Abstract
The ability of Escherichia coli to survive stress during growth in different environments is, in large part, dependent on rpoS and the genes that comprise the rpoS regulon. E. coli BJ4 and an isogenic BJ4 rpoS mutant were used to examine the influence of the rpoS gene on E. coli colonization of the streptomycin-treated mouse large intestine. Colonization experiments in which the wild-type E. coli BJ4 and its rpoS mutant were fed individually as well as simultaneously to mice suggested that E. coli BJ4 does not face prolonged periods of nutrient starvation in the mouse large intestine and that the rpoS regulon is not expressed during long-term colonization after adaptation of the bacteria to the gut environment.
It has recently been shown that leuX, which encodes tRNA5Leu (specific for the codon UUG) and which is a major tRNA at low growth rates but a minor tRNA at rapid growth rates (4), is not required for short-term growth in the streptomycin-treated mouse large intestine. However, leuX is required for long-term Escherichia coli intestinal colonization, i.e., for persistence in high numbers in the intestine for at least 2 weeks (28) and for survival of E. coli in stationary phase when it is grown in mouse intestinal mucus in vitro (4). These results suggested that the state of E. coli during long-term colonization of the mouse large intestine is akin to that found under conditions of slow growth and stationary phase in vitro. However, it has also been recently shown, using in situ hybridization with rRNA probes, that E. coli grows rapidly in mouse large intestine mucus in vivo, with doubling times ranging from 30 to 80 min for individual cells (33). Since E. coli colonizes the mouse intestine by growing in intestinal mucus (42), these results argue that the state of E. coli in the mouse large intestine during long-term colonization is similar to that found during the exponential phase in a rapidly growing culture. Collectively, these results suggest that long-term E. coli colonization of the mouse large intestine requires rapid growth but also requires that the bacterial cells express some genes that are normally associated with slow growth rates and stationary phase.
The rpoS gene is essential for the expression of a variety of stationary-phase-induced genes as well as for the expression of stationary-phase-specific stress resistance phenotypes, such as increased resistance to high temperature, high concentrations of H2O2, and high osmotic challenge (12, 13). This gene has been known as nur, katF, appR, csi-2, and abrD, a result of independent studies of different phenotypes. The rpoS sequence exhibits extensive homology to rpoD, which encodes ς70, the sigma subunit of RNA polymerase in E. coli (10). The rpoS gene encodes the sigma factor ςs. ςs is a protein of 41.5 kDa that controls a regulon of at least 30 genes which are expressed during starvation and at the transition into stationary phase (20, 24).
rpoS mutants survive poorly, compared with the wild type, under carbon and nitrogen starvation (21, 25). Furthermore, rpoS mutants fail to develop starvation-mediated cross protection to osmotic, oxidative, and heat stresses (21). Stationary-phase-induced morphological changes in E. coli are dependent on rpoS, and rpoS mutant cells appear slightly elongated and remain rod shaped in stationary phase (16).
It has been suggested that E. coli in the intestine is frequently exposed to low, growth-limiting concentrations of nutrients (29, 44). In fact, it has recently been shown that E. coli utilizes gluconate as a carbon source for growth in the mouse large intestine (39) and that gluconate is present in mouse large intestine mucus at a concentration of only about 140 μg/ml (30). Since growth in the presence of limiting nutrients in the intestine might require expression of the rpoS regulon, the aim of the present investigation was to examine whether rpoS is essential for E. coli to colonize the mouse large intestine. The investigation was performed with an E. coli wild-type strain and an isogenic rpoS mutant. The streptomycin-treated mouse model was chosen because this model can be used to simulate competitive interactions between E. coli strains and gram-positive obligate anaerobes in the natural environment of the intestine (15). Our data suggest that rpoS does not play a role in E. coli long-term colonization of the mouse large intestine, and thus that E. coli is not exposed to prolonged periods of starvation or other stress influences during growth in the intestine.
MATERIALS AND METHODS
Bacterial strains and plasmids.
E. coli BJ4, a streptomycin-resistant (Strr) rat isolate, has been previously described in detail (15). Throughout this study, a rifampin-resistant (Rifr) isolate of E. coli BJ4 Strr has been used. E. coli BJ4 Strr Rifr was made Rifr by P1 transduction from strain TC2 (kindly provided by Tove Atlung, Roskilde University Centre, Roskilde, Denmark). The rifampin-resistant isolate was found to be identical to its parent with respect to 35 biochemical reactions, ribotype, serotype, and ability to colonize the mouse gut. Strain MC4100 rpoS::Tn10 (16) was used for creating an E. coli BJ4 rpoS mutant by bacteriophage P1 transduction. E. coli BJ4 rpoS is resistant to both streptomycin and tetracycline. Bacteriophage P1v was used to ensure that the transductants were not lysogenized. Plasmid pRH320 (pBR322 harboring the wild-type rpoS gene) was kindly provided by R. Arronis (17). Bacteria were always grown in the presence of relevant antibiotics on either LB-agar plates or minimal medium (ABT supplemented with 0.2% glucose [3]) containing the relevant antibiotics, i.e., 10 μg of tetracycline hydrochloride (Tet) per ml, 50 μg of rifampin (Rif) per ml, 50 μg of ampicillin (Amp) per ml, and 100 μg of streptomycin sulfate (Str) per ml. All antibiotics were bought from Sigma.
Measurements of bacterial growth in minimal medium.
The growth rates of pure cultures under conditions of balanced growth were measured by monitoring the optical density at 450 nm (OD450).
Qualitative catalase activity.
Catalase activity was determined by adding one drop of hydrogen peroxide (30%) to bacterial colonies on an agar plate. The catalase activity was evaluated visually by the formation of bubbles.
Carbon starvation.
