Summary
T cells mediate antigen-specific immune responses to disease through the specificity and diversity of their clonotypic T-cell receptors (TCRs). Determining the spatial distributions of T cell clonotypes in tissues is essential to understanding T cell behavior, but spatial sequencing methods remain unable to profile the TCR repertoire. Here, we developed Slide-TCR-seq, a 10-μm-resolution method to sequence whole transcriptomes and TCRs within intact tissues. We confirmed the ability of Slide-TCR-seq to map the characteristic locations of T cells and their receptors in mouse spleen. In human lymphoid germinal centers, we identified spatially distinct TCR repertoires. Profiling T cells in renal cell carcinoma and melanoma specimens revealed heterogeneous immune responses: T cell states and infiltration differed intra- and inter-clonally, and adjacent tumor and immune cells exhibited distinct gene expression. Altogether, our method yields insights into the spatial relationships between clonality, neighboring cell types, and gene expression that drive T cell responses.
Keywords: spatial, transcriptomics, T-cell receptor, sequencing, clonotype, T cell, cell interactions, immune niches
eTOC
T cells’ location, clonotype, and gene expression are critical to understanding immune responses. Liu, lorgulescu, Li, and colleagues develop Slide-TCR-seq to simultaneously study spatial and transcriptomic differences between T cell clones with 10-μm-resolution and apply it to human lymphoid organs and cancer contexts.
Introduction
In many diseases, antigen-specific T cells function as the primary effector cells that mediate immune responses. The specificity of T cells is determined by their T-cell receptor (TCR) sequence – generated through recombination of genomic V(D)J segments, which, importantly, include the highly polymorphic complementarity-determining region 3 (CDR3) domains that drive recognition of cognate antigenic peptides presented in the context of major histocompatibility complex (MHC) molecules. Consequently, understanding the biological processes that underlie T cell development, immune responses, disease pathogenesis, and immunotherapy efficacy are goals that have motivated efforts to characterize TCR repertoires using bulk and single-cell approaches (Wu et al., 2020; Joshi et al., 2019; Yost et al., 2019; Thorne et al., 2014).
Although there have been remarkable advances in technologies that couple single-cell transcriptomics and TCR sequencing (Li et al., 2019; Tu et al., 2019) – which have enabled the systematic characterization of T cell clonotype repertoires – conventional strategies rely on dissociating tissues into single-cell suspensions. Consequently, valuable information is lost regarding the spatial organization of cells, thereby limiting our ability to answer how T cell clonotypes distribute in organs, interact with other T cell clones and distinct cell types during immune responses, and organize under various pathologic conditions. Conversely, current methods for spatial characterization, which primarily apply antibodies (e.g., multiplexed immunofluorescence, imaging mass cytometry) or in situ hybridization probes for mapping cell phenotypes and TCR sequences, are highly targeted, restricted in their multiplexing capability, and limited in scalability (Gohil et al., 2021). Importantly, there are currently no approaches that spatially profile TCR sequences in their tissue contexts at near single-cell resolution. As a result, an evaluation of how T cell state, clonal identity, and tissue localization are related has not been possible. Here, we developed Slide-TCR-seq, a method for sequencing whole transcriptomes and TCRs within intact tissues. To illustrate the utility of Slide-TCR-seq in identifying spatially segregated T cell niches, we identified distinct TCR repertoires in germinal centers and tertiary lymphoid structures. In renal cell carcinoma and melanoma, we found that T cell clones vary both inter- and intra-clonally in their gene expression and their tumor infiltration and that tumor and immune cells exhibited differentially expressed genes depending on the adjacent T cell clone. Overall, our method enables studying the complex relationship between clonality, localization, and gene expression.
Results
Slide-TCR-seq integrates TCR clonotype sequencing and spatial transcriptomics
To add to existing tools for studying TCRs (Table S1) and enable spatial TCR sequencing, we reasoned that Slide-seqV2 (Stickels et al., 2020), a 10-qm-resolution spatial transcriptomic approach, could be modified to capture and sequence TCR sequences—thereby allowing the integrated analysis of clonal, transcriptional, and spatial attributes of the T cell clonotype repertoire. Slide-seqV2 generates cDNA libraries from fresh-frozen tissue sections with 3’-end mRNA capture through spatially arrayed DNA-barcoded 10-qm beads: beads are affixed onto slides, in situ sequencing is performed to map the spatial bead barcode sequence to the bead’s x-y location, and then RNA can be captured onto the beads for downstream library preparation (Figure l A, Table S2). However, preparation for sequencing involves cDNA fragmentation, from which only the constant regions of TCRα and TCRβ can be detected. We recently reported rhTCRseq (Li et al., 2019) as a robust approach for targeted amplification of TCR transcripts. Hence, we hypothesized that rhTCRseq could be adapted to integrate with the Slide-seq workflow to determine the variable sequences of TCRs. rhTCRseq exploits the specificity of rhPCR (Dobosy et al., 2011), which uses 3’-b1ocked oligonucleotides with a single ribo residue cleaved by RNase H upon hybridization with DNA targets to generate functional primers extendable by Taq DNA polymerase, thereby enabling the specific multiplexed PCR needed to amplify TCR transcripts (Figure lA). We optimized rhTCRseq for Slide-seq to amplify segments that extend from the alpha or beta V region (upstream of CDR3) to the 3’ end of the TCR transcript containing the bead barcode (STAR Methods). We could then determine the variable sequences from the amplified cDNA library, which we divided into two fractions. The first fraction underwent conventional fragmentation and sequencing to determine the bead barcodes and 3’-end transcriptome information. The second fraction was processed using the modified rhTCRseq workflow and then sequenced either via short-read sequencing on a MiSeq or long-read sequencing via Oxford Nanopore (Figure SlA–B). Sequences for CDR3 regions were determined computationally (STAR Methods), and the bead barcodes were used to map back to the spatial reference. Thus, Slide-TCR-seq can generate whole transcriptome and CDR3 information with spatial context from fresh-frozen tissues at 10-μm-resolution.
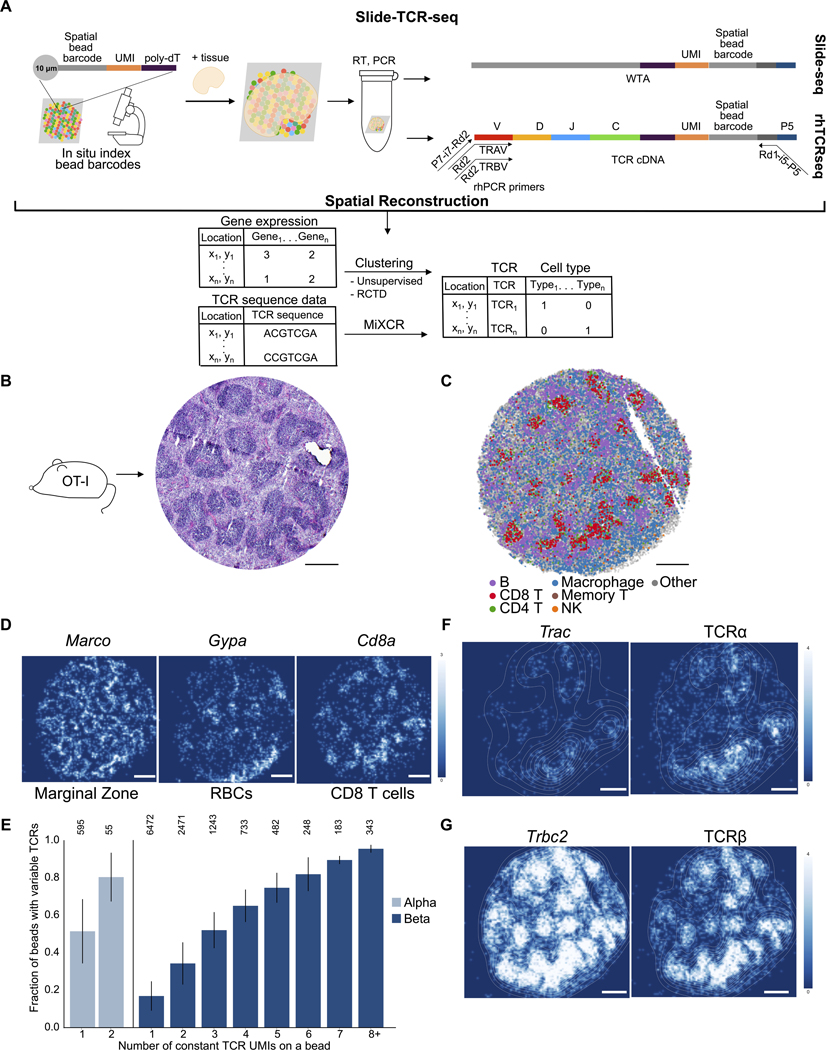
Figure 1. Slide-TCR-seq spatially localizes T cell receptors and transcriptome information.
A) Schematic of Slide-TCR-seq. First, 10 μm beads containing many DNA oligos are affixed onto a slide. The spatial bead barcode is then in situ sequenced to create a map of the bead barcodes to the spatial locations. Samples can then be placed onto the slides, and RNA captured via the poly-dT sequence on the oligos. cDNA libraries prepared with Slide-seqV2 are split before fragmentation with one portion used for targeted amplification via rhTCRseq optimized for use with Slide-seq libraries and the other portion used for whole transcriptome amplification. Slide-TCR-seq thereby provides gene expression, cell type, and clonotype information in space.
(B) Serial sections of the OT-I mouse spleen with hematoxylin and eosin stain showing characteristic architecture of red pulp and white pulp separation.
(C) Spatial reconstruction of a representative Slide-TCR-seq array from three replicates for a corresponding section of OT-I mouse spleen, with RCTD cell type assignment. Cell types are plotted individually in Figure S1C. NK = natural killer.
(D) Gene expression (in UMI counts) gaussian-filtered heatmap of a representative Slide-TCR-seq array from three replicates for visualizing the spatial distribution of gene markers for marginal zone (Marco), red blood cells (RBCs; Gypa), and CD8 T cells (Cd8a).
(E) The fraction of beads that capture CDR3 variable sequences (y-axis) when constant UMIs, UMIs corresponding to the TCR constant reads from the whole transcriptome sequencing of Slide-seq, are captured (x-axis) for TCRα (left, light blue) and TCRβ (right, dark blue), with the number of corresponding beads along the top axis (weighted average across n=3 replicates, error bars are weighted standard deviation).
(F-G) Comparing the spatial distribution of constant (left) and variable (right) sequences of a representative Slide-TCR-seq array of 3 replicates for TCRα (F) and TCRβ (G) with superimposed density plot.
UMI: unique molecular identifier. All scale bars: 500 μm. See also Figure S1.
Spatial reconstruction of the native architecture of the mouse spleen delineates CDR3 sequences and locations
To confirm the ability of Slide-TCR-seq to spatially reconstruct and quantify TCR variable region sequencing in a well-controlled setting, we first applied Slide-TCR-seq to freshly frozen spleens from OT-I transgenic mice to take advantage of the predominant expression of a single well-characterized TCR in the distinctive lymphoid architecture of spleen (Figure IB). We resolved the spatial structure of the mouse spleen using the transcriptome fraction of Slide-TCR-seq and assigned cell types to beads using robust cell type decomposition (RCTD) (Cable et al., 2021) with a murine spleen scRNA-seq reference (Figures 1C and S1C–D) (Kimmel et al., 2019). Overall cell type arrangement and marker genes for the marginal zone (Marco), red blood cells (Gypa), and CD8+ T cells (Cd8a) aligned well with the expected splenic architecture from histology and immunofluorescence staining (Figures 1B and 1D).
