Abstract
The major pore-forming outer membrane proteins (Omps) of gram-negative bacteria demonstrate numerous immunomodulating properties and are involved in the virulence of pathogenic strains. Because Escherichia coli OmpF is the best-characterized porin in terms of structural and functional characteristics, in vitro B-cell and T-cell responses to this porin in six different strains of mice were analyzed. Mice were immunized with purified OmpF trimers or overlapping synthetic polypeptides (20-mers) spanning the entire 340-amino-acid sequence of the OmpF monomer. T-cell proliferative responses and immunoglobulin G antibody responses to native OmpF and the peptide analogues were determined. For each strain, patterns of T-cell proliferation were similar regardless of whether native OmpF or synthetic peptides were inoculated, although all strains recognized one or more cryptic determinants. Mice exhibited several haplotype-specific responses, but genetically permissive epitopes were also identified. Four peptides (75-94, 265-284, 295-314, and 305-324) elicited strong T-cell proliferative responses from all strains of mice when mice were presensitized with native OmpF or a homologous peptide. In general, 10 or fewer peptides were recognized by sera from mice immunized with native OmpF or synthetic peptides, and most sera from peptide-immunized mice reacted poorly with the native protein. Four peptides spanning amino acids 45 to 64, 95 to 114, 115 to 134, and 275 to 294 were recognized by sera from all strains immunized with native OmpF but not by sera from peptide-immunized mice. Peptides 245-264 and 305-324 were universally recognized by sera from peptide-immunized mice, but these sera reacted weakly or were negative when tested against the native protein. Based on the pattern of cytokine secretion by proliferating T cells, immunization with native OmpF polarizes T helper cells toward development of a TH1 response. T-cell and B-cell responses have been investigated based on the assumption that differences in epitope specificity could influence protective or pathologic host reactions. Because of the high level of structural homology of OmpF to porins isolated from other enteric pathogens, the identification of T- and B-cell-stimulatory determinants of E. coli OmpF may have broader application.
The outer membranes of gram-negative Enterobacteriaceae contain pore-forming proteins called porins. Monomeric porin molecules associate to form stable trimeric transmembrane hydrophilic channels which facilitate the transport of various low-molecular-weight solutes. The expression of the major porin proteins of Escherichia coli, OmpC and OmpF, is regulated by environmental stimuli such as osmotic pressure, pH, and temperature. Crystallographic analysis (13) of E. coli OmpF reveals the three-dimensional structure to consist of 16 antiparallel β-strands forming a barrel which is embedded in the membrane. The external segments of the barrel consist of loop structures, seven of which are surface exposed and one (L3) which folds back inside the barrel. The trimeric structure is formed by a hydrophobic interaction between side chains of amino acid residues forming the external surfaces of adjacent barrels. Comparisons of known porin amino acid sequences demonstrate a high degree of structural and inter- and intraspecies amino acid sequence homology.
In addition to their functional properties, purified porins are immunogenic in either their trimeric or monomeric forms. Monoclonal antibodies have been used to define distinct determinants on the OmpF molecule by using mutant strains with either OmpF deletions (24), single-amino-acid substitutions (17, 50), or OmpF-OmpC hybrid porins (17). Some of these were surface-exposed epitopes, but many were also buried within the β-barrel structure. These methods, in general, have only permitted crude localization of antibody-reactive epitopes. Cross-reactions of antiporin monoclonal antibodies with porins of other Enterobacteriaceae are common (24, 28, 33, 39), indicating a high degree of antigenic similarity among porins of divergent species.
Interest in the immunological properties of porins has been fueled by their role in the pathogenesis of enteric organisms and their vaccine potential. Significant interspecies porin sequence homologies could facilitate the induction of broad-spectrum immunity to a number of pathogens following inoculation with a porin isolated from a single strain. Porin-immunized mice were protected from infection when challenged with Salmonella enterica serovar Typhimurium (49) or Salmonella enterica serovar Typhi (22, 23, 37), and mutations in the major porins of Shigella flexneri (4, 5) and S. enterica serovar Typhimurium (12) resulted in decreased virulence compared to that of the wild-type strain. As immunity to these organisms involves T-cell-mediated responses, it can be concluded that the porin molecule contains T-cell epitopes, some of which may elicit protective responses. Although Matsui and Arai (26) demonstrated that passive transfer of T cells from BALB/c mice immunized with S. enterica serovar Typhimurium porins resulted in protection against salmonellosis in naive mice, the protective epitopes were not identified. Purified porin monomers and trimers are capable of inducing T-cell proliferative responses, as measured by in vitro [3H]thymidine uptake assays (29, 46), and elicit strong delayed-type hypersensitivity reactions when inoculated into mice (26, 49). However, the difficulty of preparing purified porin which is free of lipopolysaccharide (LPS) contamination raises questions as to whether the observed responses were specific for the porin component of the inoculum.
Passive immunization of mice with monoclonal or polyclonal antiporin sera provided partial protection against subsequent challenge with S. enterica serovar Typhimurium (22, 42) or S. enterica serovar Typhi (22), with the maximum protection being afforded by monoclonal antibodies with specificity toward the porin-LPS complex. The identification of B-cell epitopes of the porin molecule responsible for the protective effects was restricted to a few strains of mice. The localization of antigenic determinants has been for the most part limited to those B-cell epitopes recognized by BALB/c (H-2d) mice, which were used to generate polyclonal antisera or monoclonal antibody panels (24, 39–41). The contribution of the H-2 haplotype to the cellular responses to the porin molecule has not previously been determined.
In order to better localize minimal B-cell and T-cell epitopes on the OmpF porin molecule and to determine the genetic restriction of responses to these epitopes, overlapping synthetic polypeptides spanning the entire sequence of E. coli OmpF were used to detect antibody responses and in vitro proliferative responses from inbred mice immunized with peptides or a native porin trimer. While the fine specificity of relatively immunodominant T-cell or B-cell epitopes varies depending on the genetic background of the mice, synthetic peptides have also proved to be extremely useful reagents in the identification of genetically permissive epitopes which may have broader application in vaccine development. Peptide-diagnostic reagents based on permissive epitopes could serve as indicators of exposure in a genetically diverse population. The present study used a synthetic-peptide approach to identify those segments of the porin molecule which are recognized by B cells and T cells, thereby eliminating the problems previously encountered with LPS-contaminated preparations. Cytokine profiles of the proliferating cell populations were also determined. The relationship of these epitopes to the three-dimensional structure of the OmpF molecule is discussed.
MATERIALS AND METHODS
Porin purification.
The CM6 strain of E. coli B/r, which produces OmpF but not OmpC (2), was the source of porin. Bacteria were grown to mid-log phase in nutrient media at 37°C. Porin was extracted by the method of Nurminen (32), followed by size exclusion chromatography on Sephacryl S-200 in the presence of 1% sodium dodecyl sulfate (SDS) (20) to reduce LPS contamination. Levels of residual LPS per microgram of porin did not exceed picogram amounts, as detected with the Limulus amebocyte lysate assay (E-Toxate kit; Sigma Chemical Company, St. Louis, Mo.). The purity of the preparation was assessed by SDS-polyacrylamide gel electrophoresis (25) using 10 to 20% linear gradient gels. Aliquots of the porin preparation were incubated with 5 μg of polymyxin B (Sigma) per ml for 1 h at room temperature to inhibit the biological activity of residual LPS (18).
Synthetic polypeptides.
Thirty-two synthetic peptides representing the entire 340-amino-acid sequence of OmpF were produced by standard 9-fluorenylmethoxycarbonyl polyamide solid-phase synthesis, using an Applied Biosystems model 430A peptide synthesizer. Peptides were synthesized as 20-mers, with adjacent peptides overlapping by 10 amino acids. Peptides corresponding to the N terminus and C terminus were synthesized as 25-mers. Syntheses proceeded on p-hydroxymethylphenoxymethyl polystyrene resins, using 2-(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate as the coupling agent. Peptides were deprotected and cleaved from the resin in 92 to 95% trifluoracetic acid (TFA)-H2O containing thioanisole, ethanedithiol, and ethylmethylsulfide as scavenger chemicals. Peptides were purified by preparative reverse-phase high-pressure liquid chromatography by using an acetonitrile gradient of 0 to 70% and 0.1% TFA as the mobile phase. Acidic peptides were purified on a reverse-phase polymeric (300-Å polystyrenedivinylbenzene) column (Vydac, Hesperia, Calif.) using 5 mM ammonium acetate, pH 8.5, as the mobile phase. Molecular weights of selected peptides were confirmed by electrospray mass spectrometry.
