Abstract
This study evaluated deep learning algorithms for semantic segmentation and quantification of intracerebral hemorrhage (ICH), perihematomal edema (PHE), and intraventricular hemorrhage (IVH) on noncontrast CT scans of patients with spontaneous ICH. Models were assessed on 1732 annotated baseline noncontrast CT scans obtained from the Tranexamic Acid for Hyperacute Primary Intracerebral Haemorrhage (ie, TICH-2) international multicenter trial (ISRCTN93732214), and different loss functions using a three-dimensional no-new-U-Net (nnU-Net) were examined to address class imbalance (30% of participants with IVH in dataset). On the test cohort (n = 174, 10% of dataset), the top-performing models achieved median Dice similarity coefficients of 0.92 (IQR, 0.89–0.94), 0.66 (0.58–0.71), and 1.00 (0.87–1.00), respectively, for ICH, PHE, and IVH segmentation. U-Net–based networks showed comparable, satisfactory performances on ICH and PHE segmentations (P > .05), but all nnU-Net variants achieved higher accuracy than the Brain Lesion Analysis and Segmentation Tool for CT (BLAST-CT) and DeepLabv3+ for all labels (P < .05). The Focal model showed improved performance in IVH segmentation compared with the Tversky, two-dimensional nnU-Net, U-Net, BLAST-CT, and DeepLabv3+ models (P < .05). Focal achieved concordance values of 0.98, 0.88, and 0.99 for ICH, PHE, and ICH volumes, respectively. The mean volumetric differences between the ground truth and prediction were 0.32 mL (95% CI: −8.35, 9.00), 1.14 mL (−9.53, 11.8), and 0.06 mL (−1.71, 1.84), respectively. In conclusion, U-Net–based networks provide accurate segmentation on CT images of spontaneous ICH, and Focal loss can address class imbalance.
International Clinical Trials Registry Platform (ICTRP) no. ISRCTN93732214
Supplemental material is available for this article.
© RSNA, 2022
Keywords: Head/Neck, Brain/Brain Stem, Hemorrhage, Segmentation, Quantification, Convolutional Neural Network (CNN), Deep Learning Algorithms, Machine Learning Algorithms
Keywords: Head/Neck, Brain/Brain Stem, Hemorrhage, Segmentation, Quantification, Convolutional Neural Network (CNN), Deep Learning Algorithms, Machine Learning Algorithms
Summary
U-Net–based networks accurately segment CT images of spontaneous intracerebral hemorrhage, with Focal loss function being used to address intraventricular hemorrhage class imbalance.
Key Points
■ A comparison of numerous deep learning networks for semantic segmentation of spontaneous intracerebral hemorrhage (ICH) showed that U-Net–based networks achieved significantly better performance than other network architectures for ICH and intraventricular hemorrhage (IVH) segmentations (P < .05).
■ A three-dimensional no-new-U-Net using the Focal loss function was able to address class imbalance in the dataset, providing significant performance improvement (P < .05) for segmentation of IVH present in approximately 30% of the training dataset (Dice score, 1.00 [IQR, 0.87–1.00]).
Introduction
Spontaneous intracerebral hemorrhage (ICH) is bleeding within the brain parenchyma in the absence of trauma or surgery, which may extend into the ventricles and subarachnoid space (1). Volumes of ICH, perihematomal edema (PHE) (2), and intraventricular hemorrhage (IVH) (3) are well-established biomarkers and are consistent independent predictors of the functional outcome and mortality of spontaneous ICH. Manual delineation and quantification of these biomarkers is labor intensive and prone to human error. Thus, an efficient automated biomarker segmentation and quantification tool could provide quantitative outcome measures for clinical trials and accelerate studies in large cohorts of patients with spontaneous ICH.
Previous studies (4–10) have trained deep neural networks to perform ICH segmentation on CT scans, but most of these works were exclusively based on ICH segmentation, PHE segmentation, or both, as accurate delineation of IVH is challenging even for an experienced radiologist (11). Additionally, previous research in this area consists of single-center studies with limited samples (12). Our work assesses the semantic segmentation and quantification of ICH, PHE, and IVH from a large multicenter dataset (from the Tranexamic Acid for Hyperacute Primary Intracerebral Haemorrhage [TICH-2] trial [13]).