To determine the spatial recovery efficiency of the CDR3 variable sequences, we measured the proportion of beads in which we both sequenced CDR3 variable regions and detected a TCR constant sequence. To extract the variable TCR repertoire from the rhTCR-seq fraction, we used MiXCR (STAR Methods) (Bolotin et al., 2015). To correct for chimeric molecules, we implemented a conservative, adaptive computational filter on the variable sequences to only consider variable sequences that had at least two reads (STAR Methods). We found that beads with constant sequences contained a variable sequence 41.7% of the time (Figure S IE). When a bead captured a single Trbc2 constant unique molecular identifier (UMI), at least one TCRβ CDR3 was sequenced on that same bead 25.8% of the time; for TCRo, the sequencing efficiency was higher, at 64.6% (Figure IE). These recovery values were comparable to that reported in recent single-cell methods for TCR sequencing (Tu et al., 2019). We also found that the targeted amplification of TCRs improved our overall T-cell detection, as 28.7% of beads containing a variable TCR were not identified in the transcriptome fraction (Figure S1E). After applying the computational filter to remove low-confidence reads, over 99% of the TCRɑ sequences matched the OT-I TCRɑ sequence, 100% of the TCRβ sequences matched the OT-I TCRβ sequence, and over 99% of the beads where both TCRɑ and TCRβ were sequenced matched the OT-I clonotype, consistent with previous tetramer studies (Table S3) (Strbo et al., 2003).
To confirm that the general architecture of the tissue was maintained in the spatial assignments of the variable CDR3 sequences, we compared their positioning with that of the constant TCR sequences and found high spatial correlation between the variable and constant sequence localizations (Pearson’s r=0.86, Figures S2F–G). Contour maps highlighting regions of high T-cell density further confirmed the spatial reconstruction of the splenic architecture (Figures IF and 1G) and revealed higher capture of Trbc2 than Trac, which reflects the higher expression of Trbc2 in T cells (Oakes et al., 2017). Slide-TCR-seq thus faithfully recovered both CDR3 sequences and their spatial locations.
Slide-TCR-seq reveals spatially distinct T cell repertoires in secondarv lymphoid tissues
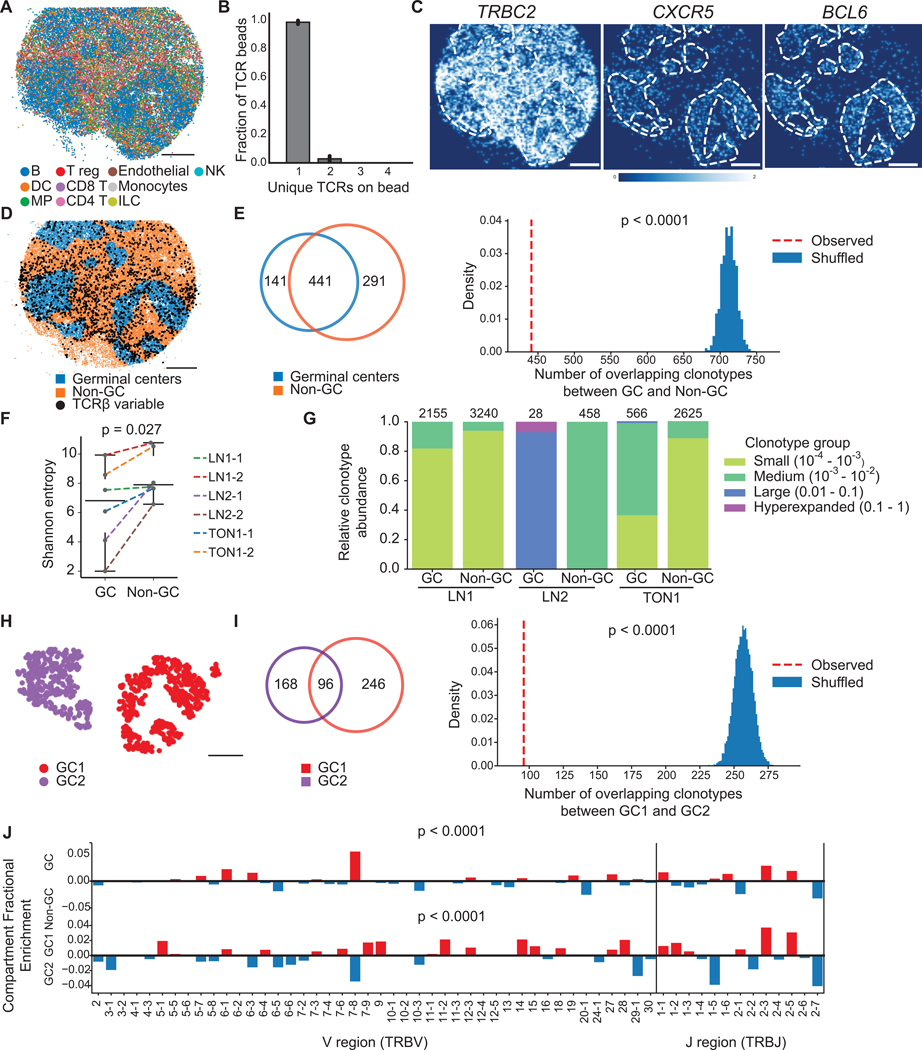
To study the native TCR repertoire organization in human secondary lymphoid tissues, we applied Slide-TCR-seq to human lymph node and tonsil samples (Figure 2). We found that, on average, 98.5% of beads with CDR3 sequences contained a single CDR3, and 2.8% contained two CDR3s (Figures 2B, S2A, S2B). This finding was consistent with our expectation that only a small fraction of beads contained multiple T cells. Germinal centers (GCs) in lymph nodes are critical locations for T follicular helper cells (Tfh) to shape B cell affinity maturation (Ansel et al., 1999; Johnston et al., 2009; Yu et al., 2009). RCTD cell type assignments showed the expected B cell-rich GCs, marked by CXCRS and BCL6 expression (Figures 2A and 2C and S2C). Since Tfh cells have been shown to travel between GCs (Shulman et al., 2013; Suan et al., 2015), we investigated whether TCR repertoires within GCs were locally distinct or shared with other tissue compartments. After assigning each bead with a TCRβ sequence to either GCs or non-GC areas, we evaluated clonotypes that were captured by at least two beads and observed a large number of clonotypes that localize in both compartments but noted that the observed clonotype overlap was lower than expected (p<0.0001, comparison to random shuffling, Figures 2D–E and S2D, STAR Methods). This observation also held in the analysis of one other reactive human lymph node and one reactive human tonsil sample (combined p<4.1×>10−10, by Fisher’s method, Figure S2E). This finding supports the notion that the distribution of T cells in the lymph node between GC and non-GC regions is non-random, suggestive of spatial segregation. The diversity of TCR clonotypes (by Shannon entropy) was higher in non-GC regions across all samples (Figure 2F) and T cells in GCs had more expanded clonotype groups (Figure 2G), further suggestive of localized TCR repertoire clonal expansion, presumably antigen-driven, in GCs (Merkenschlager et al., 2021).
Figure 2. T cell receptor repertoire differences between spatial compartments in human lymph node and tonsil.
(A) Spatial reconstruction of a representative Slide-TCR-seq array from two technical replicates a 10 μm section of human reactive lymph node, LNl, with RCTD cell type assignment. Cell types are plotted individually in Figure S2C.
(B) Characterization of the number of unique TCRs on beads that contain TCRs across lymph node and tonsil samples (n=l0, bar plots indicate means, and error bars indicate standard deviation of replicates.)
(C) Gene expression gaussian-filtered heatmap of a representative Slide-TCR-seq array from two technical replicates for visualizing the spatial distribution of gene markers for T cells (TRBC2) and germinal centers (GCs; CXCRS, BCL6).
(D) Bead designation of a representative Slide-TCR-seq array from two technical replicates into either GC regions (blue) or non-GC (orange) regions based on unsupervised clustering and GC markers on LN1. Beads with TCRβ CDR3 sequences detected are shown by black dots, but only for CDR3 sequences detected on >1 bead.
(E) For LNl, Left: For clonotypes found on >1 bead, a Venn diagram showing the clonotype overlap between GC (blue) and non-GC (orange) regions. Right: the significance of the observed number of shared clonotypes compared to the expected number from randomly shuffled assignments.
(F) Shannon entropy (i.e., a measure of diversity) of the TCR repertoire in GC vs. non-GC regions for six arrays across three human secondary lymphoid organs.
(G) Clonotypes grouped by clonotype fraction, normalized by the total number of TCRs in each compartment, in GC and non-GC regions in lymph node and tonsil, with the number of corresponding beads along the top axis.
(H) K-means clustering distinguishing two GCs in a human reactive lymph node.
(I) Left: For clonotypes detected on >l bead, a Venn diagram showing clonotype overlap between GC1 (purple) and GC2 (red) regions. Right: The significance of the observed number of shared clonotypes between GCs compared to the expected number from randomly shuffled assignments.
(J) Differential enrichment of TRBV and TRBJ sequences in GC (red) vs non-GC (blue) regions (top) and GC1 (red) vs GC2 (blue) regions (bottom). All scale bars: 500 μm. LN = lymph node. TON = tonsil. See also Figure S2.
While the GC repertoire was distinct from the non-GC repertoire, we queried whether distinct GCs had unique TCR repertoire niches for B cell maturation. We used random shuffling to estimate the expected overlap in clonotypes between two GCs that we identified in the same lymph node array. While the two GCs indeed contained many shared clonotypes, the observed clonotype overlap was lower than expected by random sampling (p<0.0001, Figures 2H–I), a result also detected upon analysis of another section from this lymph node sample (p<0.0001, Figures S2F–G) Thus, while Tfh cells can migrate between GCs, these data suggest that GCs in this lymph node sample retained locally distinct TCR environments. Furthermore, these compartmental differences extended beyond the CDR3 identity to include differences in TRBV and TRBJ distributions between compartments (p<0.0001, Figure 2I). These results reinforce the notion that in lymph nodes without ongoing antigen stimulation, TCR clonotypes are spatially segregated.
Slide-TCR-seq identifies spatial heterogeneity between clonotypes in renal cell carcinoma
Beyond examining the architecture of normal tissue, analysis of the spatial organization of individual T cell clones within the tumor microenvironment may yield insight into therapeutic response and resistance. In particular, the spatial organization of the clonal context of T cells in relation to the T cells’ tumor-infiltration patterns may contribute to response and resistance following anti-tumor immunotherapies (Lu et al., 2019). We thus applied Slide-TCR-seq to longitudinally-collected specimens from a patient with clear cell renal cell carcinoma (RCC) who developed resistance following PD-1 immune checkpoint blockade (ICB) treatment. These samples consisted of a baseline primary kidney tumor and a post-treatment therapy-refractory lung metastasis, in which cell types were assigned using RCTD with scRNA-seq data (Braun et al., 2021) from an RCC cohort that included this patient (Figures 3, S3A–C). We show that Slide-TCR-seq can facilitate the spatial dissection of transposable elements and endogenous retroviruses expression (Figures S4H–K). Confirming the recovery, we found high correlation between the clonotype fractions in Slide-TCR-seq and bulk TCR sequencing (Pearson’s r=0.79, p=9×10−15, Figure S3H). Moreover, the clonotype fractions based on variable region sequences on Slide-TCR-seq arrays obtained from consecutive serial sections were likewise correlated, confirming our expectation of relative stability in the clonal fraction of predominant clones between serial sections (Pearson’s r=0.42–0.45, all p<1×10−5; Figure S3I).
