Abstract
Background
Personalized genomic classifiers have transformed the management of prostate cancer (PCa) by identifying the most aggressive subsets of PCa. Nevertheless, the performance of genomic classifiers to risk classify African American men is thus far lacking in a prospective setting.
Methods
This is a prospective study of the Decipher genomic classifier for National Comprehensive Cancer Network low- and intermediate-risk PCa. Study-eligible non–African American men were matched to African American men. Diagnostic biopsy specimens were processed to estimate Decipher scores. Samples accrued in NCT02723734, a prospective study, were interrogated to determine the genomic risk of reclassification (GrR) between conventional clinical risk classifiers and the Decipher score.
Results
The final analysis included a clinically balanced cohort of 226 patients with complete genomic information (113 African American men and 113 non–African American men). A higher proportion of African American men with National Comprehensive Cancer Network–classified low-risk (18.2%) and favorable intermediate-risk (37.8%) PCa had a higher Decipher score than non–African American men. Self-identified African American men were twice more likely than non–African American men to experience GrR (relative risk [RR] = 2.23, 95% confidence interval [CI] = 1.02 to 4.90; P = .04). In an ancestry-determined race model, we consistently validated a higher risk of reclassification in African American men (RR = 5.26, 95% CI = 1.66 to 16.63; P = .004). Race-stratified analysis of GrR vs non-GrR tumors also revealed molecular differences in these tumor subtypes.
Conclusions
Integration of genomic classifiers with clinically based risk classification can help identify the subset of African American men with localized PCa who harbor high genomic risk of early metastatic disease. It is vital to identify and appropriately risk stratify the subset of African American men with aggressive disease who may benefit from more targeted interventions.
Prostate cancer (PCa) is one of the leading cancers and impacts millions of men in the United States and around the world, with etiologies ranging from socioeconomic factors to neighborhood characteristics and underlying genomics (1). Though PCa affects men of different races and ethnicities, African American men are disproportionately impacted by higher incidence and increased PCa-related mortality (2). However, few prospective studies maximize minority recruitment to provide an unbiased assessment of the genomic processes that underscore the disparity landscape of PCa (1). In the era of precision medicine, personalized genomic classifiers, such as the Decipher score, have transformed the management of PCa by aiding in the detection of aggressive subsets of PCa (3). The Decipher score is a validated 22-gene–based genomic classifier that predicts risk of metastasis and may offer a robust alternative to reduce disparity gaps, especially among the subset of African American men who are likely to experience increased disease severity (3-5).
Though the Decipher score was developed using a predominantly White cohort, its utility in retrospective studies has been comparable between African American men and non–African American men (4). More important, Howard et al. (4), in their recent study, showed that it had relatively similar accuracy in predicting metastasis between African American men and non–African American men. Therefore, a prospective evaluation of the Decipher score would aid in identifying aggressive African American men tumor subtypes that may benefit from targeted interventions and treatment strategies. Considering that conventional clinical risk classifiers, such as the National Comprehensive Cancer Network (NCCN) classification and the Cancer of the Prostate Risk Assessment (CAPRA), have also been shown to be suboptimal in African American men, particularly for low- and intermediate-risk PCa, the integration of conventional clinical risk classifiers with genomic classifiers may offer a robust improvement in risk stratifying and appropriately managing PCa patients (6,7). Emerging work has also shown that early PCa may harbor genomically aggressive subtypes, and these subsets are unlikely to be captured by clinical risk classifiers alone (6,8). Therefore, evaluating genomic classifiers in conjunction with clinical risk classifiers to identify patients with aggressive disease within a minority-targeted prospective study will elucidate their clinical utility across different subpopulations. Therefore, samples from the NCT02723734 prospective study were genomically interrogated to evaluate whether the integration of Decipher score with clinical risk classifiers can distinguish subsets of men with early localized PCa who may harbor aggressive disease, particularly among African American men who experience the greatest burden of PCa.
Methods
Study Design, Eligibility Criteria, and Matching
This was a multisite prospective study (NCT02723734) of the Decipher score among self-reported African American men and non–African American men. Before study activation, full institutional and local site institutional review board approval was obtained. Enrolled patients had to meet the following inclusion criteria: aged 18 years and older; Eastern Cooperative Oncology Group Performance Status 0-1 or Karnofsky Performance Status greater than 70; have pathologically and histologically confirmed NCCN-classified low- or intermediate-risk PCa (low risk defined as biopsy Gleason score 6 [3 + 3] along with pretreatment prostate-specific antigen [PSA] level <10 ng/mL and T stage T1c-T2a; intermediate risk defined as biopsy Gleason score 7 with PSA level ≥10ng/mL or <20 ng/mL, or T stage ≤T2c); and be undergoing their first line of treatment. First-line treatment included radical prostatectomy (RP) or definitive radiation therapy (RT) with or without short-term androgen deprivation therapy. Patients who were identified for active surveillance (AS) after diagnostic biopsy or on AS for more than 6 months after diagnosis were excluded from recruitment. Lastly, patients with prior PCa-related treatment were also considered ineligible. Additional details on recruitment are provided in the Supplementary Methods (available online).
