Abstract
Pseudomonas aeruginosa causes severe respiratory tract infections in patients with cystic fibrosis (CF). We have been examining nonopsonic phagocytosis of P. aeruginosa by macrophages. To study the P. aeruginosa-macrophage interaction at the molecular level, we have constructed a transposon Tn5G bank in a clinical isolate of P. aeruginosa (strain 4020) and identified mutants resistant to nonopsonic phagocytosis. Phagocytosis-resistant mutants were enriched by passaging the transposon bank over 18 macrophage monolayers. Of 900 individual mutants isolated from this enriched pool in a nonopsonic phagocytosis assay, we identified 85 putative mutants that were resistant to phagocytosis. In this study, we have characterized one of these transposon mutants, P. aeruginosa 4020 H27A, which was poorly ingested. H27A possessed a Tn5G insertion in a gene encoding a protein with homology to the MotA proteins of several species of bacteria. We have called this gene rpmA for required for phagocytosis by macrophages. RpmA is one of two MotA paralogs in P. aeruginosa. This rpmA::Tn5G mutant was motile both on agar plates and in visual examination of wet mounts. The phagocytosis defect was partially complemented by providing the rpmA gene in trans and fully complemented when both rpmA and rpmB were provided. A rpmA null mutant was ingested by macrophages similar to the H27A transposon mutant. These data suggest that the rpmA and rpmB gene products are required for the efficient ingestion of P. aeruginosa by macrophages.
Pseudomonas aeruginosa is an opportunistic pathogen that causes diverse infections in humans. It produces a wide array of both cell-associated and extracellular components that may act as virulence factors. Lung infections of cystic fibrosis (CF) patients by P. aeruginosa are particularly problematic. Once acquired, it is extremely difficult to eradicate the organism from the lungs. Colonization of the host involves initial adherence of the bacteria to host receptors on either cells or mucins. This adhesion is mediated by pili (12, 13, 43, 63), outer membrane components (9), and flagellar components (3, 50). Initial interaction of P. aeruginosa with the host immune system in the lung environment probably occurs under nonopsonic conditions. Opsonophagocytosis may be inefficient in the lungs of chronically colonized CF patients, since both host and bacterial proteases cleave complement and phagocytosis receptors (59). Initial colonizing isolates of P. aeruginosa are predominantly serum resistant, but P. aeruginosa strains recovered from chronically colonized CF patients are serum sensitive (18), indicating that both complement-mediated lysis and opsonophagocytic clearance are defective in the lungs of these patients (52). Nonopsonic phagocytosis of P. aeruginosa is a receptor-mediated event, and evidence suggests that multiple bacterial ligands are involved in the interaction of bacteria with macrophages. Several studies have demonstrated that the bacterial pilus is involved in the ingestion of P. aeruginosa by macrophages (25, 35, 54). Our laboratory has been investigating the interaction of P. aeruginosa and macrophages. The bacterium-macrophage interaction occurs in two steps. The bacteria adhere to the macrophage, followed by a signal that triggers the phagocytosis of the bacteria (53). Using isogenic mutants, we have shown that pili are involved in the interaction of P. aeruginosa with macrophages, but they are not sufficient for triggering phagocytosis (30). The flagella of P. aeruginosa, or a component associated with the flagellar filament, appear to be necessary for phagocytosis (30). Nonmotile P. aeruginosa isolates are frequently isolated from chronically colonized CF patients (28, 31), and these nonmotile strains are resistant to nonopsonic phagocytosis (31). Motility defects in these strains have been shown to be due to defects in either rpoN or algT genes (16, 31). The nonmotile phenotype may enable the bacteria to evade phagocytosis and persist in the lung. Approximately 50 genes are involved in the production and function of flagella in Escherichia coli or Salmonella enterica serovar Typhimurium (5, 29). Proteins encoded by these genes include structural components of the filament, flagellar motor components, chemotaxis components, and the export apparatus required for flagellar synthesis. Some of the genes involved in flagellar biosynthesis and function of P. aeruginosa flagella have been described (3, 24, 42, 50, 55, 60). A type III secretion system has been identified in P. aeruginosa (15, 64). Recently, it has been demonstrated that P. aeruginosa kills macrophages and macrophage-like cells via this type III secretion system (10, 20, 45). Interaction of P. aeruginosa with the macrophage is required for type III secretion system-mediated effects.
Identification of the bacterial ligands mediating the interaction of P. aeruginosa with macrophages may allow us to modulate this interaction and allow the host to eradicate P. aeruginosa more readily. In this study, we have created a transposon mutant bank in a strain of P. aeruginosa isolated from a pediatric CF patient, and we have isolated mutants resistant to nonopsonic phagocytosis. We have characterized a mutant in which the transposon had inserted into the P. aeruginosa rpmA gene. RpmA is a paralog of the MotA proteins described in enteric bacteria which comprise one component of the flagellar motor (46). Both MotA and MotB are required for torque generation (6, 8). MotA and MotB are cytoplasmic membrane proteins (41) that interact to provide conductance of protons across the membrane (7, 56). We have demonstrated that this P. aeruginosa rpmA mutant retained motility but was resistant to nonopsonic phagocytosis mediated by thioglycolate-elicited mouse peritoneal macrophages.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
P. aeruginosa 4020 is the initial strain isolated from a pediatric CF patient. Randomly amplified polymorphic DNA (RAPD) analysis (32) of subsequent isolates from this patient indicated that this strain chronically colonized this patient for several years (data not shown). All bacterial strains and plasmids are listed in Table 1. All bacterial stocks were maintained at −70°C in Luria broth (L broth) plus 8% (vol/vol) dimethyl sulfoxide (DMSO). Antibiotics were used at the following concentrations: for P. aeruginosa gentamicin (Gm) was used at 75 μg/ml for selection and at 20 μg/ml for maintenance, carbenicillin (Cb) was used at 150 μg/ml, and tetracycline (Tc) was used at 100 μg/ml and for E. coli Gm was used at 10 μg/ml, Tc was used at 20 μg/ml, ampicillin (Ap) was used at 100 μg/ml, spectinomycin was used at 50 μg/ml, and kanamycin (Km) was used at 25 μg/ml. Bacterial were routinely grown in either L broth or Minimal A salts medium (11) supplemented with 50 mM monosodium glutamate and 1% (vol/vol) glycerol. Motility was determined on L broth–0.3% agar plates.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or phenotype | Source or reference |
|---|---|---|
| Strains | ||
| P. aeruginosa | ||
| 4020 | Wild-type clinical isolate | This study |
| 4020 H27A | rpmA::Tn5G | This study |
| 4020-27 | rpmA::Tc | This study |
| 4020-27Sc | Single-crossover cointegrate of pDS27Tc in 4020 | This study |
| PAK | Wild-type laboratory strain | D. Bradley |
| PAK-N1G | PAK rpoN::Gmr | 23 |
| E. coli | ||
| DH5α | hsdR recA lacZYA Φ80 lacZ ΔM15 | Gibco-BRL |
| XL1-Blue MRF′ | Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tcr)] | Stratagene |
| Cosmid (SuperCos1) | Cosmid cloning vector | Stratagene |
| Plasmids | ||
| pRK2013::Tn5G | mob+ tra+, Kmr Gmr | S. Lory |
| pBluescript II SK and KS | Cloning vectors, Apr | Stratagene |
| pGEM5Zf(+) | Cloning vector | Promega |
| pUCP19 | Broad-host-range cloning vector, Apr | 47 |
| pUC19mob | pUC19 with mob region of broad-host-range plasmid pRP4, can be mobilized into P. aeruginosa but will not replicate | 37 |
| pPC110 | Gm cassette | 61 |
| pDSH27A | 7.5-kb SalI fragment from 4020 H27A, cloned into SalI-restricted pBluescript II SK(+) | This study |
| pDS27.1 | 0.85-kb XhoI fragment from pDSH27A, cloned into XhoI restricted pBluescript II KS(−) contains end of transposon plus flanking DNA | This study |
| pDSA4H10 | SuperCos1 cosmid vector with ∼33-kb insert containing rpmA and rpmB | This study |
| pDS27.5 | 3.3-kb SalI fragment from pDSA4H10 cloned into SalI-restricted pUCP19 rpmA+ | This study |
| pDS27.6 | Insert opposite orientation to pDS27.5 rpmA+ | This study |
| pDS27B6 | 6-kb EcoRV fragment from pDSA4H10 cloned into SmaI-restricted pUCP rpmA+ rpmB+ | This study |
| pDS27B7 | Insert opposite orientation from pDS27B6 rpmA+ rpmB+ | This study |
| pDS290 | 2-kb SacI fragment from pDS27B6 cloned into SacI-restricted pUCP19 | This study |
| pDS291 | Insert opposite orientation to pDS290 | This study |
| pDS297 | 2.8-kb SacI fragment from pDS27B7 cloned into SacI-restricted pUCP19 rpmB+ | This study |
| pDS298 | Insert opposite orientation to pDS297 rpmB+ | This study |
| pDS27Tc | SmaI-restricted Tc cassette from miniTn5Tc cloned into unique SmaI site of rpmA gene in pDS27.9 | This study |
| pDS27Tcmob | 3.5-kb PstI fragment from pDS27Tc cloned into PstI-restricted pUC19mob | This study |
Enzymes and chemicals.