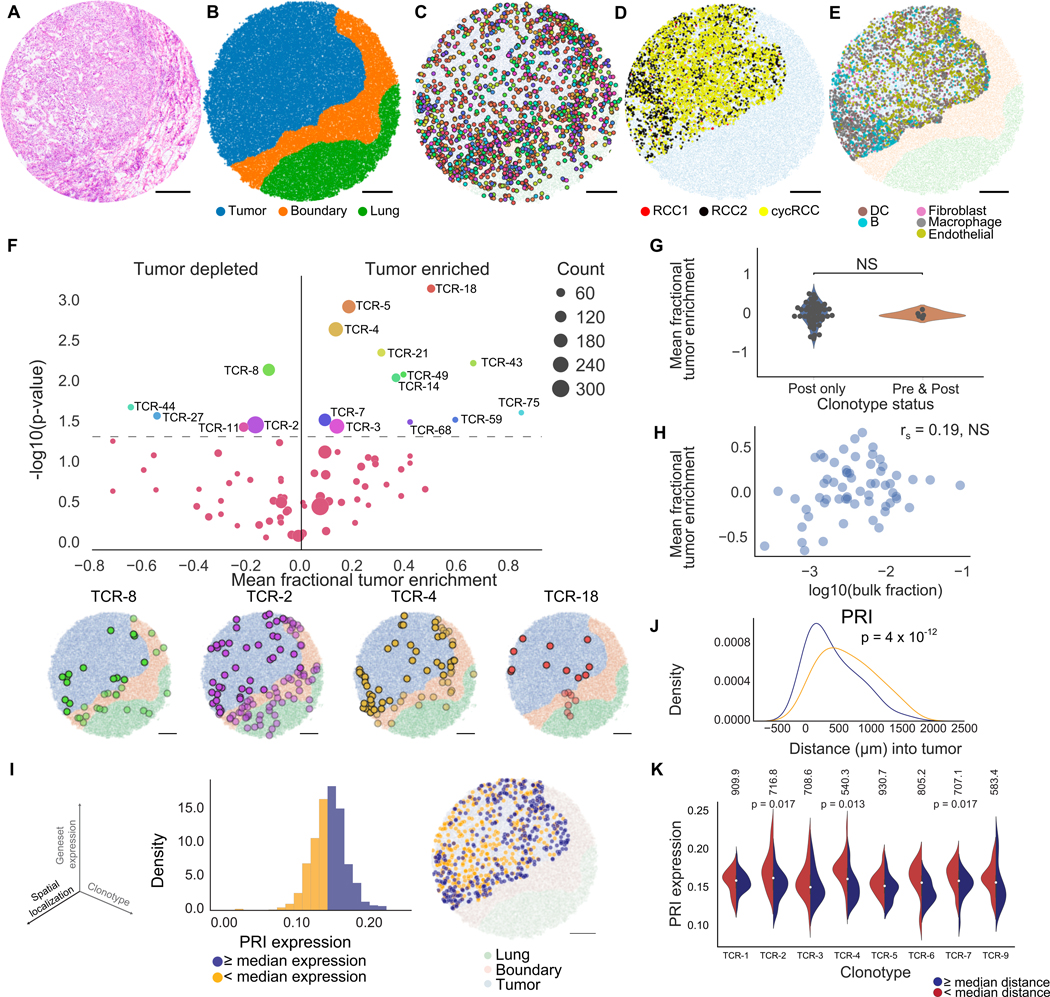
Figure 3. Slide-TCR-seq identifies spatial differences between T cell clonotypes in renal cell carcinoma.
(A) H&E stain of an RCC metastasis to the lung following treatment with a PD-1 inhibitor.
(B) Compartment assignment of the lung (green), boundary (orange), and tumor (blue) of a representative Slide-TCR-seq array from three replicates by applying K-nearest neighbors to cell types determined by unsupervised clustering.
(C) Spatial localization of T cell clonotypes (n=549 clonotypes, colored by clonotype) of a representative Slide-TCR-seq array from three replicates.
(D) Within-tumor spatial localization of 3 distinct RCC cell subtypes (STAR Methods) of a representative Slide-TCR-seq array from three replicates.
(E) Within-tumor spatial localization of a representative Slide-TCR-seq array from three replicates of the five most abundant non-tumor non-T cell types as determined by RCTD and using the combination of five scRNA-seq datasets as reference. DC = dendritic cell.
(F) Top: Significance of clonotype spatial distributions compared against all other clonotypes with at least ten beads per array from the post-treatment specimen plotted against tumor enrichment (n=3 serial arrays, two one-tailed K-S tests) with Bottom: Visualization of selected significant clonotypes, ordered by tumor enrichment, in tissue compartments for a single array (T cells within the tumor compartment are displayed as opaque, T cells within other compartments are shown as translucent).
(G) Comparison of the tumor enrichment of clonotypes that are exclusive to the post-treatment sample (n=48 clonotypes) versus clonotypes that are shared with the pre-treatment sample (n=5 clonotypes) across all serial arrays.
(H) Lack of correlation between the mean tumor enrichment of clonotypes in Slide-TCR-seq and their fraction in bulk TCR-seq from the same post-treatment specimen, by Spearman’s correlation.
(I) Left: The three axes – spatial localization, gene expression, and T cell clonotype – that Slide-TCR-seq can relate. Center: distribution of poor response to immune checkpoint inhibitor treatment (‘PRI’) gene set expression across all clonotypes in the tumor region for the post-PD-1 inhibitor RCC lung metastasis in a single array with kernel density estimation. Yellow = clonotypes with lower than median PRI expression; purple = clonotypes with PRI expression greater than or equal to the median value. Right: localization of low (yellow) and high (purple) PRI gene set expression clonotypes within the tumor region (light blue) from the Slide-TCR-seq array shows their distinct spatial separation (light blue = tumor region, orange = boundary region, green = lung region).
(J) Smoothed histograms comparing the distance infiltrated into the tumor by two-tailed K-S test comparing low (yellow) and high (purple) expression clonotypes, as dichotomized by median expression of PRI.
(K) Expression of PRI gene set across clonotypes with at least 20 beads (n=7 clonotypes), dichotomized based on each clonotype’s median distance infiltrated into the tumor from the tumor edge (red = less infiltrated, blue = more infiltrated; the median distances in μm are displayed along the top). TCR-4 exhibited higher PRI expression when localized closer to the tumor edge than farther from the edge (by two-tailed K-S test). See STAR Methods for n of each distribution. All scale bars: 500 μm. NS = nonsignificant. See also Figures S3–5.
Given the diversity of clonotypes and the presence of a tumor-lung interface, we focused on how clonotype distributions were related to their localization in the post-ICB metastasis (Figure S3J, Table S3). We used unsupervised clustering, cell markers, and the expression of CXCL9 – a chemokine facilitating T cells’ tumor infiltration (Chow et al., 2019) – to designate three compartments consisting of tumor, lung, and intervening boundary (Figures 3B, S4, STAR Methods). We examined the T cell clonality within our samples by evaluating the TCRβ CDR3 sequences and observed 1,223 unique TCRβ T cell clonotypes across the three consecutive arrays obtained from the post-ICB specimen – several with distinct spatial distributions spanning the three tissue compartments (Figures 3C and S4E, Table S3). To examine the spatial enrichment of different TCR clonotypes more deeply across tissue compartments, we focused on the most prevalent TCRβ clonotypes, each with at least ten counts per array. We tested the spatial distribution for each (based on their individual cells’ distances from the tumor edge) against the aggregated spatial distribution of all other clonotypes (across n=3 arrays, STAR Methods). We found that 17 (21.8%) had spatial distributions that differed from all other clonotypes – 12 clonotypes were tumor-enriched, while 5 were tumor-depleted (p < 0.05, Figures 3F and S4F, Table S3). Of these, TCR-4, TCR-5, and TCR-18 were the most enriched in their spatial distributions (p=0.002, p=0.001, p=0.0007, STAR Methods) and had higher tumor enrichment, a property not observed with the predominant clonotype (TCR-1) in the post-treatment specimen.
When we examined the overlap in TCR repertoire between the pre- and post-ICB RCC samples, only 10 (0.8%) of the 1,223 TCRβ clonotypes in the post-treatment arrays were also present in the pre-treatment arrays, supporting the notion that most clonotypes may be recent immigrants to the metastasis (Figures S3K–L, Table S3). Although pre-existing T cells have been reported to have limited reinvigoration capacity (Yost et al., 2019; Wu et al., 2020), we found no difference in tumor enrichment between clonotypes that were exclusive to the post-treatment specimen and clonotypes that were shared with the pre-treatment specimen (Figure 3G). Additionally, we noted the lack of correlation between the extent of clonotype tumor enrichment by Slide-TCR-seq and clonotype frequency by bulk tissue TCR-seq (Figure 3H), emphasizing the ability of Slide-TCR-seq to probe whether preexisting T cell clones exhibit distinct spatial patterning from recently infiltrated clones.
Since we observed spatial tumor enrichment differences between TCRβ clonotypes in the RCC metastasis, we asked whether we could investigate the underlying relationships between gene expression, depth of tumor infiltration, and clonotype sequence (Figure 3I, S3D–G). To evaluate how gene expression differences were associated with the depth of the tumor-infiltrating T cells (TILs) in the context of ICB resistance, we focused on the differential expression of a previously curated T cell gene set associated with poor response to ICB treatment (PRI) (Sade-Feldman et al., 2018) (Table S2). Although this geneset was originally devised from a melanoma context rather than in RCC, we found that it encompassed many known exhaustion markers (e.g., CD38, HAVCR2, CTLA4) and thus could still be useful in characterizing T cells in RCC. By comparing the PRI expression of T cells within each clonotype that were either closer to or farther from the tumor edge, we identified three clonotypes (TCR-2, TCR-4, TCR-7) for which clonotype-specific T cells more deeply infiltrated into the tumor had lower PRI expression than those closer to the tumor edge (p=0.017,0.013, and 0.017, respectively, two-tailed K-S test, Figure 3K). These data indicate that the T cells’ PRI expression was not only associated with the extent of their infiltration into the RCC metastasis but also that the effect was clonotype-specific, as different clonotypes exhibited distinct patterns of PRI expression by the depth of infiltration.