This study compares the performance of existing deep learning approaches in the semantic segmentation and quantification of ICH, PHE, and IVH. The best existing deep learning model was then refined by using different loss functions to address the class imbalance issue (unequal distribution of the lesion classes with few or no PHE or IVH pixels in a scan).
Materials and Methods
Study Patients
This retrospective analysis included baseline noncontrast CT scans from participants recruited to the prospective TICH-2 international, randomized, placebo-controlled clinical trial (ISRCTN93732214) (13,14). The trial examined the effectiveness and safety of tranexamic acid in participants with acute spontaneous ICH within 8 hours of the onset of stroke symptoms. Ethical approval was granted from the UK Health Research Authority and the relevant national or local institutional review boards (for sites not located in the United Kingdom), and written informed consent from participants or one of their relatives was obtained before enrollment. The full trial protocol is reported elsewhere (15). Our analysis included 1732 eligible patients from the previously reported cohort (16) who had valid baseline scans (ie, no incomplete or missing sections). The previous work investigated radiomics-based features, whereas this study focuses on lesion segmentation using deep learning.
Image Acquisition and Ground Truth Delineation
The noncontrast CT baseline scans were collected from 124 participating centers while complying with the local protocol. With a minimum requirement of axial image orientation, CT scans acquired by using any scanner manufacturer, settings, or section thickness were included.
The anonymized ground truth segmentations of ICH, PHE, and IVH were delineated on each scan by one of three independent trained raters (Z.K.L., vascular neurologist, with 15 years of experience; K.K., stroke physician, with 22 years of experience; A.A., CT radiographer, with 14 years of experience) by using an active-contour, semiautomated segmentation algorithm on ITK-SNAP (version 3.6.0) (17), followed by manual editing if required. Additional inter- and intraobserver details are described in Appendix E1 (supplement).
The dataset was randomly split into a training cohort (n = 1558, 90%; mean age ± SD, 69 years ± 13; 872 men) and testing cohort (n = 174, 10%; mean age ± SD, 68 years ± 14; 102 men).
Deep Neural Network Selection for Comparison
We searched the best and most relevant neural networks for brain hemorrhage segmentation from Google Scholar and PubMed and shortlisted three approaches: (a) no-new-U-Net (nnU-Net) (18), which is an automated configuration method with state-of-the-art performance in many segmentation challenges, including the Medical Segmentation Decathlon, Brain Tumor Segmentation challenge, and Kidney Tumor Segmentation challenge; (b) Brain Lesion Analysis and Segmentation Tool for CT (BLAST-CT) (19), a pipeline based on DeepMedic and a tool with top performances in the Ischemic Stroke Lesion Segmentation challenge and Brain Tumor Segmentation challenge; and (c) DeepLabv3+ (20), which is an approach that ranks highly in the semantic segmentation of general objects and can outperform notable networks such as Fully Convolutional Network, SegNet, and U-Net in the segmentation of biomedical images.
Finally, U-Net, a widely used network for general medical image segmentation, was selected as the baseline.
Network Implementation
We trained each model for 1800 epochs by using the model pipeline default parameters and tested them on the independent test cohort. Both standard U-Net and DeepLabv3+ models were implemented by using the Medical Open Network for Artificial Intelligence framework (https://github.com/Project-MONAI/MONAI). We used the Medical Open Network for Artificial Intelligence framework’s built-in, three-dimensional BasicUNet and implemented the source code for the DeepLabv3+ model (https://github.com/janetkok/MONAI-DeepLabV3plus). The open-source frameworks nnU-Net and BLAST-CT can be found online (https://github.com/MIC-DKFZ/nnUNet and https://github.com/biomedia-mira/blast-ct, respectively). General information and implementation details of these frameworks are described in Appendix E1 and Table E1 (supplement), respectively.
Refinement of the Best Existing Model through Loss Functions
We assumed that the default loss function in the best existing model—Dice and Cross-entropy (DiceCE) (Table 1)—would not be sufficiently sensitive to handle the extremely imbalanced target segmentation, low contrast, and heterogeneous appearances of PHE and IVH lesions. Inspired by previous work (21), we evaluated Tversky, Focal, FocalTversky, and DiceTopK loss using a three-dimensional nnU-Net to address the current model’s limitations (code can be found at https://github.com/JunMa11/SegLoss.git [22]). These loss functions were selected on the basis of their inherent capability to handle the class imbalance issue (see work by Ma et al [22] for a full description of loss functions).