All restriction enzymes and T4 DNA ligase were purchased from New England Biolabs. All cell culture reagents were purchased from Gibco-BRL. The random primed labeling kit was purchased from Boehringer-Mannheim Canada.
DNA manipulations.
Plasmid DNA was prepared by the method of Birnboim and Doly (4). Agarose gel electrophoresis, DNA restriction digests, DNA ligations, and Southern blots (under moderate to high stringency) were performed essentially as previously described (44). Chromosomal DNA was isolated from P. aeruginosa as described earlier (33). P. aeruginosa was electroporated using a Bio-Rad Gene Pulser (Bio-Rad, Mississauga, Ontario, Canada) by previously described methods (51).
Transposon mutagenesis of P. aeruginosa 4020.
The transposon Tn5G (36) was used to mutagenize P. aeruginosa 4020 as follows: P. aeruginosa 4020 was grown in L broth and E. coli DH5α(pRK2013::Tn5G) was grown in L broth plus Gm and Km overnight at 37°C. Prior to use in filter matings, the P. aeruginosa culture was heated to 50°C for 3 min. Aliquots (150 μl) of both donor and recipient were added to 5 ml of 10 mM MgSO4 and filtered onto 0.45-μm (pore size) nitrocellulose disks. The filters were placed on L agar and incubated overnight at 37°C. The filters were then removed from the L agar, bacteria were resuspended in 5 ml of 10 mM MgSO4, and aliquots were plated onto minimal agar containing 75 μg of Gm per ml. The colonies after overnight growth at 37°C were scraped from the plates and frozen in aliquots at −70°C in L broth plus 8% (vol/vol) DMSO. Three separate filter matings were used to derive approximately 25,000 P. aeruginosa 4020 mutants.
Enrichment for P. aeruginosa 4020 mutants resistant to nonopsonic phagocytosis.
The bank of transposon mutants was grown overnight in L broth plus 75 μg of Gm ml−1 in two conditions: either with shaking at 150 rpm or statically. The pellicle from the static culture was removed with a pipette and added to L-Gm broth and adjusted to an optical density at 600 nm (OD600) of approximately 0.6. Similarly, the OD of the shaken culture was adjusted to approximately 0.6. Each of these pools was used in parallel in the enrichment procedure described below. Thioglycolate-elicited mouse peritoneal macrophages were harvested by peritoneal lavage as described previously (31), and approximately 3.5 × 105 macrophages were added to glass coverslips in a 24-well tissue culture dish. After overnight incubation, the wells were washed three times with 2.5 ml of phosphate-buffered saline, followed by the addition of 490 μl of phagocytic medium (138 mM NaCl, 8.1 mM Na2HPO4, 1.5 mM KH2PO4, 2.7 mM KCl, 1 mM MgCl2, 0.6 mM CaCl2; pH 7.4) plus 10 mM glucose. Bacteria were added to macrophage monolayers at a multiplicity of infection (MOI) of approximately 1:10 bacteria/macrophage. The 24-well plate was then centrifuged at 2,000 × g for 10 min to facilitate interaction of the bacteria with the macrophage monolayers. After 1 h of incubation at 37°C, the supernatant containing uningested organisms was removed and added to the next uninfected macrophage monolayer. After three to five consecutive passages, depending upon the round of enrichment, the supernatant was removed and plated on Minimal A salts medium plus 75 μg of Gm per ml and grown overnight at 37°C. Colonies from each enrichment were scraped off the plates with a glass rod into L broth; DMSO was added to 8% (vol/vol), and the pooled bacteria were frozen in aliquots at −70°C. Each enrichment was performed with both shaken and statically grown cultures. Aliquots from each of the primary enrichments were inoculated into L-Gm broth and grown overnight either shaken or statically, consistent with the growth conditions prior to the primary enrichment. This enrichment procedure was repeated five times so that the uningested organisms at the end of this selection process had been passaged over 18 macrophage monolayers. Each enrichment pool, from both the shaken and the static cultures, was then plated out so individual mutants could be tested in the phagocytosis assay.
Identification of transposon mutants resistant to phagocytosis.
Individual mutants were grown with shaking at 150 rpm at 37°C overnight in L broth plus 20 μg of Gm per ml. Bacteria were diluted to an OD600 of 0.6, and 50-μl aliquots were added to macrophages adhered to glass coverslips in 24-well plates to give an MOI of approximately 100:1 bacteria/macrophage. The phagocytosis assay was performed essentially as described previously (31), except the wash volumes were increased to 2 ml. Initially, the mutants were scored visually for defects in phagocytosis. Potential mutants were then tested in triplicate and counted. All statistical analyses were performed using the Student's t test.
Identification of transposon insertions.
Chromosomal DNA isolated from putative transposon mutants was digested with restriction enzymes and electrophoretically separated in 0.8% agarose gels. Southern analysis was performed using a probe derived from the gentamicin cassette of plasmid pPC110. All probes were labeled with [α-32P]dGTP (NEN Life Science Products, Mandel Scientific, Guelph, Ontario, Canada) using a random primer labeling kit (Boehringer-Mannheim Canada), according to the manufacturer's instructions. Probes were hybridized at moderate stringency. Chromosomal DNA from 4020 H27A was restricted with SalI and ligated to SalI-restricted pBluescript II, and Gm-resistant colonies were isolated. Plasmid pDSH27A contains a 7.5-kb SalI fragment containing the entire transposon plus approximately 3.2-kb flanking DNA.
Cloning of rpmA and rpmB from P. aeruginosa 4020.