Tertiary lymphoid structures and tumor regions exhibit different degrees of clonal expansion and T cell states
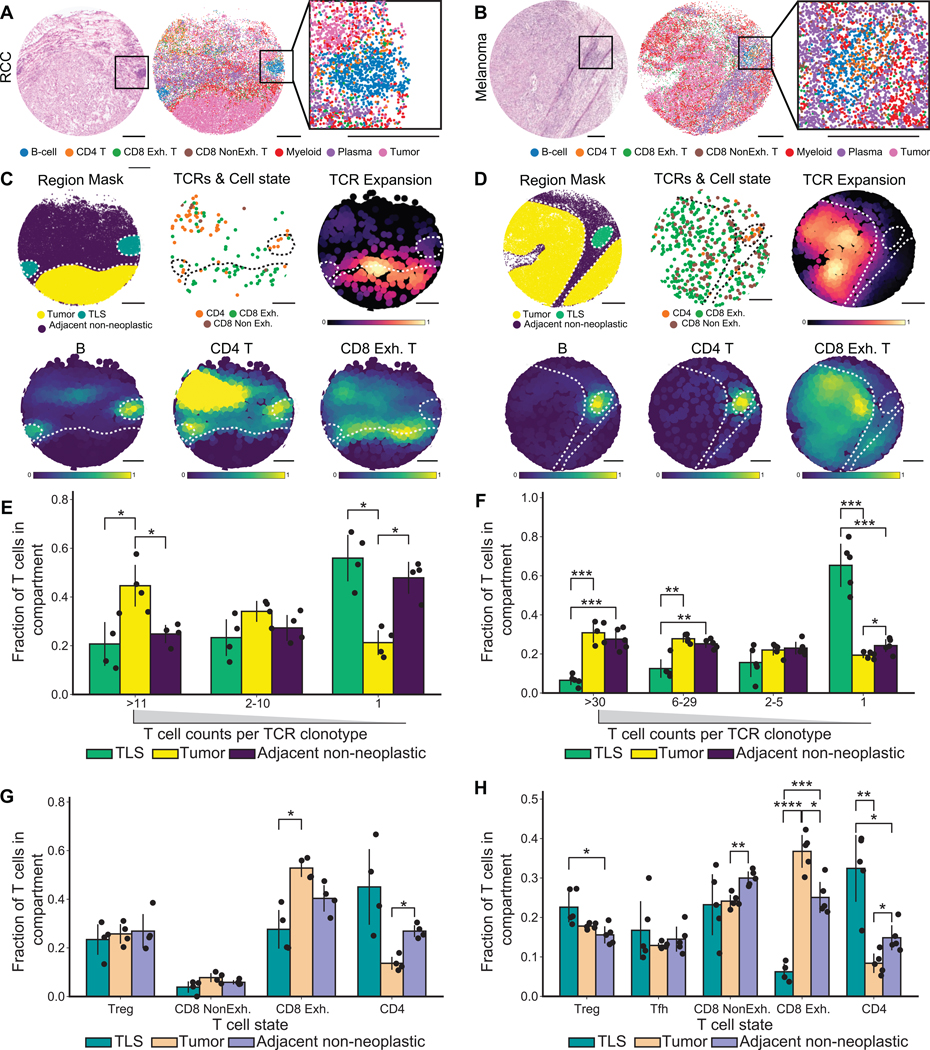
We next asked if we could study niche-level differences in T cell state and expansion across other tumor types. We focused on melanoma because of the well-characterized roles played by TCR repertoire expansion and T cell phenotype in the melanoma immune microenvironment (Sade-Feldman et al., 2018; Oliveira et al., 2021, 2022). We applied Slide-TCR-seq to multiple regions of an immunotherapy-naive axillary lymph node metastasis of malignant melanoma, which displayed T cell infiltration, a distinct multilobular spatial architecture, and interface with neighboring non-neoplastic tissue. We identified the presence of tertiary lymphoid structures (TLSs) in both the aforementioned post-treatment RCC sample and this melanoma sample (Figures 4A–B, S5). TLSs have been found in tissues with antigen persistence, including cancer, and have been hypothesized to play a role in adaptive anti-tumoral immune responses, as well as potentially associated with improved patient outcomes (Sautès-Fridman et al., 2019; Meylan et al., 2022). We observed that T cells located within the tumor regions were more clonally expanded than those in the TLSs (Figures 4C–D, S5), including across multiple arrays sampled from different regions of both the RCC and melanoma samples (Figures 4E–F, S5 n=4 arrays across two regions, n=5 arrays across three regions respectively). Furthermore, T cells in TLSs tended to be CD4+ T cells, while those infiltrating into tumor tended to be CD8+ T cells with an exhausted phenotype (p<0.05 by t-test, FDR-corrected; Figures 4C–D, 4G–H, S5). Our observations that the distinct T cell phenotype of T cell clonotypes in TLSs, that were neither actively expanding nor infiltrating into tumors, support separate roles for their presence, such as potentially facilitating B cell differentiation (Meylan et al., 2022).
Figure 4. Clonal expansion and T cell state between tertiary lymphoid structures and tumor regions in renal cell carcinoma and melanoma.
(A-B) Aligned H&E of a serial section (left), representative Slide-TCR-seq array (center) with beads colored by RCTD cell type assignments, and zoomed-in Slide-TCR-seq of tertiary lymphoid structure (TLS) (right) in RCC (A) and melanoma (B).
(C-D) Region mask of a representative Slide-TCR-seq array delineating tumor, TLS, and adjacent non-neoplastic regions (top left); beads containing both CDR3 and T cell state colored by RCTD assignment (top center); Gaussian-filtered heatmap visualizing TCRs weighted by the extent of clonal expansion (top right); Gaussian-filtered heatmap normalized to maximum values in TLSs and tumor regions, visualizing the location of B cell, CD4+ T cell, and CD8+ Exhausted (Exh) T cell types (bottom); in RCC (C) and melanoma (D).
(E-F) Grouped barplot visualizing the fraction of T cells in each compartment (TLS, tumor, and adjacent non-neoplastic) grouped by the extent of clonal expansion of the T cells in RCC (E; n=4 arrays across two regions) and melanoma (F; n=5 arrays across three regions).
(G-H) Grouped barplot visualizing the fraction of T cells in each compartment (TLS, tumor, adjacent non-neoplastic) grouped by their T cell states in RCC (G; n=4 arrays across two regions) and melanoma (H; n=5 arrays across three regions).
Barplots indicate means and error bars indicate standard deviation of replicates. All scale bars: 500 μm. All statistical tests are by t-test and corrected for false discovery rate, * p = 0.05, ** p = 0.005, *** p = 0.0005, **** p = 0.00005. Treg = regulatory T cell; Tfh = follicular T helper cell; NonExh = non-exhausted; Exh = exhausted. See also Figure S5.
Unique interactions are detectable between a T cell clone and adjacent cells in the melanoma microenvironment
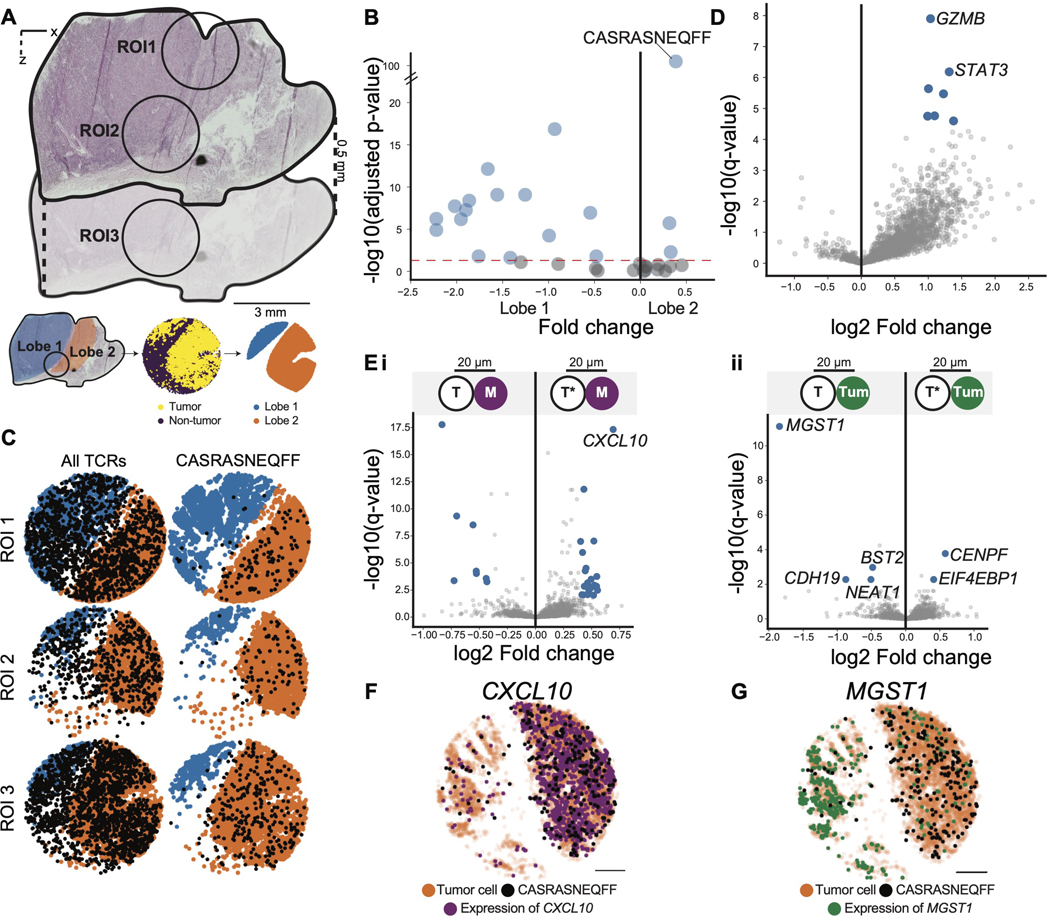
Through Slide-TCR-seq analyses of the metastatic melanoma sample, we observed that one T cell clone, with CDR3 sequence CASRASNEQFF, was preferentially enriched in one of the tumor lobes – a lobular compartment of the melanoma separated from the adjacent tumor lobes by a fibrous septum (Figures 5A–C, S5C, p = 1 10−102). Upon examining whether this T cell clone displayed cell-autonomous differences in gene expression compared to other TIL clones, we found that GZMB (associated with cytotoxic T cell function) and STAT3 (associated with enhanced survival of activated T cells (Yu et al. 2013)) were the most upregulated transcripts (Figure 5D, p = 1×10−8, p = 7×10−7, Table S4, STAR Methods).
Figure 5. Interactions between a spatially unique T cell clone and tumor cells in melanoma.
(A) H&E of melanoma tumor sample (top) and schematic illustrating the delineation of two distinct lobes (orange and blue) of the tumor across the corresponding H&E, Slide-TCR-seq arrays, and constructed region masks (bottom). ROI = region of interest.
(B) Spatial enrichment of TCRs between the two lobes, tested by chi-square test (n=7 arrays across three regions).
(C) Visualization of all TCRs (left) and the CASRASNEQFF TCR clonotype (right) across one array from each of the three regions. These visualizations are representative of the findings across the seven arrays, shown in Figure S5.
(D) Differentially expressed genes between T cells with the CASRASNEQFF clonotype (right) and all other TCRs (left) (STAR Methods), with blue dots representing statistically significant differentially-expressed genes with absolute log fold change > 0.4 (n=7 arrays across three regions).
(E) i. Differentially expressed genes between monocytes (M) within 20 μm of T cells with CASRASNEQFF (right; T*) and monocytes within 20 μm of all other TCRs (left; T), excluding any monocytes that are within 20 μm of both T cells with CASRASNEQFF and other TCRs. ii. Differentially expressed genes between tumor cells (Turn) within 20 μm of T cells with CASRASNEQFF (right; T*) and tumor cells within 20 μm of all other TCRs (left; T), excluding any tumor cells that are within 20 μm of both T cells with CASRASNEQFF and other TCRs (STAR Methods). Blue dots represent statistically significant differentially-expressed genes with absolute log fold change > 0.4 (n=7 arrays across three regions).
(F) Beads colored by CXCL10 expression (purple, counts ≥2) in one representative array of seven. arrays
(G) Beads colored by MGST1 expression (green, counts ≥1) in one representative array of seven arrays. Scale bars: 500 μm See also figure S5.