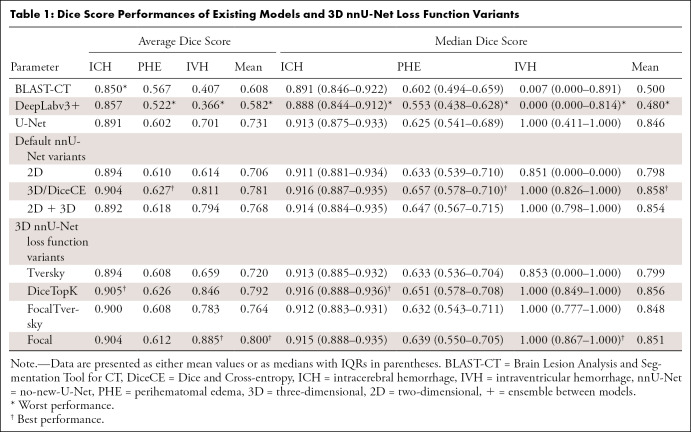
Table 1:
Dice Score Performances of Existing Models and 3D nnU-Net Loss Function Variants
Performance Measures
Quantitative performance for determining the lesion volume was measured by using automated versus human concordance and Bland-Altman plots. The accuracy overlay between the ground truth and predicted lesion was quantified by using the Dice similarity coefficient (DSC).
Statistical Analysis
Patient demographics were compared between the training and test samples by using the χ2 test or independent t test. The models’ performances were statistically compared by using the Kruskal-Wallis tests and corresponding Dunn post hoc tests with false discovery rate correction. All statistical analyses were performed by using RStudio (version 1.4.1103), and a P value less than .05 was considered to indicate significance.
Results
Patient Characteristics
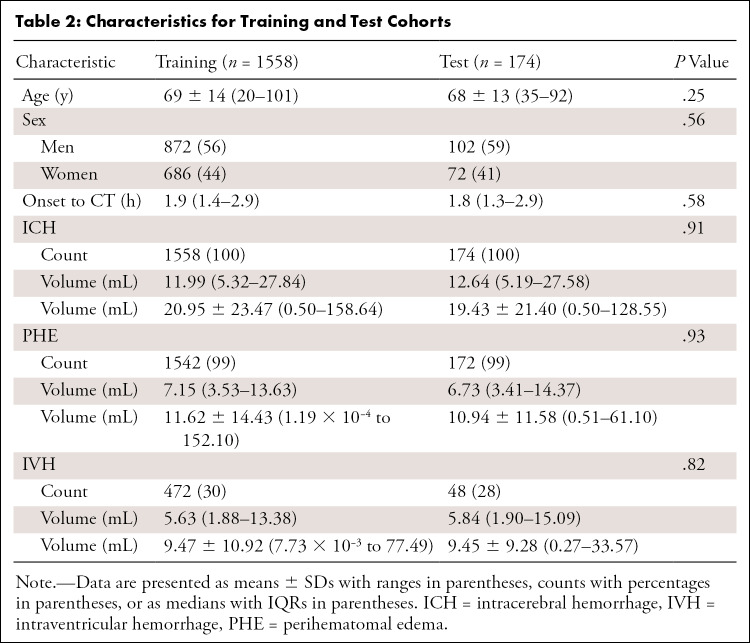
Of the 1732 included patients, we found no evidence of differences in characteristics between the training set and test set (Table 2).
Table 2:
Characteristics for Training and Test Cohorts
Lesion Segmentation Performance
Note that all three-dimensional nnU-Net loss function variants are represented as the name of their loss functions. See Appendix E1 (supplement).
Table 1 shows the DSC of various models in our experiment. Box-violin plots showing the DSC distribution of various models are presented in Figure E1 (supplement). Figures E2 and E3 (supplement) present a qualitative overview of the segmentation (best- and worst-case segmentation with respect to DiceCE) for the top-performing models: DiceCE, DiceTopK, and Focal.
