Abstract
Background
Today it is possible to profile the transcriptome of individual cells, and a key step in the analysis of these datasets is unsupervised clustering. For very large datasets, efficient algorithms are required to ensure that analyses can be conducted with reasonable time and memory requirements.
Results
Here, we present a highly efficient k-means based approach, and we demonstrate that it scales favorably with the number of cells with regards to time and memory.
Conclusions
We have demonstrated that our streaming k-means clustering algorithm gives state-of-the-art performance while resource requirements scale favorably for up to 2 million cells.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12859-022-05085-z.
Keywords: k-Means clustering, Streaming clustering, scRNAseq
Background
Technological advances have paved the way for single cell RNAseq (scRNAseq) datasets containing several million cells [1]. Such large datasets require highly efficient algorithms to enable analyses at reasonable times and hardware requirements [2]. A crucial step in single cell workflows is unsupervised clustering, which aims to delineate putative cell types or cell states based on transcriptional similarity [3]. The most popular methods for unsupervised clustering of scRNAseq data are the Louvain and Leiden algorithms. They represent cells as a neighborhood graph where densely connected modules are identified as clusters [4]. However, these methods can be biased by a poorly specified graph, running the risk of identifying structures that are not present in the data [5]. More generally, as it can be shown that no single clustering algorithm will feature all desired statistical properties and perform well for all datasets, the field would benefit from additional methodologies [6].
One of the most widely used unsupervised clustering in general is k-means clustering, and it forms the basis of several methodologies, including scCCESS [7], SCCAF [8] and the single cell consensus clustering (SC3) algorithm [9]. To achieve robust and accurate results SC3 uses a consensus approach whereby a large number of parameter combinations are evaluated and subsequently combined. However, both the k-means clustering and the consensus algorithm come at significant computational costs: both the run time and memory use scale more than quadratically with the number of cells, prohibiting application to large datasets, which are becoming increasingly commonplace with ever improving sequencing technologies.
Implementation
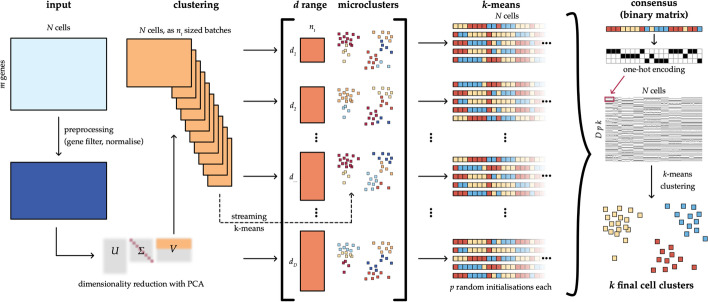
Here, we present a new version of this algorithm, single cell consensus clustering with speed (SC3s), where several steps of the original workflow have been optimized to ensure that both run time and memory usage scale linearly with the number of cells (Fig. 1; Additional file 1: Fig. S1). This is achieved by using a streaming approach for the k-means clustering [10], as implemented in the scikit-learn package [11], which makes it possible to only process a small subset of cells in each iteration. Each of the subsets can be efficiently processed at constant time and memory. In addition, as part of an intermediary step, which was not part of the original method, a large number of microclusters are calculated. The microclusters can be reused for different choices of k, and this allows substantial savings when analyzing multiple values of k, something that is very common in practice during data exploration. We have also improved the consensus step by adopting a one-hot encoding approach [12], as opposed to the original co-association based method, on which the k-means clustering algorithm could be run more efficiently (Additional file 1: Fig. S2).
Fig. 1.
