Abstract
The presence of linear amino acid motifs with the capacity to recognize the neutral lipid cholesterol, known as Cholesterol Recognition/interaction Amino acid Consensus sequence (CRAC), and its inverse or mirror image, CARC, has recently been reported in the primary sequence of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike S homotrimeric glycoprotein. These motifs also occur in the two other pathogenic coronaviruses, SARS-CoV, and Middle-East respiratory syndrome CoV (MERS-CoV), most conspicuously in the transmembrane domain, the fusion peptide, the amino-terminal domain, and the receptor binding domain of SARS-CoV-2 S protein. Here we analyze the presence of cholesterol-recognition motifs in these key regions of the spike glycoprotein in the pathogenic CoVs. We disclose the inherent pathophysiological implications of the cholesterol motifs in the virus-host cell interactions and variant infectivity.
Keywords: Cholesterol, Cholesterol recognition motifs, Viral infectivity, Cell-surface phenomena, Cholesterol-protein interactions, Fusion proteins
Graphical Abstract
1. Introduction
The appearance of ligand recognition in living organisms has occurred in various instances as a result of coevolution between interacting partners. In the case of pathogens like viruses, the interactions between their infective machinery and eukaryotic cell-surface receptors may not follow this coevolutionary mechanism [1]. This is because the virion receptors, transmembrane proteases in the case of coronaviruses (CoVs), fulfill preexisting physiological roles in the eukaryotic cells which the viruses exploit to serve their own pathophysiological cycle.
Soon after the irruption of the COVID-19 pandemic, structural work using cryo-electron microscopy (cryo-EM), and to a lesser extent X-ray diffraction and nuclear magnetic resonance (NMR) spectroscopy, produced spectacular advances in our knowledge of the causative agent, the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). These unprecedentedly vertiginous developments produced, in the course of a few months, atomistic depictions of the virus and its canonical cell-surface receptor, the metalloprotease angiotensin converting enzyme 2 (ACE2), and abundant hypotheses on the mechanistic interactions between the two (reviewed in [1]). A possible reason why ACE2 has an evolutionarily advantage in SARS-CoV-2 as a target molecule is the ubiquitous cell-surface distribution of the enzyme in many host tissues, particularly mucosal epithelia [1], [2]. The spike (S) glycoprotein of SARS-CoV-2 and other pathogenic CoVs is the one that interacts with ACE2. Binding to and fusion with the host-cell membrane appear to be sensitive to the lipid environment in which these two coupled processes occur [3].
Cholesterol plays important roles in orchestrating the biophysical properties of both proteins and lipids in the plasma membrane [4]. The observed cholesterol dependence of some viruses leads us to consider how they take advantage of cholesterol properties, in so doing optimizing viral infection or other steps of their life cycle. Several viral cholesterol-binding proteins relevant to virus entry processes have been described [5]. The amount of cholesterol in the virus envelope also apears to play a role: in members of the alphavirus genus (Chikungungya, Sindibis, Venezuelan equine encephalitis and Ross River virus), the cholesterol/phospholipid molar ratio is higher than in the host plasma membranes [6]. The higher cholesterol level in the viral particle is important in the organization of the viral envelope [7]. Cholesterol depletion in the influenza A virus envelope produces nicks and holes in the viral envelope, reducing virus infectivity [8]. Moreover, virions of influenza A virus and respiratory syncytial virus released from the cholesterol-depleted cells were less stable than those released from untreated cells [9]. The transmembrane gp41 subunit, an HIV-1 envelope protein, interacts directly with cholesterol in the viral membrane [10], [11].
Recent structural work is beginning to unravel the functional relationship between the S protein and cholesterol, present in both the host-cell membrane and the viral envelope bilayer [12]. ACE2 of Vero E6 and Caco-2 cells is recovered in the detergent-resistant phase in biochemical assays, an experimental finding that is conventionally, albeit not universally, taken as evidence that a protein resides in liquid-ordered (Lo) lipid domains, also termed “lipid rafts”. There is also substantial evidence in favor of the notion that SARS-CoV-2 entry requires the participation of cholesterol [13], [14]. In this regard, Tang et al. [15] proposed that rather than simply organizing ACE2 and SARS-CoV-2 proteins into cholesterol-rich Lo lipid domains, cholesterol might be directly involved in membrane fusion dynamics, guaranteeing the formation of the fusion intermediate. Meher et al. [16] and Pattnaik et al. [17] demonstrated that membrane cholesterol is vital for the fusogenic activity of SARS-CoVs viruses. Nardacci et al. [18] reported that the accumulation of lipids in SARS-CoV-2 infected cells, both in vitro and in the lungs of patients, could be involved in SARS-CoV-2 pathogenesis. Furthermore, membrane cholesterol depletion of ACE2-expressing HEK293T cells with methyl-β-cyclodextrin reduced SARS-CoV-2 infection, suggesting that cholesterol-rich lipid domains, as well as endosomal acidification, are essential requirements for SARS-CoV-2 infection [13]. Sanders et al. [14] showed that pre-treatment of SARS-CoV-2 with methyl-β-cyclodextrin blocked virus infection, further reinforcing the notion that cholesterol content in the viral particle is critical for infectivity.
Here we analyze the presence of cholesterol-recognition motifs in key regions of the spike glycoprotein in the pathogenic CoVs and disclose their possible pathophysiological implications in the virus-host cell interactions.