Mice.
BALB/cJ (H-2d), DBA/2J (H-2d), CBA/J (H-2k), BALB.K (H-2k), C57BL/6J (H-2b), and BALB.B (H-2b) mice were obtained from the Jackson Laboratories (Bar Harbor, Maine) and used at 6 to 8 weeks of age.
Immunization for antibody production.
B-cell epitope specificities were assessed by the presence of immunoglobulin G (IgG) serum antibodies to individual peptides. For antibody production, groups of five mice were subcutaneously immunized with 100 μg of OmpF and boosted at days 21 and 28. The first injection was given in complete Freund's adjuvant. Because of the large number of peptides to be tested, peptides were initially inoculated as pools of peptides consisting of six groups of five or six peptides each, with 200 μg of protein per injection. Individual peptides were inoculated similarly, except that only 50 μg of peptide was given per injection. Seven days later, groups of mice were bled from the tail vein, the sera were pooled, and antibody responses were determined by enzyme-linked immunosorbent assay (ELISA), using Dynatech Immulon II micro-ELISA plates coated with 0.2 μg of the respective synthetic peptides or 0.1 μg of purified porin/well. Antibody responses were determined using individual peptides which were components of the original immunizing pool. Mouse sera were diluted 1:1,000 in phosphate-buffered saline (PBS) containing 5% fetal bovine serum and 0.1% Tween 20. Bound antibodies were detected with a 1:1,000 dilution of alkaline phosphatase-conjugated goat anti-mouse IgG (Fc specific) (Jackson Immunoresearch, West Grove, Pa.) and with p-nitrophenyl phosphate as the substrate. Absorbance was read at 405 nm using a Spectramax 250 ELISA plate reader (Molecular Devices, Sunnyvale, Calif.). Results are expressed as the mean absorbance of quadruplicate wells, and the standard deviation did not exceed 10% of the mean.
Proliferative assays.
T-cell epitope specificity was determined by the ability of synthetic peptides to induce in vitro T-cell proliferation when incubated with T cells purified from porin-immunized or peptide-immunized mice. For proliferative assays, groups of five mice were immunized subcutaneously at the base of the tail or intraperitoneally with 100 μg of porin. Peptides were inoculated into groups of mice as pools of five or six adjacent peptides. For each pool, 20 μg of the individual peptides was suspended in 100 μl of sterile saline, which was emulsified with an equal volume of complete Freund's adjuvant. Mice were boosted 3 weeks later with 100 μg of porin or pooled peptides suspended in 100 μl of sterile saline. Individual peptides were similarly inoculated, except that only 50 μg of total protein was used. Ten days later, splenic T cells were purified by lysis of red blood cells with 0.17 M ammonium chloride, followed by depletion of B cells and macrophages by one panning cycle and then by passage through columns containing glass beads coated with anti-mouse Ig (R & D Systems; mouse T-cell purification columns). Postcolumn purity of T-cell suspensions was >85% Thy 1+, as assessed by flow cytometry. T cells were suspended in RPMI 1640 medium containing 10% fetal calf serum, 2 mM l-glutamine, 1% nonessential amino acids, 1 mM sodium pyruvate, 50 U of penicillin and 50 μg of streptomycin/ml, and 50 μM 2-mercaptoethanol (Gibco/BRL, Gaithersburg, Md.) and dispensed into 96-well flat-bottom microtiter plates at a concentration of 400,000 T cells/well. Antigen-presenting cells consisted of 200,000 mitomycin C-treated, T-cell-depleted normal splenocytes/well (<5% T cells). Peptides were added to a final concentration of 50 μM in a total volume of 200 μl. The optimum concentration of peptide was determined in preliminary studies using purified T cells incubated with serial dilutions of individual peptides. Additional cells were incubated with purified, polymyxin B-treated porin at concentrations ranging from 10 to 0.01 μg/well. All antigens were tested in triplicate. After 5 days, cells were pulsed with 1 μCi of [3H]thymidine/well for an additional 12 h, harvested with an automatic cell harvester, and counted in a liquid scintillation counter. Supernatants from duplicate cultures were harvested for cytokine quantitation. T cells from normal (vehicle) control mice were treated identically to those from immunized mice. Results of proliferative assays are expressed as stimulation index, i.e., the ratio of counts per minute for immune T cells plus antigen to counts per minute for control T cells plus antigen. Proliferative assays were repeated a minimum of two times for each strain of mouse, with the standard error between assays not exceeding 10%.
Cytokine quantitation.
Interleukin-2 (IL-2) and IL-4 concentrations of culture supernatants were determined by a capture ELISA assay using paired monoclonal antibodies specific for murine IL-2 and IL-4 (Pharmingen, San Diego, Calif.). Dynatech Immulon II plates were coated overnight with the capture antibody (5 μg/ml) suspended in 0.1 M carbonate-bicarbonate buffer, pH 9.6. Plates were washed three times with PBS–0.5% Tween 20, and then 100 μl of culture supernatant was added. Plates were incubated overnight at 4°C and washed four times with PBS-Tween 20, and then 100 μl of biotin-conjugated anti-murine IL-2 or IL-4 (2 μg/ml) was added. Plates were incubated for 2 h and washed six times, and then 100 μl of a 1:1,000 dilution of peroxidase-streptavidin (Jackson Immunoresearch) was added. After 30 min plates were washed eight times, and the color was developed with tetramethylbenzidine. Absorbance was read at 450 nm. Standard curves were generated by using serial dilutions of murine recombinant IL-2 or IL-4 (Genzyme, Cambridge, Mass.). Results are expressed as means of triplicate cultures, and the standard deviations did not exceed 10% of the means.
RESULTS
Mapping of linear B-cell epitopes using synthetic polypeptides.
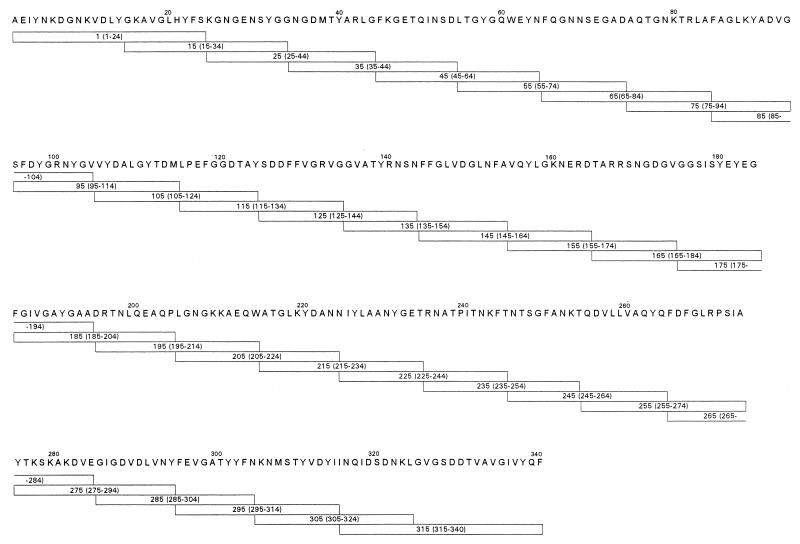
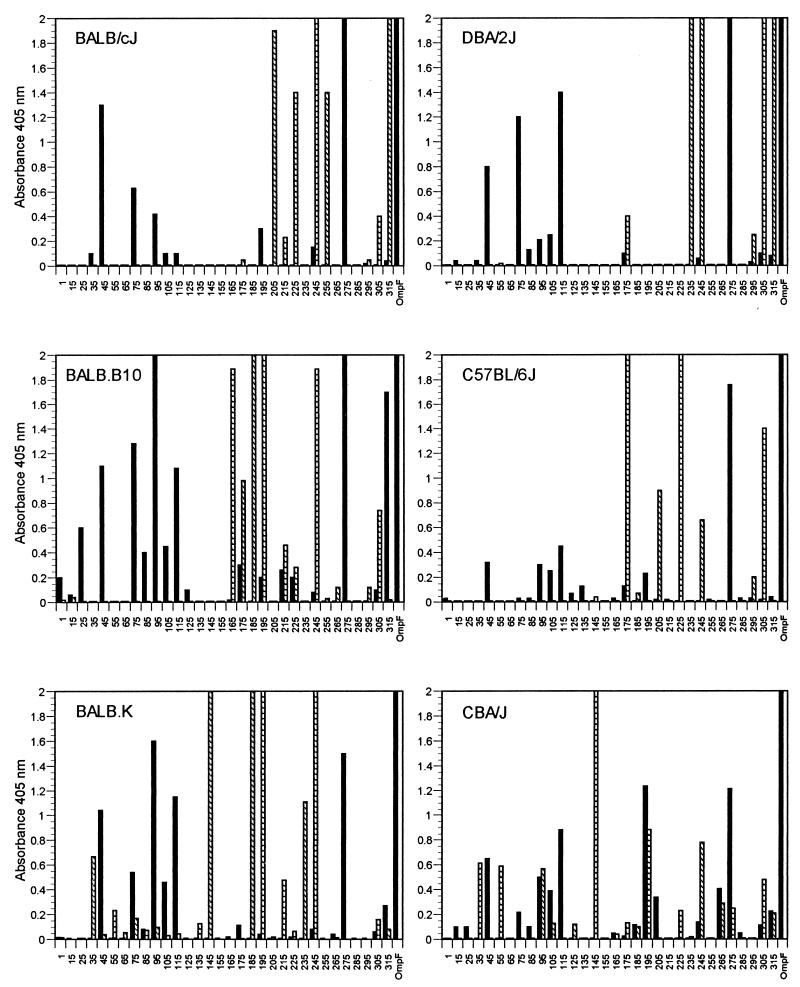
A series of polypeptides spanning the entire 36-kDa OmpF protein were synthesized as 20-mers, with the exception of peptides 1-24 and 315-340 (Fig. 1). Each peptide overlapped the adjacent peptide by 10 amino acids. Separate groups of mice were immunized with a synthetic peptide pool consisting of five or six adjacent peptides or native OmpF, and sera were assayed for IgG binding against each peptide. The IgG response against the native protein was also determined by using sera from mice immunized with native OmpF. A comparison of the reactivities of sera from the different mouse strains against synthetic peptides is shown in Fig. 2. The IgG responses of peptide-immunized mice show little correlation with those of mice immunized with native OmpF. It is also apparent that several peptides are recognized in a genetically unrestricted manner. Sera from all mouse strains immunized with native OmpF reacted with peptides 45-64, 95-114, 115-134, and 275-294. Additionally, peptides 245-264 and 305-324 elicited peptide-specific antibody responses from all strains of mice when inoculated as components of the peptide mixture.
