Abstract
The automatic segmentation of COVID-19 pneumonia from a computerized tomography (CT) scan has become a major interest for scholars in developing a powerful diagnostic framework in the Internet of Medical Things (IoMT). Federated deep learning (FDL) is considered a promising approach for efficient and cooperative training from multi-institutional image data. However, the nonindependent and identically distributed (Non-IID) data from health care remain a remarkable challenge, limiting the applicability of FDL in the real world. The variability in features incurred by different scanning protocols, scanners, or acquisition parameters produces the learning drift phenomena during the training, which impairs both the training speed and segmentation performance of the model. This paper proposes a novel FDL approach for reliable and efficient multi-institutional COVID-19 segmentation, called MIC-Net. MIC-Net consists of three main building modules: the down-sampler, context enrichment (CE) module, and up-sampler. The down-sampler was designed to effectively learn both local and global representations from input CT scans by combining the advantages of lightweight convolutional and attention modules. The contextual enrichment (CE) module is introduced to enable the network to capture the contextual representation that can be later exploited to enrich the semantic knowledge of the up-sampler through skip connections. To further tackle the inter-site heterogeneity within the model, the approach uses an adaptive and switchable normalization (ASN) to adaptively choose the best normalization strategy according to the underlying data. A novel federated periodic selection protocol (FED-PCS) is proposed to fairly select the training participants according to their resource state, data quality, and loss of a local model. The results of an experimental evaluation of MIC-Net on three publicly available data sets show its robust performance, with an average dice score of 88.90% and an average surface dice of 87.53%.
Keywords: Internet of Medical Things, Fog Computing, Deep Learning, Data Heterogeneity, COVID-19
1. Introduction
The recent COVID-19 pandemic resulting from the spread of the novel coronavirus SARS-CoV-2 has a devastating effect on the health infrastructure of many countries. Thus, there is an urgent need for time-sensitive, precise, fast, and simple technologies for efficient diagnosis in modern maritime transportation systems (MTSs). In response to these requirements, the Internet of Medical Things (IoMT) has been developed as an instantiation of the Internet of Things (IoT) to interconnect a diversity of medical and health-care platforms. The IoMT, therefore, combines a large number of computational units and IoT devices in hospitals and clinical processes to facilitate clinical diagnosis and the monitoring of patient data. The major aim of IoMT services is to computerize diagnosis tasks using intelligent techniques that can process and extract insights from medical records (e.g., medical images). Hence, developing such a service for COVID-19 diagnosis is of great importance and can significantly advance the medical diagnosis process. Based on recent studies, the diagnosis of COVID-19 should be established using the reverse transcription–polymerase chain reaction (RT-PCR) [7]. However, owing to the practical problems in sample aggregations and transportation, and the variations in testing kits, specifically at the time of the outbreak, RT-PCR testing has a high false-negative ratio. As an alternative, computerized tomography (CT) scans are a reliable tool for the diagnosis of COVID-19 infection in the medical domain.
Infection segmentation is, therefore, a primary phase of diagnosis, infection quantification, infection progression monitoring, and severity assessment. A wide variety of artificial intelligence (AI) solutions have been investigated for infection segmentation from medical images, such as fuzzy intelligence, machine learning, and mathematical modeling [27], [39]. However, given the inherent complexity of infection representations and the high volume and high-dimensionality of medical data, these approaches cannot achieve good performance. Deep learning (DL), as a subfield of machine learning, has overcome the limitations of the above methods and achieved excellent segmentation results from a different form of data, irrespective of the size and dimensionality of data. Motivated by these results, this work emphasizes studying DL to address COVID-19 segmentation in the real IoMT.
The grouping of communicating medical apparatus, devices, and applications is termed the IoMT. It includes instantaneous data, the remote nursing of patients, the situational data of patients, and diagnostic decision-making according to aggregated information. The key advantage of the online interconnections between medical appliances in the IoMT is the reduction of the time needed by specialists through offering immediate analysis of a patient’s circumstances; moreover, patient assessment can be offered at home, in a clinic, or in a hospital. However, the aggregation of a high volume of medical data is laborious and time-consuming, making it a huge problem in the IoMT environment. Moreover, relying on a single source of data limits the development of an efficient DL model [8]. Thus, it is essential to share the high volume of training data dispersed among several medical organizations. With the advancements in cloud computing of ample storage space and unrestrained computing resources, the DL approaches applied to medical data have been broadly investigated by developing IoMT applications based on cloud computing. Nevertheless, the continuous increase in the number of connections between the cloud platform and mobile individuals causes unwanted and untimely responses to user requests, along with broadcast latency. A late response to diagnostic requests can impact the health and lives of patients, particularly patients suffering from infectious or serious diseases. As a remedy, the paradigms of fog/edge computing have been introduced to provide resources positioned closer to clients and to reduce latency by making computation adjacent to clients. These computing paradigms have received increasing attention across a variety of IoT-enabled applications and have shown great potential in delivering the required response. These evolving paradigms afford edge devices, including a variety of storage space, processing, and communication technologies, efficient network bandwidth, scalability, privacy preservation, mobility, security, and short latency, thus providing a better option for time-sensitive applications.
The introduction of edge and fog computing has shifted the attention of scholars to the distributed training of DL solutions on geographically distributed devices. In this regard, federated learning (FL) was proposed to enable distributed entities to cooperatively train complex deep networks using their locally stored data by communicating and aggregating the local training parameters in a regular fashion. Contrasted with the traditional cloud-centralized learning strategies, FL solution has a design that can bring many benefits to the IoMT environment, including efficient network bandwidth, privacy preservation, and communication efficiency (in terms of low overhead and low latency). Recently, the field of medical image analysis has witnessed an active shift toward FL in a broad range of tasks, such as federated segmentation, disease prediction, image reconstruction, classification, and computational pathology. However, the research has overlooked many important design aspects, such as complexity, communication efficiency, and heterogeneity, which limit their use in real-world health-care applications.
1.1. Research gaps
Although much effort has been devoted to developing precise and reliable segmentation of COVID-19 lesions, multiple research gaps have still been uncovered in the literature. These gaps can be concisely described as follows:
-
(1)
Complex manifestations: Owing to the low contrast problem, the border between COVID-19 infection lesions and the adjacent normal tissues is frequently hazy. As a result, precise lung infection segmentation is severely hampered. Moreover, COVID-19 lung infection displays a wide range of morphological characteristics, such as size and shape, which adds to the challenge of precise segmentation.
-
(2)
Lung CT scans from various domains introduce superficial domain heterogeneity owing to changes in imaging techniques, scanning devices, and demography. Because better DL network performance often requires a well-standardized distribution of data, the discrepancies inherent in heterogeneous domain data can cause problems when compounding examples from various sources during the training.
-
(3)
Robust segmentation performance always requires complex deep networks, which makes the training and inferencing process computationally intensive, thus limiting the responsiveness of medical image analysis and diagnosis. This phenomenon represents a great barrier to applying and deploying segmentation models in real-world distributed IoMT systems.
-
(4)
The existing research has proved that the data heterogeneity of participants leads to significant drift in local and global training optimizations, slowing the convergence and reducing the stability of training, as the local training is optimized according to the local objective rather than the global one.
1.2. Main contributions
This work proposes a new multi-institutional COVID-19 segmentation framework called MIC-Net to fill in the abovementioned gaps. The proposed MIC-Net contributes to the body of knowledge as follows:
-
(1)
A novel lightweight DL technique called MIC-Net is presented for fine-tuning lung infection segmentation by exploiting heterogeneous-source CT scans aggregated in an IoMT environment. MIC-Net is integrated into a domain-adaptive FL scheme to further support the applicability of real-world distributed IoMT solutions.
-
(2)
The down-sampling module of MIC-Net is innovatively designed to effectively capture lesion features from cross-domain CT images, whereby inter-source data heterogeneity is further addressed via a new switchable domain-adaptive normalization layer.
-
(3)
In MIC-Net, a novel contextual enrichment (CE) module is introduced to empower the network to capture the combinations of static and dynamic contextual representations using a lightweight attention mechanism. The mined information CE module is later used to enrich the up-sampling path so as to accurately reconstruct the segmentation maps.
-
(4)
A novel federated periodic client selection protocol (FED-PCS) is proposed to select the training participants fairly and periodically according to multiple criteria including the resource state (time, CPU, GPU, Memory, bandwidth), data quality, and loss of a local model.