2. Material and methods
Search for the presence and localization of the consensus sequence for the "CRAC" motif, [L/V]–[X](1−5)–[Y]–[X](1−5)–[R/K], or the "CARC" motif, [R/K]–[X](1−5)–[YFW]–[X](1−5)–[L/V], was carried out on the SARS-CoV-1 (Accession: AAU04646), SARS-CoV-2 (Accession: YP_009724390), MERS-CoV (Accession: K9N5Q8), and Omicron BA.1 (Accession: 7WP9_A) spike protein sequences, available at the National Center for Biotechnology Information (NCBI). To this end, we employed the Fuzzpro application of the Jemboss software (European Molecular Biology Open Software Suite, EMBOSS) as in our original description of the onsensus motifs [19]. Molecular representations were performed with Visual Molecular Dynamics (VMD) software, and the PDB files available in the protein data bank (PDB files 6XR8 and 7WP9, for SARS-CoV-2 and Omicron BA.1 spike proteins, respectively).
3. Results and discussion
3.1. Consensus cholesterol-recognition sequences in the pathogenic CoVs
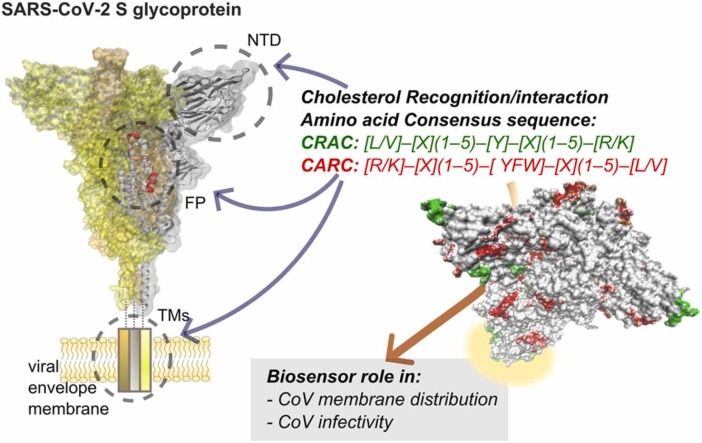
Cell-surface receptors for neurotransmitters have been instrumental in the discovery of linear amino acid sequences with the capacity to recognize cholesterol. Studies of the benzodiazepine receptor led to the identification of the sequence [L/V]–[X](1−5)–[Y]–[X](1−5)–[R/K], known as Cholesterol Recognition/interaction Amino acid Consensus sequence (CRAC) [20] ( Fig. 1A). Work on the nicotinic acetylcholine receptor (nAChR) identified the inverse or mirror image of CRAC, which we coined “CARC”: [R/K]–[X](1−5)–[YFW]–[X](1−5)–[L/V] [19] (Fig. 1A). In subsequent work, we found that these two cholesterol consensus motifs are present in a great variety of membrane proteins [21], including the superfamily of ligand-gated ion-channel (LGIC) proteins [21], other channels like the transient receptor potential (TRP) channel [22], and in the large superfamily of membrane-bound GPCR proteins [21]. CRAC/CARC sequences share a central aromatic residue, like tyrosine for CRAC, and tyrosine, phenylalanine, or tryptophan for CARC, flanked on both sides by one to five amino acid residues ending in a basic (arginine or lysine) and an apolar terminus (valine or leucine) (Fig. 1A). Both CARC and CRAC are vectorial motifs, with CARC preferentially, albeit not exclusively, located in the exofacial membrane leaflet, whereas CRAC most often occurs in the cytoplasmic-facing leaflet of the plasma membrane [21].
Fig. 1.
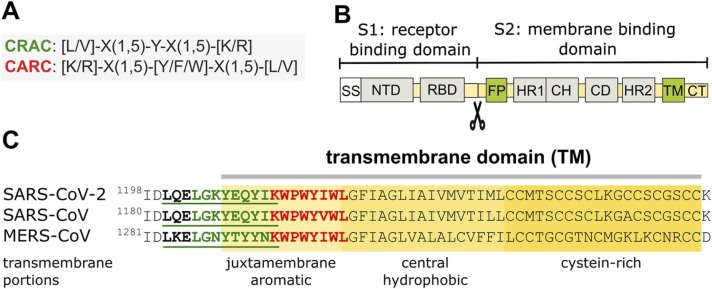
Cholesterol-recognition motifs in the TM region of spike S glycoprotein from SARS-CoV-2, SARS-CoV, and MERS-CoV. A) Sequences of the cholesterol-recognition motifs. B) Schematic diagram of SARS-CoV-2 primary structure. SS, signal sequence; NTD, N-terminal domain; RBD, receptor binding domain; FP, fusion peptide; HR1, heptad repeat 1; CH, central helix; CD, connector domain; HR2, heptad repeat 2; TM, transmembrane domain; CT, cytoplasmic tail. C) Cholesterol-recognition motifs along the linear sequences of the TMs. The TM region exhibits back-to-back mirror images of highly conserved CRAC (green) and CARC (red) motifs at the N-term juxtamembrane aromatic region.
