Abstract
Background and Objectives
Primary progressive multiple sclerosis (PPMS) displays a highly variable disease progression with a characteristic accumulation of disability, what makes difficult its diagnosis and efficient treatment. The identification of microRNAs (miRNAs)-based signature for the early detection in biological fluids could reveal promising biomarkers to provide new insights into defining MS clinical subtypes and potential therapeutic strategies. The objective of this cross-sectional study was to describe PPMS miRNA profiles in CSF and serum samples compared with other neurologic disease individuals (OND) and relapsing-remitting MS (RRMS).
Methods
First, a screening stage analyzing multiple miRNAs in few samples using OpenArray plates was performed. Second, individual quantitative polymerase chain reactions (qPCRs) were used to validate specific miRNAs in a greater number of samples.
Results
A specific profile of dysregulated circulating miRNAs (let-7b-5p and miR-143-3p) was found downregulated in PPMS CSF samples compared with OND. In addition, in serum samples, miR-20a-5p and miR-320b were dysregulated in PPMS against RRMS and OND, miR-26a-5p and miR-485-3p were downregulated in PPMS vs RRMS, and miR-142-5p was upregulated in RRMS compared with OND.
Discussion
We described a 2-miRNA signature in CSF of PPMS individuals and several dysregulated miRNAs in serum from patients with MS, which could be considered valuable candidates to be further studied to unravel their actual role in MS.
Classification of Evidence
This study provides Class II evidence that specific miRNA profiles accurately distinguish PPMS from RRMS and other neurologic disorders.
Multiple sclerosis (MS) is a chronic inflammatory and neurodegenerative disease of the CNS, the etiology of which is still unknown.1 MS is the most common nontraumatic neurologic cause of disability in young adults.2
MS is clinically heterogeneous. Approximately 85% of patients with MS are diagnosed with relapsing-remitting MS (RRMS), which is characterized by acute neurologic deficits that are followed by complete or incomplete remission.3 The remaining 15% of patients are diagnosed with a progressive disease course of neurologic dysfunction from the onset of symptoms that intensifies over time (accumulation of disability), but without the presence of clinically evident relapses, which is known as primary progressive MS (PPMS).4 The age at clinical onset might vary between RRMS and PPMS individuals because it has been established around 32 years or 42 years, respectively.5 Most current disease-modifying therapies (DMTs) are indicated in the treatment of RRMS, showing none or little efficacy in PPMS, because they modulate or suppress immune responses to reduce CNS inflammation, the number of relapses, and new inflammatory lesions in the MRI.6
MS course might be modulated by the interaction among environmental factors and genetic susceptibility through epigenetic mechanisms.7 One of these mechanisms is microRNAs (miRNAs), small single-stranded noncoding RNAs whose mature and biologically active form comprises 18–25 nucleotides long.8 miRNAs regulate gene expression posttranscriptionally through a mixture of translational inhibition and promotion of mRNA decay.9 Given that an individual miRNA is able to control many target genes, altered expression of miRNAs influences several biological processes in both immune and brain cells. Most miRNAs are located inside cells but, importantly, miRNAs could be released and circulate in different biological fluids, such as plasma, serum, urine, or CSF, where they remain stable even under really hard conditions, such as multiple freeze-thaw cycles, extreme pH values, or treatments with RNase.10,11
miRNA studies in MS have gained tremendous attention during the last few years because they might provide new insights into disease pathology, therapeutic targets, and be promising candidates to be used as biomarkers. CSF is a really interesting fluid because it directly interacts with the extracellular space in the brain, and thus, it might better reflect the level of brain damage.12 Since 2012, when the first analysis of miRNA profiles in CSF was published,13 a total of 12 studies have been released. Indeed, in different studies, miRNAs, such as miR-150 and miR-181c, have been found dysregulated in CSF of patients with MS.13-16 However, it should be mentioned that none of the studies have delved into the analysis of miRNA signatures related specifically to PPMS.
Studying the circulating miRNA profile might be a potential tool to differentiate PPMS participants from RRMS and other neurologic disease (OND) individuals. In addition, it might contribute to elucidate some of the mechanisms involved in the neurodegeneration associated with MS, without being masked by inflammatory processes. Therefore, the main objective of this study was to describe a specific miRNA signature of patients with PPMS in both CSF and serum samples compared with OND and RRMS.
Methods
Patients and Biological Samples
Patients included in this cross-sectional study were recruited at the Girona Neuroimmunology and Multiple Sclerosis Unit of Dr. Josep Trueta University Hospital (Girona, Spain), in the Immunology and Neurology Departments of Ramón y Cajal University Hospital (Madrid, Spain), and at the Neuroimmunology Unit of Hospital Universitari i Politècnic La Fe (Valencia, Spain) from 2011 to 2018. The inclusion criteria were participants older than 18 years with RRMS or PPMS diagnosis according to McDonald 2010 criteria or with OND. The list of diagnoses for the OND group is given in eTable 1, links.lww.com/NXI/A782. Those individuals diagnosed with secondary progressive MS or receiving DMTs at the moment of sample extraction were excluded. Standardized protocols, forms, and databases are used for data collection to minimize sources of bias.
Due to the multicentric nature of the study, clinical data and MRI acquisition and analysis were obtained and interpreted at the different hospitals involved by multiple specialized neurologists and radiologists, respectively. The Expanded Disability Status Scale (EDSS) score was calculated at sampling, whereas MRI assessment was obtained within a maximum time difference of 4 months.
CSF was collected at the moment of diagnosis and was centrifuged immediately after a lumbar puncture at 400g for 15 minutes to obtain cell-free CSF. Blood samples were collected in glass red top serum vacutainer tubes and centrifuged at 2,000g for 10 minutes to separate the serum. Samples were locally processed to obtain cell-free CSF and serum that were later shipped to Girona Neuroimmunology and Multiple Sclerosis Unit for further analysis.
