Abstract
An isogenic mutant of Streptococcus pyogenes Manfredo that lacks the ability to make streptococcal acid glycoprotein (SAGP) has been constructed by inserting a deletion in the sagp gene using the method of allelic exchange. An assay of cell extracts (CE) prepared from the wild-type and mutant Manfredo strains for the enzyme arginine deiminase (AD) showed that significant activity was present in wild-type CE but none could be detected in mutant CE. These findings confirm our earlier conclusion that SAGP has AD activity (B. A. Degnan, J. M. Palmer, T. Robson, C. E. D. Jones, M. Fischer, M. Glanville, G. D. Mellor, A. G. Diamond, M. A. Kehoe, and J. A. Goodacre, Infect. Immun. 66:3050–3058, 1998). Wild-type CE but not mutant CE potently inhibited human peripheral blood mononuclear cell proliferation in response to phytohemagglutinin, and this inhibition was overcome by the addition of l-arginine to proliferation assay mixtures. Invasion assays showed that the isogenic mutant organisms lacking SAGP, and thus AD activity, were between three and five times less able to enter epithelial cells (Hep-2C and A549) than were the wild-type streptococci. Both wild-type and mutant S. pyogenes bacteria were extremely sensitive to low pH. However, l-arginine (1 mM or above) significantly increased the viability of the wild type but not the isogenic mutant organisms under acidic conditions. The difference in acid susceptibility between wild-type and mutant bacteria may explain the reduced capacity of the isogenic mutant bacteria to invade and survive intracellularly.
Streptococcus pyogenes (group A Streptococcus) is an important human pathogen, infections with which can lead to acute disorders such as pharyngitis, erysipelas, otitis media, and impetigo or to pathogenic sequelae including glomerulonephritis, acute rheumatic fever and reactive arthritis (6, 7, 21, 29, 42, 53). In the last decade there has been a dramatic resurgence in the incidence of serious streptococcal infections; in particular, the number of cases of streptococcal toxic shock syndrome, bacteremia, and necrotising fasciitis has increased (11). A greater understanding of the ways in which this pathogen interacts with the host, the virulence factors that it produces, and the identification of possible targets for vaccine design are required.
During a previous study, it was observed that cell extracts (CE) prepared from a wide range of S. pyogenes strains potently inhibited antigen-, superantigen-, or phytohemagglutinin (PHA)-stimulated human peripheral blood mononuclear cell (PBMC) proliferation in vitro (16, 17). Purification of the inhibitory component present in CE of S. pyogenes Manfredo by anion-exchange chromatography followed by gel filtration chromatography yielded a single protein. When sequenced, this protein was found to have an NH2-terminal sequence identical to streptococcal acid glycoprotein (SAGP), which had been isolated from S. pyogenes Su (23, 24, 56, 57). SAGP has 31 to 39% amino acid identity to arginine deiminase (AD) from Mycoplasma hominis, Mycoplasma arginini, Pseudomonas putida, and Pseudomonas aeruginosa (3). It was subsequently shown that CE obtained from several group A streptococcal strains all contained AD activity and that a direct correlation existed between the levels of AD present in CE and their ability to inhibit T-cell proliferation (17). In addition, the inhibitory activity of S. pyogenes Manfredo CE could be overcome by the addition of l-arginine to proliferation assay mixtures in which human PBMC were stimulated with PHA. It was therefore concluded that SAGP, or its homolog, possesses AD activity and that the potent inhibition of proliferation of human T cells by streptococcal CE is due to the activity of this enzyme (17).
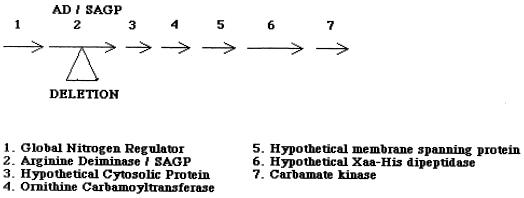
AD is one of three enzymes that comprises the arginine deiminase system, and it catalyzes the conversion of l-arginine to citrulline with the concomitant production of ammonia (13). The citrulline thus formed is then converted to ornithine and carbamoyl phosphate by the action of ornithine carbamoyltransferase. Carbamate kinase then catalyzes the breakdown of carbamoyl phosphate to yield carbon dioxide and ammonia, with the generation of 1 mol of ATP. A search of the genomic sequence of Streptococcus pyogenes (Web site http://dna1 .chem.ou.edu/strep.html) showed that SAGP is encoded by open reading frame 1429 (bp 577203 to 578438). Downstream of the SAGP gene are genes coding for a hypothetical cytosolic protein (bp 578538 to 578966), ornithine carbamoyltransferase (bp 579006 to 580018), a hypothetical membrane-spanning protein (bp 580188 to 581681), a hypothetical Xaa-His dipeptidase (bp 581698 to 583029), and carbamate kinase (bp 583049 to 583999) (Fig. 1). The close proximity of these genes on the streptococcal chromosome may suggest that they are arranged in an operon. The genes encoding the three enzymes of the AD system in P. aeruginosa have been documented as being in an operon (4, 27, 28), and expression of the three genes of the AD system in S. sanguis, Rhizobium etli, and S. faecalis is strongly interrelated (9, 18, 51).
FIG. 1.
Gene locus of S. pyogenes containing the AD/SAGP gene.
We have constructed an isogenic mutant of S. pyogenes Manfredo that lacks the ability to make SAGP by inserting a deletion in the sagp gene using the method of allelic exchange, to determine the role that AD plays in S. pyogenes. We have characterized this mutant in terms of AD production and have measured the ability of CE prepared from the wild-type and mutant Manfredo strains to inhibit human PBMC proliferation. We have also compared the pH sensitivities of the wild-type and mutant streptococci. Ammonia generated by the metabolism of l-arginine by the AD system within the bacterial cell can act to raise the pH in the cytoplasm. Extensive studies have shown that group A streptococci, once believed to be exclusively extracellular pathogens, can invade and survive within a wide range of mammalian cell types (11, 12, 19, 20, 26, 37, 40). AD may be involved in protecting streptococci from the low pH found inside cells (33, 36). To test this hypothesis, in addition to experiments to determine the pH sensitivity of the wild-type and mutant bacteria, we have carried out invasion assays using human alveolar type II epithelium-like cells derived from a lung adenocarcinoma cell line (A549) and a human laryngeal epithelial cell line (Hep-2C) to compare the abilities of the wild-type and mutant strains to survive intracellularly.
MATERIALS AND METHODS
Bacterial strains, plasmids, and tissue culture cells.
S. pyogenes Manfredo was a kind gift from the late Ed Beachey and was obtained from the Memphis VA Hospital Culture Collection, Memphis, Tenn. Streptococci were maintained routinely on Todd-Hewitt agar (Difco, Detroit, Mich.) plates containing 5% (vol/vol) horse blood (TCS Biologicals Ltd., Buckingham, United Kingdom) and were grown in Todd-Hewitt broth (THB) (Difco). Escherichia coli was grown in Luria-Bertani (LB) broth and on LB agar plates (Difco). Plasmid pG+host9 (pVE6155) is a temperature-sensitive E. coli-gram-positive shuttle vector containing an erythromycin resistance determinant. The plasmid and an E. coli host strain designated EC101-derivative which carries the wild-type plasmid repA gene in its chromosome were very kindly provided by E. Maguin, Institute National de la Recherche Agronomique, Jouy En Josas, France. Human alveolar type II epithelium-like cells (A549) were kindly provided by Ann Vernallis, Biochemistry Department, University of Birmingham, Birmingham, United Kingdom, and the human laryngeal epithelial cell line (Hep-2C) was a kind gift from Margaret Scott, Department of Microbiology and Immunology, University of Newcastle, Newcastle Upon Tyne, United Kingdom. Both cell lines were routinely grown in Dulbecco's modified Eagle's medium (DMEM) (Gibco Laboratories, Paisley, United Kingdom) supplemented with 10% fetal calf serum, penicillin (100 μg ml−1), streptomycin (100 μg ml−1), and glutamine (3 mM) and were subcultured every 3 to 4 days after being detached using 0.25% trypsin–1 mM EDTA.
