SUMMARY
Candida albicans is the most common cause of fungal infection in humans. IL-17 is critical for defense against superficial fungal infections but the role of this response in invasive disease is less understood. We show that C. albicans secretes a lipase, Lip2, that faciliates invasive disease via lipid-based suppression of the IL-17 response. Lip2 was identified as an essential virulence factor in a forward genetic screen in a mouse model of bloodstream infection. Murine infection with C. albicans strains lacking Lip2 display exaggerated IL-17 responses that lead to fungal clearance from solid organs and host survival. Both IL-17 signaling and lipase activity are required for Lip2-mediated suppression. Lip2 inhibits IL-17 production indirectly by suppressing IL-23 production by tissue resident dendritic cells. Lipase hydrolysis product palmitic acid similarly suppresses dendritic cell activation in vitro. Thus, C. albicans suppresses antifungal IL-17 defense in solid organs by altering the tissue lipid milieu.
Keywords: Candida albicans, fungal pathogenesis, virulence factor, IL-17
In Brief:
In a murine model of systemic fungal infection, Basso et al. show that C. albicans secreted lipase, Lip2, suppresses local anti-fungal IL-17 responses in infected organs by altering dendritic cell activation. Under in vitro conditions, the immunosuppressive activity of Lip2 is mimicked by palmitic acid, a product of Lip2 hydrolysis.
Graphical Abstract
INTRODUCTION
Fungal pathogens kill roughly 1.5 million people every year (Brown et al., 2012). Despite the enormous burden of fungal infectious disease, our understanding of pathogenic fungi lags well behind that of viruses, bacteria, and parasites of similar clinical importance, and we are only beginning to define rules governing interactions between fungi and mammalian hosts. This knowledge gap has contributed to a dearth of tools for timely diagnosis and treatment of fungal infections, and no vaccines are available for prevention of any fungal disease (Brown et al., 2012).
Unusually for a fungus, the yeast Candida albicans is a stable component of mammalian gut, skin, and genitourinary microbiota (Hallen-Adams and Suhr, 2017). Commensal colonization is asymptomatic, and recent studies suggest that gut colonization confers immune benefits to the host (Bacher et al., 2019; Jiang et al., 2017). In contrast, fungal overgrowth or escape into ectopic niches can produce a spectrum of noxious symptoms (Pfaller et al., 2006). The most common syndrome of “mucocutaneous candidiasis” comprises relatively superficial infections of the mouth, skin, nails, and female reproductive tract. Superficial candidiasis occurs in hosts of any age or immune status, and there are hundreds of millions of such cases every year, across the globe (Denning et al., 2018). By contrast, “systemic candidiasis” describes rarer, invasive infections that involve the bloodstream and internal organs. Unlike mucocutaneous disease, systemic infections occur most often in patients with underlying risk factors, such as immunodeficiency (either spontaneous or drug-induced), epithelial damage (such as following surgery and other invasive procedures), and microbial dysbiosis (typically following treatment with antibiotics)(Pfaller and Diekema, 2007). In keeping with the high concentration of these risk factors among hospitalized patients, Candida species are currently the fourth most common cause of bloodstream infection in US hospitals (Edmond et al., 1999), and mortality from systemic disease remains high at ~40% (Pfaller et al., 2012).
Insights into host defenses against candidiasis have emerged from studies of families with inherited predispositions to such infections. For example, the critical role played by IL-17 signaling in defense against superficial fungal infections emerged from studies of families with chronic mucocutaneous candidiasis (CMC), a syndrome of persistent or recurrent C. albicans infections of the mouth, skin, nails, and/or vulvovaginal tract (Huppler et al., 2012). CMC has been linked to independent mutations affecting IL-17F (Puel et al., 2011), a component of IL-17 cytokines, as well IL-17RA and IL-17RC (Levy et al., 2016; Ling et al., 2015; Puel et al., 2011), which together compose the heterodimeric IL-17 receptor. IL-17 signaling molecules are highly conserved, and null alleles affecting mouse orthologs also confer enhanced susceptibility to superficial C. albicans infections (Conti et al., 2009). Interestingly, however, humans with isolated CMC or treated with inhibitors of IL-17 signaling are not at higher risk for C. albicans bloodstream infections (Sparber and LeibundGut-Landmann, 2019), reinforcing the perception that IL-17 plays a less significant role controlling systemic fungal disease.
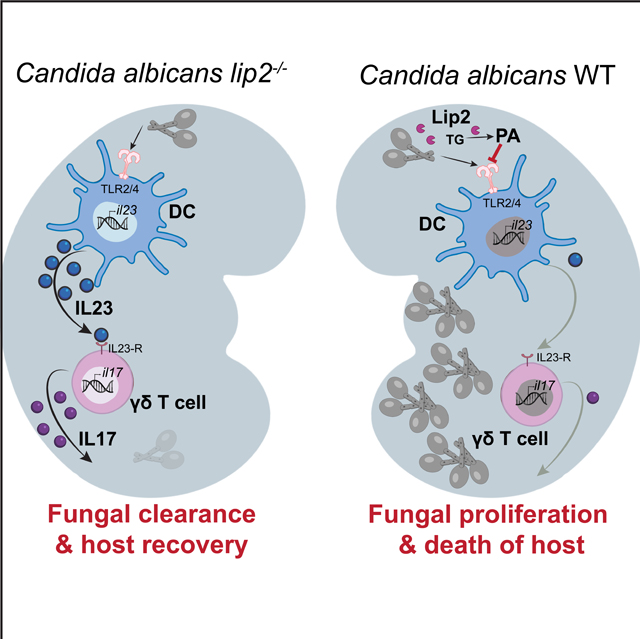
In this study, we identify a C. albicans secreted lipase, Lip2, that is required for virulence in a mouse model of systemic fungal infection. Unlike the vast majority of mutants with defects in this bloodstream infection model (Noble et al., 2010), strains that lack LIP2 undergo normal yeast-to-hypha morphogenesis, the best-studied virulence attribute of this pathogen. Rather, infection with a lip2 null mutant triggers an exaggerated IL-17A immune response in infected kidneys, which is followed by rapid fungal clearance and survival of the host. Virulence of the lip2 mutant is fully restored in Il17af−/− animals that fail to produce IL-17A and IL-17F, suggesting that IL-17 may play an unanticipated role in promoting fungal clearance from systemically infected animals. Using flow cytometry, we discovered that IL-17A production is induced in renal TCRγδ+ T cells within 6 hours of systemic infection with C. albicans, especially the lip2 mutant. Moreover, IL-23, which is known to stimulate IL-17 expression by TCRγδ+ T cells (Sutton et al., 2009), is upregulated in renal dendritic cells (DCs) with similar kinetics. To test the hypothesis that DCs are directly activated by exposure to C. albicans, we performed in vitro coculture experiments with bone marrow-derived DCs (BMDCs). Unlike WT C. albicans or a lip2+LIP2 complemented strain, the lip2 null mutant and strains expressing catalytically inactive variants of Lip2 trigger IL-23A secretion into culture supernatants. Strikingly, fungal activation of BMDCs is inhibited in the presence of 0.1 𝛍M palmitic acid, a product of Lip2 lipase activity. These findings support a model in which C. albicans utilizes Lip2 to modify the lipid milieu of infected organs during systemic infection, thereby inhibiting activation of tissue resident DCs and preventing an IL-17-dependent antifungal immune response.
RESULTS
Lip2 is critical for C. albicans pathogenicity during systemic infection.
We identified Lip2 as a candidate fungal virulence factor in a competitive screen of ~1500 C. albicans GRACE (Gene Replacement And Conditional Expression) mutants (Roemer et al., 2003) in a mouse model of bloodstream infection (Basso et al., manuscript in preparation). Each barcoded GRACE strain contains one null allele of a specific open reading frame (ORF), as well as an intact copy of the ORF whose expression is controlled by a doxycycline (DOX)-repressible promoter; note that the C. albicans genome is diploid. Strains were propagated individually in the presence of DOX (0.1 mM DOX in liquid YEPD), followed by pooling of up to 96 strains into 20 mixed inocula. 5×105 colony forming units (CFUs) of each inoculum was introduced into 5 BALB/c mice by retroorbital injection, and an aliquot was plated onto Sabouraud agar (with ampicillin and gentamicin) as the “inoculum” sample. Infected animals were maintained on DOX-containing drinking water (0.25 mg/mL) until they developed clinical morbidity criteria (body condition score ≤2, respiratory distress, and/or immobility), typically after 5–7 days of systemic infection. Kidneys (the primary target organ in this infection model) from each euthanized animal were homogenized and plated onto Sabouraud agar (with ampicillin and gentamicin) as “recovered” samples. Finally, genomic DNA from the inoculum and recovered samples was used to generate barcode sequencing libraries, and relative strain abundance was determined by Illumina sequencing, as previously described (O’Meara et al., 2015). The competitive index (CI) of each mutant in a given animal was calculated as the log2 function of its relative abundance in the recovered pool compared to its relative abundance in the inoculum.
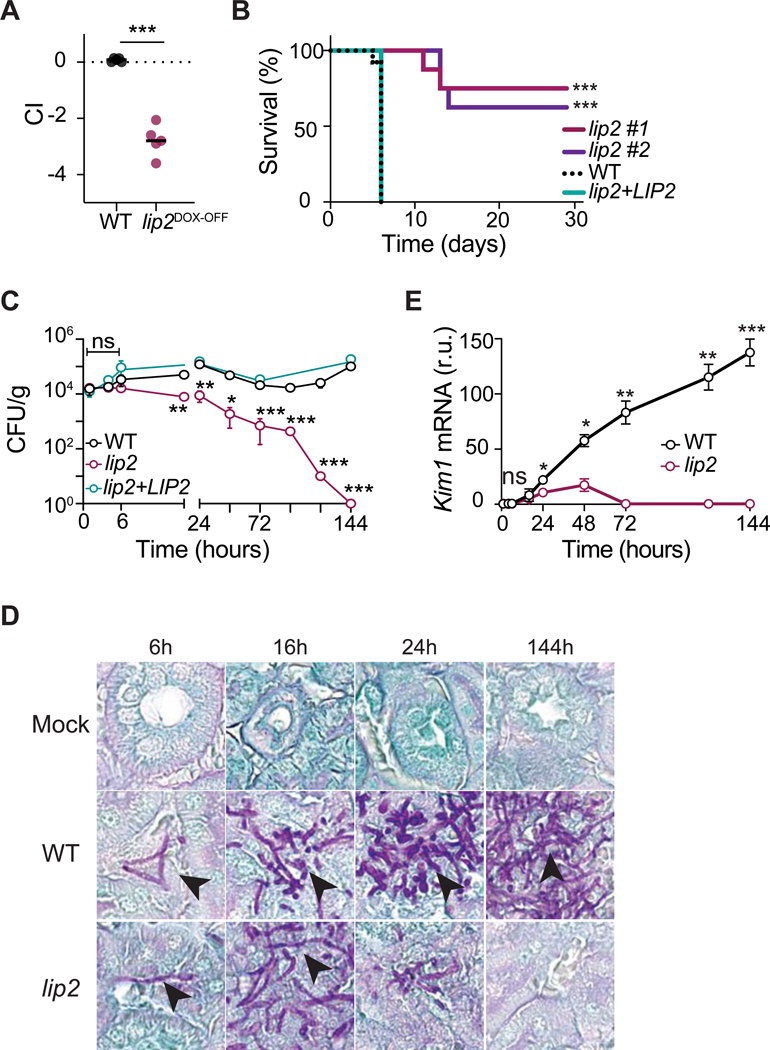
The lip2DOX-OFF mutant was one of the first loss-of-fitness mutants to be identified in the virulence screen; comprehensive results for additional virulence mutants as well as strains identified in a parallel screen for gut commensalism factors will be reported separately (Basso et al., in preparation). To validate the virulence defect of lip2DOX-OFF, we retested the GRACE strain in 1:1 competition against WT C. albicans in the presence of DOX. As shown in Figure 1A, lip2DOX-OFF was strongly outcompeted by WT in kidneys of all five systemically infected animals, similar to results obtained in the primary screen. We next compared two independent lip2 null mutants, which were constructed in a different strain background (SN152), to an isogenic WT strain and a lip2+LIP2 gene addback strain in monotypic (single strain) infections. As shown in Figure 1B, both lip2 mutants exhibited significantly reduced lethality compared to the WT and complemented strains (Figure 1B). These results establish that LIP2 is required for virulence during systemic infection.
