Abstract
Background
The conversion of lignin-derived aromatic monomers into valuable chemicals has promising potential to improve the economic competitiveness of biomass biorefineries. Pinosylvin is an attractive pharmaceutical with multiple promising biological activities.
Results
Herein, Escherichia coli was engineered to convert the lignin-derived standard model monomer cinnamic acid into pinosylvin by introducing two novel enzymes from the wood plant: stilbene synthase from Pinus pinea (PpSTS) and 4-Coumarate-CoA ligase from Populus trichocarpa (Ptr4CL4). The expression of Ptr4CL4 drastically improved the production of pinosylvin (42.5 ± 1.1 mg/L), achieving values 15.7-fold higher than that of Ptr4CL5 (another 4-Coumarate-CoA ligase from Populus trichocarpa) in the absence of cerulenin. By adjusting the expression strategy, the optimized engineered strain produced pinosylvin at 153.7 ± 2.2 mg/L with an extremely high yield of 1.20 ± 0.02 mg/mg cinnamic acid in the presence of cerulenin, which is 83.9% ± 1.17 of the theoretical yield. This is the highest reported pinosylvin yield directly from cinnamic acid to date.
Conclusion
Our work highlights the feasibility of microbial production of pinosylvin from cinnamic acid and paves the way for converting lignin-related aromatics to valuable chemicals.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13068-022-02236-5.
Keywords: Pinosylvin, Stilbene synthase, 4-Coumarate-CoA ligase, Trans-cinnamic acid, Biosynthesis
Background
As the lignocellulose volume is the largest renewable resource on earth, its biorefinery has attracted extensive attention for its sustainability [1]. Amongst the three components of lignocellulose, its carbohydrate contents (cellulose and hemicellulose) could be used for biofuel, biochemical and biomaterial production. However, lignin, accounting for 15–40%, is still difficult to degrade and utilize [2, 3]. This greatly limits the economic competitiveness of biomass biorefineries. Exploring lignin biorefineries for aromatic chemical production has promising potential to improve the lignin economic competitiveness of biomass biorefineries. According to the previous studies, many aromatic monomers from lignin can be efficiently released by pretreatment and thermochemical depolymerization [4]. These compounds mainly include various phenolic acids, such as ferulic acid, p-coumaric acid and cinnamic acid. Thus, converting these accessible lignin-related phenolic acids to value-added products contributes to realizing the value-added utilization of lignin waste. Our findings open up possibilities for the practical biosynthesis of natural pinosylvin from lignin-derived standard model monomer cinnamic acid at industrial scale.
Pinosylvin (trans-3,5-dihydroxystilbene) is an important stilbene compound that is mostly present in the heartwood of coniferous trees. Its natural function mainly protects plants against microbial and fungal decay [5]. Recent studies have shown that it has multiple promising biological activities, including anticancer, anti-cardiovascular, antioxidation, anti-inflammatory and antibacterial abilities [6–9]. Pinosylvin can be found in plant tissues, pine leaves and fruits (Pinus densiflora) [10–12]. However, several shortcomings have increased its economic and labour costs for market demand, including a low concentration (1–40 mg/g pine wood), seasonal and regional variations and the existence of structural analogues [13, 14]. In addition, from an industrial perspective, large-scale plant extraction would lead to a decrease in the vegetation coverage and damage to the ecological environment, which is contrary to the goal of sustainable development [15]. Hence, constructing engineered cells by synthetic biology and metabolic engineering technologies is desired and has been demonstrated to be a more environmentally friendly and cost-efficient platform for pinosylvin production.
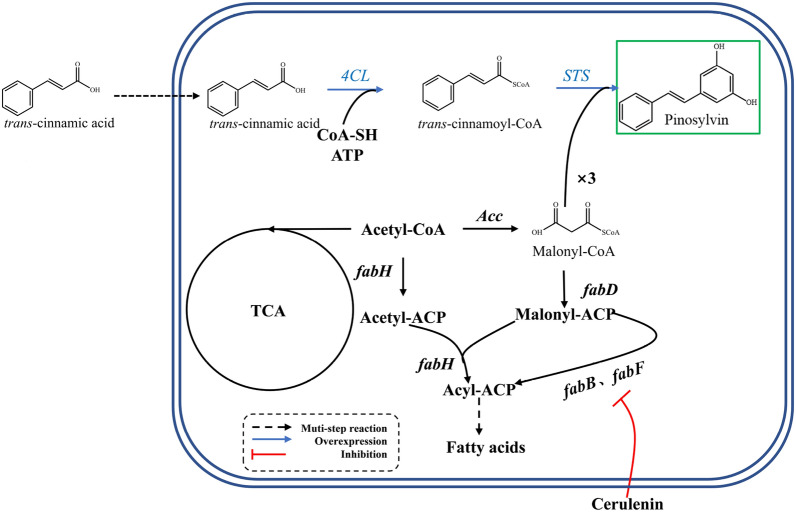
The basic skeleton of pinosylvin is composed of a B ring from a trans-cinnamoyl coenzyme A (trans-cinnamoyl-CoA) and an A ring formed by the cyclization of three malonyl coenzyme A (malonyl-CoA) molecules. Phenylalanine ammonia lyase (PAL; EC 4.3.1.24) converts l-phenylalanine synthesized by the shikimate pathway to trans-cinnamic acid. Trans-cinnamic acid is CoA-activated by a 4-coumarate-CoA ligase (4CL; EC 6.2.1.12), yielding trans-cinnamoyl-CoA. Subsequently, pinosylvin is specifically biosynthesized by stilbene synthase (STS; EC 2.3.1.146), catalysing the condensation of three malonyl-CoA molecules with trans-cinnamoyl-CoA (Fig. 1) [12]. In plants, pinosylvin is converted from trans-cinnamic acid [6, 16], a class of natural plant intermediates from the lignin metabolism pathway [17, 18]. 4CL and STS are involved in the biosynthetic pathway of pinosylvin from trans-cinnamic acid. To date, according to the plant pathway, the production of pinosylvin using engineered microorganisms has been studied [19, 20]. The supply of malonyl-CoA has been found to be critical for pinosylvin production [21, 22]. Improving the level of intracellular malonyl-CoA had a remarkable positive effect on the production of pinosylvin [23]. However, although a long and complex artificial pathway of pinosylvin production from glucose was constructed and optimized, the product titre and yield were still not satisfactory. Assembling the full pathway from glucose to pinosylvin resulted in a yield of only 56.2 mg/g glucose even through rational modular design of the metabolic network [24]. When the substrate was replaced by trans-cinnamic acid, the maximum yield of pinosylvin reached 0.64 mg/mg trans-cinnamic acid [19]. Therefore, improving the yield is of great interest for pinosylvin production.
Fig. 1.
Pinosylvin biosynthetic pathway and malonyl-CoA metabolic networks. 4CL, encoding 4-coumarate-CoA ligase; STS, encoding stilbene synthase; Acc, encoding acetyl-CoA carboxylase; fabD, encoding malonyl-CoA: ACP transacylase; fabB, encoding β-ketoacyl-ACP synthase I; fabF, encoding β-ketoacyl-ACP synthase II; fabH, encoding β-ketoacyl-ACP synthase III. The blue arrows and the red lines represent overexpression and inhibition of genes or proteins. The dotted lines indicate multistep reaction
The aim of this work was to engineer E. coli for the production of pinosylvin from trans-cinnamic acid with both high titre and high yield. To achieve this, different STSs from Pinus pinea and 4CLs from Populus trichocarpa were screened, and the most suitable combination was identified by comparing the production of pinosylvin. Based on the results, the mechanism by which the enzyme 4CL regulates the pathway was explored. Thereafter, the metabolic engineering pathway was optimized by genetic manipulation. Finally, the culture conditions for pinosylvin production in engineered E. coli were optimized. This work thus demonstrated highly efficient pinosylvin production from cinnamic acid, with the highest yield to date. The yield was 1.20 ± 0.02 mg/mg trans-cinnamic acid, corresponding to 83.9% ± 1.17 of the theoretical yield.