Standard Protocol Approvals, Registrations, and Patient Consents
The Ethics Committee and the Committee for Clinical Investigation from Dr. Josep Trueta University Hospital approved the protocol used in this study, and all participants signed written informed consent.
OCGB Detection
After quantifying serum and CSF levels of IgG using an Immage 800 nephelometer (Beckman Culter, Nyon, Switzerland), IgG bands were analyzed by isoelectric focusing and immunoblotting.17
Circulating RNA Extraction and Purification
Circulating RNA from cell-free CSF and serum was extracted using a mirVana PARIS Isolation Kit (Applied Biosystems, CA) according to the manufacturer's protocol. Briefly, 300 μL of sample were mixed with an equal amount of ×2 denaturing solution. The same volume of acid-phenol:chloroform was then added, and the upper aqueous phase was recovered after centrifugation at 17,000g for 10 minutes. This recovered phase was mixed with 100% ethanol and placed into a provided filter cartridge. After 3 washing steps, total RNA was eluted with 40 μL of nuclease-free water and stored at −80°C for its posterior use.
Circulating miRNA Retrotranscription and Preamplification
An Applied Biosystems TaqMan Advanced miRNA cDNA Synthesis kit (Applied Biosystems, CA) was used to prepare miRNA complementary DNA (cDNA) from CSF and serum samples in this study in one batch. Two microliters of RNA eluate were used to obtain miRNA cDNA following the manufacturer's instructions. Briefly, after the addition of a poly(A) tail and an adaptor, a reverse transcription reaction and miRNA amplification reaction were performed.
Circulating miRNA Quantification
To analyze circulating miRNA expression, 2 different approaches based on TaqMan Advanced miRNA Assays technology (Applied Biosystems, CA) were followed in this study. First, a screening stage analyzing multiple miRNAs in a low number of samples using OpenArray plates was performed. Those samples were selected to assure uniformity in sex and age among the groups. Second, individual quantitative polymerase chain reaction (qPCR) measurements were used to analyze specific miRNAs in a greater number of samples in the validation stage.
For the screening phase, fixed-content OpenArray plates (fc-OA) containing 754 human advanced miRNA assays were used for serum samples. CSF samples were run in triplicate using custom-configured OpenArray plates (cc-OA) specially designed to analyze CSF-enriched miRNAs.18 miRNA cDNA templates were diluted to 1:20 in tris-EDTA buffer and combined with the same volume of TaqMan OpenArray Real-Time PCR Master Mix. After loading the samples into the OpenArray plate, qPCR was performed using a QuantStudio 12K Flex Real-Time PCR System (Applied Biosystems, CA). Both types of OpenArray plates were normalized using the mean expression value of all miRNAs, known as the global normalization method, which has been validated to be used in large scale experiments.11 The delta cycle threshold method was used to calculate the relative expression.
qPCR with Individual TaqMan Advanced hydrolysis probes (Applied Biosystems, CA) was performed to determine expression levels of specific miRNAs in the validation stage. Preamplified cDNA templates were first diluted in nuclease-free water to obtain a final cDNA concentration of 1:10. Samples were run in triplicate. Individual miRNA expression was assessed in one batch with the Quant Studio 7 Flex System (Applied Biosystems, CA). Serum miRNA expression values were normalized using the mean value of 2 endogenous miRNAs (miR-15a-5p and miR-23a-3p), whereas CSF expression values were normalized using the mean value of 8 endogenous miRNAs (miR-16 + miR-24 + miR-125a + miR-30d + miR-26b + miR-145 + miR-92b + miR-23a), as shown in a previous search of suitable endogenous normalizers in these CSF samples.18 The delta cycle threshold method was also used to calculate the relative expression.
Cellular/Tissue-Enriched Source Analyses
Different public repositories, such as human miRNA tissue atlas, CNS miRNA profiles prveiously descibed, and FANTOM5, which contain data from miRNA expression profiles in diverse human tissue, CNS cells, or primary cells, respectively, were used to study potential tissue and cellular sources of CSF dysregulated miRNAs.19-21
Statistical Analysis
Demographic, clinical, and radiologic data and normalized miRNA expression levels were reported as follows: categorical variables were shown as absolute and relative frequencies, while continuous variables were represented by median and quartiles. Statistical differences were determined using nonparametric tests as the Fisher exact test and the Kruskal-Wallis test for categorical and quantitative variables, respectively. Correlation analysis of normalized serum miRNA levels and clinical and radiologic data were estimated by the Spearman rho (rs). All analyses were 2-tailed with a significance level of 5% (2-sided) and a type II error rate of 10%. Missing data were excluded. Due to the preliminary nature of this study, multiple comparison corrections were not performed. Statistical analyses were performed in the Statistical Package for the Social Sciences version 25.0 (IBM SPSS Statistics for Windows, NY). Figures were built using GraphPad PRISM v.5 (GraphPad Software, CA). The STROBE cross-sectional reporting guidelines were respected for planning, conduct, analysis, and reporting of this study.22
Data Availability
Anonymized data not published within this article will be made available by request from any qualified investigator.