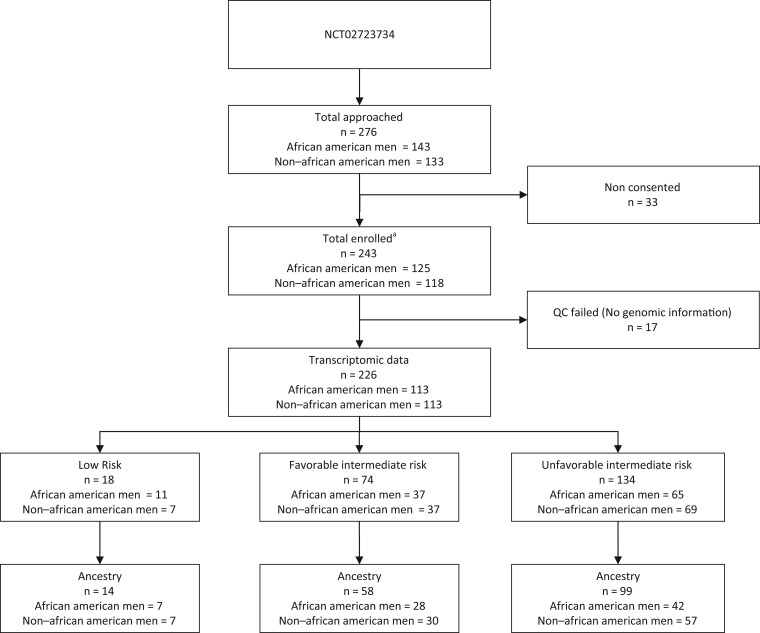
To maximize minority enrollment, study-eligible African American men were accrued first, and then clinically matched non–African American men were subsequently enrolled. Matching was performed to balance the baseline clinical difference between race groups. Non–African American men were matched to African American men on age, PSA level at diagnosis, biopsy Gleason score, clinical stage, and total percent of positive biopsy (PPB) cores. Participants were then stratified based on their treatment selection for RP vs RT with or without short-term androgen deprivation therapy. No treatment intervention beyond the standard of care was permitted, and therefore, patients were able to choose their definitive treatment option through shared decision making based on physician recommendations. The intent was to enroll early localized PCa defined as low- and intermediate-risk patients with multiple cores positive (high disease burden) to minimize accruing patients who would select AS and be excluded from study. A total of 276 patients were approached for participation in the VANDAAM study, with 243 patients (125 African American men and 118 non–African American men) enrolling in the study. The CONSORT diagram provides a detailed overview of the study cohort (see Figure 1). Though this study does not report on the primary endpoint of NCT02723734 (2-year failure rate), it presents the correlative assessment of clinical risk classifiers and the Decipher score on the samples accrued in this trial.
Figure 1.
Consort diagram of patient enrollment among the total enrolled participants (n = 243); 226 passed the quality control and had complete genomic information available. aBlood draw for ancestry genotyping was carried out for 184 of 243 enrolled patients. Of the 184 samples with ancestry information, 171 patients also had complete genomic information available. QC = Quality Control.
Data Collection
Major variables included self-identified race, age at diagnosis, pretreatment PSA, PPB (calculated by dividing cores positive by total cores evaluated), biopsy Gleason score, NCCN-defined risk categories, and CAPRA. Methods for deriving CAPRA are defined in Cooperberg et al. (9) The NCCN intermediate-risk group was subdivided as favorable intermediate risk and unfavorable intermediate risk. Favorable intermediate risk was defined as the presence of 1 intermediate risk factor alongside biopsy Gleason score (≤3 + 4) and no more than 50% of PPB, whereas unfavorable intermediate risk was defined by having more than 1 intermediate-risk factor, biopsy Gleason score (4 + 3), or more than 50% PPB. For the subset of patients who underwent RP, pathology information, including pathologic Gleason score and adverse pathologic features (extracapsular extension, surgical margins, seminal vesicle invasion, and lymph node involvement), was also collected. Additionally, Decipher scores were obtained along with the comprehensive expression-based transcriptomic signatures for each patient from the genomic resource information database repository, a database of gene expression signatures and tumor transcriptome (Veracyte Inc, California, USA). Decipher score ranges from 0 to 1 and is categorized as low (<0.45), intermediate (≥0.45-0.60), and high (>0.60) risk. Additional details on genomic signatures used in this analysis are provided in Supplementary Methods (available online). Lastly, ancestry determination was performed on the consented patients at the University of Minnesota Genomics Center (Supplementary Methods, available online) (10).
Clinical Outcome Assessment
Genomic risk of reclassification (GrR) was used as a clinical outcome to identify the subset of NCCN-classified low-risk and favorable intermediate-risk patients who harbored genomic risk of metastasis. GrR status was derived using the presence of a high Decipher score in low-risk and favorable intermediate-risk categories. Non-GrR tumors were defined as low-risk and favorable intermediate-risk patients with Decipher scores of 60 or lower.
Statistical Analysis
Baseline and clinical characteristics between race groups were compared using Pearson χ2 and Fisher exact tests. Similar statistical tests of categorical comparisons were used to assess the relationship between NCCN and CAPRA risk categories with the Decipher score. To model the GrR among low-risk and favorable intermediate-risk patients, we estimated the relative risk (RR) using a log-binomial model. We also validated preceding self-identified race model estimates by using ancestry-determined race categories. Considering the age differences between race groups, both models were also adjusted for age at diagnosis. In addition, we also evaluated the race-stratified nonlinear association of age with overall high Decipher score (>.60) and GrR tumors in a nonlinear binomial spline regression model using a B-spline function with 5 degrees of freedom. Probability of GrR tumors and high Decipher score were then estimated and plotted to compare between African American men and non–African American men. We also estimated the risk of pathologic upgrading of Gleason score using a log-binomial model within the subset of patients who received RP. Lastly, precomputed expression-based genomic signatures between race groups were compared using the Wilcoxon rank-sum test. The analyses were conducted in SAS 9.4 and R 1.3.1. All the tests were 2-sided, and cutoff values were no more than .05.
