Abstract
The B7 family ligand HERV-H LTR-associating protein 2 (HHLA2) is an attractive target for cancer immunotherapy due to its co-inhibitory function, over-expression in human cancers, and association with poor prognoses. However, the knowledge of the HHLA2 pathway is incomplete. HHLA2 has an established positive receptor transmembrane and immunoglobulin domain containing 2 (TMIGD2) but a poorly characterized negative receptor human killer cell immunoglobulin-like receptor, three Ig domains and long cytoplasmic tail (KIR3DL3). Here, KIR3DL3 and TMIGD2 simultaneously bound to different sites of HHLA2. KIR3DL3 was mainly expressed on CD56dim NK and terminally differentiated effector memory CD8+ T (CD8+ TEMRA) cells. KIR3DL3+ CD8+ TEMRA acquired an NK-like phenotype and function. HHLA2 engagement recruited KIR3DL3 to the immunological synapse and co-inhibited CD8+ T and NK cell function and killing, inducing immune evasive HHLA2+ tumors. KIR3DL3 recruited SHP-1 and SHP-2 to attenuate Vav1, ERK1/2, AKT and NF-κB signaling. HHLA2+ tumors from human kidney, lung, gallbladder and stomach were infiltrated by KIR3DL3+ immune cells. KIR3DL3 blockade inhibited tumor growth in multiple humanized mouse models. Thus, our findings elucidated the molecular and cellular basis for the inhibitory function of KIR3DL3, demonstrating that the KIR3DL3-HHLA2 pathway is a potential immunotherapeutic target for cancer.
One Sentence Summary:
Blockade of KIR3DL3 as an immunotherapy for human cancers.
INTRODUCTION
Cancer immunotherapy has fundamentally shifted the paradigm for cancer treatment from targeting the tumor itself to harnessing the host’s immune system (1). Immune checkpoint blockades (ICB) have achieved durable responses in patients with advanced-stage tumors that would have otherwise been fatal (2). Several checkpoint inhibitors targeting the cytotoxic T-lymphocyte-associated antigen 4 (CTLA-4) or programmed cell death receptor 1 (PD-1)/programmed cell death ligand 1 (PD-L1) axis have been approved for a broad spectrum of cancers (3). Although the percentage of responders to these drugs has steadily increased, the majority of cancer patients still do not benefit (3). Therefore, it is critical to characterize additional immune checkpoints and develop new therapeutic strategies.
We previously identified HHLA2 (also called B7H7/B7-H5/B7y) as a functional B7 family member that phylogenetically forms a subfamily with B7x and B7-H3 in the B7 family (4, 5). HHLA2 protein is expressed on antigen-presenting cells (APCs) (4, 6) and has a limited expression in normal human organs, whereas it is highly expressed in many human cancers (7–10). In several tumor types, HHLA2 expression is more frequent than PD-L1 and co-expression of PD-L1 and HHLA2 is rare; PD-L1 negative tumors tend to express HHLA2 (9, 11, 12). In general, elevated HHLA2 is associated with more severe pathology and worse prognosis in cancer patients (9, 13–18). However, a few studies also show that higher HHLA2 expression is associated with better survival (19–21).
This paradox could be partly explained by the dual role of HHLA2 in immune responses. HHLA2 can inhibit (4, 22) or enhance (6) immune cell function. TMIGD2 (also called IGPR-1/CD28H) is the first characterized receptor for HHLA2 (6, 7) and is primarily expressed on naïve T and NK cells, rapidly decreasing after activation (6, 23). TMIGD2 co-stimulates both T and NK cell activities (6, 23), which does not explain the inhibitory effect of HHLA2. Thus, we reasoned that there was an unidentified inhibitory receptor of HHLA2 (5, 7, 24). In 2017, we identified KIR3DL3, an orphan molecule in the killer-cell immunoglobulin-like receptor (KIR) family (25, 26), is another receptor for HHLA2 (27). Very recently, two groups perform high throughput interactome screen of immunoglobulin superfamily (IgSF) and indicate KIR3DL3 as a binding partner of HHLA2 (28, 29). During the revision of this paper, another group also independently report KIR3DL3 as the second receptor for HHLA2 and demonstrate engagement of KIR3DL3 with HHLA2 inhibits the activation of a T-cell line and the cytotoxicity of an NK-cell line (12). However, numerous key characteristics of the KIR3DL3-HHLA2 pathway remain unknown including the full expression pattern of KIR3DL3, how HHLA2 regulates immune responses through KIR3DL3, the downstream signaling pathway of KIR3DL3, the co-expression of HHLA2 and KIR3DL3 in the tumor microenvironment, and the therapeutic efficacy of KIR3DL3 blockade in human cancers.
Here, we demonstrated that KIR3DL3 was mainly expressed on terminally differentiated effector memory CD8+ T cells and CD56dim CD16+ NK cells. KIR3DL3 mediated co-inhibition of primary CD8+ T and NK cells and induced resistance of HHLA2+ tumors to killing. KIR3DL3 showed immunosuppressive activities by recruiting SHP-1 and SHP-2 to attenuate downstream Vav1, ERK1/2, AKT and NF-κB signaling in NK cells. Human HHLA2+ tumors were infiltrated by KIR3DL3+ immune cells and KIR3DL3 blockade promoted anti-tumor immunity in multiple humanized mouse models. Altogether, our study provided a detailed functional and mechanistic view of the KIR3DL3-HHLA2 pathway-mediated immunosuppression, supporting this pathway’s therapeutic application for cancer immunotherapy.
RESULTS
KIR3DL3 and TMIGD2 simultaneously bound to non-identical sites on HHLA2
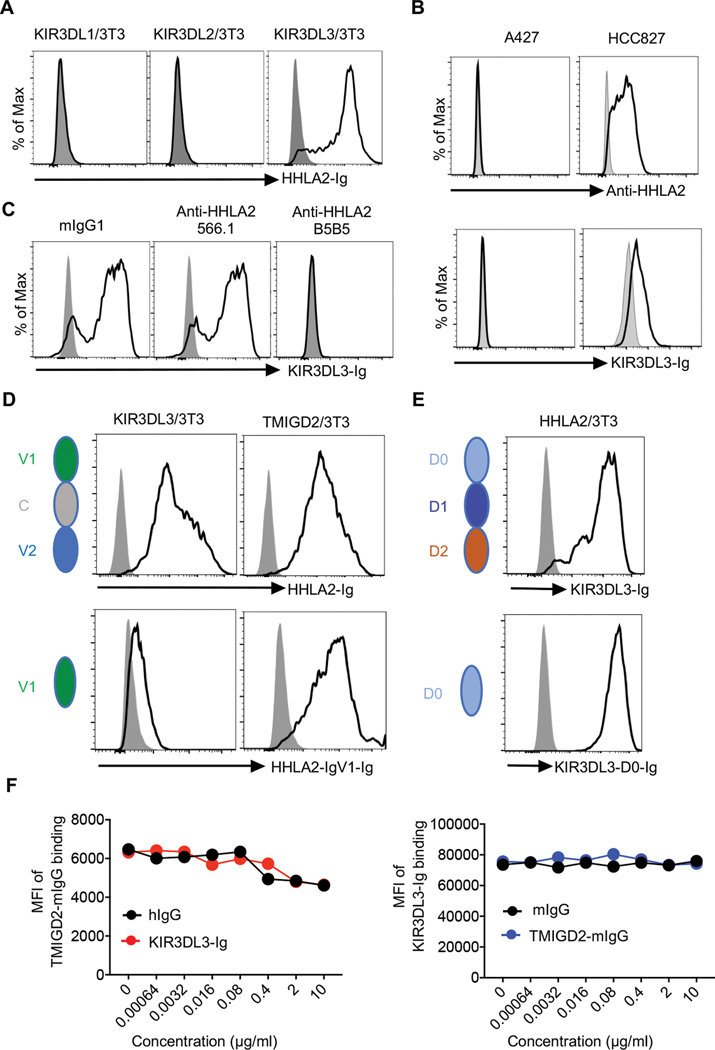
In addition to the expression of HHLA2 on APCs and tumors (4, 6, 7, 9, 10), we found its expression could also be induced on activated T and NK cells (fig. S1A). As HHLA2 is a member of IgSF and has orthologs in humans and monkeys but not in mice or rats, we screened the receptors for HHLA2 through a list of genes belonging to IgSF, exclusive to primates, and with a predicted transmembrane domain (fig. S1B). We observed that HHLA2-Ig strongly bound to cells expressing KIR3DL3, but not its closest homologs KIR3DL1 or KIR3DL2 (Fig. 1A and fig. S1C). Inversely, KIR3DL3-Ig bound to an HHLA2+ human lung cancer cell HCC827, but not an HHLA2− cell A427 (Fig. 1B). The binding of KIR3DL3-Ig to HHLA2 expressing cells was completely blocked by anti-HHLA2 monoclonal antibody (mAb) clone B5B5 (Fig. 1C). To further explore if KIR3DL3 has other binding partners, we utilized a high-throughput screening with cell microarray individually expressing 5484 full-length human membrane proteins or cell surface-tethered human secreted proteins and found that HHLA2 was the only specific binding partner for KIR3DL3 (fig. S1D–E).
Fig. 1. Characterization of KIR3DL3 as a receptor for HHLA2.
(A) Histograms showing the binding of HHLA2-Ig (open) or hIgG (shaded) to the indicated cell lines.
(B) Top: Histograms showing the HHLA2 expression on the human lung cancer cell line A427 and HCC827 examined by anti-HHLA2 mAb clone B5B5 (open) or mIgG1 (shaded). Bottom: Histograms showing the binding of KIR3DL3-Ig (open) or hIgG (shaded) to A427 and HCC827.
(C) Histograms showing the binding of KIR3DL3-Ig (open) or hIgG (shaded) to HHLA2/3T3 in the presence of anti-HHLA2 mAbs (clone B5B5 or 566.1) or mIgG1.
(D) Left: Cartoons showing the structures of HHLA2-Ig (top) and HHLA2-IgV1-Ig (bottom). Right: Histograms showing the binding of HHLA2-Ig (open, top) and HHLA2-IgV1-Ig (open, bottom) or hIgG (shaded) to KIR3DL3/3T3 and TMIGD2/3T3 cells.
(E) Left: Cartoons showing the structures of KIR3DL3-Ig (top) and KIR3DL3-D0-Ig (bottom). Right: Histograms showing the binding of KIR3DL3-Ig (open, top) and KIR3DL3-D0-Ig (open, bottom) or hIgG (shaded) to HHLA2/3T3 cells.
(F) Left: The MFI of TMIGD2-mIgG binding to HHLA2/3T3 cells. HHLA2/3T3 cells were preincubated with KIR3DL3-Ig or hIgG at the indicated concentrations and then stained by TMIGD2-mIg. Right: The MFI of KIR3DL3-Ig binding to HHLA2/3T3 cells. HHLA2/3T3 cells were preincubated with TMIGD2-mIgG or mIgG at the indicated concentrations and then stained by KIR3DL3-Ig.
In A-F, data are representative of two-three independent experiments.
HHLA2 extracellular region is composed of tandem IgV1-IgC-IgV2 domains (4). Loss of the IgC-IgV2 domains of HHLA2 largely abrogated its binding to KIR3DL3 but did not interfere with its binding to TMIGD2 (Fig. 1D). The extracellular part of KIR3DL3 is composed of tandem D0-D1-D2 domains (fig. S1F). The removal of D1-D2 domains from KIR3DL3 did not affect its binding to HHLA2 (Fig. 1E). Preincubating HHLA2/3T3 cells with KIR3DL3 did not block TMIGD2 binding, and vice versa (Fig. 1F). Taken together, our results demonstrated that KIR3DL3 and TMIGD2 bound to non-identical sites on HHLA2, with both KIR3DL3 and TMIGD2 simultaneously binding to HHLA2.