Results and discussion
Genomic mining of stilbene synthases from Pinus for pinosylvin production
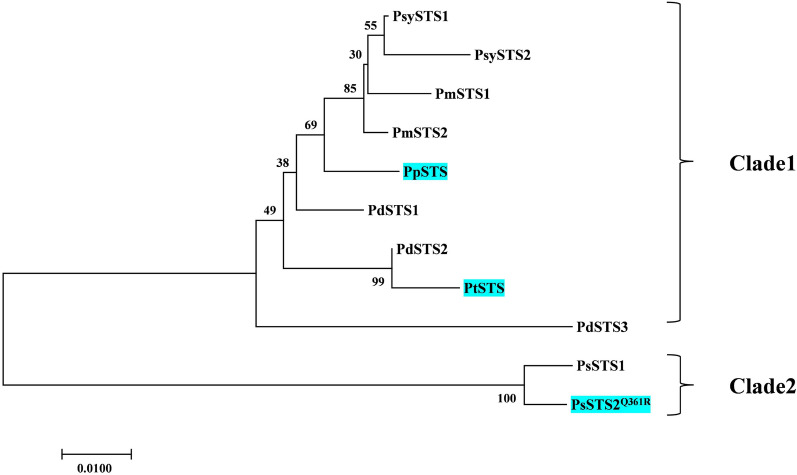
According to the biosynthetic pathway of pinosylvin in plants, both 4CL and STS are necessary for pinosylvin production from trans-cinnamic acid. Previous studies suggested that the biosynthesis of pinosylvin from trans-cinnamoyl-CoA, which is in the charge of STS, is the limiting step [25, 26]. Currently, many STSs have been characterized from plants, and most of them have been applied in the formation of resveratrol [27, 28]. However, only AhSTS (STS from Arachis hypogaea), VvSTS (STS from Vitis vinifera) and PsSTS (STS from Pinus strobus) were successfully expressed in E. coli for pinosylvin production [12, 24, 29, 30]. Pinosylvin is a stilbene that can be predominantly found in the heartwood of coniferous trees of the genus Pinus. To mine more STSs for use, 11 STS genes from six Pinus species were screened from NCBI in this study (Additional file 1: Table S1). Amongst these proteins, PsySTS1, PmSTS, PdSTS and PsSTS2 have been reported to display catalytic activities for trans-cinnamoyl-CoA [15, 16, 27, 31]. As the phylogenetic tree showed (Fig. 2), all these STSs were grouped into two clusters. PsSTS1 and PsSTS2Q361R from Pinus strobus shared the same clade. PsSTS2 was confirmed to have good activity towards trans-cinnamoyl-CoA, and its single mutant PsSTS2Q361R displayed higher activity towards trans-cinnamoyl-CoA, which would increase pinosylvin production [15, 16]. Meanwhile, PpSTS was not clearly identified in a previous study. However, it demonstrated high protein identity (> 98%) with PsySTS1, PmSTS1-2 and PdSTS1, which indicated that it might be responsible for the conversion of trans-cinnamoyl-CoA to pinosylvin. In addition, PtSTS shares a small clade with PdSTS2 which has the ability to convert trans-cinnamoyl-CoA to pinosylvin [31]. For the above reasons, we selected PsSTS2Q361R, PpSTS and PtSTS as candidates for pinosylvin synthases for further study.
Fig. 2.
Stilbene synthase sequence analysis. Phylogenetic tree constructed for the selected STS sequences from various pines based on the neighbour-joining method. All STSs used were as follows: PsySTS, STS from Pinus sylvestris; PmSTS, STS from Pinus massoniana; PpSTS, STS from Pinus pinea; PdSTS, STS from Pinus densiflora; PtSTS, STS from Pinus thunbergii; PsSTS, STS from Pinus strobus. The accession numbers of the proteins are listed in Additional file 1: Table S1
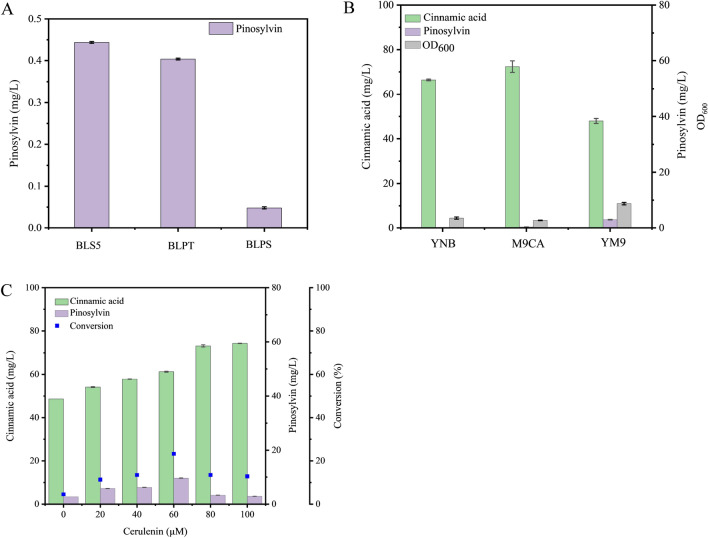
To assess the performance of the three candidate enzymes, three engineered strains, BLS5 (BL21(DE3) harbouring genes PpSTS and Ptr4CL5), BLPT (BL21 (DE3) harbouring genes PtSTS and Ptr4CL5) and BLPS (BL21 (DE3) harbouring genes PsSTSQ361R and Ptr4CL5), were constructed for pinosylvin production (Table 1). Therein, Ptr4CL5 was used for converting trans-cinnamic acid to trans-cinnamoyl-CoA, which displayed a high activity towards trans-cinnamic acid in our previous report [32]. Three strains were cultivated in M9CA medium using trans-cinnamic acid as substrates. As shown in Fig. 3A, all of them showed a certain capacity for pinosylvin production, which confirmed that Ptr4CL5 was indeed active to trans-cinnamic acid. As control, BL21(DE3) with plasmid pRSFDuet-1 did not synthesize pinosylvin. By comparison, BLS5 demonstrated the highest ability to synthesize pinosylvin, obtaining 0.44 ± 0.03 mg/L pinosylvin. Therefore, PpSTS was considered to be the best for pinosylvin production. Thereafter, we speculated that the M9CA medium (which uses glucose as a carbon source) did not provide enough nutrients for growth, which led to the trace amount of pinosylvin. Thus, YNB (which uses glucose as a carbon source) and YM9 (which uses glycerol as a carbon source and yeast extract was added) media were used (Fig. 3B). When YM9 medium was used, along with the improved growth, the concentration of product increased to 2.98 ± 0.08 mg/L, which was 571% ± 0.16 higher than that achieved with M9CA. Meanwhile, BLS5 cultivated in YM9 consumed the most trans-cinnamic acid. Nevertheless, only 46.1% of trans-cinnamic acid was consumed in YM9 medium, and the yield was obviously unsatisfactory. It was considered that the low conversion may be due to an insufficient supply of malonyl-CoA. Malonyl-CoA was reported to be a key precursor for preparing stilbene and flavone, including pinosylvin [20]. As illustrated in Fig. 1, decreasing malonyl-CoA involved in the fatty acid synthesis pathway would be an effective method for malonyl-CoA supply [33]. Owing to the inhibitory effect of cerulenin on fatty acid synthesis [34, 35], cerulenin was added to increase the intracellular level of malonyl-CoA. As expected, pinosylvin reached 9.61 ± 0.06 mg/L, which increased by 260% ± 0.45 in the presence of 60 μM cerulenin (Fig. 3C). However, the inhibition of fatty acid synthesis resulted in the poor growth of BLS5 (Additional file 1: Fig. S1), which might influence the total expression amounts of proteins in E. coli. Hence, even if cerulenin addition enhanced the supplementation of precursor malonyl-CoA, the low yield still suggested that exploring other 4CLs might be more beneficial for pinosylvin production.