Results
Baseline and Clinical Characteristics
A detailed comparison of demographic and clinical variables for all the matched evaluable patients with complete genomic information (n = 226) is presented in Table 1. There were no differences between African American men and non–African American men for pretreatment PSA (P = .26), biopsy Gleason score (P = .64), PPB (P = .46), NCCN risk (P = .60), CAPRA (P = .91), biopsy Decipher score (P = .62), or primary treatment selection (P = .76) (Table 1). Despite rigorous targeted matching, African American men enrolled in the study were relatively younger than non–African American men (aged 61 vs 65 years; P = .005).
Table 1.
Cohort characteristics (n = 226)
| Characteristic | African American men | Non–African American men, | P a |
|---|---|---|---|
| (n = 113) | (n = 113) | ||
| Median age (IQR), y | 61 (55-67) | 65 (59-70) | .005 |
| Age group, No. (%) | .07 | ||
| Younger than 50 | 12 (11) | 5 (4.4) | |
| 50 and older | 101 (89) | 108 (96) | |
| Pretreatment PSA, ng/mL | .26 | ||
| Median (IQR) | 6.9 (5.1, 8.8) | 6.3 (4.3, 8.4) | |
| PSA categories, No. (%) | .61 | ||
| 0-6 ng/mL | 44 (39) | 51 (45) | |
| >6-10 ng/mL | 45 (40) | 42 (37) | |
| >10-20 ng/mL | 24 (21) | 20 (18) | |
| Biopsy Gleason score, No. (%) | .64 | ||
| 3 + 3 | 13 (12) | 9 (8.0) | |
| 3 + 4 | 67 (59) | 68 (60) | |
| 4 + 3 | 33 (29) | 36 (32) | |
| Median No. of positive cores (IQR) | 5 (4-7) | 6 (4-8) | .11 |
| PPB category, No. (%) | .46 | ||
| <34 | 36 (32) | 31 (27) | |
| ≥34 | 77 (68) | 82 (73) | |
| NCCN risk, No. (%) | .60 | ||
| Low risk | 11 (9.7) | 7 (6.2) | |
| Favorable risk | 37 (33) | 37 (33) | |
| Unfavorable risk | 65 (58) | 69 (61) | |
| Biopsy CAPRA, No. (%) | .91 | ||
| 0-2 | 21 (19) | 20 (18) | |
| 3-5 | 70 (62) | 73 (65) | |
| 6-10 | 22 (19) | 20 (18) | |
| Decipher (biopsy) category, No. (%)b | .62 | ||
| Low risk | 47 (42) | 46 (41) | |
| Int risk | 27 (24) | 33 (29) | |
| High risk | 39 (35) | 34 (30) | |
| Treatment type, No. (%) | .76 | ||
| Prostatectomy | 41 (36) | 41 (36) | |
| Radiation (no ADT) | 41 (36) | 47 (42) | |
| Radiation (with ADT) | 25 (22) | 21 (19) | |
| NA | 6 (5.3) | 4 (3.5) |
Wilcoxon rank sum test for numeric and Pearson χ2 test for categorical variables. ADT = androgen deprivation therapy; CAPRA = Cancer of the Prostate Risk Assessment; IQR = interquartile range; NA = not available; NCCN = National Comprehensive Cancer Network; PPB = positive percentage biopsy; PSA = prostate-specific antigen.
Decipher score is categorized as low risk (<0.45), intermediate risk (≥0.45-0.60), and high risk (>0.60).
Genomic Risk of Reclassification in African American Men
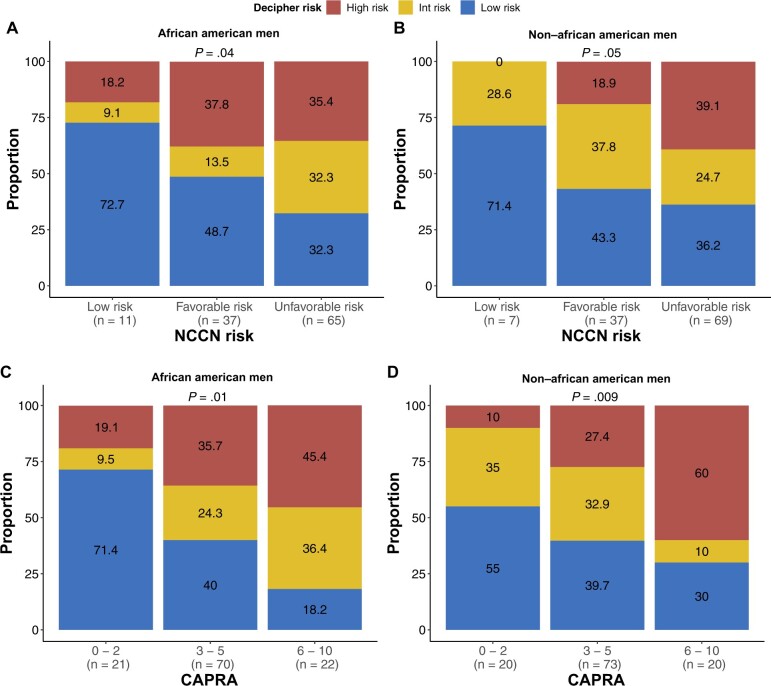
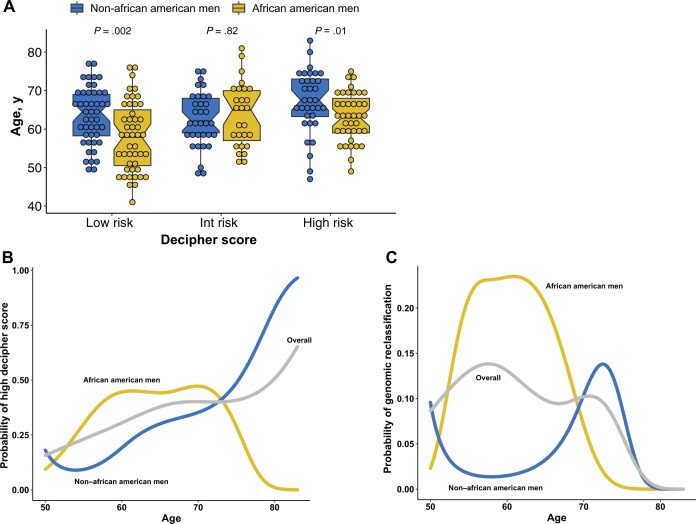
We compared GrR between clinical and genomic risk classifiers from samples with complete genomic information (113 African American men and 113 non–African American men samples). A higher proportion of African American men with NCCN-classified low risk (18.2%) and favorable intermediate risk (37.8%) harbored a higher genomic risk of metastasis than non–African American men (Figure 2, A and B). More important, among non–African American men, no low-risk patients were genomically reclassified. When we modeled the GrR from self-reported race (Table 2, model 1), African American men were more than twice as likely to experience reclassification (RR = 2.23, 95% confidence interval [CI] = 1.06 to 4.90; P = .04) (Table 2). We also compared the univariate association between baseline clinical variables and GrR and did not observe any statistically significant impact of these variables on the GrR (Supplementary Table 1, available online). Similarly, when compared with CAPRA, 19% of African American men were reclassified to a higher genomic risk subcategory within the low-risk (0-2) group (P = .01) compared with only 10% non–African American men (P = .009) (Figure 2, C and D). African American men with high genomic risk were also younger than non–African American men (P = .01) (Figure 3, A). When analyzed in the spline model, a nonlinear association between age and Decipher score was observed, with younger African American men having increased probability of a higher Decipher score (Figure 3, B). Similarly, probability of GrR was also higher among younger African American men than non–African American men (Figure 3, C). Lastly, within the subset of patients who elected for RP, African American men were also more likely to experience a Gleason score upgrade (RR = 3.25, 95% CI = 1.15 to 9.11; P = .02) (Supplementary Table 2, available online), though no differences in other pathologic features were noted (Supplementary Table 3, available online).