Figure 1. Lip2 is required for pathogenicity.
(A) lip2DOX-OFF is defective for systemic virulence in the presence of doxycycline. Female BALB/c mice were treated with 0.25 mg/ml doxycycline via drinking water for seven days prior to retro-orbital injection with 1×105 CFU of a 1:1 mixture of WT and lip2DOX-OFF; relative strain abundance in kidneys was determined using qPCR. Statistical significance was determined by a paired two-tailed t-test; *** p<0.001.
(B) lip2 mutants exhibit reduced lethality compared to WT or a lip2+LIP2 gene addback strain. Groups of female BALB/c mice were injected with 1×105 CFU WT (n=16), lip2 (isolate 1, n=8), lip2 (isolate 2, n=8) or lip2+LIP2 (n=8). Statistical significance between WT and lip2 #1 or lip2 #2 was determined by Mantel-Cox test *** p<0.001.
(C) lip2 fails to persist in infected kidneys. Groups of female BALB/c mice were infected with 1×105 CFU of WT, lip2, or lip2+LIP2, followed by euthanasia of three animals per group at the indicated time points. CFUs were determined by plating right kidney homogenates onto Sabouraud agar (supplemented gentamicin and ampicillin) and counting after two days. Data represent the means and standard deviations. Statistical significance between WT and lip2 was determined by an unpaired two-tailed t-test. ns nonsignificant, ** p<0.01; *** p<0.001. (Note that points without visible error bars displayed SEMs smaller than the circle.)
(D) lip2 forms normal-appearing hyphae in host kidneys. 400x image of PAS-stained left kidneys from experiment described in (C).
(E) lip2 causes minimal damage to kidneys. Kim1 mRNA was measured in the right kidney homogenates and normalized to Gadph. Statistical significance was determined by an unpaired two-tailed t-test. * p<0.05; ** p<0.01; *** p<0.001; **** p<0.0001. (Note that points without visible error bars displayed SEMs smaller than the circle.)
The best-known virulence attribute of C. albicans is its ability to transition from single-celled budding yeast into elongated, multicellular hyphae (Noble et al., 2017). Unlike yeast, hyphae are naturally invasive and express virulence factors such as adhesins, tissue-degrading enzymes, and the secreted toxin Candidalysin (Noble et al., 2017). The vast majority of published C. albicans virulence mutants exhibit defects in yeast-to-hypha morphogenesis (Noble et al., 2010). To determine whether LIP2 is required for hypha formation, lip2 and WT strains were profiled under multiple in vitro hypha-inducing conditions. As shown in Figure S1A, lip2 and WT exhibited equivalent patterns of filamentation in the “germ tube” clinical diagnostic assay, as well as in YEPD+5% serum, Spider, and RPMI media maintained at 37°C. Likewise, germination rates for lip2 and WT were similar throughout a 180-minute time course (Figure S1B). These results suggest that LIP2 is dispensable for morphogenesis. Exposure of WT C. albicans to hypha-promoting in vitro culture conditions results in upregulation of LIP2 gene expression (by RT-qPCR, Figure S1C), suggesting that it may encode a hypha-specific virulence factor.
To further characterize the virulence defect of the lip2 null mutant, we performed a series of monotypic, timed infections with lip2, WT, or saline (mock infection); the lip2+LIP2 strain was also tested in a subset of animals. Three BALB/c mice per comparison group were euthanized after 1, 4, 6,16, 24, 48, 72, 96, 120, and 144 hours of systemic infection, followed by analysis of kidneys for fungal burden, pattern of fungal invasion, tissue damage, and expression of selected host transcripts. As shown in Figure 1C, lip2, WT, and lip2+LIP2 were equally represented in kidneys (~104 CFU/g) after one, four, and six hours of infection. By 18 hours, however, the burden of lip2 was significantly reduced compared to the other two strains. Whereas titers of WT and the lip2+LIP2 strain remained relatively stable (~104-105 CFU/gm kidney) throughout the infectious time course, titers of lip2 progressively declined until becoming undetectable at 144 hours. Fungal titers were also determined in the liver and spleen at selected time points (Figure S2A). The three fungal strains exhibited similar abundances (~103 CFU/g) in both organs after 1 hour. In the liver, titers of WT and lip2+LIP2 remained relatively stable throughout the time course, but lip2 became undetectable by 144 hours. In contrast, all three fungal strains were rapidly cleared from the spleen. This analysis indicates that LIP2 is not necessary for the initial entry of C. albicans into solid organs, but it is required for maintenance of a stable infection in the kidney and liver.
Patterns of fungal invasion were assessed in sectioned left kidneys after staining with Periodic Acid Schiff (PAS). As shown in Figures 1D and S2B, lip2 formed normal-appearing hyphae in kidneys, suggesting that lip2 is competent for morphogenesis in the host as well as under in vitro conditions (Figure S1A and S1B). However, stark differences were apparent in the extent of renal invasion by lip2 vs. WT. By 16 hours, large clusters of WT hyphae could be visualized throughout the renal cortex (Figure S2C), a highly perfused region that receives ~25% of cardiac output (Lionakis et al., 2011; Rajendran et al., 2013). Over the remainder of the time course, the areas infiltrated by WT C. albicans enlarged and coalesced until, by 144 hours, they spanned the renal cortex, corticomedullary junction, and portions of the medulla. In contrast, lip2 formed much smaller collections within the renal cortex at 16 hours (Figure S2B and S2C), and these areas diminished in size over subsequent time points, becoming virtually undetectable by 144 hours.
Kidney damage was evaluated histologically on hematoxylin and eosin (H&E)-stained left kidney sections. Starting at 16 hours, WT-infected kidneys exhibited areas of tissue necrosis with microabscess formation at the sites of hyphal infiltration in the renal cortex (Figure S2D). These areas of injury expanded and progressed over the subsequent time course, resulting in interstitial edema, widespread acute tubular injury and parenchymal necrosis, and frank hemorrhage. By contrast, lip2-infected organs remained free of significant injury throughout the time course, despite the presence of localized microabscesses in the cortex that peaked at 24 hours and subsequently resolved (Figure S2D). We next used RT-qPCR to evaluate right kidney homogenates for Kim1 (Kidney Injury Molecule-1) mRNA, a highly sensitive marker of acute renal injury (Bonventre, 2014). As shown in Figure 1E, during infection with WT C. albicans, renal Kim1 levels were elevated throughout the time course and progressive increased until the end of observation at 144 hours. By comparison, Kim1 was briefly elevated at 24 and 48 hours in lip2-infected organs, albeit to levels that were significantly lower than in WT-infected organs, followed by a return to baseline at subsequent time points (Figure 1E). These results indicate that, consistent with its reduced titers and lesser degree of renal infiltration after ~24 hours, lip2 causes only minor, transient injury to the host.
IL-17 triggers an antifungal immune response during invasive candidiasis.
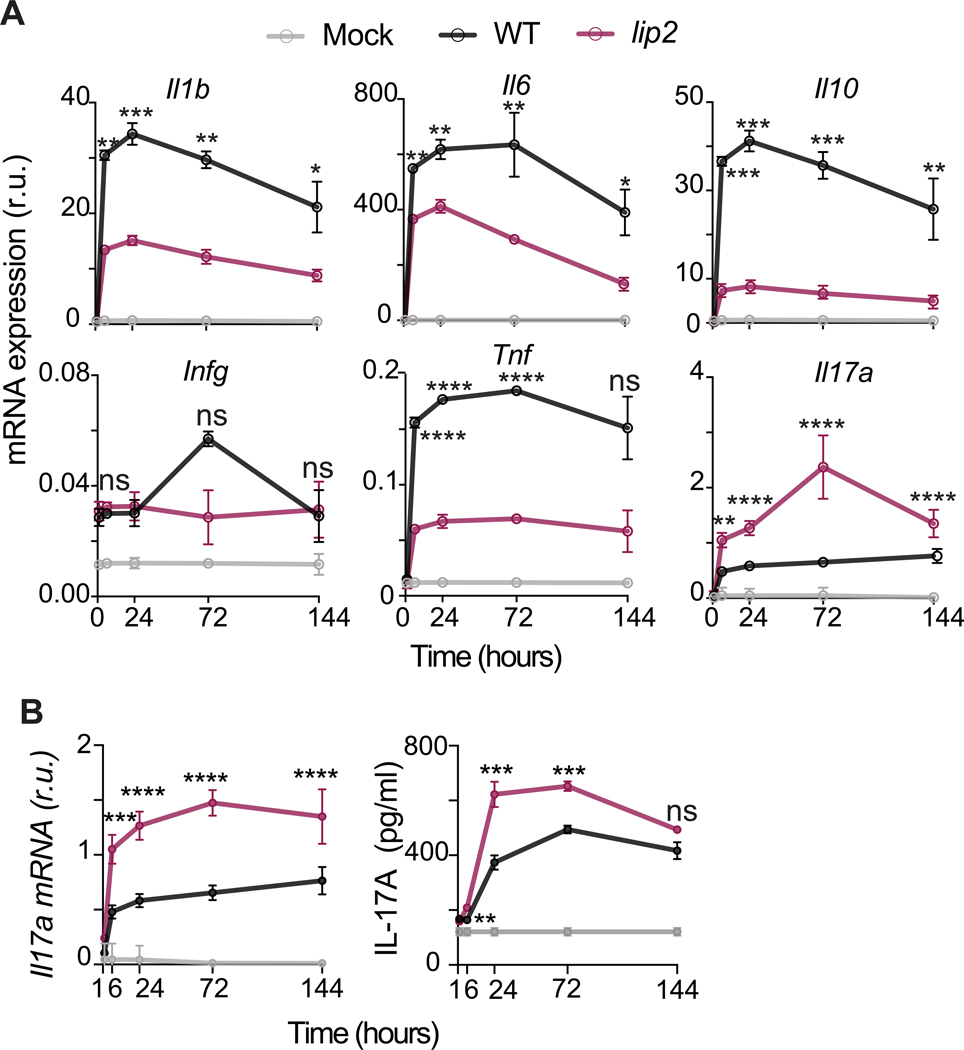
We considered two potential explanations for the persistence defect of lip2 in infected organs: The encoded lipase may be required for fungal proliferation, for example because of a role in nutrient acquisition, or it may defend against clearance by the host. To gauge the host immune response to lip2 versus WT strains in infected kidneys, we quantified transcript levels for six proinflammatory cytokines in whole kidney homogenates using RT-qPCR. As shown in Figure 2A, expression of Il1β, Il6, Il10, and Tnf was significantly higher in kidneys infected with WT compared to lip2, whereas Ifng was minimally induced by either strain. Il17a was the only cytokine to be expressed more strongly in lip2-infected organs, as early as 6 hours after infection. The latter result was confirmed in additional animals, confirming elevation of Il17a mRNA and, following a delay, IL-17A protein in kidneys infected with lip2 (Figure 2B).
Figure 2. lip2 induces an elevated IL-17 response in kidneys.