Table 1.
Strains and plasmids used in this study
| Characteristics | Source | |
|---|---|---|
| Strains | ||
| E. coli BL21 (DE3) | F− ompT hsdSB (rB− mB−) gal (λ cI857 ind1 sam7 nin5 lacI lacUV5-T7 gene1), dcm (DE3) | Transgen |
| E. coli Trans1-T1 | F− ϕ80 (lacZ) ΔM15 ΔlacX 74 hsdR(rk−mk−) ΔrecA 1398endA1tonA | Transgen |
| E. coli BL4 | BL21(DE3) carrying pE-4CL4 | Laboratory |
| E. coli BL5 | BL21(DE3) carrying pE-4CL5 | Laboratory |
| E. coli BLS5 | BL21(DE3) carrying pR-PpSTS-Ptr4CL5 | This study |
| E. coli BLPT | BL21(DE3) carrying pR-PtSTS-Ptr4CL5 | This study |
| E. coli BLPS | BL21(DE3) carrying pR-PsSTS2Q361R-Ptr4CL5 | This study |
| E. coli BR4S | BL21(DE3) carrying pR-Ptr4CL4-PpSTS | This study |
| E. coli BRS4 | BL21(DE3) carrying pR-PpSTS-Ptr4CL4 | This study |
| E. coli BE4S | BL21(DE3) carrying pE-Ptr4CL4-PpSTS | This study |
| E. coli BES4 | BL21(DE3) carrying pE-PpSTS-Ptr4CL4 | This study |
| E. coli BC4S | BL21(DE3) carrying pC-Ptr4CL4-PpSTS | This study |
| E. coli BCS4 | BL21(DE3) carrying pC-PpSTS-Ptr4CL4 | This study |
| E. coli BRT4S | BL21(DE3) carrying pRT-Ptr4CL4-PpSTS | This study |
| E. coli BRTS4 | BL21(DE3) carrying pRT-PpSTS-Ptr4CL4 | This study |
| E. coli BET4S | BL21(DE3) carrying pET-Ptr4CL4-PpSTS | This study |
| E. coli BETS4 | BL21(DE3) carrying pET-PpSTS-Ptr4CL4 | This study |
| E. coli BCT4S | BL21(DE3) carrying pCT-Ptr4CL4-PpSTS | This study |
| E. coli BCTS4 | BL21(DE3) carrying pCT-PpSTS-Ptr4CL4 | This study |
| Plasmids | ||
| pRSFDuet-1 | RSF ori with PT7; Kanr | Novagen |
| pCDFDuet-1 | CDF ori with PT7; Smr | Novagen |
| pETDuet-1 | PBR322 ori with PT7; Ampr | Novagen |
| pE-4CL4 | pETDuet-1 with Ptr4CL4 | Laboratory |
| pE-4CL5 | pETDuet-1 with Ptr4CL5 | Laboratory |
| pR-PpSTS | pRSFDuet-1 with PpSTS | Generay Biotech |
| pR-PtSTS | pRSFDuet-1 with PtSTS | Generay Biotech |
| pR-PsSTS2Q361R | pRSFDuet-1 with PsSTS2Q361R | Generay Biotech |
| pR-PpSTS-Ptr4CL5 | pRSFDuet-1 with PT7-PpSTS and PT7-Ptr4CL5 | This study |
| pR-PtSTS-Ptr4CL5 | pRSFDuet-1 with PT7-PtSTS and PT7-Ptr4CL5 | This study |
| pR-PsSTS2Q361R-Ptr4CL5 | pRSFDuet-1 with PT7-PsSTS2Q361R and PT7-Ptr4CL5 | This study |
| pR-PpSTS-Ptr4CL4 | pRSFDuet-1 with PT7-PpSTS and PT7-Ptr4CL4 | This study |
| pR-Ptr4CL4-PpSTS | pRSFDuet-1 with PT7-Ptr4CL4 and PT7-PpSTS | This study |
| pE-PpSTS-Ptr4CL4 | pETDuet-1 with PT7-PpSTS and PT7-Ptr4CL4 | This study |
| pE-Ptr4CL4-PpSTS | pETDuet-1 with PT7-Ptr4CL4 and PT7-PpSTS | This study |
| pC-PpSTS-Ptr4CL4 | pCDFDuet-1 with PT7-PpSTS and PT7-Ptr4CL4 | This study |
| pC-Ptr4CL4-PpSTS | pCDFDuet-1 with PT7-Ptr4CL4 and PT7-PpSTS | This study |
| pRT-PpSTS-Ptr4CL4 | pRSFDuet-1 with PT7-PpSTS-Ptr4CL4 | This study |
| pRT-Ptr4CL4-PpSTS | pRSFDuet-1 with PT7 -Ptr4CL4-PpSTS | This study |
| pET-PpSTS-Ptr4CL4 | pETDuet-1 with PT7-PpSTS-Ptr4CL4 | This study |
| pET-Ptr4CL4-PpSTS | pETDuet-1 with PT7 -Ptr4CL4-PpSTS | This study |
| pCT-PpSTS-Ptr4CL4 | pCDFDuet-1 with PT7-PpSTS-Ptr4CL4 | This study |
| pCT-Ptr4CL4-PpSTS | pCDFDuet-1 with PT7-Ptr4CL4-PpSTS | This study |
Fig. 3.
