Abstract
Chlamydia pneumoniae is an obligate intracellular parasite with a developmental cycle believed to be common to all members of the genus Chlamydia. We present a detailed description based on transmission and scanning electron microscopy of temporal events and inclusion structures throughout the C. pneumoniae AR-39 developmental cycle.
Chlamydia pneumoniae is a recently recognized chlamydial species causing infections of the respiratory tract. It has been estimated that C. pneumoniae infections are responsible for up to 10% of all cases of community-acquired pneumonia and 5% of bronchitis and sinusitis cases (18). C. pneumoniae infections are ubiquitous. Virtually everyone is infected at some point in life and reinfections are possible (4). Recently, C. pneumoniae has been receiving intense interest due to reports of its association with a variety of acute or chronic diseases, including atherosclerosis (reviewed in reference 4), asthma, sarcoidosis, otitis media, erythema nodosum, and Reiter's syndrome (18). Although the findings obtained so far support an association with atherosclerosis, the exact role of these bacteria in the pathogenesis of atherosclerotic lesions is unclear.
Like all chlamydiae, C. pneumoniae undergoes a developmental cycle in which two functionally and morphologically distinct cell types are recognized. The infectious cell type, which is specialized for extracellular survival and transmission, is termed the elementary body (EB). The intracellular, vegetative cell type is called the reticulate body (RB). The developmental cycle is initiated by endocytosis of an EB by a eukaryotic host cell. Chlamydiae remain within an intracellular vacuole, termed an inclusion, for their entire developmental cycle. Shortly after internalization, EBs begin to reorganize and differentiate into RBs, which then begin to multiply by binary fission. Late in the cycle, logarithmic growth ceases as RBs commence to reorganize into EBs, which are released upon lysis of the host cell.
The entry and the structure of C. pneumoniae EBs have been described previously (6, 8, 9, 20, 23, 24). However, little is known in regard to ultrastructural events throughout the C. pneumoniae developmental cycle. Indeed, many assumptions regarding C. pneumoniae development are based on analogies to C. trachomatis or C. psittaci. In this report we present a detailed analysis of the C. pneumoniae developmental cycle as characterized by transmission and scanning electron microscopy.
C. pneumoniae AR-39 (CCL 2.1) was purchased from the American Type Culture Collection and propagated in HeLa 229 cells. Confluent monolayers of HeLa 229 cells on glass coverslips in 24-well plates were infected with Renografin-purified C. pneumoniae at a multiplicity of infection of ≈1 by centrifugation at 900 × g for 1 h (19). After centrifugation, infected cells were washed twice with SPG (sucrose-phosphate-glutamate buffer) and incubated in RPMI 1640 supplemented with 10% fetal calf serum plus 10 μg of gentamicin and 0.9 μg of cycloheximide per ml. At various time points postinfection, the infected cells were fixed and processed for electron microscopy as previously described (12).
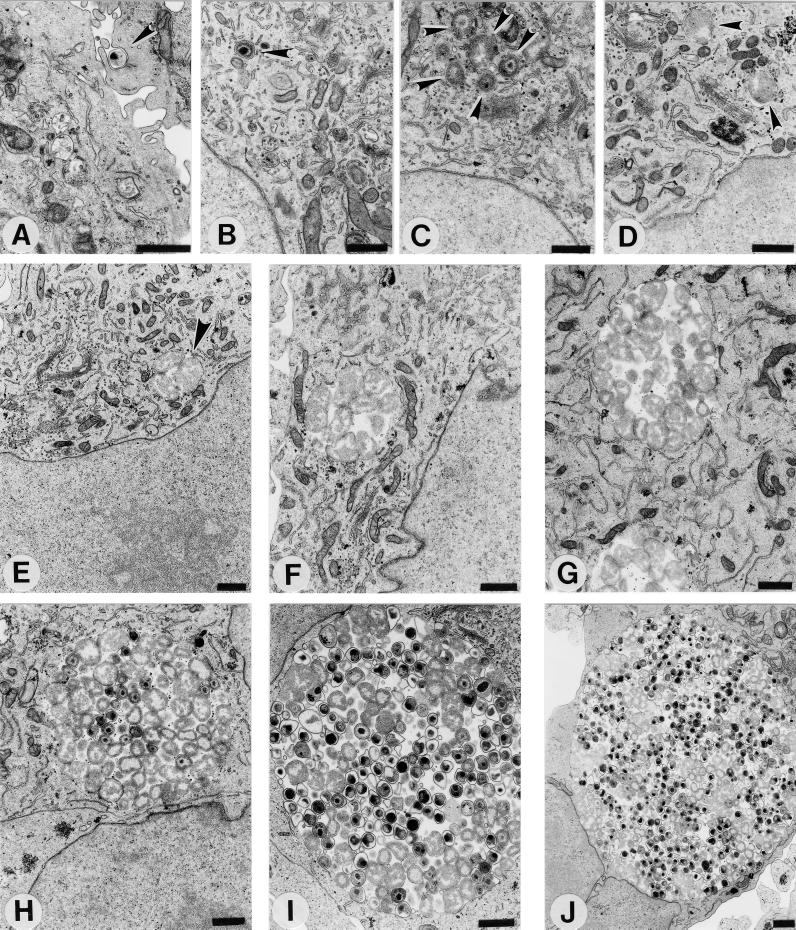