FIG. 1.
Synthetic peptides synthesized according to the E. coli OmpF protein. The amino acid sequence of OmpF (signal sequence removed) is designated by the standard one-letter code. Synthetic peptides are shown as boxes below the sequence; numbers within the boxes correspond to the amino acid residues. Peptides were synthesized as 20-mers, with the exception of peptides at the amino terminus (1-24) and carboxy terminus (315-340), which were synthesized as 25-mers. All peptides overlap by 10 amino acid residues.
FIG. 2.
Antipeptide IgG antibody response to overlapping 20-mer synthetic polypeptides following immunization of inbred mice with native OmpF (solid bars) or synthetic peptides corresponding to the OmpF sequence (hatched bars). Peptide designations (x axis) correspond to amino acid sequences identified in Fig. 1. Mice were immunized with native protein, pooled peptides, or adjuvant (vehicle control) as detailed in Materials and Methods. Results (absorbance at 405 nm) have background values (IgG response of control mice) subtracted.
Several haplotype-specific responses were also observed, mostly when peptides were used as the immunogen. Peptide-immunized BALB/cJ and DBA/2J (H-2d) mice recognized peptide 315-340, but this peptide was not recognized by OmpF-immunized mice. Similarly, peptides 35-54, 55-74, and 145-164 reacted with sera from peptide-immunized BALB.K and CBA/J mice (H-2k), and peptide 175-194 reacted strongly with sera from BALB.B10 and C57BL/6J mice (H-2b). No discernible pattern of reactivity was observed when comparing sera from the congenic strains BALB/cJ, BALB.B10, and BALB.K, indicating that any potential contribution of non-major histocompatibility complex (MHC) genes to antibody responses in these strains was either not present or was undetectable by this assay. Several strain-specific responses were also observed. Sera from peptide-immunized BALB.B10 mice recognized peptide 165-184, and peptide 265-284 was recognized by CBA/J mice when the mice were immunized with either the porin or the peptide. In several other cases, an antibody response to a peptide was observed in both peptide- and porin-immunized mice. Peptide 245-264 was recognized by sera from BALB/cJ, DBA/J, BALB.B10, and BALB.K mice, although the reactivity with sera from OmpF-immunized mice was weak. The same was true for the reactivity of sera from BALB.B10 and C57BL/6J mice using peptide 175-194. The correlation of IgG responses to peptides from mice immunized with either porin or peptide was best observed in the CBA/J strain, where 10 peptides were reactive with either antipeptide or anti-OmpF antisera. Sera from all strains of OmpF-immunized mice produced a strong IgG response against the native protein.
Antibody responses to native protein from mice inoculated with individual peptides.
Groups of mice were immunized with individual peptides to determine if an IgG antibody response against the native protein could be elicited. The individual peptides selected were those which reacted strongly against the homologous peptide when inoculated as a component of the peptide mixture (Fig. 2). A comparison of the antibodies produced by each mouse strain against the immunizing peptide and native OmpF is summarized in Table 1. Several antipeptide antisera from each strain were able to recognize epitopes on the native protein, although the absorbance values were always less than those seen against the homologous peptide. Peptides 245-264 and 305-324, which were positive with sera from all strains injected with pooled peptides, were also capable of inducing IgG in all strains tested when injected as a single peptide. In most mouse strains, these sera did not recognize native OmpF, with the exception of 245-264 antisera produced in BALB/cJ and DBA/2J mice, and 305-324 antisera produced in BALB.B10 and C57BL/6J mice. Peptide 175-194, which reacted strongly with sera from BALB.B10 and C57BL/6J mice when injected as a component of the pooled mixture, also produced antisera which recognized the native protein. The same was true for peptide 315-340 when inoculated into BALB/cJ and DBA/2J mice and peptide 145-164 when inoculated into the BALB.K and CBA/J strains. Some peptides, while strongly reactive with sera produced following inoculation with the pooled mixture, were nonreactive with the native protein or the homologous peptide when injected singly. This suggests that the T-helper epitope required for IgG synthesis was most likely present on an adjacent (or spatially distant) peptide injected as part of the pooled mixture. An unusual response was that seen in antisera from C57BL/6J mice injected with peptide 295-314. Despite the fact that this peptide reacted strongly with antisera produced when the pooled peptides were used as the immunogen, the antisera from mice immunized with 295-314 alone did not produce an IgG response against the homologous peptide. In contrast to the above situation, these antisera were strongly reactive with the native OmpF.
TABLE 1.