Results
Clinical Characteristics of the Studied Cohort
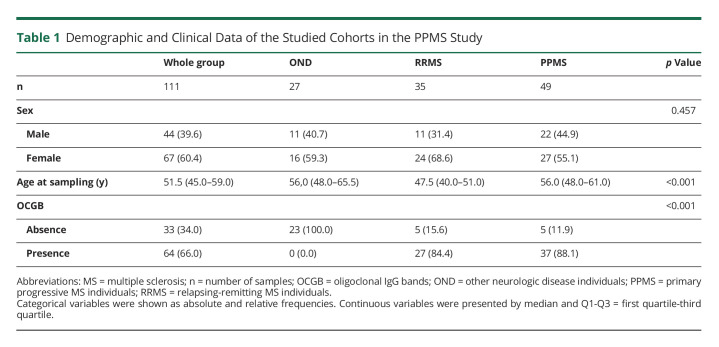
This study comprised a total of 111 individuals (60.4% women) with a median age of 51.5 years. The whole cohort distribution was the following: 49 participants belonged to the PPMS group (55.1% women), 35 to RRMS (68.6% women), and 27 were OND individuals (59.3% women). Clinical and demographic data are given in Table 1. No significant differences were observed in sex between groups. Age at sampling showed statistically significant differences between groups (p < 0.001) because RRMS individuals were younger than PPMS and OND. The intrathecal synthesis of oligoclonal IgG bands was not present in any of the OND participants (p < 0.001). Additional information regarding the different subcohorts used in this study is shown in eFigure 1, links.lww.com/NXI/A782.
Table 1.
Demographic and Clinical Data of the Studied Cohorts in the PPMS Study
miRNA Signature in CSF of Patients With PPMS
To define a characteristic miRNA profile in CSF of patients with PPMS, a screening phase was designed using cc-OA panels. A total of 41 samples (13 OND, 12 RRMS, and 16 PPMS) were used in this phase per triplicate. Twenty-seven miRNAs were detected in 70% of total samples (at least 50% of samples in each group), and their differential expression was analyzed. We identified 6 miRNAs that showed significant differences or tendency (p < 0.050 or p < 0.100, respectively) to be dysregulated in some of the groups (eTable 2, links.lww.com/NXI/A782).
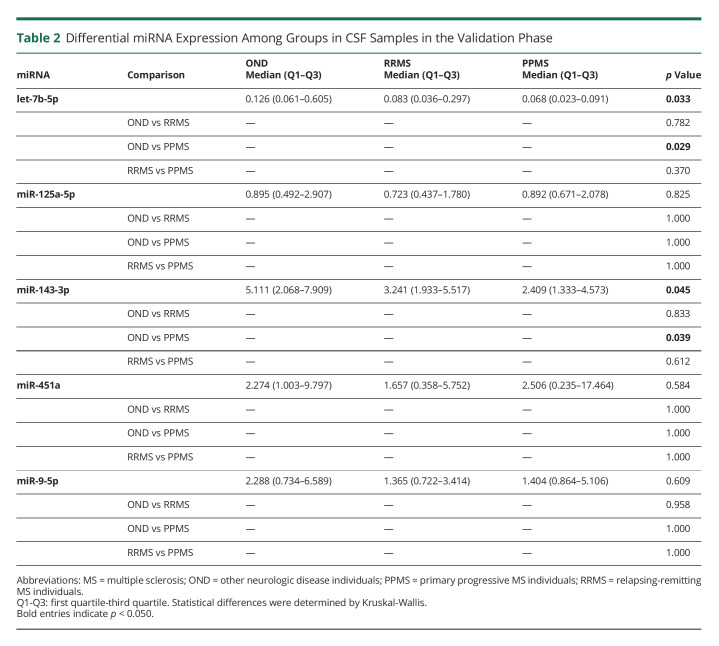
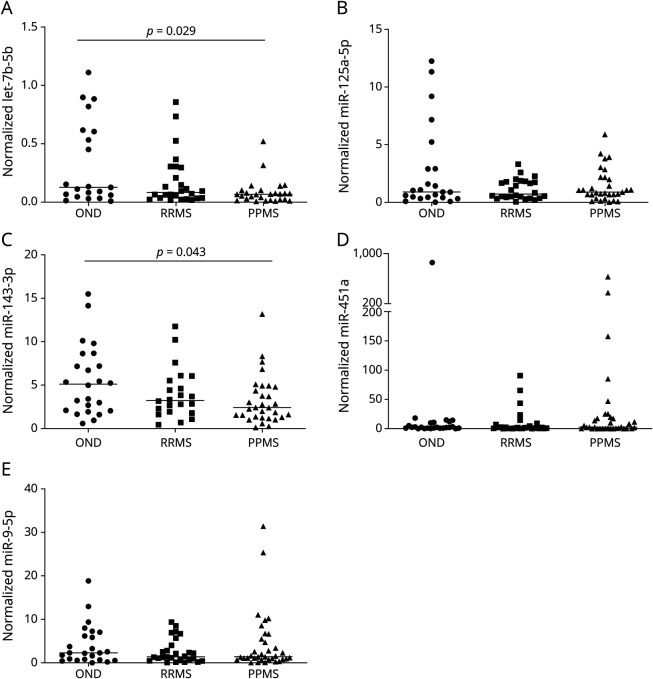
Levels of let-7b-5p, miR-125a-5p, miR-143-3p, miR-451a, and miR-9-5p were further validated in a bigger cohort of 100 samples (27 OND, 31 RRMS, and 42 PPMS) by individual qPCR. These miRNAs were selected according to the screening phase results or interesting results from previous publications. Both let-7b-5p and miR-143-3p were downregulated in PPMS samples compared with OND (Table 2 and Figure 1).
Table 2.
Differential miRNA Expression Among Groups in CSF Samples in the Validation Phase
Figure 1. Differentially Expressed miRNAs in CSF in the Validation Phase.
Dot plots for normalized values of (A) let-7b-5p, (B) miR-125a-5p, (C) miR-143-3p, (D) miR-451a, and (E) miR-9-5p in CSF samples. The dot indicates one sample, and the line indicates the median. The Kruskal-Wallis test and Mann-Whitney U test for the post hoc analysis were used to determine statistical differences among groups.
