Abstract
Accurate differentiation of intramedullary spinal cord tumors and inflammatory demyelinating lesions and their subtypes are warranted because of their overlapping characteristics at MRI but with different treatments and prognosis. The authors aimed to develop a pipeline for spinal cord lesion segmentation and classification using two-dimensional MultiResUNet and DenseNet121 networks based on T2-weighted images. A retrospective cohort of 490 patients (118 patients with astrocytoma, 130 with ependymoma, 101 with multiple sclerosis [MS], and 141 with neuromyelitis optica spectrum disorders [NMOSD]) was used for model development, and a prospective cohort of 157 patients (34 patients with astrocytoma, 45 with ependymoma, 33 with MS, and 45 with NMOSD) was used for model testing. In the test cohort, the model achieved Dice scores of 0.77, 0.80, 0.50, and 0.58 for segmentation of astrocytoma, ependymoma, MS, and NMOSD, respectively, against manual labeling. Accuracies of 96% (area under the receiver operating characteristic curve [AUC], 0.99), 82% (AUC, 0.90), and 79% (AUC, 0.85) were achieved for the classifications of tumor versus demyelinating lesion, astrocytoma versus ependymoma, and MS versus NMOSD, respectively. In a subset of radiologically difficult cases, the classifier showed an accuracy of 79%–95% (AUC, 0.78–0.97). The established deep learning pipeline for segmentation and classification of spinal cord lesions can support an accurate radiologic diagnosis.
Supplemental material is available for this article.
© RSNA, 2022
Keywords: Spinal Cord MRI, Astrocytoma, Ependymoma, Multiple Sclerosis, Neuromyelitis Optica Spectrum Disorder, Deep Learning
Keywords: Spinal Cord MRI, Astrocytoma, Ependymoma, Multiple Sclerosis, Neuromyelitis Optica Spectrum Disorder, Deep Learning
Summary
A deep learning pipeline for segmentation and classification of spinal cord lesions using T2-weighted MR images was established to support an accurate radiologic diagnosis, and it sometimes outperformed experienced neuroradiologists.
Key Points
■ The model achieved Dice scores of 0.77, 0.80, 0.50, and 0.58 for model segmentation of astrocytoma, ependymoma, multiple sclerosis (MS), and neuromyelitis optica spectrum disorders (NMOSD), respectively, against manual labeling.
■ The model achieved accuracies of 96%, 82%, and 79% for classification of tumor versus demyelinating lesion, astrocytoma versus ependymoma, and MS versus NMOSD, respectively.
■ For cases with disagreement in diagnoses by neuroradiologists, an accuracy of 79%–95% was still achieved by the classifier.
Introduction
Intramedullary spinal cord tumors and inflammatory demyelinating lesions share several MRI characteristics (eg, localization, shape, signal intensity, and contrast enhancement) (1–3), which poses a clinical challenge for accurate diagnosis. It is essential to accurately differentiate spinal cord tumors, including astrocytomas and ependymomas, from demyelinating lesions such as multiple sclerosis (MS) and neuromyelitis optica spectrum disorders (NMOSD) and to accurately classify these subtypes because they imply fundamentally different treatments and prognoses.
Substantial progress has been made in applying deep learning (DL) to diagnose brain disorders (4–6), but only a few DL studies have focused on spinal cord diseases (7,8). The limited evidence to date suggests that DL can be used to characterize and segment spinal cord tumors or demyelinating lesions (7,8), but to our knowledge no study has addressed the differential diagnosis of these lesions or their subtypes. Whereas automated pipelines for clinical diagnosis integrating lesion segmentation and differential diagnosis by DL have been reported for supratentorial lesions (eg, gliomas and white matter hyperintensities) (5,6,9), they have not yet been reported for intramedullary spinal cord lesions.
The aim of our study was to develop a DL pipeline for assisting clinical diagnosis by integrating the segmentation and classification of spinal cord tumors (astrocytoma and ependymoma) versus inflammatory demyelinating lesions (MS and NMOSD) and their subtypes. We hypothesized that such a DL pipeline could be achieved by using MRI, and therefore we conducted this study using T2-weighted images. We deliberately chose basic T2-weighted images because they are generally clinically available.