The SC3s framework for single cell consensus clustering. SC3s takes as input the gene-by-cell expression matrix, after preprocessing and dimensionality reduction via PCA using Scanpy commands. To achieve consensus clustering, SC3s attempts to combine the results of multiple clustering runs, where the number of principal components is changed (d range). All this information is then encoded into a binary matrix, which can be efficiently used to produce the final k cell clusters. The key difference from the original SC3 is that for each d, the cells are first grouped into microclusters which can be reused for multiple values of k, saving time in computation
Results
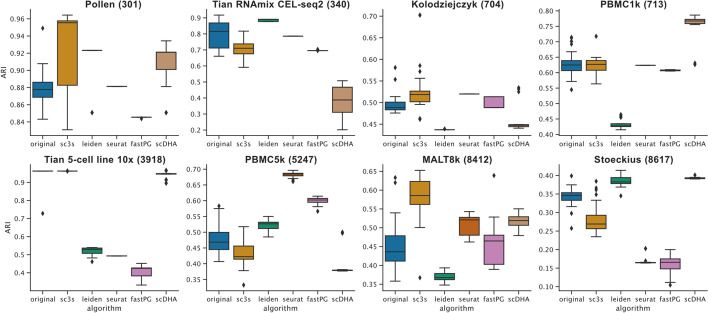
To evaluate the accuracy of SC3s we used eight datasets with < 10,000 cells where the cell labels are known or have been defined using orthogonal methods, allowing us to compare the results of the transcriptome clustering to a ground truth [9] (Additional file 1: Table S1). These benchmarks show that SC3s has an accuracy which is comparable to the original algorithm (Fig. 2), and that the performance is robust across a broad range of user-customisable parameters (Additional file 1: Figs. S3-S5). Finally, SC3s compares favorably against other clustering methodologies, such as Scanpy, Seurat, FastPG and scDHA, in terms of its accuracy, memory usage and runtime (Fig. 2; Additional file 1: Figs. S1, S6).
Fig. 2.
Clustering accuracy benchmarks on gold-standard datasets with < 10,000 cells. Boxplots show the ARI distribution across 25 realizations of each algorithm. Numbers in parentheses denote the cell count in the specified dataset. The performance of the original SC3 is shown in blue. Leiden refers to the algorithm of the same name as implemented in Scanpy. Seurat refers to its SNN modularity optimization clustering algorithm. ARI: Adjusted Rand index (ARI)
To examine the performance for large datasets, SC3s was benchmarked on the mouse organogenesis cell atlas dataset which contains 2,026,641 cells [1]. Processing, filtering and dimensionality reduction were performed as in the original publication, after which the clustering performance of SC3s was assessed. Compared to the other packages, SC3s was able to achieve both a short runtime and a low memory usage, whilst producing consistent clusters. For example, when compared to the Leiden algorithm, the peak memory usage was similar, but SC3s was ~ 18 times faster (20 min vs 6 h), even when evaluating five k values (Table 1). The slightly lower accuracy was expected because cell labels used for comparison originated from the Louvain algorithm, a method very similar to the Leiden algorithm, making them an imperfect ground truth. Visual inspection of the assigned labels also revealed that SC3s was able to capture the major structures identified by the authors (Additional file 1: Fig. S7).
Table 1.
Runtime, memory and ARI performance benchmarked on the 2 million mouse organogenesis cell atlas dataset
| Method | Runtime (hr:min:s) | Peak memory (GB) | ARI |
|---|---|---|---|
| SC3s (one k) | 00:11:12 | 22.65 | 0.408 |
| SC3s (one k) | 00:21:01 | 26.45 | 0.400 |
| Scanpy Leiden | 05:27:17 | 33.83 | 0.536 |
| SEURAT | 01:53:54 | 91.36 | 0.257 |
| FastPG | 00:15:05 | 72.75 | 0.463 |
Results are the average of five iterations
Conclusions
Overall, SC3s is a major improvement over its predecessor, and it represents a scalable and accurate alternative to the widely used neighborhood graph clustering methodologies. Moreover, it is integrated with the popular Scanpy package and utilizes the same underlying data structures [13], making it easy for users to incorporate into existing workflows and to make full use of upstream and downstream functionalities in the ecosystem. Thus, SC3s will allow researchers to analyze scRNAseq datasets as they scale to millions of cells.
Availability and requirements
Project name: SC3s. Project home page: https://github.com/hemberg-lab/sc3s/. Operating system: Platform independent. Programming language: python. License: BSD-3. Other requirements: None. Restrictions to use by non-academics: None.
Supplementary Information
Additional file 1. Contains Fig S1-S7 which provides more details about SC3s performance, and Table S1 which details the datasets used for benchmarking.