Wild-type E. coli BJ4 Strr Rifr, E. coli BJ4 rpoS Strr Tetr, and E. coli BJ4 rpoS(pRH320) Strr Tetr Ampr were grown under conditions of balanced growth in minimal medium containing glucose to an OD450 of 0.5. Each culture was centrifuged for 5 min at 3,000 rpm, and each pellet was resuspended in a flask containing warm minimal medium without glucose. Cultures were incubated at 37°C and aerated in a rotary shaking water bath (160 rpm). At various times, samples were taken from each culture. Samples were diluted and plated on LB agar plates containing streptomycin sulfate (100 μg/ml) and rifampin (50 μg/ml), on LB agar plates containing streptomycin sulfate (100 μg/ml) and tetracycline hydrochloride (10 μg/ml), and on LB agar plates containing streptomycin sulfate (100 μg/ml) and ampicillin (100 μg/ml) to determine viable counts for E. coli BJ4 Strr Rifr, E. coli BJ4 rpoS Strr Tetr, and E. coli BJ4 rpoS(pRH320) Strr Tetr Ampr, respectively. Plates were incubated at 37°C for 18 to 24 h before the colonies were counted.
Competitive growth and survival of E. coli BJ4, E. coli BJ4 rpoS, and E. coli BJ4 rpoS(pRH320) in minimal medium.
A flask containing warmed minimal medium was inoculated with about 102 CFU each of overnight cultures of E. coli BJ4 Strr Rifr and E. coli BJ4 rpoS Strr Tetr per ml. The culture was incubated at 37°C, and at various times over a period of 20 days, samples were diluted and plated on LB agar plates containing streptomycin sulfate (100 μg/ml) and rifampin (50 μg/ml) and on LB agar plates containing streptomycin sulfate (100 μg/ml) and tetracycline hydrochloride (10 μg/ml). A second flask containing warmed minimal medium was inoculated with about 102 CFU each of overnight cultures of E. coli BJ4 Strr Rifr and E. coli BJ4 rpoS(pRH320) Strr Tetr Ampr per ml. The cultures were incubated at 37°C, and at various times over a period of 20 days, samples were diluted and plated on LB agar plates containing streptomycin sulfate (100 μg/ml) and rifampin (50 μg/ml) and on LB agar plates containing streptomycin sulfate (100 μg/ml) and ampicillin (100 μg/ml). Plates were incubated at 37°C for 18 to 24 h before the colonies were counted.
Heat challenge.
E. coli BJ4 Strr Rifr, E. coli BJ4 rpoS Strr Tetr, and E. coli BJ4 rpoS(pRH320) Strr Tetr Ampr cultures grown overnight at 37°C were diluted into warmed minimal medium (57°C) without glucose to a concentration of approximately 104 cells per ml and incubated in a waterbath at 57°C. Samples from each culture were taken after 0, 2, 4, 8, 12, 18, and 30 min and plated on LB-agar plates containing streptomycin sulfate (100 μg/ml). Plates were incubated at 37°C for 18 to 24 h before the colonies were counted.
Hydrogen peroxide challenge.
Hydrogen peroxide was added to E. coli BJ4 Strr Rifr, E. coli BJ4 rpoS Strr Tetr, and E. coli BJ4 rpoS(pRH320) Strr Tetr Ampr cultures grown overnight (37°C) to a final concentration of 15 mM. The cultures were then incubated with shaking (160 rpm) at 37°C. Samples were taken at 0, 2, 5, 10, 18, 30, and 60 min and plated on LB-agar plates containing streptomycin sulfate (100 μg/ml). Plates were incubated at 37°C for 18 to 24 h before the colonies were counted.
Colonization experiments.
Six- to 8-week-old, outbred albino female mice (Ssc:CF1; Statens Serum Institut, Copenhagen, Denmark) were used for colonization experiments. The colonization experiments were performed as described previously (26). Briefly, sets of 3 mice were given sterile water containing 5 g of streptomycin sulfate per liter and fed continuously. After 24 h, 100 μl of 20% sucrose, containing either 109, 105, or 104 CFU of E. coli strains, was given to the mice orally by pipette. The strains were given either separately, for testing individual colonization abilities, or together in pairs, for determining competitive colonizing abilities. The mice were continuously given sterile water containing streptomycin during the experiment. At the indicated times, 0.5 g of feces no older than 24 h was collected, homogenized, diluted, and plated on LB-agar plates containing streptomycin sulfate (100 μg/ml) and rifampin (50 μg/ml), on LB-agar plates containing streptomycin sulfate (100 μg/ml) and tetracycline hydrochloride (10 μg/ml), and on LB-agar plates containing streptomycin sulfate (100 μg/ml) and ampicillin (100 μg/ml) to determine viable counts of E. coli BJ4 Strr Rifr, E. coli BJ4 rpoS Strr Tetr, and E. coli BJ4 rpoS(pRH320) Strr Tetr Ampr, respectively. Plates were incubated at 37°C for 18 to 24 h before the colonies were counted. During the colonization experiments, the mice were individually caged. A colonization experiment typically lasted 3 weeks, and the mice were transferred to fresh, sterile cages daily. All experiments were repeated at least twice.
RESULTS
Growth rates in minimal medium.
When cultures of E. coli BJ4 Strr, BJ4 Strr Rifr, BJ4 rpoS Strr Tetr, and BJ4 rpoS(pRH320) Strr Tetr Ampr were grown in minimal medium containing glucose as the carbon source, no differences in doubling times were discernible, i.e., in each case a doubling time of about 50 min was observed (data not shown).
Effect of rpoS mutation on in vitro carbon starvation survival.
To determine the ability of individual strains to survive during carbon starvation, the wild-type BJ4, the BJ4 rpoS mutant, and the rpoS mutant complemented with pRH320, which contains the wild-type rpoS gene, were incubated in minimal medium without glucose, and their individual abilities to survive were determined (see Materials and Methods). The BJ4 rpoS mutant had a reduced ability to survive carbon starvation relative to the wild-type BJ4 and the complemented BJ4 rpoS mutant. For example, after 5 days of carbon starvation, only about 11% of the original BJ4 rpoS mutant inoculum remained viable, whereas about 36% of the BJ4 wild-type inoculum and of the BJ4 rpoS(pRH320) inoculum survived (data not shown).
Competitive growth and survival of E. coli BJ4, E. coli BJ4 rpoS, and E. coli BJ4 rpoS(pRH320) in minimal medium.