Materials and Methods
Authors who are not employees of or consultants to BioMind, Neusoft Medical Systems, Bayer Schering, Biogen, GeNeuro, Ixico, Merck Serono, Novartis, or Roche had control of image and clinical data that might present a conflict of interest for authors Z.L., X. Guo, X. Gong, and F.B.
Our study was performed in accordance with the Declaration of Helsinki and was approved by the animal and human ethics committee of the local institution. Written informed consent was obtained from all patients.
Patients
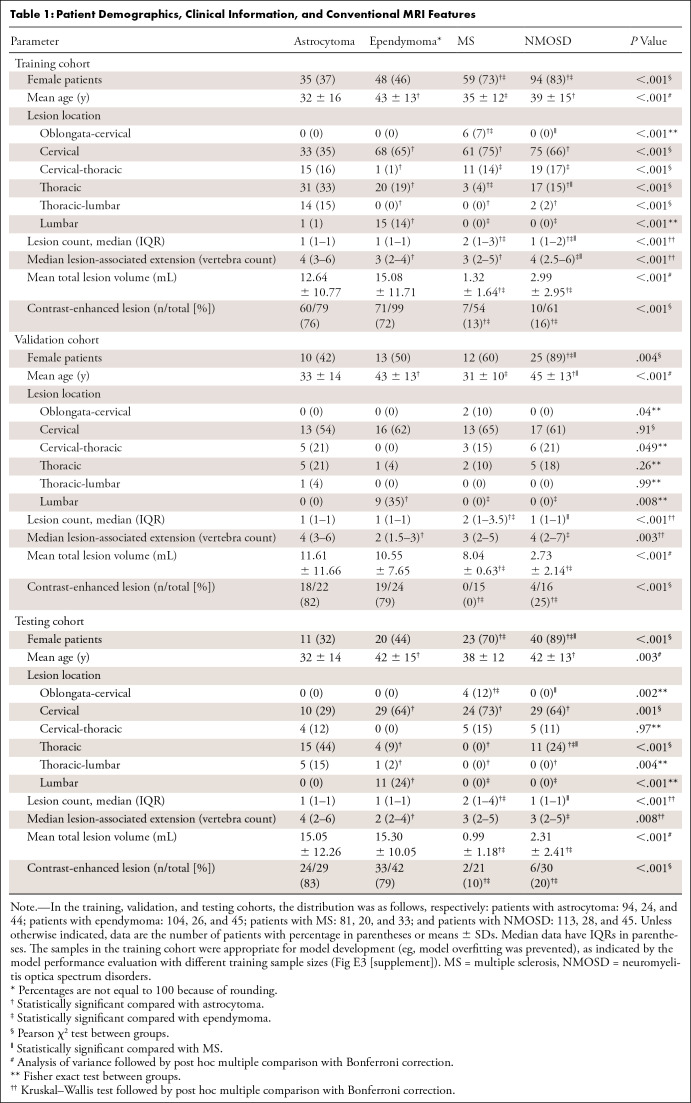
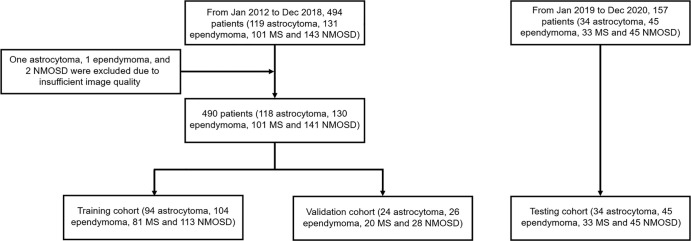
We retrospectively reviewed data collected from January 2012 to December 2018 and identified 494 patients (119 patients with astrocytoma, 131 with ependymoma, 101 with MS, and 143 with NMOSD) based on their first clinical diagnosis and before clinical treatment to train (n = 392; 80%) and validate (n = 98; 20%) the segmentation and classification models (Table 1, Appendix E1 [supplement]). Four patients (astrocytoma [n = 1], ependymoma [n = 1], and NMOSD [n = 2]) were excluded because of insufficient image quality, resulting in 490 patients (118 patients with astrocytoma, 130 with ependymoma, 101 with MS, and 141 with NMOSD) in the training and validation sets. For independent testing, 157 patients (34 astrocytoma, 45 ependymoma, 33 MS, and 45 NMOSD) were prospectively and consecutively enrolled from January 2019 to December 2020 (Table 1, Appendix E1 [supplement]). Inclusion and exclusion criteria are in Figure 1 and Appendix E1 (supplement).