Antibody response following inoculation of inbred mice with individual peptides
| Mouse strain and immunizing peptide | ELISA values of individual peptide antiseraa with:
|
|
|---|---|---|
| Homologous peptide | Native OmpF | |
| BALB/cJ | ||
| 205-224 | 0.012 ± 0.001 | 0.044 ± 0.005 |
| 225-244 | 0.954 ± 0.016 | 0.127 ± 0.007 |
| 245-264 | 1.416 ± 0.032 | 0.386 ± 0.009 |
| 255-274 | 0.031 ± 0.004 | 0.066 ± 0.006 |
| 305-324 | 1.260 ± 0.022 | 0.013 ± 0.001 |
| 315-340 | 1.460 ± 0.028 | 0.381 ± 0.016 |
| DBA/2J | ||
| 175-194 | 0.009 ± 0.000 | 0.001 ± 0.000 |
| 235-254 | 0.017 ± 0.002 | 0.024 ± 0.004 |
| 245-264 | 1.020 ± 0.028 | 0.307 ± 0.011 |
| 305-324 | 1.202 ± 0.026 | 0.021 ± 0.003 |
| 315-340 | 0.745 ± 0.019 | 0.116 ± 0.006 |
| BALB.B10 | ||
| 165-184 | 0.465 ± 0.010 | 0.001 ± 0.000 |
| 175-194 | >2.000 | 1.422 ± 0.052 |
| 185-204 | 0.018 ± 0.001 | 0.044 ± 0.005 |
| 195-214 | 0.024 ± 0.002 | 0.034 ± 0.004 |
| 245-264 | 0.757 ± 0.014 | 0.001 ± 0.000 |
| 305-324 | >2.000 | 0.352 ± 0.040 |
| C57BL/6J | ||
| 175-194 | 0.877 ± 0.021 | 0.554 ± 0.046 |
| 205-224 | 0.003 ± 0.000 | 0.045 ± 0.002 |
| 225-244 | >2.000 | 0.233 ± 0.032 |
| 245-264 | 0.565 ± 0.017 | 0.048 ± 0.006 |
| 295-314 | 0.021 ± 0.002 | 0.973 ± 0.050 |
| 305-324 | 1.214 ± 0.049 | 0.603 ± 0.043 |
| BALB.K | ||
| 35-54 | 0.018 ± 0.002 | 0.013 ± 0.001 |
| 55-74 | 0.005 ± 0.000 | 0.011 ± 0.001 |
| 145-164 | 1.677 ± 0.041 | 0.273 ± 0.013 |
| 185-204 | >2.000 | 0.682 ± 0.036 |
| 195-214 | 1.004 ± 0.043 | 0.034 ± 0.002 |
| 235-254 | 0.013 ± 0.001 | 0.011 ± 0.001 |
| 245-264 | 1.603 ± 0.046 | 0.053 ± 0.002 |
| 305-324 | 1.674 ± 0.062 | 0.067 ± 0.007 |
| CBA/J | ||
| 35-54 | 0.021 ± 0.001 | 0.010 ± 0.000 |
| 55-74 | 0.013 ± 0.001 | 0.013 ± 0.000 |
| 95-114 | 0.004 ± 0.000 | 0.010 ± 0.000 |
| 145-164 | >2.000 | 0.172 ± 0.009 |
| 195-214 | 1.603 ± 0.038 | 0.034 ± 0.002 |
| 245-264 | 0.876 ± 0.036 | 0.027 ± 0.001 |
| 305-324 | 0.683 ± 0.051 | 0.040 ± 0.003 |
Absorbance obtained when sera were tested against the homologous peptide or native OmpF. Individual peptides for inoculation were chosen based on responses obtained with sera from peptide pools. Results are mean absorbances at 405 nm of sera from four or five mice ± standard deviations, with background values subtracted. Positive responses are in boldface.
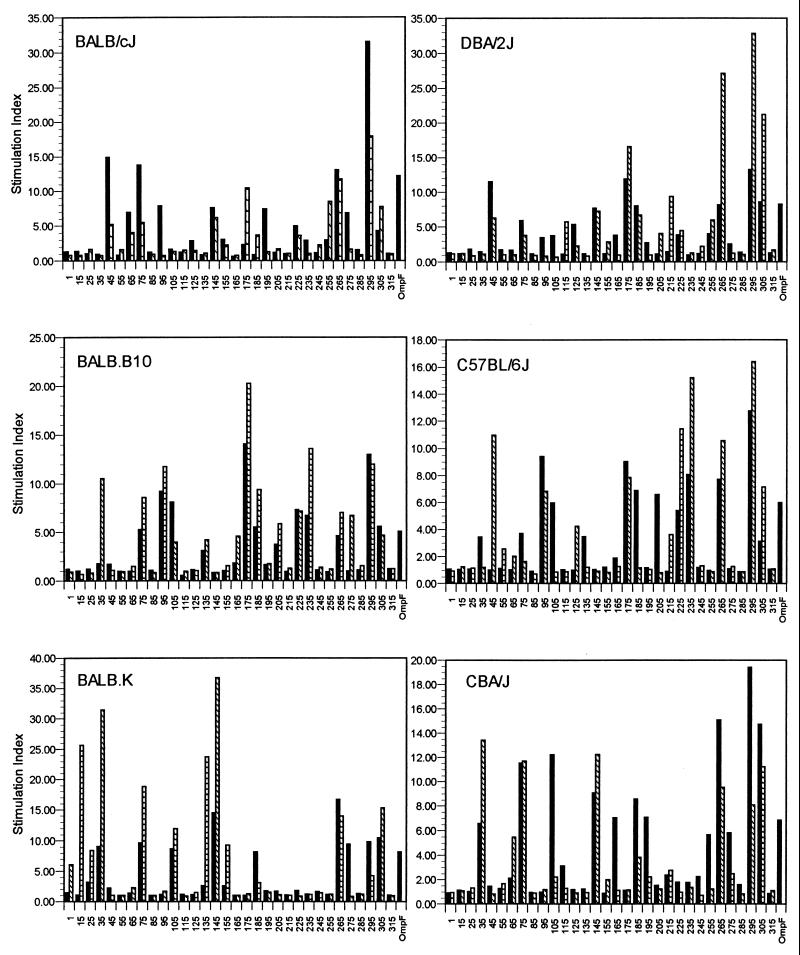
T-cell proliferative responses to synthetic peptides.
T-cell epitopes were identified by measuring the in vitro stimulatory properties of overlapping peptides on T cells primed with either native OmpF or synthetic peptides. In contrast to the IgG antibody response of immunized mice, the results presented in Fig. 3 demonstrate a high degree of correlation between the recall proliferative responses of immune T cells from mice immunized with OmpF and those from mice immunized with synthetic peptides, although a few cryptic determinants (determinants not revealed by immunization with the native protein) were identified. Additionally, several peptides are recognized in a genetically permissive manner, although haplotype-specific and strain-specific proliferative responses were also observed. Peptides 75-94, 265-284, 295-314, and 305-324 were recognized by all strains of mice when mice were immunized with either native OmpF or the homologous peptide, although there was some variability in the magnitude of response, depending on the immunogen.
FIG. 3.
Proliferative response of T cells from inbred mice immunized with native OmpF (solid bars) or synthetic peptides derived from the OmpF sequence (hatched bars) and challenged in vitro with overlapping synthetic polypeptides. Peptide designations (x axis) correspond to amino acid sequences identified in Fig. 1. [3H]thymidine incorporation was measured after 5 days in culture with 50 μM OmpF-derived 20-mers. Results of proliferative assays are expressed as stimulation index (ratio of counts per minute for immune T cells plus antigen to counts per minute for control T cells plus antigen).
In several cases, the level of response to the peptide appeared to be genetically determined. For example, peptide 95-114 induced a strong proliferative response in both peptide-immunized and OmpF-immunized BALB.B10 and C57BL/6J (H-2b) mice, but a lesser response was observed in OmpF-immunized BALB/cJ and DBA/2J (H-2d) mice, and no response was observed in BALB.K and CBA/J (H-2k) mice. Similarly, peptide 145-164 induced a strong proliferative response in peptide- and porin-immunized BALB.K and CBA/J mice, a weak response in BALB/cJ and DBA/2J mice, but no response in BALB.B10 or C57BL/6J mice. A dominant proliferative response was observed in BALB.B10 and C57BL/6J mice when they were stimulated with peptides 225-244 and 235-254, but only peptide 225-244 was able to stimulate T cells from mice bearing the H-2d haplotype. Little or no proliferative response was observed by using these peptides against T cells from mice bearing the H-2k haplotype. Peptide 35-54 induced a strong proliferative response when used to stimulate T cells from both peptide-immunized and porin-immunized BALB.K and CBA/J mice. No proliferative response was observed by using this peptide against T cells from other strains of mice, with the exception of those from peptide-immunized BALB.B10 mice. In this strain, peptide 35-54 would seem to represent a cryptic determinant. Other cryptic determinants were also strain specific. For example, peptide 175-194, while inducing a strong proliferative response in peptide- and porin-immunized BALB.B10, C57BL/6J, and DBA/2J mice, was only recognized by T cells from peptide-immunized BALB/cJ mice. No proliferative response to peptide 175-194 was observed in mice bearing the H-2k haplotype. Peptide 15-34 induced a strong proliferative response in peptide-immunized BALB.K mice, but no response was observed with the other mouse strains.
In several cases, a proliferative response to a peptide was only observed for T cells from mice immunized with native OmpF, indicating inadequate priming of T cells when the homologous peptide was inoculated or a low peptide-specific T-cell precursor frequency. For example, a proliferative response was observed only in OmpF-immunized BALB/cJ and DBA/2 mice in response to peptides 95-114 and 195-214. Similarly, peptides 105-124 and 205-224 induced a proliferative response only in C57BL/6J mice immunized with the native OmpF. All mice immunized with the native protein proliferated to similar degrees when incubated with the homologous protein.