Like those of C. trachomatis (14) and C. psittaci (15), C. pneumoniae EBs appear to interact initially with microvilli prior to endocytosis by the host cell (Fig. 1A). Once internalized, the EBs, still retaining their characteristic size and condensed nucleoid structure, remain within individual, tightly membrane-bound vesicles (Fig. 1B). By 8 h postinfection, differentiation into RBs is evidenced by the dissociation of the nucleoid and some increase in size is apparent, although EBs still containing a condensed nucleoid may be observed (Fig. 1C). At 12 h postinfection, morphologically typical RBs are present (Fig. 1D), and by 19 h postinfection, multiplication has begun (Fig. 1E). At 24 and 36 h postinfection (Fig. 1F and G), the RBs continue to multiply and accumulate within the inclusion and no EBs are yet detected. Note the morphology of the inclusions, in which RBs are observed throughout the entire volume. This morphology is distinct from that of C. trachomatis at an equivalent stage of development, since C. trachomatis RBs are typically tightly juxtaposed to the inclusion membrane, with the lumen of the inclusion being devoid of organisms (11). The developmental cycle becomes asynchronous by 48 h postinfection as the differentiation of RBs back into EBs is detected morphologically (Fig. 1H). Typical EBs of appropriate size and with a condensed nucleoid structure are observed, as are intermediate developmental forms that exhibit a condensed nucleoid but have not yet compacted to the dimensions of EBs. Although EBs and intermediate developmental forms are detected, typical RBs, some apparently in the process of binary fission, are still observed. By 60 and 72 h postinfection, the inclusion consists of RBs and an increasing percentage of EBs that take on the characteristic pear shape of C. pneumoniae EBs (Fig. 1I and J). In addition, the cytoplasmic miniature bodies, typical only of C. pneumoniae EBs, become apparent. The intracellular growth of C. pneumoniae is completed with lysis of the host cell after 84 h postinfection to release progeny EBs for subsequent rounds of infection.
FIG. 1.
The C. pneumoniae AR-39 developmental cycle in HeLa 229 cells. Infected cells were processed for transmission electron microscopy at 0 (A), 2 (B), 8 (C), 12 (D), 19 (E), 24 (F), 36 (G), 48 (H), 60 (I), and 72 (J) h postinfection. Arrowheads indicate intracellular chlamydiae. Scale bars = 1 μm.
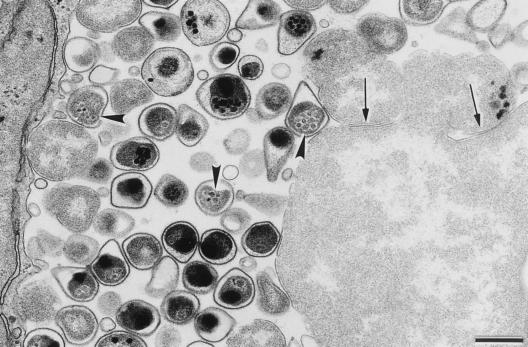
The pear shape of C. pneumoniae EBs has been proposed as a possible criterion for distinguishing C. pneumoniae EBs from EBs of other chlamydial species (6). However, the diagnostic value of the pear shape has been questioned (5, 23). Miyashita and colleagues have shown by scanning electron microscopy that some isolates, including AR-39, display the pear-like morphology but that other isolates, such as YK-41, do not (23). Thus, this particular morphology does not appear to be common to all strains of C. pneumoniae. We also observed the pear shape structure of C. pneumoniae AR-39 by both transmission and scanning electron microscopy using procedures that yield the typical spherical EB structure of C. trachomatis. Figure 2 illustrates the pear shape of C. pneumoniae EBs that appears to be largely due to a large periplasmic space and a relatively flexible and pleomorphic outer membrane. Miniature bodies, which are also considered characteristic of C. pneumoniae EBs, were also observed. However, these were observed only in the chlamydial cytoplasm and not in the periplasmic space, as shown by Chi et al. (6). Although chlamydiae typically are believed to multiply by binary fission, we occasionally observed aberrantly large RBs in the apparent process of producing additional RBs by budding.