Table 1:
Patient Demographics, Clinical Information, and Conventional MRI Features
Figure 1:
Flowcharts of patient selection for (left) training and validation and (right) testing. MS = multiple sclerosis, NMOSD = neuromyelitis optica spectrum disorders.
DL and Model Development
For integrated segmentation and classification of spinal cord tumors versus demyelinating lesions and their subtypes, three two-classification models were developed using two-dimensional MultiResUNet (10,11) and DenseNet121 networks (12): tumor versus demyelinating lesion (model 1), astrocytoma versus ependymoma (model 2), and MS versus NMOSD (model 3) based on sagittal T2-weighted images (Table E1, Appendix E1 [supplement]). Details of image preparation for DL and model development are presented in the methods section of Appendix E1 (supplement). A pipeline including segmentation and classification of spinal cord lesions is shown in Figure 2A.
Figure 2:
(A) Deep learning (DL) pipeline established for segmentation and classification of spinal cord lesions. First, the noncontrast T2-weighted (T2w) sagittal MR images (all sections; sections 1–11 herein) were used to segment the lesion, and manual verification was conducted to correct the poorly segmented lesions. Second, the sections (eg, sections 5–8) of the T2-weighted images involving lesions and lesion masks were used as the network input for classification tasks. (B) Representative cases of segmentation and classification for spinal cord tumors and demyelinating lesions in the test cohort. Diagnosis by DL and four raters (D.C. [rater 1], X.X. [rater 2], C.F. [rater 3], X.H. [rater 4]) are shown. Red areas indicate the DL segmentation, green areas indicate manual segmentation, and yellow areas indicate overlap. MS = multiple sclerosis, NMOSD = neuromyelitis optica spectrum disorders.
Data Collection
Radiologic assessments including lesion characteristics, manual lesion segmentation, and classification were performed by neuroradiologists with different levels of experience in neuroradiology (D.C. and X.X., with 2 years of experience; J.Z. and L.Q., with 3 years of experience; X.H. and C.F., with 11 years of experience; and Y.D., with 12 years of experience; Appendix E1, Table E2 [supplement]) with reference to the presentations of other available imaging sequences (eg, T1-weighted, contrast-enhanced T1-weighted, and axial T2-weighted images). Difficult cases were those with disagreement by the neuroradiologists regarding the most likely diagnoses (Appendix E1 [supplement]).
Statistical Analysis
Statistical analyses of demographics, clinical variables, and MRI variables (Appendix E1 [supplement]) were performed using software (SPSS version 22; IBM). A two-sided P value of less than .05 was considered to indicate statistical significance.
The Dice score was used to evaluate segmentation performance. Accuracy, sensitivity, specificity, positive predictive value, negative predictive value, precision, recall, and area under the receiver operating characteristic curve (AUC) were calculated to evaluate classification performance. Additionally, we conducted sensitivity analyses by using gradient-weighted class activation mapping (Grad-CAM), subgroup analyses according to patient age and sex, and additional combinations with available contrast-enhanced T1-weighted images (Appendix E1 [supplement]).
Data Availability
We made our code publicly available. It is available at https://github.com/Leezhaohui/spinalcord_classification.
