Abstract
Bacteria grown on a mixture of carbon substrates exhibit two utilization patterns: hierarchical utilization (HU) and simultaneous utilization (SU). How and why cells adopt these different behaviors remains poorly understood despite decades of research. Recent studies address various open questions from multiple viewpoints. From a mechanistic perspective, it was found that flux sensors play a central role in the regulation of substrate utilization, accounting for the known dependences on single-substrate growth rates, substrate concentrations, and the point where the substrate enters central metabolism. From a physiological perspective, several recent studies suggested HU or SU as growth-optimizing strategies through efficient allocation of essential proteome resources. However, other studies demonstrate that a significant fraction of the proteome is dedicated to functions apparently unnecessary for growth, casting doubt on explanations based on slight efficiency gains. From an ecological perspective, recent theoretical studies suggest that HU can help increase species diversity in bacterial communities.
Keywords: carbon substrate hierarchy, flux sensor, growth-rate optimization, resource allocation, resource competition
Introduction
Bacteria grown on multiple carbon substrates utilize them either hierarchically or simultaneously. Despite decades of extensive experimental and theoretical attention [1][2][3][4][5], many questions remain regarding the regulatory mechanisms, physiological roles, and ecological consequences underlying these utilization patterns. Here we review recent experimental and modeling studies that have provided fresh insights into this classical topic.
Hierarchical utilization (HU) refers to cases in which the consumption of one substrate is fully suppressed in the presence of another; simultaneous utilization (SU) to cases where multiple substrates are utilized together. In practice, however, the distinction is not clear-cut. Even if two substrates are co-utilized, the uptake of each substrate is typically reduced in the presence of the other. In Escherichia coli, this effect results from a generic negative feedback acting on carbon-substrate uptake [6][7] (Figure 1) mediated by the transcriptional regulator cAMP-Crp, which activates the expression of nearly all carbon-uptake systems. The activity of cAMP-Crp is determined by the difference between the carbon-uptake flux and the downstream biosynthesis fluxes. Hence, if carbon-uptake exceeds anabolism, further uptake is suppressed. In the presence of multiple substrates, the feedback operates on all carbon substrates, but it may not affect each equally: for instance, during growth on glucose and succinate, succinate uptake is more strongly suppressed than glucose uptake to an extent that can be quantitatively estimated based on the single-substrate growth rate [6]. Arguably, this unequal suppression amounts to a partial substrate hierarchy. Here, however, we reserve the term HU for cases where the uptake of one substrate (almost) fully suppresses the uptake of another. cAMP-Crp-mediated feedback alone cannot account for such cases [6].
Figure 1.
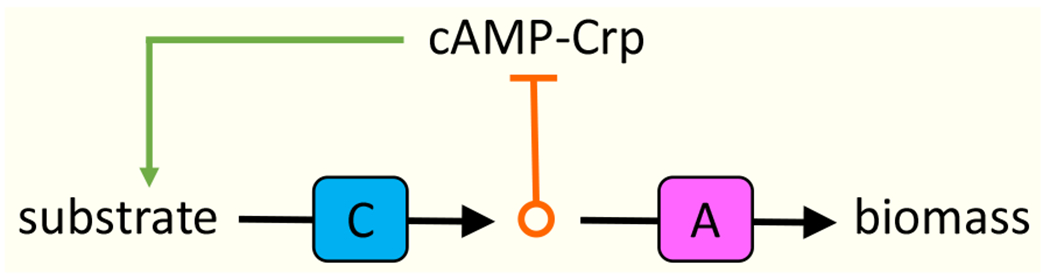
A generic negative feedback affecting the uptake of carbon substrates. A carbon substrate is taken up and catabolized through transporters and catabolic enzymes, grouped as “C”. The precursors derived from the substrate through catabolism are utilized for biosynthesis through biosynthetic enzymes including ribosomes, grouped as “A”. The difference between the fluxes through C and A is read out as the activity of cAMP-Crp, which is necessary for the expression of substrate uptake systems. If the flux through C is in excess, cAMP-Crp activity and thereby the substrate uptake is reduced [7].
Countless experiments, mostly on E. coli, have revealed several patterns in the uptake of mixed substrates. First, if two substrates give rise to HU, the preferred substrate is typically the one that supports the higher growth rate [4]. Second, if multiple substrates are present at low concentrations, as in chemostat cultures, they are co-utilized even if a hierarchy exists at higher concentrations [4][8]. Third, substrates can be classified into two categories depending on whether their degradation products merge into the “upper or middle” part of the glycolysis pathway (category A), or “lower glycolysis” or the TCA cycle (category B) (Figure 2). For E. coli, HU is typically observed only among substrates of category A [6][9]. The challenge is to explain these general observations both from the mechanistic and the physiological perspective.