For comparison of recombinant E. coli metabolites, 90 mg/L trans-cinnamic acid was supplied for pinosylvin production. A Comparison of pinosylvin produced by different recombinant E. coli BLPS, BLPT and BLS5 in M9CA medium. B Pinosylvin production, remaining trans-cinnamic acid and OD600 of E. coli BLS5 cultivated in different media for 48 h. C Pinosylvin production and remaining trans-cinnamic acid of E. coli BLS5 under different concentrations of cerulenin in YM9 medium for 48 h
Improving pinosylvin conversion from trans-cinnamic acid by altering 4-coumarate-CoA ligase
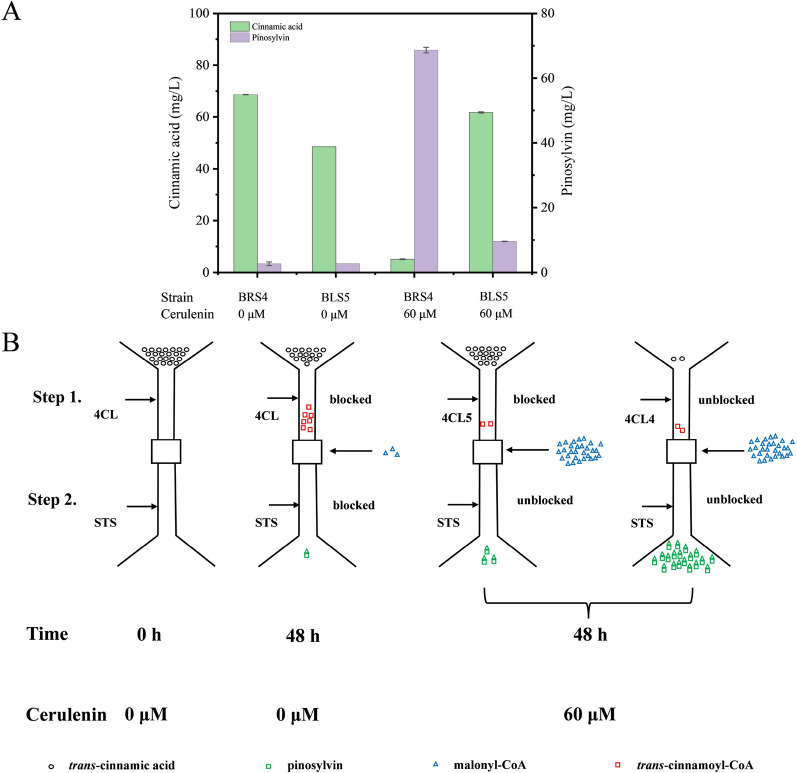
4CL is the main branch point enzyme that generates activated thioesters. Many plants have a large number of 4CLs, which connect phenylpropanoids to different product pathways. In our previous work, another 4CL from Populus trichocarpa (Ptr4CL4) was also found to be active to trans-cinnamic acid. Although it has lower activity towards trans-cinnamic acid, its primary function is confirmed to channel-activated 4-coumarate to different branch pathways of flavonoid synthesis, and its regulation is more complicated than that of Ptr4CL5 [32]. Thus, it was considered to be introduced into the pinosylvin biopathway by substituting for Ptr4CL5. As shown in Fig. 4A, E. coli BRS4 (harbouring Ptr4CL4 and PpSTS) produced 2.69 ± 0.58 mg/L pinosylvin, which did not bring an obvious change for pinosylvin production without cerulenin (p > 0.05). Moreover, the low activity of Ptr4CL4 led to more trans-cinnamic acid accumulation. However, unexpectedly, cerulenin addition resulted in a remarkable difference between Ptr4CL4 and Ptr4CL5. For Ptr4CL5, cerulenin addition resulted in less trans-cinnamic acid consumption, which might be due to the decreasing OD600 and lower protein expression level (Additional file 1: Fig. S1). However, the combination of Ptr4CL4 and PpSTS achieved the highest consumption of trans-cinnamic acid in the presence of cerulenin, with only 5.13 ± 0.50 mg/L left after fermentation of 48 h. Meanwhile, the concentration of pinosylvin for E. coli BRS4 increased dramatically to 68.64 ± 0.87 mg/L, which was approximately seven times that of E. coli BLS5.
Fig. 4.
Effects of different 4CLs on pinosylvin production. A Remaining trans-cinnamic acid and pinosylvin produced by different E. coli strains BLS5 and BRS4 under 0 and 60 μM cerulenin. The strains were cultivated at 30 °C for 48 h in YM9 medium and 90 mg/L trans-cinnamic acid was added. B The assumed regulatory mechanism involved in the biosynthesis system of pinosylvin from trans-cinnamic acid by engineered E. coli. The black circles indicate trans-cinnamic acid, the red squares indicate trans-cinnamoyl-CoA, the blue triangles indicate malonyl-CoA, the green pentagons indicate pinosylvin
The above results indicated that, in the absence of cerulenin, insufficient levels of malonyl-CoA block pinosylvin biosynthesis in the 2nd step [36–38], which might result a certain amount of trans-cinnamoyl-CoA accumulation (Fig. 4B). The addition of cerulenin had a positive effect on the supplementation of malonyl-CoA [39]. When 60 μM cerulenin was added, the 2nd step catalysis by STS was unblocked, and an increase in pinosylvin production was observed. The improved production of pinosylvin using Ptr4CL5 seemed to be only from the accumulated trans-cinnamoyl-CoA, because no more trans-cinnamic acid was consumed (Fig. 4A). In the presence of cerulenin, the combination of Ptr4CL4 and STS was more suitable. Once cerulenin was added, the overall biopathway was unobstructed, and the yield of pinosylvin increased from 0.03 mg/mg trans-cinnamic acid to 0.76 mg/mg trans-cinnamic acid.
According to the previous studies, most 4CLs do not effectively convert trans-cinnamic acid into the corresponding ester in vitro [15, 32]. Only a few 4CLs, including Sc4CL, At4CL1, Pc4CL and Ptr4CL5, were used for the biosynthesis of trans-cinnamic acid derivatives [40]. However, their results were poor [41]. Our results showed that the wood-derived gene Ptr4CL4 is more efficient than the other genes for pinosylvin production. This finding underlines the importance of altering variants of 4CL in the pathway of pinosylvin biosynthesis. In the subsequent experiment, a combination of Ptr4CL4 and PpSTS was used for pinosylvin production.
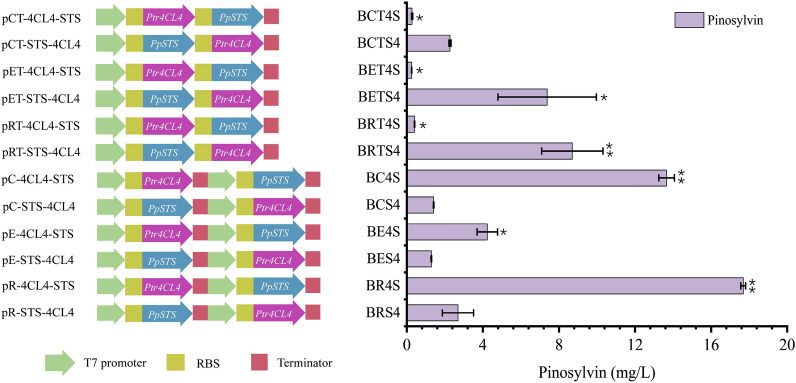
Optimization of the pinosylvin-producing strain by combinatorial engineering
Generally, basic modular optimization, such as promoter, gene copy number and gene order, is significant in reconstructing an effective metabolic pathway in microbes [42]. Thus, our strain was further engineered to balance the catalytic activities of Ptr4CL4 and PpSTS. A series of strains harbouring Ptr4CL4 and PpSTS were constructed by adjusting the expression strategy (Table 1). As shown in Fig. 5, three different plasmids, pCDFDuet-1 (CDF origin), pETDuet-1 (pBR322 origin), and pRSFDuet-1 (RSF origin), were used to regulate module expression, corresponding to gene copy numbers of 20, 40 and 100, respectively, according to previous reports [24]. When only one T7 promoter was employed, strains harbouring the PpSTS gene in front of the Ptr4CL4 gene produced more pinosylvin than those in the opposite order. This meant that the STS close to the T7 promoter was suitable for the pinosylvin pathway. Pinosylvin biosynthesis could be hampered by low heterologous pathway activity [43]. With the same T7 promoter, the production with a high-copy-number plasmid was approximately threefold greater than that with a low-copy-number plasmid (comparison between strains BRTS4 and BCTS4). However, when two T7 promoters were employed in strains, Ptr4CL4 placed in front of PpSTS tended to produce more pinosylvin. Comparing the protein expression levels of the two strains BR4S and BRS4, we found that the solubility of Ptr4CL4 and PpSTS in BRS4 was much lower than that in BR4S (Additional file 1: Fig. S2), which partly resulted in an approximately sixfold change in production. It was concluded that both the order of the two genes and the copy number of the vector play important roles in protein functional folding and expression levels, which may lead to low pinosylvin synthesis. Similar to this phenomenon, Zhou et al. produced the fusion proteins ERG20-BTS1 and BTS1-ERG20 and found that BTS1-ERG20 resulted in increased miltiradiene production compared with ERG20-BTS1 [44]. Overall, E. coli BR4S produced 17.69 ± 0.13 mg/L pinosylvin from 90 mg/L trans-cinnamic acid in the absence of cerulenin, which was the highest yield amongst these 12 constructed strains.