Results
DL Segmentation of Spinal Cord Tumors and Demyelinating Lesions
In the independent test cohort, the mean Dice scores were 0.77, 0.80, 0.50, and 0.58 for astrocytoma, ependymoma, MS, and NMOSD, respectively (Table E3 [supplement]). Representative cases are shown in Figure 2B. A subset of DL segmentations (five tumors and 16 demyelinating lesions in the validation cohort; seven tumors and 23 demyelinating lesions in the test cohort) needed further manual review and correction (Appendix E1 [supplement]).
DL Classification of Spinal Cord Tumors and Demyelinating Lesions
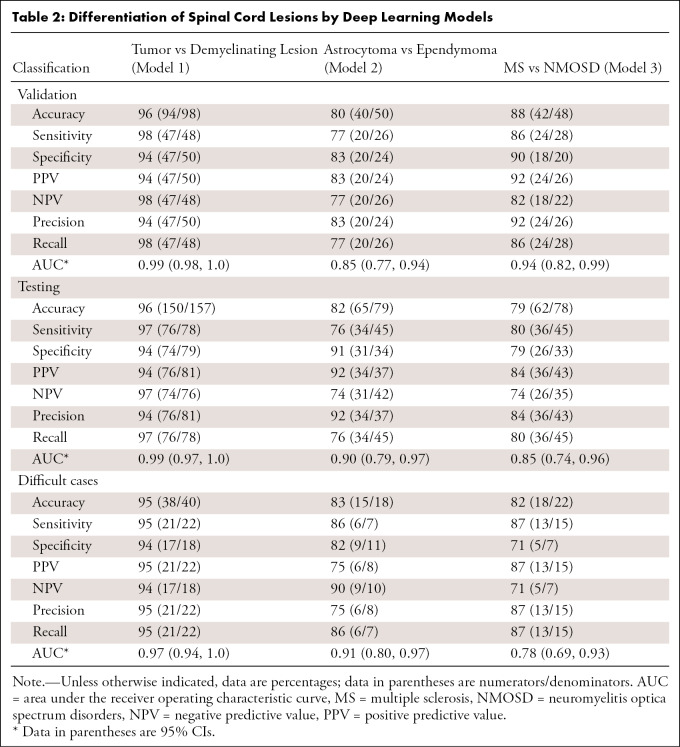
Based on model 1, accuracy of 96% (150 of 157), sensitivity of 97% (76 of 78), specificity of 94% (74 of 79), and AUC of 0.99 (95% CI: 0.97, 1.0) were achieved on the independent test cohort for the classification of tumor versus demyelination (Table 2; Fig E1, Fig E2 [supplement]), which was comparable to neuroradiologist performance (accuracy, 97% [152 of 157]; Appendix E1, Table E2 [supplement]). For classification of difficult cases, the model achieved an accuracy of 95% (38 of 40), sensitivity of 95% (21 of 22), and specificity of 94% (17 of 18).
Table 2:
Differentiation of Spinal Cord Lesions by Deep Learning Models
Based on model 2, accuracy of 82% (65 of 79), sensitivity of 76% (34 of 45), specificity of 91% (31 of 34), and AUC of 0.90 (95% CI: 0.79, 0.97) were achieved on the independent test cohort for the classification of astrocytoma versus ependymoma, which was superior to neuroradiologist performance (accuracy, 72% [57 of 79]). This performance was maintained for difficult cases, in which an accuracy of 83% (15 of 18), sensitivity of 86% (six of seven), and specificity of 82% (nine of 11) were achieved.
Based on model 3, accuracy of 79% (62 of 78), sensitivity of 80% (36 of 45), specificity of 79% (26 of 33), and AUC of 0.85 (95% CI: 0.74, 0.96) were achieved on the independent test cohort for the classification of MS and NMOSD lesions, which was superior to neuroradiologist performance (accuracy, 67%; 52 of 78). This performance was maintained for difficult cases, in which an accuracy of 82% (18 of 22), sensitivity of 87% (13 of 15), and specificity of 71% (five of seven) were achieved.
Sensitivity Analyses
The Grad-CAM showed that the main activation areas were the lesion and perilesional areas in patients with tumors or demyelinating lesions (Appendix E1, Table E4 [supplement]). Model performance degraded only in the pediatric and male subgroups, with lower sensitivity in the classification of astrocytoma versus ependymoma (Table E5 [supplement]). The contrast-enhanced T1-weighted images had no additional contribution to whole-lesion segmentation and improved only the classification accuracy of MS versus NMOSD (Table E6 [supplement]).