FIG. 2.
Transmission electron micrograph of a C. pneumoniae inclusion at 72 h postinfection showing details of the mature EBs, including the pear shape as well as the presence of miniature bodies (arrowheads) within the EBs' cytoplasms. Arrows indicate sites suggestive of septation and possible generation of C. pneumoniae RBs by budding from a giant RB located in the lower right corner. Scale bar = 0.5 μm.
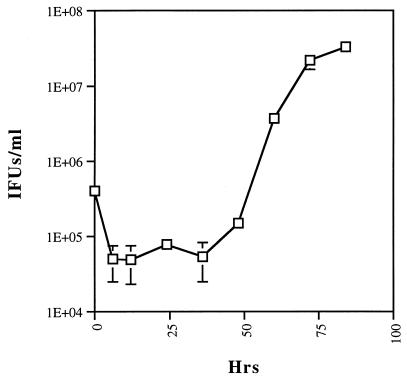
In agreement with the results of electron microscopy demonstrating the appearance of EBs by 48 h postinfection, a one-step growth curve demonstrated the production of infectious progeny EBs by 48 h and their subsequent increase in number and rapid accumulation (Fig. 3). Unlike C. trachomatis and C. psittaci EBs, which undergo a significant loss of infectivity within hours following infection, C. pneumoniae EBs display only about a log-unit decrease in recoverable infectious units. A likely explanation is that the relatively high multiplicity of infection necessary to obtain a uniformly infected culture leads to multiple EBs per cell, some of which do not differentiate into RBs or do so at prolonged intervals.
FIG. 3.
A one-step growth curve of C. pneumoniae AR-39 shows an initial 10-fold decrease in the number of infectious units and the maintenance of this level up to 36 h postinfection. The number of inclusion-forming units (IFUs) increases by 48 h postinfection, when the differentiation of RBs to EBs is initiated. Differentiation continues up to 84 h postinfection.
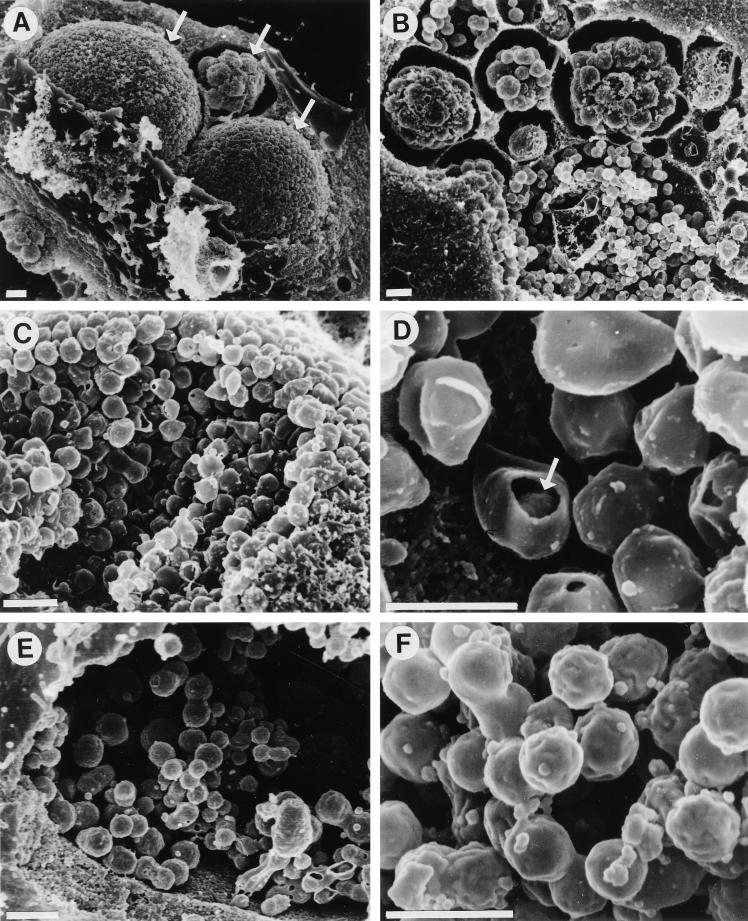
Scanning electron microscopy of C. pneumoniae AR-39-infected cells at 84 h postinfection revealed the shape of C. pneumoniae EBs as well as their distribution within the inclusion (Fig. 4), both of which are consistent with the results of transmission electron microscopy. To prepare the specimens for scanning electron microscopy, samples were fixed and dehydrated prior to critical-point drying in a Bal-Tec model cpd 030 drier (Balzers, Liechtenstein). The coverslips were then mounted on aluminum studs, and the cells were fractured by gently touching them with adhesive tape to remove their surfaces and to expose intracellular structures. Samples were then coated with 110 Å of chromium in a model IBS/TM200S ion beam sputterer (VCR Group, South San Francisco, Calif.) and viewed with a model S-4500 cold-field emission scanning electron microscope (Hitachi, Tokyo, Japan). C. pneumoniae inclusions are smaller in diameter than inclusions of C. trachomatis, but a large number of EBs with very little space between them could be seen (Fig. 4A and C). This compact structure of C. pneumoniae inclusions remained intact even after removal of the inclusion membrane. In addition, the scanning electron microscopy