(A) lip2 provokes an exaggerated Il17a response in infected kidneys. Groups of female BALB/c mice were infected with 1×105 CFU of WT or lip2, followed by euthanasia of three animals per group at the indicated time points. Il1b, Il6, Il10, Ifng, Tnf, Il17a mRNA expression was assessed by RT-qPCR of RNA recovered from kidney homogenates (expression relative to Gadph). Statistical significance of differences between WT-infected kidneys and lip2-infected kidneys was determined by an unpaired two-tailed t-test. * p<0.05; ** p<0.01; *** p<0.001; **** p<0.0001. (Note that points without visible error bars displayed SEMs smaller than the circle.)
(B) lip2 induces a strong renal IL-17A response during systemic infection. Groups of BALB/c mice were infected with 1×105 CFU WT or lip2 followed by euthanasia of three animals per group at the indicated time points. Il17a mRNA was measured by RT-qPCR in the right kidney homogenates and normalized to Gadph (left panel). IL-17A protein production was evaluated by ELISA in the left kidney homogenates (right panel). Statistical significance between WT and lip2 was determined by an unpaired two-tailed t-test. ns: p=0.1797; ** p<0.01; *** p<0.001; **** p<0.0001.
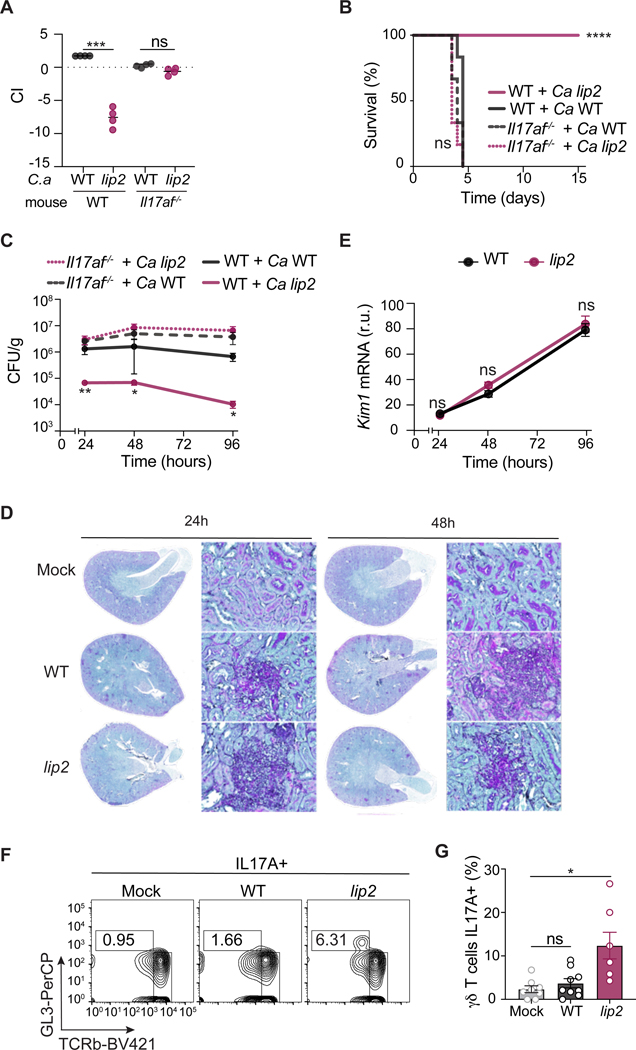
We reasoned that, if IL-17 plays an important role in eliminating lip2 from infected kidneys, then the virulence defect of the mutant may be suppressed in animals that fail to produce IL-17. Consistent with this prediction, lip2 exhibited normal competitive fitness with WT C. albicans in Il17af−/− mice, in contrast to the strong competitive defect observed in WT (Il17af+/+) littermate control animals (Figure 3A). Similarly, lip2 demonstrated similar lethality to that of WT C. albicans in monotypic infections of Il17af−/− mice, whereas the same dose of the mutant provoked no mortality in WT (Il17af+/+) littermates (Figure 3B). We confirmed that lip2 triggers elevated levels of Il17a RNA and IL-17A protein in kidneys of C57B/6J mice (the genetic background of Il17af−/− animals; Figure S3A) as previously observed in BALB/c mice (Figure 2B). To determine the impact of IL-17 on fungal clearance from infected internal organs, we compared fungal titers in kidneys of Il17af−/− vs. WT (Il17af+/+) animals infected with lip2 or WT C. albicans. As shown in Figure 3C, lip2 and WT strains exhibited nearly identical abundance (~107 CFU/gram kidney) in kidneys of Il17af−/− animals throughout a 96-hour time course, whereas the lip2 null mutant was substantially depleted at all monitored time points in WT mice. Analysis of renal histology (Figure 3D) and Kim1 mRNA levels (Figure 3E) in infected kidneys revealed that the defects of the lip2 strain in deep organ penetration and kidney damage are also suppressed in Il17af−/− animals. Finally, using RT-qPCR to evaluate the expression of multiple cytokines (IL-1β, IL-6, IL-10, and IFN𝛄) in infected kidneys, we confirmed that lip2 and WT C. albicans elicit similar cytokine responses in Il17af−/− animals (Figure S3B). These results argue that host expression of IL-17 is required for the observed fitness defects of C. albicans lip2 in a mouse model of invasive infection. Conversely, the enhanced virulence of lip2 in Il17af−/− animals suggests that IL-17 plays an important role in defending WT animals from systemic fungal infection.
Figure 3. IL-17 mediates an antifungal response during systemic infection.
(A) Competitive fitness of lip2 is restored in Il17af−/− mice. Groups of four (male and female) WT or Il17af−/− C57BL/6J mice were systemically infected with 5×105 CFU of a 1:1 mixture of WT and lip2. Mice were euthanized after four days and relative strain abundance in kidneys was determined using qPCR. Statistical significance between WT and lip2 was determined by a paired two-tailed t-test; ns: p= 0.0685; ***p < 0.001.
(B) Virulence of lip2 in monotypic infections is restored in Il17af−/− mice. Groups of WT or Il17af−/− C57BL/6 (male and female) mice were systemically infected with 5×105 CFU of WT or lip2 (n=8 per group). Statistical significance between WT and lip2 was determined by Mantel-Cox test ***p < 0.001.
(C) Clearance of lip2 from kidneys requires IL-17. Groups of three WT or Il17af−/− C57BL/6J (male and female) mice were infected with 5×105 CFU of WT or lip2. Animals were euthanized after 24,48 or 96 hours and the fungal burden in kidneys was determined by plating right kidney homogenates onto Sabouraud agar (supplemented with gentamicin and ampicillin). Statistical significance was determined by one-way ANOVA (Tukey’s multiple comparisons test); *p < 0.05; **p<0.01
(D) lip2 and WT C. albicans induce similar amounts of renal damage in Il17af−/− C57BL/6J mice (male and female were used). 400x image of PAS-stained left kidneys from the experiment described in (C).
(E) Kim1 mRNA levels in Il17af−/− C57BL/6J animals infected with lip2 or WT C. albicans. RT-qPCR was used to quantify Kim1 mRNA in right kidney homogenates, and results were normalized to Gadph. Statistical significance was determined by an unpaired two-tailed t-test ns: p=0.6130. (Note that points without visible error bars displayed SEMs smaller than the circle.) (F, G) Representative flow cytometry plots and quantification of renal TCRγδ+ T cells producing IL-17A upon infection with lip2. Groups of C57BL/6J (male and female) mice were infected with 1×106 CFU of WT or lip2. Animals were euthanized after six hours. Flow cytometry was performed on total renal cells after four hours of stimulation with PMA, ionomycin, and Golgi STOP. F) Flow cytometry plots for IL17A+ TCRγδ+ cells for one animal from each group (see Figure S3C for complete FACS gating). G) Percentage of renal TCRγδ+ T cells producing IL-17A from C. albicans–infected kidneys (gated on live CD45+ cells). Statistical significance was determined by one-way ANOVA (Tukey’s multiple comparisons test); **p < 0.01.
To probe the mechanism by which Lip2 modulates host immunity, we began by asking which cells produce IL-17 in infected kidneys. Th17 cells are generally considered to be a major source of IL-17 in mammals (Huppler et al., 2012; Sparber and LeibundGut-Landmann, 2019), and this cell type has previously been shown to protect against systemic candidiasis in mice that have been intestinally colonized with C. albicans (Shao et al., 2019). However, CD4+ T cells are poorly represented in mouse kidneys during the first 24 hours of C. albicans bloodstream infection (Lionakis et al., 2011), whereas we observe increases in renal Il17a mRNA within 6 hours (Figures 2A and 2B). We therefore focused on tissue-resident immune cells that are capable of producing IL-17, such as natural killer T cells (NKT), innate lymphoid cells (ILC), and TCRγδ+ T cells; note that TCRγδ+ T cells have previously been reported to produce IL-17 in kidneys during systemic candidiasis (Ramani et al., 2016). Kidneys were recovered from C57B/6J mice 6 and 24 hours after inoculation with the lip2 mutant, WT, or normal saline (“mock” infection), followed by cell dissociation and staining for intracellular IL-17A and leukocyte surface markers. As shown in Figures 3F, 3G, and S3C, TCRγδ+ T cells (but not ILC3s or CD4+ T cells) were identified as a major source of IL-17 in lip2-infected kidneys. Similar results were obtained using SMART17A mice (in the C57B/6J background) that display a human low-affinity nerve growth factor receptor marker on the surface of cells that express mouse Il17a (Figure S3D) (Price et al., 2012).
Lip2 suppresses the activation of renal dendritic cells.
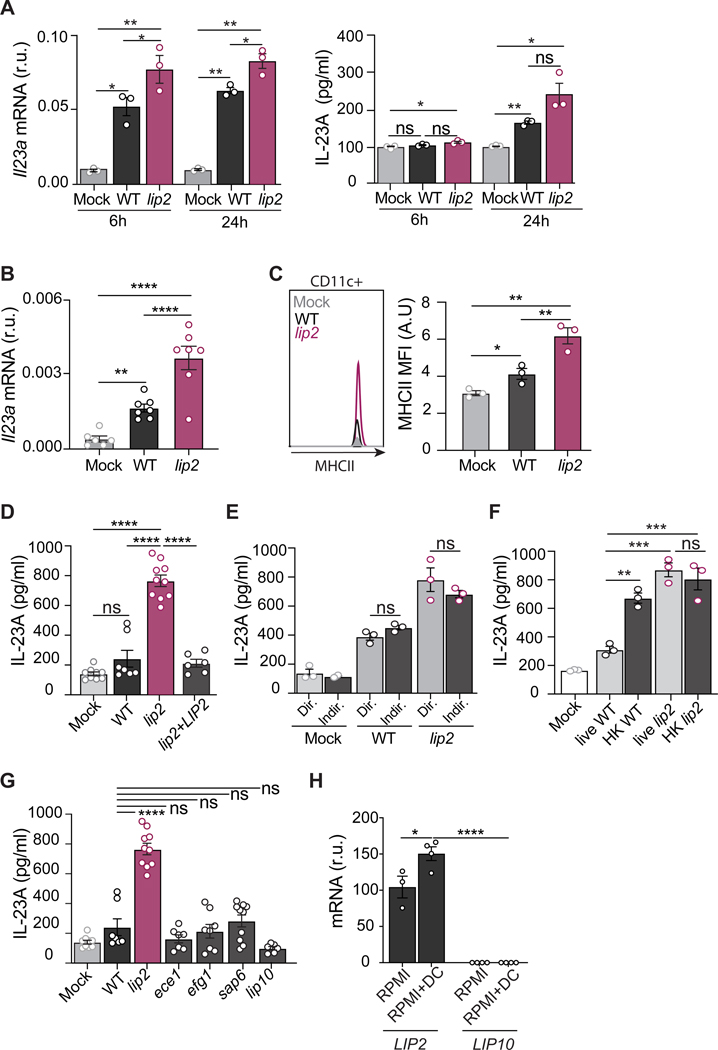
Because TCRγδ+ T cells typically upregulate IL-17 in response to an upstream cytokine, such as IL-23 (Sparber and LeibundGut-Landmann, 2019), we investigated the expression of Il23 in infected kidneys. As shown in Figure 4A, renal Il23 mRNA was significantly elevated within 6 hours of infection with WT C. albicans, and the effect was even stronger in lip2-infected organs. A similar pattern was observed with IL-23 protein, following a delay (Figure 4A). Macrophages and dendritic cells (DC) are common sources of IL-23 in solid organs. Using flow cytometry, DCs and macrophages were quantified in kidneys recovered after 6 hours of infection by WT or lip2, or in mock-infected mice. As shown in Figure S4, panels A-D, DCs were found to compose ~15% of total renal CD45+ cells, regardless of infection with C. albicans, whereas the macrophage population was too small to measure with accuracy; note that these findings are consistent with a previous report (Lionakis et al., 2011). Using RT-qPCR to quantify Il23 mRNA expression in isolated renal DCs, we observed significant upregulation after 6 hours of infection with WT C. albicans and an even stronger response to the lip2 strain (Figure 4B). In support of DC activation by C. albicans, flow cytometry revealed stepwise increases in surface expression of MHCII in DCs from organs infected with WT or lip2 (Figure 4C). Together, these results suggest that tissue-resident DCs respond to fungal invasion of the kidney by producing IL-23, a known activator of IL-17 expression. For reasons that remain unclear, this response is exaggerated in the presence of lip2.