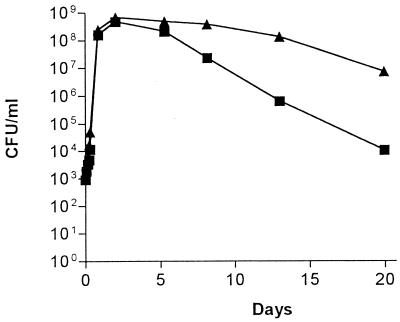
An experiment was performed to examine the ability of wild-type E. coli BJ4 and E. coli BJ4 rpoS to grow and survive in minimal medium (0.2% glucose) in direct competition after each strain was inoculated into the same culture at an initial concentration of 102 CFU/ml. The experiment was carried out for 20 days. The growth rates of the wild-type BJ4 and the BJ4 rpoS mutant were very similar during the exponential growth phase (Fig. 1). At the time of transition into stationary phase, both strains had grown to a level of approximately 5 × 108 cells/ml (Fig. 1). During extended carbon starvation, wild-type BJ4 showed a greater vitality than its rpoS mutant, such that by day 20, the wild-type BJ4 strain was present in numbers 100-fold higher than the BJ4 rpoS strain (Fig. 1). Essentially identical results were observed when E. coli BJ4 rpoS(pRH320) competed against E. coli BJ4 rpoS, i.e. by day 20 the BJ4 rpoS(pRH320) strain was present in numbers 100-fold higher than the BJ4 rpoS strain (not shown).
FIG. 1.
Competitive growth and survival in minimal medium. E. coli BJ4 (▴) and E. coli BJ4 rpoS (■) were inoculated into minimal medium (0.2% glucose) and incubated with shaking (160 rpm) at 37°C for 20 days. Viable counts (CFU per milliliter) were determined at different time points by plating on selective medium.
Sensitivity of E. coli BJ4 strains to heat and oxidative stress.
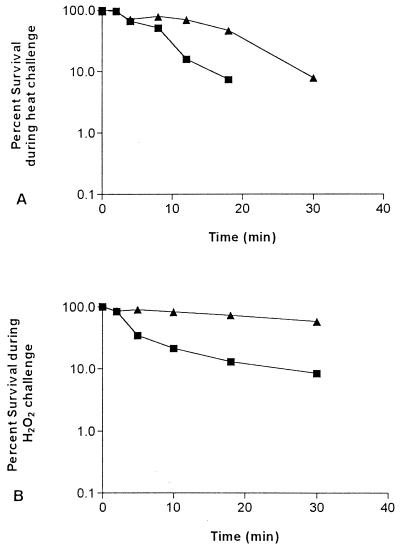
The E. coli BJ4 wild-type strain and its rpoS mutant were tested for their abilities to resist heat and oxidative stress. The BJ4 rpoS mutant exhibited a greater sensitivity than the wild type to heat challenge. After 18 min at 57°C, only 7.5% of the BJ4 rpoS mutant cells remained viable, whereas 47% of the wild-type BJ4 cells survived (Fig. 2A). Furthermore, the BJ4 rpoS mutant exhibited a greater sensitivity than the wild type to H2O2 stress. For example, after 60 min of exposure to H2O2, only 0.7% of the BJ4 rpoS mutant cells remained viable, whereas 23% of the wild-type BJ4 cells survived (Fig. 2B). The response of E. coli BJ4 rpoS(pRH320) was essentially identical to that of wild-type BJ4 (not shown).
FIG. 2.
Sensitivity to stress challenges. E. coli BJ4 (▴) and E. coli BJ4 rpoS (■) were tested for their ability to resist (A) heat (57°C) and (B) oxidative stress in the presence of 15 mM H2O2.
Individual mouse large intestine colonizing abilities.
The experiments to this point made it clear that the E. coli BJ4 rpoS mutant carried a mutation in the rpoS gene, confirmed by its response to carbon starvation, heat stress, and oxidative stress. Furthermore, the wild-type responses to carbon starvation, heat stress, and oxidative stress were reestablished in the BJ4 rpoS mutant by complementation with the wild-type rpoS gene carried on pRH230.
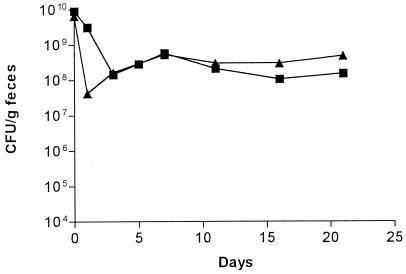
To examine the ability of the individual strains to colonize the mouse large intestine, E. coli BJ4 and E. coli BJ4 rpoS were fed separately to streptomycin-treated mice (109 CFU per mouse). Both E. coli BJ4 and its rpoS mutant colonized the large intestines of the streptomycin-treated mice at levels of about 3 × 108 CFU/g of feces (Fig. 3). One colony isolated from the day 20 postfeeding feces sample of each mouse fed wild-type BJ4 and one colony isolated from the day 20 postfeeding feces sample of each mouse fed the BJ4 rpoS mutant were tested for catalase production and for sensitivity to heat and oxidative stress. The colonies isolated from the mice fed the BJ4 rpoS mutant had a catalase-negative phenotype and retained greater sensitivity than the wild type to heat challenge and oxidative stress in vitro (not shown). The colonies isolated from the mice fed wild-type BJ4 retained the wild-type catalase activity and the wild-type sensitivities to heat and oxidative stress (not shown). Therefore, the rpoS gene is not required for long-term colonization of the streptomycin-treated mouse large intestine by E. coli BJ4.
FIG. 3.
Individual mouse large intestine colonizing abilities. One set of three mice was fed 1010 CFU of the E. coli BJ4 wild-type strain (▴), and another set of three mice was fed 1010 CFU of E. coli BJ4 rpoS (■). At the indicated times, fecal samples were plated on selective medium.
Competitive mouse large intestine colonizing abilities of E. coli BJ4 and E. coli BJ4 rpoS.