Generation and characterization of KIR3DL3 D0 domain-specific mAbs
To investigate the expression of KIR3DL3, we first generated a panel of anti-KIR3DL3 mAbs (fig. S2A). Our lead clone 26E10 bound to the KIR3DL3 D0 domain with a high affinity (Kd=0.17 nM) (fig. S2B–C) and did not cross-react with other KIRs (fig. S2A). The specificity of 26E10 was further verified by lack of reactivity with the cell microarray expressing 5484 human proteins (fig. S1D).
KIR3DL3 is a highly polymorphic gene encoding more than 100 alleles (30). The D0 domain, which our lead clone 26E10 binds to, is relatively conserved and contains about 16 amino acid variants (30). We mutated D0 domain of KIR3DL3*004 and transduced 3T3 to express the wild type and the mutant KIR3DL3*004. We then tested the binding of 26E10 with 11 KIR3DL3 D0 variants and found 9 of them, including KIR3DL3*001 and KIR3DL3*010, which are the most predominant D0 sequence in Europeans and Chinese Han respectively (30), could be detected by 26E10 (fig. S3).
KIR3DL3 protein was expressed on human innate and adaptive immune cells
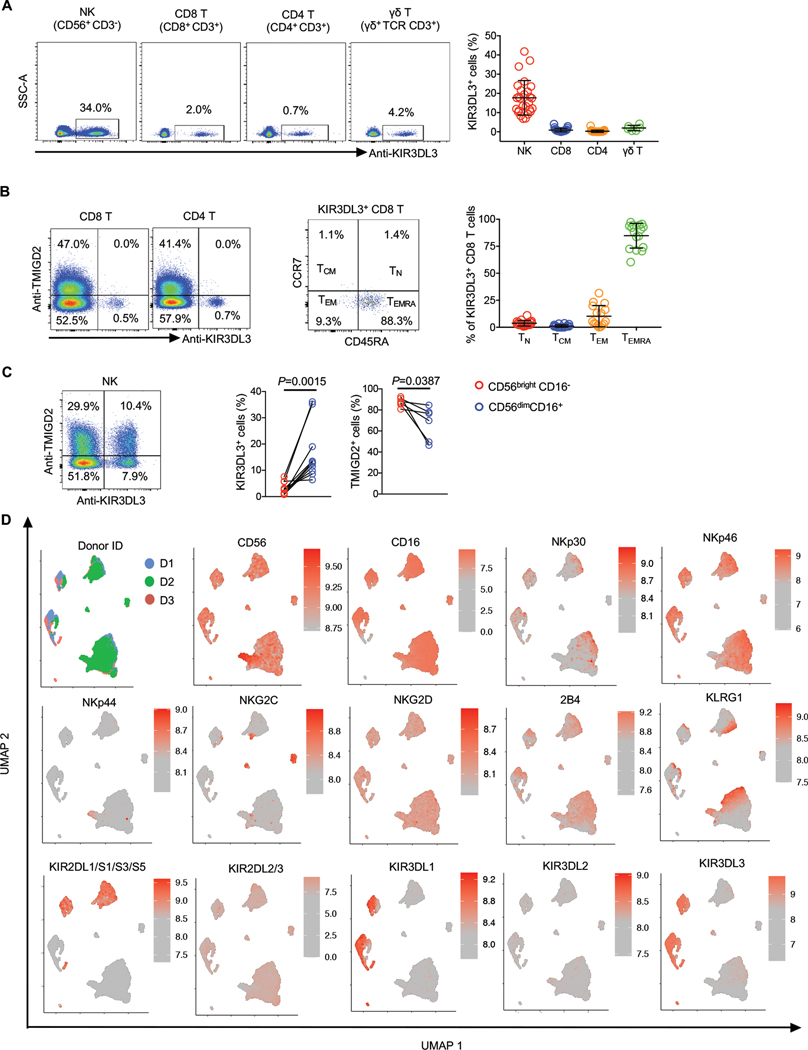
The mRNA of KIR3DL3 was undetectable in most normal human tissues but high in the blood and spleen (fig. S4A). Thus, we used 26E10 to examine KIR3DL3 protein expression on different immune cells from peripheral blood and found that NK cells, some γδ T, CD8+ and CD4+ T cells expressed KIR3DL3 (fig. S4B and Fig. 2A).
Fig. 2. KIR3DL3 was expressed on innate and adaptive immune cells.
(A) Flow cytometric analysis of KIR3DL3 expression on PBMCs. Left: KIR3DL3 expression on indicated subsets from one donor. Right: The frequencies of KIR3DL3+ cells in the indicated subsets (n=27 for NK, CD4 T, and CD8+ T; n=7 for γδ T). Data are represented as mean ± SD.
(B) Left: The expression pattern of KIR3DL3 and TMIGD2 on CD4 and CD8+ T cells. Middle: The distribution of KIR3DL3+ CD8+ T cells based on the coordinate expression of CD45RA and CCR7. Right: The summary of the distribution of KIR3DL3+ CD8+ T cells (n=16). Data are represented as mean ± SD.
(C) Left: The co-expression pattern of KIR3DL3 and TMIGD2 on NK cells. Right: The frequencies of KIR3DL3+ and TMIGD2+ cells in the indicated NK subsets (KIR3DL3: n=8; TMIGD2: n=6). P values by a two-tailed paired t-test.
(D) Human PBMCs were analyzed by spectral flow cytometry. The UMAP was generated based on flow cytometric data gated on NK cells (n=3).
In B (left) and C (left), data are representative of three independent experiments with different donors.
The expression of TMIGD2 and KIR3DL3 was mutually exclusive of CD8+ and CD4+ T cells (Fig. 2B). The major KIR3DL3 expressing CD8+ T cells were terminally differentiated (TEMRA), followed by effector memory CD8+ T cells (TEM) subset, while naive (TN) and central memory (TCM) CD8+ T cells barely expressed KIR3DL3 (Fig. 2B and fig. S4C). NK cells were the main immune cells expressing KIR3DL3 (Fig. 2A). KIR3DL3 and TMIGD2 could be co-expressed or singly expressed on an NK cell (Fig. 2C). KIR3DL3 was mainly expressed on CD56dimCD16+ NK subset, which is more differentiated and predominant in peripheral blood (31), compared to CD56birght CD16− NK subset which had higher TMIGD2 expression (Fig. 2C and fig. S4D). KIR3DL3+ NK cells expressed activating receptors, including CD16, NKG2D, NKp30, NKp46, and 2B4 (Fig. 2D). The majority of NK cells expressed at least one KIR protein and KIR3DL3 could be co-expressed with all the other KIRs (Fig. 2D).
KIR3DL3 hampered both TCR dependent and independent function of human CD8+ T cells
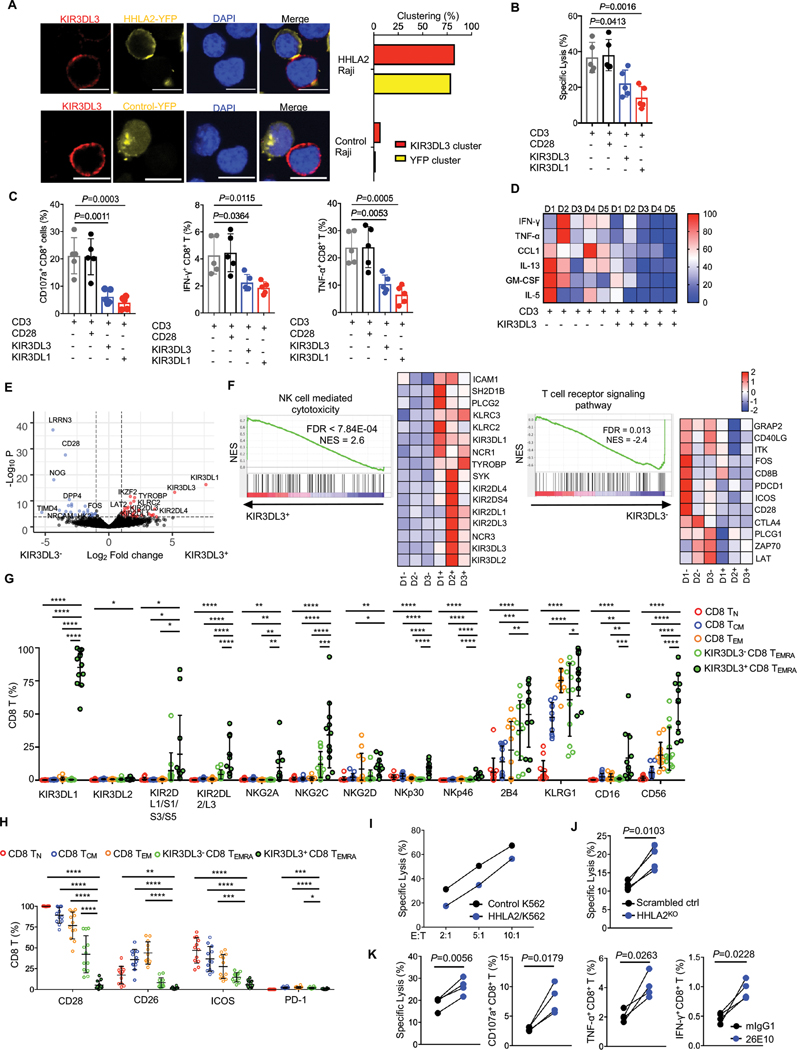
We then asked whether HHLA2 and KIR3DL3 could be recruited to T cell immunological synapses. After coculturing Jurkat cells expressing KIR3DL3 and Raji cells expressing the HHLA2-YFP (or control-YFP) fusion protein in the presence of superantigen staphylococcal enterotoxin E (SEE) (32), KIR3DL3 and HHLA2 co-localized in about 70% of the cell conjugates, whereas very little KIR3DL3 clusters were formed in the absence of HHLA2 (Fig. 3A and fig. S5A–C). We next sought to investigate the function of KIR3DL3 on T cells. FACS-purified KIR3DL3+ CD8+ T cells were expanded and the expression of KIR3DL3 was stable during the in vitro culture (fig. S5D). As the specific antigens of these CD8+ T cells are unknown, we performed a redirected cytotoxicity assay (33) (fig. S5E). After CD3 engagement, the KIR3DL3+ CD8+ T cells displayed cytolytic activity against FcR+ target cell P815, K562 and Raji (Fig 3B and fig. S5F–J). In comparison with CD3 alone, co-engagement of CD28 and CD3 did not have an additional effect (Fig. 3B–C), but co-engagement of KIR3DL3 and CD3 significantly suppressed the cytolytic function, the degranulation, cytokines (IFN-γ, TNF-α, IL-13, GM-CSF, IL-5) and chemokine (CCL1) secretion of CD8+ T cells (Fig. 3C–D and fig. S5F–I). These results suggested that KIR3DL3 hampered the TCR-dependent function of CD8+ T cells.
Fig. 3. KIR3DL3 hampered both TCR dependent and independent function of CD8+ T cells.
(A) KIR3DL3 and HHLA2 colocalized on T cell immune synapses. Left: Representative confocal images of cell conjugates acquired after Jurkat-Raji contact. Scale bars, 10 𝜇m. Right: The bar graph show the frequencies of KIR3DL3 clusters and HHLA2-YFP or control-YFP clusters in all cell conjugates (HHLA2-YFP+ Raji, n=76; Control-YFP+ Raji, n=43). Mouse Tim-3-YFP/Raji was used as the control.