P. aeruginosa 4020 chromosomal DNA was partially restricted with Sau3AI and ligated into the BamHI site of the cosmid vector SuperCos1. Cosmids were packaged using a GigaPack Gold kit (Stratagene). E. coli XL1-Blue MRF′ were transfected with the packaged cosmids. The average insert size was determined to be approximately 30 kb. Southern analysis of colony blots of the cosmid bank using a 0.85-kb XhoI fragment of DNA from pDSH27A containing the end of the transposon plus flanking DNA as a probe allowed us to identify several probe-reactive cosmids. Restriction of these cosmids followed by Southern blotting indicated that a 3.3-kb SalI fragment from cosmid pDSA4H10 hybridized with DNA flanking the transposon. This SalI fragment containing the rpmA gene was cloned into SalI restricted pUCP19 in both orientations for use in complementation experiments. The fragment was also cloned into SalI-restricted pGEM5Zf(+) to form pDS27.9 for use in sequencing. In addition, a 6-kb EcoRV fragment from pDSA4H10 was subcloned into SmaI-restricted pUCP19 in both orientations to form pDS27B6 and pDS27B7. This construct contained both the rpmA and rpmB genes. A 2.8-kb SacI fragment containing the rpmB gene from pDS27B6 was cloned into SacI-restricted pUCP19 in both orientations to form pDS297 and pDS298. Upstream sequences were cloned as a 1.7-kb SacI fragment into SacI-restricted pUCP19 in both orientations to form pDS290 and pDS291.
Sequence analysis.
A series of unidirectional nested deletions of pDS27.9 were created using the Erase-a-Base System kit (Promega). Subclones of pDS27.9 using mapped restriction sites were constructed in pBluescript vectors and also used for sequencing. All plasmid DNA used for sequencing was purified using a Qiagen plasmid kit (Qiagen). Sequencing was performed by automated PCR sequencing (Applied Biosystems 377 Automated DNA Sequencer and AmpliTaq DyeDeoxy Terminator Cycle Sequencing). Sequence data was assembled and analyzed by computer software (Lasergene for Windows; DNASTAR, Inc.). Homologs of putative proteins were identified using the BLAST algorithm (2) at the National Center for Biotechnology Information.
Construction of a rpmA null mutant.
A tetracycline cassette from miniTn5-Tc was isolated as a SmaI fragment and ligated into the unique SmaI site in the rpmA gene to form pDS27Tc. A 5.5-kb PstI fragment from pDS27Tc was cloned into pUC19mob (37) to form pDS27Tcmob. This plasmid was used for homologous recombination. P. aeruginosa 4020 was electroporated with pDS27Tcmob, and cointegrates were isolated on L-Tc plates. Putative rpmA mutants were isolated as Cb-sensitive, Tc-resistant colonies. Southern blots of chromosomal DNA of putative mutants were performed using a radiolabeled DNA fragment internal to the rpmA gene as a probe. Both a single crossover cointegrate termed P. aeruginosa 4020-27Sc and a rpmA::Tc mutant termed P. aeruginosa 4020-27 were isolated.
Detection of flagellin and pilin proteins.
Bacteria were grown overnight in L broth (plus appropriate antibiotics) with tumbling, pelleted by centrifugation, and washed one time in GET buffer (10 mM Tris-Cl, 50 mM glucose, 10 mM EDTA; pH 8.0). The cells were pelleted and resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer and boiled for 5 min prior to electrophoresis through SDS–12% PAGE gels (27). Proteins in the gel were transferred to nitrocellulose (62), and pilin protein was visualized using polyclonal antipilin antiserum (developed against P. aeruginosa PAK pilin [57]). Flagellin was visualized with monoclonal antiflagellin (developed against P. aeruginosa PAK flagellin) (49). Horseradish peroxidase-labeled labeled secondary antibody, followed by color development with 4-chloro-1-napthol, was used to identify immunoreactive components.
Pilin production was also monitored using phage PO4 sensitivity. Phage PO4 causes a lytic infection of P. aeruginosa upon binding to functional pilus organelles. Bacteria were streaked out on agar plates, and approximately 2 × 107 PFU PO4 particles in a 5-μl aliquot were spotted onto the streaked bacteria. A zone of bacterial lysis in the infected area indicated that the bacteria were producing twitching pili.
Nucleotide sequence accession number.
The sequence of pDS27.5 containing the entire rpmA gene and the first 423 nucleotides of the rpmB gene has been submitted to GenBank under accession number AF136403.
RESULTS
Identification of P. aeruginosa 4020::Tn5G mutants resistant to nonopsonic phagocytosis.
A transposon bank was constructed in P. aeruginosa 4020, the initial P. aeruginosa isolate recovered from a pediatric CF patient. RAPD analysis (32) indicated that this same strain colonized this patient for several years (data not shown). To facilitate identification of phagocytosis-resistant mutants, we grew the transposon bank either with shaking or statically, passaged each of these transposon banks in parallel over macrophages, and recovered nonphagocytosed bacteria. Initially, approximately 2% of the inoculum was recovered as noningested organisms from the phagocytic medium over the macrophage monolayers; however, after five separate enrichments, the amount recovered increased to approximately 20% of the inoculum. After this final enrichment, bacteria were plated to isolate individual mutants. Approximately 950 of these mutants (400 from the shaken pool and 550 from the statically grown pool) were examined individually in a nonopsonic phagocytosis assay. Of these mutants, 85 had a phagocytosis defect, 59 isolated from the statically grown pool and 26 isolated from the shaken pool.
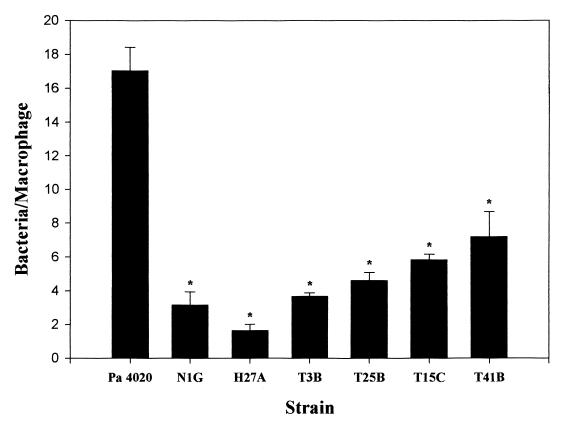
After the initial screen, Southern blots were performed with chromosomal DNA isolated and restricted from these mutants, using the Gm cassette as a probe. Several mutants were identified as siblings since they possessed the same probe-reactive fragments in the Southern blots when the DNA was restricted with multiple enzymes. In one case the same mutant was isolated from both the shaking and the statically grown pools (T25B and H7F). Subclones containing the transposon and flanking DNA on SalI fragments from several mutants were constructed in the pBluescript vectors. Table 2 lists some of these mutants and their phenotypes. Figure 1 illustrates the levels of nonopsonic phagocytosis of several P. aeruginosa 4020::Tn5G mutants by thioglycolate-elicited mouse peritoneal macrophages. All of these mutants possessed a significant resistance to nonopsonic phagocytosis compared to the parental strain (P < 0.05).
TABLE 2.
Phenotype of selected P. aeruginosa 4020::Tn5G phagocytosis-deficient mutants
| Mutanta | Characteristics
|
||
|---|---|---|---|
| Ingestionb | Motilityc | Phage PO4d | |
| H27A* (H24F) | Low | + | s |
| T3B* (T5C) | Low | + | s |
| T25B* (H7F, H8H) | Low | + | s |
| T15C* (T29F, T41F) | Low | + | s |
| T36B | Low-moderate | + | s |
| T7C | Low-moderate | + | s |
| T41B | Low-moderate | + | s |
| H39F* (H18F, H33F) | Low-moderate | + | r |
| H5E | Low-moderate | + | r |
| T13H | Low-moderate | − | s |
| T32F | Low-moderate | + | s |
| T49C | Moderate | + | s |
| H49E | Moderate | + | s |
| T2E | Moderate | − | s |
| PAK-N1G (rpoN::Gm) | Low | − | r |
An asterisk indicates that the transposon plus flanking DNA has been cloned from this mutant. Mutants in parentheses are siblings of the respective mutants (Southern blots). Mutants with the H designation were isolated from a shaking pool; mutants with the T designation were isolated from a static pool.