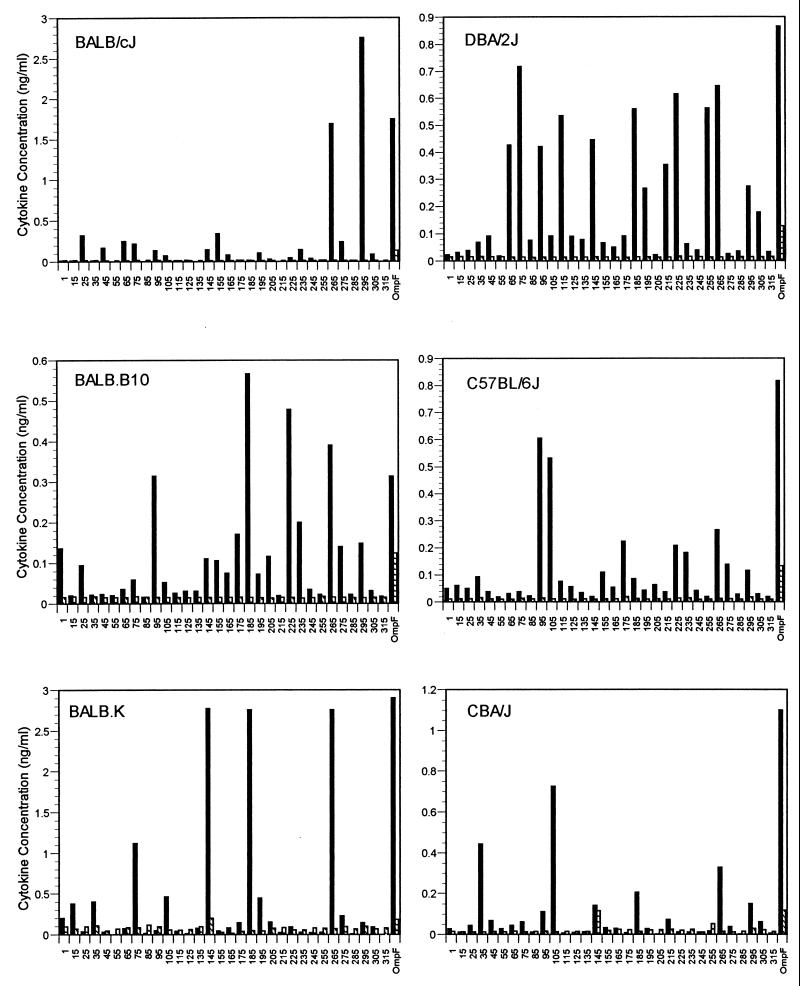
Cytokine production by proliferating T cells.
IL-2 and IL-4 production of T cells was determined by a quantitative ELISA using supernatants from duplicate proliferative cultures of T cells from OmpF-immunized mice after incubation with synthetic 20-mers and native protein. The results, summarized in Fig. 4, indicate that most proliferating T cells secrete significant levels of IL-2 into the culture supernatants, suggestive of a predominant Th1 response. In addition to IL-2 production, detectable levels of IL-4 were secreted by proliferating T cells from BALB.K and CBA/J mice incubated with peptide 145-164. T cells from all strains of mice which were incubated with native OmpF secreted high levels of IL-2 into the culture supernatant, while lower, but significant, levels of IL-4 were also detected. In many cases, the level of the proliferative response did not correlate with the level of cytokine production. This was the case with peptide 305-324 in all strains of mice. This is probably due to the fact that the peak of cytokine production of proliferating cells occurs at a different time than the peak proliferative response, and, for these assays, proliferating cells and supernatants for cytokine quantitation were harvested concurrently.
FIG. 4.
IL-2 (solid bars) and IL-4 (hatched bars) production by T cells isolated from inbred mice immunized with native OmpF and challenged in vitro with OmpF-derived overlapping 20-mer synthetic polypeptides. Peptide designations (x axis) correspond to amino acid sequences identified in Fig. 1. Cytokines were quantitated from T-cell culture supernatants following 5 days of incubation with 50 μM synthetic polypeptide or 10 ng of purified native OmpF/well.
DISCUSSION
Mapping studies to detect production of IgG to synthetic peptides in sera from mice immunized with native OmpF revealed an IgG response to relatively few of the synthetic peptides, even though all antisera contained high titers of antibodies to the native protein. This suggests that most antibodies produced after immunization with native porin are conformation dependent. Although a subpopulation of reactive antibodies could be binding to linear epitopes present on a few peptides (particularly peptides 45-64, 95-114, 115-134, and 275-294, which were recognized in a genetically unrestricted manner), these peptides may have a high conformational stability, particularly when attached to a plastic ELISA plate. This is similar to the stabilization of a folded structure when peptides are resin bound (15, 34), attached to polyethylene pins (36, 45), or conjugated to a carrier protein (8, 9). Previous studies using monoclonal antibodies generated against native OmpF trimers and denatured monomers have shown that, in almost all cases, antibodies which were generated against the native protein reacted weakly or were negative when tested against a denatured monomer (24, 55), indicating that these antibodies were also conformation dependent. Additionally, monoclonal antibodies which reacted strongly with a denatured monomer probably recognized a linear sequence, as they rarely bound to the native trimeric structure. Similarly, when synthetic OmpF peptides were used as the immunogen to generate polyclonal rabbit antisera, no reactivity was observed with native OmpF, although a strong signal was seen with the denatured monomer (38). While it is possible that antipeptide antibodies don't react with the native porin due to blockage of the epitope by LPS or O antigens (3) or inaccessibility when the reactive site is buried inside the β-barrel, it is more likely that the specificity of most antipeptide antibodies is biased toward linear, rather than conformational, epitopes.
Sera from mice immunized with peptide mixtures displayed a wide array of reactivities, few of which coincided with the patterns observed from the porin-immunized mice of the same H-2 haplotype. Two peptides, 245-264 and 305-324, while recognized universally by peptide-immune sera, were totally nonreactive with antiporin antisera. Antipeptide antisera from mice of identical H-2 phenotype exhibited similar patterns of reactivity, indicating a level of genetic restriction in the B-cell responses to synthetic peptides. For example, antisera from C57BL/6J and BALB.B mice (H-2b) recognized peptide 175-194, while sera from BALB.K and CBA mice (H-2k) reacted strongly only with peptides 35-54, 55-74, and 145-164. In some cases, strong responses to adjacent, overlapping peptides were observed, indicating that the antibody may be specific to the linear sequence of amino acids in the overlapping 10-amino-acid peptide segment.
Sera from mice immunized with single peptides as opposed to mixtures were similarly unreactive with the native OmpF trimer. Of the few individual antisera which were reactive with the native trimer, the level of reactivity was always lower than that with the homologous peptide. An exception was observed for C57BL/6J mice which were injected with peptide 295-314. In this case, while the antisera did not react with the homologous peptide, strong reactivity was observed with the native protein. An explanation for this phenomenon is that the peptide in solution (in the inoculum) could have assumed a conformation more closely resembling that of the native trimer, and one that was dissimilar to the conformation of the peptide when bound to the plastic ELISA plate. The adoption of a stable conformation in solution, and one which resembles the conformation of the native protein, has been described for other synthetic peptides (10, 14, 31).
Peptides that were recognized in a genetically unrestricted manner may have more relevance to the development of peptide-based immunodiagnostic reagents or subunit vaccines (21, 53). Because of their ability to stimulate an IgG response in mice of several H-2 haplotypes, the universally recognized peptides described in this study (45-64, 95-114, 115-134, and 275-294) are good candidate peptides for further evaluation. The segment of the porin monomer encompassed by peptide 95-114 includes a portion of the long loop L3, which contains an 8-amino-acid α-helical structure and a small segment of β-strand. Peptide 115-134 encompasses the portion of loop L3 whose secondary structure is composed entirely of turns and loops. Loop L3 is considered an internal loop, since it folds into the β-barrel, contributing to constriction and ion selectivity of the pore. The epitopes present on this loop are, therefore, probably not surface accessible. Using monoclonal antibodies specific for denatured OmpF monomers and E. coli OmpF deletion mutants, Klebba et al. (24) identified three distinct epitopes which were encompassed by residues 108 to 111, 118 to 123, and 123 to 129. Using E. coli OmpF-OmpC hybrid molecules, Yamada et al. (55) also mapped an antimonomer monoclonal antibody to residues 108 to 114. It would appear that the L3 structure contains several dominant B-cell epitopes which, as we have demonstrated, are recognized in a genetically unrestricted manner.