DNA manipulation.
Restriction endonuclease digests, DNA ligations, transformation of electrocompetent E. coli, and agarose gel electrophoresis were performed by standard techniques (47). Restriction enzymes (HpaI, EcoRI, SalI, XhoI, and Acc65I) were purchased from New England BioLabs. DNA ligase was purchased from Boehringer Mannheim. DNA fragments were purified from gels or solution using a Qiagen gel extraction kit as specified by the manufacturer.
Purification of streptococcal chromosomal DNA.
Bacteria were grown to late exponential phase at 37°C without shaking in THB. Bacterial cells were then harvested by centrifugation at room temperature for 15 min at 5,000 × g, and the bacterial pellet was washed with 50 ml of 0.2 M Tris-malate (pH 7.0) buffer before being resuspended in 2 ml of 25% (wt/vol) sucrose in 0.2 M Tris-malate (pH 7.0) buffer. Mutanolysin (0.1 mg) and lysozyme (2 mg) were then added, and the mixture was incubated at 37°C for 4 h to break down cell wall material, after which time 0.3 ml of 0.25 M EDTA, 2 ml of 0.2 M Tris-malate (pH 7.0) buffer, and 0.5 ml of 10% (wt/vol) sodium dodecyl sulfate (SDS) were added. Two phenol-chloroform extractions were then carried out, with centrifugation at 5,000 × g for 20 min between each extraction. DNase-free RNase (20 μg ml−1) was added to the top, aqueous layer, which was then incubated for 30 min at room temperature. After a further two phenol-chloroform extractions, the DNA present in the aqueous layer was precipitated with 2 volumes of absolute ethanol plus 1/10 volume of 3 M sodium acetate (pH 7.0). The precipitated DNA was removed to a clean tube before being centrifuged at 10,000 × g for 20 min. The DNA pellet was air dried and finally resuspended in 100 to 200 μl of 10 mM Tris-HCl–1 mM EDTA (pH 8.0) buffer.
Electrotransformation of S. pyogenes with pG+host9 DNA.
Streptococcal cells were transformed by the method of Simon and Ferretti (50). Briefly, transforming DNA (10 μl) was added to electrocompetent cells (100 μl) in a chilled Gene Pulser cuvette, with a 0.2-cm electrode gap (Bio-Rad Laboratories, Richmond, Calif.). A pulse was then delivered at 2.5 kV, 25 μF, and 200 Ω using a Gene Pulser (Bio-Rad Laboratories). THB (1 ml) was immediately added, and the bacteria were incubated for 3 h at 28°C. The bacteria were then pelleted by centrifugation at 12,000 × g for 2 min, resuspended in 100 μl of THB, and plated out onto THB blood agar plates containing erythromycin (1.75 μg ml−1). The plates were incubated for 36 to 48 h at 28°C.
Small-scale purification of plasmid DNA.
Minipreparations of plasmid DNA were obtained by the alkali lysis method, which is a modification of the methods of Birnboim and Doly (5) and Ish-Horowicz and Burke (22).
Gene replacement mutagenesis by allelic exchange in S. pyogenes Manfredo.
PCR fragments generated using primer pairs Middle 1 plus End 1 and Middle 2 plus End 2 with Manfredo wild-type chromosomal DNA as template were digested with Acc65I and then ligated (Table 1). This DNA fragment was then amplified by PCR using the primer pair End 1 plus End 2. Following digestion with EcoRI and SalI, the PCR fragment was ligated into plasmid pG+host9. This construct was then electroporated into E. coli (TG1 derivative), and the bacteria were plated out on LB plates containing erythromycin (300 μg ml−1), tetracycline (15 μg ml−1), and kanamycin (30 μg ml−1) and grown at 28°C. pG+host9 contains a temperature-sensitive replicon that is active at 28°C but not at 37°C. Individual bacterial colonies were picked and minipreparations of plasmid DNA were carried out as described above. Each minipreparation was screened for the presence of an insert by digestion with EcoRI and SalI followed by agarose gel electrophoresis. The desired pG+host9 construct was then electroporated into S. pyogenes Manfredo as described above, and transformants were selected following growth of colonies at 28°C on Todd-Hewitt blood agar plates containing erythromycin (1.75 μg ml−1). A single transformed colony was used to inoculate 1 ml of THB containing erythromycin (1.75 μg ml−1), and the culture was grown overnight at 28°C under stationary conditions. The overnight culture was diluted 100-fold in the same medium and was incubated as above until an optical density of 600 nm (OD600) of 0.2 to 0.3 was reached, at which point the culture was transferred to 37°C, a temperature at which the plasmid is unable to replicate. This step selects for strains in which the plasmid has integrated into the Manfredo chromosome via a single recombination event over the homologous plasmid insert and chromosome sequence. Serial 10-fold dilutions (100 μl) of this culture were plated out on Todd-Hewitt blood agar plates containing erythromycin (1.75 μg ml−1), and the plates were incubated at 37°C. A single colony containing the recombinant plasmid integrated into the chromosome was picked and used to inoculate 1 ml of THB containing erythromycin (1.75 μg ml−1), and the culture was grown at 37°C overnight, after which the culture was diluted in erythromycin-free THB and grown at 28°C. At this temperature, the plasmid is able to replicate and is excised from the chromosome via a second recombination event over the duplicated target gene sequence. The excision of the plasmid can yield the wild-type genotype or can complete the allelic exchange, resulting in an incomplete sagp gene. Serial 10-fold dilutions (100 μl) of the culture were plated out on erythromycin-free Todd-Hewitt blood agar plates and were incubated overnight at 37°C. Single colonies were then replica plated onto Todd-Hewitt blood agar plates with or without erythromycin (1.75 μg ml−1). Colonies that were erythromycin sensitive were screened by PCR to identify possible mutants.
TABLE 1.
Oligonucleotide primers used in this study
| Name | Sequence (5′ to 3′) | Location of primera | 5′-terminal restriction endonuclease site |
|---|---|---|---|
| End 1 | AGG AGT ACA CTA GGA ATA TAA GC | −81–−59 (F) | None |
| End 2 | C CCC GTC GAC CCT ATA ACC ACC TTT ACC ATA GC | 1262–1240 (R) | SalI |
| Middle 1 | TTA AGG TAC CCA CCT GTA CCG ATA GTT GCG | 523–504 (R) | Acc65I |
| Middle 2 | GAA AGG TAC CAT GTG CTT GCG GTT GGT ATT TC | 689–710 (F) | Acc65I |
| A | Same as End 1 | Same as END 1 | |
| A′ | CAA TAG AAG CAG CAT CTG TAC G | 736–715 (R) | None |
| B | GCC ATC GAC CCA ATG CCA AAC | 460–480 (F) | None |
| B′ | GTT TCT AGC GCT TCA GTG AC | 1526–1507 (R) | None |
Numbers indicate the location of the oligonucleotide primer with respect to a translational start site at position 1. F, forward primer; R, reverse primer. The accession number of SAGP in the EMBL database is X55659.
PCR.
PCR amplifications were performed by standard techniques with all primer pairs used with denaturing, annealing, and extension temperatures and times of 95°C for 1 min, 61°C for 1 min and 69°C for 2 min, respectively. Oligonucleotide primers were prepared by the Molecular Biology Unit of the University of Newcastle and are listed in Table 1. Taq DNA polymerase and deoxynucleoside triphosphates were purchased from Boehringer Mannheim.