Fig. 5.
Pinosylvin production by recombinant E. coli harbouring different modules. The strains were cultivated at 30 °C for 48 h in YM9 medium, and 90 mg/L trans-cinnamic acid was added. The data represent the means of two replicates, and error bars represent standard deviations. “*” indicates p < 0.05, “**” indicates p < 0.01
Efficient pinosylvin production by strain BR4S
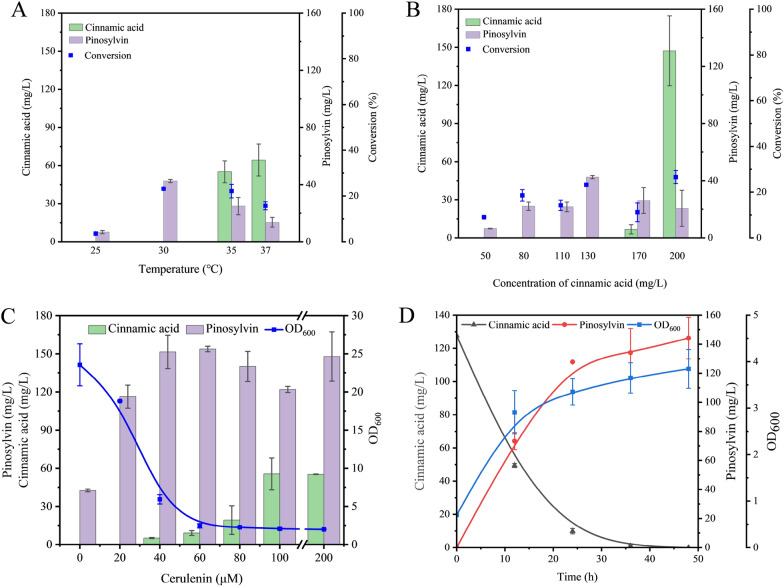
Finally, we investigated the effect of fermentation conditions on pinosylvin production efficiency, such as temperature, trans-cinnamic acid dosage and cerulenin concentration. As shown in Fig. 6A, pinosylvin production by BR4S reached the highest titre of 42.5 ± 1.1 mg/L at 30 °C. In contrast to the performance of BRS4 in Fig. 4A, the complete consumption of trans-cinnamic acid at 25 °C and 30 °C indicated that the expression of 4CL in BR4S increased greatly and that the 2nd step became the bottleneck for production by optimizing the expression strategy. As shown in Fig. 6B, when using 50–130 mg/L trans-cinnamic acid initially, no residual trans-cinnamic acid was observed after 48 h of fermentation, suggesting that the activity of Ptr4CL4 is adequate to convert trans-cinnamic acid into its thioester. The pinosylvin concentration reached a maximum of 42.5 ± 1.1 mg/L at 130 mg/L trans-cinnamic acid. However, more trans-cinnamic acid seemed to trigger cell growth inhibition, which caused less trans-cinnamic acid to be consumed (Additional file 1: Fig. S3). Especially at 200 mg/L, a large amount of trans-cinnamic acid remained after fermentation. Finally, to analyse the contribution of cerulenin to production, cultivations were performed in which cerulenin (up to 200 μM) was supplemented. Adding 20–60 μM cerulenin greatly improved pinosylvin production but also influenced cell growth and protein expression due to its toxicity [45]. When cerulenin was added up to 40 μM, the accumulation of trans-cinnamic acid was observed. The highest pinosylvin production reached 153.7 ± 2.2 mg/L at 60 μM cerulenin (Fig. 6C). Meanwhile, the yield was 1.20 ± 0.02 mg/mg trans-cinnamic acid, corresponding to 83.9% ± 1.17 of the theoretical yield. The course of BR4S with 60 μM cerulenin is shown in Fig. 6D. A total of 130 mg/L trans-cinnamic acid was consumed within 36 h, whilst the highest pinosylvin production was observed at 48 h. It is worth noting that the growth of strains was seriously inhibited in the presence of cerulenin along with the enhancement of malonyl-CoA supply. At this time, the OD600 of the strains decreased to 28.0% of the original value, and pinosylvin production increased to three times the original value. As shown in Table 2, tens of mg/L pinosylvin was produced by different optimization tools. When compared with these studies, our study resulted in a higher titre and yield of pinosylvin from trans-cinnamic acid in the presence of cerulenin.
Fig. 6.
Bioconversion of trans-cinnamic acid to pinosylvin with E. coli BR4S. A Effect of temperature on the synthesis of pinosylvin. Trans-cinnamic acid concentration was 130 mg/L and no cerulenin was added. B Effect of trans-cinnamic acid on the synthesis of pinosylvin. The temperature was 30 °C and no cerulenin was added. C Effect of cerulenin on the synthesis of pinosylvin. The temperature was 30 °C and the concentration of trans-cinnamic acid was same as (A). D The course of BR4S with 60 μM cerulenin
Table 2.
Pinosylvin production from cinnamic acid in engineering bacteria
| Host cell | Substrate concentration | Fermentation | Genetic modification | Product titre (mg/L) | Yield (mg/mg) | References |
|---|---|---|---|---|---|---|
| E. coli | 130 mg/L | + cerulenin | Ptr4CL4↑/PpSTS↑ | 153 | 1.201 | This study |
| Corynebacterium glutamicum | 740.9 mg/L | + cerulenin | AhSTS↑/Pc4CL↑ | 121 | 0.163 | [17] |
| E. coli | 74.1 mg/L | − | At4CL↑/VvSTS↑/fabD↓ | 47 | 0.674 | [19] |
| E. coli | 444.5 mg/L | − | Sc4CL↑/VvSTS↑/fabI↓ | 52 | 0.118 | [12] |
Conclusions
For the first time, the production of pinosylvin in a recombinant E. coli strain has been shown using a novel stilbene synthase PpSTS and a novel 4-coumarate-CoA ligase Ptr4CL4 from the wood plant. The choice of Ptr4CL4 is the key to increasing the yield of pinosylvin in the presence of cerulenin, which greatly benefits the biosynthesis of pinosylvin. By optimizing the expression strategy and culture conditions, we reached the highest pinosylvin production titre of 153.7 ± 2.2 mg/L at 60 μM cerulenin. To the best of our knowledge, the yield of 1.20 ± 0.02 mg/mg trans-cinnamic acid in the present study is the highest yield reported to date. Efficient biosynthesis of pinosylvin demonstrated potential application for the biosynthesis of products derived from cinnamic acid.
Methods
Strains, plasmids and media
All strains and plasmids used and constructed in this study are listed in Table 1. E. coli Trans-T1 was used as the general cloning host for plasmid construction and propagation. E. coli BL21(DE3) and its derivatives were used for enzyme expression and fermentation. The plasmids pRSFDuet-1, pETDuet-1 and pCDFDuet-1 were used as expression plasmids for recombinant plasmid construction.
All E. coli strains were grown with 90 mg/L trans-cinnamic acid in M9CA, YM9 or YNB medium for production. M9CA medium (1 L) consisted of 13.3 g M9CA broth (Sangon Biotech, Shanghai, China), 10 g glucose, 0.12 g MgSO4 and 0.5 mg VB1; YM9 medium (1 L) consisted of 11.3 g M9 salts (Sangon Biotech, Shanghai, China), 42 g MOPs (Sangon Biotech, Shanghai, China), 10 g yeast extract and 5% (v/v) glycerol [19]. For YNB medium, 100 mL of tenfold concentrated yeast nitrogen base without amino acids (Aladdin, Shanghai, China) and 10 g glucose was added to 900 mL base medium (6 g K2HPO4, 3 g KH2PO4, 10 g MOPS, pH 7.0).