-
(5)
The results of experimental simulations on real-world cross-domain COVID-19 data validate the increased efficiency and effectiveness of the proposed MIC-Net over state-of-the-art approaches in segmenting infection lesions, making the method better suited for deployment in real-world health-care systems.
1.3. Paper organization
The rest of the article is organized as follows. Section 2 reviews the recent studies relevant to this work. Section 3 details the system design. Then, a detailed explanation of the methodology of MIC-Net is presented in Section 4. Next, Section 5 describes the experimental specifications of this work. Section 6 discusses and analyzes the numerical results of our experiments. Finally, Section 7 concludes this work and lays out promising future directions.
2. Related work
This section starts by reviewing the latest research relevant to the subject matter of this work to better position the current work. The discussion of the research is decomposed into three main subsections, as described below:
2.1. COVID-19 segmentation
There have many been research efforts to use deep learning to improve the segmentation of COVID-19 by locating and segmenting the infection region in either two-dimensional (2D) or volumetric CT scans. For example, Fan et al. [6] developed a semi-supervised segmentation approach that utilizes a parallel partial decoder to fuse the complex features and produce a global map that is subsequently fed into attention modules to detect infection boundaries and improve representations. However, the reliance on generated pseudo labels limited the performance. In [9], the authors presented a dual-branch deep learning framework to concurrently accomplish two tasks of COVID-19 diagnosis: patient-based classification and lesion segmentation. In this work, a lesion attention mechanism was proposed to integrate the segmentation outcomes so as to guide the classification of the infection lesions. Moreover, one study [44] presented a convolutional network for COVID-19 segmentation that integrates a self-attention layer to extend the receptive field so as to improve the representational power by distilling valuable semantic patterns from deeper layers, with no additional training time. It also presents semi-supervised shots to ease the deficiency of annotated multi-class data and the unbalanced training data by adopting a re-weighting of the loss to precisely categorize different types of labels. Furthermore, a study [26] developed a deep segmentation network named CovTANet that integrates tri-level attention techniques to combine channel, spatial, and pixel information for precise segmentation of COVID lesions from CT scans using multistage optimization methods. In [41], the authors developed a noise-robust dice loss as a generalized variant of dice loss and mean absolute error (MAE) loss to make the segmentation network more robust against noisy COVID-19 originating from different scales and presences. They also designed a segmentation network called COPLE-Net. It is integrated into an adaptive self-ensembling architecture in which the exponential moving average (EMA) of a student network is leveraged as a teacher network that performs an adjustive update by overpowering the involvement of the student to the EMA at the time the student has a large, noise-robust dice loss. Moreover, the authors of [4] proposed a segmentation framework that first integrates the patch mechanism to extract the region of interest so as to eliminate unrelated background regions and then applies a three-dimensional (3D) network to capture spatial features from important target areas. They applied a combinatorial loss to accelerate the training convergence. Speaking of 3D networks, the work [43] presented a joint learning framework that combines classification and volumetric segmentation of COVID-19 from a large-scale CT database, whereby a 3D lesion subnetwork is applied for lesion segmentation and another subnetwork, while task-aware loss was developed to regularize the interaction between two subnetworks during the training. The most common aspect of the studies described above for COVID-19 diagnosis is that all models were trained and evaluated on homogeneous (single-source) data. Consequently, their performance degraded on unseen data from another source. Such data heterogeneity is prevalent in the IoMT environment. Additionally, all approaches exhibited high computational complexity, making them unsuitable for deployment in an IoMT environment.
2.2. Cross-Domain medical image analysis
The heterogeneity of domains in medical images has gained increasing interest in recent years owing to its considerable consequences for the efficiency of DL models. For example, in [17], the authors proposed a two-stage transfer learning approach called nCoVSegNet for efficient cross-domain segmentation of COVID-19 lesions from CT scans. nCoVSegNet was developed to extract multilevel features from the received CT scans using global context-aware modules. To improve the segmentation, nCoVSegNet adopts two-stage transfer learning methods: One uses a network trained on ImageNet, while the other conducts transfer learning based on a large public CT data set. Besides, in [19], the authors proposed a DL framework called MS-Net for enhancing the accuracy of prostate segmentation from multidomain magnetic resonance images (MRIs) by compensating for the intersite data heterogeneity via domain-related batch normalization (BN) operations. The MS-Net presents knowledge transfer to direct joint convolutional kernels to learn powerful representations from cross-domain MRIs. Similarly, the work [45] proposed explicitly addressing the problem of domain shift in cross-site COVID-19 classification by performing distinct feature normalization and applying contrastive training loss to improve the domain invariance of semantic embeddings. The aim is to improve the classification performance across different domains. Moreover, the work [42] presented a cycle-consistent framework for efficient semantic segmentation from cross domains in the medical field, which integrates consistency regularization and online diverse image translation into a single framework to promote modeling complicated relationships between domains such as many-to-many mappings. In [48], the authors presented a wholly automated machine-agnostic approach for segmenting and quantifying COVID-19 lesions from multisource CT scans. They first designed a CT scan simulator by fitting the dynamic alteration of actual patients' information captured at various time intervals, which significantly eliminates the data shortage problem; then, a deep network was proposed to decompose the volumetric segmentation into three 2D ones, thus improving the model complexity and segmentation efficiency.
2.3. IoMT
Owing to the robust performance of DL in processing and learning from large data incidents in the IoT-based system, several studies have investigated applying DL techniques in the IoMT environment for disease diagnosis. For example, in [33], the authors presented an inclusive survey of potential IoT-based solutions that can help tackle COVID-19-like pandemics by highlighting the social effects of such pandemics and recognizing the particular gaps in existing IoT systems. The authors of [31] presented an overview of the latest innovations in FL in making smart health care more intelligent and efficient by studying different federated intelligence paradigms such as resource-aware paradigms, privacy-preserved federated intelligence, personalized federated intelligence, and incentive federated intelligence. The authors of [24] proposed to detect kidney disease diagnosis using a novel heterogeneous DL segmentation approach. Furthermore, the authors of [21] discussed multiple potential deep reinforcement learning techniques for identifying and diagnosing lung cancer from CT images, and for precisely detecting tumors. Additionally, the authors of [46] used a convolutional model for the diagnosis of gallbladder stones utilizing aggregated image data in the IoMT environment. Moreover, the authors of [3] developed an integrated IoT framework to support instantaneous communication and identification of emotions from physiological signals using long-short-term memory (LSTM), where real-time health observation and distance learning provide support during epidemics. Moreover, the authors of [2] presented an IoT-empowered approach for the early evaluation of COVID-19 by applying a faster region-based CNN (Faster-RCNN) to detect the disease from chest X-rays. In [12], the authors developed a blockchain-managed FL framework in which a data normalization method is applied to lessen the impact heterogeneity of CT data originating from various hospitals. Then, a capsule network is adopted to perform the classification and segmentation of COVID-19. Blockchain technology enables the collaborative and privacy-preserved training of the model in real-life health care. Similarly, FL was used for semi-supervised training of COVID-19 from multinational CT scans; however, the cloud server takes the responsibility of coordinating the training instead of a blockchain. In [47], the authors designed a dynamic fusion-based FL framework for detecting COVID-19 from medical images through the dynamic selection of the contributing clients based on the performance of their local model while scheduling the model fusion according to the training time of participants.
In sum, although there have been many studies addressing COVID-19 segmentation from CT scans using various DL models, only a few of them have seriously considered data heterogeneity during federated training with multi-institutional data. They have not considered simulating their models physically or virtually in an IoMT context, which limits the applicability in the real-world settings with few resources.
3. System design
Real-time semantic segmentation is crucial in different IoMT tasks, such as diagnosis, patient follow-up, and detection. Generally, the application of DL requires large storage resources and powerful computing resources. Regrettably, implementing and training the segmentation model on the cloud is frequently subject to high bandwidth utilization, unanticipated latency, poor dependability, and privacy concerns. The introduction of fog and edge computing paradigms has offered a great opportunity to address the above problems by bringing the segmentation models close to the data source in the IoMT (end devices), as a supplement to the cloud.
3.1. System model
This section provides a detailed description of the system model of the proposed framework, which is composed of three main layers: the edge, fog, and cloud.