The three pathogenic CoVs, namely the severe acute respiratory syndrome (SARS-CoV), the virus that gave rise to the epidemic at the brink of the 21st century, followed by the Middle-East respiratory syndrome CoV (MERS-CoV), and the contemporary SARS-CoV-2 associated with the pandemic, possess cholesterol-recognition motifs (Fig. 1A) in various regions of the virion, and most prominently in the spike S glycoprotein. Several cholesterol-recognition motifs occur in the transmembrane (TM) domain (Fig. 1), in the fusion peptide (FP, Fig. 2), in the amino-terminal domain (NTD, Fig. 3), and the receptor binding domain (RBD, Fig. 4) of SARS-CoV-2 S protein. Fig. 1B depicts the SARS-CoV-2 spike (S) glycoprotein sequence, where the different functionally relevant regions of the virus can be identified, starting from the signal sequence (SS) in the S1 subunit, which also harbors the NTD and the important RBD. The key role of the S1 subunit is to prevent the praecox fusion of the S2 subunit to the target plasma membrane in the host cell and to bind to the mammalian cell-surface receptor, the metalloprotease angiotensin-converting enzyme-2 (ACE2). This step involves the proteolytic cleavage that activates the S protein by the host cell plasma membrane-resident transmembrane protease TMPRSS2 [23], a requisite for the corformational change of the S2 subunit [24] from a structurally disordered state into a wedge-shaped conformer [25] apt to undergo the fusion of the viral and plasma membranes via its FP.
Fig. 2.
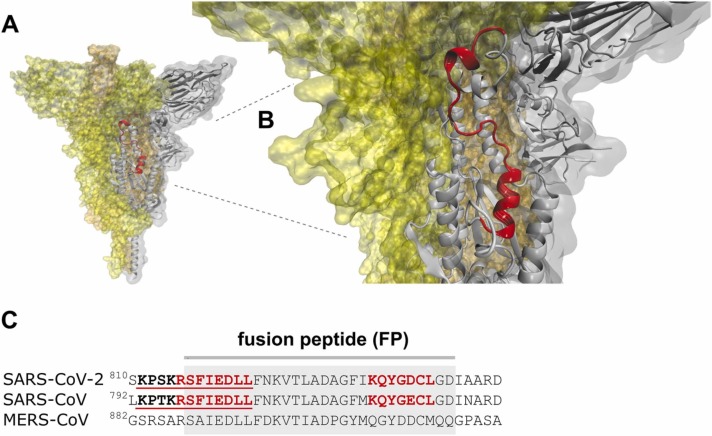
Cholesterol-recognition motifs in the fusion peptide of the spike S glycoprotein from the SARS-CoV-2, SARS-CoV, and MERS-CoV, the three highly pathogenic members of the heptad human CoVs. A) FP segment (red ribbon) in the viral envelope of the SARS-CoV-2 S glycoprotein B) Close-up view of the FP in the SARS-CoV-2 S protein. C) Cholesterol-recognition motifs along the linear sequences of the FPs.
Fig. 3.
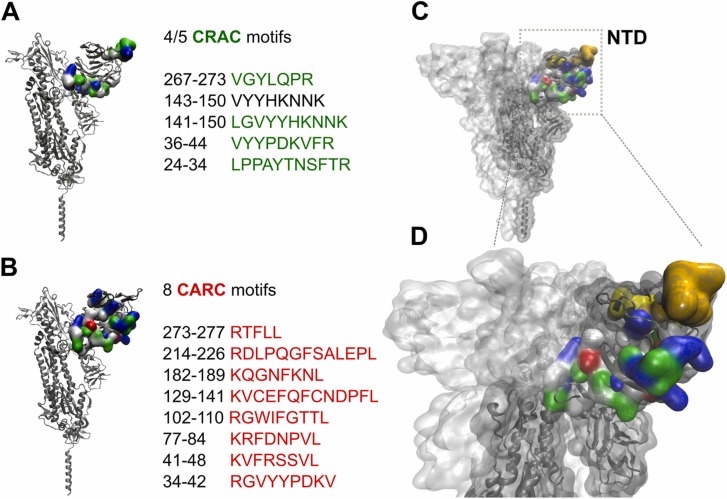
Cholesterol-recognition motifs in the N-terminal domain (NTD) region of the spike S glycoprotein from SARS-CoV-2. Sequences of the cholesterol-recognition motifs, A) CRAC and b) CARC, localized in the NTD region of SARS-CoV-2 spike protein. C) CARC and CARC motifs are colored in the NTD segment. The yellow and orange segments represent the CARC129–141 and CRAC141–150 motifs, respectively. D) Close-up view of the segments shown in C).
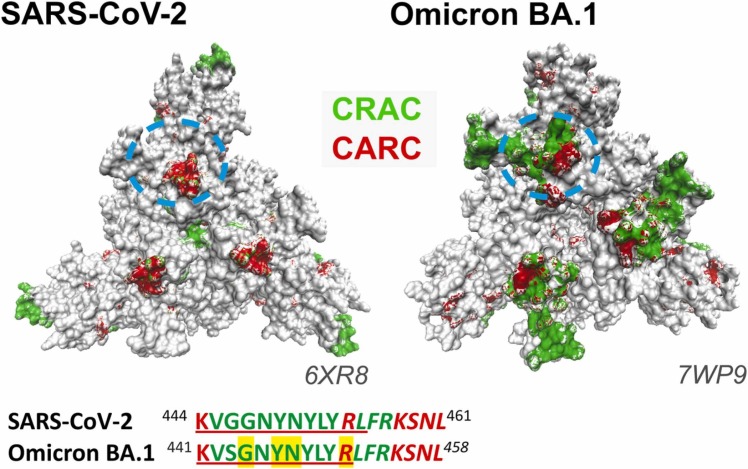
Fig. 4.
Top view of SARS-CoV-2, and Omicron BA.1 spike protein. CRAC and CARC motifs are colored green and red, respectively. The approximate position of the sequences SARS-CoV-2 444–461, and Omicron BA.1 441–458 are highlighted by a dotted light-blue circle. Letters highlighted in yellow correspond to plasma membrane-interacting residues proposed by Overduin et al. [43].