Figure 2.
Comparison between clinical risk classifier and race group in transcriptomic data (n = 226; African American men, n = 113; and non–African American men, n = 113). A) Comparison of NCCN risk groups and Decipher score in African American men. B) Comparison of NCCN risk groups and Decipher score in non–African American men. C) Comparison of CAPRA risk categories and Decipher score in African American men. D) Comparison of CAPRA risk categories and Decipher score in non–African American men. CAPRA = Cancer of the Prostate Risk Assessment; Int = intermediate; NCCN = National Comprehensive Cancer Network.
Table 2.
Log binomial regression to estimate the risk of genomic reclassification within low-favorable intermediate risk patients
| Modela | Race | RR (95% CI) | P |
|---|---|---|---|
| Model 1b | Self-reported race | ||
| African American men | 2.23 (1.02 to 4.90) | .04 | |
| Non–African American men | 1 (Referent) | ||
| Model 2c | Self-reported race | ||
| African American men | 4.13 (1.30 to 13.04) | .01 | |
| Non–African American men | 1 (Referent) | ||
| Model 3d | Ancestry-derived race | ||
| African ancestry | 5.26 (1.66 to 16.63) | .004 | |
| European ancestry | 1 (Referent) |
All models are adjusted age at diagnosis and developed in low-favorable intermediate risk patients. CI = confidence interval; RR = relative risk.
Model 1 estimates the self-reported race–based relative risk of genomic reclassification for all the patients.
Model 2 estimates the self-reported race–based relative risk of genomic reclassification for the patients with ancestry determination completed.
Model 3 with ancestry-derived race was carried out in patients who underwent ancestry determination.
Figure 3.
Impact of age on Decipher score distribution and genomic reclassification. A) Age in years is compared between race groups across Decipher score categories. B) A nonlinear binomial model with B-spline showing the probability of higher Decipher score across race groups by increasing age in years. C) A nonlinear binomial model with B-spline showing the probability of genomic risk of reclassification across race groups by increasing age in years. Int = intermediate.
Ancestry Validation of Self-Reported Race Model
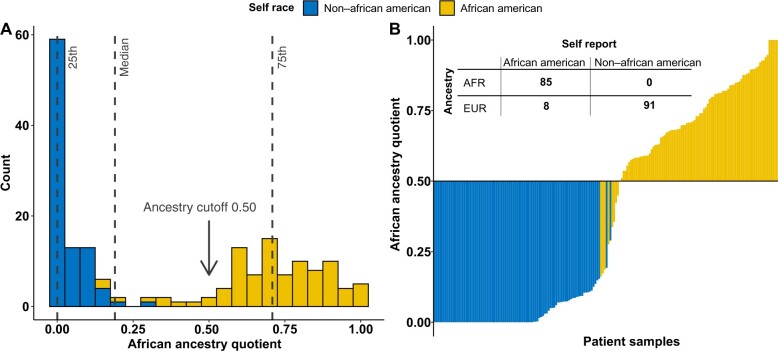
We performed ancestry evaluation on patients who consented for blood collection (n = 184). For ancestry determination, 93 self-identified African American men and 91 self-identified non–African American men were analyzed. To optimize ancestry-base race categorization, we explored overall ancestry distribution and overlayed it with self-defined race groups. Median-based ancestry cutoff captured nearly 97% (n = 90) of self-identified African American men, with only 3% (n = 3) being misclassified as having shared European ancestry (Figure 4, A). We therefore decided to apply a more stringent ancestry quotient cutoff to limit the self-identified African American men who may share borderline ancestry. Figure 4, B shows that 91.4% (n = 85) of self-identified African American men patients had more than 50% African ancestry, and 8.6% (n = 8) self-identified African American men patients were genotyped as having European ancestry. Because ancestry determination was not performed for all patients, we modified the self-identified race model 1 and subsequently developed model 2, which only included patients who had ancestry information available. This allowed for direct comparison of self-identified race estimates to ancestry-based race estimates. GrR from self-identified race model 2 was consistently higher in African American men than non–African American men (RR = 4.13, 95% CI = 1.30 to 13.04; P = .01) (Table 2, model 2). Finally, we estimated the presence of GrR using only ancestry-determined race categories in model 3 and observed nearly 5 times the risk of reclassification among low-risk and favorable intermediate-risk African American men (RR = 5.26, 95% CI = 1.66 to 16.63; P = .004) (Table 2, model 3).