U-Net–based networks achieved similarly good performance in ICH and PHE segmentations based on the average and median schemes, showing no evidence of differences between them (P > .05; Table 1). Compared with the lowest performers, BLAST-CT and DeepLabv3+, all nnU-Net variants demonstrated higher DSCs for all lesion segmentation (P < .05). The segmentation quality of ICH was satisfactory across all models (Fig E2 [supplement]), whereas PHE segmentation was not desirable, as the boundaries of the PHE label appeared to be smoothed (Fig E2B [supplement]), indicating a lack of precision.
The nnU-Net variations—DiceCE, ensemble of two-dimensional and three-dimensional nnU-Net (2D + 3D), DiceTopK, and Focal—showed significant performance improvement in IVH segmentation compared with Tversky, two-dimensional nnU-Net (2D), U-Net, BLAST-CT, and DeepLabv3+ models (P < .05). Given that the top-performing models for IVH segmentation, particularly Focal, DiceTopK, and FocalTversky, are mainly designed with an emphasis on hard samples to address high class imbalance, there was no evidence of statistical differences between the aforementioned models. Still, Focal demonstrated a notably higher average DSC and more consistent performance based on the small IQR in IVH segmentation (Table 1). In addition, based on the qualitative performance shown in Figures E2A–E2C and E3C (supplement) (note the lack of blue labels when using DiceCE and DiceTopK), Focal presented greater detection capability for small and low-contrast IVH lesions than the other top-performing models, such as DiceCE and DiceTopK. Furthermore, we observed that the DSC values of most low-performing networks in IVH segmentation have a bimodal distribution (Fig E1C [supplement]). This suggests that their segmentation results are polarized, presumably caused by differences in lesion size and intensity.
Volume Quantification and Agreement
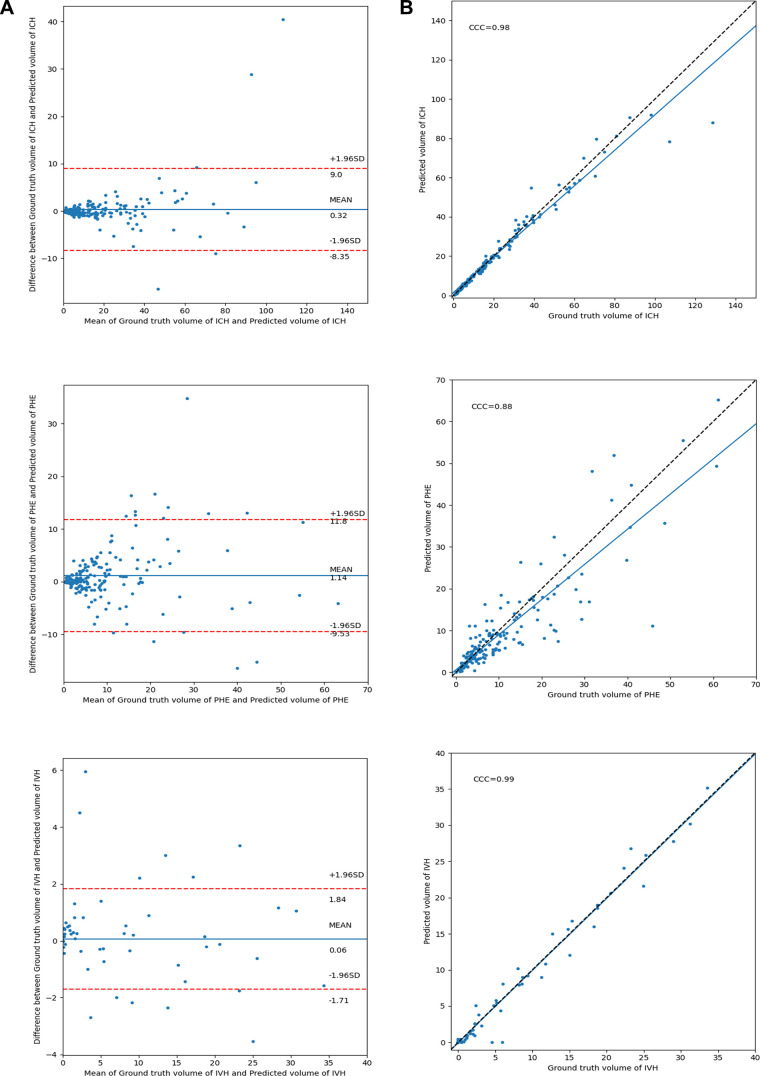
The Figure shows the concordance and Bland-Altman plot of agreements between the ground truth and predicted lesion volumes by Focal, the best overall network (based on the global mean of DSCs). Focal demonstrated high concordance and low mean difference in estimating all lesions except PHE. Additional details are presented in Appendix E1 (supplement).