3.2. CARC/CRAC cholesterol recognition motifs in the S protein transmembrane (TM) domain
The TM domain of the S protein anchors the glyoprotein in the virus envelope (Fig. 1B-C). It consists of three portions: a juxtamembrane aromatic region, a central hydrophobic region, and a cysteine-rich region [26]. As shown in Fig. 1C, the membrane-embedded regions of the S protein of the three pathogenic human CoVs share essentially the same cholesterol-recognition motifs, with a high degree of amino acid homology, especially in the two SARS virus. The CARC motif is deeper in the aromatic amino acid-rich part of the TM, with a CRAC laying immediately adjacent to it, partly embedded in the juxtamembrane portion, in a tail-to-tail disposition. We originally described this CARC-CRAC “back-to-back (or tail-to-tail)” mirror-image configuration in members of the GPCR superfamily [21]. The sequence homology observed in the TMs shown in Fig. 1 C (and fusion peptide, see below), attests to the possible functional relevance of the cholesterol-recognition motifs in CoVs [26]. The interaction between cholesterol and SARS-CoV-2 TM regions could be impaired after pre-treatment of SARS-CoV-2 with the cholesterol-depleting compound methyl-β-cyclodextrin. This cholesterol-sequestering organic compound induces a conformational change in the viral particle that could be responsible, at least in part, for the reduced virus infectivity reported by Sanders et al. [14]. Wei and coworkers recently reported the occurrence of six such tail-to-tail cholesterol mirror motifs in the SARS-CoV-2 S protein and experimentally demonstrated, using a microscale thermophoresis assay, that the S protein binds cholesterol with a half-maximum inhibitory concentration IC50 of ∼195 nM, but does not bind the cholesterol analogs campesterol or epicholesterol [27]. Contrary to expectations, the TM segment (residues 1203–1218) used in the article of Wei et al. [27] did not show interactions with cholesterol. A preliminary study using atomistic molecular dynamics simulations found that cholesterol preferred the C-terminal half of the TM segment instead of binding to the CRAC/CARC motifs in the TM region [28]. Concerning SARS-CoV 2 S protein TM segment-cholesterol interactions, the differences between live cells experiments and in silico analyses or synthetic TM-segments-cholesterol interactions may be due to the role played by the rest of the S protein in live cells, acting as a final translator that defines virus infectivity.
3.3. Cholesterol recognition motifs in the fusion peptide (FP) of the S glycoprotein
For enveloped viruses, the release of their genome into the host cell requires the fusion of their membrane bilayer to either the plasma membrane or the endocytic vesicle membranes of the target cell. The FP is a short segment (∼28 amino acid residues long) of the S2 domain that constitutes the functional fusogenic element in SARS-CoV-2 needed for direct fusion to the host lipid membrane [15] (Fig. 2A). Dacon et al. [29] have recently highlighted the potential of the FP as a target epitope to design next-generation CoV vaccines. Cholesterol affects the extent of SARS-CoV-2 binding and fusion to cellular membranes [16], [17], [30], [31]. In Fig. 2A-B, the SARS-CoV-2 S protein FP is highlighted in red in the pre-fusion conformation of the S glycoprotein. The fusion peptide is precluded from fusing to the target cell, well hidden in the S protein core.
The cholesterol-recognition motifs CARC and its mirror image CRAC of the three pathogenic viruses are shown in Fig. 2C. Interestingly, the S glycoprotein FP region contains a conserved CARC motif in the FP N-term (Fig. 2C), and an almost identical CARC motif (KQYG[D/E]CL) in SARS-CoV and SARS-CoV-2 in their C-term region. Conversely, CARC/CRAC motifs are absent in the FP C-term of another pathogenic CoV, MERS-CoV (Fig. 2C). Madu et al. [32] reported that the SARS-CoV S2 fusion peptide 798 SFIEDLLFNKVTLADAGFMKQY 819 GCGKKKK (linker), which includes ∼90% of the N-term, and ∼40% of the C-term CARC motifs, respectively, promoted a greater extent of lipid mixing in POPC-POPS-cholesterol (1:3:1) liposomes. Mahajan et al. [31], demonstrated that a 64-residue long fusion peptide (LFP) [31], [33], 758 RNTREVFAQVKQMYKTPTLKYFGGFNFSQILPSPLKPTKRSFIEDLLFNKVTLADAGFMKQYGE 821, which includes 7 CARC-motifs and a CRAC motif, exhibits a dose-dependent lipid mixing activity in DMPC liposomes, highlighting its fusogenic potential. An NMR spectroscopy study has recently shown that the FP transforms from an intrinsically disordered structure in solution into a wedge-shaped structure in lipid bicelles, with the hydrophobic, narrow end inserted in the lipid moiety [34]. The FP region that interacts more strongly with bicelles contains the CARC regions shown in Fig. 2C. Santamaria et al. [35] reported that an FP fragment containing the first CARC motif (Fig. 2C) penetrates into the hydrophobic acyl region of the host cell plasma membrane, increasing the dynamics of the fatty acid acyl tails and weakening the membrane structural integrity, possibly a requisite towards viral penetration. Importantly, the increase in membrane flexibility is more pronounced in the more rigid, Lo plasma membrane regions rich in cholesterol [35]. Shen et al. [36] have recently compared the binding modes of SARS-CoV and SARS-CoV-2 FPs using in silico molecular dynamics. They showed that SARS-CoV-2 FP binds to a synthetic POPC/POPE/cholesterol bilayer membrane more effectively than the SARS-CoV FP [36]. Although the amino acid sequences of both FPs are quite similar (Fig. 2C), the corresponding cryo-electron microscopy structures show that the helix length of SARS-CoV FP is longer than that of the SARS-CoV-2 FP [36]. The higher hydrophobicity of the SARS-CoV-2 FP may also contribute to disrupting the target cell membrane [37]. All in all, these structural differences could be responsible for the differential affinities between SARS-CoV and SARS-CoV-2 FPs [36]. The amino acids F817, I818, and L821 present in the short α-helix of the SARS-CoV-2 FP exhibited a stronger interaction with the cell membrane than those of the long α-helix from SARS-CoV [36]. The three hydrophobic amino acid residues involved are part of the N-term CARC motif present in the SARS-CoV-2 FP (Fig. 2C) and are an indispensable requirement for cholesterol binding in the CARC motif [19], [38]. In addition, SARS-CoV FP has two favorable membrane-binding modes, which overlap with the presence of CARC motifs in the N-term and C-term portions of the PF [36] (Fig. 2C). The presence of CARC motifs in the SARS-CoV and SARS-CoV-2 FP could be indicative of an absolute requirement for cholesterol-protein interactions during the early stages of the fusion of the viral particle with the host-cell plasma membrane. The occurrence of a cholesterol-recognition motif in a key region like the FP suggests a relevant biosensor role, guiding the S glycoprotein to cholesterol-rich domains in the host plasma membrane, and improving the efficiency of the virus infectivity.