The PPMS group could be further subclassified into inflammatory or noninflammatory profiles according to clinical relapse or radiologic activity (existence of Gd + lesions or increased number of T2 lesions in follow-up MRIs). It is of interest that let-7b-5p expression was also found dysregulated among the 4 groups due to the differences between OND and noninflammatory PPMS (eTable 3, links.lww.com/NXI/A782).
Analysis of Cellular/Tissue Sources of CSF Dysregulated miRNAs
The potential tissue and cellular origin of let-7b-5p and miR-143-3p were examined using different repositories. First, the human miRNA tissue atlas was established by quantifying the abundance of miRNA in tissue biopsies of 2 individuals.19 It is of interest that let-7b-5p might be found highly expressed in the spinal cord, brain, and arachnoid mater. Further investigation of miRNA expression in CNS-specific cell types showed that let-7b-5p might present an increased expression in astrocytes.20
However, miR-143-3p might not present an elevated expression in CNS organs, it could be found highly expressed in circulating cells and neutrophils, after analyzing miRNA expression across primary cells using the FANTOM5 atlas.21
miRNA Signature in Serum of Patients With PPMS
Similar to CSF samples, a screening phase with fc-OA panels was designed and 34 serum samples (9 OND, 10 RRMS, and 15 PPMS) were used in this stage. A total of 127 miRNAs were detected in 70% of samples in each group. After normalizing using the global mean value of all miRNAs, 10 miRNAs showed significant differences or tendency to be dysregulated in some of the groups (eTable 4, links.lww.com/NXI/A782).
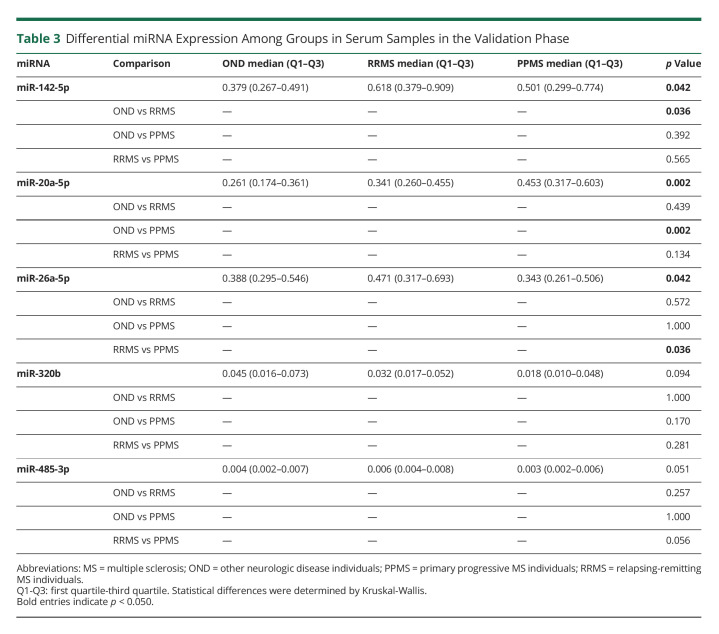
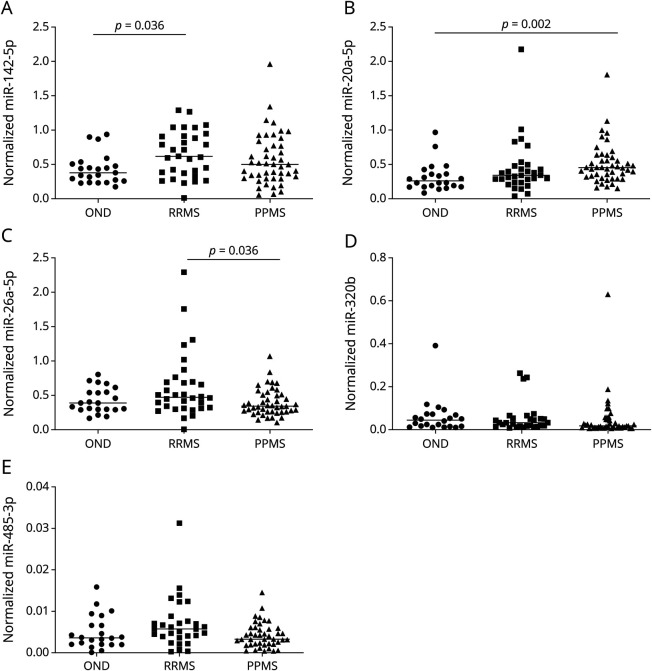
Dysregulation of these 10 miRNAs and another 2 miRNAs of interest (miR-448 and miR-485-3p) was further analyzed in a bigger cohort of 107 samples (26 OND, 32 RRMS, and 49 PPMS) by individual qPCR. We observed that miR-20a-5p and miR-320b values are dysregulated in PPMS against RRMS and OND. In addition, miR-26a-5p and miR-485-3p were downregulated in PPMS vs RRMS, and miR-142-5p was upregulated in RRMS compared with OND (Table 3 and Figure 2).
Table 3.
Differential miRNA Expression Among Groups in Serum Samples in the Validation Phase
Figure 2. Differentially Expressed miRNAs in Serum in the Validation Phase.
Dot plots for normalized values of (A) miR-142-5p, (B) miR-20a-5p, (C) miR-26a-5p, (D) miR-320b, and (E) miR-485-3p in serum samples. The dot indicates one sample, and the line indicates the median. The Kruskal-Wallis test and Mann-Whitney U test for the post hoc analysis were used to determine statistical differences among groups.
When subclassifying the PPMS cohort, miR-20a-5p levels were significantly upregulated in noninflammatory PPMS and inflammatory PPMS vs OND and miR-186-5p levels were significantly reduced in inflammatory PPMS (eTable 3, links.lww.com/NXI/A782).