(B-D) KIR3DL3+ CD8+ T redirected cytotoxicity assay against P815. (B) The lysis of P815 cells (n=5). (C) The degranulation (CD107a) and cytokine production (IFN-γ and TNF-α) of KIR3DL3+ CD8+ T cells (n=5). CD28 and KIR3DL1 served as a negative and positive control, respectively. (D) Cytokine production in the co-culture supernatant of KIR3DL3+ CD8+ T cells with indicated antibody-coated P815. The differentially secreted cytokines are shown in the heatmap (n=5). Data are represented as mean ± SD.
(E-F) RNA sequencing of KIR3DL3+ and KIR3DL3− CD8+ TEMRA (n=3). (E) The volcano plot of RNA-seq showing downregulated (blue) and upregulated (red) differentially expressed genes (DEGs). (F) Gene set enrichment analysis (GESA) in KIR3DL3+ and KIR3DL3− subset. Data shown are GESA plots and the heatmaps for the leading-edge genes.
(G-H) The T (G) and NK (H) cell surface receptor expression on CD8+ TN, TCM, TEM, KIR3DL3− and KIR3DL3+ TEMRA subsets as accessed by spectral flow cytometry (n=12). Data are represented as mean ± SD.
(I) Lysis of HHLA2/K562 or control K562 by KIR3DL3+ CD8+ T cells at indicated E:T ratios. Data are mean for duplicate measurements. Results are representative of three independent experiments.
(J) Lysis of scrambled control or HHLA2KO HCC827 by KIR3DL3+ CD8+ T cells at E:T =10:1 (n=4).
(K) The cytotoxicity, degranulation (CD107a) and cytokine production (IFN-γ and TNF-α) of KIR3DL3+ CD8+ T cell against HCC827 in the presence of anti-KIR3DL3 (26E10) or mIgG1 at E: T=10:1 (lysis) or 2:1 (CD107a, IFN-γ, and TNF-α) (n=4).
P values by a one-way ANOVA (B, C, G, H), and a two-tailed paired t-test (D, J, K). *P < 0.05, **P < 0.01, *** P < 0.001, **** P < 0.0001.
The majority (60–97%) of KIR3DL3+ CD8+ T cells were TEMRA (Fig. 2B). We performed RNA sequencing (RNA-seq) of sorted KIR3DL3+ and KIR3DL3− CD8+ TEMRA cells (Fig. 3E–F and fig. S6A–B). Gene set enrichment analysis (GESA) identified “Natural killer cell-mediated cytotoxicity” as the most significant pathway in the KIR3DL3+ subset and “T cell receptor signal pathways” as the most significant pathway in KIR3DL3− subset (Fig. 3F, Table S1–2). To verify the above results at the protein level, we performed a high-dimensional flow cytometry analysis and observed a trend of gradual acquisition of NK cell receptors and loss of T cell co-receptors from TN, TCM, TEM, to TEMRA (Fig. 3G–H). Consistent with our RNA-seq finding, KIR3DL3+ CD8+ TEMRA upregulated NK activating (NKG2C, NKp30, NKp46 and CD16) and inhibitory (KIR3DL1, KIR2DL2/3, NKG2A, KLRG1) receptors, but downregulated T cell costimulatory (CD28, ICOS) and coinhibitory (PD-1) receptors in comparison to their KIR3DL3− counterparts (Fig. 3G–H). To exclude the potential contamination of NK cells in these CD8+ T cells, we performed a redirected cytotoxicity assay by using anti-CD16 mAb. While anti-CD3 steadily activated these CD8+ T cells, we did not observe any activation induced by anti-CD16 (fig. S5J). Therefore, our results indicated that KIR3DL3+ CD8+ TEMRA cells acquired NK-like phenotypes.
KIR3DL3+ CD8+ T cells were able to lyse K562 cells without TCR engagement in an E:T ratio-dependent manner (Fig. 3I). HHLA2 expressing K562 cells were more resistant to KIR3DL3+ CD8+ T cell killing as compared to control K562 cells (Fig. 3I and fig. S6C). Gene deletion of HHLA2 sensitized HCC827 to KIR3DL3+ CD8+ T cell lysis (Fig. 3J and fig. S6D). Anti-KIR3DL3 mAb clone 26E10 was capable of completely blocking the binding of KIR3DL3 to HHLA2 (fig. S6E). As expected, 26E10 enhanced the lysis of HCC827 by KIR3DL3+ CD8+ T cells, as well as the degranulation and cytokine (TNF-α and IFN-γ) production of these T cells (Fig. 3K). Together these results indicated that KIR3DL3+ CD8+ TEMRA cells could be activated by both TCR dependent and independent signaling with KIR3DL3 inhibiting T cell function.
KIR3DL3 inhibited NK cell function and mediated HHLA2+ tumor resistance against NK cells
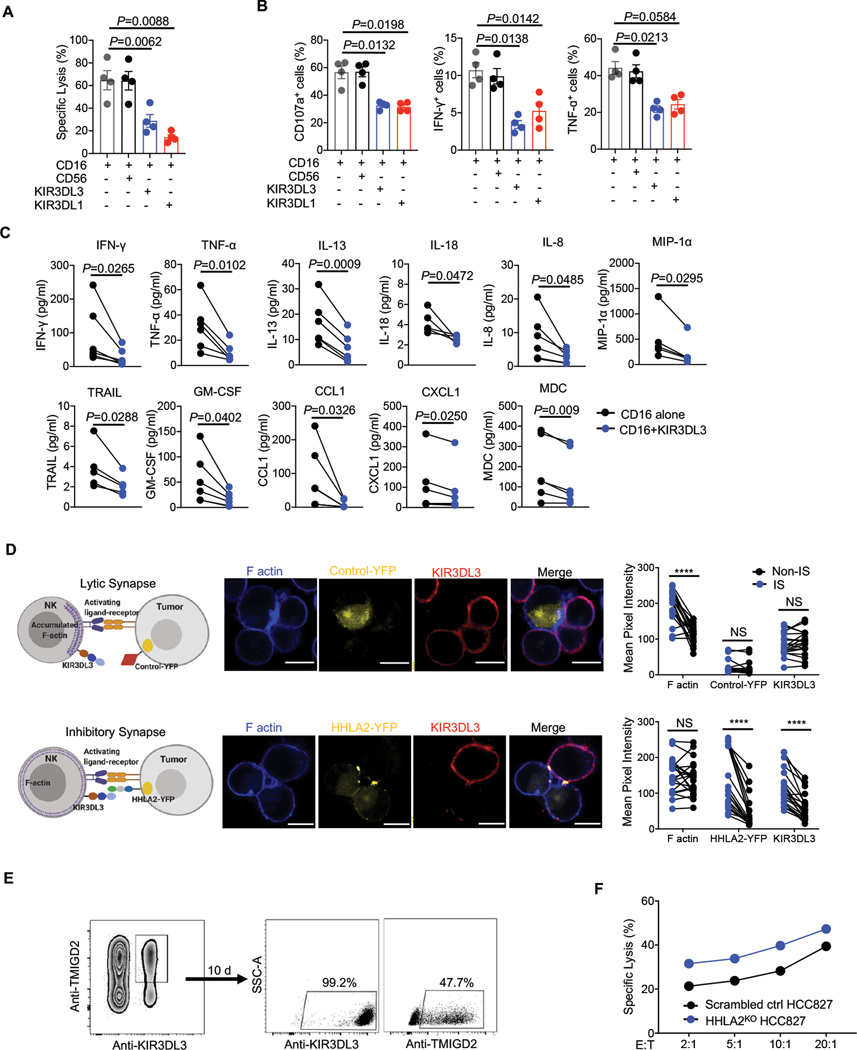
We next sought to determine the effect of KIR3DL3 on human NK cell function. We sorted out primary KIR3DL3+ NK cells and performed an NK cell-based redirected cytotoxicity assay (fig. S7A). CD16-induced lysis of P815 by primary KIR3DL3+ NK cells was significantly inhibited by the co-engagement of KIR3DL3, but was not affected by co-engagement of CD56 (Fig. 4A). Consistently, KIR3DL3 markedly decreased the degranulation, and cytokine and chemokine production of NK cells (Fig. 4B–C). Next, we explored the effect of the KIR3DL3-HHLA2 engagement on NK-mediated tumor cell lysis. The human NK cell line NK92, which were TMIGD2−, was sorted into KIR3DL3+ and KIR3DL3− populations and used as effector cells (fig. S7B). We observed that HHLA2 expressing tumor cells were more resistant to KIR3DL3+ NK92 cell-mediated killing compared to control tumor cells, but this effect was lost when KIR3DL3 was absent (fig. S7C–E).
Fig. 4. KIR3DL3 inhibited primary NK function and mediates HHLA2+ tumor immune resistance.
(A-C) Primary KIR3DL3+ NK cells redirected cytotoxicity assay against P815. (A) The lysis of P815 cells (n=4). (B) The degranulation (CD107a) and cytokine production (IFN-γ and TNF-α) of primary KIR3DL3+ NK cells (n=4). CD56 and KIR3DL1 served as a negative and positive control, respectively. (C) Cytokine production in the co-culture supernatant of primary KIR3DL3+ NK cells with indicated antibody-coated P815 (n=4–6). Data are represented as mean ± SD.
(D) HHLA2-KIR3DL3 mediated inhibitory synapses formation. Left: The cartoons depicting lytic and inhibitory synapses. Middle: Representative confocal images of cell conjugates acquired after KIR3DL3+ NK92 cell-A427 contact followed by staining with anti-KIR3DL3 and Phalloidin. Scale bars, 10 𝜇m. Right: The intensity quantification of F-actin, YFP, and KIR3DL3 at the immunological synapses (IS) and the cell surface away from synapses (Non-IS) from NK92 cell-Control/A427 (n=20) and NK92-HHLA2/A427 (n=21) conjugates. Mouse Tim-3-YFP/A427 was used as the control.
(E) Primary KIR3DL3+ TMIGD2+ NK cells were sorted and cultured in vitro. Ten days later, KIR3DL3 and TMIGD2 expression was examined by flow cytometry.
(F) Lysis of scrambled control or HHLA2KO HCC827 by primary KIR3DL3+ TMIGD2+ NK cells in (E) at indicated E:T ratios. Data are mean for duplicate measurements. Data are representative of three independent experiments with three different donors.
P values by a one-way ANOVA (A-B) or two-tailed paired (C-D). ****P < 0.0001; NS, not significant.
Similar to T cell immunological synapses, both KIR3DL3 and HHLA2 clustered at the interfaces between the KIR3DL3+ NK92 cell and HHLA2 expressing tumor cells (fig. S7F). The NK immunological synapses can be lytic or inhibitory (34, 35), thus we asked if the KIR3DL3-HHLA2 interaction could mediate inhibitory synapse formation. In the absence of HHLA2, we observed F-actin, but not KIR3DL3, polarization at the interfaces (Fig. 4D), indicating lytic synapses were formed to facilitate the tumor-killing. By contrast, in the presence of both HHLA2 and KIR3DL3, we observed HHLA2 and KIR3DL3 colocalization, but no F-actin accumulation, at the interfaces (Fig. 4D), indicating the inhibitory synapses were formed to prevent cytoskeletal reorganization.
Primary NK cells co-expressed KIR3DL3 and TMIGD2 (Fig. 2C). KIR3DL3+ TMIGD2+ NK cells were sorted and expanded (Fig. 4E). The expression of KIR3DL3 was stable, whereas the expression of TMIGD2 decreased after primary NK cell activation (Fig. 4E). HCC827 tumor cells expressed multiple activating ligands and were susceptible to primary NK killing (fig. S7G). Loss of HHLA2 sensitized HCC827 to primary NK cell killing (Fig. 4F), indicating the inhibitory KIR3DL3-HHLA2 pathway was predominant even in the presence of the activating receptor TMIGD2. Together, these results demonstrated that KIR3DL3 exerted an inhibitory effect on NK cells and mediated HHLA2+ tumor resistance to NK cell killing.