3.4. Cholesterol recognition motifs in the NTD of the S glycoprotein
Scavenger receptor class B type 1 (SR-B1) is a multifunctional membrane-bound protein mainly expressed in liver and one of the heavy-density lipoprotein (HDL) receptors [3]. Wei and coworkers [27] demonstrated that expression of SR-B1 confers susceptibility to SARS-CoV-2 infection, facilitating the attachment and entry of the virus. Since SR-B1 only enhanced viral uptake in the presence of ACE2, the authors interpreted these last results as an indication that SR-B1 is an entry cofactor of SARS-CoV-2 (reviewed in [3]). Inhibitors of SR-B1 or silencing of its expression abrogates SARS-CoV-2 infection, and the presence of HDL significantly increases viral infection. Blocking the cholesterol/HDL binding site of SARS-CoV-2 with mAb 1D2 strongly reduced HDL-enhanced SARS-CoV-2 infection [27]. The mAb 1D2 overlaps antigenic sites with mAb 48 A, a neutralizing human antibody that recognizes the NTD of the S protein [39] (Fig. 1A). Utilizing cryo-electron microscopy, Chi et al. [39] determined that mAb 4A8 mainly binds to the NTD through three complementarity-determining regions (CDRs). The amino acids Lys147, Lys150, and Tyr145, located in a CRAC motif of NTD-SARS-2-S (Fig. 3A), were identified as important antigenic sites for recognition by mAb 4A8 antibody [27], [39]. Through an in vitro binding assay, Wei et al. [27] found that three cholesterol-binding peptides encompassing the CRAC/CARC region are present in the NTD of SARS-CoV-2: amino acid residues 24–32, 129–150, and 267–277, respectively [27] (Fig. 3A-B). Interestingly, the peptide region that displays the highest interaction with cholesterol is the segment 129–150 [27]. This segment presents two cholesterol recognition motifs along its sequence, i.e., a CARC motif 129KVCEFQFCNDPFL141, and a CRAC motif 141LGVYYHKNNK150 (Fig. 3 C-D; CARC129–141 in yellow, CRAC141–150 in orange). According to Wei et al. [27], SARS-CoV-2 first binds cholesterol and high-density lipoprotein (HDL) (or one of its components). This complex is then recognized by SR-B1 as an entry cofactor, helping to increase the complex concentration in host-cell membrane regions where ACE2 is expressed and facilitating successful encounters with the receptor [3], [27].
It is intriguing how a simple lipid molecule like cholesterol can constitute a molecular target and subsequently bind to a protein motif located outside the plasma membrane. There is, however, a precedent of this molecular interaction: in the Smoothened (SMO) protein, a G protein-coupled receptor, cholesterol binds to the extracellular cysteine-rich domain, located outside the extracellular leaflet of the plasma membrane [40]. Fantini et al. [41], [42] described a ganglioside-binding domain in the NTD, which could enable the binding of SARS-CoV-2 to the plasma membrane-lipid Lo domains. Each spike protein can simultaneously bind three ganglioside molecules. This multivalent binding process could modify the membrane curvature, facilitate lipid coalescence and ACE2 receptor recruitment, thus increasing the chances of finding functional ACE2 receptor molecules in the host membrane [41].
Several mutations in the spike protein contribute to the increased transmissibility of some SARS-CoV-2 variants. The most accepted explanation is that mutations conferring higher affinity of the spike protein for the ACE2 receptor result in more transmissible virus variants. This does not necessarily account for the success of the Omicron variant, suggesting that spike ectodomains directly interact with host plasma membrane bilayers [43]. Overduin et al. [43] recently undertook a detailed in silico study of the membrane-binding surfaces of the spike protein variants, showing that membrane binding propensities increase through different versions, over time, impacting on the S protein’s affinity for cell membranes. Spike protein trimers are shown to shift from initial perpendicular stances to increasingly tilted positions that draw viral particles alongside host cell membranes before engaging ACE2 receptors. This culminates in the assembly of the fusion apparatus, enhancing membrane interactions of the more infective variants [43]. The flexibility of the S glycoprotein has been documented experimentally by cryo-electron microscopy [44] and by coarse-grained molecular dynamics studies showing that the protein can lay almost parallel to the membrane surface [45]. In an end-on view of the spike protein ectodomain, as seen from the host cell (Fig. 4), the area of CRAC and CARC exposed motifs in the NTD and RBD increase from the wild-type Wuhan SARS-CoV-2 to the highly infective Omicron BA.1 variant. The RBD segment 445–461 from SARS-CoV-2[27], which contains in its sequence the CRAC442–454 /CARC441–452 motifs exposed in the spike protein surface, has been experimentally shown to interact with cholesterol [27] (Fig. 4). Even though the segments shown in Fig. 4 are conserved in both SARS-CoV-2 444–461 and Omicron BA.1 441–458 virus variants, differences in the protein conformation may result in a better CRAC/CARC surface exposure in the Omicron variant. Higher exposure of CRAC/CARC motifs on the virus S protein binding surface could contribute to the enhanced ability of the spike protein to interact with the host plasma membrane, and consequently, augment virus infectivity.