indicated that C. pneumoniae inclusions do not fuse, as occurs in cells multiply infected with C. trachomatis (2). The protracted commitment to development shown above likely contributes to the appearance of inclusions at different stages of development observed in multiply infected cells. It is of interest that in multiply infected cells, the majority of the inclusions appear to be delayed in maturation. Although asynchronous initiation of the developmental cycle is one possible explanation for multiple inclusions at different stages of development within the same cell, the preferential maturation of one or two suggests that competition for host-derived nutrients might be an alternative mechanism. The scanning electron microscopy also confirmed the presence of a large periplasmic space between the cytoplasmic and the outer membranes of C. pneumoniae EBs as well as their pear shape (Fig. 4D) and that the shape of C. trachomatis EBs remained round (Fig. 4E and F).
FIG. 4.
Scanning electron micrographs of C. pneumoniae AR-39 and C. trachomatis L2 inclusions in fractured host cells. (A and B) Presence of several C. pneumoniae inclusions within individual infected HeLa cells (arrows). Each of these inclusions appears to be at a different stage of maturity. Scale bars = 1 μm. The remaining panels emphasize the differences in shape between EBs of C. pneumoniae (C and D) and those of C. trachomatis (E and F) and their distribution inside the inclusion, as well as the different structures of inclusions between these two chlamydial species. Scanning electron microscopy confirmed the pear shape of C. pneumoniae EBs, and a break in an EB (arrow in panel D) reveals the presence of a large periplasmic space between the cytoplasmic and the outer membranes of the EBs. Scale bars = 0.5 μm.
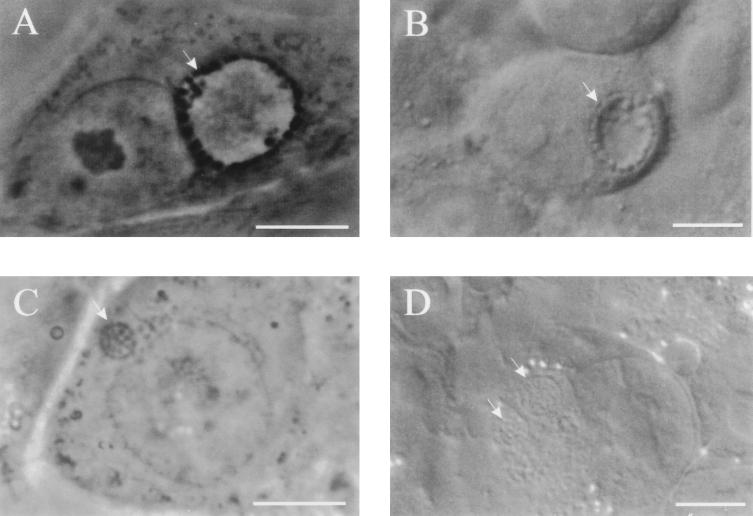
C. pneumoniae inclusions display a morphology and appearance distinct from those of C. trachomatis. Whereas C. trachomatis RBs are typically associated closely with and circumscribe the inner leaflet of the inclusion membrane (10), C. pneumoniae RBs appear to be tightly packed throughout the entire volume of the inclusion. Inclusions of C. pneumoniae viewed by phase-contrast and Nomarski differential-interference-contrast microscopy at 36 h postinfection and inclusions of C. trachomatis at 18 h postinfection demonstrate the differences in size, structure, and shape between inclusions of these two chlamydial species (Fig. 5). Both represent approximately the same stage of their development, since only dividing RBs are observed within. Because C. pneumoniae inclusions are packed with developmental forms and lack the phase-bright clear center typical of C. trachomatis inclusions, they are more difficult to visualize by standard light microscopy.