3.1.1. Cloud computing layer
In the cloud layer, the cloud server is responsible for coordinating the collaboration between the edge and fog nodes during the distributed training of segmentation models. As the collaborative training between the edge and fog layer may be considerably compromised because of local experience, the cloud layer combines different well-trained local segmentation models to achieve global learning. If the edge cannot reliably offer the service, the cloud can leverage its extensive computing resources and global expertise to help edge nodes update their DL models. The proposed solution exploits the resources of the cloud layer to avoid the problems of an overloaded fog infrastructure, latency, a larger input size, network congestion, and limited scalability. This offers extra robustness and reliability amidst heavy load requests and autonomous computation.
3.1.2. Edge/Fog computing layer
This layer contains different devices that lie in the vicinity of data owners (e.g., medical institutes, hospitals, research labs), which directly store the captured CT scans from COVID-19 patients. The data stored in this layer are usually heterogeneous because of the discrepancy in CT scanners, scanning protocols, and other conditions. Compared to the cloud layer, the edge and fog devices have far fewer resources and less computing power and storage. However, they bring the training of the DL model (i.e., MIC-Net) closer to the source of data by performing collaborative model training using the local CT data.
3.2. Resource allocation
The participation of heterogeneous devices is a common case for applying FL in the real-world IoMT. These devices can vary in the quality of their data sets, the capability of their computations, their energy states, and their opportunity to engage. Because different devices have different energy states and communication bandwidths, resource allocation needs to be optimized so that the learning process can be as effective as possible. This is necessary to make the most of the available resources. Participant selection, joint radio, computation resource management, and incentive mechanisms are common directions for addressing resource allocation in federated training scenarios. To alleviate the training traffic jam, this paper introduces a multi-criteria periodic selection protocol in federated settings. The systematic diagram of this protocol is shown in Fig. 1 , in which the cloud server acts as a coordinator of the collaborative training of the segmentation model on a heterogenous network of IoMT devices. The proposed selection protocol is operated in a periodic fashion rather than a selection per-round manner, whereas the length of the period at which the selection is performed is heuristically determined via the cloud coordinator. As shown, the proposed selection is executed as follows:
Fig. 1.
Resource allocation in the proposed system model.
Step 1: The cloud server initiates a call for a resource to acquire information about the resources of local participants that can help judge the willingness of heterogeneous health-care devices to participate in federated training.
Step 2: The Profiler of local devices is in charge of monitoring the resource consumption during the local training and generating the resource profile of its devices. The protocol uses the following metadata: time, CPU, GPU, RAM, energy, wireless channel states, bandwidth, and quality of data (e.g., number of samples, number of sources). The collected resource information is exploited to determine the highest probable number of local clients (e.g., hospitals, institutions) that can complete the training process in the preset time intervals for a later global aggregation period. Besides, the inclusion of data as selection criteria helps avoid bias toward devices with better computational resources, and enables the selection of clients hosting representative data from a population distribution.
Step 3: By choosing the highest viable number of participating clients per communication round, the segmentation performance can be easily retained and improved during the training. The cloud can regard this as a maximization problem, where greedy algorithms are widely used to select the clients with the smallest time for model uploads. Generally, the IoMT network is known to be dynamic and ambiguous, with varying conditions such as power conditions and network conditions. Thus, the proposed protocol adopts double deep q-learning (DDQL) [38] to optimize the abovementioned optimization problem, in which the server acts as the agent, the profile of client devices acts as state space, and the number of data and power items act as action space. The collected data, power usage, and training time are adopted as a reward function. Typically, the cloud coordinator seeks to reach the optimal resource allocation that maximizes training speed and simultaneously minimizes the usage of client resources.
Step 4: Given the selected clients, the cloud server starts broadcasting the parameters of the global model to those clients to perform local training and share their local updates to the cloud server so as to aggregate their acquired knowledge from the distributed cross-domain devices.
Unlike previous protocols designed and evaluated for simple deep models [32], the proposed selection protocol is suitable for segmentation models that are relatively more complex than previous ones. Another difference is that our method factors in GPU acceleration resources. Another problem to encounter is fraudulent resource allocation, which means that the FL is overrepresented by the distribution of data held by clients possessing higher computing resources. To address this problem, inspired by previous research [13], fairness of selection is regarded as an extra objective in DDQL, which is actually defined as the difference in the performance of the segmentation model (MIC-Net) across different clients. If the variation in the segmentation performance is high, it implies the existence of high bias or less equality, as the learned model may be extremely precise for particular applicants and less so for other underrepresented participants.
3.3. System security
Although FL avoids the need to communicate training data with a remote cloud by communicating only the learned parameters, it is still subject to a broad range of attacks, such as model and data poisoning, wherein a malevolent client can deliver inaccurate parameters or degraded models to forge the learning procedure throughout global aggregation. Therefore, the vulnerability of FL to such attacks can corrupt the entire learning system. Moreover, FL still suffers from critical privacy challenges, which can be caused by a malicious client trying to infer sensitive information about clients. Security and privacy are major concerns in the integration of FL in health care; however, these topics are out of the scope of this work. Hence, the design of our system assumes that honest clients participate in training and an honest cloud is involved in the coordination for training. To imitate a real scenario, the security administrator module is settled on the cloud server to manage the security mode of the federated training process and decide the best security and/or privacy-preservation mechanisms in dishonest and semi-honest scenarios.
4. MIC-Net
In this section, we provide a detailed explanation of MIC-Net. For convenience, the architecture is illustrated in Fig. 2 . As a pre-training step for the MIC-Net, an intelligent harmonization mechanism is applied to lessen the impact of distribution shifts in the training data. As shown, the MIC-Net has a U-shaped structure, which is a common design for segmentation networks. In particular, the MIC-Net consists of three main building blocks: the encoding path, decoding path, and contextual enrichment (CE) module. Unlike previous approaches, our method has an encoding path consisting of new down-sampler modules instead of conventional commotional encoders. To avoid losing the contextual representations between the encoding and decoding path, the CE module is integrated to enable the network to capture both static and dynamic representations from multi-distribution data. Similarly, instead of a de-convolutional decoder, the decoding path was designed with new up-sampler modules to precisely reconstruct the segmentation maps from the learning solution. In the following subsections, we detail each building module.
Fig. 2.
System model of the proposed federated MIC-Net in the IoMT environment.
4.1. Feature encoding path (Down-Samplers)
The structure of the down-sampler module is composed of two parallel modules, the lightweight attention encoding (LAE) module and lightweight convolutional encoding (LCE) module. These modules bring two advantages to the encoding: the robust feature extraction capabilities of convolution and the ability to capture global context with attention.
The design of the LEA module is largely inspired by vision transformers [20], albeit with reduced complexity (See Fig. 3 ). Assume that we have as the input map of the LCE block, with denoting the number of channels, height, and width, respectively. The LAE passes input to a point-wise convolution (), as follows:
| (1) |
Fig. 3.
Architecture of the down-sampler module.
Then, the generated map is passed to the sparse self-attention layer (SSA), whose input is decomposed into three paths: input , key , and value . A linear layer is used to map the input () to the d-dimensional token into a scalar value. This layer comes with a weight matrix () that acts as the latent node (). The output of this linear projection is a k-dimensional vector representing the calculated distance between input and . These k-dimensional vectors are passed to a SoftMax function to generate attention scores (). Unlike standard vision transformers, the SSA calculates the attention score for every token in regard to rather than all tokens (). This, in turn, implies linear time computation, as demonstrated by [29]. The context scores () are then adopted to calculate a context vector (). In particular, input is linearly encoded into a d-dimensional representation through and weighted by the parameters to generate an outcome (). Follow, the context vector () is calculated as a weighted combination of , as follows:
| (2) |
Similar to the attention matrix, encapsulates the knowledge from all tokens, but in a computationally cheap manner. The encoded knowledge in is later combined with all tokens in input by linearly projecting input to a d-dimensional representation utilizing weighted by the parameters , then passed to the function to generate . The context representations in are later transmitted to through transmitted Hadamard products. The obtained outcome is later passed to one more linear projection operation weighted by the parameters to generate the ultimate outcome (). In mathematical terms, the SSA can be designated separately, as follows:
| (3) |
where * and denote the are broadcast-capable Hadamard product and summation functions, respectively. At the end of the LAE, a point-wise convolution is applied, followed by an adaptive and switchable normalization (ASN) layer, as follows:
| (4) |
The ASN is described in more detail later in this section.