Figure 2.
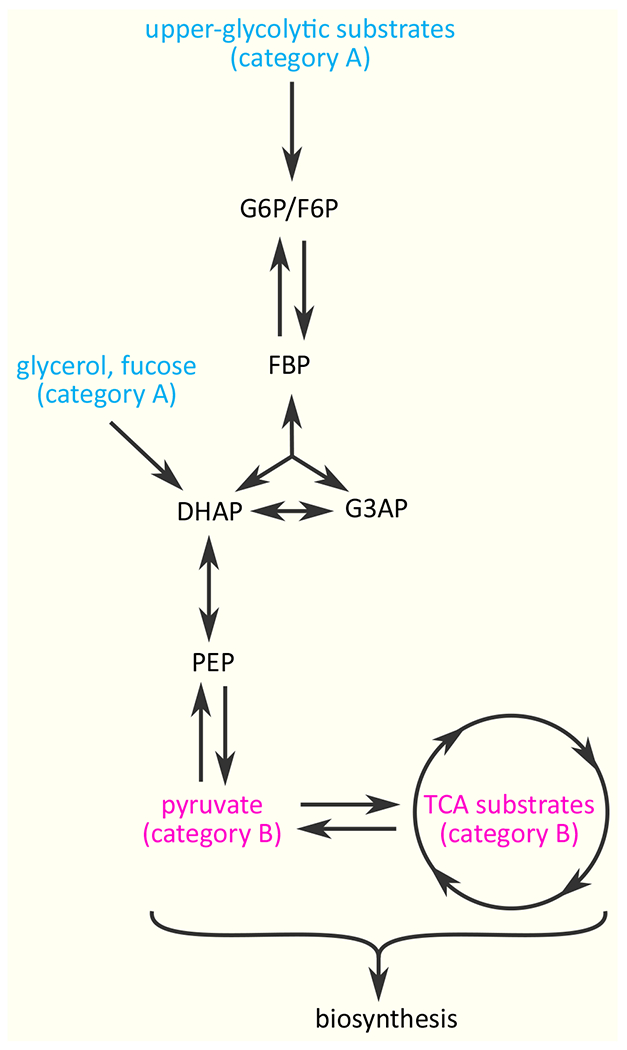
Category A and B substrates. Category A substrates, shown in cyan, are the substrates entering glycolysis either at the upper part (e.g., D-glucose, D-glucose-6-phosphate, D-gluconate, D-mannitol, D-fructose, D-sorbitol, D-mannose, lactose, melibiose, L-arabinose, and D-xylose) or in the middle part (e.g., glycerol and L-fucose). Category B substrates, shown in pink, include pyruvate and TCA substrates such as succinate, fumarate, malate, and oxaloacetate. As a rule, hierarchical utilization is observed only among substrates of category A.
Mechanistic insights
A priori, HU could be implemented by regulators that directly repress the uptake of one substrate in the presence of another. An example is the HU of arabinose and xylose in E. coli [10][11][12]. Few such direct interactions, however, have been identified. This may be because establishing a hierarchy among multiple substrates using direct interactions is impractical: the uptake system of each substrate would have to be repressed by many transcription factors – one for each preferred substrate [13]. Indeed, the preference for lactose over arabinose or xylose appears to be mediated by the generic cAMP-Crp signal rather than direct interactions [14]. Hence, the question becomes how a substrate hierarchy can be implemented using generic rather than specific molecular signals.
A recent study on HU between lactose and glycerol suggests a possible scheme [15]. This study used engineered E. coli cells for which the uptake of lactose in batch culture can be tuned via an inducer. If the lactose uptake flux exceeds a particular threshold, glycerol uptake is fully suppressed (HU). Strikingly, if lactose uptake is below this threshold, glycerol uptake is ‘supplemented’ (i.e., SU) such that the total carbon uptake flux is maintained at the threshold value. Such observations indicate that glycerol uptake responds to the lactose uptake flux rather than to the substrate itself.
How can cells sense fluxes? Within metabolic pathways, particular network motifs can result in correlations between the flux through a pathway and the concentration of certain related molecules[16]. If such molecules engage in regulatory interactions, they can act as flux sensors, allowing the cell to respond to the corresponding fluxes [16][17].
In fact, glycerol uptake responds to two signals: transcriptional activator cAMP-Crp and metabolite fructose-1,6-bisphosphate (FBP), which inhibits the activity of the glycerol kinase GlpK [18]. Both signals are flux sensors: the activity of cAMP-Crp correlates (negatively) with the total carbon-uptake flux [7] (Figure 1) whereas the concentration of FBP correlates with the flux through upper glycolysis [15][19]. A large lactose flux reduces both transcription of the glycerol uptake system (through cAMP-Crp) and GlpK activity (through FBP). These signals are amplified by another positive feedback loop, mediated by the glycerol-specific regulator GlpR, which shuts down glycerol uptake when lactose uptake exceeds the threshold flux value [15].