The initial screen of ingestion was graded: low, 25% of the parental P. aeruginosa 4020 strain; low-moderate, 25 to 40% of the parental strain; and moderate, ∼50% of the parental strain.
Motility was determined on L broth–0.3% agar plates.
s, sensitive to PO4 infection; r, resistant to PO4.
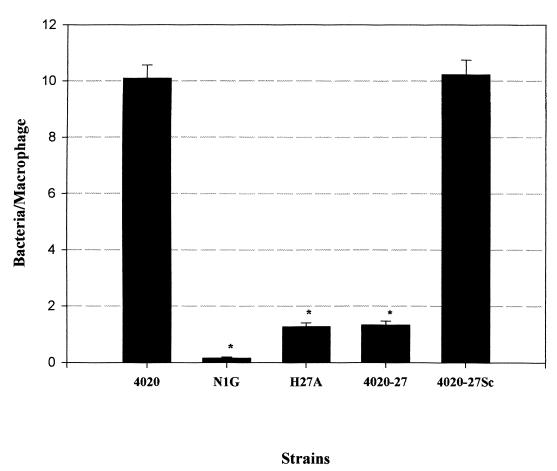
FIG. 1.
Nonopsonic phagocytosis of P. aeruginosa 4020 and selected Tn5G mutants by mouse thioglycolate-elicited murine peritoneal macrophages. Phagocytosis assays were performed as described in Materials and Methods. The y axis represents the mean number of bacteria ingested per macrophage. Error bars represent the standard error of three separate experiments. An asterisk indicates that the number of bacteria phagocytosed was significantly lower than with P. aeruginosa 4020 (P < 0.05).
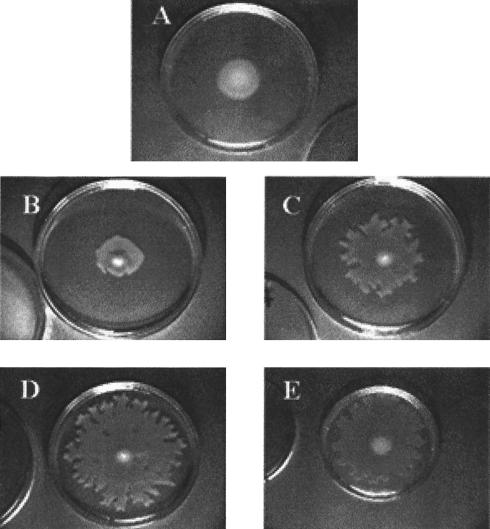
Since flagella and pili are involved in the interaction of P. aeruginosa with macrophages, we examined both the motility and the pilus production of the parental strain and selected mutants. Motility of the mutants was determined on 0.3% agar, and pilus production was monitored by phage PO4 sensitivity. The normal pattern of motility, in which bacteria swim through the agar matrix, results in the formation of concentric circles centered about the inoculation site. P. aeruginosa PAK exhibits this pattern (Fig. 2A). Motility exhibited by P. aeruginosa 4020 initially resembled this phenotype, but after the colony of bacteria reached approximately 15 mm in diameter (which occurred after 14 h of incubation at 37°C), rapid spreading upon the surface of the agar commenced (Fig. 2B to D). This phenotype was lost after repeated subcultures of P. aeruginosa 4020 on L agar. Several nonmotile mutants, such as T13H, and mutants that exhibited a normal motility pattern, such as H39F, were isolated, but the majority of mutants exhibited the unusual motility pattern. The majority of phagocytosis-resistant mutants were phage PO4 sensitive, indicating that they produced an intact functional pilus. We isolated several phage PO4-resistant mutants, although further molecular characterization indicated that many of these were siblings. We selected one of the phagocytosis-deficient transposon mutants, P. aeruginosa 4020 H27A for further study.
FIG. 2.
Motility of P. aeruginosa 4020 and phagocytosis-deficient mutants. P. aeruginosa 4020 strains were stabbed into 0.3% L agar plates and incubated for the indicated times at 37°C. Panels: A, P. aeruginosa PAK, 18 h of incubation; B, C, and D, and P. aeruginosa 4020 after 14, 16, or 18 h of incubation, respectively; E, P. aeruginosa 4020 H27A after 16 h of incubation at 37°C.
Characterization of P. aeruginosa 4020 H27A.
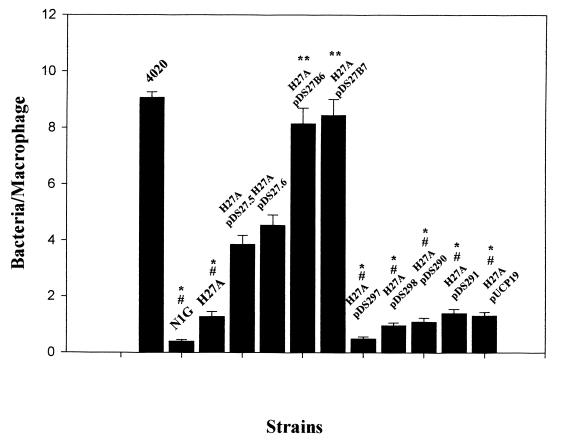
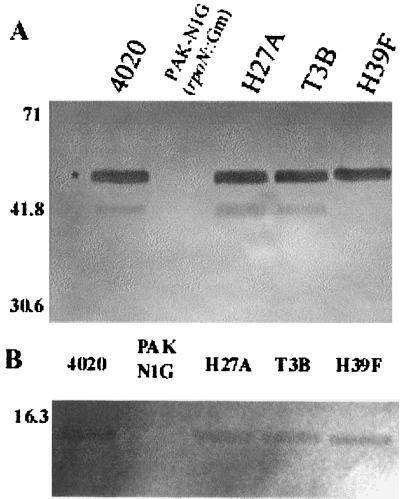
P. aeruginosa 4020 H27A was identified as the most resistant to nonopsonic phagocytosis of the mutants that were isolated using our screen (Fig. 1). This mutant was ingested at a level similar to that of an RpoN mutant (PAK-N1G). The ingestion of H27A was similar whether or not the bacteria had been centrifuged onto the monolayer (compare Fig. 1, where bacteria were centrifuged, versus Fig. 5, where bacteria were not centrifuged). H27A exhibited the parental motility phenotype (Fig. 2E), and it was also sensitive to phage PO4 infection. Visual examination of wet mounts of P. aeruginosa 4020 H27A also indicated that the mutant was motile. Western blots of both whole cells and sheared extracts from cells indicated that H27A produced both flagellin (Fig. 3A) and pilin proteins (Fig. 3B).
FIG. 5.
Phagocytosis of P. aeruginosa 4020 H27A containing rpmA and rpmB derivatives in trans. Phagocytosis assays were performed as described in the text. Bacteria were not centrifuged onto the monolayer. Plasmid-encoded proteins were induced with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside). The numbers represent the mean, and the error bars represent the standard error for triplicate experiments. ∗, Number of bacteria phagocytosed significantly lower than with P. aeruginosa 4020 H27A (pDS27.6) (P < 0.05); ∗∗, number of bacteria phagocytosed significantly higher than with P. aeruginosa 4020 H27A (pDS27.6) (P < 0.01); #, number of bacteria phagocytosed significantly lower than with P. aeruginosa 4020 (P < 0.001).
FIG. 3.
Detection of flagellin or pilin in selected mutants. Strains are labeled at the top of each lane. PAK-N1G (rpoN::Gm) was used as a negative control (i.e., no pilin, no flagellin). Samples were electrophoresed in SDS–12% PAGE gels and transferred to nitrocellulose. Immunoreactive components were visualized with either monoclonal anti-PAK flagellin (A) or anti-PAK pilin polyclonal antibody (B), followed by horseradish peroxidase-labeled secondary antibody. The asterisk in panel A indicates flagellin. Numbers on the left refer to the molecular mass in kilodaltons.