(A) Bland-Altman and (B) concordance plot of agreements between ground truth and predicted lesion volumes in the test cohort by using the Focal model. CCC = concordance correlation coefficient, ICH = intracerebral hemorrhage, IVH = intraventricular hemorrhage, PHE = perihematomal edema.
Discussion
We compared the segmentation performance of existing deep learning networks on a large dataset from the TICH-2 trial. The analysis demonstrated that U-Net–based networks have immense potential in segmenting targeted lesions. We investigated how the application of a selected range of loss functions could be a feasible technique to address the issue of class imbalance. We showed that Focal can address this problem and significantly improve IVH segmentation.
Based on the median DSC of ICH, three-dimensional nnU-Net variant performance was similar to that found in the single-center study by Zhao et al (12) and in the multicenter study by Sharrock et al (23). Our reported average DSC for the best ICH segmentation was lower than that previously found with Ψ-Net (0.95) (24), but it should be noted that this prior work had lower CT variability and a smaller dataset.
Regarding PHE segmentation, studies by Ironside et al (10) and Zhao et al (12) reported higher performance levels than those shown in our results. Nevertheless, our performance results remain adequate considering the variability of our multicenter dataset and smaller PHE volume (larger lesion volumes are positively correlated with higher DSC [12,19]).
The best model for IVH segmentation (Focal) outperformed that in the single-center study by Zhao et al (12). Of note, our initial assumption that PHE segmentation performance would be superior to that of IVH did not hold, albeit PHE was much larger in volume and was found in 99% of the training dataset. A possible explanation is that PHE demands the resolution of indistinct, low-contrast edges, whereas IVH typically shows up as areas of high attenuation and often has sharp edges where the IVH contacts the ventricle wall.
This study had limitations. First, a scan not containing a lesion class but was mislabeled by the model had a DSC of 0. In the same setting, but with no error made, the DSC was counted as 1 to include patient scans with no target lesions that were correctly predicted. Thus, the metric tends to favor nonexistent lesions and can result in misrepresentation of model performance. To address this issue, we included two other metrics, volume intersection and false-positive results, in Table E2 (supplement) for better performance comparison. Additionally, we are aware that we only performed a single split, and the performance evaluation could have been strengthened by using k-fold cross-validation. In addition, we acknowledge that our ground truth segmentations contained segmentation errors in a few cases, as supported by the high but imperfect rates of inter- and intrarater agreement. Inspection of the “worst-case” segmentation in Figure E3 (supplement) shows that, in retrospect, network-based segmentations were more accurate than the manually edited, semiautomated segmentations. That being said, the study would be enhanced if a radiologist had been among the reviewers.
In summary, we compared numerous deep learning approaches for the segmentation and quantification of ICH, PHE, and IVH in spontaneous ICH from a large-scale, international, multicenter dataset. We showed that U-Net–based networks remain robust in medical imaging segmentation, demonstrating similarly high performances for both ICH and PHE lesions. We also investigated a selected range of loss functions on the three-dimensional nnU-Net, but none of the networks had the best result in every lesion; however, Focal can address class imbalance and showed greater detection capability with significant performance improvement in IVH segmentation, a prominent yet rarely investigated lesion because of its complexity and scarcity. We believe that the future development of a fully accurate and automated deep learning–based segmentation model could potentially eliminate human error in manual segmentation and provide early prediction of hematoma expansion and clinical outcome when combined with quantitative radiomic analysis (16).
Acknowledgments
Acknowledgments
We thank all the participants, staff, and centers who took part in the Tranexamic Acid for Intracerebral Haemorrhage (TICH-2) trial.
This study included data from participants recruited to the Tranexamic Acid for Intracerebral Haemorrhage (TICH-2) international randomized, placebo-controlled clinical trial (ISRCTN93732214). The TICH-2 trial was funded in part by a grant from the UK National Institute for Health Research Health Technology Assessment Programme (project code 11_129_109).