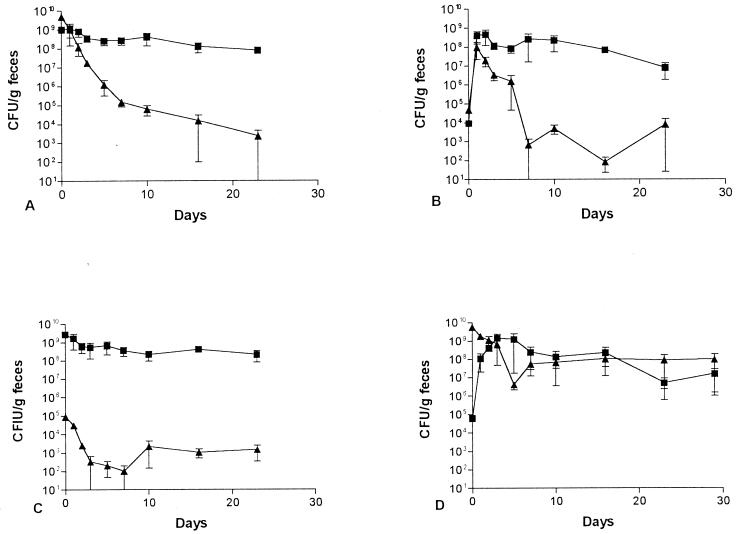
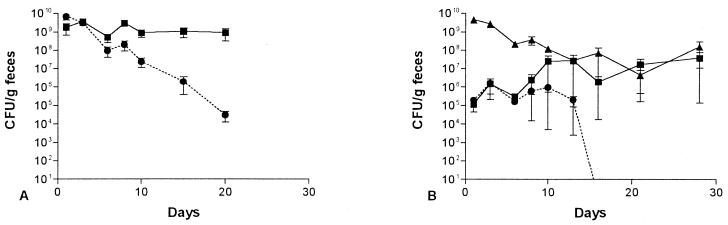
The ability of the E. coli BJ4 rpoS mutant to colonize the mouse intestine in competition with wild-type BJ4 was examined. Surprisingly, when each strain was fed to the same mice at 109 CFU per mouse, the wild-type BJ4 strain was initially rapidly eliminated from the intestine (Fig. 4A). Several days later, wild-type BJ4 stabilized at a level of about 104 CFU/g of feces, i.e., it was not completely eliminated (Fig. 4A). In contrast, the BJ4 rpoS mutant colonized at a level of about 108 CFU/g of feces (Fig. 4A). One colony of the wild-type BJ4 and one colony of the BJ4 rpoS mutant isolated from the feces of each mouse on the last day of the experiment were tested for catalase and heat and oxidative stress. The wild-type BJ4 colonies acted like wild-type BJ4 and the BJ4 rpoS mutant colonies acted like rpoS mutants.
FIG. 4.
Competitive mouse large intestine colonizing abilities. E. coli BJ4 (▴) and E. coli BJ4 rpoS (■) were fed simultaneously to four sets of three mice as follows: (A) both at 109 CFU; (B) both at 104 CFU; (C) BJ4 at 105 CFU and BJ4 rpoS at 109 CFU; (D) BJ4 at 109 CFU and BJ4 rpoS at 105 CFU. Error bars represent the standard errors of the geometric means.
When mice were fed 104 CFU each of wild-type BJ4 and the BJ4 rpoS mutant, both strains grew rapidly to about 108 CFU/g of feces (Fig. 4B). Thereafter, however, the wild-type BJ4 strain was eliminated rapidly and colonized at a level of only 104 CFU/g of feces, whereas the rpoS mutant remained at a level of about 108 CFU/g of feces for the duration of the experiment (Fig. 4B). One colony of the wild-type BJ4 and one colony of the BJ4 rpoS mutant isolated from the feces of each mouse on the last day of the experiment were tested for catalase and heat and oxidative stress. The wild-type BJ4 colonies acted like the wild type and the BJ4 rpoS mutant colonies acted like rpoS mutants.
When mice were fed 109 CFU of E. coli BJ4 rpoS and 105 CFU of wild-type BJ4, the BJ4 rpoS mutant colonized at high levels, i.e., 109 CFU/g of feces, whereas the wild-type BJ4 colonized the mouse large intestine at levels of only 102 to 103 CFU/g of feces (Fig. 4C). However, when mice were fed 109 CFU of wild-type BJ4 and 104 CFU of the BJ4 rpoS mutant, the BJ4 rpoS mutant grew to the level of the wild-type BJ4 within a few days and then both strains cocolonized the large intestine at levels of about 108 CFU/g of feces for the duration of the experiment (Fig. 4D). One colony of the wild-type BJ4 and one colony of the BJ4 rpoS mutant isolated from the feces of each mouse on the last day of the experiment were tested for catalase and heat and oxidative stress. The wild-type BJ4 colonies acted like the wild type and the BJ4 rpoS mutant colonies acted like rpoS mutants. It therefore appears that if given enough time to adapt to the intestine in the absence of high levels of the BJ4 rpoS mutant, wild-type BJ4 can subsequently compete well in the presence of high levels of the BJ4 rpoS mutant.
Competitive mouse large intestine colonizing abilities of E. coli BJ4 and E. coli BJ4 rpoS(pRH320).
To this point, the colonization experiments suggested that the BJ4 rpoS mutant is initially a better colonizer of the mouse large intestine than the wild-type BJ4. Therefore, experiments were performed to determine whether complementing the BJ4 rpoS mutant with the wild-type rpoS gene in pRH320 would eliminate its advantage. Plasmid pRH320 consists of the rpoS gene in pBR322. It has been shown that pBR322 as such has no effect on the colonization ability of E. coli (2) and that some plasmids are known to segregate from E. coli in the mouse intestine (2). The degree to which pRH320 segregates in the mouse intestine was determined by feeding streptomycin-treated mice 109 CFU of E. coli BJ4 rpoS(pRH320) (Fig. 5A). For 7 days postfeeding, the counts of viable bacteria in feces samples cultivated on plates containing streptomycin and tetracycline (109 BJ4 rpoS) were essentially identical to the counts on plates containing ampicillin [5 × 108 BJ4 rpoS(pRH320)]. Thereafter, pRH320 segregated from BJ4 rpoS(pRH320) in the intestine so that by day 15 postfeeding, only 5 in 104 fecal CFU retained the plasmid (Fig. 5A). However, since pRH320 was stable in BJ4 rpoS in the mouse intestine for at least 1 week, we were able to determine the relative colonizing abilities of wild-type BJ4 and BJ4 rpoS(pRH320).
FIG. 5.