Complementation of the H27A mutation.
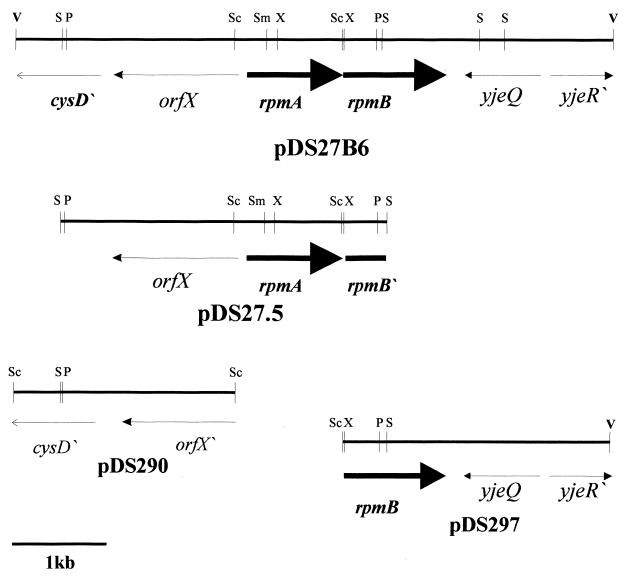
The transposon plus flanking DNA was cloned as a 7.5-kb SalI fragment into pBluescript II SK(+), giving pDSH27A. A 0.85-kb XhoI fragment from pDSH27A encompassing the end of Tn5G plus approximately 400 bp of flanking DNA (termed pDS27.1) was used as a probe to screen a P. aeruginosa 4020 cosmid bank. Five probe-positive cosmids were identified. Cosmid DNA was restricted with several enzymes, and a Southern blot was performed using the 0.85-kb XhoI fragment from pDS27.1 as a probe. Several probe-reactive fragments were cloned from the cosmid into pUCP19 (50), which can be electroporated into P. aeruginosa. A 3.3-kb SalI fragment from the cosmid was cloned into pUCP19 in both orientations to form pDS27.5 and pDS27.6, respectively (Fig. 4, only pDS27.5 is shown). P. aeruginosa 4020 H27A containing either pDS27.5 or pDS27.6 was phagocytosed at a level approximately half that of the parent but twofold greater than the mutant H27A (Fig. 5). This is significantly higher than either H27A alone or H27A carrying the vector (pUCP19) (P < 0.05). Since the orientation of the fragment played no role in complementation, the genes present in the insert were likely being transcribed from their own promoters. A 6-kb EcoRV probe-reactive fragment from the cosmid was cloned into pUCP19 in both orientations to form pDS27B6 (Fig. 4) and pDS27B7, and H27A containing either of these constructs were phagocytosed at a level similar to that of the parental P. aeruginosa 4020 strain (Fig. 5). Thus, genes present on pDS27B6 were required in trans to fully complement the phagocytosis defect of H27A. Plasmids pDS290 (Fig. 4) and pDS291, which contain a portion of the pDS27.5 sequence plus upstream sequences, did not complement the phagocytosis defect when conjugated into H27A (Fig. 5) (P < 0.005). Similarly, plasmids pDS297 (Fig. 4) and pDS298, which contain a small portion of pDS27.5 sequence and downstream sequences, had no effect on the H27A phagocytosis defect (Fig. 5) (P < 0.005).
FIG. 4.
Mapping of the rpmA region. Plasmids are indicated in boldface. Only one orientation of each insert is shown. Upstream and in an opposite orientation to the rpmA gene are orfX, which codes for a protein of unknown function, and the 5′ region of a cysD homolog encoding a putative N-acetylhomocysteine sulfhydrylase. The yjeQ gene and a partial yjeR (orn) gene are present on pDS27B6 downstream of the rpmB gene. Various subclones of pDS27B6 described in the text are shown. Restriction enzyme abbreviations: P, PstI; S, SalI; Sc, SacI; Sm, SmaI; V, EcoRV; X, XhoI. The map is drawn to scale (bar = 1 kb).
Genes identified as rpmA and rpmB.
The site of insertion of the transposon in H27A was identified by sequencing pDS27.1. Sequence analysis indicated that the DNA flanking the transposon encoded a paralog of the MotA proteins present in a number of species of bacteria. To identify the entire gene, plasmid pDS27.5 was sequenced in its entirety using a series of nested deletions. The insert was 3,315 nucleotides in length and contained an open reading frame (ORF) encoding a paralog of the MotA protein; we have called this gene rpmA for required for phagocytosis by macrophages. The putative ATG initiation and stop codons for rpmA are present at nucleotide positions 2007 and 2858, respectively (data not shown). This ORF would encode a 283-amino-acid protein. There are several other potential initiation sites that would produce smaller proteins. A consensus ς28 recognition site (TAA-N15-GCCGATAA) was located 58 bases upstream from the putative ATG initiation codon (at position 2007) of the rpmA gene. There are 20 nucleotides separating the stop codon of rpmA from the ATG initiation codon of a putative rpmB gene. The first 423 nucleotides of rpmB, which encode the first 141 amino acids of the RpmB protein, are present on pDS27.5. Partial complementation of phagocytosis of H27A was observed when only the rpmA gene was present in trans. Both the rpmA and rpmB genes were present on the plasmid pDS27B6 which, when introduced in trans, restored phagocytosis to parental levels. There were two ORFs present on pDS27.5 upstream of the rpmA gene. Plasmid pDS290, which contained these genes, did not complement the phagocytosis defect. The gene immediately upstream of rpmA has been termed orfX since it possessed little homology to any component in the databases, while a partial cysD homolog encoding an O-acetylhomoserine sulfhydrylase was present downstream of the orfX gene (Fig. 4). Plasmid pDS297, which contained the entire rpmB gene as well as a gene encoding a homolog of the YjeQ protein from E. coli, had no effect on phagocytosis of H27A when provided in trans. This arrangement of genes is in agreement with that of the P. aeruginosa PAO1 genome (http://www.pseudomonas.com). Sequencing of pDS27.1 indicated that the transposon had inserted between nucleotides 2397 and 2398 of pDS27.5 corresponding to insertion between R130 and I131 of the putative RpmA protein.
Construction of a rpmA null mutant.
A Tc cassette from mini-Tn5Tc was ligated into a unique SmaI site in the rpmA gene. This was mobilized into P. aeruginosa 4020 as a construct in pUC19mob. A mutant in the rpmA gene was isolated (4020-27) and utilized in a nonopsonic phagocytosis assay. The 4020-27 mutant was as resistant as H27A to nonopsonic phagocytosis (Fig. 6). A single crossover cointegrate (4020-27Sc) was phagocytosed as well as the parental strain.
FIG. 6.
Nonopsonic phagocytosis of a P. aeruginosa 4020 rpmA::Tc (4020-27) mutant by mouse peritoneal thioglycolate-elicited macrophages. Phagocytosis assays were performed as described in Materials and Methods. The numbers represent the mean plus the standard error for triplicate experiments. An asterisk indicates the number of bacteria phagocytosed was significantly lower than with P. aeruginosa 4020 (P < 0.0001).
Analysis of RpmA.