Although the factorization of a convolution operation into a point-wise convolution and a depthwise separable convolution (DwS-Conv) can considerably decrease the number of learning parameters, and thereby the computing complexity, it generally results in a performance drop. Motivated by the reality that successful multi-scale learning has a significant role in enhancing the segmentation performance [1], [10], we introduce a lightweight convolutional encoding (LCE) module for efficient and effective multiscale extraction of lesion features using dilated versions of DwS-Conv layers. Each DwS-Conv layer is denoted as , where represents the kernel size and denotes the dilation rate. For convenience, when , the layer can be denoted as. Given the aforementioned definitions, the structural design of the LCE block can be illustrated as shown in Fig. 2. Given as the input map of the LCE block, with denoting the number of channels, height, and width, respectively, the LCE block generates the output map , whereas designates the function of the transformation of the input. follow the above definition, but for an output map.
Let us dive into the working At an early stage, the number of channels is reduced to by passing input to a point-wise convolution (), in which is the number of concurrent paths (see Fig. 1). This layer can be mathematically expressed as follows:
| (5) |
The output maps () from the above operation are passed to parallel dilated DwS-Conv, whereby the output of each path is added to the input of the next path such that
| (6) |
whereby the dilation factor grows in an exponential manner to expand the receptive field. These parallel convolutions shape the core of multi-scale representational, as high dilation factors enable extracting large-scale representations, while small-scale representations can be extracted by low dilation factors. In the above operation, the generated output maps are down-sampled by setting a stride of 2 for each layer. In the LCE block, the breaks are handled in a multi-scale manner, which enables the network to capture both the local and global representations from multi-distribution inputs. The output of each DwS-Conv layer in each path is passed to the point-wise convolution preceded by adaptive normalization, as follows:
| (7) |
The Gaussian error linear unit (GELU) activation is applied after the above convolution to obtain smooth activation. Next, a spatial squeeze and excite (sSE) [36] is applied to enable the LCE block to automatedly learn to emphasize target patterns of different scales. Moreover, the sSE acts as an attention method that can also learn to overturn unrelated representations at certain scales and focus on important representations on other scales. The sSE enables each scale to express itself to determine which one it contributes to in the multi-scale learning procedure. The transformation incurred by the sSE can be expressed as
| (8) |
where is a sigmoid function and denotes the Hadamard product.
By the end, information from various scales is fused by concatenating the output of all paths and processing them with point-wise convolution, as follows:
| (9) |
| (10) |
The convolution in the above formula is a convolution generating a feature map with channels, which acts as a fuser of features from parallel paths with DwS-Conv layers. In the next LCE blocks, the fusion can be performed via point-wise convolution at the beginning. The design of the LCE block can considerably decrease the number of learning parameters by times when contrasted with the approach of applying a standard convolution. Notably, increasing the number of paths is beneficial for reducing the number of parameters of LCE blocks. To maintain a better balance of computing cost and segmentation precision, we set the value of to . Thus, LCE block brings advantages. One is a significant reduction in the number of learning parameters, and the other is the ability to efficiently learn effective multi-scale features. Thus, it can precisely address the variable size COVID-19 lesions.
The ASN layer can be described as follows. Generally, an input instance () to a specific normalization layer is shaped into a four-dimensional tensor , representing the number of examples per batch, number of input channels, height, and width, respectively. In this context, represents the pixel value before normalization, while represents the corresponding normalized pixel value, which is computed as follows:
| (11) |
where , , , and . and denote a scale and a shifting parameter, respectively. denotes the tiny constant that maintains numerical constancy, while and represent the mean and standard deviation statistics, respectively. The above formula is the same for instance normalization (IN), layer normalization (LN), and BN; however, they adopt diverse groups of pixels to approximate and . This can be generally represented as
| (12) |
| (13) |
where is adopted to differentiate diverse normalization layers methods.
In this formulation, signifies the number of pixels. In particular, , , and denote the group of pixels applied to calculate statistics in various layers.
Inspired by switchable normalization (SN) [23], the ASN layer was designed based on adaptive IN (AdaIN) [11], [16], in which the mean and variance of the input features can be aligned with those of the style features. The normalization of the pixel value can be formulated as follows:
| (14) |
where represents the statistics calculated from integrated normalization methods. However, this formulation exhibits highly repetitive calculations. To avoid this problem, the dependency between statistics can be exploited to reuse computations, as follows:
| (15) |
| (16) |
| (17) |
This design leads to a computing cost of . Moreover, and denote the significance fractions applied to calculate the weighted average of and , respectively. Both and are designated as a scalar instance that is commonly used in each channel. Similar to SN, the ASN layer contains a total of significance weights, while is calculated as follows:
| (18) |
where each is calculated with the SoftMax function. , , and are the monitor parameters to be updated during the training. The definition of is the same as that of , with three control parameters: , , and . In ASN, weight standardization (WS) [34] is applied to standardize the weights of the encoding layers with the aim of smoothing the landscape. Specifically, weight standardization alleviates the concern about transmitting smoothing outcomes from activations to weights. The traditional convolutional layer is.
| (19) |
where represents the weight matrix, and designates the convolution layer. and denote the number of the output channels and input channels, respectively. In this setting, rather than explicitly optimizing the loss with the original weight’s matrix (, the network reparametrizes the weights as follows:
| (20) |
| (21) |
whereas,
| (22) |
In the way that BN manipulates the first and second moments of the weights in convolutional layers for each individual output channel, WS does the same for each output channel. Keep in mind that the weights are of ten initialized in a consistent way across multiple initialization strategies. When compared to these other methods, WS attempts to normalize gradients through back-propagation by standardizing the weights in a differentiable way. The absence of affine transformations in is important to keep in mind. It is expected that subsequent normalization layers, such as a BN or group normalization (GN), can re-normalize the output of this convolutional layer. Thus, including an affine transformation generates confusion and slows down the training process.
4.2. CE module
A popular shortcoming of encoder–decoder networks is that successive convolution striding or pooling layers lead to a considerable reduction in feature resolution during the learning of encoded feature representation, resulting in loss of contextual information. The CE module is introduced to encode the contextual representations from multi-scale features fused from the down-samplers with no more learning weights. In particular, the CE module was designed to extract semantic representations with an intelligent multi-headed window-based attention layer, aiming to enrich the up-samplers with contextual information necessary for improving the accuracy of segmentation results. The CE equips MIC-Net with an elegant method for improving the segmentation performance through better exploitation of the abundance of contexts amongst input keys over multi-scale 2D encoding maps. The design of the CE module combines both contexts digging between keys and SSA over feature maps from down-samplers, thus preventing the creation of another path for context fusion (See Fig. 4 ). Technologically, the CE module first contextualizes the representation of the key via applying to adjacent keys inside the grid. In mathematical terms, we have feature map with query , key , and value . Rather than encoding each key with point-wise convolution in traditional SA, the CE module applies DwS-Conv to generate a contextualized key representation, as follows:
| (23) |
Fig. 4.
Architecture of CE module.
The obtained contextualized keys () inherently indicate the static contextual information between local neighboring keys, so can be considered as the static contextual mapping of multi-scale input .
Next, these contextualized key maps and input query are concatenated and passed to two successive point-wise convolutions. The aim is to calculate the attention scores by exploiting the joint relationships between query and latent keys (as described in SSA) under the supervision of the static context.
| (24) |
For each attention head, the local attention score for every position of is learned according to the query information and the context key information instead of the solitary query-key couples. This improves the SSA with the extra supervision from the fused static context . Later, matrix is adopted to compute attention feature map by the fusion of , as in SSA:
| (25) |
This way, the calculated attention scores can be exploited to fuse all input elements and learn a contextual dynamic representation of encoding maps to represent the dynamic context. Thus, the output of the CE module is calculated as a combination of the dynamic contextual representation () and the static contextual representation. As shown in Fig. 5 , the output of CE is scaled to various resolutions such that . Each scaled output is later exploited to enrich the semantic knowledge transferred from the down-sampler to the up-sampler through the skip connections.
| (26) |
Fig. 5.
Systematic diagram of the proposed MIC-Net with three main constituents: the down-sampling path, CE module, and up-sampling path.