Discussion
In this study, a DL pipeline for spinal cord lesion segmentation and classification was developed using T2-weighted images, the most widely available type of MR images. In the test cohort, the model achieved Dice scores of 0.77, 0.80, 0.50, and 0.58 for segmentation of astrocytoma, ependymoma, MS, and NMOSD, respectively, against manual labeling. Accuracy of 96%, 82%, and 79% was achieved for the classifications of tumor versus demyelinating lesion, astrocytoma versus ependymoma, and MS versus NMOSD, respectively, which was comparable to or better than that achieved by neuroradiologists (accuracy of 97%, 72%, and 67%, respectively). In a subset of radiologically difficult cases, an accuracy of 79%–95% was achieved by the classifier. This DL pipeline could both benefit patients without available contrast-enhanced T1-weighted images and facilitate fast clinical translation with robust performance across different subpopulations. For the differentiation of demyelinating lesions, contrast-enhanced T1-weighted imaging is recommended to achieve higher classification performance (accuracy, 90%).
Few studies have focused on spinal cord lesion segmentation by using DL (8). Spinal cord tumor segmentation benefits from a relatively high tumor intensity compared with surrounding normal spinal cord tissue (13). Our DL model showed promising segmentation performance comparable to that of a previous study in which a Dice score of 0.77 was reported (8). For demyelinating lesions, DL segmentation achieved a slightly lower performance (even combining contrast-enhanced T1-weighted images) because of the smaller volume of disseminated lesions and lower contrast enhancement in the lesion and surrounding tissue (7); these factors also pose a challenge in manual delineation. Although the current automatic segmentation of demyelinating lesions requires manual review and frequent modification, it may still aid efficient lesion segmentation.
Our study classifies spinal cord tumors and demyelinates lesions and their subtypes, a clinically relevant and sometimes challenging task, using DL. Our model showed an excellent differentiation of spinal cord tumors versus demyelinating lesions using only T2-weighted images, acquired by using standard of care, without the need for advanced MRI sequences or other modalities, which benefits a fast and general clinical application. The model performance was comparable to that of neuroradiologists. Our model may benefit from the different intensity contrast and morphologic characteristics (eg, orientation, shape, size, and count, as shown in Grad-CAM) (14,15). In addition, cysts, necrosis, cavities, and hemorrhages, which are specific to tumors and typically are absent in demyelinating lesions, may also contribute to the final classification (1,14–16). Even though the differentiation of brain tumors has been widely reported in previous studies, with accuracies greater than 80% (5,17), studies on the differentiation of spinal cord tumors are lacking. The differentiation within spinal cord tumors using DL in the current study was superior to neuroradiologists’ diagnostic performance and was comparable to that reported in previous brain tumor studies (5,17). The accuracy of the differentiation of demyelinating lesions (MS vs NMOSD) using DL was higher than that of neuroradiologists, of which performance could be further improved by combining contrast-enhanced T1-weighted images. The contribution of the entire demyelination lesion and/or lesion central area and perilesional areas along the lesion margin revealed by Grad-CAM indicated potential distinct underlying pathologic causes, which has potential value for radiologic diagnosis. Good to excellent performance was achieved using DL for clinically difficult cases (ie, conflicting diagnoses from neuroradiologists), which suggests that it may be useful in clinically difficult spinal cord cases.
Our study had some limitations. First, only spinal cord T2-weighted images were used. Use of multimodal spinal cord MRI scans and available brain MRI scans, which would provide complementary profiles, could be considered in further studies. An additionally performed analysis comparing the performance of radiologist assessment of T2-weighted images only versus all available MRI sequences found no relevant difference in accuracy (Table E2, Appendix E1 [supplement]). Second, lesion segmentation by DL may have been suboptimal, particularly for demyelinating lesions. Additionally, the whole lesion on the T2-weighted image was segmented, and different tumor components (eg, cyst, edema, and hemorrhage) may improve the classification model performance. Third, a prospective study with more types of spinal cord lesions (eg, spinal cord infarction) and external testing is warranted to validate the established pipeline and extend the model to other spinal cord diseases.