DNA sequencing.
DNA sequencing was performed on an automatic sequencer (Perkin-Elmer Biosystems Model 877), using the dideoxynucleotide chain termination method with fluorescent tags, by the Molecular Biology Unit of the University of Newcastle.
Southern hybridization.
Streptococcal chromosomal DNA was digested overnight at 37°C with HpaI and XhoI. The DNA fragments were then separated by agarose gel electrophoresis on a 0.7% agarose gel at 4 V/cm before being transferred overnight by capillary transfer by the method of Southern (52) to Hybond N+ blotting membrane (Amersham Life Science). Fluorescein-labeled DNA molecular weight markers (1-kbp ladder) were also separated on the agarose gel. The membrane was dried at 80°C for 2 h and then incubated at 60°C for at least 30 min with hybridization buffer (5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.1% [wt/vol] SDS, 5.0% [wt/vol] dextran sulfate). Denatured fluorescein-labeled probe was then added to the hybridization buffer, and the membrane was incubated at 60°C overnight. A PCR product generated with the primers End 1 plus Middle 1 was used as the probe. DNA molecular weight markers and the probe were labeled with fluorescein using the Gene Images random-primer labeling module purchased from Amersham Life Science. Following two high-stringency washes of the labeled membrane at 60°C with 1× SSC–0.1% (wt/vol) SDS and 0.5× SSC–0.1% (wt/vol) SDS for 15 min each, fluorescein-labeled DNA fragments were detected using the Gene Images CDP-Star detection module obtained from Amersham Life Science. The membrane was then developed.
Inhibition of T-cell proliferation by streptococcal CE.
Bacterial CE were prepared and the proliferation assays using human PBMC were performed as described previously (17).
Assay of AD activity.
AD activity was assayed by measuring the rate of conversion of l-arginine to citrulline by the method of Oginsky (35).
Growth curves.
THB (50 ml) and THB (50 ml) supplemented with yeast extract (0.5%, wt/vol) were inoculated with a loopful of S. pyogenes taken from blood agar plates, and the bacteria were grown at 37°C under stationary conditions. After overnight growth, 50 ml of fresh medium was inoculated with 0.5 ml of each culture and incubated as above. Samples (2.0 ml) were removed at hourly intervals to determine the OD600.
Comparison of the pH sensitivity of S. pyogenes Manfredo wild type and mutant.
S. pyogenes was grown under stationary conditions at 37°C in THB to an OD600 of 0.7 to 0.8. The bacteria were then harvested by centrifugation at 5,000 × g for 15 min and resuspended in 20 mM Na2HPO4/NaH2PO4–1 mM MgCl2 (pH 7.0) buffer to give approximately 109 to 1010 CFU ml−1. Equal numbers of bacteria were removed and added to 20 mM Na2HPO4/NaH2PO4–1 mM MgCl2 buffer at pH 4, 5, 6, or 7 supplemented with either l-arginine or citrulline. Aliquots (100 μl) were removed at various time intervals, and the number of viable bacteria remaining was determined by plating out 10-fold dilutions on Todd-Hewitt blood agar plates.
Invasion assays.
Invasion assays were carried out as described by LaPenta et al. (26). Hep-2C or A549 cells (5 × 105 cells/well) were incubated for 24 h in a humidified atmosphere of 5% CO2 at 37°C in antibiotic-free DMEM containing 10% fetal calf serum and glutamine (3 mM) in 24-well plates. The confluent monolayers were then washed three times with fresh antibiotic-free DMEM before being infected with approximately 105 to 107 CFU of streptococci. The S. pyogenes inoculum was grown overnight in THB (50 ml), harvested by centrifugation at 5,000 × g for 10 min and washed three times in phosphate-buffered saline (PBS). The infected monolayers were incubated for 90 min and then washed three times with PBS, and DMEM containing gentamicin (200 μg ml−1) was added. After a further 90-min incubation, the cells were washed three times and dispersed using 0.25% trypsin–1 mM EDTA (200 μl). Intracellular bacteria were released by adding sterile ice-cold distilled H2O (800 μl) to the cells and counted by plating out on Todd-Hewitt blood agar plates.
Statistical analysis.
Results are expressed as means ± standard error of the mean (SEM). Statistical differences were determined by Student's t test.
Other reagents.
Unless otherwise stated, all chemicals were purchased from Sigma Chemical Co., Poole, United Kingdom.
RESULTS
Generation of an S. pyogenes Manfredo SAGP-deficient isogenic mutant.
An isogenic mutant of S. pyogenes Manfredo impaired in SAGP production was constructed by replacing the wild-type sagp gene with a disrupted gene containing a 165-bp deletion (bp 523 to 689, where bp 1 is the translational start point) by the method of allelic exchange. The deletion, together with the insertion of 6 bp (GGTACC) from the Acc65I restriction endonuclease site of primers Middle 1 and Middle 2, resulted in an in-frame mutation and did not create any termination codons. PCR analyses were carried out to screen potential mutants for the presence of the disrupted gene. Chromosomal DNA preparations from wild-type Manfredo, a mutant bacterium in which the cloned gene and plasmid had been incorporated into the chromosome by homologous recombination, and potential mutants that were generated by excision of the plasmid from the chromosome by a second homologous recombination step were used as templates with the primer pairs B plus End 2, B plus B′, A′ plus End 1, and End 1 plus B. After agarose gel electrophoresis of the PCR products, a mutant was identified by the fact that each PCR product from this colony was approximately 165 bp smaller than the PCR products generated from the wild-type DNA (results not shown). To verify that the reduction in size of the PCR products was due to the required deletion, a PCR-amplified product covering region −81 to 1507, using primers End 1 plus B′, was sequenced and found to have the desired sequence (data not shown). Southern hybridization of HpaI- and XhoI-digested chromosomal DNA prepared from the mutant and wild-type Manfredo strains with a fluorescein-labeled probe generated by PCR amplification of the region from −81 to 523) yielded a single band in each case with approximate sizes of 1,100 and 1,300 bp, respectively (results not shown), showing that initial integration of the plasmid containing the cloned sagp gene into the Manfredo chromosome by homologous recombination was unlikely to take place at any site on the chromosome other than at the sagp gene.
No differences were observed in the growth rates of the mutant and wild-type bacteria in THB or THB supplemented with 0.5% yeast extract (results not shown). CE prepared from the wild-type and mutant bacteria were assayed for AD activity, and it was shown that whereas wild-type CE possessed AD activity (43.7 μM citrulline produced/h/μg of protein), no AD activity was detected in mutant CE, endorsing our view that SAGP has AD activity.
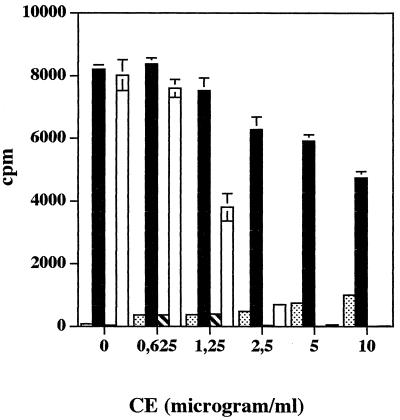
Effect of CE prepared from S. pyogenes Manfredo wild type and isogenic mutant on PHA-stimulated proliferation of human PBMC.
To determine whether it is solely the action of AD that is responsible for inhibiting T-cell proliferation, human PBMC were incubated with a range of concentrations of CE prepared from the wild-type and isogenic mutant Manfredo strains in either the presence or absence of PHA (1.0 μg ml−1). Figure 2 shows that the proliferative response to PHA was significantly inhibited by the addition of wild-type CE at 1.25 μg ml−1 or above (P < 0.005). In the presence of 5 or 10 μg of wild-type CE per ml, proliferation of human PBMC was completely abolished (P < 0.0001). In contrast, strong PHA-stimulated proliferative responses of human PBMC were observed in the presence of all concentrations of mutant CE tested, although significant reduction in proliferation was elicited by 5 or 10 μg of mutant CE per ml (P < 0.001). No significant proliferation of human PBMC was observed in the presence of either wild-type or mutant CE alone after 3 days.