4.3. Feature decoding path (Up-Sampler)
Given that the last feature map in the encoding path comes with the scale of the network input, it is not ideal to model COVID-19 infection lesions explicitly because of the loss of fine-grained information. As an alternative, the decoding path is introduced to re-establish the complex semantic multi-scale features captured by the feature down-sampling modules. Skip linking is employed to transmit detailed information from the encoder modules to the corresponding decoder modules to avoid losing semantic information. The decoding path structure consists of up-sampler modules that progressively up-sample and aggregate the learned representational map at every down-sampling stage. An up-sampler module is in charge of aggregating learned features. We denote the transformation function of the up-sampler as
| (27) |
| (28) |
where , applies the point-wise convolution to adapt the number of channels. The up-sampling feature map of the decoder is . In the first stem of the decoding path, we have
| (29) |
Thereby, is calculated as follows:
| (30) |
| (31) |
where denotes the up-sampling of the feature map with scale applying bilinear interpolation. Thus, the decoding path can fuse the fine-grained semantics to enable the network to perform precise segmentation of COVID-19 infection lesions. Given , the dense estimation map is computed with a point-wise convolution, as follows:
| (32) |
The predicted output map () is calculated with two channels indicative of two classes: background (black) and COVID-19 infections lesion (white). Hence, the final segmentation map can be designated with .
5. Experimental design
In this section, a detailed explanation of the experimental design is given to enable reproducing the experiments in terms of the implementation setup, data set description, preprocessing, and performance metrics.
5.1. Implementation setup
The overall implementation of MIC-Net is conducted with the TensorFlow library using three NVIDIA Quadro GPUs, one for every data source. Table 1 summarizes the best hyper-parameters of our simulation model. Notably, the optimal values for each parameter are selected after an exhaustive experiment with different possible sets of parameters.
Table 1.
Hyper-parameters of the proposed MIC-NET.
| Hyper-parameter | Optimal values |
|---|---|
| 0.9 | |
| 0.999 | |
| 0.6 | |
| 0.0001 | |
| Number of iterations | 25,000 |
| Dropout | 0.1 |
| Optimizer | Adam |
| Learning rate | 0.0001 |
| Total clients | 100 |
| Aggregation | q-FedAVG [13] |
| Local Epochs | 50 |
| Batch-size | 4 |
| Communication rounds | 200 |
5.2. Data sets
In order to assess the performance of the proposed MIC-Net on heterogeneous multi-source data, we selected three publicly available COVID-19 CT data sets. The first is the COVID-19-CT-Seg data set [25], which comprises 20 public COVID-19 CT volumes from the Coronacases Initiative and Radiopaedia, with more than 1,800 annotated slices. In our experiments, we refer to this data set as D1. Second, we use the 50 CT volumes published in the MosMedData data set [30], which was aggregated from municipal hospitals in Moscow, Russia. In our experiments, we refer to this data set as D2. The third is a larger data set, MedSeg [28], which consists of nine CT volumes that comprise a total of 829 slices, with 373 slices confirmed as positive. We refer to this data set as D3. The specification of each data set is reported in corresponding studies.
5.3. Data preparation
Following [19], all three data sets were prepared by slicing whole volumes into 2D images and applying some standard augmentation (i.e., cropping, rotating, and scaling) to alleviate the observed data imbalances. Then, we resized all the generated CT slices to in order to minimize the discrepancy of intensity among CT scans from diverse sources. We allocated 80 % and 20 % of the data sets into training and testing sets, respectively. Once again motivated by [45], three types of intensity normalization were adopted: bias field correction, noise filtering, and whitening. Moreover, harmonization mechanisms (HMs) [18] have been demonstrated as an effective way of lessening inter-site heterogeneity through continuous frequency space interpolation. Thus, an HM was applied to prepare the data before training. All experiments of the proposed framework and competing 2D segmentation models are performed on an axial view of a CT scan. For every CT image, the intensity score is stabilized to exhibit unit variance and a zero average before being fed into MIC-Net.
5.4. Evaluation metrics
Inspired by the extensively used approaches for evaluating segmentation techniques, we assess the segmentation performance of MIC-Net using two complementary metrics: the normalized surface dice (NSD) and the dice similarity coefficient (DSC). Let denote the GT and model outcomes, respectively. The two measures are given as follows:
| (33) |
| (34) |
where designate the border area of the GT and segmentation surface at tolerance, respectively, and are expressed as and , respectively. Herein, is set to 1 mm and 3 mm for segmenting the lung and lesions, respectively. The acceptance is calculated by estimating the difference between two diverse radiologists.
6. Results and analysis
This section comprehensively discusses and analyzes the findings obtained from the experimental evaluation of the proposed framework. The details of each experiment and the corresponding results can be found in the following subsections.
6.1. Domain shift analysis
To begin, MIC-Net experiments are done under two scenarios. One trains MIC-Net with merged multi-source CT scans (mixed), and the other scenario emphasizes training MIC-Net on single-source data (independent). To experiment with data from different domains in the IoMT, it is important to numerically investigate the data heterogeneity from different domains. Motivated by a recent paradigm for analyzing domain shift [23], we perform cross-source validation among D1, D2, and D3 by training independent networks on each separately, and then evaluating the networks using different samples from the three data sets. It is obvious from Table 2 that the independent models achieve better performance when tested on samples from the same data set, and their performance degrades when tested using samples from another domain. Conversely, the mixed model realizes a slight performance improvement compared to the independent model on D1 and has comparable performance to the independent model on D2. In some cases, the mixed training may lead to lower segmentation performance than that achieved under independent settings.
Table 2.
Performance comparison between independent and mixed approach, trained and evaluated on different data sets (mean ± standard deviation).
| Methods | D1 | D2 | D3 |
|---|---|---|---|
| DSC | |||
| Independent (D1) | 79.3 ± 13.1 | 69.5 ± 10.04 | 64.8 ± 12.3 |
| Independent (D2) | 72.9 ± 11.40 | 80.7 ± 08.91 | 72.2 ± 11.18 |
| Independent (D3) | 76.67 ± 9.17 | 70.8 ± 12.50 | 81.3 ± 08.19 |
| Mixed | 81.8 ± 10.60* | 79.6 ± 13.10 | 81.1 ± 09.13 |
| NSD | |||
| Independent (D1) | 78.6 ± 8.66 | 67.5 ± 5.33 | 61.8 ± 9.46 |
| Independent (D2) | 72.5 ± 9.38 | 76.8 ± 8.01 | 72.4 ± 7.15 |
| Independent (D3) | 74.6 ± 6.89 | 70.3 ± 8.78 | 78.3 ± 6.68 |
| Mixed | 79.8 ± 7.14* | 79.6 ± 10.29* | 77.8 ± 7.71* |
6.2. Comparative analysis
As a common research practice, the MIC-Net is compared with state-of-the-art segmentation methods to evaluate its competitive capabilities under the same experimental settings. The numerical results obtained from these experiments are displayed in Table 3 . Specifically, the MIC-Net is compared with the common 2D segmentation models U-Net [35], FCN-8 [22], and Inf-net [6]. They are also in compared with —the 3D segmentation models 3D U-Net [5] and 3D V-Net [33], with patch size dimensions of , and 2.5-dimensional-based approaches H-DUnet [14] and MultiPlanar UNet (MPUnet) [37], which perform view accumulation from 2D patch architectures. The inclusion of models with different dimensionalities in comparative experiments is beneficial to evaluate the competitiveness of the proposed solution. For multi-site approaches, the proposed framework is compared with MS-Net [19], MS-Fed [15], and FED-DG [18].
Table 3.