In conclusion, we developed and validated a DL framework for the segmentation and classification of spinal cord lesions including tumors (astrocytoma and ependymoma) and demyelinating diseases (MS and NMOSD), the performance of which sometimes exceeded that of radiologists.
Acknowledgments
Acknowledgments
We acknowledge the colleagues who helped with patient recruitment and MRI.
Z.Z. and J.Z. contributed equally to this work.
C.Y. and Y.L. are co–senior authors.
Supported by the National Science Foundation of China (nos. 81870958 and 81571631), the Beijing Municipal Natural Science Foundation for Distinguished Young Scholars (no. JQ20035), the Special Fund of the Pediatric Medical Coordinated Development Center of Beijing Hospitals Authority (no. XTYB201831), and the ECTRIMS-MAGNMIS Fellowship from ECTRIMS (Y.L.).
Data sharing: Data generated or analyzed during the study are available from the corresponding author by request.
Disclosures of conflicts of interest: Z.Z. No relevant relationships. J.Z. No relevant relationships. Y.D. No relevant relationships. L.Q. No relevant relationships. C.F. No relevant relationships. X.H. No relevant relationships. D.C. No relevant relationships. X.X. No relevant relationships. T.S. No relevant relationships. Z.L. No relevant relationships. X. Guo Employee of Neusoft Group who contributed to segmentation model design. X. Gong Employee of Neusoft Group who contributed to segmentation model design. Y.W. No relevant relationships. W.J. No relevant relationships. D.T. No relevant relationships. X.Z. No relevant relationships. F.S. No relevant relationships. S.H. No relevant relationships. F.B. Consultant for Bayer-Schering, Biogen-Idec, GeNeuro, Ixico, Merck-Serono, Novartis and Roche. He has received grants, or grants are pending, from the Amyloid Imaging to Prevent Alzheimer’s Disease (AMYPAD) initiative, the Biomedical Research Centre at University College London Hospitals, the Dutch MS Society, ECTRIMS–MAGNIMS, EU-H2020, the Dutch Research Council (NWO), the UK MS Society, and the National Institute for Health Research, University College London. He has received payments for the development of educational presentations from Ixico and his institution from Biogen-Idec and Merck. He is on the editorial board of Radiology, Brain, European Radiology, Multiple Sclerosis Journal, and Neurology. C.Y. No relevant relationships. Y.L. No relevant relationships.
Abbreviations:
- AUC
- area under the receiver operating characteristic curve
- DL
- deep learning
- Grad-CAM
- gradient-weighted class activation mapping
- MS
- multiple sclerosis
- NMOSD
- neuromyelitis optica spectrum disorders
References
- 1. Abul-Kasim K , Thurnher MM , McKeever P , Sundgren PC . Intradural spinal tumors: current classification and MRI features . Neuroradiology 2008. ; 50 ( 4 ): 301 – 314 . [DOI] [PubMed] [Google Scholar]
- 2. Karussis D . The diagnosis of multiple sclerosis and the various related demyelinating syndromes: a critical review . J Autoimmun 2014. ; 48-49 : 134 – 142 . [DOI] [PubMed] [Google Scholar]
- 3. Kim HJ , Paul F , Lana-Peixoto MA , et al. MRI characteristics of neuromyelitis optica spectrum disorder: an international update . Neurology 2015. ; 84 ( 11 ): 1165 – 1173 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zeng C , Gu L , Liu Z , Zhao S . Review of Deep Learning Approaches for the Segmentation of Multiple Sclerosis Lesions on Brain MRI . Front Neuroinform 2020. ; 14 : 610967 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shaver MM , Kohanteb PA , Chiou C , et al. Optimizing Neuro-Oncology Imaging: A Review of Deep Learning Approaches for Glioma Imaging . Cancers (Basel) 2019. ; 11 ( 6 ): 829 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zlochower A , Chow DS , Chang P , Khatri D , Boockvar JA , Filippi CG . Deep Learning AI Applications in the Imaging of Glioma . Top Magn Reson Imaging 2020. ; 29 ( 2 ): 115 . [DOI] [PubMed] [Google Scholar]