Data sharing: Data generated or analyzed during the study are available upon written request to the TICH-2 trial CI Nikola Sprigg (Nikola.Sprigg@nottingham.ac.uk). Proposals will be assessed by the CI (with advice from the TICH-2 trial Steering Committee if required), and a Data Transfer Agreement will be established before any data are shared.
Disclosures of conflicts of interest: Y.E.K. No relevant relationships. S.P. No relevant relationships. Z.K.L. Research grants from National University of Malaysia; speakers honoraria from Bayer and Boehringer Ingelheim; support to attend meetings from Bayer and Boehringer Ingelheim; member of World Stroke Organisation Future Stroke Leaders Programme and an executive committee member of the Malaysian Stroke Council. A.A. No relevant relationships. K.K. No relevant relationships. P.M.B. NIHR HTA funding to institution; funding for SAVANNAS study from British Heart Foundation; funding for PhEAST trial from NIHR HTA; consulting fees/advisory committee from DiaMedica, Moleac, Phagenesis, and Roche; Chair Data Safety Monitoring Board for AVERT trial and ECST-2 trial; co-chair for Industry Committee World Stroke Organisation; stock/stock options in DiaMedica. N.S. No relevant relationships. R.A.D. Funding for the TICH-2 Trial (grant reference 11/129/109) from United Kingdom National Institute of Health and Care Research (NIHR), Health Technology Assessment (HTA) Programme. A.P.F. No relevant relationships.
Abbreviations:
- BLAST-CT
- Brain Lesion Analysis and Segmentation Tool for CT
- DiceCE
- Dice and Cross-entropy
- DSC
- Dice similarity coefficient
- ICH
- intracerebral hemorrhage
- IVH
- intraventricular hemorrhage
- nnU-Net
- no-new-U-Net
- PHE
- perihematomal edema
- TICH-2
- Tranexamic Acid for Hyperacute Primary Intracerebral Haemorrhage
References
- 1. Aguilar MI , Freeman WD . Spontaneous intracerebral hemorrhage . Semin Neurol 2010. ; 30 ( 5 ): 555 – 564 . [DOI] [PubMed] [Google Scholar]
- 2. Appelboom G , Bruce SS , Hickman ZL , et al. Volume-dependent effect of perihaematomal oedema on outcome for spontaneous intracerebral haemorrhages . J Neurol Neurosurg Psychiatry 2013. ; 84 ( 5 ): 488 – 493 . [DOI] [PubMed] [Google Scholar]
- 3. Trifan G , Arshi B , Testai FD . Intraventricular hemorrhage severity as a predictor of outcome in intracerebral hemorrhage . Front Neurol 2019. ; 10 : 217 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rajat D , Falcone GJ , Chen Y , et al. Deep learning for automated measurement of hemorrhage and perihematomal edema in supratentorial intracerebral hemorrhage . Stroke 2020. ; 51 ( 2 ): 648 – 651 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Islam M , Sanghani P , See AAQ , James ML , King NKK , Ren H . ICHNet: intracerebral hemorrhage (ICH) segmentation using deep learning . In: Crimi A , Bakas S , Kuijf H , Keyvan F , Reyes M , van Walsum T , eds . Brainlesion: glioma, multiple sclerosis, stroke and traumatic brain injuries. Lecture Notes in Computer Science, Vol 11383 . Cham, Switzerland: : Springer; , 2019. ; 456 – 463 . [Google Scholar]
- 6. Chang PD , Kuoy E , Grinband J , et al. Hybrid 3D/2D convolutional neural network for hemorrhage evaluation on head CT . AJNR Am J Neuroradiol 2018. ; 39 ( 9 ): 1609 – 1616 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Guo D , Wei H , Zhao P , et al. Simultaneous classification and segmentation of intracranial hemorrhage using a fully convolutional neural network . In: 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI) . Piscataway, NJ: : Institute of Electrical and Electronics Engineers; , 2020. ; 118 – 121 . [Google Scholar]
- 8. Hu K , Chen K , He X , et al. Automatic segmentation of intracerebral hemorrhage in CT images using encoder–decoder convolutional neural network . Inf Process Manage 2020. ; 57 ( 6 ): 102352 . [Google Scholar]