Sequence analysis
All protein coding sequences were obtained from the National Center for Biotechnology Information (NCBI) and BRENDA. The protein accession numbers used in this study are listed in Additional file 1: Table S1. Multiple sequence alignment was performed using Espript 3.0 (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi). The phylogenetic tree was constructed by MEGA 7.0 software based on the neighbour-joining method.
Pathway construction
The Ptr4CL4 (GenBank accession number EEF00197.1) and Ptr4CL5 (GenBank accession number EEE79804.2) sequences were codon optimized and synthesized by Generay Biotech. The gene products were excised via BamHI and HindIII and then ligated into pETDuet-1 to yield pE-Ptr4CL5 and pE-Ptr4CL4, respectively.
PsSTS2Q361R (the variant of PsSTS2 (GenBank accession number P48408.1), PtSTS (GenBank accession number AHK13302.1) and PpSTS (GenBank accession number ALN42233.1) sequences were codon optimized and synthesized by Generay Biotech. To construct the pinosylvin synthetic pathway, the high-copy-number vector pRSFDuet-1 was employed, generating plasmids pR-PsSTS2Q361R, pR-PtSTS and pR-PpSTS. Thereafter, Ptr4CL5 was amplified by PCR using the primer pair 4CL5-f/4CL5-r and cloned into pR-PsSTS2Q361R, pR-PtSTS and pR-PpSTS through the NdeI and XhoI sites to generate plasmids pR-PsSTS2Q361R-Ptr4CL5, pR-PtSTS-Ptr4CL5 and pR-PpSTS-Ptr4CL5, respectively. In addition, the Ptr4CL4 gene was amplified by PCR using the primer pair 4CL4-P-f/4CL4-P-r and cloned into pR-PpSTS through the NdeI and XhoI sites to generate the plasmid pR-PpSTS-Ptr4CL4.
To optimize modular expression, the PpSTS gene in plasmid pRSFDuet-1 was digested and cloned into plasmids pETDuet-1 and pCDFDuet-1 through the BamHI and HindIII sites, yielding the intermediate expression plasmids pE-PpSTS and pC-PpSTS. The Ptr4CL4 gene in plasmid pR-PpSTS-Ptr4CL4 was digested and cloned into plasmids pE-PpSTS and pC-PpSTS through the NdeI and XhoI sites, yielding the expression plasmids pE-PpSTS-Ptr4CL4 and pC-PpSTS-Ptr4CL4. Ptr4CL4 was digested from plasmid pE-Ptr4CL4 through BamHI and HindIII and cloned into plasmids pRSFDuet-1 and pCDFDuet-1, generating plasmids pR-Ptr4CL4 and pC-Ptr4CL4, respectively. The PpSTS gene was amplified by PCR using the primer pair STS-f/STS-r and ligated with the linear DNAs pE-Ptr4CL4, pR-Ptr4CL4 and pC-Ptr4CL4, which were amplified by PCR using the primer pair P-4-f/P-4-r, yielding the plasmids pE-Ptr4CL4-PpSTS, pR-Ptr4CL4-PpSTS and pC-Ptr4CL4-PpSTS, respectively. Plasmids pR-PpSTS-Ptr4CL4, pE-PpSTS and pC-PpSTS were amplified by PCR using primer pair plasmids-STS-f/plasmids-STS-r, yielding the linear DNAs pR-PpSTS, pE-PpSTS and pC-PpSTS. The Ptr4CL4 gene was amplified by PCR using the primer pair RBS-4CL4-f/RBS-4CL4-r and ligated with the linear DNAs pR-PpSTS, pE-PpSTS and pC-PpSTS, generating plasmids pRT-PpSTS-Ptr4CL4, pET-PpSTS-Ptr4CL4 and pCT-PpSTS-Ptr4CL4. The PpSTS gene was amplified by PCR using the primer pair RBS-STS-f/RBS-STS-r, and the plasmids pR-Ptr4CL4, pE-Ptr4CL4 and pC-Ptr4CL4 were amplified by PCR using the primer pair plasmids-4CL4-f/plasmids-4CL4-r. PpSTS was ligated with the linear DNAs pR-Ptr4CL4, pE-Ptr4CL4 and pC-Ptr4CL4, yielding plasmids pRT-Ptr4CL4-PpSTS, pET-Ptr4CL4-PpSTS and pCT-Ptr4CL4-PpSTS, respectively. All primers used in this study are listed in Additional file 1: Table S2.
Microbial pinosylvin production
The recombinant strain was first cultivated in 50 mL fresh LB medium in 250 mL flasks for 12 h at 37 °C and 200 rpm. Then, these cells were diluted to an OD600 of 0.7 in 10 mL of M9CA, YM9 or YNB medium in 50 mL flasks at 200 rpm, and gene expression was induced with 1 mM IPTG. The cultivation was continued for 48 h at 30 °C, and 90 mg/L trans-cinnamic acid was added. After 48 h of fermentation, the fermentation broth was collected for analysis.
To test the effect of temperature, cultivation was performed with 1.0 mM IPTG and 130 mg/L trans-cinnamic acid. The effect of trans-cinnamic acid concentration was explored at 1.0 mM IPTG and 30 °C. To determine the effect of cerulenin on pinosylvin production, cultivation was carried out at 30 °C with 1.0 mM IPTG, 130 mg/L trans-cinnamic acid and different concentrations of cerulenin ranging from 0 to 100 μM. After 48 h of fermentation, the fermentation broth was collected for analysis.
For 48 h of optimized fermentation, samples were taken at 12, 24, 36 and 48 h intervals for subsequent analysis, and cultivation was carried out at 30 °C with 1.0 mM IPTG, 130 mg/L trans-cinnamic acid and 60 μM cerulenin. At least two duplicates were used for all experiments.
Detection and quantification
Before HPLC analysis for compounds, the cell optical density (OD) of the culture was measured on a 752S spectrophotometer at 600 nm. Thereafter, the culture was centrifuged to remove the cell debris and further filtered through a 0.22 μm filter. The supernatant sample was used for trans-cinnamic acid analysis.
To analyse the amount of pinosylvin, 500 μL of supernatant was mixed with the same volume of ethyl acetate. The mixture was vortexed for 5 min and centrifuged at 13,400×g for 2 min. Then, 400 μL of the top organic layer was transferred to a new microtube and volatilized to dryness. Subsequently, the samples were resolubilized in 400 μL methanol for HPLC analysis.
HPLC analysis was carried out on an HPLC (Agilent 1260 Series, USA) using an Eclipse XDB-C18 column (250 mm × 4.6 mm, 5 μm). The UV‒Vis detector was set at 294 nm. The mobile phase consisted of 40% solvent A and 60% solvent B, where A was acetonitrile and B was water/acetic acid (98.5:1.5, v/v). The flow rate was set at 1.0 mL min−1. The column temperature was set to 30 ℃.
The calculation of the pinosylvin yield is shown below.
Cp is the concentration of pinosylvin in the medium; Ct is the initial concentration of trans-cinnamic acid. The unit of Cp and Ct is mg/L, and the theoretical yield was 1.43 mg/mg.
Statistical analyses
All experiments were performed at least in two duplicates, and the results were expressed as means ± standard deviation (SD). The statistical analysis was performed using SPSS Statistics 20.0 (SPSS Inc., Chicago, IL, USA) with independent samples t test. Figures were drawn using OriginPro 2021(OriginLab, Northampton, MA, USA).