Figure 4.
Ancestry determination in VANDAAM samples. A) Figure presents the African ancestry quotient on the x-axis to show its distribution across all the patients who underwent genotyping (n = 184). We overlayed the ancestry distribution with the self-defined race group to determine optimal cut point for African ancestry quotient to dichotomize race groups. At 0.50 and above, nearly all the self-reported African American patients shared African ancestry and were categorized as AFR, whereas others were grouped as EUR. B) Waterfall figure shows patient samples on y-axis and their ancestry quotient on x-axis. Samples were sorted based on their quotients and were overlayed with self-reported race group. The right side represents all the self-reported African American men with ancestry quotient above 0.50. Whereas on the left side, 8 self-reported African American men were considered as being misclassified and were grouped as EUR in ancestry table. The ancestry table presents the overlap of the patients between self-reported race and ancestry-based race categories. At African ancestry quotient of 0.50, no misclassification in non–African American men was noted, whereas only 8 self-reported African American men were misclassified out of 93. AAM = African American men; AFR = African ancestry; EUR = European ancestry; NAAM = non–African American men.
Molecular Basis of GrR in African American Men
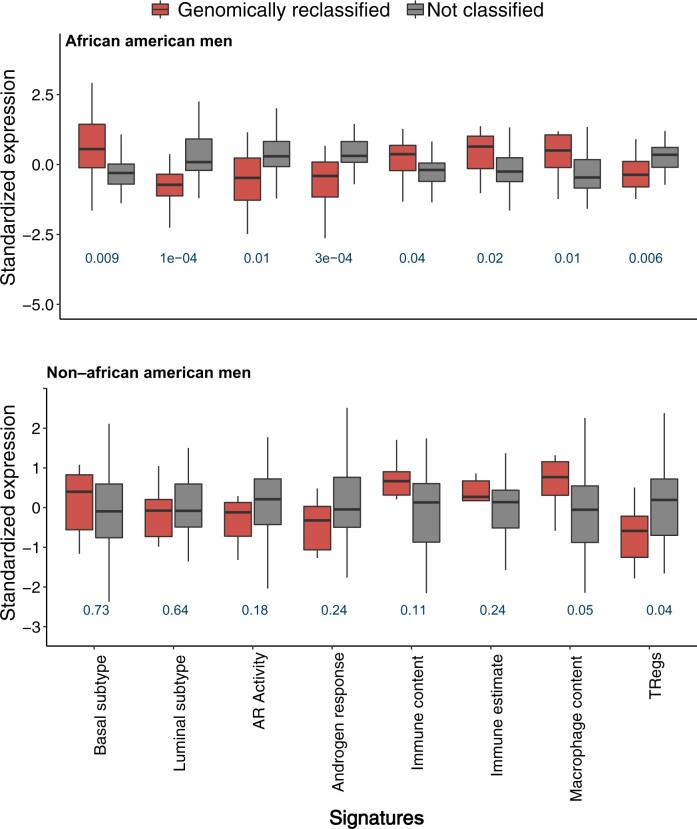
We evaluated the molecular underpinnings of GrR within the low-risk and favorable intermediate-risk subset of African American men. We used precomputed gene expression signatures available within the genomic resource information database repository and evaluated their distribution between GrR and non-GrR African American men tumors. Tumors from African American men who experienced reclassification exhibited higher basal-like characteristics and had lower rates of androgen receptor (AR) activity. We applied Zhang et al. (11) gene expression signature for classification of basal-like tumors and found that genomically reclassified tumors within African American men had higher basal-like gene expression (P = .009; Figure 5). Consistently, GrR tumors also had a lower score for luminal tumor subtypes (P < .001; Figure 5) (12). To assess AR activity, we used the hallmark AR signature from Liberzon et al. (13) and AR activity score from Spratt et al. (14) and showed that GrR tumors in African American men consistently had lower AR activity than non-GrR tumors of African American men (Figure 5). Lastly, we evaluated the enrichment of immune content in African American men tumors with GrR and observed that these tumor subsets were enriched for 190 immune-mediated gene expression markers (P = .04) (15). Furthermore, we also used an estimate-based signature of immune estimate and showed that GrR tumors from African American men indeed had higher immune enrichment (Figure 5) (16). Considering that the proportion of GrR tumors were much lower in non–African American men, reclassified tumors from non–African American men did not reveal similar molecular features to those seen in African American men tumors. Finally, reclassified tumors among African American men and non–African American men had higher expression of macrophage signatures; however, both groups consistently had lower expression of the regulatory T-cell signature (Figure 5) (17,18).