FIG. 2.
Effect of CE prepared from S. pyogenes Manfredo wild-type and isogenic mutant bacteria on the PHA-stimulated proliferation of human PBMC. Human PBMC (2 × 105/well) were incubated in 96-well round-bottom plates in the presence of either wild-type streptococcal CE alone ( ), isogenic mutant CE alone (
), isogenic mutant CE alone ( ), wild-type streptococcal CE plus PHA (□), or isogenic mutant CE plus PHA (■). A range of concentrations of CE was used, while PHA was added to give a final concentration of 1.0 μg ml−1. After 3 days, the cells were pulsed with tritiated thymidine for the final 6 h of culture and the incorporated radioactivity was measured. Results show mean [3H]thymidine uptake and SEM for triplicate cultures.
), wild-type streptococcal CE plus PHA (□), or isogenic mutant CE plus PHA (■). A range of concentrations of CE was used, while PHA was added to give a final concentration of 1.0 μg ml−1. After 3 days, the cells were pulsed with tritiated thymidine for the final 6 h of culture and the incorporated radioactivity was measured. Results show mean [3H]thymidine uptake and SEM for triplicate cultures.
To further confirm that the inhibitory activity of wild-type CE was mainly due to AD, l-arginine was titrated into proliferation assay mixtures in which human PBMC were stimulated with PHA (1.0 μg ml−1) in the presence of an inhibitory concentration of wild-type CE (2.5 μg ml−1). In the absence of exogenous l-arginine, T-cell proliferation was significantly reduced (P < 0.0002), but this was restored in a dose-dependent manner by the addition of exogenous l-arginine. No inhibition due to wild-type CE was seen in the presence of 4, 8, or 16 mM l-arginine, although at these concentrations the amino acid itself was inhibitory (data not shown).
Thus, SAGP has AD activity, and the inhibitory action of Manfredo CE on proliferation of human PBMC can be largely attributed to the action of this enzyme.
pH sensitivity of wild-type and isogenic mutant S. pyogenes Manfredo bacteria.
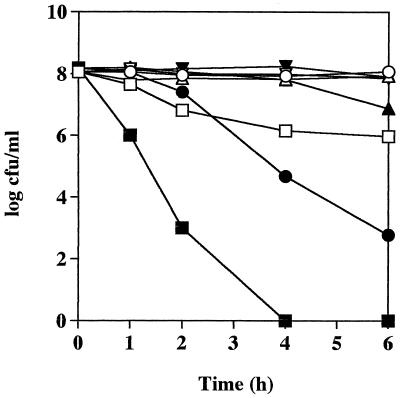
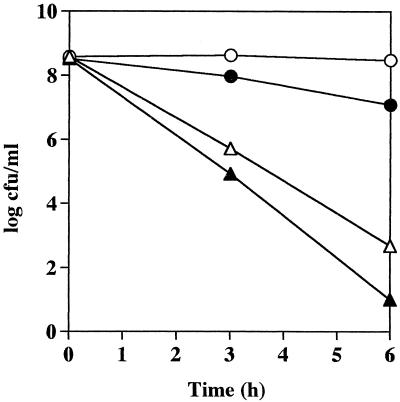
To compare the pH sensitivity of the wild-type and isogenic mutant streptococci, equal numbers of bacteria were added to 20 mM Na2HPO4/NaH2PO4–1 mM MgCl2 buffer containing 50 mM l-arginine at pH 4, 5, 6, or 7 and incubated at 37°C without shaking. Samples were removed at various time intervals, and the number of viable bacteria remaining was measured. Figure 3 shows a significant difference between the survival rates of the isogenic mutant and wild-type bacteria at pH 4. Although the numbers of wild-type bacteria did decrease at pH 4 after 1 h of incubation, they stabilized after this initial reduction and 9.43 × 105 CFU ml−1 of the original inoculum (1.12 × 108 CFU ml−1) was still viable after 6 h incubation. However, isogenic mutant bacteria were rapidly killed at pH 4, and after 4 h no viable streptococci were detected. A dramatic difference was also seen in the survival rates of the isogenic mutant and wild-type bacteria at pH 5. After 2 h of incubation, the number of viable isogenic mutant bacteria decreased rapidly with time, and after 6 h of incubation, only 6 × 102 CFU ml−1 of the initial inoculum (1.53 × 108 CFU ml−1) was still viable. In contrast, the numbers of wild-type bacteria remained unchanged during the 6-h incubation at pH 5. At pH 6, the viability of both groups of bacteria did not vary with time until after 4 h, when there was a slight reduction in the numbers of isogenic mutant bacteria compared to wild-type S. pyogenes Manfredo. At pH 7, no difference was seen between the viability of the wild-type and isogenic mutant organisms and the numbers of both types of bacteria remained constant over the 6-h incubation (Fig. 3).
FIG. 3.
Effect of pH on the survival of S. pyogenes Manfredo wild-type and isogenic mutant bacteria. Either wild-type (open symbols) or isogenic mutant (solid symbols) streptococci were harvested from a late-exponential culture, and equal numbers were suspended in 20 mM Na2HPO4/NaH2PO4–1 mM MgCl2 buffer containing 50 mM l-arginine at pH 4 (□, ■), pH 5 (○, ●), pH 6 (▵, ▴), or pH 7 (▿), ▾). The bacteria were incubated under stationary conditions at 37°C, and aliquots (100 μl) were removed at various time intervals to determine the number of remaining viable bacteria by plating out serial dilutions on Todd-Hewitt blood agar plates. Results are representative of three separate experiments.
Effect of l-arginine on the pH sensitivity of S. pyogenes Manfredo.
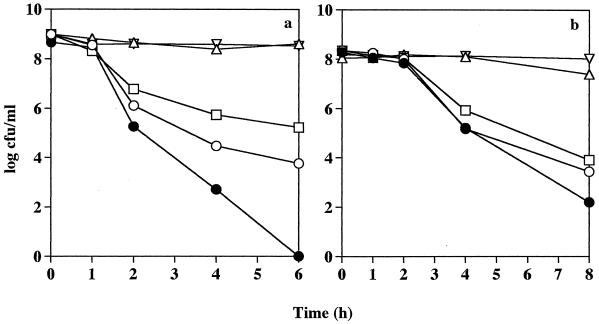
Having shown that S. pyogenes Manfredo isogenic mutant bacteria deficient in SAGP production were more susceptible to acidic conditions than were the wild-type Manfredo bacteria in the presence of 50 mM l-arginine, experiments were done to confirm that l-arginine was protecting the wild-type bacteria from the acid environment and also to determine the minimum concentration of l-arginine needed to be protective. Wild-type bacteria were suspended in 20 mM Na2HPO4/NaH2PO4–1 mM MgCl2 buffer at pH 4, 5, 6, and 7 supplemented with 0, 0.1, 1.0, or 10 mM l-arginine. Isogenic mutant organisms were suspended in buffer supplemented with 10 mM l-arginine only, since it has already been shown that 50 mM was not protective. Samples were removed at various time intervals to determine bacterial viability. Figure 4a indicates that at pH 4 in the presence of either 1.0 or 10 mM l-arginine, the numbers of wild-type bacteria remained constant over a 6-h incubation, but bacterial survival was significantly diminished when a lower concentration of l-arginine was used (0 or 0.1 mM), with 5.89 × 103 and 1.70 × 105 CFU ml−1 of the original inoculum (9.67 × 108 CFU ml−1), respectively, remaining viable after 6 h. The wild-type bacteria in the absence of exogenous l-arginine still appeared to have a survival advantage over the isogenic mutant organisms, with the latter being rapidly killed at pH 4 despite the presence of 10 mM l-arginine (Fig. 4a). Similar survival patterns were observed at pH 5. Supplementation of the buffer with either 1.0 or 10 mM l-arginine completely protected the wild-type bacteria at pH 5, but survival of wild-type streptococci closely resembled that of the isogenic mutant organism in the presence of 0 or 0.1 mM l-arginine, with numbers of viable bacteria severely decreasing after 2 h of incubation (Fig. 4b). A similar but less pronounced trend was also seen at pH 6 (data not shown). At pH 7, the numbers of wild-type and isogenic mutant bacteria both remained constant over the incubation period, with no detectable effect due to the presence of l-arginine (results not shown). Thus, S. pyogenes Manfredo is highly sensitive to acidic conditions but l-arginine at a minimum concentration of 1 mM greatly enhances bacterial survival at low pH. Conversely, a SAGP-deficient mutant is not given a survival advantage by the presence of l-arginine.