Figure 4. Lip2 suppresses the activation of renal dendritic cells.
(A) lip2 induces IL-23A production in kidneys. Groups of female C57BL/6J mice were infected with 1×106 CFU of WT or lip2 followed by euthanasia of three animals per group at the indicated time points. Il23a mRNA was measured by RT-qPCR in right kidney homogenates and normalized to Gadph (left panel). IL-23A protein production was evaluated by ELISA in the left kidney homogenates (right panel). Statistical significance was determined by one-way ANOVA (Tukey’s multiple comparisons test); * p< 0.05; ** p< 0.01.
(B) Kidney resident DCs respond to C. albicans invasion. Groups C57BL/6J mice were infected 1×106 CFU of WT or lip2 for six hours. Renal DCs were isolated and Il23a mRNA levels were measured by RT-qPCR and normalized to Gadph. Statistical significance was determined by one-way ANOVA (Tukey’s multiple comparisons test); ** p< 0.01; *** p<0.001; **** p<0.0001.
(C) Infection with lip2 is associated with increased cell surface expression of MCHII by renal DCs. Groups C57BL/6J mice were infected 1×106 CFU of WT or lip2 for six hours. DCs were analyzed by flow cytometry for MCHII. Statistical significance was determined by one-way ANOVA (Tukey’s multiple comparisons test); * p<0.05; ** p< 0.01.
(D) lip2 stimulates IL-23 production by BMDCs. BMDCs from C57BL/6J mice were cultured with WT, lip2, or lip2+LIP2 at an MOI of 1 for two hours followed by measurement of IL-23A in cell supernatants by ELISA. Statistical significance was determined by one-way ANOVA (Tukey’s multiple comparisons test); ns: p= 0.6247; **** p<0.0001.
(E) lip2-stimulated activation of BMDCs does not require cell-cell contact. BMDCs from C57BL/6J mice were cultured in the same well with WT or lip2 (Dir) or separately using a transwell system (Indir) at an MOI of 1 for two hours followed by measurement of IL-23A in cell supernatants by ELISA. Statistical significance between Direct vs Indirect in WT or lip2 condition was determined by an unpaired two-tailed t-test; ns: p= 0.6247.
(F) Cell viability is required to repress BMDC activation. BMDCs from C57BL/6J mice were cocultured with live or heat-killed (HK) forms of WT and lip2 at an MOI of 1 for two hours followed by measurement of IL-23A in cell supernatants by ELISA. Statistical significance between the different groups was determined by one-way ANOVA (Tukey’s multiple comparisons test); ns: p>0.5 ; ** p<0.01; *** p<0.001.
(G) The ability to activate BMDCs is specific to lip2. Coculture assays were performed with lip2, three other virulence-defective mutants (efg1, ece1, sap6), and lip2 (lip10) as described in (D). Statistical significance between WT and the different mutants was determined by one-way ANOVA (Tukey’s multiple comparisons test); ns WT vs ece1: 0.8937; ns WT vs efg1: 0.9999, ns WT vs sap6: 0.9977, ns WT vs lip10: 0.3074: WT vs lip2: **** p<0.0001
(H) LIP2 but not LIP10 is expressed under co-coculture conditions. WT C. albicans was propagated alone or in co-culture with BMDCs for two hours, followed by assessment of LIP2 and LIP10 mRNAs by RT-qPCR; reported values were to ACT1. Statistical significance between RPMI and RPMI+DC was determined by an unpaired two-tailed t-test; * p<0.05; **** p<0.0001.
To test whether C. albicans can directly activate IL-23 production by DCs, coculture experiments were performed with bone marrow-derived dendritic cells (BMDCs) prepared from C57B/6J mice. BMDCs were incubated at an MOI of 1 with WT, lip2, a lip2+LIP2 complemented strain, or with cell medium alone (mock) for two hours, followed by quantitation of IL-23 in the culture supernatants. As shown in Figure 4D, exposure to lip2 but not WT or lip2+LIP2 induced a strong IL-23 response from cocultured BMDCs. To determine whether direct cell-cell contact is required for BMDC activation by C. albicans, we repeated the experiment, this time varying whether cells were mixed directly in the same wells (Dir) or indirectly (Indir) in compartments separated by a porous membrane. As shown in Figure 4E, C. albicans triggered similar IL-23 responses from BMDCs regardless of whether the cells were in direct physical contact, suggesting that a soluble fungal signal may be responsible for the activation of BMDCs. Finally, we asked whether cell viability is required for the ability of WT C. albicans to suppress activation of BMDCs. As shown in Figure 4F, coincubation of BMDCs with heat-killed (HK) but not viable (live) WT C. albicans resulted in secretion of IL-23, similar to the effect of coincubation with live or heat-killed lip2. Together, these results suggest that a soluble fungal signal can activate IL-23 production by BMDCs, but live yeasts are able to block immune activation in a LIP2-dependent fashion.
To determine whether BMDC activation is specific to lip2, we tested three additional virulence mutants and a mutant defective in the closest paralog to Lip2 in the coculture assay. As shown in Figure 4G, coculture of BMDCs with mutants defective in Candidalysin psignificantly reduced lipase activity was associated with the point mutant roduction (ece1) (Moyes et al., 2016), yeast-to-hypha morphogenesis (efg1) (Stoldt et al., 1997), a secreted aspartyl protease (sap6) (Jackson et al., 2007), or Lip10 (lip10) failed to elicit IL-23 production beyond the level observed during coculture with WT. In contrast to LIP2, ECE1, EFG1, and SAP6 (Figure S4E), LIP10 is not detectably expressed by WT C. albicans under the coculture assay conditions (Figures 4H and S4E); therefore, the lack of a lip10 phenotype may reflect the absence of gene expression rather than a lack of immunomodulatory activity pf the encoded enzyme. This caveat notwithstanding, the ability to activate BMDCs in the coculture assay appears to be unique to lip2, raising the possibility that the virulence-promoting activity of Lip2 is to dampen immune activation of tissue-resident DCs in kidneys and other infected organs.
Lipase activity is required for the immunomodulatory role of Lip2.
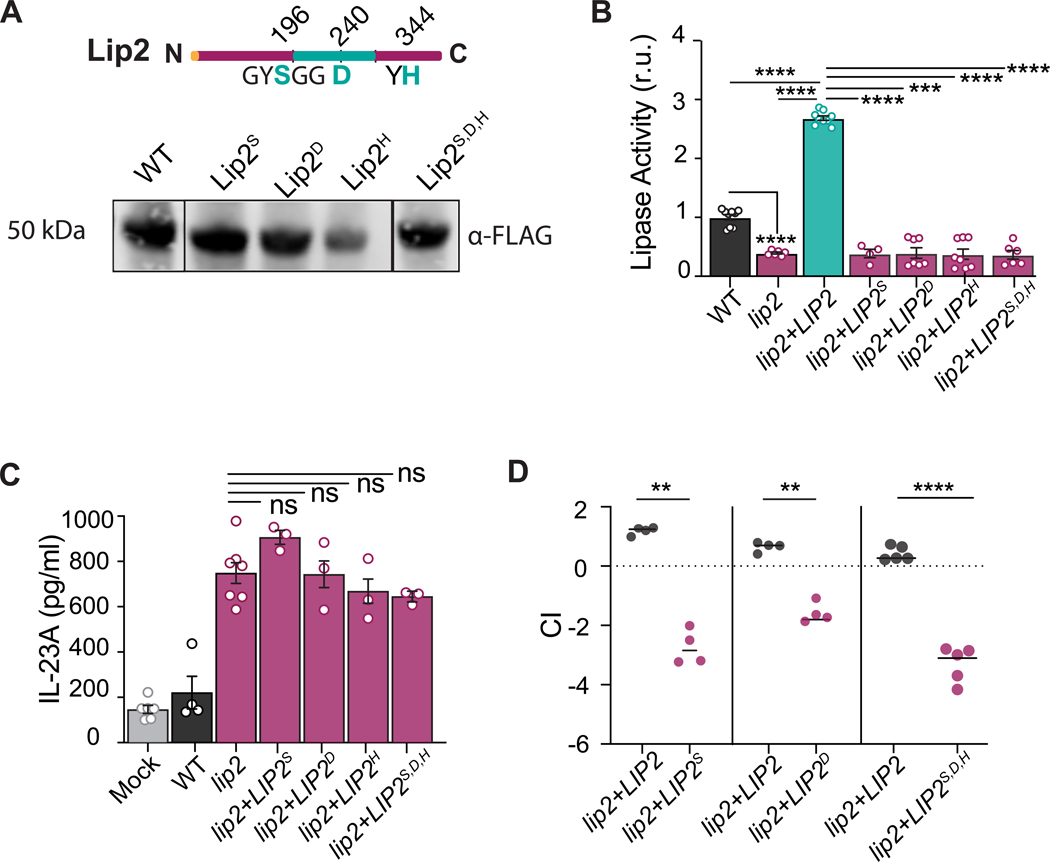
LIP2 encodes a secreted lipase with nine paralogs in C. albicans (Hube et al., 2000). To determine whether lipase activity is required for the immune dampening function of Lip2, we engineered four LIP2 point mutants (lip2S196A, lip2D240A, lip2H344A, and lip2S196A,D240A,H344A) that target conserved residues in the predicted lipase domain (Figure 5A). FLAG-tagged versions of each mutant allele and wild-type LIP2 were individually introduced into a lip2 null strain, and protein expression and secretion were confirmed by immunoblotting of the respective cell supernatants with anti-FLAG antibodies (Figure 5A). Cell supernatants from the point mutant strains were next assessed for in vitro lipase activity. Compared to WT C. albicans or lip2+LIP2, significantly reduced lipase activity was associated with the point mutant strains, similar to lip2 null mutant (Figure 5B), suggesting that the encoded proteins are catalytically inactive. Further, in coculture assays with BMDCs, all four point mutant strains activated IL-23 production to a similar degree as lip2 (Figure 5C). Finally, competitive fitness of the lip2+LIP2S196A, lip2+LIP2D240A, and lip2+LIP2S196A,D240A,H344A strains was assessed in 1:1 competitions against WT in the mouse systemic infection model. All three mutants exhibited significantly reduced fitness in kidneys (Figure 5D), similar to the phenotype of lip2. Together, these experiments suggest that the lipase activity of Lip2 is required for suppression of the BMDC response to C. albicans and for virulence in the host.
Figure 5. Predicted catalytic residues of Lip2 is required for its immunomodulatory role.