The molecular mass of the predicted 283 amino acid RpmA protein is 30,743 Da. The P. aeruginosa 4020 RpmA protein is 95% identical to the predicted PAO1 RpmA protein. Analysis of the predicted amino acid sequence indicates that P. aeruginosa RpmA likely has six putative transmembrane helices. Overall, there was 41% identity and 64% similarity between P. aeruginosa RpmA and its most closely related homolog, S. enterica serovar Typhimurium MotA. Other closely related homologs include: E. coli MotA, Vibrio parahaemolyticus LafT, Agrobacterium tumefaciens MotA, and Sinorhizobium meliloti MotA.
DISCUSSION
The interaction of P. aeruginosa with macrophages in the absence of opsonins is multifactorial. Both the bacterial flagellar filament and the pilus have been shown to be required for the initial interaction of the bacterium with the macrophage. In this study, we have mutagenized a clinical strain of P. aeruginosa and identified mutants with defects in nonopsonic phagocytosis mediated by mouse peritoneal macrophages. Since bacterial organelles involved in motility are implicated in the bacterium-macrophage interaction, we devised specific selection and enrichment procedures to decrease the likelihood of isolating these types of mutations. The rationale for growing the mutant pool under static conditions and utilizing the pellicle as a source of organisms was to decrease our chances that pilin and flagellin mutants would be isolated. These classes of mutants would not be well represented in the pellicle of a static culture. After testing 950 individual mutants, we identified 85 mutants that had a phagocytic defect. Several mutants had a number of readily identifiable phenotypic defects in addition to the phagocytosis defect. Despite the screening procedures, we did identify mutants in both pilus and flagellar biosynthetic pathways, including flagellin-deficient mutants (T13H) and phage PO4-resistant mutants (pilus defective; H39F).
The parental strain used for the mutagenesis exhibited a motility pattern unusual for P. aeruginosa. It resembled the swarming phenotype exhibited by other bacteria, including E. coli, Serratia marcescens, and S. enterica serovar Typhimurium (1, 19). This swarming phenotype has also recently been described in Pseudomonas syringae B728a (26). Swarming motility exhibited by P. aeruginosa has recently been observed in other laboratories (48). We are currently characterizing the nature of this swarming phenotype, but it appears that it is not required for nonopsonic phagocytosis since H27A displays the same motility pattern as the parent.
We characterized in depth one P. aeruginosa mutant which had a transposon insertion in a gene that encoded a paralog of enteric MotA proteins, which we have called rpmA. This mutant was poorly ingested compared to the parental strain. With the sequencing of the P. aeruginosa PAO1 genome it is apparent that P. aeruginosa possesses two MotA homologs in separate regions of the chromosome. Recently, Kato et al. (24) constructed a mutant in orf1 of P. aeruginosa PAO1 which encodes a MotA homolog and demonstrated that this mutant (PM1) does not swim in agar motility plates but is motile when viewed microscopically. This suggests that orf1 likely encodes the functional equivalent of enteric MotA proteins. Furthermore, the arrangement of genes surrounding orf1 includes a number of chemotaxis genes, a finding which is similar to the clusters of chemotaxis genes surrounding enteric motA genes. The orf1 gene would encode a 246-amino-acid protein of a predicted molecular mass of 25,962 Da. In this study, we have identified the other paralog of MotA, RpmA. Analysis of the region surrounding the P. aeruginosa 4020 rpmA gene revealed a high degree of identity with the PAO1 genome. A MotB paralog termed RpmB is immediately downstream of RpmA. There are no apparent chemotaxis or motility genes surrounding rpmA or rpmB. This contrasts with other MotA homologs. The predicted PAO1 Orf1/MotA and 4020 RpmA proteins are 24.7% identical and 59.6% similar over a 246-amino-acid overlap, which suggests that they have some structural similarity but are likely functionally distinct. RpmA is longer than PAO1 Orf1/MotA (283 versus 246 amino acids) and is structurally more similar to enteric MotA proteins than to PAO1 MotA. The only other organisms that have had two MotA homologs described are Bacillus subtilis and B. megaterium (17, 21). In B. subtilis, one of these homologs termed MotA plays a role in motility (34), while the function of the second homolog (ORF1) is unknown but unrelated to motility (17, 22). This ORF1 is present downstream of the ccpA gene, which is involved in catabolite repression, and immediately upstream of a MotB homolog, ORF2.
The phagocytic defect of H27A (rpmA::Tn5G) was only partially complemented when the rpmA gene was provided in trans, possibly due to a dominant-negative effect of the rpmA mutation. Multiple copies of MotA form a complex with other proteins to form the flagellar motor (56, 58). It is possible that RpmA also forms a complex with other components. It is more likely, however, that the transposon insertion into the rpmA gene in H27A had a polar effect on the rpmB gene, since providing both genes in trans fully abrogated the ingestion defect. Furthermore, a rpmA::Tc mutant (4020-27) was as resistant as the transposon mutant H27A to nonopsonic phagocytosis.
Our results suggest that RpmA of P. aeruginosa may have different or additional functions than the MotA of other species. E. coli and S. enterica serovar Typhimurium motA and motB null mutants produce flagella that are paralyzed and so do not swim in motility plates. P. aeruginosa H27A is motile in the absence of a functional RpmA (and possibly RpmB), and this motility was indistinguishable from that of the parental strain. This indicated that RpmA was not involved in the unusual motility exhibited by P. aeruginosa 4020. This swarming motility is not involved in nonopsonic phagocytosis, since the phagocytosis-resistant mutant H27A (rpmA::Tn5G) exhibits the same motility as the parental strain. Flagellar rotation in P. aeruginosa 4020 appears to be unrelated to RpmA function, since H27A remains motile. However, it was possible that the unusual motility expressed by P. aeruginosa 4020 masks our ability to detect a defect in flagellar motility of the rpmA strain.
A consensus ς28 recognition site upstream of the rpmA gene suggests that this gene is regulated via the flagellum regulatory hierarchy. Since rpmA was not involved in the unusual motility pattern, it is possible that a second flagellum-dependent motility system involving RpmA exists. The phagocytosis defect exhibited by H27A (rpmA mutant phenotype) suggests that some subtle aspect of genes regulated by the flagellar cascade are involved in phagocytosis. It has been well established that motility is involved in bacterial interactions with cells (reviewed in reference 39). Flagella have been shown to play a role in certain types of infections caused by P. aeruginosa (14). Flagellum-mediated motility is involved in the formation of biofilms on abiotic surfaces by both P. aeruginosa and E. coli (38, 40). The flagellar cap protein, FliD, interacts with mucin (3), and other flagellar components are involved in the interaction of P. aeruginosa with eukaryotic cells (50). We believe that it is unlikely that the RpmA protein is directly involved in the interaction of P. aeruginosa with macrophages, but the function of RpmA is required for ingestion.
This study indicates that a functional RpmA protein (and to a lesser extent RpmB) is required for efficient phagocytosis of P. aeruginosa 4020 by thioglycolate-elicited murine peritoneal macrophages. Two paralogs of enteric MotA proteins exist in P. aeruginosa, and these appear to be functionally distinct. P. aeruginosa PAO1 Orf1/MotA mutants are not motile in agar swim plates (24), while P. aeruginosa 4020 rpmA mutants are still motile. Thus, the function of RpmA in P. aeruginosa is different from that of MotA in enteric bacteria. Ongoing characterization of RpmA should allow us to delineate the precise role it plays in nonopsonic phagocytosis.
ACKNOWLEDGMENTS
We thank Eshwar Mahenthiralingam and members of the Speert lab for helpful comments, Jacqueline Chung for technical assistance, Marguerite Cervin for careful review of the manuscript, and Steve Lory for strains.
This work was supported by a grant to D.P.S. from the Medical Research Council of Canada.