- 7. Gros C , De Leener B , Badji A , et al. Automatic segmentation of the spinal cord and intramedullary multiple sclerosis lesions with convolutional neural networks . Neuroimage 2019. ; 184 : 901 – 915 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lemay A , Gros C , Zhuo Z , et al. Automatic multiclass intramedullary spinal cord tumor segmentation on MRI with deep learning . Neuroimage Clin 2021. ; 31 : 102766 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ye Z , George A , Wu AT , et al. Deep learning with diffusion basis spectrum imaging for classification of multiple sclerosis lesions . Ann Clin Transl Neurol 2020. ; 7 ( 5 ): 695 – 706 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ibtehaz N , Rahman MS . MultiResUNet : Rethinking the U-Net architecture for multimodal biomedical image segmentation . Neural Netw 2020. ; 121 : 74 – 87 . [DOI] [PubMed] [Google Scholar]
- 11. Ramamurthy M , Krishnamurthi I , Vimal S , Robinson YH . Deep learning based genome analysis and NGS-RNA LL identification with a novel hybrid model . Biosystems 2020. ; 197 : 104211 . [DOI] [PubMed] [Google Scholar]
- 12. Zhang X , Hu Y , Chen W , Huang G , Nie S . 3D brain glioma segmentation in MRI through integrating multiple densely connected 2D convolutional neural networks . J Zhejiang Univ Sci B 2021. ; 22 ( 6 ): 462 – 475 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jung JS , Choi YS , Ahn SS , Yi S , Kim SH , Lee SK . Differentiation between spinal cord diffuse midline glioma with histone H3 K27M mutation and wild type: comparative magnetic resonance imaging . Neuroradiology 2019. ; 61 ( 3 ): 313 – 322 . [DOI] [PubMed] [Google Scholar]
- 14. Ogunlade J , Wiginton JG 4th , Elia C , Odell T , Rao SC . Primary Spinal Astrocytomas: A Literature Review . Cureus 2019. ; 11 ( 7 ): e5247 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wu J , Armstrong TS , Gilbert MR . Biology and management of ependymomas . Neuro Oncol 2016. ; 18 ( 7 ): 902 – 913 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dauleac C , Messerer R , Obadia-Andre N , Afathi M , Barrey CY . Cysts associated with intramedullary ependymomas of the spinal cord: clinical, MRI and oncological features . J Neurooncol 2019. ; 144 ( 2 ): 385 – 391 . [DOI] [PubMed] [Google Scholar]
- 17. Bi WL , Hosny A , Schabath MB , et al. Artificial intelligence in cancer imaging: Clinical challenges and applications . CA Cancer J Clin 2019. ; 69 ( 2 ): 127 – 157 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
We made our code publicly available. It is available at https://github.com/Leezhaohui/spinalcord_classification.



![(A) Deep learning (DL) pipeline established for segmentation and classification of spinal cord lesions. First, the noncontrast T2-weighted (T2w) sagittal MR images (all sections; sections 1–11 herein) were used to segment the lesion, and manual verification was conducted to correct the poorly segmented lesions. Second, the sections (eg, sections 5–8) of the T2-weighted images involving lesions and lesion masks were used as the network input for classification tasks. (B) Representative cases of segmentation and classification for spinal cord tumors and demyelinating lesions in the test cohort. Diagnosis by DL and four raters (D.C. [rater 1], X.X. [rater 2], C.F. [rater 3], X.H. [rater 4]) are shown. Red areas indicate the DL segmentation, green areas indicate manual segmentation, and yellow areas indicate overlap. MS = multiple sclerosis, NMOSD = neuromyelitis optica spectrum disorders.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/146a/9745442/bbca289c145a/ryai.210292.fig2.jpg)