Colonizing abilities of the complemented mutant BJ4 rpoS(pRH320). (A) Mice were fed 109 CFU of E. coli BJ4 rpoS(pRH320). Symbols: ●, E. coli BJ4 rpoS(pRH320); ■, E. coli BJ4 rpoS segregants, i.e., without the plasmid. (B) Mice were fed simultaneously with E. coli BJ4 (▴) at 109 CFU and E. coli BJ4 rpoS(pRH320) at 105 CFU (●). ■, E. coli BJ4 rpoS segregants, i.e., without the plasmid. Fecal samples were plated on selective medium on the indicated days. Error bars represent the standard errors of the geometric means.
Streptomycin-treated mice were simultaneously fed 109 CFU of the wild-type BJ4 and 109 CFU of rpoS(pRH320). Wild-type BJ4 and BJ4 rpoS(pRH320) cocolonized at levels ranging between 108 and 109 CFU/g of feces for 7 days, i.e., wild-type BJ4 was not eliminated (not shown). Thereafter, pRH320 rapidly segregated from BJ4 rpoS(pRH320) (data not shown). Therefore, complementing BJ4 rpoS with the wild-type rpoS gene prevented the initial elimination of the wild-type BJ4. In a second experiment, streptomycin-treated mice were simultaneously fed 109 CFU of BJ4 and 105 CFU of BJ4 rpoS(pRH320) (Fig. 5B). For the duration of the experiment, wild-type BJ4 colonized at a level of between 108 and 109 CFU/g of feces. The complemented mutant BJ4 rpoS(pRH320) colonized at a level of between 105 and 106 CFU/g of feces for 13 days, i.e., complementing BJ4 rpoS with the wild-type rpoS gene prevented it from growing in the intestine in the presence of high numbers of the wild-type BJ4 strain. However, beginning at about 6 days postfeeding, BJ4 rpoS cells free of pRH320 appeared, and these cells grew rapidly to the level of BJ4 over the next 4 days and cocolonized at the level of BJ4 (about 108 CFU/g of feces) thereafter. By day 15 postfeeding, BJ4 rpoS(pRH320) was undetectable, presumably due to segregation of the plasmid (Fig. 5B).
DISCUSSION
The large intestine of the mouse consists of the cecum and the colon, both of which contain the mucosa and the luminal contents. Two components of the mucosa are the layer of epithelial cells on the intestinal wall and the mucus layer covering the epithelial cells. The relatively thick (up to 400 μm) mucus layer consists of mucin, a 2-MDa gel-forming glycoprotein, and a large number of smaller glycoproteins, proteins, glycolipids, and lipids (1, 7, 14, 31, 34, 37, 41). Presumably, shed epithelial cells are a source of many of the smaller mucus components (31, 34). The mucus layer itself is in a dynamic state, constantly being synthesized and secreted by specialized goblet cells and, to a large extent, degraded by the indigenous intestinal microbes (11, 27). Degraded mucus components are shed into the intestinal lumen, forming a part of the luminal contents, and are excreted in feces (11).
The prevalent theory as to how bacteria colonize the mammalian gut is that all species can coexist as long as each member of the flora is able to utilize one or a few limited nutrients better than all the others and that its growth rate during the colonization process is at least equal to the washout rate from the intestine (9). The growth rate of a particular bacterium in the intestine is determined by the nature of the limited nutrient that it utilizes, and the density to which it grows is determined by the available concentration of that nutrient. It is also possible for a species that does not compete well for limited nutrients to avoid washout and colonize if it is able to adhere to the intestinal wall (8).
In recent studies, the location of E. coli BJ4 in the streptomycin-treated mouse large intestine has been examined using in situ hybridization with species-specific rRNA probes (32). E. coli BJ4 was found dispersed in the intestinal mucus and the luminal contents but was not found associated with epithelial cells (15, 32). Moreover, we have shown that oral streptomycin treatment does not affect the essentially constant growth rate of E. coli in the large intestine of germ-free, conventional, and streptomycin-treated mice (36). Finally, a large body of experimental evidence strongly suggests that E. coli grows rapidly in intestinal mucus both in vitro and in vivo but does not grow or grows poorly in luminal contents (19, 33, 42). It is therefore very likely that the ability of an E. coli strain to continuously grow rapidly in intestinal mucus (because of mucus turnover) using nutrients that are present there plays a critical role in its ability to colonize the intestine. One other facet in the ability of enteric bacteria to colonize deserves mention. It has been shown that the ability of E. coli and Salmonella typhimurium to colonize the streptomycin-treated mouse large intestine is dependent on their ability to rapidly penetrate deeply into the mucus layer (18, 22, 23).
In the present investigation, we examined the influence of the rpoS gene on the ability of E. coli BJ4 to grow in and colonize the streptomycin-treated mouse large intestine. Both the wild-type BJ4 strain and BJ4 rpoS colonized equally well at levels of about 108 CFU/g of feces when fed individually to mice (Fig. 3). Since the BJ4 rpoS mutant was significantly more sensitive to carbon starvation in vitro than both wild-type BJ4 and the BJ4 rpoS mutant complemented with the wild-type rpoS gene, the colonization experiments suggest that E. coli BJ4 is not starved for carbon for prolonged periods in the streptomycin-treated mouse large intestine, despite the presence of an essentially normal gram-positive anaerobic population consisting of a myriad of species, each competing for limited nutrients (8).
Surprisingly, in two different types of colonization experiments involving competition between the strains, the BJ4 rpoS mutant was found to be the better colonizer. In the first instance, mice were fed 109 CFU of overnight cultures of the wild-type BJ4 and the BJ4 rpoS mutant. For the first several days, the rpoS mutant outcompeted the wild-type BJ4 strain (Fig. 4A). Thereafter, each strain remained at a relatively stable level, i.e., the wild-type BJ4 strain stabilized at a level 4 orders of magnitude lower than the BJ4 rpoS mutant (Fig. 4A). The data therefore suggest that in the initial stages of the colonization process, the ability to express the genes of the rpoS regulon is a disadvantage. In support of this view, the advantage of the BJ4 rpoS mutant was eliminated when it was complemented with the wild-type rpoS gene.