4. Conclusions
From the data presented here, we propose two functional attributes of the cholesterol-recognition motifs in the S glycoprotein of the SARS-CoV-2: 1) the presence of CARC and CRAC motifs in the surface of the S trimer (NTD and RBD) that docks onto the host membrane surface is indicative of actual S protein-cholesterol interactions occurring at the earliest stage of virus infection, that is, during the landing of the virion on the host cell surface; 2) the presence of cholesterol-recognition motifs in the FP segment strongly suggests their involvement in the subsequent fusion step. Thus, the presence of the cholesterol consensus regions provides the structural basis for the sequential i) reduction of dimensionality of the virion binding step, i.e., from the random walk in the 3D space to the 2D of the membrane surface, ii) the accelerated diffusion of the S trimer in the target plasma membrane to iii) dock in cholesterol-rich domains containing the ACE2 receptor. The combination of these cholesterol-dependent steps appears to bear direct relevance to the higher infectivity of the Omicron SARS-CoV-2 variant.
Funding
Project PIP 222021-2023 GI to FJB.
CRediT authorship contribution statement
Carlos Javier Baier: Data curation, Conceptualization, Investigation, Methodology, Visualization, Writing - review & editing. Francisco J. Barrantes: Data curation, Conceptualization, Investigation, Methodology, Writing - original draft, Writing - review & editing.
Author contributions
Conceptualization: FJB, CJB; Methodology: FJB, CJB; Investigation: FJB, CJB; Visualization: CJB; Writing: original draft: FJB; review & editing: FJB, CJB.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Data Availability
Data will be made available on request.
References
- 1.Barrantes F.J. The contribution of biophysics and structural biology to current advances in COVID-19. Annu. Rev. Biophys. 2021;50:493–523. doi: 10.1146/annurev-biophys-102620-080956. [DOI] [PubMed] [Google Scholar]
- 2.Barrantes F.J. The unfolding palette of COVID-19 multisystemic syndrome and its neurological manifestations. Brain Behav. Immun. Health. 2021;14 doi: 10.1016/j.bbih.2021.100251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrantes F.J. The constellation of cholesterol-dependent processes associated with SARS-CoV-2 infection. Prog. Lipid Res. 2022;87 doi: 10.1016/j.plipres.2022.101166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maxfield F.R., van Meer G. Cholesterol, the central lipid of mammalian cells. Curr. Opin. Cell Biol. 2010;22:422–429. doi: 10.1016/j.ceb.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schroeder C. Cholesterol-binding viral proteins in virus entry and morphogenesis. Subcell. Biochem. 2010;51:77–108. doi: 10.1007/978-90-481-8622-8_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sousa I.P., Jr., Carvalho C.A.M., Gomes A.M.O. Current understanding of the role of cholesterol in the life cycle of alphaviruses. Viruses. 2020;13 doi: 10.3390/v13010035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sousa I.P., Jr., Carvalho C.A., Ferreira D.F., Weissmuller G., Rocha G.M., Silva J.L., Gomes A.M. Envelope lipid-packing as a critical factor for the biological activity and stability of alphavirus particles isolated from mammalian and mosquito cells. J. Biol. Chem. 2011;286:1730–1736. doi: 10.1074/jbc.M110.198002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barman S., Nayak D.P. Lipid raft disruption by cholesterol depletion enhances influenza A virus budding from MDCK cells. J. Virol. 2007;81:12169–12178. doi: 10.1128/JVI.00835-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bajimaya S., Frankl T., Hayashi T., Takimoto T. Cholesterol is required for stability and infectivity of influenza A and respiratory syncytial viruses. Virology. 2017;510:234–241. doi: 10.1016/j.virol.2017.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nieto-Garai J.A., Arboleya A., Otaegi S., Chojnacki J., Casas J., Fabrias G., Contreras F.X., Krausslich H.G., Lorizate M. Cholesterol in the viral membrane is a molecular switch governing HIV-1 Env clustering. Adv. Sci. 2021;8 doi: 10.1002/advs.202003468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Epand R.F., Thomas A., Brasseur R., Vishwanathan S.A., Hunter E., Epand R.M. Juxtamembrane protein segments that contribute to recruitment of cholesterol into domains. Biochemistry. 