Peptide 45-64 encompasses a segment of the OmpF monomer which is embedded in the transmembrane portion of the barrel structure and makes up a portion of the subunit (monomer) contact region. Peptide 275-294 is composed of amino acids which form the exposed loop L7. Previous studies using monoclonal or polyclonal antisera have not mapped these determinants, but a smaller peptide fragment of the L7 region (275-285) induced polyclonal activation of BALB/c splenocytes and induced B-lymphocyte differentiation into Ig-secreting cells (52). In addition, short peptides (11-mers) derived from these segments were immunogenic when inoculated into rabbits (38).
Using monoclonal antibodies generated against E. coli OmpF and S. enterica serovar Typhimurium OmpC and OmpD (24, 39, 40), other investigators have shown that the transmembrane portion of the barrel structure is conserved among the family of porin molecules from numerous species of Enterobacteriaceae, both structurally and antigenically, while the exposed loops are antigenically diverse. Our studies have not addressed the genetic restriction of response to these phylogenically conserved (or diverse) structures, but it would be interesting to determine if the universal B-cell epitopes identified in this study are conserved in porin molecules from other enterobacterial species. Preliminary data using monoclonal antibodies specific for the peptide composed of amino acids 275 to 294 indicate that the identified epitope is present on porin molecules isolated from numerous species of Enterobacteriaceae despite the external location of this amino acid segment (data not shown).
Results of T-cell proliferative responses revealed that the OmpF protein contains a limited number of dominant T-cell-stimulatory epitopes. While some of the responses were strain specific, several peptides were recognized in a genetically permissive manner. In contrast to the situation with antibody production, T cells isolated from mice immunized with the native protein or synthetic peptides demonstrated similar patterns of proliferation when stimulated with a peptide. This is probably due to the fact that T cells recognize segments of the protein that are processed into smaller peptide fragments. Although the conformation of the native protein may affect how it is processed by antigen-presenting cells, the peptide ultimately presented to the T cell will, most often, depend on the linear amino acid motif recognized by MHC class I (MHC-I) or MHC-II molecules. In several cases, overlapping peptides elicited proliferative responses, indicating that the minimal T-cell epitope may be contained in the shared amino acid segments.
Four peptides (75-94, 265-284, 295-314, and 305-324) elicited strong proliferative responses in all strains tested, whether the mice were immunized with native OmpF or the peptide. Three of these universal T-cell epitopes (75-94, 265-284, and 295-314) were localized to the transmembrane region of the native OmpF protein, while one epitope (305-324) encompasses a portion of a surface-exposed loop (L8). Haplotype- and strain-specific epitopes were localized to other transmembrane and loop regions of the native protein, indicating that both the conserved (transmembrane) and heterogeneous (loop) portions of the porin molecule are composed of multiple T-cell-stimulatory determinants. Based on our results, a general conclusion cannot be made with regard to genetic restriction of the T-cell response in relation to the structure of the native OmpF protein.
Several cryptic epitopes were identified in each mouse strain tested. This would indicate that there are structural differences between synthetic peptides and those peptide fragments resulting from naturally processed antigens. Naturally processed peptides could lack the necessary amino acid residues for stable MHC binding or T-cell activation. Another possibility is that when the native protein is used as the immunogen, there is competition among naturally processed peptides for binding to the MHC molecules, with the result being that only the peptides with the highest affinity to the MHC molecule are finally presented to T cells. This competition would not be observed when a single peptide is inoculated. On the other hand, some peptides were recognized after injection with the native protein but not after injection with the peptide itself. In these cases the processing of the whole antigen may result in a peptide with different flanking amino acids or a different conformation, which would impart a higher affinity for MHC than that imparted by the synthetic peptide. Alternatively, the synthetic peptide could be more sensitive to destruction by proteases involved in antigen processing. Mapping studies using peptides of shorter length or which conform to known MHC binding motifs will be required to identify minimal epitopes.
Protective immunity to enteric pathogens is partially dependent on the activation of cellular defense mechanisms by T cells. Intracellular bacteria and parasites stimulate MHC-II-restricted CD4+ T-helper cells, which consist of distinct subsets, designated TH1 and TH2 cells, based on their cytokine secretion profiles and functional characteristics (27). TH1 cells secrete IL-2, gamma interferon (IFN-γ), tumor necrosis factor alpha (TNF-α), TNF-β, and granulocyte/macrophage colony-stimulating factor and are associated with cell-mediated immunity and an Ig class switch to IgG2a. TH2 cells secrete IL-4, IL-5, and IL-10 and promote humoral immunity by activation of B cells and an Ig class switch to IgG1 and IgE. TH1 cells promote recovery and resistance to intracellular pathogens by activation of antimicrobial effector functions of macrophages through their secreted cytokines. But TH1 cells also exert a protective role through direct cytotoxic activity against infected macrophages. Native porin isolated from S. enterica serovar Typhimurium induced the secretion of IL-2 from BALB/c splenocytes (26) and IFN-γ from BALB/cByJ splenocytes (43) levels of which were not reduced by coculturing with polymyxin B (not attributable to LPS). With purified porin preparations of S. enterica serovar Typhi, pretreatment of mice with porin prior to inoculation with an unrelated antigen not only enhanced antigen-specific cellular immunity as demonstrated by a positive delayed-type hypersensitivity response but also enhanced the level of antigen-specific serum IgG2a (1). Although the segment of the porin molecule responsible for the enhanced response was not determined, these data provide further evidence that exposure to porin molecules may provide immunomodulatory or protective effects by favoring a TH1-type response, possibly by inducing the secretion of IFN-γ by T cells or NK cells or of IL-12 by macrophages or dendritic cells. Based on the level of IL-2 secretion by porin-immune T cells in response to specific peptides, our data demonstrate that inoculation with native OmpF induces an overwhelming TH1-type response in mice of several MHC haplotypes, although lower, but significant, levels of IL-4 were detected in T cells restimulated with the porin itself. Since supernatants were harvested at only one time point following restimulation with the antigen, we were unable to determine if the relative levels of IL-2 and IL-4 vary over time as precursor TH cells (TH0) become more polarized toward a TH1 response.
This study identifies the dominant B-cell and T-cell epitopes of E. coli OmpF using a synthetic peptide approach. The pattern of responses to 20-mer peptides demonstrates the diversity of epitopes represented on the OmpF protein. The repertoire of response is, in some cases, specific to the MHC haplotype of the mouse, but in many cases a promiscuous response to peptides was observed. In several cases the B-cell and T-cell epitopes were localized to identical peptides; however, in many cases the sites of antibody recognition and proliferative responses were spatially distinct. The relationship between those peptides containing T-cell epitopes capable of providing B-cell help and the resultant location of the site of antibody recognition is not well understood. Conformational differences between the native protein and the corresponding peptide clearly affect the processing and/or presentation of an antigen to T cells, and it is reasonable to assume that T-cell or B-cell responses to isolated porins or synthetic peptides derived thereof could differ from responses observed during a natural infection, where the site of initial exposure and the presence of conjugated proteins and other complex bacterial structures could affect antigen processing. Indeed, other investigators have found that chimeric peptides synthesized with B-cell and T-cell epitopes as tandem linear or branched repeating sequences may be more effective immunogens (11, 16, 35, 44). If the goal of epitope mapping in different mouse strains is to develop model peptide-based vaccines, it is clear that more than one B-cell or T-cell determinant may be necessary to elicit a protective response in a genetically diverse population, and it will be necessary to select individual immunogenic peptides from a multideterminant antigen which displays degeneracy in binding to diverse MHC-encoded alleles.