KIR3DL3-induced inhibitory signaling in NK cells
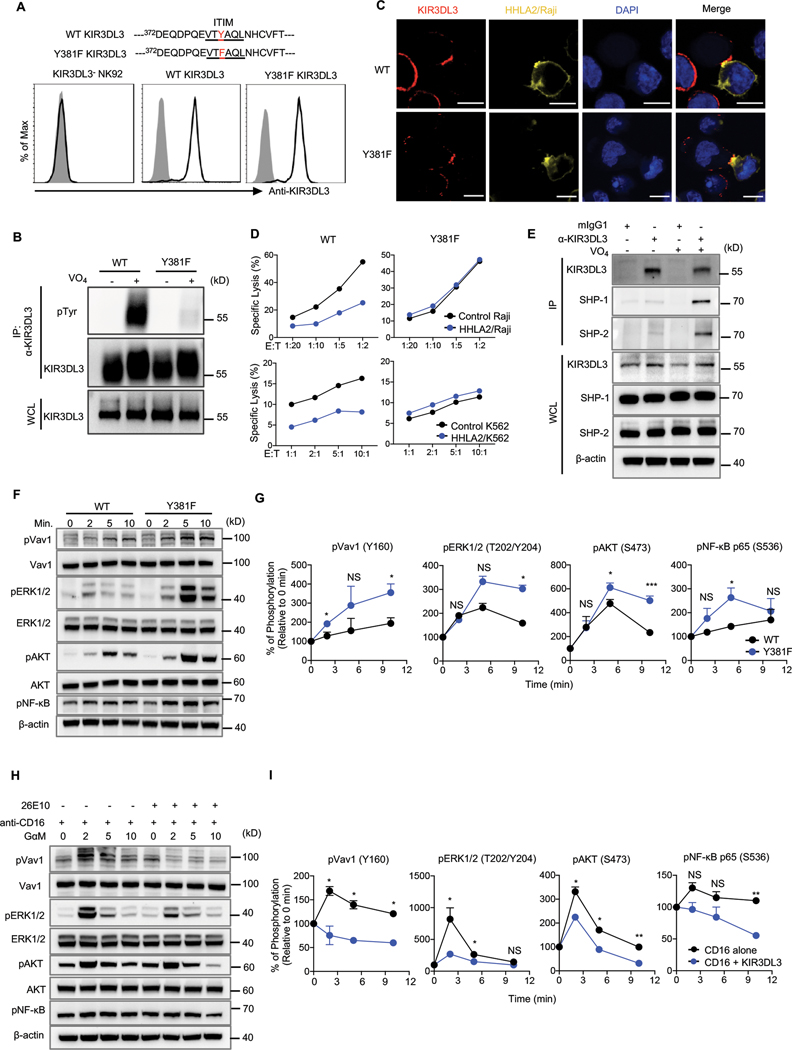
The intracellular tail of KIR3DL3 contained one immunoreceptor tyrosine-based inhibition motif (ITIM). We mutated the tyrosine381 in this ITIM to phenylalanine (Y381F) and introduced wild type (WT) KIR3DL3 or a Y381F mutant into the KIR3DL3− NK92 cell line (Fig. 5A). After treatment with the tyrosine phosphatase inhibitor pervanadate (36), WT KIR3DL3, but not Y381F mutant, displayed tyrosine phosphorylation (Fig. 5B and fig. S8A). Y381F mutation did not affect the recruitment of KIR3DL3 by HHLA2 to immunologic synapses (Fig. 5C) but abolished its inhibitory effect on the NK92 cell line (Fig. 5D).
Fig. 5. KIR3DL3 ITIM mediated NK-cell suppression through recruiting SHP-1 and SHP-2 to attenuate downstream signaling.
(A) Tyrosine (Y) in ITIM of KIR3DL3 was mutated to phenylalanine (F). The KIR3DL3− NK92 cell line was then transduced to express a similar level of WT KIR3DL3 or Y381F mutant.
(B) Tyrosine phosphorylation in WT or Y381F KIR3DL3/NK92 cells cultured in the presence or absence of pervanadate (VO4). IP, immunoprecipitation; WCL, whole-cell lysates.
(C) Images of cell conjugates acquired after indicated NK92 cell line and HHLA2/Raji contact followed by staining with anti-KIR3DL3 and DAPI. Scale bars, 10 𝜇m.
(D) Lysis of Raji (top) or K562 (bottom) by WT or Y381F KIR3DL3/NK92 cell line at the indicated E:T ratios. Data are mean for duplicate measurements.
(E) SHP-1 and SHP-2 were co-immunoprecipitated with KIR3DL3 in primary KIR3DL3+ NK cells.
(F-G) Expression and phosphorylation of Vav1, ERK1/2, AKT, and NF-κB in WT or Y381F KIR3DL3/NK92 cells stimulated by HHLA2/Raji for the indicated time (F). Quantification of immunoblotting (G). Data are mean ± SD from two independent experiments.
(H-I) Expression and phosphorylation of Vav1, ERK1/2, AKT, and NF-κB in primary KIR3DL3+ NK cells after crosslinking with indicated mAbs at the indicated time (H). Quantification of immunoblotting (I). Data are mean ± SD from two independent experiments.
In A-F and H, data are representative of two or three independent experiments. P values by an unpaired Student’s t-test (G, I). *P < 0.05, **P < 0.01, ***P < 0.001; NS, not significant.
ITIMs recruit the SH2 domain-containing tyrosine phosphatase, such as SHP-1 or SHP-2, inhibiting signaling induced by activating receptors (37, 38). Co-immunoprecipitation showed that KIR3DL3 recruited both SHP-1 and SHP-2 in primary NK cells (Fig. 5E and fig. S8B). We thus investigated KIR3DL3/SHP-1/2-induced downstream pathways. WT KIR3DL3 or Y381F mutant transduced NK92 cells were cocultured with HHLA2+ Raji cells, to initiate signaling, and then subjected to analysis with human phospho-kinase arrays. Compared with WT KIR3DL3, the Y381F mutant displayed a higher phosphorylation level of multiple kinases, including ERK1/2 and AKT (fig. S8C–D). Further immunoblot analysis showed enhanced activation of Vav1, ERK1/2, AKT and the downstream transcription factor NF-κB upon Y381F mutation (Fig. 5F–G and fig. S8E). Consistently, upon KIR3DL3 signaling initiation, a significantly reduced activation of Vav1, ERK1/2, AKT and NF-κB was observed in CD16-stimulated KIR3DL3+ primary NK cells (Fig. 5H–I and fig. S8F). These results suggested that the ITIM in KIR3DL3 was essential for KIR3DL3-mediated NK cell suppression by recruiting SHP-1/2, which inhibited Vav1, ERK1/2 and AKT activation and downstream NF-κB signaling.
KIR3DL3+ immune cells infiltrated human HHLA2+ tumors
To further explore the KIR3DL3-HHLA2 pathway in human cancers, we analyzed multiple databases through the Gene Expression Omnibus. KIR3DL3 mRNA was upregulated in several human solid and hematopoietic malignancies as compared to the normal tissues (fig. S9A). HHLA2 mRNA was also significantly upregulated in renal cell carcinoma, whereas TMIGD2 mRNA was downregulated in the tumor (fig. S9A), revealing potential immunosuppressive roles of the KIR3DL3-HHLA2 pathway in the tumor microenvironment. Consistent with the above mRNA data, we observed higher KIR3DL3 and lower TMIGD2 expression on both CD4+ and CD8+ T cells of adult T cell leukemia/lymphoma patients compared to healthy donors (fig. S9B–E).
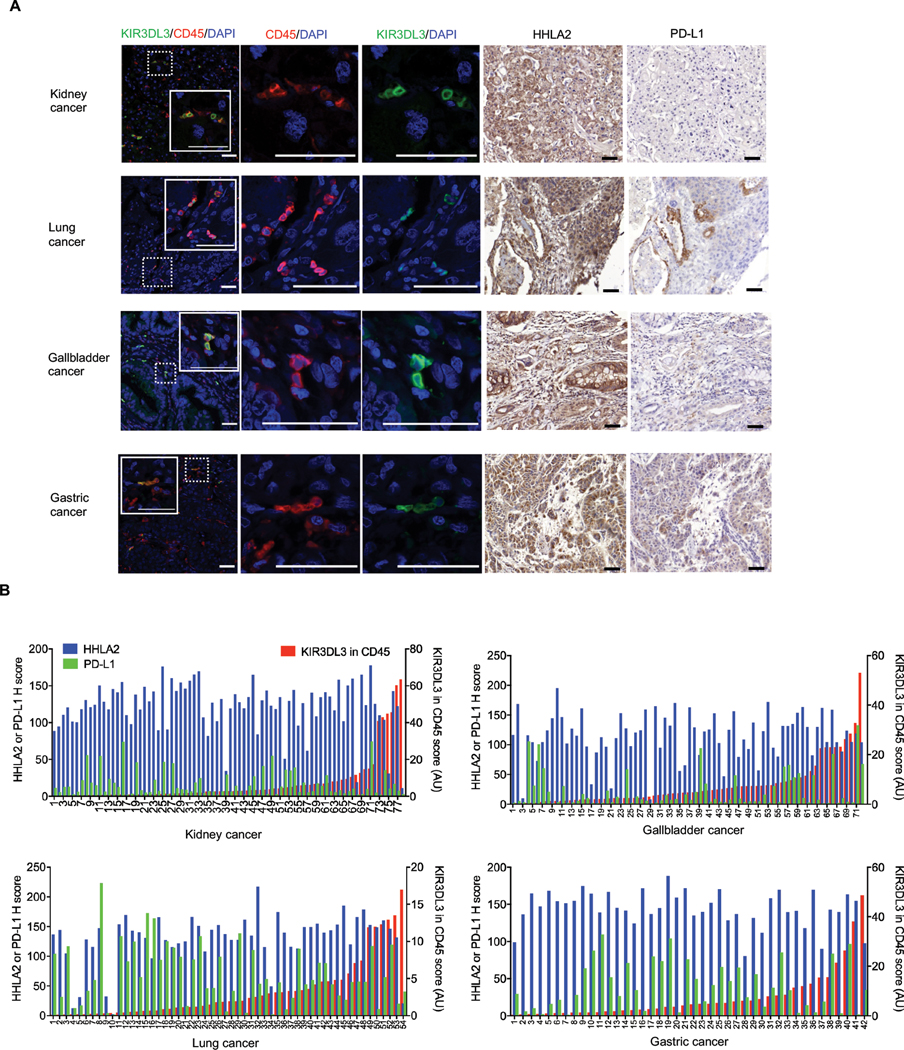
We then used quantitative multiplex immunofluorescence to determine KIR3DL3 and CD45 expression in four tumor microarray (TMA) cohorts, including renal cell carcinoma, lung cancer, gastric cancer, and gallbladder cancer. We observed KIR3DL3+ CD45+ cells infiltrated in all four tumors types (Fig. 6A–B, fig. S9F and table S3). Immunohistochemistry (IHC) staining showed HHLA2 was strongly and widely expressed in these cancers (Fig. 6A–B). By contrast, high PD-L1 expression was limited to a small portion of the lung, gastric, and gallbladder cancer, while it was barely expressed in renal cell carcinoma (Fig. 6A–B). These results suggested that HHLA2+ tumors were infiltrated by KIR3DL3+ immune cells and the KIR3DL3-HHLA2 interaction may represent an immune checkpoint pathway in these cancer patients.
