) Raw counts for ATAC-Seq of chicken neural crest time course.
Supplemental Table 4 (Related to Figure 2) Peak to gene correlation analysis for all genes and neural crest GRN genes (galGal6).
Supplemental Table 5 (Related to Figure 2) DiffTF results classifying transcription factors as activators or repressors during neural crest development.
Supplemental Table 6 (Related to Figure 3) Shared (~50 bp) foci of SOX2 and OCT4 binding from EcHO analysis (galGal6).
Supplemental Table 7 (Related to Figure 6) AlphaFold modeling of human TFAP2A and SOX2 in pdb format.