FIG. 5.
Phase- and Nomarski differential-interference-contrast images of C. trachomatis (A and B, respectively) and C. pneumoniae (C and D) inclusions. In comparison to inclusions of C. trachomatis obtained 18 h postinfection, C. pneumoniae inclusions at 36 h postinfection differ in shape and size, although both chlamydiae reach similar stages of their development at these time points postinfection, characterized by the presence of dividing RBs. This difference is most apparent in the presence of clear fluid-filled centers in C. trachomatis inclusions, since C. pneumoniae inclusions are filled with RBs. Scale bars = 10 μm.
The presence of persistent C. pneumoniae is believed to be a common sequel of acute respiratory infections with C. pneumoniae in humans (13), and persistent chlamydial infection is thought to play a role in the development of chronic chlamydial diseases. In many studies demonstrating an association of C. pneumoniae with heart disease, electron microscopy, PCR, and immunocytochemical staining are used individually or in combination to detect organisms in atherosclerotic lesions (reviewed in reference 3). There are, however, few examples of C. pneumoniae isolation by culture from atherosclerotic lesion homogenates of patients with severe coronary artery disease (16, 25). These observations suggest that C. pneumoniae may survive under certain circumstances in a condition in which infectious EBs are not produced. Events triggering such an alteration of chlamydial development may include activation of an immune response against chlamydiae. As described for C. trachomatis, the presence of gamma interferon in the culture medium leads to formation of abnormal RBs (1). However, there are a variety of other factors causing growth inhibition, including the use of β-lactam antibiotics and d-cycloserine and deviations in the levels of essential nutrients, that can trigger aberrant chlamydial development (7, 17, 21, 22). These aberrant RBs can recover to generate infectious EBs upon removal of the growth inhibitory factor.
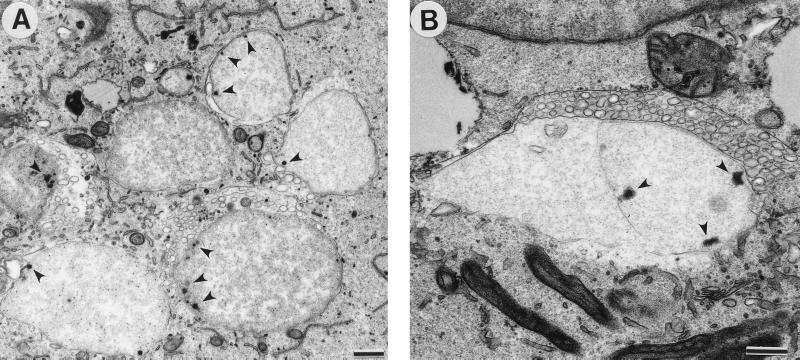
Because chlamydiae in a persistent state may not display typical morphology, we induced aberrant C. pneumoniae development by culture in the presence of ampicillin to visualize the morphology of C. pneumoniae in a nonproductive infection but one which maintains the chlamydiae in a viable state. C. pneumoniae-infected HeLa cells were incubated for 48 h in the presence of ampicillin (50 μg/ml) to induce the development of abnormal RBs. Abnormal RBs are much larger than normal RBs and undergo little or no cell division. Numerous vesicles of unknown origin and composition are present in the inclusions surrounding some giant RBs (Fig. 6).
FIG. 6.
Transmission electron micrograph of C. pneumoniae-infected cells treated with ampicillin. (A) Large, abnormal RBs displaying single-cell or no cell division and multiple electron-dense sites, possibly representing nucleoid condensation, near the chlamydial cell wall (arrowheads). (B) Vesicles of unknown function and origin surrounded some giant RBs. Scale bars = 0.5 μm.