Importantly, because the glycerol system responds to fluxes rather than to a specific substrate, it is also repressed by other glycolytic substrates, such as glucose, provided their flux exceeds the threshold [15]. Thus, a single mechanism establishes glycerol’s position in the hierarchy.
Such mechanisms can explain several substrate utilization patterns described above. By sensing the total carbon flux, cells switch from HU to SU as the concentration of a preferred substrate – and hence its uptake flux – is reduced, as observed in chemostats [4][8]. Also, by sensing the upper-glycolytic flux, substrates of categories A and B can be discriminated, consistent with their differences in utilization patterns [6]. Moreover, for co-utilized category A and B substrates, a model incorporating only the total-flux-sensor-based feedback by cAMP-Crp can successfully predict the two-substrate growth rate based on the growth rates on each substrate alone [6].
Detailed studies on substrates beyond glycerol are needed to assess how widely flux sensors are employed. That said, other examples of flux sensors involved in the implementation of HU have been identified in the phosphoenolpyruvate:sugar phosphotransferase system (PTS). In various bacterial species, multiple carbon substrates are transported and phosphorylated via this system [3][20]. Early theoretical work already argued that the HU observed among PTS substrates could result from an inherent negative feedback of the total uptake flux of PTS substrates (i.e., the total phosphoryl flux) on the uptake of each individual PTS substrate, and that this total flux would be reflected in the phosphorylated fraction of the PTS components [21]. Indeed, these fraction correlate with the uptake fluxes of both PTS and non-PTS substrates [22][23]. Thus, these components can be regarded as flux sensors. Classical studies have demonstrated that one PTS component, EIIAGlc, is involved in the HU of glucose over non-PTS sugars through inducer exclusion [3][24][25][26]. Recent studies demonstrate that the preference of E. coli for glucose over mannitol – both PTS sugars – involves the dephosphorylated form of another PTS component, HPr [27][28].
While most work has focused on minimal media containing mixtures of carbohydrates, HU and SU also occur in complex media containing mixtures of amino acids [29][30][31][32]. Because most amino acids are degraded into category-B substrates, SU of category A and B substrates may be related to SU of category A substrates and amino acids [31]. Future studies shall reveal the mechanism underlying utilization pattern of amino acids.
Physiological roles
While some consumption patterns can be understood from a mechanistic perspective, a different question is whether we can rationalize them from a physiological perspective.
Many attempts have been made to explain consumption patterns as the result of natural selection acting to optimize the performance of the cell. From this point of view, the task is to find a reasonable objective function, plus relevant constraints, such that the predicted optimal behavior matches the observed patterns. Often the specific growth rate is taken as a proxy for bacterial fitness and hence as the objective.
Many studies assume that the amount of enzyme a cell can produce or contain is constrained due to some limiting resource, be it nutrients, ribosomal capacity, cytosolic or membrane space [13][33][34]. This results in a resource allocation problem: given a choice of two substrates, what fraction of the resource should be invested in the uptake of either substrate? Under a wide range of model assumptions, if the return on investment (ROI), i.e. the carbon-uptake flux per unit of resource invested, differs among substrates, HU is optimal [13][33]: the cell only consumes the substrate providing the largest ROI. If this ROI also dictates the specific growth rate on substrates, this naturally explains that the observed rank order of substrates typically follows their growth rate. However, this argument does not readily explain SU. One possibility is that SU results from nonlinear cost/benefit functions [13], but no specific mechanisms have been identified.
Recent mathematical work provides more general insight by studying Elementary Growth Modes (EGMs): minimal sets of enzymes that can support balanced growth under given conditions [35]. In this framework, SU can be optimal in two ways. First, if no specific constraints are imposed, optimal growth is characterized by a single EGM. SU can then be optimal only if each substrate plays a distinct role in biomass production. Second, if additional constraints are imposed, optimal growth may involve multiple EGMs consuming different substrates, also resulting in SU. However, it is difficult for this theory to explain transitions from HU to SU observed when substrate concentrations are reduced. Also, detailed information, such as rate constants of the metabolic enzymes, is needed to generate quantitative predictions.