FIG. 4.
Effect of l-arginine on the pH sensitivity of S. pyogenes Manfredo wild-type and isogenic mutant bacteria. Streptococci were harvested from a late-exponential culture, and equal numbers were suspended in 20 mM Na2HPO4/NaH2PO4–1 mM MgCl2 buffer at pH 4 (a) or pH 5 (b). The buffer was supplemented with 0 mM (○), 0.1 mM (□), 1.0 mM (▵), or 10.0 mM (▿) l-arginine for the wild-type bacteria and 10 mM (●) l-arginine for the isogenic mutant organisms. The cultures were incubated as described in the legend to Fig. 3, and the number of viable streptococci was determined at various time intervals. Results are representative of two separate experiments.
Effect of citrulline on the viability of mutant and wild-type S. pyogenes.
Experiments were done to determine whether citrulline could protect isogenic mutant bacteria from acidic conditions. Citrulline, the product of AD, is metabolized further via the AD system to yield ATP, NH3, and CO2. Therefore, if the expression of ornithine carbamoyltransferase and carbamate kinase was not affected in the isogenic mutant, citrulline should protect the wild-type and mutant bacteria equally well at a low pH through the production of NH3.
Equal numbers of wild-type and mutant bacteria were suspended in 20 mM Na2HPO4/NaH2PO4–1 mM MgCl2 buffer at pH 4 with or without 50 mM citrulline, and samples were removed for viable-count determination at certain time intervals. Figure 5 shows that citrulline protected the wild-type and isogenic mutant bacteria equally well at pH 4, with the numbers of viable bacteria remaining relatively constant over time. In the absence of citrulline, both wild-type and isogenic mutant bacteria were rapidly killed. Therefore, the isogenic mutant bacteria were able to metabolize citrulline as efficiently as were the wild-type organisms, indicating that expression of ornithine carbamoyltransferase and carbamate kinase was not affected in the S. pyogenes mutant bacteria.
FIG. 5.
Effect of citrulline on the survival of S. pyogenes Manfredo wild-type (open symbols) and isogenic mutant (solid symbols) bacteria at pH 4.0. Streptococci were harvested and incubated in buffer in the presence (○, ●) or absence (▵, ▴) of 50 mM citrulline as described in the legend to Fig. 3. The number of viable streptococci remaining at various time intervals was determined. Results are representative of three separate experiments.
Invasion of epithelial cells by S. pyogenes Manfredo wild-type and isogenic mutant bacteria.
Gentamicin protection assays were done to compare the ability of the wild-type and isogenic mutant bacteria to enter and/or survive in Hep-2C and A549 cells. Initially, epithelial monolayers (5 × 105 cells per well) were incubated for 90 min with either wild-type or isogenic mutant streptococci to give a bacterium-to-cell ratio of 100:1, 10:1, or 1:1. The numbers of intracellular bacteria were then determined after a further 90-min incubation in DMEM containing gentamicin (200 μg ml−1). The results consistently showed little difference between the numbers of wild-type and isogenic mutant streptococci recovered from the monolayers at the highest inoculum used, but between three- and fivefold fewer isogenic mutant bacteria were recovered from the monolayers compared to the wild type at the two lower infective doses, irrespective of the nature of the monolayer used (results not shown).
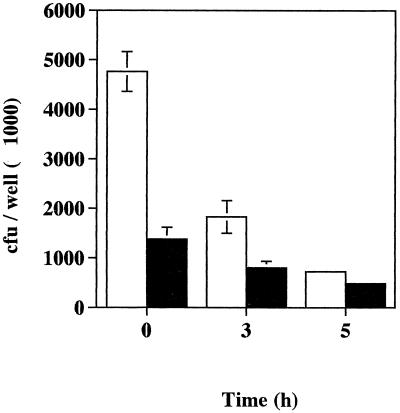
Further experiments were then done to compare the recovery of the bacteria over a longer incubation period (after a further 3 and 5 h of incubation). Hep-2C cells were incubated for 90 min with streptococci to give a bacterium-to-cell ratio of approximately 10:1. After 90 min of incubation with 200 μg of gentamicin per ml (T = 0), the monolayers were washed thoroughly with PBS and either lysed or incubated for a further 3 and 5 h in fresh DMEM supplemented with a reduced concentration of gentamicin (5.0 μg ml−1). At the required time intervals, viable intracellular streptococci were counted. Approximately threefold fewer isogenic mutant organisms were recovered from the Hep-2C cells compared to wild-type streptococci at T = 0 (Fig. 6). The invasion of wild-type and isogenic mutant bacteria was 55.7 and 27.1%, respectively, where these percentages are the number of bacteria recovered from the monolayer expressed as a percentage of the total number of bacteria added to each well. The numbers of both wild-type and isogenic mutant bacteria recovered from the cells decreased with time, and wild-type bacterial counts appeared to decrease more sharply than those of the mutant organisms. Therefore, wild-type Manfredo streptococci appear to have an enhanced capacity to invade and/or survive intracellularly during the early stages of infection (within the first 3 h) compared to the SAGP-deficient mutant, but this advantage is lost over time (after a further 3 or 5 h of incubation).
FIG. 6.
Intracellular survival of S. pyogenes Manfredo wild-type (□) and isogenic mutant (■) bacteria in Hep-2C cells. Epithelial cells (5 × 105 cells/well) were incubated for 24 h in 24-well plates in antibiotic-free DMEM containing 10% fetal calf serum and 3 mM glutamine. The resulting monolayers were washed three times with fresh medium before streptococci were added. Infected monolayers were incubated for 90 min at 37°C in a humidified atmosphere of 5% CO2 and then washed three times with PBS to remove extracellular bacteria. Medium supplemented with gentamicin (200 μg ml−1) was added, and after a further 90 min of incubation (T = 0), the cells were washed three times with PBS and then either trypsinized and lysed with ice-cold sterile distilled H2O or incubated for a further 3 or 5 h in fresh DMEM plus gentamicin (5 μg ml−1) before being lysed. Numbers of released viable bacteria were determined by plating out serial dilutions. Control wells with no monolayers were set up and treated in the same way. Results show the mean number of viable streptococci recovered from the monolayers at each time point and the SEM and are representative of results obtained in at least two separate experiments.
DISCUSSION
In this study we have constructed an isogenic mutant of S. pyogenes Manfredo deficient in SAGP production by inserting a deletion in the sagp gene using the method of allelic exchange. Characterization of this mutant showed that in contrast to the wild-type streptococci, it did not possess AD activity; CE prepared from the isogenic mutant did not inhibit the proliferation of human PBMC; the mutant was less easily able to enter epithelial cells or to survive intracellularly during the early stages of cell infection; and isogenic mutant bacteria were not protected from acidic environments by l-arginine and were thus rapidly killed at a low pH.