(A) Lip2 point mutant proteins are expressed. Cartoon of Lip2 protein domains (top). The lipase domain is represented by the green central region, with targeted residues of the predicted catalytic site also colored green. Immunoblot (bottom) of WT and point mutant alleles recovered from cell culture supernatants. Lip2 proteins are fused at the C-terminus to 6-His-FLAG (HHHHHHGGDYKDDDDK), and immunoblots were probed with anti-FLAG.
(B) Lip2 point mutants exhibit reduced lipase activity. Strains expressing the indicated allele of LIP2 were propagated to mid log growth (OD600=1) in liquid YEPD. Lipase assays were performed following a 10-minute incubation of supernatants with mixed lipids. Statistical significance between the different groups was determined by one-way ANOVA (Tukey’s multiple comparisons test); *** p<0.001; **** p<0.0001.
(C) Lip2 lipase activity is required to suppress the activation of BMDCs by C. albicans. Co-culture assays were performed with yeasts expressing WT Lip2 or the indicated Lip2 point mutants as in Figure 4D. Statistical significance between lip2 and the point-mutation mutants was determined by one-way ANOVA (Tukey’s multiple comparisons test); ns: 0.1964
(D) Lip2 lipase activity is required for virulence. BALB/c mice were infected with 1×105 CFU of a 1:1 mixture between lip2+LIP2 and either lip2+LIP2S196A, lip2+LIP2D240A, or lip2+LIP2S196A,D240A,H344A. Relative strain abundance in kidneys was determined by qPCR. Statistical significance between lip2+LIP2 and mutants was determined by a paired two-tailed t-test; **p < 0.01, ****p < 0.0001.
Palmitic acid suppresses the activation of dendritic cells
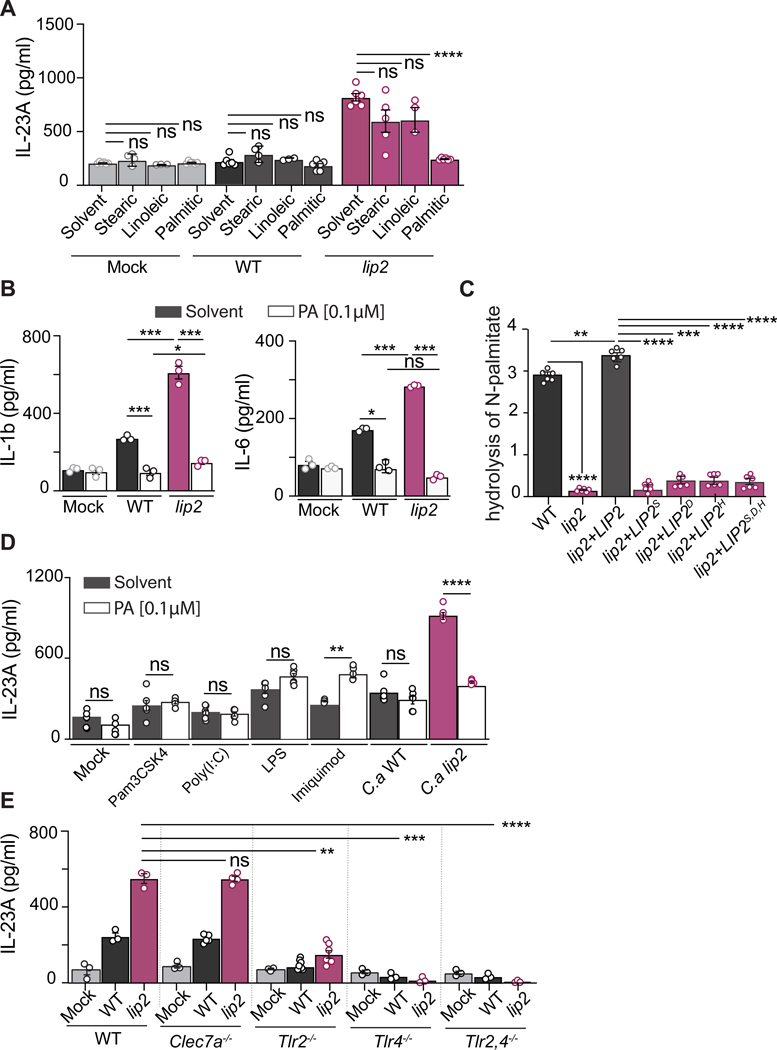
Lipases are enzymes that catalyze the hydrolysis of triglycerides to fatty acids and glycerol. We hypothesized that, within the host, Lip2 may modulate DC reactivity by increasing the local concentration of one or more fatty acids. To test this hypothesis, we assayed the ability of several common fatty acids to suppress IL-23 production by BMDCs exposed to lip2. As shown in Figures 6A, S5A, and S5B, 0.1 μM palmitic acid but not stearic acid. linoleic acid, or solvent alone suppressed the IL-23 response of BMDCs to lip2 to the level observed in unexposed BMDCs (or WT-exposed BMDCs). In addition to IL-23, cytokines such as IL-1β and IL-6 are capable of driving IL-17 production by TCRγδ+ T cells. To determine whether palmitic acid affects the production of IL-1β and/or IL-6 following exposure of BMDCs to C. albicans, we assessed the levels of these cytokines following two-hour coculture experiments performed in the presence or absence of palmitic acid. Interestingly, in the absence of palmitic acid, lip2 and, to a lesser extent, WT C. albicans triggered enhanced production of both IL-1β and IL-6 by BMDCs; however, both responses were blocked in the presence of 0.1 μM palmitic acid (Figure 6B). As shown in Figure S5C, palmitic acid also suppresses BMDC activation (enhanced expression of IL-23) by strains expressing catalytically inactive alleles of LIP2. Note that fungal morphogenesis in the coculture assays (Figure S5D) and expression of LIP2 by WT C. albicans (Figure S5E) are unaffected by palmitate. Finally, we confirmed that strains expressing WT LIP2 but not a lip2 null mutant or strains expressing catalytically inactive LIP2 point mutants are able to hydrolyze a pNp-palmitate substrate (Figure 6C). Together, these results indicate that Lip2 liberates palmitic acid from suitable substrates and that palmitic acid suppresses BMDC activation by C. albicans.
Figure 6. The immune dampening defect of lip2 is complemented by palmitic acid.
(A) Palmitic acid suppresses the IL-23 response of BMDCs to C. albicans. BMDCs from C57BL/6J mice were incubated with 0.1 μM of fatty acids (palmitic acid, stearic acid or linoleic acid) or solvent alone (chloroform) with or without WT C. albicans or lip2 at an MOI of 1 for two hours, followed by measurement of IL-23A in cell supernatants by ELISA. Statistical significance between solvent control and fatty acid was determined by one-way ANOVA (Tukey’s multiple comparisons test); ns: p> 0.05; **** p<0.0001.
(B) Palmitic acid also suppresses the IL1b and IL-6 responses of BMDCs to lip2. BMDCs from C57BL/6J mice were incubated with 0.1 μM palmitic acid or solvent alone with or without WT C. albicans or lip2 at an MOI of 1 for two hours, followed by measurement of IL-1b or IL-6 in culture supernatants by ELISA. Statistical significance between the different conditions was determined by one-way ANOVA (Tukey’s multiple comparisons test); ns: p> 0.05; * p=0.05; *** p<0.001.
(C) Lip2 hydrolyzes pNp-palmitate to palmitic acid. WT C. albicans, lip2, lip2+LIP2, and strains expressing catalytically inactive Lip2 point mutants were propagated in BMDC culture medium to OD600=1, supernatants were incubated with pNp-palmitate, and hydrolysis was assessed by spectrophotometry. Statistical significance between the different mutants was determined by one-way ANOVA (Tukey’s multiple comparisons test); ** p<0.01; *** p<0.001; **** p<0.0001.
(D) Palmitic acid dampens the BMDC response to lip2 but enhances the response to imiquimod. BMDCs from C57BL/6J mice were incubated for two hours with solvent (dark bars) or 0.1 μM palmitic acid (white bars) with or without TLR ligands (Pam3CSK4: TLR1/2; Poly(I:C): TLR3; LPS: TLR4; imiquimod: TLR7/8) or C. albicans (WT or lip2 at a MOI of 1), followed by measurement of IL-23A in cell supernatants by ELISA. Statistical significance between the solvent control and the PA conditions was determined by an unpaired two-tailed t-test; ns: p> 0.05; ** p<0.001; **** p<0.0001.
(E) TLR2 and TLR4 are required for the BMDC activation by C. albicans. BMDCs from C57BL/6J mice were incubated with 0.1 μM fatty acids (palmitic acid, stearic acid or linoleic acid) or solvent alone with or without WT C. albicans or lip2 at an MOI of 1 for two hours, followed by measurement of IL-23A in cell supernatants by ELISA. Statistical significance between the different DC infected with lip2 was determined by one-way ANOVA (Tukey’s multiple comparisons test); ns: p> 0.05; **** p<0.0001.
The immune dampening effect of palmitic acid on DC activation by C. albicans was somewhat surprising in light of published reports that palmitic acid enhances the pro-inflammatory responses of macrophages and DCs to other pathogen-associated molecular patterns (PAMPs), thereby contributing to the morbidity of chronic diseases, such as obesity and type 2 diabetes mellitus, that are associated with elevated serum palmitate (Korbecki and Bajdak-Rusinek, 2019). In particular, one study reported that palmitic acid enhances BMDC activation (IL-23A production) in response to TLR agonists, including Pam3CSK4 (TLR1/TLR2 agonist), poly(I:C) (TLR3 homodimer agonist), LPS (TLR4 homodimer agonist), and imiquimod (TLR7/TLR8 agonist) (Mogilenko et al., 2019). We hypothesized that the distinct effect of palmitic acid on BMDCs in response to C. albicans reflects either a difference in our experimental assay or a difference in BMDC responses to live lip2 yeasts compared to specific TLR agonists. To distinguish between these possibilities, we repeated our coculture experiments using both sets of assay conditions and included the published panel of TLR agonists as controls. Under our assay conditions, imiquimod was the only one of four tested TLR agonists to induce significant IL-23A production by BMDCs, and this response was substantially enhanced in the presence of 0.1 𝛍M palmitic acid (Figure 6D). By comparison, coincubation with lip2 yeasts triggered an even larger IL-23A response, and this response was suppressed by 0.1 𝛍M palmitic acid (Figure 6D). Under the previously published assay conditions, which notably includes a 24-hour incubation period (compared to 2 hours in our assay), LPS and imiquimod triggered significant IL-23A responses, and both responses were significantly enhanced in the presence of 500 𝛍M palmitic acid (Figure S5F), as previously reported (Mogilenko et al., 2019). Coincubation with lip2 also triggered IL-23A production under these assay conditions, and the response was suppressed by 500 𝛍M palmitic acid (Figure S5F). These results confirm that BMDC activation by lip2 occurs under both sets of assay conditions and suggest that the immune modulatory activity of palmitic acid can vary with the nature of the immune stimulus (pro-inflammatory with purified TLR agonists vs. anti-inflammatory with whole yeast).
TLR2, TLR4, and Dectin-1 are cell surface-associated pattern recognition receptors that have previously been implicated in host immune recognition of C. albicans (Netea et al., 2008). TLR2 and TLR4 are thought to recognize O-linked mannose residues while Dectin-1 recognizes β−1,3-linked glycan residues of the fungal cell wall. To determine whether any of these receptors plays a role in the IL-23 response to lip2, we performed coculture assays with C. albicans and BMDCs prepared from clec7a−/− (Dectin-1 deficient), tlr2−/−, tlr4−/−, or tlr2/tlr4−/− mice. As shown in Figure 6E, Clec7a was found to be dispensable for BMDC activation by lip2, but the IL-23 response was significantly diminished in cells lacking Tlr2 or Tlr4 and virtually eliminated in cells lacking both Tlr2 and Tlr4. These results suggest that TLR2 and TLR4 play nonredundant roles in BMDC activation by C. albicans.