A recent study on substrate consumption patterns reveals the importance of the topology of the metabolic network [34]. The production of biomass requires multiple precursors that are derived from various metabolite pools. Some of these pools are more efficiently supplied by glycolytic substrates, others by gluconeogenic substrates. Importantly, SU of category A and B substrates can hence be rationalized as the most efficient way to supply all pools. However, it is difficult to judge whether the modest improvements in ROI thus obtained account for the observed 20 – 50% differences between single-substrate and mixed-substrate growth rates [6][34]. To validate growth-optimization theories, quantitative predictions on growth rate differences between HU and SU, would be valuable.
While many studies consider growth rate as the objective function supplemented with a resource allocation constraint, several facts known for E. coli are hard to square with this framework. First, glucose is preferentially utilized over lactose, and yet in several laboratory strains the growth rate on lactose is indistinguishable from that on glucose and possibly slightly higher [15][36]. Second, the regulation of several uptake/catabolic systems seems suboptimal. For example, GlpK mutations that abolish allosteric inhibition by FBP increase the growth rate on glycerol by over 20% [37][38] and such mutants co-utilize glucose and glycerol without a measurable reduction in growth rate (Okano, unpublished). Third, mass spectrometry shows that the transporter systems of quite a few substrates, such as DppA, RbsB, and MglB, are expressed regardless of the presence of these substrate, each occupying ~0.5% of the proteome, comparable to the glycerol uptake system expressed on glycerol (1.15% for GlpK and 0.25% for GlpD [39][40]. If the consumption patterns are to be explained by ROI gained through such modest fractions of the proteome, it is hard to see why other uptake systems are expressed when they are clearly not required. Indeed, growth increased with deletion of these useless transporters [41]. Generally, a large fraction of proteomes is allocated towards proteins that are unnecessary for growth (but presumably required for other functions, such as motility and chemotaxis) [39][42][43]. Moreover, even the abundances of proteins essential for metabolism and growth are not necessarily set optimal [44]. Deletion of transcriptional regulator Cra, which represses the expression of glycolytic enzymes, increases the growth rate on fructose and mannose by at least 10% [45]. Additionally, recent work combining proteomic and metabolomic analysis of E. coli suggests that the abundances of most metabolic enzymes far exceed what it takes to maintain desired metabolic fluxes [46].
Together, such facts suggest that objectives beyond growth rate and constraints other than resource allocation should be considered. Examples of other potential constraints include an upper bound on the Gibbs free energy dissipation [47] and toxicity of metabolic intermediates [48][49][50][51]. As an alternative objective function one might also consider the average growth rate in a fluctuating environment [45][52]. Adaptation of growth after an environmental switch usually involves a lag time. A recent study demonstrates that this lag time increases with the preshift growth rate[45]. Thus, if substrate availability fluctuates, a larger growth rate on one substrate might result in a smaller growth rate on average.
Ecological consequences
Nutrient utilization strategies adopted by bacterial species may also significantly influence the emergent properties of microbial communities. For example, a model inspired by the stable marriage problem of Game Theory predicts that multi-stability emerges in microbial populations composed of species adopting HU [53].
In some natural habits, such as rivers or intestines, bacteria may encounter growth conditions that resemble chemostat cultures. In chemostat cultures, nutrients are supplied at a fixed low rate and the concentration of a limiting nutrient changes until the cellular growth rate matches the dilution rate. Recent theoretical studies have assessed the effects of substrate utilization strategies on the biodiversity of microbial communities under chemostat-like conditions.
Classical competition theory predicts that the number of coexisting species cannot exceed the number of limiting nutrients – the “competitive exclusion principle” [54][55]. In reality, however, many bacterial species have been found to coexist on a few limiting nutrients [56][57]. This puzzle, known as “the paradox of the plankton” [58], has been studied through consumer-resource models [59][60][61][62][56][63][64]. Recent studies introduced trade-offs in allocating cellular resources into such consumer-resource models under chemostat-like conditions [62][63][64]. Posfai et al [62] showed that, on p substitutable substrates, an arbitrary number of bacterial species can coexist irrespective of their strategies of substrate utilization, provided substrate supply fluxes are located within the p-dimensional space defined by strategies of “keystone” species that preferentially consume different substrates. In the context of substrate utilization strategies, if a population includes p species each adopting a different HU strategy, then those species would act as keystone species that maximize the p-dimensional space and thereby allow any species to coexist in the population at any supply fluxes. Under the assumption of optimal allocation of essential resources, species adopting a distinct HU strategy can invade the preexisting bacterial communities [65], thereby supporting coexistence of more species with different utilization strategies. More recently, Pacciani-Mori et al [63][64] showed that if each species can dynamically adjust their utilization strategy to optimize its own growth rate, then the keystone species of Ref. [62] are not even necessary because the system can “self-organize” to find the right conditions for coexistence.