Figure 5.
Relative difference in precomputed gene expression–based genomic signatures between GrR and non-GrR tumors in a race-stratified comparison. All statistical tests were 2-sided. AR = androgen receptor; Treg = regulatory T cell.
Discussion
This prospective study used a clinically balanced cohort of African American men and non–African American men and showed that a subset of African American men harbor genomically aggressive PCa, which may not be detected with conventional clinical classifiers. Therefore, results from this study support the integration of personalized biomarkers with conventional clinical risk classifiers, particularly for African American men, to optimize timely detection of genomically aggressive PCa and guide appropriate treatment recommendations.
Timely detection of aggressive tumor subtypes portends favorable prognoses and is associated with improved outcomes (7). Considering that clinical risk classifiers, primarily the NCCN risk groups, are a mainstay in PCa risk stratification and management, their ability to discern these tumor subtypes across racial subgroups is vital (7). However, clinical risk classifiers alone do not offer optimal detection of these aggressive subtypes because they exclusively rely on clinicopathologic features and do not account for the genomic diversity of prostate tumors (6,7). An earlier study by Spratt et al. (6) demonstrated that integration of a genomic classifier into clinical risk classifier outperformed both NCCN and CAPRA classifications in the prediction of long-term outcomes. This is highly relevant to African American men whose tumors exhibit high genomic variability (8,19,20). Prior retrospective studies have revealed that PCa in African American men with low-risk disease may often show higher Decipher scores and are more likely to experience pathological upgrading compared with non–African American men (8,21,22). An alternative approach is to use a multiparametric magnetic resonance imaging (mp-MRI) of the prostate and pelvis, which offers enhanced staging accuracy for African American men and non–African American men, but its utilization for PCa remains disparate, with African American men less likely to undergo mp-MRI (23,24). Additionally, mp-MRI–based risk classification of PCa does not account for genomic distinctions in tumor characteristics. Therefore, genomic classification may offer a robust alternative to identify specific subsets of patients with disease that is likely to metastasize despite being classified as low risk or favorable intermediate risk by conventional clinical risk classifiers. Recently, Feng et al. (25) validated genomic classifiers in a prospective randomized trial (RTOG 9601) to predict androgen deprivation therapy response among patients. Their findings suggested that genomic classifiers may help personalize shared decision making to weigh the absolute benefit of androgen deprivation therapy in addition to primary therapy. Therefore, using genomic-based precision biomarkers alongside clinical risk classifiers may also aid clinicians to further personalize treatment decision making and identify patients who may benefit from additional therapeutic interventions.
Considering that African American men are known to experience a higher burden of PCa at a much earlier age, we further evaluated age-specific differences in Decipher scores and GrR tumors (1). Our nonlinear spline model suggested that younger African American men harbored higher genomic risk of metastasis, which may underlie the presence of GrR tumors in this group. These results may inform the clinical recommendations pertaining to early detection of aggressive PCa and support the use of additional diagnostic tools to appropriately stage patients for optimal care.
To understand the genomic characteristics of GrR tumors, we investigated the expression-based signature that may be enriched within these tumors. In a race-stratified analysis of GrR vs non-GrR tumors, we observed a relative difference in the precomputed signatures that underlie AR biology, basal and luminal cell lineages, and overall immune cell enrichment. AR activity plays a vital role in tumorigenesis and regulation of DNA repair pathways (26). Considering that DNA repair and AR biology can also modulate response to RT, our results may offer insights into RT response of GrR tumors from African American men (26). Furthermore, GrR tumors among African American men also exhibited higher enrichment of a signature estimating basal cell lineage than non-GrR tumors, which is consistent with the earlier work of Zhang et al. (11). Lastly, we also identified a higher immune cell abundance within GrR tumors from African American men, signifying the role of the immune repertoire within the prostate tumor microenvironment of genomically aggressive subtypes. Consistently, a number of studies in PCa have shown that higher immune content enrichment in PCa is associated with an elevated risk of poor outcomes (27,28). Additionally, immune content can be evaluated alongside the Decipher score to identify subtypes that are harbingers of lethal outcomes (27). Emerging research has revealed the presence of a distinct immune microenvironment in PCa of African American men that is enriched in inflammatory markers (19). Whether such immune repertoire within prostate tumors of African American men could underlie observed higher prevalence of GrR tumors in this subset remains unexplored. This study does not suggest that observed differences in aforementioned signatures drive disparities in African American men; instead, it shows that GrR tumors of African American men are genomically distinct and that further validation may be warranted to assess their race-specific functional role in the progression of these genomically aggressive tumor subtypes.
This prospective study reports that African American men may benefit from the integration of precision biomarkers and clinical risk classifiers to optimize risk stratification. Detection of these tumor subtypes within early localized PCa may help clinicians select the subset of patients who are likely to benefit from additional therapeutic interventions. This work has a few limitations. First, like other clinical trials, the African American men enrolled in this study may be less representative of minorities across the United States; African American men participants may have higher health-care–seeking behavior, socioeconomic status, and overall willingness to participate in clinical trials compared with the general population (1). Second, despite rigorous matching, a significant age gap persisted between race groups, emphasizing the role of age in studying minority populations. However, we attempted to achieve a clinically balanced cohort to facilitate a first-of-its-kind genomic study in a prospective manner. Additionally, using ancestry-informed markers of race further strengthens our findings from a self-identified race model and obviates any resulting biases. Overall, our findings show that conventional clinical risk classifiers models may be suboptimal in assessing true disease severity in a subset of African American men who are likely to have a high genomic risk for distant metastasis.