DISCUSSION
Candida albicans is the primary fungus of the human gut microbiota and is among only a handful of fungal species to colonize humans. The perpetual association of C. albicans with mammalian hosts, paired with the absence of a known environmental reservoir, implies the existence of extensive coevolution between host and fungus. Indeed, it can be argued that C. albicans and humans enjoy a largely mutualistic relationship, with the fungus benefitting from a nutrient-rich, stable environment, and the host receiving certain immune benefits. Latter benefits include the stimulation of Th17-based immunity to other fungal pathogens, as well as a poorly understood, generalized resilience to fungi, bacteria, and viruses in diverse niches (Bacher et al., 2019; Jiang et al., 2017; Shao et al., 2019). Nevertheless, C. albicans remains a perpetual threat with the ability to cause serious disease, particularly in hosts with risk factors for systemic infection.
Our screen of DOX-repressible GRACE mutants identified Lip2 as a fungal effector that is required for competitive fitness in kidneys (Figure 1A) and overall lethality (Figure 1B) in a murine model of systemic infection. Unlike most C. albicans mutants with defects in this model, a lip2 null mutant is competent for yeast-to-hypha morphogenesis, both under laboratory conditions (Figure S1A and S1B) and in the host (Figures 1D and S1B). Further, after one hour of infection, equal titers of lip2 and WT can be recovered from kidneys, livers, and spleens, suggesting that the mutant is proficient in egress from the vasculature and penetration into target organs (Figures 1C and S2A). Instead, the first detectable deviation from standard bloodstream infections occurs at 6 hours, when unusually high levels of pro-inflammatory cytokines (IL-23 and IL-17; Figures 2A, 2B, 4A and S3A) are seen in lip2-infected kidneys. At subsequent time points, lip2 displays progressive defects in persistence (Figures 1C and S2B–D) and damage to (Figures 1E and S2D) target organs, leading to eradication of the infection (Figure 1C) and host survival (Figure 1B) in most cases.
Prior to this study, there were several reports of enhanced susceptibility to bloodstream candidiasis among mice with defects in IL-17 signaling (Huang et al., 2004; Ramani et al., 2018; Saijo et al., 2010). IL-17 was proposed to protect the host by increasing the resilience of kidneys during infection. Specifically, IL-17 was shown to activate the renal protective Kallikrein-Kinin proteolytic cascade and to exert direct anti-apoptotic activity on renal tubular epithelial cells (Ramani et al., 2016; Ramani et al., 2018). Our finding that lip2 is virulent in Il17af−/− animals but highly attenuated in wild-type littermates (Figure 3A–E) suggests that IL-17 plays an additional role in elimination of fungal pathogens from systemically infected animals. Medzhitov, Schneider, and Soares have hypothesized that injury from infectious disease is the result of pathogen proliferation plus host intolerance of the pathogen (Medzhitov et al., 2012). Viewed through this lens, our findings complement the published findings on renal tolerance (Ramani et al., 2016; Ramani et al., 2018) and suggest that IL-17 antagonizes both pathways of infection-related injury.
Following our identification of renal DCs as a source of IL-23A (Figure 4B) and renal TCRγδ+ T cells as a source of IL-17A (Figures 3F, 3G, and S3C) in lip2-infected kidneys, we tested whether lip2 directly activates BMDCs in coculture experiments. The finding that IL-23A production is stimulated by strains lacking LIP2 (Figure 4D) or expressing catalytically inactive LIP2 point mutants (Figure 5C) but not WT C. albicans suggested that a Lip2-dependent metabolite might suppress the activation of BMDCs. Consistent with this model, exposure of cocultured cells to 0.1 𝛍M palmitic acid suppresses induction of IL-23A (as well as IL-1β and IL-6; Figure 6A–B). Interestingly, the immune dampening effect of palmitic acid on BMDCs in response to a fungal stimulus is opposite to its published pro-inflammatory effects in response to multiple TLR agonists (Mogilenko et al., 2019). Although we have yet to define which features of the lip2 mutant (or heat-killed WT cells, Figure 4F) are sensed by DCs, TLR2 and TLR4 appear to be required for the full response (Figure 6E).
In sum, our results support a model in which Lip2 promotes fungal virulence by suppressing the host IL-17 response. We propose that, in organs such as the kidney, Lip2 increases the local concentration of immune modulatory fatty acids such as palmitic acid, thereby blunting TLR2- and TLR4-dependent activation of tissue resident DCs. The immune dampening effect may be mediated directly on DCs or may be indirect, for example because of decreased expression of TLR2/TLR4 ligands on the C. albicans cell surface. In contrast, during infections with C. albicans mutants that lack LIP2 or express catalytically inactive versions of the enzyme, DCs are activated to release IL-23, leading to IL-17 expression by tissue resident TCRγδ+ T cells. The lipid-mediated suppression of IL-17 that we observe in solid organs is apparently not active in skin (Kashem et al., 2015), where IL-17 is known to play an active role in suppressing the proliferation of C. albicans. This difference may be related to the observation that, in skin, C. albicans is sensed by nociceptive sensory fibers, which leads to DC activation and IL-17 production by dermal TCRγδ+ T cells (Kashem et al., 2015). This neuro-immune pathway may bypass the Lip2-mediated DC suppression that we observe in solid organs. Future work will be needed to clarify whether Lip2 plays a role in mucocutaneous candidiasis.
STAR METHODS
Resource availability
Lead contact
Further information and requests for reagents may be directed to and will be fulfilled by the Lead Contact, Suzanne M. Noble (suzanne.noble@ucsf.edu).
Materials availability
C. albicans strains in this study will be made available on request, but may require payment and/or a completed Materials Transfer Agreement if there is potential for commercial application.
Data and code availability
All data reported in this paper will be shared by the lead contact upon request..
This paper does not report original code.
Any additional information required to reanalyze the data reported in this work is available from the Lead Contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Yeast manipulations
Yeast strains and primers used for this study are described in Key Resources Table, and Table S1. Growth assays were performed with strains that were freshly streaked from frozen glycerol stocks onto YPD agar and incubated for two days at 30ºC. A single colony was suspended in sterile distilled water to an optical density at 600 nm (OD600) of 1 and diluted as appropriate for the assay. New strains were constructed as described in (Noble and Johnson, 2005).
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Brilliant Violet 785 anti-mouse CD11b (M1/70) | Biolegend | 101243 |
| PE-Cy7 anti-mouse CD11c (N418) | Biolegend | 117317 |
| Pacific Blue anti-mouse CD45.2 (104) | Biolegend | 109819 |
| Brilliant Violet 785 anti-mouse CD90.2 (30-H12) | Biolegend | 105331 |
| Brilliant Violet 605 anti-mouse CD45R/B220 (RA3-6B2) | Biolegend | 103243 |
| PE anti-mouse IL-17A (TC11-18H10.1) | Biolegend | 506903 |
| PerCP/Cyanine5.5 anti-mouse TCR gamma delta (GL3) | Biolegend | 118117 |
| PE anti-mouse MHCII (I-A/I-E) (M5/114.15.2) | Biolegend | 50-5321-U025 |
| FITC anti-mouse CD4 (GK1.5) | Biolegend | 100405 |
| Brilliant Violet 785 anti-mouse CD11b (M1/70) | Biolegend | 101243 |
| PE-Cy7 anti-mouse CD11c (N418) | Biolegend | 117317 |
| Pacific Blue anti-mouse CD45.2 (104) | Biolegend | 109819 |
| anti-FLAG | Thermo Fisher Scientific | MA1-142-A488 |
| Chemicals, peptides, and recombinant proteins | ||
| Acid phenol chloroform | Ambion | AM9722 |
| Phenol chloroform isoamyl alcohol | Ambion | AM9732 |
| Recombinant mouse GM-CSF | Prepotech | Cat# 315-03 |
| IL-4 | Gibco | Cat# PMC0045 |
| Pam3CSK4 | InvivoGen | Cat# tlrl-pms |
| Poly(I:C) | InvivoGen | Cat# tlrl-pic |
| LPS from E.coli | Sigma | Cat# L3024 |
| imiquimod | Calbiochem | Cat# CAS 99001-02-6 |
| Palmitic acid | Sigma | Cat# P0500 |
| Stearic acid | Sigma | Cat# S4751 |
| Linoleic acid | Sigma | Cat #L1376 |
| RPMI media | UCSF Media Production | Cat# CCFAE001 |
| FBS | UCSF Media Production | Cat# CCFAQ009 |
| Bovine serum Albumin | Sigma | Cat# A6003 |
| Percoll | Cytivia | Cat# 17089101 |
| PMA | Sigma | Cat# P1585 |
| ionomycin | Sigma | Cat# I9657 |
| GolgiStop | Fisher Scientific | Cat# BDB555029 |
| BD Cytofix buffer | Fisher Scientific | Cat# BDB554655 |
| Perm/Wash reagent | BD Biosciences | Cat# BDB554723 |
| Critical commercial assays | ||
| Collagenase I | Thermo Fisher Scientific | Cat# 17100017 |
| DNAse I | Thermo Fisher Scientific | Cat# EN0521 |
| RNeasy Plus mini kit | Qiagen | Cat# 74134 |
| DuoSet ELISA kit IL-23 | R&D Systems | Cat# DY1887 |
| DuoSet ELISA kit IL-17 | R&D Systems | Cat# DY421 |
| DuoSet ELISA kit IL-1 | R&D Systems | Cat# DY401 |
| DuoSet ELISA kit IL-6 | R&D Systems | Cat# DY406 |
| Lipase Assay Kit | Sigma | Cat# MAK046 |
| pNp-palmitate | Sigma | Cat# N2752 |
| Trizol reagent | ThermoFisher | Cat# 15596026 |
| SYBR Green Master Mix | Biorad | Cat# 1725275 |
| Superscript III | Invitrogen | Cat# 18080 |
| Deposited data | ||
| Experimental models: Cell lines | ||
| Bone marrow-derived dendritic cells differentiated in the presence of GM-CSF and IL-4 | This paper | N/A |
| Experimental models: Organisms/strains | ||
| Mouse: BALB/c | Charles River Laboratories | Stock No: 028 |
| Mouse: C57BL/6J | Jackson Laboratories | Stock no. 000664 |
| Mouse: il17af−/− | Jackson Laboratories | Stock no. 034140 |
| SN2019 | this study | WT control strain for lip2DOX-OFF |
| SN2464 | Roemer et al. (2003) | lip2 DOX-OFF |
| SN250 | Noble et al. (2010) | leu2Δ::C.d.LEU2/leu2Δ::C.m.HIS1, his1Δ/his1Δ, arg4Δ/arg4Δ, ura3Δ/URA3, iro1/IRO1 |
| SN2461 | Noble et al. (2010) | lip2Δ::C.d.LEU2/lip2Δ::C.m.HIS1, his1Δ/his1Δ, arg4Δ/arg4Δ, ura3Δ/URA3, iro1/IRO1 |
| SN2242 | This study | LIP2/lip2Δ, leu2Δ::C.d.LEU2/leu2Δ::C.m.HIS1, his1Δ/his1Δ, arg4Δ::C.d.ARG4/arg4Δ, ura3Δ/URA3, iro1/IRO1 |
| SN2243 | This study | LIP2S196A/lip2Δ, leu2Δ::C.d.LEU2/leu2Δ::C.m.HIS1, his1Δ/his1Δ, arg4Δ::C.d.ARG4/arg4Δ, ura3Δ/URA3, iro1/IRO1 |
| SN2246 | This study | LIP2D240A/lip2Δ, leu2Δ::C.d.LEU2/leu2Δ::C.m.HIS1, his1Δ/his1Δ, arg4Δ::C.d.ARG4/arg4Δ, ura3Δ/URA3, iro1/IRO1 |
| SN2250 | This study | LIP2H344A/lip2Δ, leu2Δ::C.d.LEU2/leu2Δ::C.m.HIS1, his1Δ/his1Δ, arg4Δ::C.d.ARG4/arg4Δ, ura3Δ/URA3, iro1/IRO1 |
| SN2253 | This study | LIP2S196A,D240A,H344A/lip2Δ, leu2Δ::C.d.LEU2/leu2Δ::C.m.HIS1, his1Δ/his1Δ, arg4Δ::C.d.ARG4/arg4Δ, ura3Δ/URA3, iro1/IRO1 |
| SN119 | Noble and Johnson (2005) | efg1Δ::C.d.HIS1/efg1Δ::C.m.LEU2, leu2Δ/leu2Δ, his1Δ/his1Δ, ura3Δ/URA3, iro1/IRO1 |
| SN2462 | Noble et al. (2010) | ece1Δ::C.d.LEU2/ece1Δ::C.m.HIS, leu2Δ/leu2Δ, his1Δ/his1Δ, arg4Δ/arg4Δ, ura3Δ/URA3, iro1/IRO1 |
| SN1664 | Noble et al. (2010) | sap6Δ::C.d.LEU2/sap6Δ::C.m.HIS, leu2Δ/leu2Δ, his1Δ/his1Δ, arg4Δ/arg4Δ, ura3Δ/URA3, iro1/IRO1 |
| SN2463 | Noble et al. (2010) | lip10Δ::C.d.LEU2/lip10Δ::C.m.HIS, leu2Δ/leu2Δ, his1Δ/his1Δ, arg4Δ/arg4Δ, ura3Δ/URA3, iro1/IRO1 |
| Oligonucleotides | ||
| Oligonucleotides are described in Table S1 | N/A | N/A |
| Software and algorithms | ||
| Other | ||
Studies in animals
All procedures involving animals were approved by the Institutional Animal Care and Use Committee at the University of California San Francisco and were carried out according to the National Institute of Health (NIH) guidelines for the ethical treatment of animals. Experiments were performed with 8–10 week female BALB/c (no.028) mice from Charles River Laboratories or eight-week-old (male and female) C57BL/6J (no. 000664) WT and il17af−/− (no. 034140) mice from Jackson Laboratories and bred in-house. Systemic infection was performed by inoculation of 1×105 CFUs (BALB/C), 1×106 CFUs (WT C57BL/6J) or 5×105 CFUs (il17af−/− C57BL/6J) of mid-log phase yeasts into the retro bulbar sinus of animals, under isoflurane anesthesia. Animals were monitored closely and euthanized at the experimentally determined endpoints or upon development of signs of clinical morbidity (defined as BCS≤2, hunched posture, decreased motor activity), whichever was first. Kidneys were then recovered for CFU analysis, histology, RNA analysis, and/or analysis of competitive fitness. Female BALB/C infected with the lip2(DOX-OFF) strain were additionally treated with 0.25 mg/mL of doxycycline via drinking water beginning seven days prior to infection and continued throughout the experiment.