Currently, experimental validation of these intriguing predictions is lacking. A critical requirement for the model of Posfai et al to support broad coexistence is that each coexisting species has an identical constraint on total resource. This constraint is somewhat relaxed in the model of Pacciani-Mori et al, which provides an explicit mapping [63][64] of the model components to a model of proteome allocation established for E. coli [66]. However, currently it is unknown whether such resource constraints and variability exist across microbes.
Conclusion
While substrate utilization patterns have been studied extensively for over half a century, recent developments reviewed above demonstrate that this subject is far from settled.
The notion of a flux sensor provides a new mechanistic perspective. Flux-sensor-based regulation can serve as a general strategy to implement HU, readily rationalizing the observed hierarchical order of utilization according to single-substrate growth rates, as well as the switch to SU at low substrate concentrations, and the lack of HU between category A and B substrates. While the involvement of flux sensors in the regulation of several uptake systems has been demonstrated, more work is needed to survey the generality of these mechanisms.
The proposed physiological functions of HU and SU have mainly been based on growth-rate optimization achieved through the efficient allocation of essential resources. Recent developments have widened the scope of such arguments considerably. However, several experimental observations, including those from proteomic and metabolomic studies, show that the specific growth rate is often not optimal and gene expression often not streamlined, casting doubt on the notion that the adoption of HU or SU is based on small gains in ROI. Possibly, the identification of additional constraints and/or the application of different (multi-)objective functions can help explain the discrepancies. The dependence of HU and SU on fluxes suggests that such constraints and/or objective functions are condition dependent. Also, further studies will benefit greatly from incorporating elements from the environmental and ecological context of bacterial cells.
Acknowledgement
We are grateful to helpful discussions with Leonardo Pucciani-Mori. This work is supported by the National Institutes of Health through Grant R01GM109069 to TH.
Footnotes
Declaration of interest: none
References
•• of outstanding interest
• of special interest
- 1.Monod J: Recherches sur la croissance des cultures bacteriennes. Hermann & Cie; 1942. [Google Scholar]
- 2.Monod J: The phenomenon of enzymatic adaptation and its bearings on problems of genetic and cellular differentiation. Growth 1947, 11:223–289. [Google Scholar]
- 3.Deutscher J, Francke C, Postma PW: How Phosphotransferase System-Related Protein Phosphorylation Regulates Carbohydrate Metabolism in Bacteria. Microbiology and Molecular Biology Reviews 2006, 70:939–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harder W, Dijkhuizen L: Strategies of mixed substrate utilization in microorganisms. Philosophical transactions of the Royal Society of London Series B, Biological sciences 1982, 297:459–480. [DOI] [PubMed] [Google Scholar]
- 5.Narang A, Pilyugin SS: Bacterial gene regulation in diauxic and non-diauxic growth. Journal of theoretical biology 2007, 244:326–348. [DOI] [PubMed] [Google Scholar]
- 6.Hermsen R, Okano H, You C, Werner N, Hwa T: A growth-rate composition formula for the growth of E. coli on co-utilized carbon substrates. Molecular Systems Biology 2015, 11:801. [DOI] [PMC free article] [PubMed] [Google Scholar]; •This study derives a formula that predict the growth rate of E. coli on two carbon substrates from the growth rates on individual substrates for cases where the uptake of the two substrates is subject only to total-flux feedback by cAMP-Crp.
- 7.You C, Okano H, Hui S, Zhang Z, Kim M, Gunderson CW, Wang YP, Lenz P, Yan D, Hwa T: Coordination of bacterial proteome with metabolism by cyclic AMP signalling. Nature 2013, 500:301–306. [DOI] [PMC free article] [PubMed] [Google Scholar]; •This study identifies the total-flux feedback through cAMP-Crp signaling.
- 8.Lendenmann U, Snozzi M, Egli T: Kinetics of the simultaneous utilization of sugar mixtures by Escherichia coli in continuous culture. Applied and Environmental Microbiology 1996, 62:1493–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Narang A, Konopka A, Ramkrishna D: New patterns of mixed-substrate utilization during batch growth of Escherichia coli K12. Biotechnology and Bioengineering 1997, 55:747–757. [DOI] [PubMed] [Google Scholar]
- 10.Kang HY, Song S, Park C: Priority of Pentose Utilization at the Level of Transcription: Arabinose, Xylose, and Ribose Operons. Molecules and Cells 1998, 8:318–323. [PubMed] [Google Scholar]
- 11.Desai TA, Rao CV.: Regulation of arabinose and xylose metabolism in Escherichia coli. Applied and Environmental Microbiology 2010, 76:1524–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koirala S, Wang X, Rao CV.: Reciprocal regulation of L-arabinose and D-xylose metabolism in Escherichia coli. Journal of Bacteriology 2016, 198:386–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aidelberg G, Towbin BD, Rothschild D, Dekel E, Bren A, Alon U: Hierarchy of non-glucose sugars in Escherichia coli. BMC Systems Biology 2014, doi: 10.1186/s12918-014-0133-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ammar EM, Wang X, Rao CV.: Regulation of metabolism in Escherichia coli during growth on mixtures of the non-glucose sugars: Arabinose, lactose, and xylose. Scientific Reports 2018, 8:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okano H, Hermsen R, Kochanowski K, Hwa T: Regulation underlying hierarchical and simultaneous utilization of carbon substrates by flux sensors in Escherichia coli. Nature Microbiology 2020, 5:206–215. [DOI] [PMC free article] [PubMed] [Google Scholar]; ••Focusing on the glycerol uptake system as a model system, this study shows that the regulation of substrate utilization is not based on the identity of the available substrates but rather on the uptake fluxes they produce.