In the second instance, mice were fed 104 CFU each of the wild-type BJ4 and the BJ4 rpoS mutant. Neither strain had an advantage during an initial stage of extremely rapid growth, i.e., under conditions in which genes of the rpoS regulon are minimally expressed (16). However, thereafter the BJ4 rpoS mutant eliminated wild-type BJ4 until both strains once again reached relatively stable population levels, the wild-type BJ4 again stabilizing at a level about 4 orders of magnitude lower than the rpoS mutant (Fig. 4B). These data suggest that after the initial rapid rate of growth in mucus, wild-type BJ4 entered stationary phase, maximally expressed the genes of the rpoS regulon (16), and reduced its rate of growth relative to the rpoS mutant. This scenario explains why the BJ4 rpoS mutant eliminated wild-type BJ4 (Fig. 4B). Alternatively, it is possible that the BJ4 rpoS mutant was better able to penetrate the mucus layer than its parent and thereby eliminated its parent from the intestine.
As stated above, the prevailing evidence suggests that E. coli grows both continuously and rapidly (30 to 80 min for individual cells [33]) as it colonizes the intestine to avoid washout as mucus is sloughed. Remaining in stationary phase at a slow growth rate as described above would prove disastrous for E. coli BJ4 in the intestine. It would appear more reasonable that for long-term colonization, BJ4 maintains a rapid growth rate, a condition in which the rpoS regulon is minimally expressed (16). In support of this view, when the wild-type BJ4 was fed to mice in high numbers (109 CFU per mouse) and the BJ4 rpoS mutant was fed in low numbers (104 CFU per mouse), after the BJ4 rpoS mutant grew to the level of the wild-type BJ4, both strains colonized at about 108 CFU/g of feces (Fig. 4D). In other words, it is possible that after a few days in the intestine, the wild-type BJ4 resumed a higher rate of growth, reduced its expression of genes of the rpoS regulon, and was therefore able to cocolonize with the BJ4 rpoS mutant.
It is of great interest that in the large intestine, small numbers of the BJ4 rpoS mutant grew in the presence of large numbers of the wild-type BJ4 strain (Fig. 4D and 5B), which was not true of small numbers of wild-type BJ4 in the presence of large numbers of the BJ4 rpoS mutant (Fig. 4C). A recent report demonstrated that when small numbers of a nalidixic acid-resistant E. coli strain were fed to streptomycin-treated mice simultaneously with large numbers of its isogenic nalidixic acid-sensitive parent, the resistant strain did not grow in the intestine relative to its parent, but the colonization ratio of the two strains was the same as the input ratio, i.e., the amounts of each fed to the mice (39). In contrast, small numbers of an E. coli strain better able to transport gluconate than its otherwise isogenic parental strain were able to grow in the intestine in the presence of large numbers of its parent (39). In the present context, it may be that the BJ4 rpoS mutant can utilize a limiting nutrient in the intestine better than the wild-type BJ4 strain and therefore can grow in the presence of high numbers of the wild type. It is known that RpoS can negatively regulate the expression of some genes (6), and it may be that the small amount of RpoS that is present in exponentially growing cells (17) reduces the ability of the wild-type BJ4 to utilize a specific limiting nutrient relative to the BJ4 rpoS mutant. Furthermore, the reduction in expression of the rpoS regulon in vivo might be necessary in order to accommodate a chemostat-like type of growth in which mucus is constantly being synthesized and sloughed into feces. Experiments designed to test this hypothesis are in progress.
If the inability to express the rpoS regulon allows small numbers of BJ4 rpoS to grow in the presence of large numbers of BJ4 in the intestine, it would be expected that complementing the BJ4 rpoS mutant with the wild-type rpoS gene would eliminate its growth advantage. This is indeed what happened, i.e., when small numbers of BJ4 rpoS(pRH320) were fed to mice along with high numbers of the wild-type BJ4, the mutant initially remained at a constant low level in the intestine relative to the wild-type BJ4 strain (Fig. 5B). However, when BJ4 rpoS(pRH320) began to segregate the plasmid in the intestine, the BJ4 rpoS segregants grew rapidly to the level of the wild-type BJ4, and thereafter both strains cocolonized at that level (Fig. 5B), as expected if loss of the wild-type rpoS gene allowed it to grow more rapidly than the wild-type BJ4 strain.
A major problem encountered in studying colonization of the mammalian intestine is what is commonly referred to as colonization resistance, in which all intestinal niches are occupied in a balanced ecosystem and most ingested microorganisms fail to colonize due to lack of an available niche (40). Colonization resistance is, in fact, the reason why we use streptomycin, i.e., to clear the mouse intestine of facultative microorganisms and create an available niche for E. coli strains. Streptomycin treatment selectively reduces the facultative microflora, but the anaerobic population in the large intestine remains largely intact, and large numbers of different species coexist (36).
The present results suggest that the rpoS gene may be involved in colonization resistance. That is, if, during long-term colonization, E. coli cells express genes of the rpoS regulon minimally, then when small numbers of an invading strain enter the large intestine expressing the rpoS regulon maximally, either because they were exposed to conditions of poor nutrient availability in their natural environment or because of the acid environment of the stomach (38), the invading strain may not be able to compete with the colonizing strain and, if so, will be eliminated from the intestine.
It should be emphasized that we do not intend to imply that the rpoS gene is dispensable in animals. In fact, it has been shown that an S. typhimurium rpoS mutant is attenuated for virulence in mice (5, 38). However, it is possible that the rpoS gene is not required for S. typhimurium survival in the intestine but is critical for its survival intracellularly in the stressful environments of extraintestinal cells (e.g., M cells, macrophages, etc.).
The results presented here are in agreement with those in which low but equal numbers of a wild-type Vibrio cholerae strain and its isogenic rpoS mutant were administered by gavage into the intestines of infant mice (43). At the end of 20 h, the mice were sacrificed and equal numbers of both strains survived (43), showing that the defect in rpoS was not disadvantageous.
In summary, the data presented here are consistent with the hypothesis that E. coli BJ4 expresses the rpoS regulon during the initial stages of growth in the mucus layer as it enters stationary phase, but within a few days thereafter expression of the rpoS regulon is reduced.
ACKNOWLEDGMENTS
Leo Eberl, Technical University of Munich, Munich, Germany, is thanked for participating with stimulating suggestions and for constructing the E. coli BJ4 rpoS mutant.