Quantitative results (mean ± standard deviation) for COVID-19 lesion segmentation on test sets.
| DSC↑ |
NSD↑ |
# Parameters |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Methods | D1 | D2 | D3 | Average | D1 | D2 | D3 | Average | |
| 2D-U-Net [21] | 82.1 ± 8.9 | 86.1 ± 9.9 | 87.1 ± 8.9 | 85.10 ± 9.23 | 80.9 ± 10.3 | 84.9 ± 11.1 | 87.1 ± 11.5 | 84.3 ± 10.97 | 7.8530 M |
| 2D-FCN-8[11] | 82.2 ± 9.1 | 86.2 ± 9.1 | 86.9 ± 10.1 | 85.10 ± 9.43 | 80.3 ± 11.2 | 84.4 ± 10.3 | 86.9 ± 10.3 | 83.9 ± 10.60 | 41.530 M |
| 3D-U-Net [16] | 80.8 ± 11.4 | 84.0 ± 8.4 | 85.9 ± 14.3 | 83.57 ± 11.37 | 79.7 ± 14.5 | 83.5 ± 9.8 | 85.2 ± 13.4 | 82.8 ± 12.57 | 22.577 M |
| 3D-V-Net [33] | 80.1 ± 11.2 | 84.7 ± 9.2 | 85.3 ± 15.1 | 83.37 ± 11.83 | 79.9 ± 13.1 | 84.6 ± 10.7 | 85.3 ± 13.3 | 83.2 ± 12.37 | 46.048 M |
| H-DUnet [34] | 81.2 ± 9.2 | 81.9 ± 7.2 | 86.1 ± 9.7 | 83.07 ± 8.70 | 80.3 ± 8.7 | 80.1 ± 9.3 | 86.1 ± 10.5 | 82.2 ± 9.50 | 45.082 M |
| MPUnet [40] | 80.4 ± 10.2 | 81.1 ± 9.7 | 84.7 ± 13.3 | 82.07 ± 11.07 | 79.9 ± 9.3 | 79.1 ± 12.2 | 84.7 ± 9.7 | 81.2 ± 10.40 | 62.001 M |
| Inf-Net [9] | 81.4 ± 7.2 | 85.6 ± 10.3 | 86.0 ± 11.1 | 84.99 ± 9.77 | 81.9 ± 7.9 | 85.8 ± 6.3 | 87.9 ± 7.16 | 85.1 ± 8.44 | 33.122 M |
| MS-Net [9] | 83.8 ± 6.5 | 87.8 ± 6.4 | 88.5 ± 5.9 | 86.70 ± 6.27 | 82.9 ± 9.4 | 86.6 ± 8.7 | 87.9 ± 6.5 | 85.9 ± 8.20 | 18.841 M |
| MS-Fed [15] | 81.6 ± 4.7 | 85.1 ± 7.1 | 84.9 ± 3.8 | 83.87 ± 5.20 | 81.8 ± 3.7 | 87.6 ± 4.3 | 86.2 ± 7.4 | 85.20 ± 5.13 | 7.8530 M |
| FEDDG [18] | 84.7 ± 9.1 | 87.3 ± 8.8 | 89.2 ± 6.6 | 87.06 ± 8.17 | 83.3 ± 5.6 | 85.6 ± 9.2 | 88.3 ± 8.8 | 85.73 ± 7.87 | 18.841 M |
| * MIC-Net | 85.8 ± 7.8 | 90.1 ± 7.3 | 90.8 ± 5.6 | 88.90 ± 6.90 | 85.1 ± 6.7 | 88.8 ± 7.8 | 88.7 ± 9.2 | 87.53 ± 7.90 | 980.131 K |
Table 3 presents the quantitative results of the proposed MIC-Net against existing approaches for COVID-19 lesion segmentation. We observe that MPUnet achieves the lowest performance, with DSCs of 82.07 and 81.23. 3D-U-Net and 3D-VNet attain around 2 % and 1 % reductions in DSC and NSD, respectively, in comparison with the 2D models and realize comparable NSD. It is also noted that the recently proposed DSBN and MS-Net attain better performance, with 86.93 and 86.70 DSCs, respectively; furthermore, they yield the highest NSDs, of 85.87 and 86.87, respectively. Compared to the MS-Net, MIC-Net achieves a 2.3 % and 1.6 % improvement on DSC and NSD, respectively. Compared to the recent FL approaches, the proposed MIC-Net achieves a 1 %–2% improvement across different metrics. The results demonstrate the superiority of MIC-Net in the supervised segmentation of pneumonia lesions from heterogeneous CT scans; its better generalization performance qualifies it for integration in the IoMT environment.
6.3. Statistical & visual analysis
To further validate the significance of results obtained in the comparative analysis, a statistical significance test is performed to assess how the obtained results significantly differ from those of the previous one. The p-value obtained from this test is shown in Table 4 . In this setting, the significance threshold is set to be 0.05. As shown, the majority of statistical results are beyond the significance threshold. This, in turn, demonstrates the competing advantages of MIC-Net.
Table 4.
Results of the statistical significance test (p-value) for comparing MIC-Net with competing methods.
| DSC↑ |
NSD↑ |
|||||
|---|---|---|---|---|---|---|
| Method | D1 | D2 | D3 | D1 | D2 | D3 |
| MIC-Net vs 2D-U-Net | 8.112E-05 | 6.930E-04 | 3.490E-04 | 3.114E-03 | 8.975E-06 | 8.820E-04 |
| MIC-Net vs Inf-Net | 8.846E-02 | 6.263E-05 | 1.912E-03 | 2.323E-03 | 2.342E-03 | 2.586E-03 |
| MIC-Net vs MS-Net | 2.155E-03 | 2.827E-03 | 8.014E-02 | 1.509E-03 | 2.832E-03 | 3.285E-04 |
| MIC-Net vs MS-Fed | 1.762E-02 | 2.464E-03 | 6.680E-04 | 9.858E-02 | 2.454E-03 | 1.745E-03 |
| MIC-Net vs FEDDG | 2.549E-03 | 2.459E-02 | 3.425E-05 | 1.709E-02 | 2.405E-03 | 2.111E-03 |
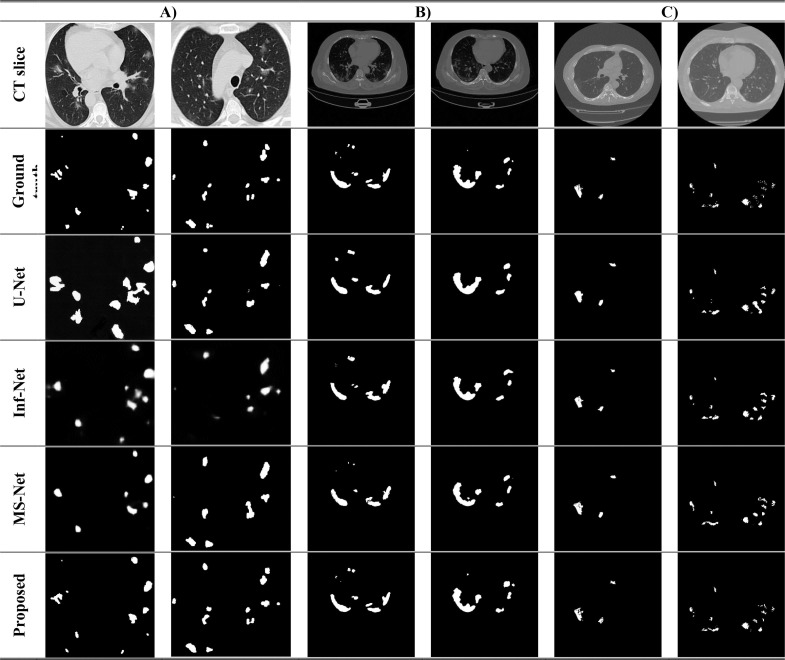
Moreover, Fig. 6 provides a visual comparison of the segmentation outcomes from different competing methods on samples belonging to different domains. The illustration provides an obvious indication of the precision of the segmentation maps compared to that of competing methods. Notably, MIC-Net achieves accurate segmentation for various lesions of different shapes and sizes.
Fig. 6.
Comparison of the segmentation outcomes from different methods against the ground-truth label.
6.4. Ablation analysis
This section analyzes and discusses the ablation experiments to understand the contribution of different building blocks to the final segmentation performance. In this scenario, common U-Net is chosen as the baseline method. Table 5 shows the results of ablation experiments across different performance metrics. For convenience, a nickname is given to each ablation experiment in each row of Table 5. Obviously, the harmonization of training CT images shows considerable performance improvements (DSC: NSD:). Speaking of building blocks, note that redesigning the encoder path with the proposed down-sampler module improves the ability of the network to learn from cross-domain CT slices. This can be attributed to the ability of the ASN to adaptively lessen the impact of domain shift during the training process. To validate this justification, we implement the down-sampler without ASN (See V4), which leads to a significant drop in the performance of MIC-Net. Moreover, eliminating the SSA from the down-sampler (See V5) also results in a significant drop in segmentation performance, which indicates that the attention enables the network to effectively encode lesion features under distribution shifts. In addition, the inclusion of the CE module improves the performance over the baseline (See V6), and similarly, when combined with a down-sampler or up-sampler, it shows notable improvements in all data sets. This can be attributed to the fusion of static and dynamic contextual features by the CE module, enabling the network captures the features of lesions from different size and shapes in CT images from varied distribution. Finally, the up-sampler module is elegantly designed to reconstruct the segmentation maps given the down-sampling maps as well as contextual representations from the CE module, leading to notable improvements compared with the standard deconvolution in the baseline.