- 16.Kotte O, Zaugg JB, Heinemann M: Bacterial adaptation through distributed sensing of metabolic fluxes. Molecular Systems Biology 2010, 6:355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Litsios A, Ortega ÁD, Wit EC, Heinemann M: Metabolic-flux dependent regulation of microbial physiology. Current Opinion in Microbiology 2018, 42:71–78. [DOI] [PubMed] [Google Scholar]
- 18.Lin EC: Glycerol dissimilation and its regulation in bacteria. Annual review of microbiology 1976, 30:535–578. [DOI] [PubMed] [Google Scholar]
- 19.Kochanowski K, Volkmer B, Gerosa L, Van Rijsewijk BRH, Schmidt A, Heinemann M: Functioning of a metabolic flux sensor in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America 2013, 110:1130–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Postma PW, Lengeler JW, Jacobson GR: Phosphoenolpyruvate: Carbohydrate phosphotransferase systems of bacteria. Microbiological Reviews 1993, 57:543–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thattai M, Shraiman BI: Metabolic switching in the sugar phosphotransferase system of Escherichia coli. Biophysical Journal 2003, 85:744–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bettenbrock K, Sauter T, Jahreis K, Kremling A, Lengeler JW, Gilles ED: Correlation between growth rates, EIIACrr phosphorylation, and intracellular cyclic AMP levels in Escherichia coli K-12. Journal of Bacteriology 2007, 189:6891–6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Somavanshi R, Ghosh B, Sourjik V: Sugar Influx Sensing by the Phosphotransferase System of Escherichia coli. PLoS Biology 2016, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saier MH, Straud H, Massman LS, Judice JJ, Newman MJ, Feucht BU: Permease-Specific Mutations in Salmonella typhimurium and Escherichia coli That Release the Glycerol, Maltose, Melibiose, and Lactose Transport Systems from Regulation by the Phosphoenolpyruvate:Sugar Phosphotransferase System Downloaded from. Journal of Bacteriology 1978, 133:1358–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inada T, Kimata K, Aiba H: Mechanism responsible for glucose-lactose diauxie in Escherichia coli: Challenge to the cAMP model. Genes to Cells 1996, 1:293–301. [DOI] [PubMed] [Google Scholar]
- 26.Okada T, Ueyama K, Niiya S, Kanazawa H, Futai M, Tsuchiya T: Role of inducer exclusion in preferential utilization of glucose over melibiose in diauxie growth of Escherichia coli. Journal of Bacteriology 1981, 146:1030–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choe M, Park YH, Lee CR, Kim YR, Seok YJ: The general PTS component HPr determines the preference for glucose over mannitol. Scientific Reports 2017, 7:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choe M, Min H, Park YH, Kim YR, Woo JS, Seok YJ: Structural insight into glucose repression of the mannitol operon. Scientific Reports 2019, 9:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prub BM, Nelms JM, Park C, Wolfe AJ: Mutations in NADH: Ubiquinone oxidoreductase of Escherichia coli affect growth on mixed amino acids. Journal of Bacteriology 1994, 176:2143–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sezonov G, Joseleau-Petit D, D’Ari R: Escherichia coli physiology in Luria-Bertani broth. Journal of Bacteriology 2007, 189:8746–8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zampieri M, Hörl M, Hotz F, Müller NF, Sauer U: Regulatory mechanisms underlying coordination of amino acid and glucose catabolism in Escherichia coli. Nature Communications 2019, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]; •This study demonstrates that E. coli cells grown in glucose medium supplemented with casamino acid co-utilize several amino acids together with glucose. In particular, serine, glycine, aspartate, glutamate, and threonine, all of which are low-cost amino acids, are consumed at much higher rates than required as building blocks for protein synthesis.