REFERENCES
- 1.Allan A. Structure and function of gastrointestinal mucus. In: Johnson L R, editor. Physiology of the gastrointestinal tract. New York: Raven Press; 1981. pp. 637–639. [Google Scholar]
- 2.Burghoff R L, Laux D C, Cohen P S. Construction of stable cloning vectors that do not segregate from a human fecal Escherichia coli strain in the streptomycin-treated mouse large intestine. Infect Immun. 1990;58:1141–1145. doi: 10.1128/iai.58.5.1141-1145.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clark D J, Maaløe O. DNA replication and the division cycle in E. coli J. Mol Biol. 1967;23:99–112. [Google Scholar]
- 4.Dobrint U, Cohen P S, Utley M, Mühldorfer I, Hacker J. The leuX-encoded tRNA5Leu but not the pathogenicity islands I and II influence the survival of the uropathogenic Escherichia coli strain 53 in CD-1 mouse bladder mucus in the stationary phase. FEMS Microbiol Lett. 1998;162:135–141. doi: 10.1111/j.1574-6968.1998.tb12990.x. [DOI] [PubMed] [Google Scholar]
- 5.Fang F C, Libby S J, Budhmeier N A, Loewen P C, Switala J, Harwood J, Guiney D. The alternative sigma factor KatF (RpoS) regulates Salmonella virulence. Proc Natl Acad Sci USA. 1993;90:3511–3515. doi: 10.1073/pnas.89.24.11978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farewell A, Kvint K, Nystrom T. Negative regulation by RpoS: a case of sigma factor competition. Mol Microbiol. 1998;29:1039–1051. doi: 10.1046/j.1365-2958.1998.00990.x. [DOI] [PubMed] [Google Scholar]
- 7.Forstner G G. [1-14C]glucosamine incorporation by subcellular fractions of small intestine mucosa. J Biol Chem. 1970;245:3584–3592. [PubMed] [Google Scholar]
- 8.Freter R, Brickner H, Fekete J, Vickerman M M, Carey K E. Survival and implantation of Escherichia coli in the intestinal tract. Infect Immun. 1983;39:686–703. doi: 10.1128/iai.39.2.686-703.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freter R, Brickner H, Botney M, Cleven D, Arankl A. Mechanisms that control bacterial populations in continuous-flow culture models or mouse large intestinal flora. Infect Immun. 1983;39:676–685. doi: 10.1128/iai.39.2.676-685.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hengge-Aronis R, Klein W, Lange R, Rimmele M, Boss W. Trehalose synthesis genes are controlled by the putative sigma factor encoded by rpoS and are involved in stationary-phase thermotolerance in Escherichia coli. J Bacteriol. 1991;173:7918–7924. doi: 10.1128/jb.173.24.7918-7924.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoskins L. Mucin degradation by enteric bacteria: ecological aspects and implications for bacterial attachment to gut mucosa. In: Boedecker E C, editor. Attachment of organisms to the gut mucosa. II. Boca Raton, Fla: CRC Press, Inc.; 1984. pp. 51–65. [Google Scholar]
- 12.Jenkins D E, Schultz J E, Matin A. Starvation-induced cross protection against heat or H2O2 challenge in Escherichia coli. J Bacteriol. 1988;170:3910–3914. doi: 10.1128/jb.170.9.3910-3914.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jenkins D E, Chaisson S A, Martin A. Starvation-induced cross protection against osmotic challenge in Escherichia coli. J Bacteriol. 1990;172:2779–2781. doi: 10.1128/jb.172.5.2779-2781.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim Y S, Morita A, Miura S, Siddiqul B. Structure of glycoconjugates of intestinal mucosal membranes: role of bacterial adherence. In: Boedecker E C, editor. Attachment of organisms to the gut mucosa. II. Boca Raton, Fla: CRC Press, Inc.; 1984. pp. 99–109. [Google Scholar]
- 15.Krogfelt K A, Poulsen L K, Molin S. Identification of coccoid Escherichia coli BJ4 cells in the large intestine of streptomycin-treated mice. Infect Immun. 1993;61:5029–5034. doi: 10.1128/iai.61.12.5029-5034.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lange R, Hengge-Aronis R. Growth phase-regulated expression of bolA and morphology of stationary-phase Escherichia coli cells are controlled by the novel sigma factor ςs. J Bacteriol. 1991;173:4474–4481. doi: 10.1128/jb.173.14.4474-4481.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lange R, Hengge-Aronis R. The cellular concentration of the ςs subunit of RNA polymerase in Escherichia coli is controlled at the levels of transcription, translation, and protein stability. Genes Dev. 1994;8:1600–1612. doi: 10.1101/gad.8.13.1600. [DOI] [PubMed] [Google Scholar]
- 18.Licht R T, Krogfelt K A, Cohen P S, Kongsbak Poulsen L, Urbance J, Molin S. Role of lipopolysaccharide in colonization of the mouse intestine by Salmonella typhimurium studied by in situ hybridization. Infect Immun. 1996;64:3811–3817. doi: 10.1128/iai.64.9.3811-3817.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Licht T R, Tolker-Nielsen T, Holmstrøm K, Krogfelt K A, Molin S. Inhibition of Escherichia coli precursor 16S rRNA processing by mouse intestinal contents. Environ Microbiol. 1999;1:23–32. doi: 10.1046/j.1462-2920.1999.00001.x. [DOI] [PubMed] [Google Scholar]
- 20.Loewen P C, Hengge-Aronis R. The role of the sigma factor ςs (KatF) in bacterial global regulation. Annu Rev Microbiol. 1994;48:53–80. doi: 10.1146/annurev.mi.48.100194.000413. [DOI] [PubMed] [Google Scholar]
- 21.McCann M P, Kidwell J P, Matin A. The putative ς factor KatF has a central role in development of starvation-mediated general resistance in Escherichia coli. J Bacteriol. 1991;173:4188–4194. doi: 10.1128/jb.173.13.4188-4194.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCormick B A, Klemm P, Krogfelt K A, Burghoff R L, Pallesen L, Laux D C, Cohen P S. Escherichia coli F-18 phase locked “ON” for expression of type 1 fimbriae is a poor colonizer of the streptomycin-treated mouse large intestine. Microb Pathog. 1993;14:33–43. doi: 10.1006/mpat.1993.1004. [DOI] [PubMed] [Google Scholar]