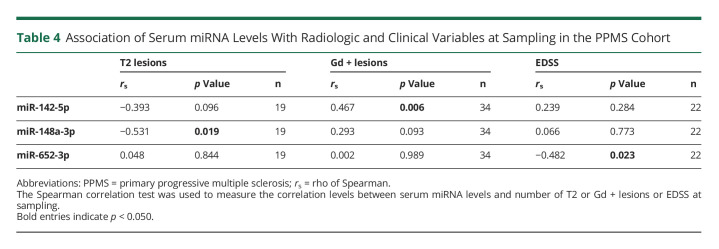
Correlation of Serum miRNA Levels With Radiologic and Clinical Variables
The correlation in the PPMS cohort between serum miRNA levels in the validation stage and the number of T2 and Gd+ lesions in MRI and EDSS at sampling was studied. Lower levels of miR-142-5p and miR-148a-3p were associated with higher number of T2 lesions (rs = −0.393, p = 0.096; rs = −0.531, p = 0.019, respectively). At the same time, increased miR-142-5p levels correlated with higher number of Gd+ lesions (rs = 0.467, p = 0.006). Last, lower EDSS scores and higher serum levels of miR-652-3p were associated (rs = −0.482, p = 0.023) (Table 4).
Table 4.
Association of Serum miRNA Levels With Radiologic and Clinical Variables at Sampling in the PPMS Cohort
Classification of Evidence Assignment
Circulating miRNA signatures might be potential biomarkers in MS to discriminate subtypes and MS individuals from OND participants. This study provides Class II evidence that miRNA profiles accurately distinguish PPMS from RRMS and OND individuals. Specifically in serum samples, miR-20a-5p and miR-320b might differentiate PPMS from RRMS and OND, miR-26a-5p and miR-485-3p might show differential expression in PPMS compared with RRMS, and higher miR-142-5p levels might be characteristic of RRMS compared with OND.
Discussion
In this study, we investigated the potential role of circulating miRNAs to differentiate PPMS from OND and RRMS. We identified downregulation in let-7b-5p and miR-143-3p in PPMS CSF samples compared with OND. In addition, differential expression in miR-20a-5p, miR-320b, miR-26a-5p, and miR-485-3p was found in serum of PPMS individuals by comparing with RRMS and OND. Taken together, our findings provide possible miRNAs candidates to differentiate PPMS and potential targetable biological pathways involved in PPMS.
RRMS is mainly associated with an inflammatory burden, whereas PPMS is characterized by demyelination and disability from the onset.23 Even if the pathologic mechanisms causing tissue damage in both RRMS and PPMS overlap, some characteristics differ enormously. Although focal inflammatory lesions with an accumulation of T and B cells and blood-brain barrier leakage are the main pathologic indicators of RRMS, the gradual accumulation of disability characteristic of PPMS individuals might be the result of diffuse immune mechanisms and neurodegeneration.23 Most current DMTs are largely only suitable for RRMS because they decrease relapse frequency and new lesion formation. Recently, some clinical trials have shown the efficacy of B-cell depletion therapy in PPMS individuals.24 Several studies have also suggested the benefits of an early treatment with high efficacy DMTs to delay neurologic damage.25 However, one unmet need might be finding remyelinating therapies that could restore damage and function, preventing neurodegeneration.26
miRNA studies in MS have gained attention recently due to the new insights they can offer.27 Beyond their role as biomarkers, miRNA studies might also provide new concepts into disease pathology because MS etiology is still unknown,1 and deep into therapeutic targets,27 to address new challenges as limiting progressive degeneration or restoring function.28 The therapeutic potential for MS of several miRNAs such as miR-155, miR-326, miR-20a, or miR-219a by targeting its inhibition or overexpression has already been explored in mouse models.27,29,30
Although most studies have been focused on analyzing miRNA profiles in blood cells27 or even in postmortem brain lesions of patients with MS,31 circulating miRNAs in serum present lots of advantages to be considered promising biomarkers.32 In addition, as CSF is in direct contact with CNS organs and it might mirror neurologic events, miRNA CSF studies might be really valuable to deepen the knowledge of MS pathology.33 However, some challenges need to be solved to fully exploit miRNA potential in this field and overcome the lack of replication among studies and conflicting results such as methodological heterogeneity, lack of controls, reduced samples sizes, poor experimental designs, or handling missing data.34
Since the first analysis of circulating miRNAs in the CSF of patients with MS was published,13 a total of 12 MS studies analyzing miRNA expression in CSF samples have been reported. Four of them performed a preliminary screening phase using high-throughput platforms as TaqMan Low Density Arrays15,16 or arrays,13,35 whereas the rest only analyzed the expression of specific individual miRNAs. Three studies used CSF samples from PPMS, and while 2 of them only analyzed specific miRNAs,14,36 Haghikia et al.13 could not find any statistical change during their profiling in PPMS probably due to the small size of the cohort (n = 6). However, none of these CSF MS studies aimed to specifically uncover miRNA signatures in PPMS individuals. Therefore, we aimed to characterize for the first time a specific miRNA profile of patients with PPMS compared with OND and RRMS in both CSF and serum samples.