- 32.Perrin E, Ghini V, Giovannini M, Di Patti F, Cardazzo B, Carraro L, Fagorzi C, Turano P, Fani R, Fondi M: Diauxie and co-utilization of carbon sources can coexist during bacterial growth in nutritionally complex environments. Nature Communications 2020, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dhurjati P, Ramkrishna D, Flickinger MC, Tsao GT: A cybernetic view of microbial growth: Modeling of cells as optimal strategists. Biotechnology and Bioengineering 1985, 27:1–9. [DOI] [PubMed] [Google Scholar]
- 34.Wang X, Xia K, Yang X, Tang C: Growth strategy of microbes on mixed carbon sources. Nature Communications 2019, 10:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]; ••This work seeks to explain the conditions for HU and SU from the perspective of optimal resource allocation by incorporating the topological structure of E. coil’s metabolic network. Thus, it is shown that SU can be the optimal strategy for combinations of category A and B substrates if some metabolite pools are more efficiently produced from the former, and others from the latter.
- 35.de Groot DH, Hulshof J, Teusink B, Bruggeman FJ, Planqué R: Elementary Growth Modes provide a molecular description of cellular self-fabrication. PLOS Computational Biology 2020, 16:e1007559. [DOI] [PMC free article] [PubMed] [Google Scholar]; •This paper develops a mathematical framework to describe all molecular states of a cell that produce balanced exponential cell growth. It introduces Elementary Growth Modes (EGMs) as minimal subsets of enzymes that can support exponential growth, and proves that, within the assumptions of the framework, the state that optimizes the growth rate cannot involve more EGMs than the number of linear constraints imposed plus one.
- 36.Erickson DW, Schink SJ, Patsalo V, Williamson JR, Gerland U, Hwa T: A global resource allocation strategy governs growth transition kinetics of Escherichia coli. Nature 2017, 551:119–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Applebee MK, Joyce AR, Conrad TM, Pettigrew DW, Palsson B: Functional and metabolic effects of adaptive glycerol kinase (GLPK) mutants in Escherichia coli. Journal of Biological Chemistry 2011, 286:23150–23159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zwaig N, Kistler WS, Lin EC: Glycerol kinase, the pacemaker for the dissimilation of glycerol in Escherichia coli. Journal of Bacteriology 1970, 102:753–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmidt A, Kochanowski K, Vedelaar S, Ahrné E, Volkmer B, Callipo L, Knoops K, Bauer M, Aebersold R, Heinemann M: The quantitative and condition-dependent Escherichia coli proteome. Nature Biotechnology 2016, 34:104–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mori M, Zhang Z, Banaei-Esfahani A, Lalanne J, Okano H, Collins BC, Schmidt A, Schubert OT, Lee D, Li G, et al. : From coarse to fine: the absolute Escherichia coli proteome under diverse growth conditions. Molecular Systems Biology 2021, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Balakrishnan R, Hwa T, Cremer J: Suboptimal proteome allocation during changing environments constrains bacterial response and growth recovery. bioRxiv 2021, doi: 10.1101/2021.04.28.441780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Price MN, Deutschbauer AM, Skerker JM, Wetmore KM, Ruths T, Mar JS, Kuehl JV., Shao W, Arkin AP: Indirect and suboptimal control of gene expression is widespread in bacteria. Molecular Systems Biology 2013, 9:660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hui S, Silverman JM, Chen SS, Erickson DW, Basan M, Wang J, Hwa T, Williamson JR: Quantitative proteomic analysis reveals a simple strategy of global resource allocation in bacteria. Molecular Systems Biology 2015, 11:784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Towbin BD, Korem Y, Bren A, Doron S, Sorek R, Alon U: Optimality and sub-optimality in a bacterial growth law. Nature Communications 2017, 8:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Basan M, Honda T, Christodoulou D, Hürl M, Chang YF, Leoncini E, Mukherjee A, Okano H, Taylor BR, Silverman JM, et al. : A universal trade-off between growth and lag in fluctuating environments. Nature 2020, 584:470–474. [DOI] [PMC free article] [PubMed] [Google Scholar]; •E. coli exhibits multi-hour lag times when the carbon substrate is switched from a glycolytic to a gluconeogenic substrates. This study demonstrates that the lag times increases with the preshift growth rate, with an apparent divergence at a critical growth rate. This trade-off may help explain that growth on glycolytic substrates is not always optimized for speed.