Fig. 6. Human HHLA2+ tumors were infiltrated by KIR3DL3+ immune cells.
(A) Representative images from the indicated cancer type (taken from the patients in B) showed co-expression of KIR3DL3 and CD45 detected by immunofluorescence (left), expression of HHLA2 and PD-L1 detected by immunohistochemistry (right). Scale bars, 50 𝜇m.
(B) Quantitative analysis of KIR3DL3 (QIF score, red, right Y-axis), HHLA2 (H score, blue, left Y-axis) and PD-L1 (H score, green, left Y-axis) expression on kidney (n=78), gallbladder (n=72), lung (n=54), and gastric (n=42) tumors from cancer patients.
KIR3DL3 blockade promoted NK cell-based anti-tumor immunity in vitro and in vivo
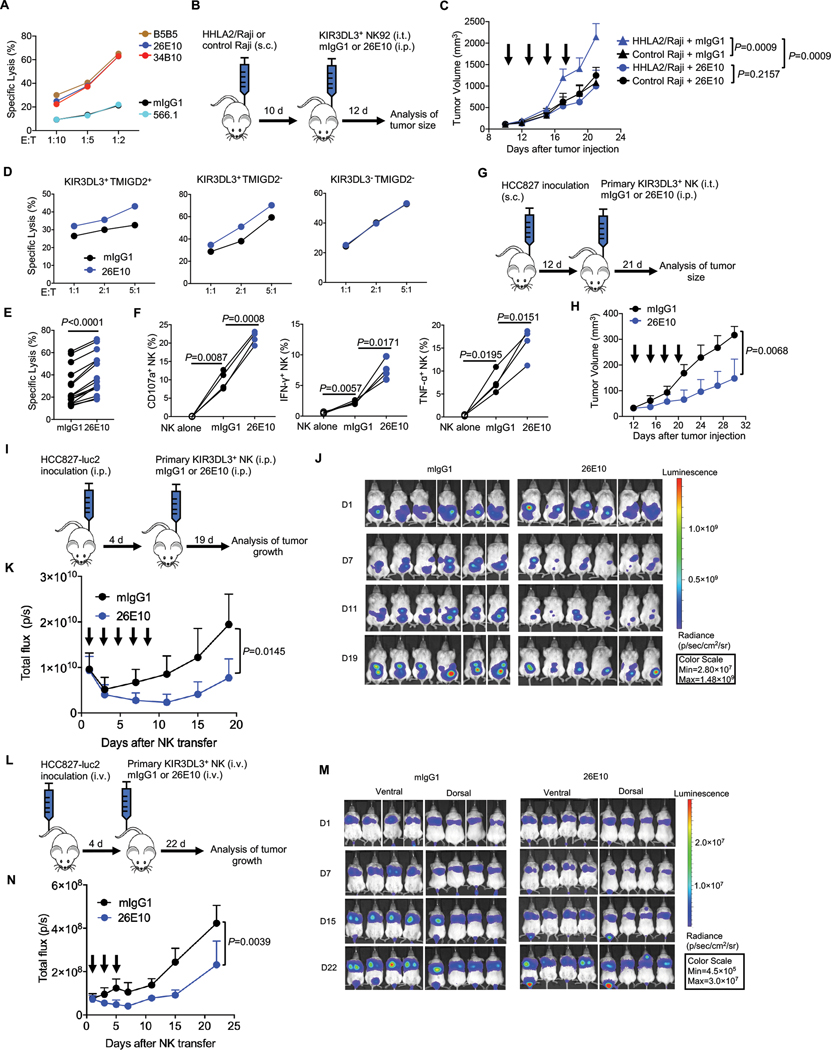
We next explored if the KIR3DL3-HHLA2 blockade could enhance NK-mediated lysis of tumors. Treatment with either anti-KIR3DL3 (clone 26E10 or 34B10) or anti-HHLA2 (clone B5B5) blocking mAbs restored the cytotoxicity of the KIR3DL3+ NK92 cell line against HHLA2+ K562 cells (Fig. 7A). To test this concept in vivo, we subcutaneously engrafted NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NSG) mice with HHLA2+ Raji or control HHLA2− Raji cells and then treated them with KIR3DL3+ NK92 cell line, together with 26E10 or mIgG1 (Fig. 7B). With the treatment of the KIR3DL3+ NK92 cell line alone, the HHLA2+ tumors grew more aggressively than the control tumors (Fig. 7C). Compared to mIgG1, 26E10 significantly suppressed tumor growth in HHLA2+ tumor-bearing mice (Fig. 7C).
Fig. 7. KIR3DL3 blockade promoted NK cell anti-tumor immunity in vitro and in vivo.
(A) The lysis of HHLA2/K562 by KIR3DL3+ NK92 cells in the presence of anti-KIR3DL3 mAbs (clone 26E10 and 34B10) or anti-HHLA2 mAbs (clone B5B5 and 566.1) at an E: T=5:1. Data are mean for duplicate measurements.
(B-C) Subcutaneous Raji mouse model treated with NK92 cells. (B) Outline of the experiment procedure. (C) The tumor growth in the treatment of 26E10 or mIgG1 (n=4). Each arrow indicates treatment with NK92 cells and the antibody. Mean tumor volumes ± SD are shown.
(D) The indicated primary NK cells cytotoxicity against HCC827 at indicated E:T ratios in the presence of 26E10 or mIgG1. Data are mean for duplicate measurements.
(E-F) The cytotoxicity (E) (n=15), degranulation (CD107a) and cytokine production (IFN-γ and TNF-α) (F) (n=4) of primary KIR3DL3+ NK cells against HCC827 in the presence of anti-KIR3DL3 (26E10) or mIgG1 at an E: T=10:1.
(G-H) Subcutaneous HCC827 mouse model treated with primary KIR3DL3+ NK cells. (G) Outline of the procedure. (H) The tumor growth in treatment of the 26E10 or mIgG1 (n=4). Each arrow indicates treatment with primary KIR3DL3+ NK cells and the antibody. Mean tumor volumes ± SD are shown.
(I-K) Intraperitoneal HCC827 tumor model treated with primary KIR3DL3+ NK cells. (I) Outline of the procedure. (J) Bioluminescence images by ventral imaging of each mouse at indicated days. (K) Tumor growth in the 26E10 or mIgG1 treatment group (n=6). Each arrow indicates treatment with primary KIR3DL3+ NK cells and the antibody. Data are mean ± SD of the total flux from ventral imaging for each mouse.
(L-N) HCC827 lung metastasis mouse model treated with primary KIR3DL3+ NK cells. (L) Outline of the experiment procedure. (M) Bioluminescence images by dorsal and ventral imaging of each mouse at indicated days. (N) Tumor growth in the treatment of 26E10 or mIgG1 group (n=4). Each arrow indicates a treatment with primary KIR3DL3+ NK cells and the antibody. Data are mean ± SD of the average of total flux from dorsal and ventral imaging for each mouse.
Data are representative of three (A, D) or two (C, H, K, N) independent experiments. P values by a two-way ANOVA (C, H, K, N), a two-tailed paired t-test (E) and a one-way ANOVA (F).
We next tested the effect of KIR3DL3 blockade on primary NK cells. 26E10 mAb significantly enhanced the lysis of HCC827 by primary KIR3DL3+ NK cells regardless of the presence of TMIGD2 or not, but it showed no effect on primary KIR3DL3− NK cells (Fig. 7D–E). Also, 26E10 significantly improved primary KIR3DL3+ NK cells’ degranulation as well as IFN-γ and TNF-α production (Fig. 7F). We then tested the therapeutic efficacy of KIR3DL3 blockade with primary NK cells in vivo. NSG mice were subcutaneously engrafted with HCC827 and then treated with expanded primary KIR3DL3+ NK cells intratumorally, as well as 26E10 or mIgG1 (Fig. 7G). 26E10 significantly reduced tumor growth when compared with mIgG1 (Fig. 7H). We next intraperitoneally engrafted the luciferase-transduced HCC827 (HCC827-luc2) into NSG mice. The mice were then treated with primary KIR3DL3+ NK cells and 26E10 or mIgG1 intraperitoneally (Fig. 7I). Recombinant human IL-2 (rhIL-2) and rhIL-15 were injected to support primary NK cell survival in vivo. Tumor growth was significantly suppressed in 26E10 treatment group compared with control group (Fig. 7J–K). Using a more physiologically relevant human lung cancer model, mice were inoculated with HCC827-luc2 cells intravenously and then were reconstituted with primary KIR3DL3+ NK cells and treated with 26E10 or mIgG1, rhIL-2, and rhIL-15 (Fig. 7L). Tumor growth was significantly inhibited in the mice treated with 26E10 compared with mIgG1 (Fig. 7M–N). As a control, the therapeutic efficacy of 26E10 was lost when the mice were transferred with primary KIR3DL3− NK cells (fig. S10A–C). Altogether, our results demonstrated that the KIR3DL3-HHLA2 blockade restored the effector function of the NK cells and promoted anti-tumor immunity.
DISCUSSION
HHLA2 plays dual roles in immune response (4, 6, 22, 23), making the HHLA2 blockade approach for cancer immunotherapy a challenge. In this study, we reported that KIR3DL3 mediated HHLA2 co-inhibition on primary CD8+ T and NK cells, which provided a cellular and molecular mechanistic basis for the inhibitory function of HHLA2. We further showed that the KIR3DL3 blockade, which reinvigorated immune responses and enhanced anti-tumor immunity without interrupting the costimulatory function of HHLA2, is a potential therapeutic strategy for cancers.
We demonstrated that our anti-KIR3DL3 clone 26E10 could recognize the majority of KIR3DL3 allelic variants. Although KIR3DL3 is a highly polymorphic gene, KIR3DL3 D0 domain is relatively conserved and bound to HHLA2, indicating that targeting KIR3DL3 D0 domain is a rational strategy for KIR3DL3-HHLA2 blockade. Different KIR3DL1 alleles display different surface expression levels, binding affinity with HLA-Bw4 ligands, and inhibition on NK cells (39–41). Further studies will be needed to determine if KIR3DL3 polymorphisms affect its cell surface expression, its binding to HHLA2, its inhibitory signaling capacities and its association to cancer risk.
The expression of TMIGD2 and KIR3DL3 on T cells was mutually exclusive, suggesting that they function independently in the different stages of T cell differentiation. Naïve CD8+ T cells constitutively express TMIGD2, which transmits a costimulatory signal to T cells (6). Repetitive stimulation downregulates TMIGD2 expression, thus the majority of CD8+ TEMRA lose their TMIGD2 expression. By contrast, KIR3DL3 was mainly expressed on CD8+ TEMRA, but not naïve CD8+ T cells, suggesting the acquisition of KIR3DL3 may require long-term differentiation of some antigen-experienced T cells. CD8+ TEMRA cells, which accumulate during aging, downregulate TCR signaling molecules and costimulatory molecules but upregulate NK receptors (42). This switch enables them to broaden their capacity for immune surveillance by different recognition systems. We showed that the KIR3DL3+ CD8+ TEMRA cells expressed NK receptors and acquired NK-like function. These KIR3DL3+ CD8+ T cells were capable of immediately lysing NK-sensitive K562 targets and less sensitive solid tumor cells, indicating they may exhibit important immune surveillance function by sensing ligands on stressed and transformed cells.