SAGP was originally isolated from S. pyogenes Su and was shown to inhibit the proliferation of a number of cell lines including murine embryonic cells (BALB/3T3), murine leukemic L1210 cells, murine fibrosarcoma Meth A cells, and human HL60 cells (23, 24, 56, 57). Its precise mode of action, however, was not elucidated, although it was postulated that -SH groups may be involved, since thiol-reactive agents such as cystamine and 5,5′-dithiobis(2-nitrobenzoic acid) diminished its activity (56). Evidence was also put forward for the involvement of inhibitory guanine nucleotide-binding (Gi) proteins (57). However, we later showed that SAGP is most probably the group A streptococcal equivalent or homolog of AD (17). The generation of the isogenic mutant in this present study further endorses this view. The isogenic mutant was made by introducing a deletion in the sagp gene using the S. pyogenes Su sagp gene sequence as a template for all oligonucleotide primer design, and the resulting mutant bacteria lacked AD activity. CE derived from the isogenic mutant has severely reduced inhibitory activity against the proliferation of human PBMC (Fig. 2). This shows that the inhibition elicited by wild-type CE can be attributed mainly to AD activity rather than to more than one CE component.
The enzyme AD has been described previously in S. pyogenes (17, 43), but to our knowledge the exact molecular organization of the genes encoding the enzymes of the full AD system has not. As mentioned in the introduction, the close proximity of the genes for AD, ornithine carbamoyltransferase, and carbamate kinase on the streptococcal chromosome suggests that they may be arranged in an operon. If this is true, it was important that the method we used to create the SAGP-deficient mutant had no polar effects on expression of other genes. The deletion that we have introduced in the sagp gene resulted in an in-frame mutation, and no termination codons were formed; therefore, transcription of the gene and any genes downstream should not be altered. We have also shown here that citrulline protected both the isogenic mutant and wild-type bacteria equally well at pH 4.0 (Fig. 5), showing that expression of ornithine carbamoyltransferase and carbamate kinase was not affected by the mutation. Citrulline can be broken down by the action of these two enzymes to yield NH3, which will protect the bacteria at low pH. Figure 1 shows that the gene encoding carbamate kinase is found downstream of the genes encoding SAGP and AD and the three hypothetical proteins that may also form part of the operon; therefore, although we could not assay for the expression of these hypothetical proteins, our results would suggest that their expression also has not been affected by the mutation.
AD and the AD system for metabolizing l-arginine is found in a range of prokaryotic organisms including Mycoplasma (31, 32, 48, 55), P. aeruginosa (4, 27, 28), P. putida (49), S. faecalis, S. faecium (51), oral streptococci such as S. sanguis, S. milleri, and S. rattus (9, 10, 14), and the protozoan Giardia intestinalis (25), but it is not found in mammalian species. It has been proposed to have three possible functions (10, 13, 14): first, the system can provide ATP for growth of organisms (e.g., Mycoplasma is able to grow on l-arginine as the sole source of carbon and energy) (13, 48); second, the system may play a biosynthetic role (e.g., the reaction catalyzed by carbamate kinase can work in a noncatabolic direction to produce carbamoyl phosphate for synthesis of citrulline or pyrimidines); and third, the system can protect bacteria against damage caused by acidic environments through the production of NH3. We set out to determine what role AD may play in S. pyogenes. Several groups have previously shown that group A streptococci can grow in chemically defined media, but group A streptococci are fastidious feeders and the medium must be supplemented with over 18 amino acids, a range of B vitamins, salts, and glucose to achieve growth rates comparable to those attained in complex media (15, 30, 34, 44). Therefore, it seems unlikely that the main function of the AD system in group A streptococci is to provide energy and metabolic intermediates. Studies were therefore done to determine whether the system was involved in the acid-base physiology of the bacteria. The AD system is known to play a crucial role in protecting oral streptococci such as S. rattus, S. sanguis, and S. milleri from the acidic environment found in dental plaque (10, 14). The NH3 produced within the cell combines with protons to yield NH4+, and this raises the cytoplasmic pH value to protect sensitive cytoplasmic structures against acid damage. S. sanguis can be protected from killing at pH 4.0 by addition of as little as 2.9 mM l-arginine (10). The results presented here show that S. pyogenes Manfredo wild-type bacteria were very sensitive to low pH; for example, at pH 4 to 5, cell viability decreased rapidly, but l-arginine at a concentration as low as 1 mM protected the organisms from such acidic environments (Fig. 4). In contrast, even at concentrations of 50 mM, l-arginine had no effect on the survival of isogenic mutant organisms at low pH (Fig. 3). Therefore, it appears that the AD system plays a major role in the pH tolerance of S. pyogenes.
Invasion assays were set up to compare the ability of wild-type and SAGP-deficient isogenic mutant bacteria to enter and survive within epithelial cells. The main sites of infection for group A streptococci are the skin and soft tissues (2). It was found that the wild-type streptococci had a survival advantage over the isogenic mutant bacteria during the initial stages of invasion and that this was lost over longer periods (Fig. 6). Further experiments are needed to determine the precise mechanism by which AD enhances cell invasion and/or intracellular survival and to determine whether AD simply protects group A streptococci from the low pH found inside cells. In lysosomes, the pH can reach values as low as 4.7 (33, 36), and in a recent paper it has been shown that S. pyogenes is taken up by HeLa cells by a zipper-like mechanism and that streptococci can be found in lysosomes 2 h after infection (19). The invasion results presented in the present paper compare the recovery of viable wild-type and isogenic mutant bacteria from epithelial cells (Hep-2C and A549) at what we have designated T = 0, but at this time point the streptococci have been in contact with the cells for 3 h (90 min without gentamicin and 90 min in the presence of gentamicin) and it is therefore difficult to distinguish whether the isogenic mutant bacteria were either less able to enter the cells, e.g., less able to adhere to the cells, or more readily killed than the wild-type organisms once inside the cells during the early stages of invasion, i.e., during the first 3 h. Intracellular survival of the wild-type bacteria was not significantly greater than that of the isogenic mutant organisms after longer incubation periods, i.e., after an extra 3 or 5 h (Fig. 6). One possible explanation for this may be that l-arginine was depleted from the tissue culture media by the action of AD relatively early and therefore any advantage conferred by AD would be lost with time due to a lack of available substrate.
The physiological concentration of free l-arginine found in inflammatory lesions is thought to be <0.1 mM (1). This concentration did not fully protect S. pyogenes from the damaging effects of acidic conditions in vitro (Fig. 4). However, S. pyogenes may be able to specifically cleave l-arginine from peptides at inflamed sites to increase the available concentration of free amino acid. One source of l-arginine for oral streptococci is sialin (H2N-Gly-Gly-Lys-Arg-COOH), which is also known as the pH rise peptide (10). Oral streptococci grow poorly in a chemically defined medium containing amino acids and dipeptides but were able to utilize large peptides, and it is thought that the trypsin-like activity shown by some dental plaque bacteria would provide peptides that have C-terminal arginine residues (45).
If AD enhances the intracellular survival of S. pyogenes in vivo, this may have very important consequences in determining the outcome of an infection. Group A streptococci are highly sensitive to antibiotics such as penicillin, but in 10 to 15% of cases of streptococcal infections there is a relapse of the disease after the end of a course of antibiotic therapy (41, 54). It has been hypothesized that this is due to small numbers of streptococci residing inside cells, such as laryngeal cells, where penicillin is unable to penetrate and that these organisms reinitiate the infection when antibiotic treatment has finished (37, 38, 39). Indeed, it has been shown that treatment of patients with recurrent pharyngotonsillitis using antibiotics that are able to enter cells, for example clindamycin and rifampin, reduces the incidence of relapsing infections (8, 46). Persistence of streptococcal infections may lead to increased exposure of bacterial products to the immune system and enhance the potential for developing autoimmunity. With the use of the isogenic mutant deficient in SAGP and AD, we intend to make a more detailed study of the role this enzyme plays in intracellular survival of the bacteria.