Supplementary Information
Additional file 1: Table S1. GenBank IDs and sequences of stilbene synthases used in this study. Table S2. Primers used in this study. Figure S1. Growth curves of E. coli BLS5 cultured in YM9. Figure S2. SDS-PAGE of Ptr4CL4 and PpSTS in BRS4 and BR4S. Figure S3. The OD600 of BR4S at 48 h under different concentrations of trans-cinnamic acid.
Acknowledgements
The work was founded by the National Key Research and Development Program of China (2017YFD0600205). We also kindly acknowledge partial support from the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Abbreviations
- PAL
Phenylalanine ammonia lyase
- 4CL
4-Coumarate-CoA ligase
- STS
Stilbene synthase
- CoA
Coenzyme A
- Acc
Acetyl-CoA carboxylase encoding gene
- fabD
Malonyl-CoA: ACP transacylase encoding gene
- fabB
β-Ketoacyl-ACP synthase I encoding gene
- fabF
β-Ketoacyl-ACP synthase II encoding gene
- fabH
β-Ketoacyl-ACP synthase III encoding gene
Author contributions
Y.H.: Conceptualization, Data curation, Methodology, Investigation, Validation, Software, Visualization, Writing-original draft Writing-review and editing. C.Z.: Data curation, Methodology, Investigation, Validation. L.Z.: Investigation, Software. Z.Z.: conceptualization, Supervision, Writing-review and editing. J.OY.: Methodology, Project administration, Funding acquisition, Supervision, Writing-review and editing.
Availability of data and materials
The data supporting the conclusions of this article are included with the article and its supplementary material.
Declarations
Competing interests
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Shen X, Sun R. Recent advances in lignocellulose prior-fractionation for biomaterials, biochemicals, and bioenergy. Carbohydr Polym. 2021;261:117884. doi: 10.1016/j.carbpol.2021.117884. [DOI] [PubMed] [Google Scholar]
- 2.Garlapati VK, Chandel AK, Kumar SPJ, et al. Circular economy aspects of lignin: towards a lignocellulose biorefinery. Renew Sustain Energy Rev. 2020;130:109977. doi: 10.1016/j.rser.2020.109977. [DOI] [Google Scholar]
- 3.Li J, Yue C, Wei W, et al. Construction of a p-coumaric and ferulic acid auto-regulatory system in Pseudomonas putida KT2440 for protocatechuate production from lignin-derived aromatics. Bioresour Technol. 2022;344:126221. doi: 10.1016/j.biortech.2021.126221. [DOI] [PubMed] [Google Scholar]
- 4.Azubuike CC, Allemann MN, Michener JK. Microbial assimilation of lignin-derived aromatic compounds and conversion to value-added products. Curr Opin Microbiol. 2022;65:64–72. doi: 10.1016/j.mib.2021.10.014. [DOI] [PubMed] [Google Scholar]
- 5.Lieutier F, Sauvard D, Brignolas F, et al. Changes in phenolic metabolites of Scots-pine phloem induced by Ophiostoma brunneo-ciliatum, a bark-beetle-associated fungus. Eur J For Res. 1996;26:145–158. [Google Scholar]
- 6.Hwang H-S, Han JY, Choi YE. Enhanced accumulation of pinosylvin stilbenes and related gene expression in Pinus strobus after infection of pine wood nematode. Tree Physiol. 2021;41(10):1972–1987. doi: 10.1093/treephys/tpab053. [DOI] [PubMed] [Google Scholar]
- 7.Kivimäki K, Leppänen T, Hämäläinen M, Vuolteenaho K, Moilanen E. Pinosylvin shifts macrophage polarization to support resolution of inflammation. Molecules. 2021;26(9):2772. doi: 10.3390/molecules26092772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koo HB, Hwang H-S, Han JY, et al. Enhanced production of pinosylvin stilbene with aging of Pinus strobus callus and nematicidal activity of callus extracts against pinewood nematodes. Sci Rep. 2022;12(1):770. doi: 10.1038/s41598-022-04843-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song J, Seo Y, Park H. Pinosylvin enhances leukemia cell death via down-regulation of AMPKα expression. Phytother Res. 2018;32(10):2097–2104. doi: 10.1002/ptr.6156. [DOI] [PubMed] [Google Scholar]
- 10.Rivière C, Pawlus AD, Mérillon JM. Natural stilbenoids: distribution in the plant kingdom and chemotaxonomic interest in vitaceae. Nat Prod Rep. 2012;29:1317–1333. doi: 10.1039/c2np20049j. [DOI] [PubMed] [Google Scholar]
- 11.Bakrim S, Machate H, Benali T, et al. Natural sources and pharmacological properties of pinosylvin. Plants. 2022;11(12):1541. doi: 10.3390/plants11121541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salas-Navarrete C, Hernández-Chávez G, Flores N, et al. Increasing pinosylvin production in Escherichia coli by reducing the expression level of the gene fabI-encoded enoyl-acyl carrier protein reductase. Electron J Biotechnol. 2018;33:11–16. doi: 10.1016/j.ejbt.2018.03.001. [DOI] [Google Scholar]
- 13.Chong J, Poutaraud A, Hugueney P. Metabolism and roles of stilbenes in plants. Plant Sci. 2009;177(3):143–155. doi: 10.1016/j.plantsci.2009.05.012. [DOI] [Google Scholar]
- 14.Conde E, Fang W, Hemming J, et al. Recovery of bioactive compounds from Pinus pinaster wood by consecutive extraction stages. Wood Sci Technol. 2014;48(2):311–323. doi: 10.1007/s00226-013-0604-1. [DOI] [Google Scholar]
- 15.van Summeren-Wesenhagen PV, Marienhagen J, Pettinari MJ. Metabolic engineering of Escherichia coli for the synthesis of the plant polyphenol pinosylvin. Appl Environ Microbiol. 2015;81(3):840–849. doi: 10.1128/AEM.02966-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raiber S, Schröder G, Schröder J. Molecular and enzymatic characterization of two stilbene synthases from Eastern white pine (Pinus strobus). A single Arg/His difference determines the activity and the pH dependence of the enzymes. FEBS Lett. 1995; 361(2):299–302. [DOI] [PubMed]
- 17.Adisakwattana S, Moonsan P, Yibchok-anun S. Insulin-Releasing properties of a series of cinnamic acid derivatives in vitro and in vivo. J Agric Food Chem. 2008;56(17):7838–7844. doi: 10.1021/jf801208t. [DOI] [PubMed] [Google Scholar]
- 18.Wang S, Chen J, Shi J, et al. Novel cinnamic acid derivatives containing the 1,3,4-oxadiazole moiety: design, synthesis, antibacterial activities, and mechanisms. J Agric Food Chem. 2021;69(40):11804–11815. doi: 10.1021/acs.jafc.1c03087. [DOI] [PubMed] [Google Scholar]
- 19.Liang J-l, Guo L-q, Lin J-f, et al. A novel process for obtaining pinosylvin using combinatorial bioengineering in Escherichia coli. World J Microbiol Biotechnol. 2016;32(6):102. doi: 10.1007/s11274-016-2062-z. [DOI] [PubMed] [Google Scholar]
- 20.Kallscheuer N, Vogt M, Stenzel A, et al. Construction of a Corynebacterium glutamicum platform strain for the production of stilbenes and (2S)-flavanones. Metab Eng. 2016;38:47–55. doi: 10.1016/j.ymben.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 21.Hong J, Im D-K, Oh M-K. Investigating E. coli coculture for resveratrol production with 13C metabolic flux analysis. J Agric Food Chem. 2020; 68(11):3466–73. [DOI] [PubMed]