The recovery of infectious EBs from C. pneumoniae and C. trachomatis abnormal RBs was assessed by blind passaging 24, 48, and 72 h after removal of ampicillin from the culture media. Neither C. trachomatis nor C. pneumoniae regenerated infectious EBs by 24 h after removal of ampicillin. However, 33.3% of the original C. trachomatis inoculum but only 0.13% of the original C. pneumoniae inoculum could be recovered by 48 h of incubation in the absence of the antibiotic. Eighty-six and seven-tenths percent of the C. trachomatis and 0.22% of the C. pneumoniae original inocula were obtained by 72 h after removal of ampicillin. Attempts to recover EBs after incubation for longer than 72 h after removal of ampicillin were unsuccessful due to death of the host cells. Ampicillin-induced C. pneumoniae aberrant RBs thus appear to recover less efficiently than C. trachomatis RBs upon removal of the antibiotic. This result may be due to a delayed ability to initiate the developmental cycle of C. pneumoniae abberant RBs following release from inhibition. Alternatively, this may be due to a higher eradication rate of C. pneumoniae abnormal RBs by host cells. The low numbers of EBs recovered after release from growth inhibition may contribute to the difficulty of cultivating C. pneumoniae organisms from patient samples.
In this work we have emphasized the unique attributes of the C. pneumoniae developmental cycle, which is otherwise similar to those of other chlamydial species. This information may help to overcome some difficulties of successful growth of C. pneumoniae in vitro and improve recognition of C. pneumoniae in vivo by documenting the distinctive morphologies of both normal inclusions and growth-inhibited aberrant developmental forms.
Acknowledgments
We thank H. Caldwell, G. McClarty, M. Scidmore, R. Carabeo, E. Shaw, and K. Fields for critical review of the manuscript.
REFERENCES
- 1.Beatty W L, Byrne G I, Morrison R P. Morphological and antigenic characterization of interferon-γ mediated persistent Chlamydia trachomatis infection in vitro. Proc Natl Acad Sci USA. 1993;90:3998–4002. doi: 10.1073/pnas.90.9.3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blyth W A, Taverne J. Some consequences of the multiple infection of cell cultures by TRIC organisms. J Hyg. 1972;70:33–37. doi: 10.1017/s0022172400022063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boman J, Gaydos C A, Quinn T C. Molecular diagnosis of Chlamydia pneumoniae infection. J Clin Microbiol. 1999;37:3791–3799. doi: 10.1128/jcm.37.12.3791-3799.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell L A, Kuo C-C, Grayston J T. Chlamydia pneumoniae and cardiovascular disease. Emerg Infect Dis. 1998;4:571–579. doi: 10.3201/eid0404.980407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carter M W, Al-Mahdawi S A H, Giles I G, Treharne J D, Ward M E, Clarke I N. Nucleotide sequence and taxonomic value of the major outer membrane protein gene of Chlamydia pneumoniae IOL-207. J Gen Microbiol. 1991;137:465–475. doi: 10.1099/00221287-137-3-465. [DOI] [PubMed] [Google Scholar]
- 6.Chi E Y, Kuo C-C, Grayston J T. Unique ultrastructure in the elementary body of Chlamydia sp. strain TWAR. J Bacteriol. 1987;169:3757–3763. doi: 10.1128/jb.169.8.3757-3763.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coles A M, Reynolds D J, Harper A, Devitt A, Pearce J H. Low-nutrient induction of abnormal chlamydial development: a novel component of chlamydial pathogenesis? FEMS Microbiol Lett. 1993;106:193–200. doi: 10.1111/j.1574-6968.1993.tb05958.x. [DOI] [PubMed] [Google Scholar]
- 8.Grayston J T, Kuo C-C, Campbell L A, Wang S-P. Chlamydia pneumoniae sp. nov. for Chlamydia sp. strain TWAR. Int J Syst Bacteriol. 1989;39:88–90. [Google Scholar]
- 9.Grayston J T. Infections caused by Chlamydia pneumoniae strain TWAR. Clin Infect Dis. 1992;15:757–763. doi: 10.1093/clind/15.5.757. [DOI] [PubMed] [Google Scholar]