ACKNOWLEDGMENTS
We thank Andrew Allen for his help with the manuscript.
This work was funded by project grant D0547 from the Arthritis Research Campaign awarded to J. A. Goodacre and B. A. Degnan and by Wellcome Trust grant 045692 to M. A. Kehoe.
REFERENCES
- 1.Albina J E, Mills C D, Caldwell M D. Alterations in macrophage physiology associated with the metabolism of l-arginine through the oxidative l-arginine deiminase pathway. In: Moncada S, Higgs E A, editors. Nitric oxide from l-arginine: a bioregulatory system. Amsterdam, The Netherlands: Elsevier Science Publishers B.V.; 1990. pp. 243–248. [Google Scholar]
- 2.Ashbaugh C D, Warren H B, Carey V J, Wessels M R. Molecular analysis of the role of the group A streptococcal cysteine protease, hyaluronic acid capsule, and M protein in a murine model of human invasive soft-tissue infection. J Clin Investig. 1998;102:550–560. doi: 10.1172/JCI3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bairoch A, Apweiler R. The SWISS-PROT protein sequence data bank and its new supplement TrEMBL. Nucleic Acids Res. 1996;24:21–25. doi: 10.1093/nar/24.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baur H, Luethi E, Stalon V, Mercenier A, Haas D. Sequence analysis and expression of the arginine-deiminase and carbamate-kinase genes of Pseudomonas aeruginosa. Eur J Biochem. 1989;179:53–60. doi: 10.1111/j.1432-1033.1989.tb14520.x. [DOI] [PubMed] [Google Scholar]
- 5.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bisno A L. Group A streptococcal infections and rheumatic fever. N Engl J Med. 1991;325:783–793. doi: 10.1056/NEJM199109123251106. [DOI] [PubMed] [Google Scholar]
- 7.Bisno A L. Acute rheumatic fever: a present-day perspective. Medicine. 1993;72:278–283. [Google Scholar]
- 8.Brook I, Leyva F. The treatment of the carrier state of group A beta-hemolytic streptococci with clindamycin. Chemotherapy. 1981;27:360–367. doi: 10.1159/000238005. [DOI] [PubMed] [Google Scholar]
- 9.Burne R A, Parsons D T, Marquis R E. Cloning and expression in Escherichia coli of the genes of the arginine deiminase system of Streptococcus sanguis NCTC 10904. Infect Immun. 1989;57:3540–3548. doi: 10.1128/iai.57.11.3540-3548.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casiano-Colon A, Marquis R E. Role of arginine deiminase system in protecting oral bacteria and an enzymatic basis for acid tolerance. Appl Environ Microbiol. 1988;54:1318–1324. doi: 10.1128/aem.54.6.1318-1324.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cue D R, Cleary P P. High-frequency invasion of epithelial cells by Streptococcus pyogenes can be activated by fibrinogen and peptides containing the sequence RGD. Infect Immun. 1997;65:2759–2764. doi: 10.1128/iai.65.7.2759-2764.1997. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Cue D, Dombek P E, Lam H, Cleary P P. Streptococcus pyogenes serotype M1 encodes multiple pathways for entry into human epithelial cells. Infect Immun. 1998;66:4593–4601. doi: 10.1128/iai.66.10.4593-4601.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cunin R, Glansdorff N, Pierard A, Stalon V. Biosynthesis and metabolism of arginine in bacteria. Microbiol Rev. 1986;50:314–352. doi: 10.1128/mr.50.3.314-352.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Curran T M, Lieou J, Marquis R E. Arginine deiminase system and acid adaptation of oral streptococci. Appl Environ Microbiol. 1995;61:4494–4496. doi: 10.1128/aem.61.12.4494-4496.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dassy B, Alouf J E. Growth of Streptococcus pyogenes and streptolysin O production in complex and synthetic media. J Gen Microbiol. 1983;129:643–651. doi: 10.1099/00221287-129-3-643. [DOI] [PubMed] [Google Scholar]
- 16.Degnan B A, Kehoe M A, Goodacre J A. Analysis of human T cell responses to group A streptococci using fractionated Streptococcus pyogenes proteins. FEMS Immunol Med Microbiol. 1997;17:161–170. doi: 10.1111/j.1574-695X.1997.tb01009.x. [DOI] [PubMed] [Google Scholar]
- 17.Degnan B A, Palmer J M, Robson T, Jones C E D, Fischer M, Glanville M, Mellor G D, Diamond A G, Kehoe M A, Goodacre J A. Inhibition of human peripheral blood mononuclear cell proliferation by Streptococcus pyogenes cell extract is associated with arginine deiminase activity. Infect Immun. 1998;66:3050–3058. doi: 10.1128/iai.66.7.3050-3058.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D'Hooghe I, Wauven C V, Michiels J, Tricot C, De Wilde P, Vanderleyden J, Stalon V. The arginine deiminase pathway in Rhizobium etli: DNA sequence analysis and functional study of the arcABC genes. J Bacteriol. 1997;179:7403–7409. doi: 10.1128/jb.179.23.7403-7409.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dombek P E, Cue D, Sedgewick J, Lam H, Ruschkowski S, Finlay B B, Cleary P P. High-frequency intracellular invasion of epithelial cells by serotype M1 group A streptococci: M1 protein-mediated invasion and cytoskeletal rearrangements. Mol Microbiol. 1999;31:859–870. doi: 10.1046/j.1365-2958.1999.01223.x. [DOI] [PubMed] [Google Scholar]
- 20.Fluckiger U, Jones K F, Fischetti V A. Immunoglobulins to group A streptococcal surface molecules decrease adherence to and invasion of human pharyngeal cells. Infect Immun. 1998;66:974–979. doi: 10.1128/iai.66.3.974-979.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ginsburg I. Mechanisms of cell and tissue injury induced by group A streptococci: relation to poststreptococcal sequelae. J Infect Dis. 1972;126:294–340. doi: 10.1093/infdis/126.3.294. [DOI] [PubMed] [Google Scholar]
- 22.Ish-Horowicz D, Burke J F. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981;9:2989–2998. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanaoka M, Kawanaka C, Negoro T, Fukita Y, Taya K, Agui H. Cloning and expression of the antitumour glycoprotein gene of Streptococcus pyogenes Su in Escherichia coli. Agric Biol Chem. 1987;51:2641–2648. [Google Scholar]
- 24.Kanaoka M, Negoro T, Kawanaka C, Agui H, Nabeshima S. Streptococcal antitumour protein: expression in Escherichia coli cells and properties of the recombinant protein. Agric Biol Chem. 1991;55:743–750. [PubMed] [Google Scholar]
- 25.Knodler L A, Schofield P J, Gooley A A, Edwards M R. Giardia intestinalis: purification and partial amino acid sequence of arginine deiminase. Exp Parasitol. 1997;85:77–80. doi: 10.1006/expr.1996.4099. [DOI] [PubMed] [Google Scholar]
- 26.LaPenta D, Rubens C, Chi E, Cleary P P. Group A streptococci efficiently invade human respiratory epithelial cells. Proc Natl Acad Sci USA. 1994;91:12115–12119. doi: 10.1073/pnas.91.25.12115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luthi E, Baur H, Gamper M, Brunner F, Villeval D, Mercenier A, Haas D. The arc operon for anaerobic arginine catabolism in Pseudomonas aeruginosa contains an additional gene, arcD, encoding a membrane protein. Gene. 1990;87:37–43. doi: 10.1016/0378-1119(90)90493-b. [DOI] [PubMed] [Google Scholar]