METHOD DETAILS
Determination of competitive index
Competitive index (CI) of strains in 1:1 competitive infections of the systemic infection model was determined as in (Noble et al., 2010). Strain abundance was determined by qPCR of genomic DNA prepared from colonies plated on Sabouraud agar recovered from the inoculum and infected kidneys, using strain-specific primers (Table S2) and a Roche LightCycler 480 instrument. The significance of observed differences was determined using the paired Student’s t-test.
Histology
Kidneys were fixed in 10% formalin for 24 hours and washed in 80% ethanol. Preparation of paraffin blocks and 4 μm sections as well as staining with PAS and H&E were performed by Nationwide Histology (MT).
Kidney dissociation and recovery of renal dendritic cells
Kidneys were minced and digested with Collagenase I (0.125mg/mL, Thermo Fisher Scientific), DNAse I (0.2 mg/mL, Millipore) in RPMI with 10% of FBS for 30 min at 37ºC, prior to filtration through a 100-micron strainer (Thermo Fisher Scientific) and washing with RPMI + 2% FBS + 5 mM EDTA. Cells were suspended in FACS buffer (PBS, 2% FBS, 1 mM EDTA) and purified over a discontinuous gradient of 70% and 30% Percoll (Cytivia). Cells collected from the 70%−30% interface were washed in FACS buffer for further analysis.
Isolation of renal DCs: Cells recovered from the Percoll gradient were suspended in MACS buffer and incubated with CD11c-Ab beads for 15 min at 4ºC. CD11c+ DCs were isolated using MS MACS columns per the manufacturer’s instructions. Quantitation was performed with a hemocytometer, and total RNA was extracted using an RNeasy Plus mini kit (Qiagen).
Flow cytometry
Cells were stained with antibodies against TCRgd (GL3), CD4 (GK1.5), B220 (RA3–6B2), CD45 (30-F11), IL-17A (TC11–18H10.1), hNGFR (ME20.4), CD90.2 (53–2.1), CD11b (M1/70) Ly6G (1A8), CD11c (N418), F4/80 (BM8), MHCII (M5/114.15.2) (from Biolegend, BD Biosciences or eBiosciences). To determine the source of IL-17A in kidneys, dissociated cells were incubated for two hours in MACS buffer supplemented with PMA (50 ng/ml) and ionomycin (500 ng/ml) with GolgiStop (1000X). To detect intracellular IL-17A, cells were treated with BD Cytofix buffer and Perm/Wash reagent (BD Biosciences) and then stained with anti-IL-17A (C57BL/6J) or anti-hNGFR (SMART17 C57BL/6J) in Perm/wash buffer. Samples were analyzed by FACS (BD), and data were analyzed with FlowJo software (Version 10, BD, https://www.flowjo.com/).
RNA isolation and RT-qPCR
Isolation of total RNA from kidneys or dendritic cells was performed using TriZol or RNeasy Plus Mini kits per the manufacturer’s protocols. First-strand complementary DNA (cDNA) was synthesized from 1 μg of total RNA using the Superscript III cDNA Reverse Transcription Kit (Thermo Fisher Scientific). Quantitative PCR was performed with specific primers described in Table S1. We employed the ΔΔCt method (Livak and Schmittgen, 2001); results were normalized to the housekeeping genes Gadph or ACT1 for LIP2 and LIP10 expression.
Preparation of BMDCs
Bone-marrow-derived dendritic cells (BMDC) were isolated from femurs and tibias of 6- to 8-week old (male and female) C57BL/6J mice (WT, tlr2−/−, tlr4−/−, tlr2−/−4−/−, clec7a−/−). Bone marrow was eluted from bones with RPMI + 5% FBS and filtered through a cell strainer (70 μm). The cell suspension was centrifuged at 250 × g for 5 min at 4°C, and the pellet was re-suspended in 2 ml of RPMI + 5% FBS and 1 mL/mouse of ACK erythrocyte lysis buffer (Ammonium-Chloride-Potassium Lysing Buffer, Thermo Fisher Scientific). After incubation for 7 min one ice, 20 mL PBS was added before centrifugation, and pellets were washed once in RPMI. Cells were plated onto 100 mm non-tissue culture treated culture plate in complete RPMI (10% FCS, 10 μM 2-mercaptoethanol, 25 mM HEPES, Glutamax 100X, 100 U/mL penicillin, 100 μg/mL of streptomycin sulfate and 20 ng/mL GM-CSF (Prepotech)) for three days. On day three, cells were treated with ten mL of complete RPMI. On days six, eight, and ten, adherent cells were collected, washed, and plated with a Complete RPMI supplemented with 10 ng/mL of IL-4 (Gibco). Mature DCs were used on day 12.
In vitro activation of dendritic cells
Murine BMDCs were seeded into 24-well tissue culture-treated plates (BD) at 1×105 cells per well in one ml complete RPMI (without penicillin and streptomycin) and cultured O/N at 37ºC in 5% CO2. Activation by specific C. albicans strains was assessed after coincubation for two hours at MOI 1, followed by measurement of IL-23 in cell supernatants by ELISA.
Free fatty acid (FFA) supplementation. FFA (in chloroform) was added to BMDCs in culture medium to a final concentration of 1, 0.1, 0.01 μM for two hours.
Activation of DCs by TLR ligands. 5×105 cells/mL were incubated in complete RPMI and treated with 100 ng/mL Pam3CSK4, 10 μg/mL Poly(I:C), 100 ng/mL LPS, or 3 μg/mL imiquimod with or without palmitic acid conjugated with BSA when mentioned. Cells were activated for 24 hours or 2 hours.
Measurement of cytokine production
Cytokine production was measured by enzyme-linked immunoassay using a DuoSet ELISA kit (R&D Systems) following the manufacturer’s protocol. Briefly, for detection of IL-17A, and IL-23A, 200 μL of kidney homogenate was used. For in vitro coculture experiments with BMDCs, 100 μL of cell supernatant was used to detect IL-23A, IL1 β, and IL6.
Lipase activity assay
Lipase activity was assessed with a Lipase Assay Kit (Sigma). Briefly, strains were propagated in liquid YPD to an OD600 of 1. One mL of each culture was pelleted, and 100 μL of the supernatant was used for the assay.
pNp-Palmitate Hydrolase Activity Assay
Lipase activity was assayed spectrophotometrically using pNp-palmitate as a substrate. The reaction buffer was composed of 50 mM Tris-HCl pH 8, 1 mg/mL Arabic gum, 0.005% of Triton X-100, and 3.9 mM pNp-palmitate (Sigma). Strains were grown in dendritic cell media to A600 of 1. 5 mL of supernatant was concentrated using concentrator tubes (Thermo Fisher Scientific) for a final volume of one ml. Samples were incubated at 30ºC in the dark for 15 min, and absorbance was read at 410 nm.
Immunoblotting
Strains were propagated in liquid YPD at 30ºC O/N. Supernatants from 10 mL of pelleted cultures were concentrated using protein concentrator tubes (Pierce), denatured in Laemmli buffer, and boiled at 100ºC for 10 min. 15 μL of each sample was analyzed by SDS-PAGE and immunoblotted with anti-FLAG (Thermo Fisher Scientific).
QUANTIFICATION AND STATISTICAL ANALYSIS
Competitive fitness assays (competitive index of mutant vs. WT in the systemic infection model) are presented with a single dot representing the CI of each C. albicans strain in each mouse (biological) replicate, based on the average of three technical replicates. A bar represents the median CI. Fungal burden in infected organs (CFU of C. albicans/gram organ) are presented as the mean of three independent biological replicates. RNA expression is presented as the mean of three biological replicates, each based on the average of three technical replicates. ELISA assays (cytokine protein levels) are presented as the mean of at least three biological replicates; in some figures the results of individual biological replicates are indicated as dots. Error bars depict the standard error of the mean (SEM)
Statistical analyses were performed using GraphPad Prism 9.2 (Graphpad Software). Statistical significance was analyzed by the Student’s t-test (CFU analysis, mRNA expression, cytokine levels, competitive fitness), One-way ANOVA (with Tukey’s correction for multiple comparisons; CFU analysis, mRNA expression, flow cytometry measurements), or the Mantel-Cox test (survival curve), as indicated in the figure legends. No methods were used to determine whether the data met assumptions of the statistical approaches. Significant differences are indicated as * p<0.05; ** p <0.01; *** p<0.001; **** p<0.0001.
Supplementary Material
Table S1. Primers used in this study (related to the Star Methods)
Highlights.