In this study, let-7b-5p and miR-143-3p were found downregulated in CSF samples of PPMS compared with OND. Although let-7b-5p did not present statistical significance in the screening phase, it was chosen to be analyzed in the posterior validation stage because it has been described as dysregulated in different MS studies. In line with our results, it has been recently described that CSF let-7b-5p levels were significantly reduced in patients with PPMS and it was found negatively correlated with clinical disability at disease onset and after a follow-up period.37 let-7b-5p has also been found upregulated in peripheral blood of pediatric MS cases compared with healthy controls,38 and its expression changes during interferon-β treatment.39 Indeed, it has been suggested as a protective factor for MS course, in both inflammation and clinical disability, although further experiments are needed to elucidate this aspect. Regarding miR-143-3p, it was found downregulated in different studies focused on MS. Its downregulation was evident in astrocytes from active MS lesions.40 In experimental ischemic injury, inhibition of miR-143-3p has been related to glucose metabolism.41 Since glucose is the main nutrient used by the brain and neurotransmission processes are high energy-consuming, deepening into metabolic pathways involving miR-143-3p would be of interest to elucidate possible therapeutic targets for MS.
Despite the observed differences, we could not find dysregulated miRNAs that differentiate the PPMS group from both RRMS and OND in CSF samples. On the contrary, miR-20a-5p and miR-320b in serum might assume that role because their levels were increased and decreased in PPMS, respectively. In this regard, miR-20 has been described as a contributor to the immune cell signature observed in MS involving regulation of T-cell activation.42 Thus, these miRNAs may provide useful targets for new therapeutic approaches. In addition, other changes in serum samples were observed in this study because miR-26a-5p and miR-485-3p levels were downregulated in PPMS vs RRMS, whereas miR-142-5p was found upregulated in RRMS against OND. It is of interest that upregulation of miR-142-5p has been found in white matter lesions and in normal-appearing white matter, which might also modulate inflammatory processes.43
There are 2 published studies whose aims were to associate miRNA expression in serum samples with MRI measures and phenotypes in MS.44,45 Correlations between miR-142-5p and miR-148a-3p and the number of T2 lesions and Gd + lesions, respectively, in the PPMS cohort have also been described in this study. Only miR-142-5p levels were found associated with the T1:T2 ratio in one of the studies.45
Regarding clinical variables in the studied cohort, 1 statistically significant difference was observed. Although we tried to match samples of similar ages among the 3 studied groups in the screening phase when increasing the cohort for the validation phase, the RRMS group was statistically younger than PPMS and OND. It has already been established that the age at clinical onset in PPMS is higher than the one observed in RRMS.46 For this reason, at least, an OND cohort comparable with PPMS in age was used to be able to avoid possible miRNA dysregulation due to aging.
In conclusion, this study was focused on describing miRNA profiles in CSF of PPMS individuals. The fact that the validation cohort of this study comprised samples coming from 3 different centers strengthened the miRNA expression results with a global character. However, this specific characteristic provided at the same time one of the main limitations in this study, the existence of multiple missing data regarding radiologic and clinical information from patients with MS. In addition, we were unable to find a CSF miRNA that distinguished PPMS from OND and RRMS. Sample size should be increased, and some other potential miRNAs such as miR-125b-5p or miR-29a-3p might be analyzed. Total RNA extracted from cell-free CSF was analyzed in this study instead of focusing on exosomes, small vesicles containing miRNAs that might provide information about their cellular origin.47 Despite the mentioned deficiencies and the preliminary nature of this work, let-7b-5p and miR-143-3p are firm candidates to be analyzed in further functional studies to unravel their potential biological role in PPMS and serum dysregulated miRNAs might be potential candidates that might differentiate MS subtypes.
Acknowledgment
The authors want to particularly acknowledge the patients and the IDIBGI Biobank (Biobanc IDIBGI, B. 0000872), integrated in the Spanish National Biobank Network, for their collaboration.
Glossary
- cc-OA
custom-configured OpenArray plates
- cDNA
complementary DNA
- EDSS
Expanded Disability Status Scale
- DMTs
disease-modifying therapies
- miRNAs
microRNAs
- MS
multiple sclerosis
- OND
other neurologic disease
- PPMS
primary progressive MS
- qPCR
quantitative polymerase chain reaction
- RRMS
relapsing-remitting MS
Appendix. Authors

Footnotes
Class of Evidence: NPub.org/coe
Study Funding
The author(s) disclose the receipt of the following financial support for the research, authorship, and/or publication of this article: this study has been funded by grants from Instituto de Salud Carlos III—Red Española de Esclerosi múltiple (REEM) through the projects PI13/01782 and RD16/0015/004 (co-funded by ERDF/ESF, “A way to make Europe”/“Investing in your future”) and Asociación Esclerosis Múltiple (EME; EME-REEM 2020). M. Muñoz-San Martín was supported by a FI-DGR 2016 from AGAUR. A. Miguela was supported by a grant from Granés Fundació.
Disclosure
All authors report no disclosures. Go to Neurology.org/NN for full disclosure.