We next investigated the cell non-autonomous mechanisms unique to the CASRASNEQFF clone by comparing the transcriptional profiles of the cells adjacent to the CASRASNEQFF T cells against those of the cells neighboring all other TIL clonotypes (STAR Methods). In particular, we found that the monocytes neighboring CASRASNEQFF T cells exhibited elevated expression of the chemokine CCCL10 (p = 5×10−21, Figure 5Ei, Table S4), which has been shown to recruit tumor-reactive effector T cells (Spranger et al., 2017). Notably, monocytic expression of CXCL10 was higher in the same lobe enriched with CASRASNEQFF T cells (Figure 5F), implicating a preferential interaction between CASRASNEQFF T cells and tumor-infiltrating monocytes. Of note, we also found that the tumor cells neighboring the CASRASNEQFF T cells displayed several differentially expressed genes, including lower expression of MGST1 (p=3×10−15, Figure 5Eii, 5G, Table S4), a tumor-associated gene that has been shown to repress ferroptosis (Kuang et al., 2021; Wang et al., 2019). The spatial differences highlighted by this clone and the gene expression differences between cell types in distinct regions of melanoma emphasize the need for further study into the spatial-transcriptomic heterogeneity of T cells and the tumor microenvironment.
Discussion
Slide-TCR-seq is a straightforward methodology that can be applied to existing, routine frozen tissue samples without the need for specialized processing and produces data outputs that can be processed with commonly used single-cell and spatial analytical software packages. As a result, our approach can be readily used to answer a broad spectrum of important questions in immunology, including investigations of T cell biology and TCR repertoires in primary and secondary lymphoid organ development, infectious disease, autoimmunity, cancer immunology, and the effects of immunomodulatory and immunotherapeutic agents. For example, by employing untargeted poly(A) capture, Slide-TCR-seq could also enable the detection of polyadenylated viral transcripts, which could enable the study of how antiviral T cell clonotypes are spatially and transcriptionally related to virus-infected cells across multiple viral settings (e.g., HIV, HPV, SARS-CoV-2) (Golumbeanu et al., 2018; Steuerman et al., 2018; Bost et al., 2020). Likewise, Slide-TCR-seq can facilitate the spatial dissection of transposable elements and endogenous retroviruses expression and how they relate to immune responses. Furthermore, because our method relies on the capture and sequencing of nucleotide sequences, it is also potentially compatible with oligonucleotide-barcoded antibody reagents for protein staining (Stoeckius et al., 2017) – enabling the simultaneous interrogation of T cell clonotypes’ transcriptional and protein expression. Finally, because Slide-TCR-seq’s modified rhPCR utilizes readily available primer sets (Li et al., 2019), we envision that our method can be extended to the spatial mapping of additional targets in immunology where longer sequence read information is needed, such as B-cell receptors in B lymphocytes and mutation-derived neoantigens in tumor cells.
In summary, we have reported Slide-TCR-seq, a method that enables sequencing transcriptomes and TCRs at 10-μm spatial resolution. Slide-TCR-seq facilitates the study of the complex spatial relationships between T cell clonotypes and other cell types within tissues across healthy and diseased settings – as illustrated by our analyses of a transgenic TCR mouse model’s native splenic architecture, compartmentalization of TCRs in human lymph nodes, spatially-defined transcriptional differences between TILs of the same clonotype, and distinct clonotypic repertoires in TILs vs. tumor-associated TLSs. Altogether, our findings highlight Slide-TCR-seq’s ability to integrate T cell transcriptional profiles, localization, and clonotypic identity, thereby enabling users to dissect inter- and intra-clonotype spatial transcriptional heterogeneity. We anticipate that Slide-TCR-seq will be a valuable technology for studying the complex spatial relationships between T cell clonotypes, neighboring cell types, and gene expression among diverse immunological and immunotherapeutic contexts, yielding insights into the spatially-organized mechanisms that underlie T cell responses.
Limitations of Study
Slide-TCR-seq is limited by the capture rates inherent in barcoded beads. As bead capture reagents and capabilities advance in molecular biology, we anticipate that analysis of paired TCR alpha-beta clonotypes will also improve. We also expect that future experimental work with amplification alternatives to PCR will enhance the fidelity of the amplification process and permit the use of less stringent computational filters. Additionally, although Slide-TCR-seq was initially developed for research use with fresh-frozen tissue, ongoing refinement of this technology will enable the analysis of formalin-fixed paraffin-embedded tissues.
STAR Methods
Lead Contact
Correspondence should be directed to the Lead Contact: Fei Chen (chenf@broadinstitute.org)
Materials Availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Fei Chen (chenf@broadinstitute.org)
Data and Code Availability
All raw Slide-TCR-seq data are available at the Broad Institute Single Cell Portal at: https://sing1ecell.broadinstitute.org/single_cell/study/SCP1348/. Arrays with cell type prediction have also been uploaded to https://cel1xgene.cziscience.com/co1lections/02b0l703-bflb-48de-b99a-23bef8cccc81 for visualization and browsing. Single-cell RNA sequencing data for RCC were used from dbGaP (dbGaP:phs002252.vl.p1 at https://www.ncbi.nlm.nih.gov/gap/). Single-cell RNA sequencing data for mouse spleen were used from NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE132901. Single-cell RNA sequencing data for melanoma were used from European Genome-Phenome Archive (EGA), under the accession code EGAS0000l 00511. The code used can be found at https://github.com/MacoskoLab/slideseq-tools and https://github.com/soph-liu/Slide-TCR-seq.
Sample information and processing
All procedures involving animals at the Broad Institute were conducted in accordance with the US National Institutes of Health Guide for the Care and Use of Laboratory Animals under protocol number 0211–06-18. Male C57BL/6-Tg OT-I mice (The Jackson Laboratory) were housed in a 12-h light-12-h dark cycle with ad libitum access to food and water. At 5–7 weeks, they were anesthetized with 3% isofluorane in a gas chamber until a negative foot pinch response. They were then moved to nose-cone isofluorane and transcardially perfused with ice-cold lX pH 7.4 PBS to remove blood. Following a color change in the liver, spleens were harvested, embedded in optimal cutting temperature (OCT) compound, and flash-frozen for one minute in liquid nitrogen chilled isopentane and stored at −80°C. RCC specimens were acquired with informed consent from a patient who underwent nephrectomy for AJCC pT2b cM1 histologic grade 4 clear cell RCC (i.e., the pre-treatment specimen) and then received approximately 11 months of a PD-1 inhibitor and VEGF inhibitor until resection of progressing lung metastasis (i.e., the post-treatment specimen) (Braun et al., 2021). Melanoma specimens were acquired from a patient who underwent axillary lymphadenectomy for metastatic BRAF-mutant melanoma prior to starting PD-1 inhibitor. Human RCC, melanoma, lymph node, and tonsil specimens were embedded in OCT, snap-frozen following surgery, and stored at −80°C. Institutional Review Board approval was obtained from Dana-Farber Cancer Institute, the Broad Institute, and Brigham and Women’s Hospital.
Histological processing
Serial 5–10 μm thickness sections from the frozen tissue samples were mounted on glass slides. For hematoxylin & eosin (H&E), sections were fixed in either 10% neutral-buffered formalin or methanol, processed through a graded ethanol series, stained with H&E, dehydrated, coverslipped, and imaged.
Bulk TCR sequencing
Five 30 μM sections of each frozen sample were taken, and RNA was extracted using the RNeasy Micro kit (Qiagen) as per the manufacturer’s protocol. Alpha and beta TCR repertoire analysis in bulk RNA samples was performed using an adapted rhTCRseq protocol published previously (Li et al., 2019). Specifically, 10 ng bulk RNA was used in each RT reaction, and four replicates were done for each sample. Excess RT primers were eliminated by exonuclease digestion, and then rhPCR was done based on the published protocol (Li et al., 2019). After the sequencing library was made, it was sequenced using MiSeq 300 cycle Reagent Kit v2 on the Illumina sequencing system according to the manufacturer’s protocol with 248-nt read 1, 48-nt read 2, 8-nt index 1, and 8-nt index 2. The sequencing data analysis was done based on the method published previously (Li et al., 2019).
In situ transcriptome processing via Slide-seq
Slide-seq arrays were prepared and spatial bead barcodes sequenced following Slide-seqV2 (Stickels et al., 2020) protocol using arrays created with custom synthesized barcoded beads (5’-TTT_PC_GCCGGTAATACGACTCACTATAGGGCTACACGACGCTCTTCCGATCTJJJJJJJJTCTTCAGCGTTCCCGAGAJJJJJJNNNNNNNVVT30-3’) with a photocleavable linker (PC), a bead barcode sequence (J, 14 bp), a UMI sequence (NNNNNNNVV, 9 bp), and a poly dT tail.
OCT-embedded frozen tissue samples were warmed to −20°C in a cryostat (Leica CM3050S) and serially sectioned at a 10 μm thickness (2–3 Slide-seq array replicates per sample), with consecutive sections used for hematoxylin and eosin staining and immunofluorescence staining. Each tissue section was affixed to an array and moved into a 1.5 mL Eppendorf tube for downstream processing. The sample library was prepared as previously described (Stickels et al., 2020) except with the following modifications. Library amplification before tagmentation for the human samples included an additional two cycles, for a PCR program of 1 cycle of 98°C for 2 min, 4 cycles of 98°C for 20 s, 65°C for 45 s, 72°C for 3 min, 11 cycles of 98°C for 20 s, 67°C for 20 s, 72°C for 3 min, and 1 cycle of 72°C for 5 min. The PCR was performed in a final volume of 200 μL of PCR mix, divided into 4 PCR tubes.
Libraries were sequenced using the following read structure on a NovaSeq (S2; Illumina): Readl: 42 bp; Read2: 41 bp; Index1: 8 bp, and sequences were processed as previously described (Stickels et al., 2020) using the pipeline available at https://github.com/MacoskoLab/slideseq-tools.
TCR clonotype enrichment via rhTCRseq
As previously described (Li et al., 2019), several solutions were prepared for the first rhPCR reaction. 20× rhPCR buffer was made (final concentration: 300 mM Tris-HCl, pH 8.4; 500 Mm KCl; 80 mM MgCl2). RNase H2 was diluted to 20 mU/μL using RNase H2 Dilution Buffer (IDT). For the human TCRs, 69 rhPCR primers specific for the V segments of human alpha and beta TCR genes, Rd2.AV.xl/Rd2.BV.xl (Table S2) were combined at a concentration of 5 μM each was made by mixing 5 μL of each primer at 500 μM with 155 μL of TE buffer. For the mouse TCRs, 56 rhPCR primers specific for the V segments of mouse alpha and beta TCR genes were used (Table S2).
The reaction was prepared as follows: 6 μL of Slide-seq cDNA library (0.5ng/μL); 8μL of 6μM each of P5.IDTxxx.Rd1x.x1 and P7.IDTxxx.Rd2x.x1 primers, same as in the earlier publication (Li et al., 2019), except that we added eight additional new P5x.IDTxxx.Rd1x.x2 primers (Table S2). 10 μL of rhPCR master mix (final concentration: 1× rhPCR Buffer, 400 μM dNTPs, 50 nM of Rd2.AV.x1 and Rd2.BV.xl primers, 0.5 mU/μL RNase H2, 0.2 units/μL One Taq hot-start DNA polymerase [New England BioLabs M0481L]). The following program was used on the thermal cycler: 1 cycle of 95°C for 5 min, 18 cycles of 96°C for 20 s, 60°C for 4 min, and 72°C for 2 min, and then hold at 4°C. Then 19.2 μL of ProNex beads (0.8× clean-up; Promega NG2003) were added for clean-up following the manufacturer’s instructions and eluted into 20 μL of ProNex Elution Buffer.