Acknowledgements
We would like to thank the Cellular Genetics Informatics team at the Wellcome Trust Sanger Institute for providing compute resources, particularly Simon Murray for helping package SC3s.
Abbreviations
- scRNAseq
Single cell RNAseq
- SC3
Single cell consensus clustering
- SC3s
Single cell consensus clustering with speed
Author contributions
The project was conceived by FXQ and MH. FXQ wrote the code and analyzed the data. MH supervised the research. FXQ and MH wrote the manuscript. All authors read and approved the final manuscript.
Funding
FXQ was supported by a Wellcome Trust PhD studentship. MH was funded by a core grant from the Wellcome Trust. The funder did not play any role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
All datasets used for benchmarking are available publically, and they are listed in Additional file 1: Table S1. The Python code for SC3s is licensed under a BSD-3 Clause License. Instructions to install from pip and conda channels are available on GitHub: https://github.com/hemberg-lab/sc3s.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
None.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Cao J, Spielmann M, Qiu X, Huang X, Ibrahim DM, Hill AJ, et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 2019;566:496–502. doi: 10.1038/s41586-019-0969-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Melsted P, Booeshaghi AS, Liu L, Gao F, Lu L, Min KHJ, et al. Modular, efficient and constant-memory single-cell RNA-seq preprocessing. Nat Biotechnol. 2021 doi: 10.1038/s41587-021-00870-2. [DOI] [PubMed] [Google Scholar]
- 3.Kiselev VY, Andrews TS, Hemberg M. Challenges in unsupervised clustering of single-cell RNA-seq data. Nat Rev Genet. 2019;20:273–282. doi: 10.1038/s41576-018-0088-9. [DOI] [PubMed] [Google Scholar]
- 4.Traag VA, Waltman L, van Eck NJ. From Louvain to Leiden: guaranteeing well-connected communities. Sci Rep. 2019;9:5233. doi: 10.1038/s41598-019-41695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pasta MQ, Zaidi F. Topology of complex networks and performance limitations of community detection algorithms. IEEE Access. 2017;5:10901–10914. doi: 10.1109/ACCESS.2017.2714018. [DOI] [Google Scholar]
- 6.Sun S, Zhu J, Ma Y, Zhou X. Accuracy, robustness and scalability of dimensionality reduction methods for single-cell RNA-seq analysis. Genome Biol. 2019;20:269. doi: 10.1186/s13059-019-1898-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geddes TA, Kim T, Nan L, Burchfield JG, Yang JYH, Tao D, et al. Autoencoder-based cluster ensembles for single-cell RNA-seq data analysis. BMC Bioinform. 2019;20(Suppl 19):660. doi: 10.1186/s12859-019-3179-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miao Z, Moreno P, Huang N, Papatheodorou I, Brazma A, Teichmann SA. Putative cell type discovery from single-cell gene expression data. Nat Methods. 2020;17:621–628. doi: 10.1038/s41592-020-0825-9. [DOI] [PubMed] [Google Scholar]
- 9.Kiselev VY, Kirschner K, Schaub MT, Andrews T, Yiu A, Chandra T, et al. SC3: consensus clustering of single-cell RNA-seq data. Nat Methods. 2017;14:483–486. doi: 10.1038/nmeth.4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sculley D. Web-scale k-means clustering. In: Proceedings of the 19th international conference on World wide web. New York: Association for Computing Machinery; 2010. p. 1177–8.
- 11.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, et al. Scikit-learn: machine learning in python. J Mach Learn Res. 2011;12:2825–2830. [Google Scholar]
- 12.Liu H, Liu T, Wu J, Tao D, Fu Y. Spectral ensemble clustering. In: Proceedings of the 21th ACM SIGKDD international conference on knowledge discovery and data mining. ACM; 2015. p. 715–24.
- 13.Wolf FA, Angerer P, Theis FJ. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19:15. doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Contains Fig S1-S7 which provides more details about SC3s performance, and Table S1 which details the datasets used for benchmarking.
Data Availability Statement
All datasets used for benchmarking are available publically, and they are listed in Additional file 1: Table S1. The Python code for SC3s is licensed under a BSD-3 Clause License. Instructions to install from pip and conda channels are available on GitHub: https://github.com/hemberg-lab/sc3s.