References
- 1.Thompson AJ, Baranzini SE, Geurts J, Hemmer B, Ciccarelli O. Multiple sclerosis. Lancet. 2018;391(10130):1622-1636. doi: 10.1016/S0140-6736(18)30481-1. [DOI] [PubMed] [Google Scholar]
- 2.Hollen CW, Paz Soldán MM, Rinker JR, Spain RI. The future of progressive multiple sclerosis therapies. Fed Pract. 2020;37(suppl 1):S43-S49. Accessed October 14, 2020. ncbi.nlm.nih.gov/pubmed/32341636. [PMC free article] [PubMed] [Google Scholar]
- 3.Gross HJ, Watson C. Characteristics, burden of illness, and physical functioning of patients with relapsing-remitting and secondary progressive multiple sclerosis: a cross-sectional US survey. Neuropsychiatr Dis Treat. 2017;13:1349-1357. doi: 10.2147/NDT.S132079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olek MJ. Multiple sclerosis. Ann Intern Med. 2021;174(6):ITC81-ITC96. doi: 10.7326/AITC202106150 [DOI] [PubMed] [Google Scholar]
- 5.Rommer PS, Ellenberger D, Hellwig K, et al. Relapsing and progressive MS: the sex-specific perspective. Ther Adv Neurol Disord. 2020;13:175628642095649. doi: 10.1177/1756286420956495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ciotti JR, Valtcheva MV, Cross AH. Effects of MS disease-modifying therapies on responses to vaccinations: a review. Mult Scler Relat Disord. 2020;45:102439. doi: 10.1016/j.msard.2020.102439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huynh JL, Casaccia P. Epigenetic mechanisms in multiple sclerosis: implications for pathogenesis and treatment. Lancet Neurol. 2013;12(2):195-206. doi: 10.1016/S1474-4422(12)70309-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhaskaran M, Mohan M. MicroRNAs: history, biogenesis, and their evolving role in animal development and disease. Vet Pathol. 2014;51(4):759-774. doi: 10.1177/0300985813502820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dexheimer PJ, Cochella L. MicroRNAs: from mechanism to organism. Front Cell Dev Biol. 2020;8:409. doi: 10.3389/fcell.2020.00409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang L, Ding H, Zhang Y, Wang Y, Zhu W, Li P. Circulating MicroRNAs: biogenesis and clinical significance in acute myocardial infarction. Front Physiol. 2020;11:1088. doi: 10.3389/fphys.2020.01088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marabita F, de Candia P, Torri A, Tegnér J, Abrignani S, Rossi RL. Normalization of circulating microRNA expression data obtained by quantitative real-time RT-PCR. Brief Bioinform. 2016;17(2):204-212. doi: 10.1093/bib/bbv056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jagot F, Davoust N. Is it worth considering circulating microRNAs in multiple sclerosis? Front Immunol. 2016;7:129. doi: 10.3389/fimmu.2016.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haghikia A, Haghikia A, Hellwig K, et al. Regulated microRNAs in the CSF of patients with multiple sclerosis: a case-control study. Neurology. 2012;79(22):2166-2170. doi: 10.1212/WNL.0b013e3182759621. [DOI] [PubMed] [Google Scholar]
- 14.Kramer S, Haghikia A, Bang C, et al. Elevated levels of miR-181c and miR-633 in the CSF of patients with MS: a validation study. Neurol Neuroimmunol Neuroinflammation. 2019;6(6):e623. doi: 10.1212/NXI.0000000000000623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bergman P, Piket E, Khademi M, et al. Circulating miR-150 in CSF is a novel candidate biomarker for multiple sclerosis. Neurol Neuroimmunol Neuroinflamm. 2016;3(3):e219. doi: 10.1212/NXI.0000000000000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quintana E, Ortega FJ, Robles-Cedeño R, et al. miRNAs in cerebrospinal fluid identify patients with MS and specifically those with lipid-specific oligoclonal IgM bands. Mult Scler J. 2017;23(13):1716-1726. doi: 10.1177/1352458516684213. [DOI] [PubMed] [Google Scholar]
- 17.Sádaba MC, González Porqué P, Masjuan J, Alvarez-Cermeño JC, Bootello A, Villar LM. An ultrasensitive method for the detection of oligoclonal IgG bands. J Immunol Methods. 2004;284(1-2):141-145. doi: 10.1016/j.jim.2003.09.018. [DOI] [PubMed] [Google Scholar]
- 18.Muñoz-San Martín M, Gomez I, Miguela A, et al. Description of a CSF-enriched miRNA panel for the study of neurological diseases. Life (Basel). 2021;11(7):594. doi: 10.3390/life11070594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ludwig N, Leidinger P, Becker K, et al. Distribution of miRNA expression across human tissues. Nucleic Acids Res. 2016;44(8):3865-3877. doi: 10.1093/nar/gkw116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoye ML, Koval ED, Wegener AJ, et al. MicroRNA profiling reveals marker of motor neuron disease in ALS models. J Neurosci. 2017;37(22):5574-5586. doi: 10.1523/JNEUROSCI.3582-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lizio M, Harshbarger J, Shimoji H, et al. Gateways to the FANTOM5 promoter level mammalian expression atlas. Genome Biol. 2015;16(1):22. doi: 10.1186/s13059-014-0560-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.von Elm E, Altman DG, Egger M, Pocock SJ, Gøtzsche PC, Vandenbroucke JP. Strengthening the reporting of observational studies in epidemiology (STROBE) statement: guidelines for reporting observational studies. BMJ. 2007;18(6):800-804. doi: 10.1136/BMJ.39335.541782. [DOI] [PubMed] [Google Scholar]
- 23.Ontaneda D. Progressive multiple sclerosis. Contin Lifelong Learn Neurol. 2019;25(3):736-752. doi: 10.1212/CON.0000000000000727. [DOI] [PubMed] [Google Scholar]
- 24.Miyazaki Y, Niino M. B-cell depletion therapy for multiple sclerosis. Immunol Med. 2021;45(2):54-62. doi: 10.1080/25785826.2021.1952543 [DOI] [PubMed] [Google Scholar]