Funding
This research was supported by the George Edgecomb Society; Cancer Center Support grant P30-CA076292 to the Moffitt Cancer Center; and P20‐CA233255 (to KY).
Notes
Role of the funder: Funding sources had no role in the design, study commencement, analysis, results interpretation and writing of this manuscript, or decision to submit the manuscript for publication.
Disclosures: Corresponding author KY is on the advisory committee on health equity for Janssen Research & Development LLC, Flatiron Health Inc, and MyCareGorithm, LLC. Other authors do not report any conflict of interest related to the publication of this paper.
Author contributions: Conceptualization: KY. Data curation: S., YL, ED, AH, AD, AKF. Formal Analysis: SA. Funding acquisition: KY. Investigation: All authors. Supervision: KY, GDG, JTR, PASJ, JPS, JD, MP, RL, BM, DF, AN, KG, EK; RJB, AL, CEE, JDP, NV, CD, APD, WK Writing—Original draft: KY, SA, GDG. Writing—review & editing: All authors.
Acknowledgements: Decipher genomic testing was covered by Veracyte Inc, formally known as Decipher Biosciences, Inc. Editorial assistance was provided by the Moffitt Cancer Center’s Office of Scientific Publishing by Daley Drucker and Gerard Hebert. No compensation was given beyond their regular salaries.
Prior presentation: This was presented at ASCO 2021 as podium presentation. Abstract title: A prospective ValiDation of the genomic classifier defines high-metastasis risk in a subset of African American men with early localized prostate cancer: VanDAAM study. Online conference, June 8, 2021.
Supplementary Material
Contributor Information
Shivanshu Awasthi, H. Lee Moffitt Cancer Center & Research Institute, Tampa, FL, USA.
G Daniel Grass, H. Lee Moffitt Cancer Center & Research Institute, Tampa, FL, USA.
Javier Torres-Roca, H. Lee Moffitt Cancer Center & Research Institute, Tampa, FL, USA.
Peter A S Johnstone, H. Lee Moffitt Cancer Center & Research Institute, Tampa, FL, USA.
Julio Pow-Sang, H. Lee Moffitt Cancer Center & Research Institute, Tampa, FL, USA.
Jasreman Dhillon, H. Lee Moffitt Cancer Center & Research Institute, Tampa, FL, USA.
Jong Park, H. Lee Moffitt Cancer Center & Research Institute, Tampa, FL, USA.
Robert J Rounbehler, H. Lee Moffitt Cancer Center & Research Institute, Tampa, FL, USA.
Elai Davicioni, Veracyte Inc, South San Francisco, CA, USA.
Alex Hakansson, Veracyte Inc, South San Francisco, CA, USA.
Yang Liu, Veracyte Inc, South San Francisco, CA, USA.
Angelina K Fink, H. Lee Moffitt Cancer Center & Research Institute, Tampa, FL, USA.
Amanda DeRenzis, H. Lee Moffitt Cancer Center & Research Institute, Tampa, FL, USA.
Jordan H Creed, H. Lee Moffitt Cancer Center & Research Institute, Tampa, FL, USA.
Michael Poch, H. Lee Moffitt Cancer Center & Research Institute, Tampa, FL, USA.
Roger Li, H. Lee Moffitt Cancer Center & Research Institute, Tampa, FL, USA.
Brandon Manley, H. Lee Moffitt Cancer Center & Research Institute, Tampa, FL, USA.
Daniel Fernandez, H. Lee Moffitt Cancer Center & Research Institute, Tampa, FL, USA.
Arash Naghavi, H. Lee Moffitt Cancer Center & Research Institute, Tampa, FL, USA.
Kenneth Gage, H. Lee Moffitt Cancer Center & Research Institute, Tampa, FL, USA.
Grace Lu-Yao, Sidney Kimmel Cancer Center at Thomas Jefferson University, Philadelphia, PA, USA.
Evangelia Katsoulakis, James A. Haley Veterans Hospital, Tampa, FL, USA.
Ryan J Burri, Bay Pines VA Healthcare System, Tampa, FL, USA.
Andrew Leone, Bay Pines VA Healthcare System, Tampa, FL, USA.
Cesar E Ercole, James A. Haley Veterans Hospital, Tampa, FL, USA.
Joshua D Palmer, The James Cancer Hospital at Ohio State University, Columbus, OH, USA.
Neha Vapiwala, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA.
Curtiland Deville, Johns Hopkins University, Baltimore, MD, USA.
Timothy R Rebbeck, Harvard T.H. Chan School of Public Health, Boston, MA, USA.
Adam P Dicker, Sidney Kimmel Cancer Center at Thomas Jefferson University, Philadelphia, PA, USA.
William Kelly, Sidney Kimmel Cancer Center at Thomas Jefferson University, Philadelphia, PA, USA.
Kosj Yamoah, H. Lee Moffitt Cancer Center & Research Institute, Tampa, FL, USA.
Data Availability
The data discussed in this work have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO series accession number GSE209954. (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE209954).