Table 5.
Results of ablation experiments for the proposed MIC-NET.
| Variants | Baseline | +HM | Down-sampler |
CE | Up-sampler | DSC↑ |
NSD↑ |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| All | w/o ASN | w/o SSA | D1 | D2 | D3 | D1 | D2 | D3 | |||||
| V1 | 82.1 ± 8.9 | 86.1 ± 9.9 | 87.1 ± 8.9 | 80.9 ± 10.3 | 84.9 ± 11.1 | 87.1 ± 11.5 | |||||||
| V2 | 83.3 ± 8.2 | 87.3 ± 10.3 | 87.3 ± 9.3 | 82.1 ± 7.2 | 86.3 ± 10.9 | 87.7 ± 8.4 | |||||||
| V3 | 84.6 ± 11.3 | 88.3 ± 9.1 | 88.5 ± 8.2 | 83.4 ± 5.9 | 86.9 ± 6.2 | 87.3 ± 3.7 | |||||||
| V4 | 83.8 ± 9.2 | 87.8 ± 8.8 | 87.31 ± 7.7 | 83.1 ± 6.8 | 86.6 ± 4.7 | 87.4 ± 6.3 | |||||||
| V5 | 83.9 ± 8.5 | 87.8 ± 10.2 | 87.9 ± 9.4 | 82.9 ± 9.1 | 86.7 ± 5.4 | 87.1 ± 5.7 | |||||||
| V6 | 84.4 ± 10.1 | 88.1 ± 9.9 | 88.2 ± 8.1 | 83.1 ± 8.4 | 86.6 ± 8.2 | 87.2 ± 7.2 | |||||||
| V7 | 85.4 ± 9.7 | 89.1 ± 6.8 | 89.3 ± 4.9 | 84.2 ± 7.5 | 87.6 ± 6.6 | 87.5 ± 4.6 | |||||||
| V8 | 84.9 ± 8.8 | 88.5 ± 8.1 | 88.4 ± 6.3 | 83.8 ± 4.9 | 87.2 ± 7.4 | 87.4 ± 8.1 | |||||||
| V9 | 85.8 ± 7.8 | 90.1 ± 7.3 | 90.8 ± 5.6 | 85.1 ± 6.71 | 88.8 ± 7.81 | 88.7 + 9.2 | |||||||
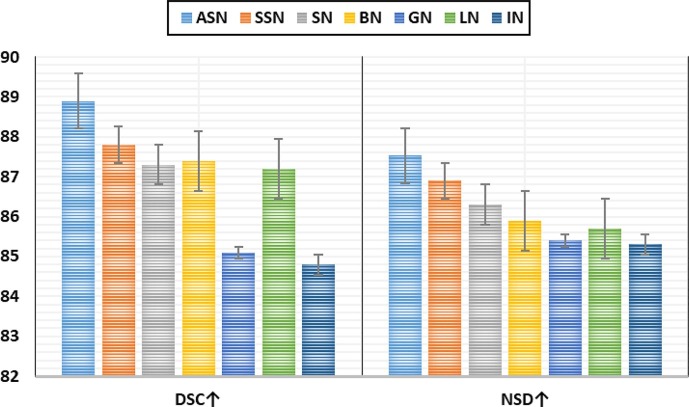
Given the notable performance gain achieved by the ASN, an additional experimental analysis is performed to analyze and compare the performance of MIC-Net when implemented using different normalization layers such as BN, IN, LN, GN, SN, and sparse switchable normalization (SSN). The numerical results obtained from these experiments are shown in Fig. 7 . Notably, the proposed ASN is conducive to improving the segmentation performance under distribution shifts owing to its ability to perform adjustive normalization according to the underlying distribution.
Fig. 7.
Comparison of the performance of MIC-Net under different normalization layers.
6.5. Computational analysis
The framework setup for the proposed IoMT framework assessment and the employed computer hardware configurations are summarized in Table 6 .
Table 6.
Hardware setup for the proposed COVID-19-Fog.
| Fog Component | Configuration Setup |
|---|---|
| Gateway Device | Samsung J4, 4 GB RAM, 16G storage, android 8 |
| Broker | Laptop: Dell inspiron 3500, with Intel(R) Core (TM) i5-7200 CPU @ 1.7 GHZ, 8.00 GB DDR4 RAM, 64-bit system bus, and Windows 10. Apache HTTP Server 2.4.34 utilized for deployment |
| Worker Node | Laptop: Toshiba PS582E-002002AR, with Intel(R)Core (TM) i7-7200 CPU @ 2.5 GHZ, 8.00 GB, 32-bit system bus, and Windows 8.1. Apache HTTP Server 2.4.34. |
| CDC | Microsoft Azure Engine, 2 GB SSD, 1vCPU, 2 GB RAM, Windows Server 2016. |
Latency analysis. Latency is a major concern for the health-care community, especially when it comes to IoMT-aided diagnosis. Thus, the latency of the proposed framework is compared under different FL schemes, namely vanilla FL, FED-CS [32], FED-MCCS, q-FFL, and FED-PCS, computed by combining the queueing delay and communication delay. Note that the FED-PCS can achieve the lowest latency compared to the other approaches, as shown in Fig. 8 . This may be because the FED-PCS can adaptively and fairly select the clients according to their resources and data statistics. In general, the lightweight nature of the MIC-Net enables attaining low latency during the training under different FL schemes, thereby leading to significant differences in latency performance.
Fig. 8.
Latency of the MIC-Net under different resource allocation protocols.
Execution time analysis. When it comes to model training in the real-world IoMT, the Execution time is essential to judging the ability of the model to generate real-time segmentation of the uploaded COVID-19 CT scan. Thus, the Execution time of MIC-Net is calculated and compared under different resource allocation protocols, as shown in Fig. 9 . As anticipated, the FED-PCS protocol configuration exhibits the shortest Execution time (188.3 ms) owing to its powerful resource accessibility. However, the average inference times at edge devices are relatively larger, but it is still acceptable to be considered a real-time response.
Fig. 9.
Execution time of the MIC-Net under different resource allocation protocols.
Network Bandwidth Analysis. As shown in Fig. 10 , the average network bandwidth consumption is compared under different FL allocation protocols. The bandwidth consumption reaches the highest degree when MIC-Net is trained with vanilla FL (46.3 kbps). However, the FED-PCS protocol achieves relatively lower bandwidth consumption, as the parameters are communicated intelligently based on the profile state considered during the client selection. The intelligent FED-PCS achieves the lowest consumption, as it adaptively selects participants according to the resource profile of the clients.
Fig. 10.
The bandwidth consumption of the MIC-Net under different resource allocation protocols.
Power Consumption Analysis: Investigating power/energy utilization is important for any IoT framework. Our analysis for the power proposed IoMT framework is shown in Fig. 11 . It can be seen that training MIC-Net under the FED-PCS protocol results in the lowest power consumption (14.3 W) compared to the other competing methods. Furthermore, the lightweight nature of MIC-Net helps it maintain low power consumption under different FL allocation protocols.
Fig. 11.
The power consumption of the MIC-Net under different resource allocation protocols.
7. Conclusions and future work
This work presented a framework, MIC-Net, for federated segmentation from multi-domain CT data. The concept of drift phenomena is tackled through two stages: one is the attentive learning of cross-domain images, and the other is the adaptive normalization layer. A multi-criteria selection FL protocol is presented to train the MIC-Net in such a way that we can preserve the resources and maximize the training efficiency in the IoMT environment. The experimental findings demonstrate the efficiency and effectiveness of our solution over recent centralized and federated segmentation approaches. The lightweight nature of MIC-Net enables low resource consumption (in terms of power, CPU, memory, and communication bandwidth), making it a robust candidate to be deployed as a distributed pneumonia diagnosis tool, especially in resource-constrained IoMT environments.