- 28.Mercenier A, Stalon V, Simon J-P, Haas D. Mapping of the arginine deiminase gene in Pseudomonas aeruginosa. J Bacteriol. 1982;149:787–788. doi: 10.1128/jb.149.2.787-788.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Michie C, Scott A, Cheesbrough J, Beverley P, Pasvol G. Streptococcal toxic shock-like syndrome: evidence of superantigen activity and its effects on T lymphocyte subsets in vivo. Clin Exp Immunol. 1994;98:140–144. doi: 10.1111/j.1365-2249.1994.tb06620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mickelson M N. Chemically defined medium for growth of Streptococcus pyogenes. J Bacteriol. 1964;88:158–164. doi: 10.1128/jb.88.1.158-164.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Misawa S, Aoshima M, Takaku H, Matsumoto M, Hayashi H. High-level expression of Mycoplasma arginine deiminase in Escherichia coli and its efficient renaturation as an anti-tumor enzyme. J Biotechnol. 1994;36:145–155. doi: 10.1016/0168-1656(94)90050-7. [DOI] [PubMed] [Google Scholar]
- 32.Miyazaki K, Takaku H, Umeda M, Fujita T, Huang W, Kimura T, Yamashita J, Horio T. Potent growth inhibition of human tumor cells in culture by arginine deiminase purified from a culture medium of a Mycoplasma-infected cell line. Cancer Res. 1990;50:4522–4527. [PubMed] [Google Scholar]
- 33.Myers B M, Tietz P S, Tarara J E, LaRusso N F. Dynamic measurements of the acute and chronic effects of lysosomotropic agents on hepatocyte lysosomal pH using flow cytometry. Hepatology. 1995;22:1519–1526. [PubMed] [Google Scholar]
- 34.Ogburn C A, Harris T N, Harris S. Extracellular antigens in steady-state cultures of the hemolytic Streptococcus: production of proteinase at low pH. J Bacteriol. 1958;76:142–151. doi: 10.1128/jb.76.2.142-151.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oginsky E L. Isolation and determination of arginine and citrulline. Methods Enzymol. 1957;3:639–643. [Google Scholar]
- 36.Ohkuma S, Poole B. Fluorescence probe measurement of the intralysosomal pH in living cells and the perturbation of pH by various agents. Proc Natl Acad Sci USA. 1978;75:3327–3331. doi: 10.1073/pnas.75.7.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Osterlund A, Engstrand L. Intracellular penetration and survival of Streptococcus pyogenes in respiratory epithelial cells in vitro. Acta Otolaryngol. 1995;115:685–688. doi: 10.3109/00016489509139387. [DOI] [PubMed] [Google Scholar]
- 38.Osterlund A, Engstrand L. An intracellular sanctuary for Streptococcus pyogenes in human tonsillar epithelium: studies of asymptomatic carriers and in vitro cultured biopsies. Acta Otolaryngol. 1997;117:883–888. doi: 10.3109/00016489709114219. [DOI] [PubMed] [Google Scholar]
- 39.Osterlund A, Popa R, Nikkila T, Scheynius A, Engstrand L. Intracellular reservoir of Streptococcus pyogenes in vivo: a possible explanation for recurrent pharyngotonsillitis. Laryngoscope. 1997;107:640–647. doi: 10.1097/00005537-199705000-00016. [DOI] [PubMed] [Google Scholar]
- 40.Ozeri V, Rosenshine I, Mosher D F, Fassler R, Hanski E. Roles of integrins and fibronectin in the entry of Streptococcus pyogenes into cells via protein F1. Mol Microbiol. 1998;30:625–637. doi: 10.1046/j.1365-2958.1998.01097.x. [DOI] [PubMed] [Google Scholar]
- 41.Pichichero M E. Cephalosporins are superior to penicillin for treatment of streptococcal tonsillopharyngitis: is the difference worth it? Pediatr Infect Dis J. 1993;12:268–274. doi: 10.1097/00006454-199304000-00002. [DOI] [PubMed] [Google Scholar]
- 42.Rammelkamp C H, Weaver R S. Acute glomerulonephritis. The significance of the variation in the incidence of the disease. J Clin Investig. 1953;320:345–358. doi: 10.1172/JCI102745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reizer J, Novotny M J, Panos C, Saier M H., Jr Mechanism of inducer expulsion in Streptococcus pyogenes: a two-step process activated by ATP. J Bacteriol. 1983;156:354–361. doi: 10.1128/jb.156.1.354-361.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van de Rijn I, Kessler R E. Growth characteristics of group A streptococci in a new chemically defined medium. Infect Immun. 1980;27:444–448. doi: 10.1128/iai.27.2.444-448.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rogers A H, Zilm P S, Gully N J, Pfennig A L. Response of a Streptococcus sanguis strain to arginine-containing peptides. Infect Immun. 1988;56:687–692. doi: 10.1128/iai.56.3.687-692.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roos K, Grahn E, Holm S E, Johansson H, Lind L. Interfering alpha-streptococci as a protection against recurrent streptococcal tonsillitis in children. Int J Pediatr Otorhinolaryngol. 1993;25:141–148. doi: 10.1016/0165-5876(93)90047-7. [DOI] [PubMed] [Google Scholar]
- 47.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 48.Schimke R T, Berlin C M, Sweeney E W, Carroll W R. The generation of energy by the arginine dihydrolase pathway in Mycoplasma hominis 07. J Biol Chem. 1966;241:2228–2236. [PubMed] [Google Scholar]
- 49.Shibatani T, Kakimoto T, Chibata I. Crystallization and properties of l-arginine deiminase of Pseudomonas putida. J Biol Chem. 1975;250:4580–4583. [PubMed] [Google Scholar]
- 50.Simon D, Ferretti J J. Electrotransformation of Streptococcus pyogenes with plasmid and linear DNA. FEMS Microbiol Lett. 1991;82:219–224. doi: 10.1016/0378-1097(91)90336-9. [DOI] [PubMed] [Google Scholar]
- 51.Simon J-P, Wargnies B, Stalon V. Control of enzyme synthesis in the arginine deiminase pathway of Streptococcus faecalis. J Bacteriol. 1982;150:1085–1090. doi: 10.1128/jb.150.3.1085-1090.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–507. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 53.Stollerman G H. Rheumatogenic streptococci and autoimmunity. Clin Immunol Immunopathol. 1991;61:131–142. doi: 10.1016/s0090-1229(05)80019-4. [DOI] [PubMed] [Google Scholar]
- 54.Stromberg A, Schwan A, Cars O. Five versus ten days treatment of group A streptococcal pharynotonsillitis: a randomized controlled clinical trial with phenoxymethylpenicillin and cefadroxil. Scand J Infect Dis. 1988;20:37–46. doi: 10.3109/00365548809117215. [DOI] [PubMed] [Google Scholar]
- 55.Weickmann J L, Fahrney D E. Arginine deiminase from Mycoplasma arthritidis. Evidence for multiple forms. J Biol Chem. 1977;252:2615–2620. [PubMed] [Google Scholar]
- 56.Yoshida J, Takamura S, Suzuki S. Evidence for the involvement of sulfhydryl groups in the expression of antitumor activity of streptococcal acid glycoprotein (SAGP) purified from crude extract of Streptococcus pyogenes. Anticancer Res. 1994;14:1833–1838. [PubMed] [Google Scholar]
- 57.Yoshida J, Takamura S, Suzuki S, Nishio M. Streptococcal glycoprotein-induced tumour cell growth inhibition involves the modulation of a pertussis toxin-sensitive G protein. Br J Cancer. 1996;73:917–923. doi: 10.1038/bjc.1996.182. [DOI] [PMC free article] [PubMed] [Google Scholar]