- 25.Filippi M, Danesi R, Derfuss T, et al. Early and unrestricted access to high-efficacy disease-modifying therapies: a consensus to optimize benefits for people living with multiple sclerosis. J Neurol. 2022;269(3):1670-1677. doi: 10.1007/S00415-021-10836-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cunniffe N, Coles A. Promoting remyelination in multiple sclerosis. J Neurol. 2021;268(1):30-44. doi: 10.1007/S00415-019-09421-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piket E, Zheleznyakova GY, Kular L, Jagodic M. Small non-coding RNAs as important players, biomarkers and therapeutic targets in multiple sclerosis: a comprehensive overview. J Autoimmun. 2019;101:17-25. doi: 10.1016/j.jaut.2019.04.002. [DOI] [PubMed] [Google Scholar]
- 28.Franklin RJM, Ffrench-Constant C. Regenerating CNS myelin–from mechanisms to experimental medicines. Nat Rev Neurosci. 2017;18(12):753-769. doi: 10.1038/nrn.2017.136. [DOI] [PubMed] [Google Scholar]
- 29.Osorio-Querejeta I, Carregal-Romero S, Ayerdi-Izquierdo A, et al. MiR-219a-5p enriched extracellular vesicles induce OPC differentiation and EAE improvement more efficiently than liposomes and polymeric nanoparticles. Pharmaceutics. 2020;12(2):186. doi: 10.3390/pharmaceutics12020186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang H, Moyano AL, Ma ZZ, et al. miR-219 cooperates with miR-338 in myelination and promotes myelin repair in the CNS. Dev Cell. 2017;40(6):566-582.e5. doi: 10.1016/j.devcel.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fritsche L, Teuber-Hanselmann S, Soub D, Harnisch K, Mairinger F, Junker A. MicroRNA profiles of MS gray matter lesions identify modulators of the synaptic protein synaptotagmin-7. Brain Pathol. 2020;30(3):524-540. doi: 10.1111/bpa.12800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bär C, Thum T, De Gonzalo-Calvo D. Circulating miRNAs as mediators in cell-to-cell communication. Epigenomics. 2019;11(2):111-113. doi: 10.2217/epi-2018-0183. [DOI] [PubMed] [Google Scholar]
- 33.Vernau W, Vernau KA, Sue Bailey C. Cerebrospinal fluid. In: Clinical Biochemistry of Domestic Animals. Elsevier Inc; 2008:769-819. doi: 10.1016/B978-0-12-370491-7.00026-X. [DOI] [Google Scholar]
- 34.de Gonzalo-Calvo D, Pérez-Boza J, Curado J, Devaux Y, EU-CardioRNA COST Action CA17129. Challenges of microRNA-based biomarkers in clinical application for cardiovascular diseases. Clin Transl Med. 2022;12(2):e585. doi: 10.1002/CTM2.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perdaens O, Dang HA, D'Auria L, van Pesch V. CSF microRNAs discriminate MS activity and share similarity to other neuroinflammatory disorders. Neurol Neuroimmunol Neuroinflammation. 2020;7(2):673. doi: 10.1212/NXI.0000000000000673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bruinsma IB, van Dijk M, Bridel C, et al. Regulator of oligodendrocyte maturation, miR-219, a potential biomarker for MS. J Neuroinflammation. 2017;14(1):235. doi: 10.1186/s12974-017-1006-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mandolesi G, Rizzo FR, Balletta S, et al. The microRNA let-7b-5p is negatively associated with inflammation and disease severity in multiple sclerosis. Cells. 2021;10(2):330-422. doi: 10.3390/CELLS10020330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liguori M, Nuzziello N, Licciulli F, et al. Combined microRNA and mRNA expression analysis in pediatric multiple sclerosis: an integrated approach to uncover novel pathogenic mechanisms of the disease. Hum Mol Genet. 2018;27(1):66-79. doi: 10.1093/hmg/ddx385. [DOI] [PubMed] [Google Scholar]
- 39.Hecker M, Thamilarasan M, Koczan D, et al. MicroRNA expression changes during interferon-beta treatment in the peripheral blood of multiple sclerosis patients. Int J Mol Sci. 2013;14(8):16087-16110. doi: 10.3390/ijms140816087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rao VTS, Fuh SC, Karamchandani JR, et al. Astrocytes in the pathogenesis of multiple sclerosis: an in situ microRNA study. J Neuropathol Exp Neurol. 2019;78(12):1130-1146. doi: 10.1093/jnen/nlz098. [DOI] [PubMed] [Google Scholar]
- 41.Zeng X, Liu N, Zhang J, et al. Inhibition of miR-143 during ischemia cerebral injury protects neurones through recovery of the hexokinase 2-mediated glucose uptake. Biosci Rep. 2017;37(4):BSR20170216. doi: 10.1042/BSR20170216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cox MB, Cairns MJ, Gandhi KS, et al. MicroRNAs miR-17 and miR-20a inhibit T cell activation genes and are under-expressed in MS whole blood. PLoS One. 2010;5(8):e12132. doi: 10.1371/JOURNAL.PONE.0012132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Teuber-Hanselmann S, Meinl E, Junker A. MicroRNAs in gray and white matter multiple sclerosis lesions: impact on pathophysiology. J Pathol. 2020;250(5):496-509. doi: 10.1002/PATH.5399. [DOI] [PubMed] [Google Scholar]
- 44.Hemond CC, Healy BC, Tauhid S, et al. MRI phenotypes in MS: longitudinal changes and miRNA signatures. Neurol Neuroimmunol Neuroinflammation. 2019;6(2):e530. doi: 10.1212/NXI.0000000000000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Regev K, Healy BC, Khalid F, et al. Association between serum MicroRNAs and magnetic resonance imaging measures of multiple sclerosis severity. JAMA Neurol. 2017;74(3):275-285. doi: 10.1001/jamaneurol.2016.5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moreno-Torres I, Sabín-Muñoz J, García-Merino A. Chapter 1: multiple sclerosis: epidemiology, genetics, symptoms, and unmet needs. In: RSC Drug Discovery Series, Vol 2019-January. Royal Society of Chemistry; 2019:3-32. doi: 10.1039/9781788016070-00001. [DOI] [Google Scholar]
- 47.Mycko MP, Baranzini SE. microRNA and exosome profiling in multiple sclerosis. Mult Scler J. 2020;26(5):599-604. doi: 10.1177/1352458519879303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Anonymized data not published within this article will be made available by request from any qualified investigator.