From this, 5 μL was taken for a subsequent PCR with P5 and P7 primers. This was added to 45 μL of reaction mix (final concentration: l× Q5 Ultra II PCR Master Mix [New England BioLabs M0544L], 500 nM P5 primer, 500 nM P7 primer). The following program was used on the thermal cycler: 1 cycle of 98°C for 30 s, 12 cycles of 98°C for 10 s, 62°C for 1 min, 75°C for 1 min, 1 cycle of 75°C for 2 min, and hold at 4°C. Then, 40 μL of ProNex beads (0.8× clean-up; Promega NG2003) were added following the manufacturer’s instructions and eluted into 20 μL of ProNex Elution Buffer. 1 μL of each library was used to confirm the fragment size distribution and yield using an Agilent High Sensitivity DNA Kit (Agilent Technologies) on an Agilent 2100 bioanalyzer. Libraries were loaded at 4 pM and sequenced using the following read structure on a MiSeq 300 cycle kit (V2; Illumina): Read1: 48 nt, Read2: 248 nt, Index1: 8 nt, Index2: 8 nt.
To see whether a long-read sequencing platform can increase the quality of the TCRseq data, we adapted the Oxford Nanopore system. Specifically, after TCRseq libraries were prepared, we added the nanopore adaptors using the Nanopore ligation kit (Nanopore, SQL-LSKl 12) and purified and eluted them. The final sequencing libraries were loaded onto R10 flow cells (FLO-MINl12) in the Nanopore MinION MklC system with 10 fmol library input.
Processing TCR sequencing data
We identified variable TCR sequences using MiXCR (v3.0.13) (Bolotin et al., 2015) to align to the human or mouse genomes. For Miseq data to account for sequencing errors-introduced diversity, we corrected the CDR3 amino acid sequences with a Hamming distance of 1 and collapsed to the most predominant sequence. We then identified the bead barcode and UMI sequence, removing all reads that did not have a match within a Hamming distance of 1 to an in situ sequenced bead barcode. For sequencing data generated by Oxford Nanopore, raw signals were base called using the super-accurate model in guppy 6.1.2. Reads were aligned to constant alpha and beta sequences of either human or OT-1 mouse using minimap2 (Li, 2018).
Alignments were filtered to cover at least 95% of the constant region as a pre-filtering step to reduce chimeras. 150 bases from the 5’ soft-clipped part of the alignments were extracted to assign clonotypes with MiXCR. 300 bases from the 3’ soft-clipped part of the alignments were searched for the linker sequence in slide-seq barcodes within one edit distance. The candidate barcodes were then corrected against in situ barcodes within one edit distance. The clonotypes were collapsed to 95% identity at the nucleotide level of the CDR3 region by increasing mutational probability in MiXCR to 5%. For each bead, a clonotype was assigned only if 95% or more of the remaining reads came from one clonotype. Otherwise, the bead was removed from further analysis. For both MiSeq and Oxford Nanopore data, we generated a knee plot, ranking the beads based on number of reads aligning to a TCR CDR3, and identified the inflection point (which highlights the point at which it becomes difficult to distinguish genuine TCRs from sequencing or amplification-introduced chimeras, and is a built-in control for sequencing depth) and set that as the cutoff to only consider high-confidence reads with a low-likelihood for sequence chimerism for subsequent analyses; a minimum cutoff was at least 2 UMIs per bead or 2 reads per UMI. For samples at the minimum cutoff, we then added back any variable TCR reads that had a bead barcode and UMI sequence that matched exactly with a barcode-UMI mapping to Trac, TRAC, Trbc2 or TRBC2 sequenced in Slide-seq.
Cell type and compartment identification
Robust cell type decomposition (RCTD; v1.1.0) (Cable et al., 2021) was performed on spleen, lymph node, tonsil, and RCC samples, using published scRNA-seq datasets as cell type references. For the spleen sample, we added two classes of immune cell identity: CD4 T cell, CD8 T cell, and memory T cells, all mapping to T cells, and CD8 macrophage and CD4 macrophage mapping to macrophage, as annotated from a published spleen scRNA-seq dataset (Kimmel et al., 2019). RCTD initially tries to make cell type assignments using the broad cell type (e.g., T cell, macrophage) and then reports the most likely subtype (e.g., CD4, CD8, or memory for T cell). Broad cell types were annotated for the lymph node sample from a published thymus scRNA-seq dataset (Park et al., 2020). For the RCC specimens, broad cell types of tumor, myeloid, NK, T cell, and B cell populations were annotated from a published RCC scRNA-seq dataset that included this patient (Braun et al., 2021).
To identify cellular compartments, unsupervised clustering using Seurat v3 (v3.2.3) (Stuart et al. 2019) was performed on the RCC specimens. For each specimen, replicate arrays were merged into a single object, transformed using sctransform (v0.3.2) (Hafemeister and Satija, 2019), and a Uniform Manifold Approximation and Projection (UMAP) (McInnes, Healy and Melville, 2018) plot was generated using the first 30 principal components. In the post-therapy RCC specimen, we performed unsupervised clustering and identified a tumor cell cluster, a non-neoplastic lung cluster, and a tumor-lung ‘boundary’ cluster by differential expression of marker genes (Figure S4), which aligned with the tissue compartments identified in the corresponding H&E. The intervening boundary cluster was marked by high expression of the CXCL9 chemokine gene and closely corresponded spatially with the band of myeloid cells identified in RCTD analysis. Cell type clusters were used to delineate tissue compartments in each array by first plotting the location of beads from the tumor, lung, and boundary clusters and then assigning all other beads to one of those three clusters by using a k-nearest neighbors classifier (KNeighborsClassifier, from Scikit-learn (Pedregosa et al., 2011)). In the pre-treatment RCC specimen, no non-neoplastic tissue compartment was identified by either unsupervised clustering or histological examination.
For the melanoma samples, a publicly available scRNA-seq dataset (Wu et al., 2021) was re-clustered, and subtypes of T-cells were annotated into five groups, consisting of naive/memory CD4+ T cells, T follicular helper cells, T regulatory cells, exhausted CD8+ T cells, memory/cytotoxic CD8+ T cells. These subtypes were then mapped to the Slide-TCR-seq data using cell2location (Kleshchevnikov et al., 2022) using the scRNA-seq dataset as a reference.
TCR variable sequence recoverv and spatial concordance with TCR constant sequences
For each bead barcode on the spleen array, we determined the number of UMIs for each Trac or Trbc2 constant sequence using the Slide-seq data. For the bead barcodes with n UMIs for Trac or Trbc2, we then ascertained whether a variable CDR3 sequence was also detected and calculated F, the fraction of all bead barcodes at n counts of constant UMIs that had at least one variable sequence for Figure IE.
In Figure 1E, the height of each bar is the weighted average fraction, Fc, of beads with c constant TCR UMIs that also have variable TCR UMIs. For each replicate, i, we went through c, each number of constant TCR UMIs, and calculated Fi, the fraction of beads with variable TCRs measured from each replicate. We then weighed Fi by ni, the number of beads for each replicate, and averaged the weighted values. A weighted average better controls for variation in sequencing depth between replicates.
Similarly, the weighted standard deviation was calculated by the following using the DescrStatsW() function from statsmodels (Seabold et al., 2010).
To determine spatial concordance for the constant and variable sequences in Figure S1, we generated two heatmaps of the spleen array with bin sizes of 50 pixels (i.e., 32.5 μm). The first heatmap displayed beads with Tcra or Trbc2 constant sequences, and the second heatmap showed beads with TCRɑ or TCRβ variable sequences. We then determined the Pearson’s correlation between the number of beads in each bin between the two heatmaps.
Benchmarking Slide-TCR-seq against bulk TCR sequencing
For Figure S3H, we filtered for clonotypes that had at least ten beads across all post-treatment RCC arrays in Slide-TCR-seq and at least ten counts in the post-treatment bulk TCR sequencing to determine the Pearson’s correlation. Clonotype fractional values for Slide-TCR-seq were determined by dividing the total number of beads for each clonotype across all three arrays by the total number of beads with TCR CDR3 sequences.
Clonotype spatial enrichment analysis
In the post-treatment RCC specimen, after identifying the tumor, lung, and boundary compartments in each array, we then filtered for clonotypes composed of at least ten beads to be able to analyze their spatial distributions robustly. Clonotype enrichment in each compartment was calculated as follows, where Ec = clonotype enrichment for the compartment c; Cc = number of beads for a clonotype in the compartment c; Call = total number of beads for a clonotype (across all compartments); Ac = number of beads for all other clonotypes in the compartment c; and Aall = total number of beads for all other clonotypes (across all compartments):
We determined the tumor edge by identifying beads in the KNN-assigned tumor compartment with less than 90% of neighboring beads (within a 50 pixel [i.e., 32.5 μm] distance) mapping to the tumor compartment. The spatial distribution for each clonotype was evaluated by measuring the distance from each bead with that clonotype to the nearest point on the tumor edge. The aggregate distribution for each clonotype was generated by measuring the distances from all other beads to their closest point on the tumor edge. Each clonotype had two one-sided K-S tests performed between the clonotype’s spatial distribution and the aggregate spatial distribution. Plots in Figures 3F and S4F show the results of the “greater” K-S test (where the alternative hypothesis was that the spatial distribution of the aggregate was less than the clonotype distribution) for all clonotypes that had positive tumor enrichments and the results of the “lesser” K-S test (where the alternative hypothesis was that the spatial distribution of the aggregate was greater than the clonotype distribution) for all clonotypes that had negative tumor enrichment, with p values combined across all three post-treatment arrays using Fisher’s method (Fisher, 1992).
Clonotype gene set analysis
In the post-treatment RCC specimen, for each bead in the tumor compartment, the UMIs for all the genes in the PRI gene set were summed and normalized by total counts to determine the gene set expression. The gene set expression distribution for all beads containing variable TCR reads was created and plotted as histograms with a kernel density estimation (bandwidth=0.3) in Figure 3I. All beads containing TCR variable sequences in the tumor region were then dichotomized into high expression and low PRI expression groups based on the median gene expression, and spatial distributions were generated from the distance of each bead in the tumor compartment to the closest point on the tumor edge. Depth of tumor infiltration distances was plotted as kernel density estimations (bandwidth=0.5) separately for high and low PRI expression beads in Figure 3J. Two-sided K-S tests were performed to assess if the distributions of tumor infiltration distance differed between high and low expression beads, with correction of p values using the Benjamini-Hochberg method.
In Figure 3K, the analysis applied for all clonotypes that had at least 20 counts overall, or at least ten counts in each of the two groups dichotomized by the depth of infiltration into the tumor. Two-sided K-S tests were performed comparing these groups’ different PRI gene set expression. The number of individuals for each clonotype (displayed as n>median : n<median) were: TCR-1 26:26, TCR-2 20:20, TCR-3 23:23, TCR-4 22:21, TCR-5 16:15, TCR-6 17:16, TCR-7 18:17, and TCR-9 11:10.
The correlation cluster map in Figure S3G between T cell clonotypes was then generated by l) using a heatmap of the effect size (Cohen’s d) of the PRI gene set – comparing expression for each clonotype with at least ten beads compared against the aggregate gene expression of all other clonotypes 2) showing the Pearson’s correlation between each clonotype and 3) clustering by hierarchical clustering.