The non-competitive binding of KIR3DL3 and TMIGD2 to HHLA2 and the co-expression pattern of both receptors on NK cells suggested that both receptors can function simultaneously. NK cells are regulated by the balance of signaling from numerous activating and inhibitory receptors (43). These receptors with opposing effects can share the same ligand, such as PVR: DNAM1/TIGIT; HLA-E: NKG2C/NKG2A (43). The immune response is fine-tuned by these paired receptors. Among them, the inhibitory receptors usually dominantly bind to the ligand by a greater affinity to compete with the activating receptor (43). However, KIR3DL3 and TMIGD2 did not compete for the binding to HHLA2. Instead, during NK cell activation, TMIGD2 expression is downregulated (23), while KIR3DL3 expression was stable and thus became predominant. Through this spatial and temporal divergent expression, the NK immune response maintains homeostasis.
Although the biological function of these negative regulators is to limit immune responses, they can be hijacked by tumor cells to evade immunologic destruction. Ectopic expression of HHLA2 on tumor cells was sufficient to make them resistant to human NK and CD8+ T cell killing. Meanwhile, gene deletion of HHLA2 from endogenous HHLA2+ tumors sensitized them to the killing of primary NK and CD8+ T cells. Tumor cells expressing HLA-C could evade KIR2DL1/2/3+-induced NK killing (44). HLA-E also contributes to protecting tumor cells from killing by NKG2A+ NK cells (45, 46). However, downregulation or even complete loss of HLA I molecules occurs in human tumors (47). Tumor cells with impaired expression of HLA I become resistant to CD8+ T cell killing (48), whereas these tumors are highly sensitive to NK cell killing because of “missing-self recognition” (49). Therefore, the selective pressure may drive tumors to exploit the KIR3DL3-HHLA2 pathway to evade NK cell surveillance.
The presence of KIR3DL3+ immune cells in HHLA2+ human tumors and the enhanced anti-tumor immunity by KIR3DL3 blockade were other key findings of our study. We showed that KIR3DL3+ immune cells infiltrated the tumor beds of kidney, lung, gastric, and gallbladder cancers. Its ligand HHLA2 was also widely and highly expressed on these tumors. HHLA2 on cancer could suppress the function of KIR3DL3+ immune cells in the tumor microenvironment leading to immune evasion and poor clinical outcomes in these patients. Indeed, multiple independent studies demonstrate that elevated HHLA2 in cancers is significantly associated with poor prognosis in various cancer patients (9, 10, 13–18). Finally, we showed that KIR3DL3 blockade effectively suppressed tumor growth in multiple humanized mouse models. Subsequent humanization of these anti-KIR3DL3 mAbs and testing in clinical settings will provide further evidence for this new therapeutic application in cancer patients.
MATERIALS AND METHODS
More details on these methods and the cell lines used can be found in the supplementary material.
Study design
This study was designed to elucidate the HHLA2-KIR3DL3 pathway and the therapeutic potential. We utilized multiple approaches to demonstrate KIR3DL3 as a receptor for HHLA2, and used human PBMCs to study the expression pattern, the function and the downstream signaling of KIR3DL3. We then used imaging methods to inspect HHLA2 and KIR3DL3 in the immunological synapse and in the tumor microenvironments. We further utilized humanized NSG mice to test the therapeutic efficacy of KIR3DL3 blockade. Group sizes were typically n =4 or 5 or as indicated in the figure legends, and experiments were repeated at least twice. Investigators were not blinded to group identity during experimentation.
Mice
BALB/c and NSG (NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ) mice at 6–8 weeks old were purchased from Charles River Laboratory and Jackson Laboratory, respectively. All mice were kept in the specific pathogen-free animal facility. All mouse protocols followed NIH guidelines and were approved by the Animal Care and Use Committee (IACUC) of Albert Einstein College of Medicine.
Human primary cells
Human peripheral blood mononuclear cells (PBMCs) were isolated from buffy coats of healthy donors purchased from New York Blood Center, using Ficoll-Hypaque (GE Healthcare) density gradient separation. Human peripheral blood samples of Adult T-Cell Leukemia/Lymphoma patients were obtained at the time of hospitalization or clinic visit in Montefiore Medical Center under IRB approved protocol and with written informed consent from each patient. PBMCs were separated via Ficoll-Hypaque Plus (GE Healthcare).
Human tumor tissues
Formalin-fixed paraffin-embedded (FFPE) commercial human cancer tissue micro-arrays (TMAs) were purchased from USBiomax. There were four different cohorts, including kidney, lung, gastric, and gallbladder cancer, used in this study.
The human protein cell microarray system
The cell microarray technology platform (Retrogenix, https://www.retrogenix.com/the-technology/) was used to identify new binding partners for KIR3DL3 and to examine the specificity of mAb 26E10.
Human phospho-kinase arrays
The phosphorylation profiles of kinases downstream of KIR3DL3-HHLA2 pathway were determined by using a human phospho-kinase array (R&D, Cat# ARY003B) following the manufacturing instructions.
Production and purification of fusion proteins
KIR3DL3-Ig, KIR3DL3-D0-Ig, HHLA2-IgV1-Ig, and TMIGD2-mIgG proteins were generated in an inducible secreted serum-free Drosophila expression system as described previously (4).
Cell binding and blocking assays
In the fusion protein-cell binding assay, 5–20 μg/ml (or indicated concentration) HHLA2-IgG (R&D system), HHLA2-IgV1-Ig, KIR3DL3-Ig, KIR3DL3-D0-Ig or hIgG (R&D system) were incubated with corresponding NIH 3T3 cells on ice for 45 min, followed by incubation with APC or PE conjugated anti-human IgG Fc antibody (1:100, Biolegend, clone HP6017) on ice for 30 min. Then the cells were acquired on an LSR II flow cytometer (BD Biosciences).
Generation of monoclonal antibodies against KIR3DL3 and HHLA2
Mouse anti-KIR3DL3 monoclonal antibodies were generated by standard hybridoma techniques (4).
Biolayer interferometry (BLI)
The affinities of anti-KIR3DL3 mAbs were assayed by biolayer interferometry, using an Octet Red96 system (ForteBio, Pall LLC).
Flow cytometry
Human PBMCs were pre-incubated with FcR blocking reagents (Miltenyi Biotec) before staining. For surface marker staining, cells were incubated with specific antibodies for 30–45 min at 4°C. For intracellular cytokine and CD107a staining, cells were cultured with Protein Transport Inhibitor Cocktail (eBioscience) and anti-CD107a (clone H4A3) for 5 hr, followed by staining with Live/Dead Ghost Dye UV 450 (Tonbo biosciences). Cells were then fixed and permeabilized using Fixation/Permeabilization Solution Kit (BD Biosciences) according to the manufacturer’s guidelines, followed by staining with intracellular antibodies for 30–45 min at 4°C.
For the spectral flow cytometry, PBMCs were stained with a fixable blue dead cell stain kit (Invitrogen) and then incubated with FcR blocking reagents (Miltenyi Biotec). The cells were then stained by anti-KIR3DL3 clone 26E10 on ice for 30 min, followed by goat anti-mouse IgG PE (Biolegend). After blocking with normal mouse serum, the cells were then stained by a mixture of surface markers diluted in Brilliant stain buffer (BD Biosciences) on ice for 30 min. The cells were then acquired on Aurora (Cytek).
Jurkat-Raji and NK92-tumor conjugation assay
For the Jurkat-Raji conjugation assay, Raji cells were pre-pulsed with 100 ng/ml SEE (Toxin Technology) in RPMI medium for 30 min at 37°C. The cells were then washed twice and resuspended in RPMI medium. 4×105 SEE-loaded Raji and 4×105 Jurkat cells were pre-cooled on ice and then mixed in a 96-well round-bottom plate. The plate was centrifuged at 290×g for 1 minute at 4°C and then incubated for 10 min at 37°C. The mixture of cells was resuspended and loaded onto Poly-l-lysine pre-coated slides at RT for 40 min. The cells were fixed with 4% formaldehyde at RT for 15 min and then blocked by 5% normal goat serum at RT for 1 hr, followed by staining with 20 μg/ml anti-KIR3DL3 antibodies (a mixture of clone 3B7, 8G7, 14F8, 15D2, 26E10, 29H7, 34B10 and 51C3) at 4°C overnight and then with goat anti-mIgG (H+L) Alexa Flour 647 (Invitrogen) at RT for 2 hr. The slides were then mounted by Gold Antifade Mountant with DAPI (Life technologies).
For NK92-tumor Conjugation Assay, 5×105 NK92 and 5×105 A427 (or Raji) cells were mixed in a 50 ml Falcon tube centrifuged at 1500 rpm for 2 min at RT. The tube was then incubated at 37°C for 40 min. The cells were then loaded onto Poly-l-lysine pre-coated slides and then stained as described above.
For F actin staining, after staining with primary and secondary antibodies as described above, the cells were permeabilized by 0.1% Triton X-100 at RT for 15 min after staining with primary and secondary antibodies, as described above. The permeabilized cells were then stained with Alexa Flour Plus 405 Phalloidin (Life technologies) for 1 hr at RT. The slides were then mounted by Gold Antifade Mountant without DAPI (Life technologies). Images were acquired by Leica SP8 confocal microscope and processed by ImageJ.
Isolation and culture of human NK and CD8+ T cells
KIR3DL3+ CD8+ T cells were purified by FACS and then cultured with allogeneic PBMCs as feeder cells (irradiated at 3000 rads, feeder cells: T cells=10:1) in OpTimizer (Invitrogen) supplemented with 5% human AB serum (Sigma-Aldrich), 1% L-glutamine, 100 U/mL penicillin, 100 μg/mL streptomycin and anti-CD3/CD28 Dynabeads (Gibco), recombinant human IL-2 (100 ng/mL, Biolegend).
Human primary NK cell subpopulations or NK92 cell subpopulations (KIR3DL3+ or KIR3DL3− NK92 cells) were sorted by FACS based on KIR3LD3 and/or TMIGD2 expression. Primary NK cell subpopulations were then expanded by culturing with autologous PBMCs as feeder cells (irradiated at 3000 rads, feeder cells: NK cells=20:1) in OpTimizer (Invitrogen) supplemented with 5% human AB serum (Sigma-Aldrich), 1% L-glutamine, 100 U/mL penicillin, 100 μg/mL streptomycin and anti-CD3 OKT3 (10 ng/mL, Biolegend), recombinant human IL-2 (40 ng/mL, Biolegend) and IL-15 (10 ng/mL, Biolegend). Five days later, NK cells were further expanded in the same medium without anti-CD3 and feeder cells.
Cytotoxicity assay
Cytotoxicity assays were performed through a flow-based assay. Briefly, target cells were labeled with PKH26 (Sigma-Aldrich) for 2 min at 37°C. Prelabelled target cells were co-incubated with effector cells (NK92, primary NK or CD8+ T cells) in a 96-well round-bottom plate at indicated effector to target (E:T) ratios for 4–6 hr at 37°C. 7-AAD was used to differentiate dead cells from live cells. Supernatants were collected after 24 hr co-culture for Human cytokine 65-Plex Assay (Eve Technologies). The standard formula of 100 × PKH26+7-AAD+ cells/ PKH26+ cells % was used to calculate specific lysis percentages. For KIR3DL3 or HHLA2 blocking, NK cells or target cells were pre-incubated with 20 μg/ml anti-KIR3DL3 or anti-HHLA2 mAbs for 30 min, respectively, before mixing.
CRISPR/Cas9-mediated knockout of HHLA2
The scramble control sgRNA and HHLA2 targeting sgRNA, generated by GPP sgRNA Designer (https://portals.broadinstitute.org/gpp/public/analysis-tools/sgrna-design), was cloned into lentiCRISPR v2 (Addgene 52961). All these constructs are not predicted to target any known sequences in the human genome. The lentiviruses were produced as described above. HCC827 were transduced with viral supernatant and then selected by puromycin (2 μg/ml) for three days. Stable knockout of HHLA2 (HHLA2KO) was confirmed by flow cytometry analysis.