- 9. Ironside N , Chen CJ , Mutasa S , et al. Fully automated segmentation algorithm for hematoma volumetric analysis in spontaneous intracerebral hemorrhage . Stroke 2019. ; 50 ( 12 ): 3416 – 3423 . [DOI] [PubMed] [Google Scholar]
- 10. Ironside N , Chen CJ , Mutasa S , et al. Fully automated segmentation algorithm for perihematomal edema volumetry after spontaneous intracerebral hemorrhage . Stroke 2020. ; 51 ( 3 ): 815 – 823 . [DOI] [PubMed] [Google Scholar]
- 11. Dowlatshahi D , Kosior JC , Idris S , et al. Planimetric hematoma measurement in patients with intraventricular hemorrhage: is total volume a preferred target for reliable analysis? Stroke 2012. ; 43 ( 7 ): 1961 – 1963 . [DOI] [PubMed] [Google Scholar]
- 12. Zhao X , Chen K , Wu G , et al. Deep learning shows good reliability for automatic segmentation and volume measurement of brain hemorrhage, intraventricular extension, and peripheral edema . Eur Radiol 2021. ; 31 ( 7 ): 5012 – 5020 . [DOI] [PubMed] [Google Scholar]
- 13. Sprigg N , Flaherty K , Appleton JP , et al. Tranexamic acid for hyperacute primary IntraCerebral Haemorrhage (TICH-2): an international randomised, placebo-controlled, phase 3 superiority trial . Lancet 2018. ; 391 ( 10135 ): 2107 – 2115 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sprigg N , Flaherty K , Appleton JP , et al. Tranexamic acid to improve functional status in adults with spontaneous intracerebral haemorrhage: the TICH-2 RCT . Health Technol Assess 2019. ; 23 ( 35 ): 1 – 48 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sprigg N , Robson K , Bath P , et al. Intravenous tranexamic acid for hyperacute primary intracerebral hemorrhage: protocol for a randomized, placebo-controlled trial . Int J Stroke 2016. ; 11 ( 6 ): 683 – 694 . [DOI] [PubMed] [Google Scholar]
- 16. Pszczolkowski S , Manzano-Patrón JP , Law ZK , et al. Quantitative CT radiomics-based models for prediction of haematoma expansion and poor functional outcome in primary intracerebral haemorrhage . Eur Radiol 2021. ; 31 ( 10 ): 7945 – 7959 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yushkevich PA , Piven J , Hazlett HC , et al. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability . Neuroimage 2006. ; 31 ( 3 ): 1116 – 1128 . [DOI] [PubMed] [Google Scholar]
- 18. Isensee F , Jaeger PF , Kohl SAA , Petersen J , Maier-Hein KH . nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation . Nat Methods 2021. ; 18 ( 2 ): 203 – 211 . [DOI] [PubMed] [Google Scholar]
- 19. Monteiro M , Newcombe VFJ , Mathieu F , et al. Multiclass semantic segmentation and quantification of traumatic brain injury lesions on head CT using deep learning: an algorithm development and multicentre validation study . Lancet Digit Health 2020. ; 2 ( 6 ): e314 – e322 . [DOI] [PubMed] [Google Scholar]
- 20. Chen LC , Zhu Y , Papandreou G , Schroff F , Adam H . Encoder-decoder with atrous separable convolution for semantic image segmentation . ArXiv 1802.02611 [preprint] https://arxiv.org/abs/1802.02611. Posted February 7, 2018. Accessed November 29, 2020 .
- 21. Ma J . Loss ensembles for extremely imbalanced segmentation . ArXiv 2101.10815 [preprint] https://arxiv.org/abs/2101.10815. Posted December 31, 2020. Accessed March 7, 2021 .
- 22. Ma J , Chen J , Ng M , et al. Loss odyssey in medical image segmentation . Med Image Anal 2021. ; 71 : 102035 . [DOI] [PubMed] [Google Scholar]
- 23. Sharrock MF , Mould WA , Ali H , et al. 3D deep neural network segmentation of intracerebral hemorrhage: development and validation for clinical trials . Neuroinformatics 2021. ; 19 ( 3 ): 403 – 415 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kuang Z , Deng X , Yu L , Wang H , Li T , Wang S . Ψ-Net: focusing on the border areas of intracerebral hemorrhage on CT images . Comput Methods Programs Biomed 2020. ; 194 : 105546 . [DOI] [PubMed] [Google Scholar]