T-cell clonotype clustering
For the correlation map in Figure S3G, genes from several published T-cell gene sets (Tirosh et al., 2016; Pace et al., 2018; Sade-Feldman et al., 2018; Zemmour et al., 2018; Miller et al., 2019; Hwang et al., 2020) were combined to generate a list of common T-cell genes (Table S2). Non-negative matrix factorization (NMF from Scikit-learn (Pedregosa et al., 2011)) was applied to a matrix containing the bead-normalized expression of these genes across all TCR clonotypes with variable TCR sequence on at least ten beads across three arrays to identify ten different factors. Then, the effect size of the cell loadings of the beads for each clonotype vs. all other clonotypes was calculated by computing the Cohen’s d. This process resulted in a matrix with the dimensions of clonotypes by factors containing values for the effect size of each factor’s cell loading on a clonotype. This matrix was then clustered via hierarchical clustering.
Endogenous Retrovirus (ERV) expression identification
ERV expression in RCC Slide-seq arrays was estimated using Telescope (vl.0.3.1) (Bendall et al., 2019). Genome-aligned BAM files from each RCC Slide-seq array were used as inputs to Telescope, accompanied by the corresponding GTF annotation file containing 60 ERV families across 14,896 loci to quantify the expression of ERVs. The default settings were used except for reassign_mode, where ‘unique’ was selected to include only uniquely aligned reads. The updated BAM files were subsequently used to extract the reads for each ERV feature (using tag ZF) along with the corresponding bead barcodes and UMIs. A count matrix was constructed to facilitate the downstream analysis.
Analysis of spatial cell interactions between TCR clonotypes and tumor features
To assess the degree of colocalization of each clonotype to a feature in the tumor, including subtypes of cancer cells and other cells present within the tumor region, we calculated the proportion of each clonotype’s beads that had at least one bead with the feature within a 13 μm radius. This radius was selected for spatial interaction analyses based on the observation that, in the array’s expression data, adjacent beads were predominantly located within 20 pixels (i.e., 13 μm) of each other. One-tailed Fisher’s exact test was used to test whether the colocalization of the TCR clonotype with the feature was significantly higher than the colocalization displayed by other clonotypes by constructing the contingency table with the following elements: number of colocalized beads of the TCR, number of non-colocalized beads of the TCR, number of colocalized beads of the other TCRs, number of non-colocalized beads of the different TCRs. For the identification of RCC subtypes in Figure 3D, we used the scRNA-seq dataset from Bi et al. as a reference for RCTD (Bi et al., 2021). To improve the non-tumor non-T cell type assignments specific for the microenvironment neighborhood analysis, we incorporated two additional published RCC scRNA-seq datasets (Young et al., 2018; Bi et al., 2021; Krishna et al., 2021) and a lung scRNA-seq dataset (Travaglini et al., 2020) as references for RCTD – all with a less stringent doublet search parameter delta of 5. We then used a voting scheme across the five different RCTD references to arrive at consensus cell type assignments for each bead: first, the bead was assigned as tumor cell if predicted to be tumor type by >1 reference; among the remaining non-tumor cell beads, the bead was assigned as lung epithelial cell if predicted to be either lung or non-neoplastic epithelial types by any reference; finally, microenvironmental cell types (i.e., macrophage, dendritic cell, endothelial cell, B cell, T cell, fibroblast) were assigned to beads if predicted to be the corresponding cell type for that bead by ˃l reference; with multiple cell type assignments to a single bead permitted to account for the physical overlap of multiple cells over the same bead.
Analysis of differential expression in spatial transcriptomic data
To study the differential expression between the CASRASNEQFF clonotype and all other TCR clones in the melanoma samples, we used the Slide-TCR-seq CDR3 data to build an explanatory variable with l = CASRASNEQFF and 0 = all other TCR clones. We processed all data with RCTD (Cable et al., 2021) using a single-cell melanoma reference (Wu et al., 2021) and applied C-SIDE (Cable et al., 2022) in batch mode on all replicates to perform population-level statistical inference. DE outputs report the differentially expressed genes in RCTD-defined T cells over the CDR3-defined explanatory variable.
To study differences in gene expression between cells that were adjacent to CASRASNEQFF clones vs. other T cell clones, we built an explanatory variable that reported 1 for any bead that was within a 30-pixel distance of a CASRASNEQFF T cell and a 0 for any bead that was within a 30-pixel distance of all other TCR clones, excluding any beads that were within a 30-pixel distance of both. We then used C-SIDE (Cable et al., 2022) in batch mode to identify the differentially expressed genes in monocytes and tumor cells over the explanatory variable.
Supplementary Material
Table S1. Comparison of technologies. Related to Figure 1.
Table S2. Oligonucleotides and genesets used. Related to STAR Methods.
Table S3. Bulk and Slide-TCR-seq clonotype sequencing results in OT-I spleen and pre- and post-treatment RCC samples. Related to Figure 3.
Table S4. Differentially expressed genes in spatial transcriptomics data. Related to Figure 5.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| Raw and analyzed data | This paper |
https://singlecell.broadinstitute.org/single_cell/study/SCP1348/
https://cellxgene.cziscience.com/collections/02b01703-bf1b-48de-b99a-23bef8cccc81 |
| Single-cell RNA sequencing data for RCC | dbGaP | dbGaP:phs002252.v1.p1 at https://www.ncbi.nlm.nih.gov/gap/ |
| Single-cell RNA sequencing data for mouse spleen | NCBI Gene Expression Omnibus | GEO:GSE132901 |
| Single-cell RNA sequencing data for melanoma | European Genome-Phenome Archive | EGAS0000100511 |
| Software and algorithms | ||
| Slide-seq-tools for processing spatial arrays | ||
| Robust decomposition of cell type mixtures | Cable et al., 2021 | doi: 10.1038/s41587-021-00830-w. |
| MiXCR: software for comprehensive adaptive immunity profiling | Bolotin et al., 2015 | doi: 10.1038/nmeth.3364 |
| Minimap2: pairwise alignment for nucleotide sequences | Li et al., 2018 | doi: 10.1093/bioinformatics/bty191 |
| Seurat v3 | Stuart et al., 2019 | doi: 10.1016/j.cell.2019.05.031 |
| sctransform | Hafemeister et al. 2019 | doi: 10.1186/s13059-019-1874-1 |
| UMAP | McInnes et al. 2018 | doi: 10.48550/arXiv.1802.03426 |
| Cell2location | Kleshchevnikov et al., 2022 | doi: 10.1038/s41587-021-01139-4 |
| C-SIDE | Cable et al., 2020 | doi: 10.1101/2021.12.26.474183. |
| Other | ||
| Code and analyses | This paper | https://github.com/soph-liu/Slide-TCR-seq |
Highlights.
Slide-TCR-seq measures the location, clonotype, and gene expression of T cells
Germinal centers in human lymph nodes and tonsil have spatially segregated T clones
Cancers can show spatial and transcriptomic inter- & intra-clonotype heterogeneity
Expression profiles differ between T clones in neighboring monocytes and tumor cells
Acknowledgments
We thank Jamie L. Marshall and Wandi Zhang for expert technical assistance, Sabina Signoretti and Toni Choueiri for RCC biobanking, and Aziz Al’Khafaji, Hans T. Bergal, Ravi Uppaluri, Hattie Chung, Haiqi Chen, Zack Chiang, Dylan M. Cable, Lucas Pomerance, and Giacomo Oliveira for helpful feedback. This work was supported in part by the DFCI Glioblastoma Tiger Team, the NIH U19AI082630, and the Dana-Farber/Harvard Cancer Center Kidney Cancer SPORE (NCI P50CA101942). S.Liu is supported by NIH Molecular Biophysics Training Grant (NIH/NIGMS T32GM008313) and the National Science Foundation Graduate Research Fellowship. J.B.I gratefully acknowledges support from the NIH/NCI (K12CA090354) and the Conquer Cancer Foundation/Sontag Foundation. S.Li is supported by the NCI Research Specialist Award (R50CA251956). D.A.B. acknowledges support from the DF/HCC Kidney Cancer SPORE Career Enhancement Program (NCI P50CA l 01942-15) and the DOD Academy of Kidney Cancer Investigators (KC190128). C.J.W. acknowledges support from NIH/NCI R01CA155010 and NIH/NCI U24CA224331 and is supported in part by the Parker Institute for Cancer Immunotherapy. F.C. acknowledges support from NIH Early Independence Award (1DP5OD024583), the NHGRI (R0l HG010647), the Burroughs Wellcome Fund CASI award, and the Merkin Institute.