The secreted lipase Lip2 is required for C. albicans virulence in the bloodstream
Lip2 suppresses an antifungal IL-17 response in infected internal organs
Lip2 indirectly impacts IL-17 by suppressing IL-23 production by dendritic cells
Lipase product palmitic acid mimics the activity of Lip2 on cultured dendritic cells
ACKNOWLEDGMENTS
We are grateful to Prof. Gregory Barton (UC Berkeley) for the gift of tlr2−/−, tlr4−/− and tlr2,4−/− femurs and to Prof. Richard Locksley (UCSF) for the gift of SMART17 mice. We thank Allison Cohen for technical advice on the preparation of BMDCs. Finally, we are indebted to Prof. Anita Sil (UCSF), Prof. Sarah L. Gaffen (U. Pittsburg), and Prof. Partha S. Biswas (U. Pittsburg) for illuminating discussions and advice. We thank Merck and Genome Canada for making the original C. albicans GRACE mutant collections available. This work was funded by R01AI127375 to SMN and LEC and R01AI00272 and R01 AI165541 to HDM. EVD was supported by a Damon Runyon Postdoctoral Fellowship. LEC is a Canada Research Chair (Tier 1) in Microbial Genomics & Infectious Disease and co-Director of the CIFAR Fungal Kingdom: Threats & Opportunities program. LEC is a co-founder and shareholder in Bright Angel Therapeutics, a platform company for development of novel antifungal therapeutics.
Footnotes
DECLARATION OF INTERESTS
Competing interests: Authors declare that they have no competing interests.
Data and materials availability: All data are available in the main text or the supplementary materials.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bacher P, Hohnstein T, Beerbaum E, Rocker M, Blango MG, Kaufmann S, Rohmel J, Eschenhagen P, Grehn C, Seidel K, et al. (2019). Human Anti-fungal Th17 Immunity and Pathology Rely on Cross-Reactivity against Candida albicans. Cell 176, 1340–1355 e1315. 10.1016/j.cell.2019.01.041. [DOI] [PubMed] [Google Scholar]
- Bonventre JV (2014). Kidney injury molecule-1: a translational journey. Trans Am Clin Climatol Assoc 125, 293–299; discussion 299. [PMC free article] [PubMed] [Google Scholar]
- Brown GD, Denning DW, Gow NA, Levitz SM, Netea MG, and White TC (2012). Hidden killers: human fungal infections. Sci Transl Med 4, 165rv113. 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- Conti HR, Shen F, Nayyar N, Stocum E, Sun JN, Lindemann MJ, Ho AW, Hai JH, Yu JJ, Jung JW, et al. (2009). Th17 cells and IL-17 receptor signaling are essential for mucosal host defense against oral candidiasis. J Exp Med 206, 299–311. 10.1084/jem.20081463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning DW, Kneale M, Sobel JD, and Rautemaa-Richardson R. (2018). Global burden of recurrent vulvovaginal candidiasis: a systematic review. Lancet Infect Dis 18, e339–e347. 10.1016/S1473-3099(18)30103-8. [DOI] [PubMed] [Google Scholar]
- Edmond MB, Wallace SE, McClish DK, Pfaller MA, Jones RN, and Wenzel RP (1999). Nosocomial bloodstream infections in United States hospitals: a three-year analysis. Clin Infect Dis 29, 239–244. [DOI] [PubMed] [Google Scholar]
- Hallen-Adams HE, and Suhr MJ (2017). Fungi in the healthy human gastrointestinal tract. Virulence 8, 352–358. 10.1080/21505594.2016.1247140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Na L, Fidel PL, and Schwarzenberger P. (2004). Requirement of interleukin-17A for systemic anti-Candida albicans host defense in mice. J Infect Dis 190, 624–631. 10.1086/422329. [DOI] [PubMed] [Google Scholar]
- Hube B, Stehr F, Bossenz M, Mazur A, Kretschmar M, and Schafer W. (2000). Secreted lipases of Candida albicans: cloning, characterisation and expression analysis of a new gene family with at least ten members. Arch Microbiol 174, 362–374. 10.1007/s002030000218. [DOI] [PubMed] [Google Scholar]
- Huppler AR, Bishu S, and Gaffen SL (2012). Mucocutaneous candidiasis: the IL-17 pathway and implications for targeted immunotherapy. Arthritis Res Ther 14, 217. 10.1186/ar3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson BE, Wilhelmus KR, and Hube B. (2007). The role of secreted aspartyl proteinases in Candida albicans keratitis. Invest Ophthalmol Vis Sci 48, 3559–3565. 48/8/3559 [pii] 10.1167/iovs.07-0114. [DOI] [PubMed] [Google Scholar]
- Jiang TT, Shao TY, Ang WXG, Kinder JM, Turner LH, Pham G, Whitt J, Alenghat T, and Way SS (2017). Commensal Fungi Recapitulate the Protective Benefits of Intestinal Bacteria. Cell Host Microbe 22, 809–816 e804. 10.1016/j.chom.2017.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashem SW, Riedl MS, Yao C, Honda CN, Vulchanova L, and Kaplan DH (2015). Nociceptive Sensory Fibers Drive Interleukin-23 Production from CD301b+ Dermal Dendritic Cells and Drive Protective Cutaneous Immunity. Immunity 43, 515–526. 10.1016/j.immuni.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbecki J, and Bajdak-Rusinek K. (2019). The effect of palmitic acid on inflammatory response in macrophages: an overview of molecular mechanisms. Inflamm Res 68, 915–932. 10.1007/s00011-019-01273-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy R, Okada S, Beziat V, Moriya K, Liu C, Chai LY, Migaud M, Hauck F, Al Ali A, Cyrus C, et al. (2016). Genetic, immunological, and clinical features of patients with bacterial and fungal infections due to inherited IL-17RA deficiency. Proc Natl Acad Sci U S A 113, E8277–E8285. 10.1073/pnas.1618300114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling Y, Cypowyj S, Aytekin C, Galicchio M, Camcioglu Y, Nepesov S, Ikinciogullari A, Dogu F, Belkadi A, Levy R, et al. (2015). Inherited IL-17RC deficiency in patients with chronic mucocutaneous candidiasis. J Exp Med 212, 619–631. 10.1084/jem.20141065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionakis MS, Lim JK, Lee CC, and Murphy PM (2011). Organ-specific innate immune responses in a mouse model of invasive candidiasis. J Innate Immun 3, 180–199. 10.1159/000321157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, and Schmittgen TD (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Medzhitov R, Schneider DS, and Soares MP (2012). Disease tolerance as a defense strategy. Science 335, 936–941. 10.1126/science.1214935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogilenko DA, Haas JT, L’Homme L, Fleury S, Quemener S, Levavasseur M, Becquart C, Wartelle J, Bogomolova A, Pineau L, et al. (2019). Metabolic and Innate Immune Cues Merge into a Specific Inflammatory Response via the UPR. Cell 178, 263. 10.1016/j.cell.2019.06.017. [DOI] [PubMed] [Google Scholar]
- Moyes DL, Wilson D, Richardson JP, Mogavero S, Tang SX, Wernecke J, Hofs S, Gratacap RL, Robbins J, Runglall M, et al. (2016). Candidalysin is a fungal peptide toxin critical for mucosal infection. Nature 532, 64–68. 10.1038/nature17625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea MG, Brown GD, Kullberg BJ, and Gow NA (2008). An integrated model of the recognition of Candida albicans by the innate immune system. Nat Rev Microbiol 6, 67–78. [DOI] [PubMed] [Google Scholar]
- Noble SM, French S, Kohn LA, Chen V, and Johnson AD (2010). Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat Genet 42, 590–598. ng.605 [pii] 10.1038/ng.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble SM, Gianetti BA, and Witchley JN (2017). Candida albicans cell-type switching and functional plasticity in the mammalian host. Nat Rev Microbiol 15, 96–108. 10.1038/nrmicro.2016.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble SM, and Johnson AD (2005). Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot Cell 4, 298–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Meara TR, Veri AO, Ketela T, Jiang B, Roemer T, and Cowen LE (2015). Global analysis of fungal morphology exposes mechanisms of host cell escape. Nat Commun 6, 6741. 10.1038/ncomms7741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller M, Neofytos D, Diekema D, Azie N, Meier-Kriesche HU, Quan SP, and Horn D. (2012). Epidemiology and outcomes of candidemia in 3648 patients: data from the Prospective Antifungal Therapy (PATH Alliance(R)) registry, 2004–2008. Diagn Microbiol Infect Dis 74, 323-331. 10.1016/j.diagmicrobio.2012.10.003. [DOI] [PubMed] [Google Scholar]
- Pfaller MA, and Diekema DJ (2007). Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev 20, 133–163. 20/1/133 [pii] 10.1128/CMR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller MA, Pappas PG, and Wingard JR (2006). Invasive Fungal Pathogens: Current Epidemiological Trends. Clin Infect Dis 43 (1), S3–14. [Google Scholar]
- Price AE, Reinhardt RL, Liang HE, and Locksley RM (2012). Marking and quantifying IL-17A-producing cells in vivo. PLoS One 7, e39750. 10.1371/journal.pone.0039750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puel A, Cypowyj S, Bustamante J, Wright JF, Liu L, Lim HK, Migaud M, Israel L, Chrabieh M, Audry M, et al. (2011). Chronic mucocutaneous candidiasis in humans with inborn errors of interleukin-17 immunity. Science 332, 65–68. 10.1126/science.1200439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendran R, Lew SK, Yong CX, Tan J, Wang DJ, and Chuang KH (2013). Quantitative mouse renal perfusion using arterial spin labeling. NMR Biomed 26, 1225–1232. 10.1002/nbm.2939. [DOI] [PubMed] [Google Scholar]
- Ramani K, Garg AV, Jawale CV, Conti HR, Whibley N, Jackson EK, Shiva SS, Horne W, Kolls JK, Gaffen SL, and Biswas PS (2016). The Kallikrein-Kinin System: A Novel Mediator of IL-17-Driven Anti-Candida Immunity in the Kidney. PLoS Pathog 12, e1005952. 10.1371/journal.ppat.1005952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramani K, Jawale CV, Verma AH, Coleman BM, Kolls JK, and Biswas PS (2018). Unexpected kidney-restricted role for IL-17 receptor signaling in defense against systemic Candida albicans infection. JCI Insight 3. 10.1172/jci.insight.98241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roemer T, Jiang B, Davison J, Ketela T, Veillette K, Breton A, Tandia F, Linteau A, Sillaots S, Marta C, et al. (2003). Large-scale essential gene identification in Candida albicans and applications to antifungal drug discovery. Mol Microbiol 50, 167–181. [DOI] [PubMed] [Google Scholar]
- Saijo S, Ikeda S, Yamabe K, Kakuta S, Ishigame H, Akitsu A, Fujikado N, Kusaka T, Kubo S, Chung SH, et al. (2010). Dectin-2 recognition of alpha-mannans and induction of Th17 cell differentiation is essential for host defense against Candida albicans. Immunity 32, 681–691. 10.1016/j.immuni.2010.05.001. [DOI] [PubMed] [Google Scholar]
- Shao TY, Ang WXG, Jiang TT, Huang FS, Andersen H, Kinder JM, Pham G, Burg AR, Ruff B, Gonzalez T, et al. (2019). Commensal Candida albicans Positively Calibrates Systemic Th17 Immunological Responses. Cell Host Microbe 25, 404–417 e406. 10.1016/j.chom.2019.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparber F, and LeibundGut-Landmann S. (2019). Interleukin-17 in Antifungal Immunity. Pathogens 8. 10.3390/pathogens8020054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoldt VR, Sonneborn A, Leuker CE, and Ernst JF (1997). Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J 16, 1982–1991. 10.1093/emboj/16.8.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton CE, Lalor SJ, Sweeney CM, Brereton CF, Lavelle EC, and Mills KH (2009). Interleukin-1 and IL-23 induce innate IL-17 production from gammadelta T cells, amplifying Th17 responses and autoimmunity. Immunity 31, 331–341. 10.1016/j.immuni.2009.08.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Primers used in this study (related to the Star Methods)
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request..
This paper does not report original code.
Any additional information required to reanalyze the data reported in this work is available from the Lead Contact upon request.