- 22.Yang Y, Lin Y, Li L, Linhardt RJ, Yan Y. Regulating malonyl-CoA metabolism via synthetic antisense RNAs for enhanced biosynthesis of natural products. Metab Eng. 2015;29:217–226. doi: 10.1016/j.ymben.2015.03.018. [DOI] [PubMed] [Google Scholar]
- 23.Milke L, Aschenbrenner J, Marienhagen J, Kallscheuer N. Production of plant-derived polyphenols in microorganisms: current state and perspectives. Appl Microbiol Biotechnol. 2018;102(4):1575–1585. doi: 10.1007/s00253-018-8747-5. [DOI] [PubMed] [Google Scholar]
- 24.Wu J, Zhang X, Zhu Y, et al. Rational modular design of metabolic network for efficient production of plant polyphenol pinosylvin. Sci Rep. 2017;7(1):1459. doi: 10.1038/s41598-017-01700-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeandet P, Sobarzo-Sánchez E, Clément C, et al. Engineering stilbene metabolic pathways in microbial cells. Biotechnol Adv. 2018;36(8):2264–2283. doi: 10.1016/j.biotechadv.2018.11.002. [DOI] [PubMed] [Google Scholar]
- 26.Villa-Ruano N, Rivera A, Rubio-Rosas E, et al. Comparative activity of six recombinant stilbene synthases in yeast for resveratrol production. Appl Sci. 2020;10(14):4847. doi: 10.3390/app10144847. [DOI] [Google Scholar]
- 27.Schanz S, Schröder G, Schröder J. Stilbene synthase from Scots pine (Pinus sylvestris) FEBS Lett. 1992;313(1):71–74. doi: 10.1016/0014-5793(92)81187-Q. [DOI] [PubMed] [Google Scholar]
- 28.Wang C, Zhi S, Liu C, et al. Characterization of stilbene synthase genes in Mulberry (Morus atropurpurea) and metabolic engineering for the production of resveratrol in Escherichia coli. J Agric Food Chem. 2017;65(8):1659–1668. doi: 10.1021/acs.jafc.6b05212. [DOI] [PubMed] [Google Scholar]
- 29.Cotner M, Zhan J, Zhang Z. A computational metabolic model for engineered production of resveratrol in Escherichia coli. ACS Synth Biol. 2021;10(8):1992–2001. doi: 10.1021/acssynbio.1c00163. [DOI] [PubMed] [Google Scholar]
- 30.Xu J-Y, Xu Y, Chu X, Tan M, Ye B-C. Protein acylation affects the artificial biosynthetic pathway for pinosylvin production in engineered E. coli. ACS Chem Biol 2018; 13(5):1200–8. [DOI] [PubMed]
- 31.Kodan A, Kuroda H, Sakai F. A stilbene synthase from Japanese red pine (Pinus densiflora): Implications for phytoalexin accumulation and down-regulation of flavonoid biosynthesis. Proc Natl Acad Sci USA. 2002;99(5):3335–3339. doi: 10.1073/pnas.042698899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang C, Zang Y, Liu P, Zheng Z, Ouyang J. Characterization, functional analysis and application of 4-Coumarate: CoA ligase genes from Populus trichocarpa. J Biotechnol. 2019;302:92–100. doi: 10.1016/j.jbiotec.2019.06.300. [DOI] [PubMed] [Google Scholar]
- 33.Zhao Y, Wu B-H, Liu Z-N, Qiao J, Zhao G-R. Combinatorial optimization of resveratrol production in engineered E. coli. J Agric Food Chem. 2018; 66(51):13444–53. [DOI] [PubMed]
- 34.Funabashi H, Kawaguchi A, Tomoda H, et al. Binding site of cerulenin in fatty acid synthetase. J Biochem. 1989;105(5):751–755. doi: 10.1093/oxfordjournals.jbchem.a122739. [DOI] [PubMed] [Google Scholar]
- 35.Lim Chin G, Fowler Zachary L, Hueller T, Schaffer S, Koffas Mattheos AG. High-yield resveratrol production in engineered Escherichia coli. Appl Environ Microbiol. 2011;77(10):3451–3460. doi: 10.1128/AEM.02186-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miyahisa I, Kaneko M, Funa N, et al. Efficient production of (2S)-flavanones by Escherichia coli containing an artificial biosynthetic gene cluster. Appl Microbiol Biotechnol. 2005;68:498–504. doi: 10.1007/s00253-005-1916-3. [DOI] [PubMed] [Google Scholar]
- 37.Leonard E, Lim KH, Saw PN, Koffas MAG. Engineering central metabolic pathways for high-level flavonoid production in Escherichia coli. Appl Environ Microbiol. 2007;73:3877–3886. doi: 10.1128/AEM.00200-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson O, Gonzalez-Villanueva M, Wong L, et al. Design and application of genetically-encoded malonyl-CoA biosensors for metabolic engineering of microbial cell factories. Metab Eng. 2017;44:253–264. doi: 10.1016/j.ymben.2017.10.011. [DOI] [PubMed] [Google Scholar]
- 39.Wen J, Tian L, Xu M, Zhou X, Zhang Y, Cai M. A synthetic malonyl-CoA metabolic oscillator in Komagataellaphaffii. ACS Synth Biol. 2020;9:1059–1068. doi: 10.1021/acssynbio.9b00378. [DOI] [PubMed] [Google Scholar]
- 40.Mark R, Lyu X, Ng KR, Chen WN. Gene source screening as a tool for naringenin production in engineered Saccharomyces cerevisiae. ACS Omega. 2019;4(7):12872–12879. doi: 10.1021/acsomega.9b00364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H, Lyv Y, Zhou S, Yu S, Zhou J. Microbial cell factories for the production of flavonoids–barriers and opportunities. Bioresour Technol. 2022;360:127538. doi: 10.1016/j.biortech.2022.127538. [DOI] [PubMed] [Google Scholar]
- 42.Li S, Liang C, Liu G, et al. De novo biosynthesis of chlorogenic acid using an artificial microbial community. J Agric Food Chem. 2021;69(9):2816–2825. doi: 10.1021/acs.jafc.0c07588. [DOI] [PubMed] [Google Scholar]
- 43.Wei W, Zhang P, Shang Y, Zhou Y, Ye B-C. Metabolically engineering of Yarrowia lipolytica for the biosynthesis of naringenin from a mixture of glucose and xylose. Bioresour Technol. 2020;314:123726. doi: 10.1016/j.biortech.2020.123726. [DOI] [PubMed] [Google Scholar]
- 44.Zhou YJ, Gao W, Rong Q, et al. Modular pathway engineering of diterpenoid synthases and the mevalonic acid pathway for miltiradiene production. J Am Chem Soc. 2012;134(6):3234–3241. doi: 10.1021/ja2114486. [DOI] [PubMed] [Google Scholar]
- 45.Zhou S, Lama S, Jiang J, Sankaranarayanan M, Park S. Use of acetate for the production of 3-hydroxypropionic acid by metabolically-engineered Pseudomonas denitrificans. Bioresour Technol. 2020;307:123194. doi: 10.1016/j.biortech.2020.123194. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. GenBank IDs and sequences of stilbene synthases used in this study. Table S2. Primers used in this study. Figure S1. Growth curves of E. coli BLS5 cultured in YM9. Figure S2. SDS-PAGE of Ptr4CL4 and PpSTS in BRS4 and BR4S. Figure S3. The OD600 of BR4S at 48 h under different concentrations of trans-cinnamic acid.
Data Availability Statement
The data supporting the conclusions of this article are included with the article and its supplementary material.