REFERENCES
- 1.Alberti L, Harshey R M. Differentiation of Serratia marcescens 274 into swimmer and swarmer cells. J Bacteriol. 1990;172:4322–4328. doi: 10.1128/jb.172.8.4322-4328.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T F, Schaffer A A, Zhang J, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arora S K, Ritchings B W, Almira E C, Lory S, Ramphal R. The Pseudomonas aeruginosa flagellar cap protein, FliD, is responsible for mucin adhesion. Infect Immun. 1998;66:1000–1007. doi: 10.1128/iai.66.3.1000-1007.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blair D F. How bacteria sense and swim. Annu Rev Microbiol. 1995;49:489–522. doi: 10.1146/annurev.mi.49.100195.002421. [DOI] [PubMed] [Google Scholar]
- 6.Blair D F, Berg H C. Restoration of torque in defective flagellar motors. Science. 1988;242:1678–1681. doi: 10.1126/science.2849208. [DOI] [PubMed] [Google Scholar]
- 7.Blair D F, Berg H C. The MotA protein of Escherichia coli is a proton-conducting component of the flagellar motor. Cell. 1990;60:439–449. doi: 10.1016/0092-8674(90)90595-6. [DOI] [PubMed] [Google Scholar]
- 8.Block S M, Berg H C. Successive incorporation of force generating units in the bacterial rotary motor. Nature. 1984;309:470–472. doi: 10.1038/309470a0. [DOI] [PubMed] [Google Scholar]
- 9.Carnoy C, Scharfman A, Van B E, Lamblin G, Ramphal R, Roussel P. Pseudomonas aeruginosa outer membrane adhesins for human respiratory mucus glycoproteins. Infect Immun. 1994;62:1896–1900. doi: 10.1128/iai.62.5.1896-1900.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coburn J, Frank D W. Macrophages and epithelial cells respond differently to the Pseudomonas aeruginosa type III secretion system. Infect Immun. 1999;67:3151–3154. doi: 10.1128/iai.67.6.3151-3154.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis B D, Mingioli E S. Mutants of Escherichia coli requiring vitamin B-12. J Bacteriol. 1950;60:17–28. doi: 10.1128/jb.60.1.17-28.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doig P, Todd T, Sastry P A, Lee K K, Hodges R S, Paranchych W, Irvin R T. Role of pili in adhesion of Pseudomonas aeruginosa to human respiratory epithelial cells. Infect Immun. 1988;56:1641–1646. doi: 10.1128/iai.56.6.1641-1646.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doig P, Sastry P A, Hodges R S, Lee K K, Paranchych W, Irvin R T. Inhibition of pilus-mediated adhesion of Pseudomonas aeruginosa to human buccal epithelial cells by monoclonal antibodies directed against pili. Infect Immun. 1990;58:124–130. doi: 10.1128/iai.58.1.124-130.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drake D, Montie T C. Flagella, motility, and invasive virulence of Pseudomonas aeruginosa. J Gen Microbiol. 1988;134:43–52. doi: 10.1099/00221287-134-1-43. [DOI] [PubMed] [Google Scholar]
- 15.Frank D. The exoenzyme S regulon of Pseudomonas aeruginosa. Mol Microbiol. 1997;26:621–629. doi: 10.1046/j.1365-2958.1997.6251991.x. [DOI] [PubMed] [Google Scholar]
- 16.Garrett E S, Perlegas D, Wozniak D J. Negative control of flagellum synthesis in Pseudomonas aeruginosa is modulated by the alternative sigma factor AlgT (AlgU) J Bacteriol. 1999;181:7401–7404. doi: 10.1128/jb.181.23.7401-7404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grundy F J, Waters D A, Takova T Y, Henkin T M. Identification of genes involved in utilization of acetate and acetoin in Bacillus subtilis. Mol Microbiol. 1993;10:259–271. doi: 10.1111/j.1365-2958.1993.tb01952.x. [DOI] [PubMed] [Google Scholar]
- 18.Hancock R E W, Mutharia L M, Chan L, Darveau R P, Speert D P, Pier G B. Pseudomonas aeruginosa isolates from patients with cystic fibrosis: a class of serum sensitive, nontypable strains deficient in lipopolysaccharide O side chains. Infect Immun. 1983;42:170–177. doi: 10.1128/iai.42.1.170-177.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harshey R M, Matsuyama T. Dimorphic transition in Escherichia coli and Salmonella typhimurium: surface-induced differentiation into hyperflagellate swarmer cells. Proc Natl Acad Sci USA. 1994;91:8631–8635. doi: 10.1073/pnas.91.18.8631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hauser A R, Engel J E. Pseudomonas aeruginosa induces type-III-secretion-mediated apoptosis of macrophages and epithelial cells. Infect Immun. 1999;67:5530–5537. doi: 10.1128/iai.67.10.5530-5537.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hueck C J, Kraus A, Hillen W. Sequences of ccpA and two downstream Bacillus megaterium genes with homology to the motAB operon from Bacillus subtilis. Gene. 1994;143:147–148. doi: 10.1016/0378-1119(94)90621-1. [DOI] [PubMed] [Google Scholar]
- 22.Hueck C J, Kraus A, Schmiedel D, Hillen W. Cloning, expression and functional analyses of the catabolite control protein CcpA from Bacillus megaterium. Mol Microbiol. 1995;16:855–864. doi: 10.1111/j.1365-2958.1995.tb02313.x. [DOI] [PubMed] [Google Scholar]
- 23.Ishimoto K S, Lory S. Identification of pilR, which encodes a transcriptional activator of the Pseudomonas aeruginosa pilin gene. J Bacteriol. 1992;174:3514–3521. doi: 10.1128/jb.174.11.3514-3521.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kato J, Nakamura T, Kuroda A, Ohtake H. Cloning and characterization of chemotaxis genes in Pseudomonas aeruginosa. Biosci Biotechnol Biochem. 1999;63:155–161. doi: 10.1271/bbb.63.155. [DOI] [PubMed] [Google Scholar]
- 25.Kelly N M, Kluftinger J, Pasloske B L, Paranchych W, Hancock R E W. Pseudomonas aeruginosa pili as ligands for nonopsonic phagocytosis by fibronectin-stimulated macrophages. Infect Immun. 1989;57:3841–3845. doi: 10.1128/iai.57.12.3841-3845.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kinscherf T G, Willis D K. Swarming by Pseudomonas syringae B728a requires gacS (lemA) and gacA but not the acyl-homoserine lactone biosynthetic gene ahlI. J Bacteriol. 1999;181:4133–4136. doi: 10.1128/jb.181.13.4133-4136.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 28.Luzar M A, Thomassen M J, Montie T C. Flagella and motility alterations in Pseudomonas aeruginosa strains from patients with cystic fibrosis: relationship to patient clinical condition. Infect Immun. 1985;50:577–582. doi: 10.1128/iai.50.2.577-582.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Macnab R M. Genetics and biogenesis of bacterial flagella. Annu Rev Genet. 1992;26:131–158. doi: 10.1146/annurev.ge.26.120192.001023. [DOI] [PubMed] [Google Scholar]