- 10.Hackstadt T, Rockey D D, Heinzen R A, Scidmore M A. Chlamydia trachomatis interrupts an exocytic pathway to acquire endogenously synthesized sphingomyelin in transit from the Golgi apparatus to the plasma membrane. EMBO J. 1996;15:964–977. [PMC free article] [PubMed] [Google Scholar]
- 11.Hackstadt T, Fischer E R, Scidmore M A, Rockey D D, Heinzen R A. Origins and functions of the chlamydial inclusion. Trends Microbiol. 1997;5:288–293. doi: 10.1016/S0966-842X(97)01061-5. [DOI] [PubMed] [Google Scholar]
- 12.Hackstadt T, Scidmore-Carlson M, Shaw E, Fischer E. The Chlamydia trachomatis IncA protein is required for homotypic vesicle fusion. Cell Microbiol. 1999;1:119–130. doi: 10.1046/j.1462-5822.1999.00012.x. [DOI] [PubMed] [Google Scholar]
- 13.Hammerschlag R M, Chirgwin K, Roblin P M, Gelling M, Dumornay W, Mandel L, Smith P, Schachter J. Persistent infection with Chlamydia pneumoniae following acute respiratory illness. Clin Infect Dis. 1992;14:178–182. doi: 10.1093/clinids/14.1.178. [DOI] [PubMed] [Google Scholar]
- 14.Hodinka R L, Davis C H, Choong J, Wyrick P B. Ultrastructural study of endocytosis of Chlamydia trachomatis by McCoy cells. Infect Immun. 1988;56:1456–1463. doi: 10.1128/iai.56.6.1456-1463.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hodinka R L, Wyrick P B. Ultrastructural study of mode of entry of Chlamydia psittaci into L-929 cells. Infect Immun. 1986;54:855–863. doi: 10.1128/iai.54.3.855-863.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jackson L A, Campbell L A, Kuo C-C, Lee A, Grayston J T. Isolation of Chlamydia pneumoniae from a carotid endarterectomy specimen. J Infect Dis. 1997;176:292–295. doi: 10.1086/517270. [DOI] [PubMed] [Google Scholar]
- 17.Kramer M J, Gordon F B. Ultrastructural analysis of the effects of penicillin and chlortetracycline on the development of a genital tract Chlamydia. Infect Immun. 1971;3:333–341. doi: 10.1128/iai.3.2.333-341.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuo C-C, Jackson L A, Campbell L A, Grayston J T. Chlamydia pneumoniae. Clin Microbiol Rev. 1995;8:451–461. doi: 10.1128/cmr.8.4.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuo C-C, Grayston J T. Factors affecting viability and growth in HeLa 229 cells of Chlamydia sp. strain TWAR. J Clin Microbiol. 1988;26:812–815. doi: 10.1128/jcm.26.5.812-815.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuo C-C, Chi E Y, Grayston J T. Ultrastructural study of entry of Chlamydia strain TWAR into HeLa cells. Infect Immun. 1988;56:1668–1672. doi: 10.1128/iai.56.6.1668-1672.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin H-S, Moulder J W. Patterns of response to sulfadiazine, d-cycloserine, and d-alanine in the members in the psittacosis group. J Infect Dis. 1966;116:372–376. doi: 10.1093/infdis/116.3.372. [DOI] [PubMed] [Google Scholar]
- 22.Matsumoto A, Manire G P. Electron microscopic observations on the effects of penicillin on the morphology of C. psittaci. J Bacteriol. 1970;101:278–285. doi: 10.1128/jb.101.1.278-285.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miyashita N, Kamanoto Y, Matsumoto A. The morphology of Chlamydia pneumoniae. J Med Microbiol. 1993;38:418–425. doi: 10.1099/00222615-38-6-418. [DOI] [PubMed] [Google Scholar]
- 24.Popov V L, Shatkin A A, Pankratova V N, Smirnova N S, von Bonsdorff C-H, Ekman M-R, Morttinen A, Saikku P. Ultrastructure of Chlamydia pneumoniae in cell culture. FEMS Microbiol Lett. 1991;84:129–134. doi: 10.1016/0378-1097(91)90115-q. [DOI] [PubMed] [Google Scholar]
- 25.Ramirez J, Ahkee A, Ganzel B L, Ogden L L, Gaydos C A, Quinn T C, et al. Isolation of Chlamydia pneumoniae (C pn) from the coronary artery of a patient with coronary atherosclerosis. Ann Intern Med. 1996;125:979–982. doi: 10.7326/0003-4819-125-12-199612150-00008. [DOI] [PubMed] [Google Scholar]