2006;45:6105–6114. doi: 10.1021/bi060245+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glende J., Schwegmann-Wessels C., Al-Falah M., Pfefferle S., Qu X., Deng H., Drosten C., Naim H.Y., Herrler G. Importance of cholesterol-rich membrane microdomains in the interaction of the S protein of SARS-coronavirus with the cellular receptor angiotensin-converting enzyme 2. Virology. 2008;381:215–221. doi: 10.1016/j.virol.2008.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li X., Zhu W., Fan M., Zhang J., Peng Y., Huang F., Wang N., He L., Zhang L., Holmdahl R., Meng L., Lu S. Dependence of SARS-CoV-2 infection on cholesterol-rich lipid raft and endosomal acidification. Comput. Struct. Biotechnol. J. 2021;19:1933–1943. doi: 10.1016/j.csbj.2021.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanders D.W., Jumper C.C., Ackerman P.J., Bracha D., Donlic A., Kim H., Kenney D., Castello-Serrano I., Suzuki S., Tamura T., Tavares A.H., Saeed M., Holehouse A.S., Ploss A., Levental I., Douam F., Padera R.F., Levy B.D., Brangwynne C.P. SARS-CoV-2 requires cholesterol for viral entry and pathological syncytia formation. Elife. 2021;10 doi: 10.7554/eLife.65962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang T., Bidon M., Jaimes J.A., Whittaker G.R., Daniel S. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antivir. Res. 2020;178 doi: 10.1016/j.antiviral.2020.104792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meher G., Bhattacharjya S., Chakraborty H. Membrane cholesterol modulates oligomeric status and peptide-membrane interaction of severe acute respiratory syndrome coronavirus fusion peptide. J. Phys. Chem. B. 2019;123:10654–10662. doi: 10.1021/acs.jpcb.9b08455. [DOI] [PubMed] [Google Scholar]
- 17.Pattnaik G.P., Bhattacharjya S., Chakraborty H. Enhanced cholesterol-dependent hemifusion by internal fusion peptide 1 of SARS coronavirus-2 compared to its N-terminal counterpart. Biochemistry. 2021;60:559–562. doi: 10.1021/acs.biochem.1c00046. [DOI] [PubMed] [Google Scholar]
- 18.Nardacci R., Colavita F., Castilletti C., Lapa D., Matusali G., Meschi S., Del Nonno F., Colombo D., Capobianchi M.R., Zumla A., Ippolito G., Piacentini M., Falasca L. Evidences for lipid involvement in SARS-CoV-2 cytopathogenesis. Cell Death Dis. 2021;12:263. doi: 10.1038/s41419-021-03527-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baier C.J., Fantini J., Barrantes F.J. Disclosure of cholesterol recognition motifs in transmembrane domains of the human nicotinic acetylcholine receptor. Sci. Rep. 2011;1:69. doi: 10.1038/srep00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H., Papadopoulos V. Peripheral-type benzodiazepine receptor function in cholesterol transport. Identification of a putative cholesterol recognition/interaction amino acid sequence and consensus pattern. Endocrinology. 1998;139:4991–4997. doi: 10.1210/endo.139.12.6390. [DOI] [PubMed] [Google Scholar]
- 21.Fantini J., Epand R.M., Barrantes F.J. Cholesterol-recognition motifs in membrane proteins. Adv. Exp. Med. Biol. 2019;1135:3–25. doi: 10.1007/978-3-030-14265-0_1. [DOI] [PubMed] [Google Scholar]
- 22.Lee A.G. Interfacial binding sites for cholesterol on TRP ion channels. Biophys. J. 2019;117:2020–2033. doi: 10.1016/j.bpj.2019.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181 doi: 10.1016/j.cell.2020.02.052. 271-280.e278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cai Y., Zhang J., Xiao T., Peng H., Sterling S.M., Walsh R.M., Rawson S., Rits-Volloch S., Chen B. Distinct conformational states of SARS-CoV-2 spike protein. Science. 2020;369:1586–1592. doi: 10.1126/science.abd4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koppisetti R.K., Fulcher Y.G., Van Doren S.R. Fusion peptide of SARS-CoV-2 spike rearranges into a wedge inserted in bilayered micelles. J. Am. Chem. Soc. 2021 doi: 10.1021/jacs.1c05435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.X. Xia, Domains and Functions of Spike Protein in SARS-Cov-2 in the Context of Vaccine Design, 13 (2021) 109. [DOI] [PMC free article] [PubMed]
- 27.Wei C., Wan L., Yan Q., Wang X., Zhang J., Yang X., Zhang Y., Fan C., Li D., Deng Y., Sun J., Gong J., Yang X., Wang Y., Wang X., Li J., Yang H., Li H., Zhang Z., Wang R., Du P., Zong Y., Yin F., Zhang W., Wang N., Peng Y., Lin H., Feng J., Qin C., Chen W., Gao Q., Zhang R., Cao Y., Zhong H. HDL-scavenger receptor B type 1 facilitates SARS-CoV-2 entry. Nat. Metab. 2020;2:1391–1400. doi: 10.1038/s42255-020-00324-0. [DOI] [PubMed] [Google Scholar]
- 28.S. Lall, P. Balaram, S. Gosavi, M.K. Mathew, Dynamics and self-assembly of the SARS-CoV-2 spike transmembrane domain, (2021) 2021.2006.2007.447334.