- 46.Kochanowski K, Okano H, Patsalo V, Williamson J, Sauer U, Hwa T: Global coordination of metabolic pathways in Escherichia coli by active and passive regulation. Molecular Systems Biology 2021, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Niebel B, Leupold S, Heinemann M: An upper limit on Gibbs energy dissipation governs cellular metabolism. Nature Metabolism 2019, 1:125–132. [DOI] [PubMed] [Google Scholar]; •This computational study on metabolism in budding yeast and E. coli presents a combined thermodynamic and stoichiometric model by adding to a stoichiometric metabolic network model a Gibbs energy balance and suggests an upper limit on the total cellular Gibbs energy dissipation rate.
- 48.Ferguson GP, Tütemeyer S, MacLean MJ, Booth IR: Methylglyoxal production in bacteria: Suicide or survival? Archives of Microbiology 1998, 170:209–218. [DOI] [PubMed] [Google Scholar]
- 49.Booth IR, Ferguson GP, Miller S, Li C, Gunasekera B, Kinghorn S: Bacterial production of methylglyoxal: A survival strategy or death by misadventure? Biochemical Society Transactions 2003, 31:1406–1408. [DOI] [PubMed] [Google Scholar]
- 50.Kadner RJ, Murphy GP, Stephens CM: Two mechanisms for growth inhibition by elevated transport of sugar phosphates in Escherichia coli. Journal of General Microbiology 1992, 138:2007–2014. [DOI] [PubMed] [Google Scholar]
- 51.Bobrovskyy M, Vanderpool CK: The small RNA SgrS: Roles in metabolism and pathogenesis of enteric bacteria. Frontiers in Cellular and Infection Microbiology 2014, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mori M, Schink S, Erickson DW, Gerland U, Hwa T: Quantifying the benefit of a proteome reserve in fluctuating environments. Nature Communications 2017, doi: 10.1038/s41467-017-01242-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goyal A, Dubinkina V, Maslov S: Multiple stable states in microbial communities explained by the stable marriage problem. ISME Journal 2018, 12:2823–2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hardin G: The competitive exclusion principle. Science 1960, 131:1292–1297. [DOI] [PubMed] [Google Scholar]
- 55.MacArthur R: Species packing and competitive equilibrium for many species. Theoretical Population Biology 1970, 1:1–11. [DOI] [PubMed] [Google Scholar]
- 56.Goldford JE, Lu N, Baji;ć D, Estrela S, Tikhonov M, Sanchez-Gorostiaga A, Segré D, Mehta P, Sanchez A: Emergent simplicity in microbial community assembly. Science 2018, 361:469–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bello MD, Lee H, Goyal A, Gore J: A simple linear relationship between resource availability and microbial community diversity. bioRxiv 2020, doi: 10.1101/2020.09.12.294660. [DOI] [Google Scholar]
- 58.Hutchinson GE: The Paradox of the Plankton. The American Naturalist 1961, 95:137–145. [Google Scholar]
- 59.Huisman J, Weissing J: Biodiversity of plankton by species oscilations and chaos. Nature 1999, 402:407–410. [Google Scholar]
- 60.Pfeiffer T, Bonhoeffer S: Evolution of cross-feeding in microbial populations. The American naturalist 2004, 163. [DOI] [PubMed] [Google Scholar]
- 61.Beardmore RE, Gudelj I, Lipson DA, Hurst LD: Metabolic trade-offs and the maintenance of the fittest and the flattest. Nature 2011, 472:342–346. [DOI] [PubMed] [Google Scholar]
- 62.Posfai A, Taillefumier T, Wingreen NS: Metabolic Trade-Offs Promote Diversity in a Model Ecosystem. Physical Review Letters 2017, 118:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]; •This study shows that an unlimited number of species can coexist in an environment with a finite number of resources by introducing trade-offs and constraint into a consumer-resource model.
- 63.Pacciani-Mori L, Giometto A, Suweis S, Maritan A: Dynamic metabolic adaptation can promote species coexistence in competitive microbial communities. PLoS Computational Biology 2020, 16:e1007896. [DOI] [PMC free article] [PubMed] [Google Scholar]; ••This work shows that by dynamically adapting the carbon utilization strategies under an overall resource constraint, bacteria species can “self-organize” their strategies such that an unlimited number of species can coexist in environments with a limited number of nutrients.
- 64.Pacciani-Mori L, Suweis S, Maritan A, Giometto A: Constrained proteome allocation affects coexistence in models of competitive microbial communities. The ISME Journal 2021, doi: 10.1038/s41396-020-00863-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li Z, Liu B, Li SHJ, King CG, Gitai Z, Wingreen NS: Modeling microbial metabolic trade-offs in a chemostat. PLoS Computational Biology 2020, 16:1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Scott M, Gunderson CW, Mateescu EM, Zhang Z, Hwa T: Interdependence of cell growth and gene expression: Origins and consequences. Science 2010, 330:1099–1102. [DOI] [PubMed] [Google Scholar]