- 30.Mahenthiralingam E, Speert D P. Nonopsonic phagocytosis of Pseudomonas aeruginosa by macrophages and polymorphonuclear leukocytes requires the presence of the bacterial flagellum. Infect Immun. 1995;63:4519–4523. doi: 10.1128/iai.63.11.4519-4523.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mahenthiralingam E, Campbell M E, Speert D P. Nonmotility and phagocytic resistance of Pseudomonas aeruginosa isolates from chronically colonized patients with cystic fibrosis. Infect Immun. 1994;62:596–605. doi: 10.1128/iai.62.2.596-605.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mahenthiralingam E, Campbell M E, Foster J, Lam J S, Speert D P. Random amplified polymorphic DNA typing of Pseudomonas aeruginosa isolates recovered from patients with cystic fibrosis. J Clin Microbiol. 1996;34:1129–1135. doi: 10.1128/jcm.34.5.1129-1135.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mahenthiralingam E, Simpson D A, Speert D P. Identification and characterization of a novel DNA marker associated with epidemic Burkholderia cepacia strains recovered from patients with cystic fibrosis. J Clin Microbiol. 1997;35:808–816. doi: 10.1128/jcm.35.4.808-816.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mirel D B, Lustre V M, Chamberlin M J. An operon of Bacillus subtilis motility genes transcribed by the ςD form of RNA polymerase. J Bacteriol. 1992;174:4197–4204. doi: 10.1128/jb.174.13.4197-4204.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mork T, Hancock R E W. Mechanisms of nonopsonic phagocytosis of Pseudomonas aeruginosa. Infect Immun. 1993;61:3287–3293. doi: 10.1128/iai.61.8.3287-3293.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nunn D N, Lory S. Components of the protein-excretion apparatus of Pseudomonas aeruginosa are processed by the type IV prepilin peptidase. Proc Natl Acad Sci USA. 1992;89:47–51. doi: 10.1073/pnas.89.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nunn D N, Bergman S, Lory S. Product of the three accessory genes, pilB, pilC, and pilD, are required for biogenesis of Pseudomonas aeruginosa pili. J Bacteriol. 1990;172:2911–2919. doi: 10.1128/jb.172.6.2911-2919.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Toole G A, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol. 1998;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 39.Ottemann K M, Miller J F. Roles for motility in bacterial-host interactions. Mol Microbiol. 1997;24:1109–1117. doi: 10.1046/j.1365-2958.1997.4281787.x. [DOI] [PubMed] [Google Scholar]
- 40.Pratt L A, Kolter R. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol. 1998;30:285–293. doi: 10.1046/j.1365-2958.1998.01061.x. [DOI] [PubMed] [Google Scholar]
- 41.Ridgway H F, Silverman M, Simon M I. Localization of proteins controlling motility and chemotaxis in Escherichia coli. J Bacteriol. 1977;132:657–665. doi: 10.1128/jb.132.2.657-665.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ritchings B W, Almira E C, Lory S, Ramphal R. Cloning and phenotypic characterization of fleS and fleR, new response regulators of Pseudomonas aeruginosa which regulate motility and adhesion to mucin. Infect Immun. 1995;63:4868–4876. doi: 10.1128/iai.63.12.4868-4876.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saiman L, Ishimoto K, Lory S, Prince A. The effect of piliation and exoproduct expression on the adherence of Pseudomonas aeruginosa to respiratory epithelial monolayers. J Infect Dis. 1990;161:541–548. doi: 10.1093/infdis/161.3.541. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1989. [Google Scholar]
- 45.Sawa T, Yahr T L, Ohara M, Kurahashi K, Gropper M A, Wiener-Kronish J P, Frank D W. Active and passive immunization with the Pseudomonas V antigen protects against type III intoxication and lung injury. Nat Med. 1999;5:392–398. doi: 10.1038/7391. [DOI] [PubMed] [Google Scholar]
- 46.Schuster S C, Khan S. The bacterial flagellar motor. Annu Rev Biophys Biomol Struct. 1994;23:509–539. doi: 10.1146/annurev.bb.23.060194.002453. [DOI] [PubMed] [Google Scholar]
- 47.Schweizer H P. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene. 1991;97:109–121. doi: 10.1016/0378-1119(91)90016-5. [DOI] [PubMed] [Google Scholar]
- 48.Semmler A B, Whitchurch C B, Mattick J S. A re-examination of twitching motility in Pseudomonas aeruginosa. Microbiology. 1999;145:2863–2873. doi: 10.1099/00221287-145-10-2863. [DOI] [PubMed] [Google Scholar]
- 49.Simpson D A, Ramphal R, Lory S. Genetic analysis of Pseudomonas aeruginosa adherence: distinct genetic loci control attachment to epithelial cells and mucins. Infect Immun. 1992;60:3771–3779. doi: 10.1128/iai.60.9.3771-3779.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simpson D A, Ramphal R, Lory S. Characterization of Pseudomonas aeruginosa fliO, a gene involved in flagellar biosynthesis and adherence. Infect Immun. 1995;63:2950–2957. doi: 10.1128/iai.63.8.2950-2957.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith A W, Iglewski B H. Transformation of Pseudomonas aeruginosa by electroporation. Nucleic Acids Res. 1989;17:10509. doi: 10.1093/nar/17.24.10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Speert D P. Pseudomonas aeruginosa infections in patients with cystic fibrosis. In: Baltch A L, Smith R P, editors. Pseudomonas aeruginosa infections and treatment. New York, N.Y: Marcel Dekker, Inc.; 1994. pp. 183–235. [Google Scholar]
- 53.Speert D P, Gordon S. Phagocytosis of unopsonized Pseudomonas aeruginosa by murine macrophages is a two-step process requiring glucose. J Clin Investig. 1992;90:1085–1092. doi: 10.1172/JCI115924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Speert D P, Loh B A, Cabral D A, Salit I E. Nonopsonic phagocytosis of nonmucoid Pseudomonas aeruginosa by human neutrophils and monocyte-derived macrophages is correlated with bacterial piliation and hydrophobicity. Infect Immun. 1986;53:207–212. doi: 10.1128/iai.53.1.207-212.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Starnbach M N, Lory S. The fliA (rpoF) gene of Pseudomonas aeruginosa encodes an alternative sigma factor required for flagellin synthesis. Mol Microbiol. 1992;6:459–469. doi: 10.1111/j.1365-2958.1992.tb01490.x. [DOI] [PubMed] [Google Scholar]
- 56.Stolz B, Berg H C. Evidence for interactions between MotA and MotB, torque-generating elements of the flagellar motor of Escherichia coli. J Bacteriol. 1991;173:7033–7037. doi: 10.1128/jb.173.21.7033-7037.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Strom M S, Lory S. Cloning and expression of the pilin gene of Pseudomonas aeruginosa PAK in Escherichia coli. J Bacteriol. 1986;165:367–372. doi: 10.1128/jb.165.2.367-372.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tang H, Braun T F, Blair D F. Motility protein complexes in the bacterial flagellar motor. J Mol Biol. 1996;261:209–221. doi: 10.1006/jmbi.1996.0453. [DOI] [PubMed] [Google Scholar]
- 59.Tosi M F, Zakem H, Berger M. Neutrophil elastase cleaves C3Bi on opsonized Pseudomonas as well as CR1 on neutrophils to create a functionally important opsonin receptor mismatch. J Clin Investig. 1988;86:300–308. doi: 10.1172/JCI114699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Totten P A, Lory S. Characterization of the type a flagellin gene from Pseudomonas aeruginosa PAK. J Bacteriol. 1990;172:7188–7199. doi: 10.1128/jb.172.12.7188-7199.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Totten P A, Lara J C, Lory S. The rpoN gene product of Pseudomonas aeruginosa is required for expression of diverse genes, including the flagellin gene. J Bacteriol. 1990;172:389–396. doi: 10.1128/jb.172.1.389-396.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Woods D E, Straus D C, Johanson W J, Berry V K, Bass J A. Role of pili in adherence of Pseudomonas aeruginosa to mammalian buccal epithelial cells. Infect Immun. 1980;29:1146–1151. doi: 10.1128/iai.29.3.1146-1151.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yahr T L, Barbieri J T, Frank D W. Exoenzyme S of Pseudomonas aeruginosa is secreted by a type III pathway. Mol Microbiol. 1996;22:991–1003. doi: 10.1046/j.1365-2958.1996.01554.x. [DOI] [PubMed] [Google Scholar]