RNA-seq data analysis
RNA-seq reads were aligned to the human genome (hg38) using STAR (version 2.6.1b) (52). The number of RNA-seq fragments mapped to each gene in the reference gene annotation (downloaded from the UCSC genome browser in 11/2019) was then counted using HTseq (version 0.6.1) (53). Genes were considered expressed if their average expression counts were greater than or equal to 1 in either of the two sample groups, and selected for differential expression analysis and principal component analysis (PCA) by DESeq2 (version 3.11) (54). To perform gene set enrichment analysis (GSEA), we used KEGG database and ranked all genes by -log10(p-value) and multiplied by the sign of the log2(fold change) (55). The ranks were then used for GSEA to determine gene sets that were enriched among the genes expressed greater in either KIR3DL3+ or KIR3DL3− samples (55, 56).
Co-immunoprecipitation and immunoblotting
NK92 cells or primary NK cells pretreated with 1 mM pervanadate (NEB) for 2 min at 37°C or not treated were lysed in Pierce IP Lysis Buffer supplemented with protease and phosphatase inhibitor cocktail (Thermo Fisher). Proteins from whole cell lysis were further incubated with anti-KIR3DL3 antibodies and Dynabeads protein G (Thermo Fisher) for further immunoprecipitation. To analyze phosphorylation status, after mixing and receptor crosslinking, cells were lysed in RIPA lysis buffer (50 mM Tris-HCl pH 7.5, 0.15 M NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) supplemented with protease and phosphatase inhibitor cocktail. Samples were separated on SDS-PAGE gels (Genscript) and transferred onto nitrocellulose membranes (Bio-Rad) for protein detection.
KIR3DL3 crosslinking and cell mixing experiments
For mAb-mediated crosslinking of KIR3DL3, KIR3DL3+ NK cells were preincubated with 10 μg/ml isotype control or anti-KIR3DL3 mAbs (clone 26E10) in the presence of anti-CD16 (5 μg/ml) for 30 min on ice. After washing with medium, NK cells were crosslinked with 25 μg/ml goat anti-mouse IgG (minimal x-reactivity) (Biolegend) at 37°C water bath for the indicated times. For cell mixing, pre-chilled NK92 cells and target cells were mixed at a ratio of 2:1 on ice and then transferred to a 37°C water bath for the indicated time. Cells were moved to ice and then lysed for further immunoblotting analysis.
Immunohistochemistry staining
Immunohistochemistry staining were performed as described before (7).
Immunofluorescence staining
FFPE human cancer TMAs (USBiomax) were deparaffinized, followed by antigen retrieval with EDTA unmasking buffer (CST) in a steamer for 20 min at a sub-boiling temperature (95–98°C). Slides were then blocked by 3% hydrogen peroxidase solution at RT for 10 min and subsequently by 5% normal goat serum at RT for 1 hr. A rabbit anti-KIR3DL3 (clone 1136B, R&D) mAb was used at a dilution of 1:3500; a mouse anti-CD45 mAb (clone: 2B11 + PD7/26, Novus Biologicals) was used at a dilution of 1:75 for overnight incubation. Then a goat anti-mIgG (H+L) Alexa Fluor 647 (Invitrogen) and a goat anti-Rabbit IgG (H+L) Alexa Fluor 488 (Invitrogen) were used both at 1:2000 dilution in RT for 1 hr. The slides were then mounted by Gold Antifade Mountant with DAPI (Life technologies). Positive and negative controls (FFPE cell blocks) were included in each staining.
In vivo mouse models of human cancers and NK cells
Six to eight weeks old NSG mice were used in all tumor models. For the subcutaneous Raji tumor model, HHLA2/Raji cells or control Raji cells (1×106) were implanted into NSG mice subcutaneously (s.c.). Once the tumor size reached around 150 mm3, control tumor or HHLA2+ tumor-bearing mice were allocated to two groups (n= 4). Mice were then treated with KIR3DL3+ NK92 cells (2×106) intratumorally (i.t.) and 200 μg anti-KIR3DL3 mAb (clone 26E10) or isotype control (mIgG1) intraperitoneally (i.p.) every two days for four total treatments. For the subcutaneous HCC827 tumor model, 3×106 HCC827 were s.c. injected. After two weeks, the mice were divided into two groups (n=4 for each group) and received KIR3DL3+ NK cell (i.t., 1×107) and mAb (i.p., 200 μg) treatment every two days for four times. The tumor volume was calculated as (width2×length)/2.
For the intraperitoneal tumor mouse model, 4×106 HCC827-Luc2 cells were i.p. injected into NSG mice. KIR3DL3+NK cells were expanded as described before. Four days later, the mice were imaged and then allocated to 26E10 or mIgG1 group based on baseline bioluminescence to ensure sure each group had an equivalent tumor burden (n=6 for each group). Each mouse then received 1×107 KIR3DL3+ NK cells together with 1 μg rhIL-2, 1 μg rhIL-15, and 200 μg 26E10 (or mIgG1) i.p. every other day (5 times in total).
For the intravenous tumor mouse model, 5×105 HCC827-Luc2 cells were intravenously (i.v.) injected into NSG mice. Four or five days later, the mice were allocated to 26E10 or mIgG1 group based on baseline bioluminescence to ensure each group had an equivalent tumor burden (n=4–5 for each group). Each mouse then received 7×106 expanded KIR3DL3+ NK or 4×106 expanded KIR3DL3− NK cells together with 1 μg rhIL-2, 1 μg rhIL-15, and 200 μg 26E10 (or mIgG1) i.v. every other day (3 times in total). Bioluminescence images were acquired by dorsal and/or ventral imaging of each mouse by the caliper life science IVIS spectrum in vivo imaging system beginning 10–18 min after i.p. injection of D-Luciferin (150 μg/g body weight, Gold Biotechnology). The data were analyzed with Living Image 3.0 software.
Quantification and statistical analysis
Statistical analysis was performed with Prism GraphPad software v8.0.2. The error bars in the figures represent the standard deviation (SD). Statistical significance was determined with the paired or unpaired Student’s t-test (two-tailed), one-way or two-way ANOVA, or Fisher’s exact test. The statistical test used, the sample size n, and the P values are stated in the corresponding figure legend. N in the figure represents the biological replicate unless indicated. All in vitro experiments with cell lines were done with at least two replicates per condition and repeated at least twice. In vitro experiments with primary human cells were performed with at least 4 independent donors. All in vivo experiments were performed with at least 4 mice per group and repeated at least twice.
Supplementary Material
Acknowledgments:
We thank Huan Huang for the IVIS Spectrum Imaging System, Hillary Guzik for the 3DHistec Pannoramic 250 Flash II slide scanner and Jinghang Zhang for the Cytek Aurora.
Funding:
Flow Cytometry Core Facility and Analytical Imaging Facility are supported by the National Institutes of Health (NIH) P30CA013330 (Einstein Cancer Center) and P30DK020541 (Einstein-Mount Sinai Diabetes Research Center). The 3DHistec Pannoramic 250 Flash II slide scanner is supported by SIG 1S10OD019961-01. Phillip M. Galbo Jr is supported by NIH 5TL1TR002557.
This work was supported by NIH R01CA175495 and R01DK100525, Department of Defense BC190403, and Cancer Research Institute awarded to Xingxing Zang.
Footnotes
Competing interests:
Yao Wei, Xiaoxin Ren, Scott Moerdler and Xingxing Zang are inventors on a pending patent (KIR3DL3 is an inhibitory receptor of the immune system and uses thereof); Bijan Etemad-Gilbertson is an employee and Xingxing Zang is a co-founder of NextPoint Therapeutics, Inc.
Data and materials availability:
The RNA-seq data have been uploaded to the Gene Expression Omnibus database with accession number GSE175550. All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
References and Notes
- 1.Yang Y, Cancer immunotherapy: harnessing the immune system to battle cancer. J Clin Invest 125, 3335–3337 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Waldman AD, Fritz JM, Lenardo MJ, A guide to cancer immunotherapy: from T cell basic science to clinical practice. Nat Rev Immunol 20, 651–668 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haslam A, Gill J, Prasad V, Estimation of the percentage of US patients with cancer Who are eligible for immune checkpoint inhibitor drugs. JAMA Netw Open 3, e200423 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao R, Chinai JM, Buhl S, Scandiuzzi L, Ray A, Jeon H, Ohaegbulam KC, Ghosh K, Zhao A, Scharff MD, Zang X, HHLA2 is a member of the B7 family and inhibits human CD4 and CD8+ T-cell function. Proc Natl Acad Sci U S A 110, 9879–9884 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Janakiram M, Shah UA, Liu W, Zhao A, Schoenberg MP, Zang X, The third group of the B7-CD28 immune checkpoint family: HHLA2, TMIGD2, B7x, and B7-H3. Immunol Rev 276, 26–39 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu Y, Yao S, Iliopoulou BP, Han X, Augustine MM, Xu H, Phennicie RT, Flies SJ, Broadwater M, Ruff W, Taube JM, Zheng L, Luo L, Zhu G, Chen J, Chen L, B7-H5 costimulates human T cells via CD28H. Nat Commun 4, 2043 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Janakiram M, Chinai JM, Fineberg S, Fiser A, Montagna C, Medavarapu R, Castano E, Jeon H, Ohaegbulam KC, Zhao R, Zhao A, Almo SC, Sparano JA, Zang X, Expression, clinical significance, and receptor identification of the newest B7 family member HHLA2 protein. Clin Cancer Res 21, 2359–2366 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng H, Janakiram M, Borczuk A, Lin J, Qiu W, Liu H, Chinai JM, Halmos B, Perez-Soler R, Zang X, HHLA2, a new immune checkpoint member of the B7 Family, is widely expressed in human lung cancer and associated with EGFR mutational status. Clin Cancer Res 23, 825–832 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jing CY, Fu YP, Yi Y, Zhang MX, Zheng SS, Huang JL, Gan W, Xu X, Lin JJ, Zhang J, Qiu SJ, Zhang BH, HHLA2 in intrahepatic cholangiocarcinoma: an immune checkpoint with prognostic significance and wider expression compared with PD-L1. J Immunother Cancer 7, 77 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou QH, Li KW, Chen X, He HX, Peng SM, Peng SR, Wang Q, Li ZA, Tao YR, Cai WL, Liu RY, Huang H, HHLA2 and PD-L1 co-expression predicts poor prognosis in patients with clear cell renal cell carcinoma. J Immunother Cancer 8, e000157 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng H, Borczuk A, Janakiram M, Ren X, Lin J, Assal A, Halmos B, Perez-Soler R, Zang X, Wide expression and significance of alternative immune checkpoint molecules, B7x and HHLA2, in PD-L1-negative human lung cancers. Clin Cancer Res 24, 1954–1964 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhatt RS, Berjis A, Konge JC, Mahoney KM, Klee AN, Freeman SS, Chen CH, Jegede OA, Catalano PJ, Pignon JC, Sticco-Ivins M, Zhu B, Hua P, Soden J, Zhu J, McDermott DF, Arulanandam AR, Signoretti S, Freeman GJ, KIR3DL3 is an inhibitory receptor for HHLA2 that mediates an alternative immunoinhibitory pathway to PD1. Cancer Immunol Res 9, 156–169 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.. Chen D, Chen W, Xu Y, Zhu M, Xiao Y, Shen Y, Zhu S, Cao C, Xu X, Upregulated immune checkpoint HHLA2 in clear cell renal cell carcinoma: a novel prognostic biomarker and potential therapeutic target. J Med Genet 56, 43–49 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Koirala P, Roth ME, Gill