The question also remains as to whether recognition by human B cells and T cells will reflect epitope specificities similar to those recognized by inbred mice. Several studies have demonstrated the proinflammatory and immunomodulatory effects of porin proteins on human monocytes and lymphocytes in vitro (6, 19, 47, 54), but the fine specificity of stimulatory sequences was not determined. Acute and convalescent sera from patients with typhoid fever (30, 51) or gram-negative bacteremia (7) contain high titers of antibodies to purified porins from the implicated organisms, but these sera also contain cross-reacting antibodies to porins isolated from other species of Enterobacteriaceae. The ideal peptide-based diagnostic reagent must (i) contain structures which will permit recognition by antisera produced following exposure to the native protein (dominant epitopes), (ii) contain one or more epitopes recognized by sera produced from a population with diverse HLA phenotypes (universal epitopes), and (iii) contain epitopes common to all isolates of the strain or species to be detected, but with sufficient diversity from related species which may cross-react serologically. We have identified epitopes of the OmpF protein which are potential candidates for such reagents, and future investigations will focus on the evaluation of panels of human sera to determine if these peptides or peptides synthesized from the corresponding sequences of porins from other Enterobacteriaceae would be useful diagnostic reagents.
REFERENCES
- 1.Alurkar V, Kamat R. Immunomodulatory properties of porins of some members of the family Enterobacteriaceae. Infect Immun. 1997;65:2382–2388. doi: 10.1128/iai.65.6.2382-2388.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bavoil P, Nikaido H, von Meyenburg K. Pleiotropic transport mutants of Escherichia coli lack porin, a major outer membrane protein. Mol Gen Genet. 1977;158:23–33. doi: 10.1007/BF00455116. [DOI] [PubMed] [Google Scholar]
- 3.Bentley A T, Klebba P E. Effect of lipopolysaccharide structure on reactivity of antiporin monoclonal antibodies with the bacterial cell surface. J Bacteriol. 1988;170:1063–1068. doi: 10.1128/jb.170.3.1063-1068.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernardi M L, Fontaine A, Sansonetti P J. The two-component regulatory system OmpR-EnvZ controls the virulence of Shigella flexneri. J Bacteriol. 1990;172:6274–6281. doi: 10.1128/jb.172.11.6274-6281.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernardini M L, Sanna M G, Fontaine A, Sansonetti P J. OmpC is involved in invasion of epithelial cells by Shigella flexneri. Infect Immun. 1993;61:3625–3635. doi: 10.1128/iai.61.9.3625-3635.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blanco F, Isibasi A, Gonzalez C R, Ortiz V, Paniagua J, Arreguin C, Kumate J. Human cell-mediated immunity to porins from Salmonella typhi. Scand J Infect Dis. 1993;25:73–80. [PubMed] [Google Scholar]
- 7.Brauner A, Kallenius G, Wrangsell G, Wretlind B, Svenson S B. Antibody responses to Escherichia coli J5 lipopolysaccharide and to Salmonella porin in patients with bacteremia. Microb Pathog. 1986;1:475–481. doi: 10.1016/0882-4010(86)90009-4. [DOI] [PubMed] [Google Scholar]
- 8.Briand J P, Muller S, van Regenmortel M H. Synthetic peptides as antigens: pitfalls of conjugation methods. J Immunol Methods. 1985;78:59–69. doi: 10.1016/0022-1759(85)90329-1. [DOI] [PubMed] [Google Scholar]
- 9.Briand J P, Barin C, van Regenmortel M H V, Muller S. Application and limitations of the multiple antigen peptide (MAP) system in the production and evaluation of anti-peptide and anti-protein antibodies. J Immunol Methods. 1992;156:255–265. doi: 10.1016/0022-1759(92)90033-p. [DOI] [PubMed] [Google Scholar]
- 10.Campbell A P, Sykes B D, Norrby E, Assa-Munt N, Dyson H J. Solution conformation of an immunogenic peptide derived from the principal neutralizing determinant of the HIV-2 envelope glycoprotein gp125. Fold Des. 1996;1:157–165. doi: 10.1016/S1359-0278(96)00024-7. [DOI] [PubMed] [Google Scholar]
- 11.Chai S K, Clavijo P, Tam J P, Zavala F. Immunogenic properties of multiple antigen peptide systems containing defined T and B epitopes. J Immunol. 1992;149:2385–2390. [PubMed] [Google Scholar]
- 12.Chatfield S M, Dorman C J, Hayward C, Dougan G. Role of ompR-dependent genes in Salmonella typhimurium virulence: mutants deficient in both OmpC and OmpF are attenuated in vivo. Infect Immun. 1991;59:449–452. doi: 10.1128/iai.59.1.449-452.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cowan S W, Schirmer T, Rummel G, Steiert M, Ghosh R, Pauptit R A, Jansonium J N, Rosenbusch J P. Crystal structures explain functional properties of two E. coli porins. Nature. 1992;358:727–733. doi: 10.1038/358727a0. [DOI] [PubMed] [Google Scholar]
- 14.Cuniasse P, Thomas A, Smith J C, Thanh J L, Leonetti M, Menez A. Structural basis of antibody cross-reactivity: solution conformation of an immunogenic peptide fragment containing both T and B epitopes. Biochemistry. 1995;34:12782–12789. doi: 10.1021/bi00039a039. [DOI] [PubMed] [Google Scholar]
- 15.Fisher P M, Comis A, Howden M E H. Direct immunization with synthetic peptidyl-polyamide resin. J Immunol Methods. 1989;118:111–123. doi: 10.1016/0022-1759(89)90061-6. [DOI] [PubMed] [Google Scholar]
- 16.Fitzmaurice C J, Brown L E, McInerney T L, Jackson D C. The assembly and immunological properties of non-linear synthetic immunogens containing T-cell and B-cell determinants. Vaccine. 1996;14:553–560. doi: 10.1016/0264-410x(95)00217-o. [DOI] [PubMed] [Google Scholar]
- 17.Fourel D, Mizushima S, Bernadac A, Pages J. Specific regions of Escherichia coli OmpF protein involved in antigenic and colicin receptor sites and in stable trimerization. J Bacteriol. 1993;175:2754–2757. doi: 10.1128/jb.175.9.2754-2757.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galdiero F, Cipollaro de L'Ero G, Benedetto N, Galdiero M, Tufano M A. Release of cytokines induced by Salmonella typhimurium porins. Infect Immun. 1993;61:155–161. doi: 10.1128/iai.61.1.155-161.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galdiero M, Cipollaro de L'Ero G, Donnarumma G, Marcatili A, Galdiero F. Interleukin-1 and interleukin-6 gene expression in human monocytes stimulated with Salmonella typhimurium porins. Immunology. 1995;86:612–619. [PMC free article] [PubMed] [Google Scholar]
- 20.Gehring K B, Nikaido H. Existence and purification of porin heterotrimers of Escherichia coli K12 OmpC, OmpF and PhoE proteins. J Biol Chem. 1989;264:2810–2815. [PubMed] [Google Scholar]
- 21.Herrera S, Escobar P, DePlata C, Avila G I, Corradin G, Herrera M A. Human recognition of T cell epitopes on the Plasmodium vivax circumsporozoite protein. J Immunol. 1992;148:3986–3990. [PubMed] [Google Scholar]
- 22.Isibasi A, Ortiz V, Vargas M, Paniagua J, González C, Moreno J, Kumate J. Protection against Salmonella typhi infection in mice after immunization with outer membrane proteins isolated from Salmonella typhi 9,12,d,Vi. Infect Immun. 1988;56:2953–2959. doi: 10.1128/iai.56.11.2953-2959.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Isibasi A, Ortiz-Navarrete V, Paniagua J, Pelayo R, Gonzalez C R, Garcia J A, Kumate J. Active protection of mice against Salmonella typhi by immunization with strain-specific porins. Vaccine. 1992;10:811–813. doi: 10.1016/0264-410x(92)90041-h. [DOI] [PubMed] [Google Scholar]
- 24.Klebba P E, Benson S A, Bala S, Abdullah T, Reid J, Singh S P, Nikaido H. Determinants of OmpF porin antigenicity and structure. J Biol Chem. 1990;265:6800–6810. [PubMed] [Google Scholar]