- 29.Dacon C., Tucker C., Peng L., Lee C.D., Lin T.H., Yuan M., Cong Y., Wang L., Purser L., Williams J.K., Pyo C.W., Kosik I., Hu Z., Zhao M., Mohan D., Cooper A.J.R., Peterson M., Skinner J., Dixit S., Kollins E., Huzella L., Perry D., Byrum R., Lembirik S., Drawbaugh D., Eaton B., Zhang Y., Yang E.S., Chen M., Leung K., Weinberg R.S., Pegu A., Geraghty D.E., Davidson E., Douagi I., Moir S., Yewdell J.W., Schmaljohn C., Crompton P.D., Holbrook M.R., Nemazee D., Mascola J.R., Drawbaugh I.A., Tan J. Broadly neutralizing antibodies target the coronavirus fusion peptide. Science. 2022 doi: 10.1126/science.abq3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Correa Y., Waldie S., Thepaut M., Micciula S., Moulin M., Fieschi F., Pichler H., Trevor Forsyth V., Haertlein M., Cardenas M. SARS-CoV-2 spike protein removes lipids from model membranes and interferes with the capacity of high density lipoprotein to exchange lipids. J. Colloid Interface Sci. 2021;602:732–739. doi: 10.1016/j.jcis.2021.06.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mahajan M., Chatterjee D., Bhuvaneswari K., Pillay S., Bhattacharjya S. NMR structure and localization of a large fragment of the SARS-CoV fusion protein: Implications in viral cell fusion. Biochim. Biophys. Acta Biomembr. 2018;1860:407–415. doi: 10.1016/j.bbamem.2017.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Madu I.G., Roth S.L., Belouzard S., Whittaker G.R. Characterization of a highly conserved domain within the severe acute respiratory syndrome coronavirus spike protein S2 domain with characteristics of a viral fusion peptide. J. Virol. 2009;83:7411–7421. doi: 10.1128/JVI.00079-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mahajan M., Bhattacharjya S. NMR structures and localization of the potential fusion peptides and the pre-transmembrane region of SARS-CoV: Implications in membrane fusion. Biochim. Biophys. Acta. 2015;1848:721–730. doi: 10.1016/j.bbamem.2014.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koppisetti R.K., Fulcher Y.G., Van Doren S.R. Fusion peptide of SARS-CoV-2 spike rearranges into a wedge inserted in bilayered micelles. J. Am. Chem. Soc. 2021;143:13205–13211. doi: 10.1021/jacs.1c05435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Santamaria A., Batchu K.C., Matsarskaia O., Prevost S.F., Russo D., Natali F., Seydel T., Hoffmann I., Laux V., Haertlein M., Darwish T.A., Russell R.A., Corucci G., Fragneto G., Maestro A., Zaccai N.R. Strikingly different roles of SARS-CoV-2 fusion peptides uncovered by neutron scattering. J. Am. Chem. Soc. 2022;144:2968–2979. doi: 10.1021/jacs.1c09856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen H., Wu Z., Chen L. Different binding modes of SARS-CoV-1 and SARS-CoV-2 fusion peptides to cell membranes: the influence of peptide helix length. J. Phys. Chem. B. 2022 doi: 10.1021/acs.jpcb.2c01295. [DOI] [PubMed] [Google Scholar]
- 37.Lai A.L., Freed J.H. SARS-CoV-2 fusion peptide has a greater membrane perturbating effect than SARS-CoV with highly specific dependence on Ca2+ J. Mol. Biol. 2021;433 doi: 10.1016/j.jmb.2021.166946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Di Scala C., Baier C.J., Evans L.S., Williamson P.T.F., Fantini J., Barrantes F.J. Relevance of CARC and CRAC cholesterol-recognition motifs in the nicotinic acetylcholine receptor and other membrane-bound receptors. Curr. Top. Membr. 2017;80:3–23. doi: 10.1016/bs.ctm.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 39.Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M., Zhang Z., Fan P., Dong Y., Yang Y., Chen Z., Guo Y., Zhang J., Li Y., Song X., Chen Y., Xia L., Fu L., Hou L., Xu J., Yu C., Li J., Zhou Q., Chen W. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kinnebrew M., Woolley R.E., Ansell T.B., Byrne E.F.X., Frigui S., Luchetti G., Sircar R., Nachtergaele S., Mydock-McGrane L., Krishnan K., Newstead S., Sansom M.S.P., Covey D.F., Siebold C., Rohatgi R. Patched 1 regulates Smoothened by controlling sterol binding to its extracellular cysteine-rich domain. Sci. Adv. 2022;8 doi: 10.1126/sciadv.abm5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fantini J., Chahinian H., Yahi N. Leveraging coronavirus binding to gangliosides for innovative vaccine and therapeutic strategies against COVID-19. Biochem. Biophys. Res. Commun. 2021;538:132–136. doi: 10.1016/j.bbrc.2020.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fantini J., Chahinian H., Yahi N. Synergistic antiviral effect of hydroxychloroquine and azithromycin in combination against SARS-CoV-2: What molecular dynamics studies of virus-host interactions reveal. Int. J. Antimicrob. Agents. 2020;56 doi: 10.1016/j.ijantimicag.2020.106020. [DOI] [PubMed] [Google Scholar]
- 43.Overduin M., Kervin T.A., Tran A. Progressive membrane-binding mechanism of SARS-CoV-2 variant spike proteins. iScience. 2022;25 doi: 10.1016/j.isci.2022.104722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.B. Turoňová, M. Sikora, C. Schürmann, W.J.H. Hagen, S. Welsch, F.E.C. Blanc, S. von Bülow, M. Gecht, K. Bagola, C. Hörner, G. van Zandbergen, J. Landry, N.T.D. de Azevedo, S. Mosalaganti, A. Schwarz, R. Covino, M.D. Mühlebach, G. Hummer, J. Krijnse Locker, M. Beck, In situ structural analysis of SARS-CoV-2 spike reveals flexibility mediated by three hinges, (2020) eabd5223. [DOI] [PMC free article] [PubMed]
- 45.Wang B., Zhong C., Tieleman D.P. Supramolecular organization of SARS-CoV and SARS-CoV-2 virions revealed by coarse-grained models of intact virus envelopes. J. Chem. Inf. Model. 2022;62:176–186. doi: 10.1021/acs.jcim.1c01240. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.