References
- 1. Mahal BA, Gerke T, Awasthi S, et al. Prostate cancer racial disparities: a systematic review by the prostate cancer foundation panel. Eur Urol Oncol. 2021;5(1):18-29. [DOI] [PubMed] [Google Scholar]
- 2. DeSantis CE, Miller KD, Goding Sauer A, Jemal A, Siegel RL.. Cancer statistics for African Americans, 2019. CA A Cancer J Clin. 2019;69(3):211-233. [DOI] [PubMed] [Google Scholar]
- 3. Jairath NK, Dal Pra A, Vince R Jr, et al. A systematic review of the evidence for the decipher genomic classifier in prostate cancer. Eur Urol. 2021;79(3):374-383. [DOI] [PubMed] [Google Scholar]
- 4. Howard LE, Zhang J, Fishbane N, et al. Validation of a genomic classifier for prediction of metastasis and prostate cancer-specific mortality in African-American men following radical prostatectomy in an equal access healthcare setting. Prostate Cancer Prostatic Dis. 2020;23(3):419-428. [DOI] [PubMed] [Google Scholar]
- 5. Feng FY, Huang HC, Spratt DE, et al. Validation of a 22-gene genomic classifier in patients with recurrent prostate cancer: an ancillary study of the NRG/RTOG 9601 randomized clinical trial. JAMA Oncol. 2021;7(4):544-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Spratt DE, Zhang J, Santiago-Jiménez M, et al. Development and validation of a novel integrated clinical-genomic risk group classification for localized prostate cancer. J Clin Oncol. 2018;36(6):581-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Herlemann A, Huang HC, Alam R, et al. Decipher identifies men with otherwise clinically favorable-intermediate risk disease who may not be good candidates for active surveillance. Prostate Cancer Prostatic Dis. 2020;23(1):136-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mahal BA, Alshalalfa M, Spratt DE, et al. Prostate cancer genomic-risk differences between African-American and White men across Gleason scores. Eur Urol. 2019;75(6):1038-1040. [DOI] [PubMed] [Google Scholar]
- 9. Cooperberg MR, Pasta DJ, Elkin EP, et al. The University of California, San Francisco Cancer of the Prostate Risk Assessment score: a straightforward and reliable preoperative predictor of disease recurrence after radical prostatectomy. J Urol. 2005;173(6):1938-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Alexander DH, Novembre J, Lange K.. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19(9):1655-1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang D, Park D, Zhong Y, et al. Stem cell and neurogenic gene-expression profiles link prostate basal cells to aggressive prostate cancer. Nat Commun. 2016;7:10798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Seiler R, Ashab HAD, Erho N, et al. Impact of molecular subtypes in muscle-invasive bladder cancer on predicting response and survival after neoadjuvant chemotherapy. Eur Urol. 2017;72(4):544-554. [DOI] [PubMed] [Google Scholar]
- 13. Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P.. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1(6):417-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Spratt D, Dess R, Hartman H, et al. Androgen receptor activity and radiotherapeutic sensitivity in African-American men with prostate cancer: a large scale gene expression analysis and meta-analysis of RTOG trials. Int J Radiat Oncol Biol Phys. 2018;102(3):S3. [Google Scholar]
- 15. Korpal M, Puyang X, Jeremy Wu Z, et al. Evasion of immunosurveillance by genomic alterations of PPARγ/RXRα in bladder cancer. Nat Commun. 2017;8(1):103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yoshihara K, Shahmoradgoli M, Martínez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ricketts CJ, De Cubas AA, Fan H, et al. ; for the Cancer Genome Atlas Research Network. The Cancer Genome Atlas comprehensive molecular characterization of renal cell carcinoma. Cell Rep. 2018;23(1):313-326.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Charoentong P, Finotello F, Angelova M, et al. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18(1):248-262. [DOI] [PubMed] [Google Scholar]
- 19. Awasthi S, Berglund A, Abraham-Miranda J, et al. Comparative genomics reveals distinct immune-oncologic pathways in African American men with prostate cancer. Clin Cancer Res. 2021;27(1):320-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Echevarria MI, Awasthi S, Cheng CH, et al. African American specific gene panel predictive of poor prostate cancer outcome. J Urol. 2019;202(2):247-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sundi D, Ross AE, Humphreys EB, et al. African American men with very low-risk prostate cancer exhibit adverse oncologic outcomes after radical prostatectomy: should active surveillance still be an option for them? J Clin Oncol. 2013;31(24):2991-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Maurice MJ, Sundi D, Schaeffer EM, Abouassaly R.. Risk of pathological upgrading and up staging among men with low risk prostate cancer varies by race: results from the national cancer database. J Urol. 2017;197(3, pt 1):627-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Falagario UG, Ratnani P, Lantz A, et al. Staging accuracy of multiparametric magnetic resonance imaging in Caucasian and African American men undergoing radical prostatectomy. J Urol. 2020;204(1):82-90. [DOI] [PubMed] [Google Scholar]
- 24. Abashidze N, Stecher C, Rosenkrantz AB, Duszak R Jr, Hughes DR.. Racial and ethnic disparities in the use of prostate magnetic resonance imaging following an elevated prostate-specific antigen test. JAMA Netw Open. 2021;4(11):e2132388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Feng FY, Sandler HM, Huang H-C, et al. Transcriptome profiling of NRG Oncology/RTOG 9601: validation of a prognostic genomic classifier in salvage radiotherapy prostate cancer patients from a prospective randomized trial. J Clin Oncol. 2020;38(suppl 6):276. [Google Scholar]
- 26. Polkinghorn WR, Parker JS, Lee MX, et al. Androgen receptor signaling regulates DNA repair in prostate cancers. Cancer Discov. 2013;3(11):1245-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yamoah K, Johnson MH, Choeurng V, et al. Novel biomarker signature that may predict aggressive disease in African American men with prostate cancer. J Clin Oncol. 2015;33(25):2789-2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhao SG, Lehrer J, Chang SL, et al. The immune landscape of prostate cancer and nomination of PD-L2 as a potential therapeutic target. J Natl Cancer Inst. 2019;111(3):301-310. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data discussed in this work have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO series accession number GSE209954. (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE209954).