Declaration of interests
F.C., C.J.W., S.L., J.B.I., K.J.L., S.Li., have filed a patent on this work. D.A.B. reports nonfinancial support from Bristol Myers Squibb, honoraria from LM Education/Exchange Services, advisory board fees from Exelixis and AVEO, personal fees from Charles River Associates, Schlesinger Associates, Imprint Science, Insight Strategy, Trinity Group, Cancer Expert Now, Adnovate Strategies, MDedge, CancerNetwork, Catenion, OncLive, Cello Health BioConsulting, PWW Consulting, Haymarket Medical Network, Aptitude Health, and AbbVie, and research support from Exelixis, outside of the submitted work. C.J.W. hold equity in BioNTech, Inc. K.J.L. holds equity in Standard BioTools Inc. (formerly Fluidigm Corporation). F.C. is a paid consultant for Atlas Bio.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ansel KM et al. (1999) ‘In vivo-activated CD4 T cells upregulate CXC chemokine receptor 5 and reprogram their response to lymphoid chemokines’, The Journal of experimental medicine, 190(8), pp. 1123–1134. doi: 10.1084/jem.190.8.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendall ML et al. (2019) ‘Telescope: Characterization of the retrotranscriptome by accurate estimation of transposable element expression’, PLoS computational biology, 15(9), p. e1006453. doi: 10.1371/journal.pcbi.1006453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi K et al. (2021) ‘Tumor and immune reprogramming during immunotherapy in advanced renal cell carcinoma’, Cancer cell, 39(5), pp. 649–661.e5. doi: 10.1016/j.ccel1.2021.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin DA et al. (2015) ‘MiXCR: software for comprehensive adaptive immunity profiling’, Nature methods, 12(5), pp. 380–381. doi: 10.1038/nmeth.3364. [DOI] [PubMed] [Google Scholar]
- Bost P et al. (2020) ‘Host-Viral Infection Maps Reveal Signatures of Severe COVID-19 Patients’, Cell, 181(7), pp. 1475–1488.e12. doi: 10.1016/j.ce1l.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun DA et al. (2021) ‘Progressive immune dysfunction with advancing disease stage in renal cell carcinoma’, Cancer cell [Preprint]. doi: 10.1016/j.ccell.2021.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cable DM et al. (2021) ‘Robust decomposition of cell type mixtures in spatial transcriptomics’, Nature biotechnology [Preprint]. doi: 10.1038/s41587-021-00830-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cable DM et al. (2022) ‘Cell type-specific inference of differential expression in spatial transcriptomics’, bioRxiv. doi: 10.1101/2021.12.26.474183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow MT et al. (2019) Intratumoral Activity of the CXCR3 Chemokine System Is Required for the Efficacy of Anti-PD- l Therapy’, Immunity, 50(6), pp. 1498–1512.e5. doi: 10.1016/j.immuni.2019.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobosy JR et al. (2011) ‘RNase H-dependent PCR (rhPCR): improved specificity and single nucleotide polymorphism detection using blocked cleavable primers’, BMC biotechnology, 11, p. 80. doi: 10.1186/1472-6750-11-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA (1992) ‘Statistical Methods for Research Workers’, in Kotz S and Johnson NL (eds) Breakthroughs in Statistics: Methodology and Distribution. New York, NY: Springer New York, pp. 66–70. doi: 10.1007/978-1-4612-4380-9_6. [DOI] [Google Scholar]
- Gohil SH et al. (2021) ‘Applying high-dimensional single-cell technologies to the analysis of cancer immunotherapy’, Nature reviews. Clinical oncology, 18(4), pp. 244–256. doi: 10.1038/s4157l-020-00449-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golumbeanu M et al. (2018) Single-Cell RNA-Seq Reveals Transcriptional Heterogeneity in Latent and Reactivated HIV-Infected Cells’, Cell reports, 23(4), pp. 942–950. doi: 10.1016/j.celrep.2018.03.102. [DOI] [PubMed] [Google Scholar]
- Hafemeister C and Satija R (2019) ‘Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression’, Genome biology, 20(l), p. 296. doi: 10.1186/sl3059-0l9-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang S et al. (2020) ‘Immune gene signatures for predicting durable clinical benefit of anti-PD-1 immunotherapy in patients with non-small cell lung cancer’, Scientific reports, 10(1), p. 643. doi: 10.1038/s4l598-0l9-572l8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston RJ et al. (2009) ‘Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation’, Science, 325(5943), pp. 1006–1010. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi K et al. (2019) ‘Spatial heterogeneity of the T cell receptor repertoire reflects the mutational landscape in lung cancer’, Nature medicine, 25(10), pp. 1549–1559. doi: 10.1038/s4l59l-019-0592-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel JC et al. (2019) ‘Murine single-cell RNA-seq reveals cell-identity- and tissue-specific trajectories of aging’, Genome research, 29(12), pp. 2088–2103. doi: 10.1101/gr.253880.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleshchevnikov V et al. (2022) ‘Cell2location maps fine-grained cell types in spatial transcriptomics’, Nature biotechnology, 40(5), pp. 661–671. doi: 10.1038/s4l587-021-01l39-4. [DOI] [PubMed] [Google Scholar]
- Krishna C et al. (2021) ‘Single-cell sequencing links multiregional immune landscapes and tissue-resident T cells in ccRCC to tumor topology and therapy efficacy’, Cancer cell, 39(5), pp. 662–677.e6. doi: 10.1016/j.ccell.2021.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang F et al. (2021) ‘MGST1 is a redox-sensitive repressor of ferroptosis in pancreatic cancer cells’, Cell chemical biology, 28(6), pp. 765–775.e5. doi: 10.1016/j.chembiol.2021.01.006. [DOI] [PubMed] [Google Scholar]
- Li H (2018) ‘Minimap2: pairwise alignment for nucleotide sequences’, Bioinformatics, 34(18), pp. 3094–3100. doi: 10.1093/bioinformatics/btyl9l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S et al. (2019) ‘RNase H-dependent PCR-enabled T-cell receptor sequencing for highly specific and efficient targeted sequencing of T-cell receptor mRNA for single-cell and repertoire analysis’, Nature protocols, 14(8), pp. 2571–2594. doi: 10.1038/s4l596-019-0195-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S et al. (2019) ‘Comparison of Biomarker Modalities for Predicting Response to PD-1/PD-L1 Checkpoint Blockade: A Systematic Review and Meta-analysis’, JAMA oncology, 5(8), pp. 1195–1204. doi: 10.1001/jamaoncol.2019.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes L, Healy J and Melville J (2018) ‘UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction’, arXiv [stat.ML]. Available at: http://arxiv.org/abs/1802.03426. [Google Scholar]
- Merkenschlager J et al. (2021) ‘Dynamic regulation of TFH selection during the germinal centre reaction’, Nature, 591(7850), pp. 458–463. doi: 10.1038/s41586-02l-03l87-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan M et al. (2022) ‘Tertiary lymphoid structures generate and propagate anti-tumor antibody-producing plasma cells in renal cell cancer’, Immunity, 55(3), pp. 527–541.e5. doi: 10.1016/j.immuni.2022.02.001. [DOI] [PubMed] [Google Scholar]
- Miller BC et al. (2019) ‘Subsets of exhausted CD8+ T cells differentially mediate tumor control and respond to checkpoint blockade’, Nature immunology, 20(3), pp. 326–336. doi: 10.1038/s41590-0l9-03l2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes T et al. (2017) ‘Quantitative Characterization of the T Cell Receptor Repertoire of Naïve and Memory Subsets Using an Integrated Experimental and Computational Pipeline Which Is Robust, Economical, and Versatile’, Frontiers in immunology, 8, p. 1267. doi: 10.3389/fimmu.2017.01267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira G et al. (2021) ‘Phenotype, specificity and acidity of antitumour CD8+ T cells in melanoma’, Nature [Preprint]. doi: 10.1038/s41586-021-03704-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira G et al. (2022) ‘Landscape of helper and regulatory antitumour CD4+ T cells in melanoma’, Nature, 605(7910), pp. 532–538. doi: 10.1038/s41586-022-04682-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace L et al. (2018) ‘The epigenetic control of stemness in CD8+ T cell fate commitment’, Science, 359(6372), pp. 177–186. doi: 10.1126/science.aah6499. [DOI] [PubMed] [Google Scholar]
- Park J-E et al. (2020) ‘A cell atlas of human thymic development defines T cell repertoire formation’, Science, 367(6480). doi: 10.1126/science.aay3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedregosa F et al. (2011) ‘Scikit-learn: Machine learning in Python’, the Journal of machine Learning research, 12, pp. 2825–2830. Available at: https://www.jm1r.org/papers/volumel2/pedregosa1la/pedregosalla.pdf?source=post_age-----------------------. [Google Scholar]
- Sade-Feldman M et al. (2018) ‘Defining T Cell States Associated with Response to Checkpoint Immunotherapy in Melanoma’, Cell, 175(4), pp. 998–1013.e20. doi: 10.1016/j.cell.2018.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sautès-Fridman C et al. (2019) ‘Tertiary lymphoid structures in the era of cancer immunotherapy’, Nature reviews. Cancer, 19(6), pp. 307–325. doi: 10.1038/s41568-019-0144-6. [DOI] [PubMed] [Google Scholar]
- Seabold S & Perktold J, 2010. statsmodels: Econometric and statistical modeling with python. In 9th Python in Science Conference. [Google Scholar]
- Shulman Z et al. (2013) ‘T follicular helper cell dynamics in germinal centers’, Science, 341(6146), pp. 673–677. doi: 10.1126/science.1241680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spranger S et al. (2017) ‘Tumor-Residing Batf3 Dendritic Cells Are Required for Effector T Cell Trafficking and Adoptive T Cell Therapy’, Cancer cell, 31(5), pp. 711–723.e4. doi: 10.1016/j.ccell.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steuerman Y et al. (2018) ‘Dissection of Influenza Infection In Vivo by Single-Cell RNA Sequencing’, Cell systems, 6(6), pp. 679–691.e4. doi: 10.1016/j.cels.2018.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stickels RR et al. (2020) ‘Highly sensitive spatial transcriptomics at near-cellular resolution with Slide-seqV2’, Nature biotechnology [Preprint]. doi: 10.1038/s41587-020-0739-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeckius M et al. (2017) ‘Simultaneous epitope and transcriptome measurement in single cells’, Nature methods, 14(9), pp. 865–868. doi: 10.1038/nmeth.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strbo N et al. (2003) ‘Perforin is required for innate and adaptive immunity induced by heat shock protein gp96’, Immunity, 18(3), pp. 381–390. doi: 10.1016/sl074-76l3(03)00056-6. [DOI] [PubMed] [Google Scholar]
- Stuart T et al. (2019) ‘Comprehensive Integration of Single-Cell Data’, Cell, 177(7), pp. 1888–1902.e2l. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suan D et al. (2015) ‘T follicular helper cells have distinct modes of migration and molecular signatures in naive and memory immune responses’, Immunity, 42(4), pp. 704–718. doi: 10.1016/j.immuni.2015.03.002. [DOI] [PubMed] [Google Scholar]
- Thorne JJC et al. (2014) ‘Spatial map of human T cell compartmentalization and maintenance over decades of life’, Cell, 159(4), pp. 814–828. doi: 10.1016/j.cell.2014.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I et al. (2016) ‘Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma’, Nature, 539(7628), pp. 309–313. doi: 10.1038/nature20l23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travaglini KJ et al. (2020) ‘A molecular cell atlas of the human lung from single-cell RNA sequencing’, Nature, 587(7835), pp. 619–625. doi: 10.1038/s41586-020-2922-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu AA et al. (2019) ‘TCR sequencing paired with massively parallel 3′ RNA-seq reveals clonotypic T cell signatures’, Nature immunology, 20(12), pp. 1692–1699. doi: 10.1038/s41590-019-0544-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W et al. (2019) ‘CD8+ T cells regulate tumour ferroptosis during cancer immunotherapy’, Nature, 569(7755), pp. 270–274. doi: 10.1038/s4l586-019-1170-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SZ et al. (2021) ‘Cryopreservation of human cancers conserves tumour heterogeneity for single-cell multi-omics analysis’, Genome medicine, 13(1), p. 81. doi: 10.1186/sl3073-021-00885-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TD et al. (2020) ‘Peripheral T cell expansion predicts tumour infiltration and clinical response’, Nature, 579(7798), pp. 274–278. doi: 10.1038/s41586-020-2056-8. [DOI] [PubMed] [Google Scholar]
- Yost KE et al. (2019) ‘Clonal replacement of tumor-specific T cells following PD-1 blockade’, Nature medicine, 25(8), pp. 1251–1259. doi: 10.1038/s4l59l-019-0522-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MD et al. (2018) ‘Single-cell transcriptomes from human kidneys reveal the cellular identity of renal tumors’, Science, 361(6402), pp. 594–599. doi: 10.1126/science.aat1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu CR, Dambuza IM, Lee YJ, Frank GM, Egwuagu CE (2013). STAT3 Regulates Proliferation and Survival of CD8+ T Cells: Enhances Effector Responses to HSV-1 Infection, and Inhibits IL-10+ Regulatory CD8+ T Cells in Autoimmune Uveitis. Mediat. Inflamm. 2013:359674. doi: 10.1155/2013/359674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D et al. (2009) ‘The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment’, Immunity, 31(3), pp. 457W68. doi: 10.1016/j.immuni.2009.07.002. [DOI] [PubMed] [Google Scholar]
- Zemmour D et al. (2018) ‘Single-cell gene expression reveals a landscape of regulatory T cell phenotypes shaped by the TCR’, Nature immunology, 19(3), pp. 291–301. doi: 10.1038/s41590-018-0051-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Comparison of technologies. Related to Figure 1.
Table S2. Oligonucleotides and genesets used. Related to STAR Methods.
Table S3. Bulk and Slide-TCR-seq clonotype sequencing results in OT-I spleen and pre- and post-treatment RCC samples. Related to Figure 3.
Table S4. Differentially expressed genes in spatial transcriptomics data. Related to Figure 5.
Data Availability Statement
All raw Slide-TCR-seq data are available at the Broad Institute Single Cell Portal at: https://sing1ecell.broadinstitute.org/single_cell/study/SCP1348/. Arrays with cell type prediction have also been uploaded to https://cel1xgene.cziscience.com/co1lections/02b0l703-bflb-48de-b99a-23bef8cccc81 for visualization and browsing. Single-cell RNA sequencing data for RCC were used from dbGaP (dbGaP:phs002252.vl.p1 at https://www.ncbi.nlm.nih.gov/gap/). Single-cell RNA sequencing data for mouse spleen were used from NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE132901. Single-cell RNA sequencing data for melanoma were used from European Genome-Phenome Archive (EGA), under the accession code EGAS0000l 00511. The code used can be found at https://github.com/MacoskoLab/slideseq-tools and https://github.com/soph-liu/Slide-TCR-seq.