- 23.McCormick B A, Stocker B A D, Laux D C, Cohen P S. The role of motility, chemotaxis, penetration through, and growth in intestinal mucus in the ability of an avirulent strain of Salmonella typhimurium to colonize the large intestines of streptomycin-treated mice. Infect Immun. 1988;56:2209–2217. doi: 10.1128/iai.56.9.2209-2217.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mulvey M R, Loewen P C. Nucleotide sequence of katF of Escherichia coli suggests KatF protein is a novel ς transcription factor. Nucleic Acids Res. 1989;17:9979–9991. doi: 10.1093/nar/17.23.9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mulvey M R, Switala J, Borys A, Loewen P C. Regulation of transcription of katE and katF in Escherichia coli. J Bacteriol. 1990;172:6713–6720. doi: 10.1128/jb.172.12.6713-6720.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Myhal M L, Laux D C, Cohen P S. Relative colonizing abilities of human fecal and K12 strains of Escherichia coli in the large intestines of streptomycin-treated mice. Eur J Clin Microbiol. 1982;1:186–192. doi: 10.1007/BF02019621. [DOI] [PubMed] [Google Scholar]
- 27.Neutra M R. The mechanism of intestinal mucous secretion. In: Boedecker E C, editor. Attachment of organisms to the gut mucosa. II. Boca Raton, Fla: CRC Press, Inc.; 1984. pp. 33–41. [Google Scholar]
- 28.Newman J V, Kolter R, Laux D C, Cohen P S. Role of leuX in Escherichia coli colonization of the streptomycin-treated mouse large intestine. Microb Pathog. 1994;17:301–311. doi: 10.1006/mpat.1994.1076. [DOI] [PubMed] [Google Scholar]
- 29.Notley L, Ferenci T. Induction of RpoS-dependent functions in glucose-limited continuous culture: what level of nutrient limitation induces the stationary phase of Escherichia coli. J Bacteriol. 1996;178:1465–1468. doi: 10.1128/jb.178.5.1465-1468.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peekhaus N, Conway T. What's for dinner?: Entner-Doudoroff metabolism in Escherichia coli. J Bacteriol. 1998;180:3495–3502. doi: 10.1128/jb.180.14.3495-3502.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Potten C S, Allen T D. Ultrastructure of cell loss in intestinal mucosa. J Ultrastruct Res. 1977;60:272–277. doi: 10.1016/s0022-5320(77)80071-3. [DOI] [PubMed] [Google Scholar]
- 32.Poulsen L K, Lan F, Kristensen C S, Hobolth P, Molin S, Krogfelt K A. Spatial distribution of Escherichia coli in the mouse large intestine from rRNA in situ hybridization. Infect Immun. 1994;62:5191–5194. doi: 10.1128/iai.62.11.5191-5194.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poulsen L K, Licht T R, Rang C, Krogfelt K A, Molin S. The physiological state of Escherichia coli BJ4 growing in the large intestine of streptomycin treated mice. J Bacteriol. 1995;177:5840–5845. doi: 10.1128/jb.177.20.5840-5845.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quastler H, Sherman F G. Cell population in the intestinal epithelium of the mouse. Exp Cell Res. 1959;17:420–438. doi: 10.1016/0014-4827(59)90063-1. [DOI] [PubMed] [Google Scholar]
- 35.Que J U, Casey S W, Hentges D J. Factors responsible for increased susceptibility of mice to intestinal colonization after treatment with streptomycin. Infect Immun. 1986;53:116–123. doi: 10.1128/iai.53.1.116-123.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rang C U, Licht T R, Krogfelt K A, Conway P, Midtvedt T, Molin S. Estimation of growth rates of Escherichia coli BJ4 in streptomycin-treated and previously germfree mice by in situ rRNA hybridization. Clin Diagn Lab Immunol. 1999;6:434–436. doi: 10.1128/cdli.6.3.434-436.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Slomlany A, Yano S, Slomlany B I, Glass G B J. Lipid composition of the gastric mucus barrier in the rat. J Biol Chem. 1978;253:3785–3791. [PubMed] [Google Scholar]
- 38.Small P, Blankenhorn D, Welty D, Zinser F, Slonczewski J L. Acid and base resistance in Escherichia coli and Shigella flexneri: role of RpoS and growth pH. J Bacteriol. 1994;176:1729–1737. doi: 10.1128/jb.176.6.1729-1737.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sweeney N J, Klemm P, McCarmich B A, Moller-Nielsen E, Utley M, Schembri M A, Laux D C, Cohen P S. The Escherichia coli K-12 gntP gene allows E. coli F-18 to occupy a distinct nutritional niche in the streptomycin-treated mouse large intestine. Infect Immun. 1996;64:3497–3503. doi: 10.1128/iai.64.9.3497-3503.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van der Waaij D, Berghuis-de Vries J M, Lekkerkerk-van der Wees J E C. Colonization resistance of the digestive tract in conventional and antibiotic treated mice. J Hyg. 1971;69:405–411. doi: 10.1017/s0022172400021653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vercellotti J R, Salyers A A, Bullard W S, Wilkins T D. Breakdown of mucins and plant polysaccharides in the human colon. Can J Biochem. 1977;55:1190–1196. doi: 10.1139/o77-178. [DOI] [PubMed] [Google Scholar]
- 42.Wadolkowski E A, Laux D C, Cohen P S. Colonization of the streptomycin-treated mouse large intestine by a human fecal Escherichia coli strain: role of growth in mucus. Infect Immun. 1988;56:1030–1035. doi: 10.1128/iai.56.5.1030-1035.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yildiz F H, Schoolnik G K. Role of rpoS in stress survival and virulence of Vibrio cholerae. J Bacteriol. 1998;180:773–784. doi: 10.1128/jb.180.4.773-784.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zambrano M M, Siegele D A, Almirón M, Tormo A, Kolter R. Microbial competition: Escherichia coli mutants that take over stationary phase cultures. Science. 1993;259:1757–1760. doi: 10.1126/science.7681219. [DOI] [PubMed] [Google Scholar]