- 25.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 26.Matsui K, Arai T. Protective immunity induced by porin in experimental mouse salmonellosis. Microbiol Immunol. 1989;33:699–708. doi: 10.1111/j.1348-0421.1989.tb00957.x. [DOI] [PubMed] [Google Scholar]
- 27.Mosmann T R, Coffman R L. TH1 and TH2 cells: different patterns of lymphokine secretion lead to different functional properties. Annu Rev Immunol. 1989;7:145–173. doi: 10.1146/annurev.iy.07.040189.001045. [DOI] [PubMed] [Google Scholar]
- 28.Muthukkaruppan V R, Nandakumar K S, Palanivel V. Monoclonal antibodies against Salmonella porins: generation and characterization. Immunol Lett. 1992;33:201–206. doi: 10.1016/0165-2478(92)90047-r. [DOI] [PubMed] [Google Scholar]
- 29.Muthukkumar S, Muthukkaruppan V R. Mechanism of protective immunity induced by porin-lipopolysaccharide against murine salmonellosis. Infect Immun. 1993;61:3017–3025. doi: 10.1128/iai.61.7.3017-3025.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nandakumar K S, Palanievl V, Muthukkaruppan V R. Diagnosis of typhoid fever: detection of Salmonella typhi porins-specific antibodies by inhibition ELISA. Clin Exp Immunol. 1993;94:317–321. doi: 10.1111/j.1365-2249.1993.tb03450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niman H L, Houghten R A, Walker L E, Reisfeld R A, Wilson I A, Hogle J M, Lerner R A. Generation of protein-reactive antibodies by short peptides is an event of high frequency: implications for the structural basis of immune recognition. Proc Natl Acad Sci USA. 1983;80:4949–4953. doi: 10.1073/pnas.80.16.4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nurminen M. A mild procedure to isolate the 34K, 35K, and 36K porins of the outer membrane of Salmonella typhimurium. FEMS Microbiol Lett. 1978;3:331–334. [Google Scholar]
- 33.Pai S R, Upshaw Y, Singh S P. Characterization of monoclonal antibodies to the outer membrane protein (OmpD) of Salmonella typhimurium. Can J Microbiol. 1992;38:1102–1107. doi: 10.1139/m92-181. [DOI] [PubMed] [Google Scholar]
- 34.Paniagua-Solis J, Sanchez J, Ortiz-Navarrete V, Gonzalez C R, Isibasi A. Construction of CTB fusion proteins for screening of monoclonal antibodies against Salmonella typhi OmpC peptide loops. FEMS Microbiol Lett. 1996;141:31–36. doi: 10.1111/j.1574-6968.1996.tb08359.x. [DOI] [PubMed] [Google Scholar]
- 35.Peterson E M, Cheng X, Qu Z, de la Maza L M. The effect of orientation within a chimeric peptide on the immunogenicity of Chlamydia trachomatis epitopes. Mol Immunol. 1996;33:335–339. doi: 10.1016/0161-5890(95)00157-3. [DOI] [PubMed] [Google Scholar]
- 36.Savoca R, Schwab C, Bosshard H R. Epitope mapping employing immobilized synthetic peptides. How specific is the reactivity of these peptides with antiserum raised against the parent protein? J Immunol Methods. 1991;141:245–252. doi: 10.1016/0022-1759(91)90151-5. [DOI] [PubMed] [Google Scholar]
- 37.Sharma P, Ganguly N K, Sharma B K, Sharma S, Rawal I J, Saxena S N, Sehgal R. Humoral and cell mediated immune responses to porins of Salmonella typhi. Jpn J Exp Med. 1989;89:73–77. [PubMed] [Google Scholar]
- 38.Simonet V, Mallea M, Fourel D, Bolla J, Pages J. Crucial domains are conserved in Enterobacteriaceae porins. FEMS Microbiol Lett. 1996;136:91–97. doi: 10.1111/j.1574-6968.1996.tb08030.x. [DOI] [PubMed] [Google Scholar]
- 39.Singh S P, Upshaw Y, Abdullah T, Singh S R, Klebba P E. Structural relatedness of enteric bacterial porins assessed with monoclonal antibodies to Salmonella typhimurium OmpD and OmpC. J Bacteriol. 1992;174:1265–1973. doi: 10.1128/jb.174.6.1965-1973.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singh S P, Miller S, Williams Y U, Rudd K E, Nikaido H. Immunochemical structure of the OmpD porin from Salmonella typhimurium. Microbiology. 1996;142:3201–3210. doi: 10.1099/13500872-142-11-3201. [DOI] [PubMed] [Google Scholar]
- 41.Singh S P, Singh S R, Williams Y U, Jones L, Abdullah T. Antigenic determinants of the OmpC porin from Salmonella typhimurium. Infect Immun. 1995;63:4600–4605. doi: 10.1128/iai.63.12.4600-4605.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singh S P, Williams Y U, Benjamin W H, Klebba P E, Boyd D. Immunoprotection by monoclonal antibodies to the porins and lipopolysaccharide of Salmonella typhimurium. Microb Pathog. 1996;21:249–263. doi: 10.1006/mpat.1996.0059. [DOI] [PubMed] [Google Scholar]
- 43.Sommese L, Donnarumma G, Cipollaro de L'ero G, Marcatili A, Vitiello M, Galdiero M. Growth hormone modulates IL-1α and IFN-γ release by murine splenocytes activated by LPS or porins of Salmonella typhimurium. J Med Microbiol. 1996;45:40–47. doi: 10.1099/00222615-45-1-40. [DOI] [PubMed] [Google Scholar]
- 44.Steward M W, Partidos C D, D'Mello F, Howard C R. Specificity of antibodies reactive with hepatitis B surface antigen following immunization with synthetic peptides. Vaccine. 1993;11:1405–1414. doi: 10.1016/0264-410x(93)90169-x. [DOI] [PubMed] [Google Scholar]
- 45.Trifilieff E, Dubs M C, van Regenmortel M H V. Antigenic cross-reactivity potential of synthetic peptides immobilized on polyethylene rods. Mol Immunol. 1991;28:889–894. doi: 10.1016/0161-5890(91)90053-m. [DOI] [PubMed] [Google Scholar]
- 46.Tufano M A, Berlingieri M T, Sommese L, Galdiero F. Immune response in mice and effects on cells by outer membrane porins from Salmonella typhimurium. Microbiologica. 1984;7:353–366. [PubMed] [Google Scholar]
- 47.Tufano M A, Tetta C, Biancone L, Iorio E L, Baroni A, Giovane A, Camussi G. Salmonella typhimurium porins stimulate platelet-activating factor synthesis by human polymorphonuclear neutrophils. J Immunol. 1992;149:1023–1030. [PubMed] [Google Scholar]
- 48.Udhayakumar V, Muthukkaruppan V R. Protective immunity induced by outer membrane proteins of Salmonella typhimurium in mice. Infect Immun. 1987;55:816–821. doi: 10.1128/iai.55.3.816-821.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Udhayakumar V, Muthukkaruppan V R. An outer membrane protein (porin) as an eliciting antigen for delayed-type hypersensitivity in murine salmonellosis. Infect Immun. 1987;55:822–824. doi: 10.1128/iai.55.3.822-824.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van der Ley P, Struyve M, Tommassen J. Topology of outer membrane pore protein PhoE of Escherichia coli. J Biol Chem. 1986;261:12222–12226. [PubMed] [Google Scholar]
- 51.Verdugo-Rodriguez A, Lopez-Vidal Y, Puente J L, Ruiz-Palacios G M, Calva E. Early diagnosis of typhoid fever by an enzyme immunoassay using Salmonella typhi outer membrane protein preparations. Eur J Clin Microbiol Infect Dis. 1993;12:248–254. doi: 10.1007/BF01967254. [DOI] [PubMed] [Google Scholar]
- 52.Vordermeier H M, Hoffmann P, Gombert F O, Jung G, Bessler W G. Synthetic peptide segments from the Escherichia coli porin OmpF constitute leukocyte activators. Infect Immun. 1990;58:2719–2724. doi: 10.1128/iai.58.8.2719-2724.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vordermeier H M, Harris D P, Roman E, Lathigra R, Moreno C, Ivanyi J. Identification of T cell stimulatory peptides from the 38-kDa protein of Mycobacterium tuberculosis. J Immunol. 1991;147:1023–1029. [PubMed] [Google Scholar]
- 54.Vordermeier H M, Drexler H, Bessler W G. Polyclonal activation of human peripheral blood lymphocytes by bacterial porins and defined porin fragments. Immunol Lett. 1987;15:121–126. doi: 10.1016/0165-2478(87)90042-3. [DOI] [PubMed] [Google Scholar]
- 55.Yamada H, Oshima N, Mizuno T, Matsui H, Kai Y, Noguchi H, Mizushima S. Use of a series of OmpF-OmpC chimeric proteins for locating antigenic determinants recognized by monoclonal antibodies against the OmpC and OmpF proteins of the Escherichia coli outer membrane. J Biochem. 1987;102:455–464. doi: 10.1093/oxfordjournals.jbchem.a122076. [DOI] [PubMed] [Google Scholar]