J, Chinai JM, Ewart MR, Piperdi S, Geller DS, Hoang BH, Fatakhova YV, Ghorpade M, Zang X, Gorlick R, HHLA2, a member of the B7 family, is expressed in human osteosarcoma and is associated with metastases and worse survival. Sci Rep 6, 31154 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin G, Ye H, Wang J, Chen S, Chen X, Zhang C, Immune checkpoint human endogenous retrovirus-H long terminal repeat-associating protein 2 is upregulated and independently predicts unfavorable prognosis in bladder urothelial carcinoma. Nephron 141, 256–264 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Wei L, Tang L, Chang H, Huo S, Li Y, HHLA2 overexpression is a novel biomarker of malignant status and poor prognosis in gastric cancer. Hum Cell 33, 116–122 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Xiao Y, Li H, Yang LL, Mao L, Wu CC, Zhang WF, Sun ZJ, The expression patterns and associated clinical parameters of human endogenous retrovirus-H long terminal repeat-associating protein 2 and transmembrane and immunoglobulin domain containing 2 in oral squamous cell carcinoma. Dis Markers 2019, 5421985 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu Z, Dong W, Overexpression of HHLA2, a member of the B7 family, is associated with worse survival in human colorectal carcinoma. Onco Targets Ther 11, 1563–1570 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boor PPC, Sideras K, Biermann K, Hosein Aziz M, Levink IJM, Mancham S, Erler NS, Tang X, van Eijck CH, Bruno MJ, Sprengers D, Zang X, Kwekkeboom J, HHLA2 is expressed in pancreatic and ampullary cancers and increased expression is associated with better post-surgical prognosis. Br J Cancer 122, 1211–1218 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan H, Qiu W, Koehne de Gonzalez AK, Wei JS, Tu M, Xi CH, Yang YR, Peng YP, Tsai WY, Remotti HE, Miao Y, Su GH, HHLA2 is a novel immune checkpoint protein in pancreatic ductal adenocarcinoma and predicts post-surgical survival. Cancer Lett 442, 333–340 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Z, Liu J, Zhang C, Li F, Li L, Wang D, Chand D, Guan F, Zang X, Zhang Y, Over-expression and prognostic significance of HHLA2, a new immune checkpoint molecule, in human clear cell renal cell carcinoma. Front Cell Dev Biol 8, 280 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rieder SA, Wang J, White N, Qadri A, Menard C, Stephens G, Karnell JL, Rudd CE, Kolbeck R, B7-H7 (HHLA2) inhibits T-cell activation and proliferation in the presence of TCR and CD28 signaling. Cell Mol Immunol, doi: 10.1038/s41423-020-0361-7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhuang X, Long EO, CD28 Homolog is a strong activator of natural killer cells for lysis of B7H7(+) tumor cells. Cancer Immunol Res 7, 939–951 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Janakiram M, Chinai JM, Zhao A, Sparano JA, Zang X, HHLA2 and TMIGD2: new immunotherapeutic targets of the B7 and CD28 families. Oncoimmunology 4, e1026534 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trundley AE, Hiby SE, Chang C, Sharkey AM, Santourlidis S, Uhrberg M, Trowsdale J, Moffett A, Molecular characterization of KIR3DL3. Immunogenetics 57, 904–916 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Beziat V, Hilton HG, Norman PJ, Traherne JA, Deciphering the killer-cell immunoglobulin-like receptor system at super-resolution for natural killer and T-cell biology. Immunology 150, 248–264 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zang X, New immune checkpoint pathways: HHLA2 and its receptors including TMIGD2. Cold Spring Harbor Asia Conference on Precision Cancer Biology: From Targeted to Immune Therapies, Suzhou, China, 18 to 22 September 2017. [Google Scholar]
- 28.Wojtowicz WM, Vielmetter J, Fernandes RA, Siepe DH, Eastman CL, Chisholm GB, Cox S, Klock H, Anderson PW, Rue SM, Miller JJ, Glaser SM, Bragstad ML, Vance J, Lam AW, Lesley SA, Zinn K, Garcia KC, A human IgSF cell-surface interactome reveals a complex network of protein-protein interactions. Cell 182, 1027–1043 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Verschueren E, Husain B, Yuen K, Sun Y, Paduchuri S, Senbabaoglu Y, Lehoux I, Arena TA, Wilson B, Lianoglou S, Bakalarski C, Franke Y, Chan P, Wong AW, Gonzalez LC, Mariathasan S, Turley SJ, Lill JR, Martinez-Martin N, The immunoglobulin superfamily receptome defines cancer-relevant networks associated with clinical outcome. Cell 182, 329–344 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Leaton LA, Shortt J, Kichula KM, Tao S, Nemat-Gorgani N, Mentzer AJ, Oppenheimer SJ, Deng Z, Hollenbach JA, Gignoux CR, Guethlein LA, Parham P, Carrington M, Norman PJ, Conservation, extensive heterozygosity, and convergence of signaling potential all indicate a critical role for KIR3DL3 in higher primates. Front Immunol 10, 24 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moretta L, Dissecting CD56dim human NK cells. Blood 116, 3689–3691 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Zhao Y, Lee CK, Lin CH, Gassen RB, Xu X, Huang Z, Xiao C, Bonorino C, Lu LF, Bui JD, Hui E, PD-L1:CD80 cis-heterodimer triggers the co-stimulatory receptor CD28 while repressing the inhibitory PD-1 and CTLA-4 pathways. Immunity 51, 1059–1073 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saverino D, Tenca C, Zarcone D, Merlo A, Megiovanni AM, Valle MT, Manca F, Grossi CE, Ciccone E, CTLA-4 (CD152) inhibits the specific lysis mediated by human cytolytic T lymphocytes in a clonally distributed fashion. J Immunol 162, 651–658 (1999). [PubMed] [Google Scholar]
- 34.Mace EM, Orange JS, Multiple distinct NK-cell synapses. Blood 118, 6475–6476 (2011). [DOI] [PubMed] [Google Scholar]
- 35.Orange JS, Formation and function of the lytic NK-cell immunological synapse. Nat Rev Immunol 8, 713–725 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huyer G, Liu S, Kelly J, Moffat J, Payette P, Kennedy B, Tsaprailis G, Gresser MJ, Ramachandran C, Mechanism of inhibition of protein-tyrosine phosphatases by vanadate and pervanadate. J Biol Chem 272, 843–851 (1997). [DOI] [PubMed] [Google Scholar]
- 37.Daeron M, Jaeger S, Du Pasquier L, Vivier E, Immunoreceptor tyrosine-based inhibition motifs: a quest in the past and future. Immunol Rev 224, 11–43 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Yokoyama WM, Plougastel BF, Immune functions encoded by the natural killer gene complex. Nat Rev Immunol 3, 304–316 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Gardiner CM, Guethlein LA, Shilling HG, Pando M, Carr WH, Rajalingam R, Vilches C, Parham P, Different NK cell surface phenotypes defined by the DX9 antibody are due to KIR3DL1 gene polymorphism. J Immunol 166, 2992–3001 (2001). [DOI] [PubMed] [Google Scholar]
- 40.Vivian JP, Duncan RC, Berry R, O’Connor GM, Reid HH, Beddoe T, Gras S, Saunders PM, Olshina MA, Widjaja JM, Harpur CM, Lin J, Maloveste SM, Price DA, Lafont BA, McVicar DW, Clements CS, Brooks AG, Rossjohn J, Killer cell immunoglobulin-like receptor 3DL1-mediated recognition of human leukocyte antigen B. Nature 479, 401–405 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carr WH, Pando MJ, Parham P, KIR3DL1 polymorphisms that affect NK cell inhibition by HLA-Bw4 ligand. J Immunol 175, 5222–5229 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Pereira BI, De Maeyer RPH, Covre LP, Nehar-Belaid D, Lanna A, Ward S, Marches R, Chambers ES, Gomes DCO, Riddell NE, Maini MK, Teixeira VH, Janes SM, Gilroy DW, Larbi A, Mabbott NA, Ucar D, Kuchel GA, Henson SM, Strid J, Lee JH, Banchereau J, Akbar AN, Sestrins induce natural killer function in senescent-like CD8+ T cells. Nat Immunol 21, 684–694 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Levi-Schaffer F, Mandelboim O, Inhibitory and coactivating receptors recognising the same Ligand: immune homeostasis exploited by pathogens and tumours. Trends Immunol 39, 112–122 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Romagne F, Andre P, Spee P, Zahn S, Anfossi N, Gauthier L, Capanni M, Ruggeri L, Benson DM Jr., Blaser BW, Della Chiesa M, Moretta A, Vivier E, Caligiuri MA, Velardi A, Wagtmann N, Preclinical characterization of 1–7F9, a novel human anti-KIR receptor therapeutic antibody that augments natural killer-mediated killing of tumor cells. Blood 114, 2667–2677 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lo Monaco E, Tremante E, Cerboni C, Melucci E, Sibilio L, Zingoni A, Nicotra MR, Natali PG, Giacomini P, Human leukocyte antigen E contributes to protect tumor cells from lysis by natural killer cells. Neoplasia 13, 822–830 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kamiya T, Seow SV, Wong D, Robinson M, Campana D, Blocking expression of inhibitory receptor NKG2A overcomes tumor resistance to NK cells. J Clin Invest 129, 2094–2106 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leone P, Shin EC, Perosa F, Vacca A, Dammacco F, Racanelli V, MHC class I antigen processing and presenting machinery: organization, function, and defects in tumor cells. J Natl Cancer Inst 105, 1172–1187 (2013). [DOI] [PubMed] [Google Scholar]
- 48.Sade-Feldman M, Jiao YJ, Chen JH, Rooney MS, Barzily-Rokni M, Eliane JP, Bjorgaard SL, Hammond MR, Vitzthum H, Blackmon SM, Frederick DT, Hazar-Rethinam M, Nadres BA, Van Seventer EE, Shukla SA, Yizhak K, Ray JP, Rosebrock D, Livitz D, Adalsteinsson V, Getz G, Duncan LM, Li B, Corcoran RB, Lawrence DP, Stemmer-Rachamimov A, Boland GM, Landau DA, Flaherty KT, Sullivan RJ, Hacohen N, Resistance to checkpoint blockade therapy through inactivation of antigen presentation. Nat Commun 8, 1136 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Karre K, Ljunggren HG, Piontek G, Kiessling R, Selective rejection of H-2-deficient lymphoma variants suggests alternative immune defence strategy. Nature 319, 675–678 (1986). [DOI] [PubMed] [Google Scholar]
- 50.Stewart SA, Dykxhoorn DM, Palliser D, Mizuno H, Yu EY, An DS, Sabatini DM, Chen IS, Hahn WC, Sharp PA, Weinberg RA, Novina CD, Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA 9, 493–501 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Welm BE, Dijkgraaf GJ, Bledau AS, Welm AL, Werb Z, Lentiviral transduction of mammary stem cells for analysis of gene function during development and cancer. Cell Stem Cell 2, 90–102 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR, STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anders S, Pyl PT, Huber W, HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K, KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 45, D353–D361 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP, Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gettinger SN, Choi J, Mani N, Sanmamed MF, Datar I, Sowell R, Du VY, Kaftan E, Goldberg S, Dong W, Zelterman D, Politi K, Kavathas P, Kaech S, Yu X, Zhao H, Schlessinger J, Lifton R, Rimm DL, Chen L, Herbst RS, Schalper KA, A dormant TIL phenotype defines non-small cell lung carcinomas sensitive to immune checkpoint blockers. Nat Commun 9, 3196 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data have been uploaded to the Gene Expression Omnibus database with accession number GSE175550. All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.