In our plans for future works, the proposed framework will be extended to support a multitask diagnosis of COVID-19 patients by including extra tasks such as automatic classification, multi-class segmentation, severity assessment, and other follow-up functionalities. Responsible AI is currently regarded as the most interesting topic for both academia and industry; hence, this work will be updated to satisfy the principles of responsibility in terms of security against adversaries, precision, interpretability, fairness, and privacy preservation. Moreover, the efficiency-cost tradeoff is still a major concern for IoMT application; thus, extending the proposed framework to be more lightweight and efficient will be an important step in our future work. Furthermore, estimating uncertainty in the decisions generated by DL has become a major concern in the majority of IoMT applications; thus, type-2 and type-3 fuzzy sets will be investigated to expand the proposed model to include this functionality.
CRediT authorship contribution statement
Weiping Ding: Conceptualization, Methodology, Investigation, Supervision, Writing-Reviewing and Editing, and Funding acquisition. Mohamed Abdel-Basset: Writing-Original draft preparation, Formal analysis, Data curation, Visualization, Writing-Reviewing and Editing. Hossam Hawash: Writing-Original draft preparation,Writing-Reviewing and Editing. Witold Pedrycz: Writing-Reviewing and Editing, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors would like to express sincere appreciation to the editor and anonymous reviewers for their insightful comments, which greatly improved the quality of this paper. This work is in part by the National Natural Science Foundation of China under Grants 61300167 and 61976120, the Natural Science Foundation of Jiangsu Province under Grant BK20191445, and the Natural Science Key Foundation of Jiangsu Education Department under Grant 21KJA510004.
Data availability
No data was used for the research described in the article.
References
- 1.Abdel-Basset M., Chang V., Hawash H., Chakrabortty R.K., Ryan M. FSS-2019-nCov: A deep learning architecture for semi-supervised few-shot segmentation of COVID-19 infection. Knowl.-Based Syst. 2021;212 doi: 10.1016/j.knosys.2020.106647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmed I., Ahmad A., Jeon G. An IoT-Based Deep Learning Framework for Early Assessment of COVID-19. IEEE Internet Things J. 2020;8(21):15855–15862. doi: 10.1109/JIOT.2020.3034074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Awais M., Raza M., Singh N., Manzoor U., Islam S.U., Rodrigues J.J.P.C. LSTM-based emotion detection using physiological signals: IoT framework for healthcare and distance learning in COVID-19. IEEE Internet Things J. 2020;8(23):16863–16871. doi: 10.1109/JIOT.2020.3044031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen C., Zhou K.N., Zha M.X., Qu X.Y., Guo X.Y., Chen H.Y., Wang Z., Xiao R. An effective deep neural network for lung lesions segmentation from COVID-19 CT images. IEEE Trans. Ind. Inf. 2021;17(9):6528–6538. doi: 10.1109/TII.2021.3059023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Çiçek Ö., Abdulkadir A., Lienkamp S.S., Brox T., Ronneberger O. International Conference on Medical Image Computing and Computer-assisted Intervention. Springer; Cham: 2016. 3D U-Net: learning dense volumetric segmentation from sparse annotation; pp. 424–432. [Google Scholar]
- 6.Fan D.P., Zhou T., Ji G.P., Zhou Y., Chen G., Fu H.Z., Shen J.B., Shao L. Inf-net: Automatic covid-19 lung infection segmentation from ct images. IEEE Trans. Med. Imaging. 2020;39(8):2626–2637. doi: 10.1109/TMI.2020.2996645. [DOI] [PubMed] [Google Scholar]
- 7.Fang Y.C., Zhang H.Q., Xie J.C., Lin M.J., Ying L.J., Pang P.P., Ji W.J. Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology. 2020;296:E115–E117. doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ganesh P., Chen Y., Lou X., Khan M.A., Yang Y., Sajjad H., Nakov P., Chen D.M., Winslett M. Compressing large-scale transformer-based models: A case study on bert, Transactions of the Association for. Comput. Linguist. 2021;9:1061–1080. [Google Scholar]
- 9.Gao K., Su J.P., Jiang Z.B., Zeng L.L., Feng Z.C., Shen H., Rong P.F., Xu X., Qin J., Yang Y.X., Wang W., Hu D.W. Dual-branch combination network (DCN): Towards accurate diagnosis and lesion segmentation of COVID-19 using CT images. Med. Image Anal. 2021;67 doi: 10.1016/j.media.2020.101836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao S.H., Cheng M.M., Zhao K., Zhang X.Y., Yang M.H., Torr P. Res2Net: A New Multi-Scale Backbone Architecture. IEEE Trans. Pattern Anal. Mach. Intell. 2019;43(2):652–662. doi: 10.1109/TPAMI.2019.2938758. [DOI] [PubMed] [Google Scholar]
- 11.Huang X., Belongie S. Proceedings of the IEEE International Conference on Computer Vision. 2017. Arbitrary style transfer in real-time with adaptive instance normalization; pp. 1501–1510. [Google Scholar]
- 12.Kumar R., Khan A.A., Kumar J., Zakria N.A., Golilarz S.M., Zhang Y., Ting C.Y., Zheng W.Y.W. Blockchain-federated-learning and deep learning models for covid-19 detection using ct imaging. IEEE Sens. J. 2021;21(14):16301–16314. doi: 10.1109/JSEN.2021.3076767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li T., Sanjabi M., Beirami A., Smith V. Fair Resource Allocation in Federated Learning, International Conference on Learning. Representations. 2020:1–13. [Google Scholar]
- 14.Li X.M., Chen H., Qi X.J., Dou Q., Fu C.W., Heng P.A. H-DenseUNet: Hybrid Densely Connected UNet for Liver and Tumor Segmentation from CT Volumes. IEEE Trans. Med. Imaging. 2018;37(12):2663–2674. doi: 10.1109/TMI.2018.2845918. [DOI] [PubMed] [Google Scholar]
- 15.Li X.X., Gu Y.F., Dvornek N., Staib L.H., Ventola P., Duncan J.S. Multi-site fMRI analysis using privacy-preserving federated learning and domain adaptation: ABIDE results. Med. Image Anal. 2020;65 doi: 10.1016/j.media.2020.101765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ling J., Xue H., Song L., Xie R., Gu X. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. 2021. Region-aware adaptive instance normalization for image harmonization; pp. 9361–9370. [Google Scholar]
- 17.Liu J.N., Dong B., Wang S., Cui H., Fan D.P., Ma J.Q., Chen G. COVID-19 lung infection segmentation with a novel two-stage cross-domain transfer learning framework. Med. Image Anal. 2021;74 doi: 10.1016/j.media.2021.102205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Q.D., Chen C., Qin J., Dou Q., Heng P.A. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. 2021. Feddg: Federated domain generalization on medical image segmentation via episodic learning in continuous frequency space; pp. 1013–1023. [Google Scholar]
- 19.Liu Q.D., Dou Q., Yu L.Q., Heng P.A. MS-Net: Multi-Site Network for Improving Prostate Segmentation with Heterogeneous MRI Data. IEEE Trans. Med. Imaging. 2020;39(9):2713–2724. doi: 10.1109/TMI.2020.2974574. [DOI] [PubMed] [Google Scholar]
- 20.Liu Z., Lin Y.T., Cao Y., Hu H., Wei Y.X., Zhang Z., Lin S., Guo B.N. Proceedings of the IEEE/CVF International Conference on Computer Vision. 2021. Swin transformer: Hierarchical vision transformer using shifted windows; pp. 10012–10022. [Google Scholar]
- 21.Liu Z., Yao C.H., Yu H., Wu T.H. Deep reinforcement learning with its application for lung cancer detection in medical Internet of Things. Futur. Gener. Comput. Syst. 2019;97:1–9. [Google Scholar]
- 22.Long J., Shelhamer E., Darrell T. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2015. Fully convolutional networks for semantic segmentation; pp. 3431–3440. [DOI] [PubMed] [Google Scholar]
- 23.Luo P., Zhang R.M., Ren J.M., Peng Z.L., Li J.Y. Switchable Normalization for Learning-to-Normalize Deep Representation. IEEE Trans. Pattern Anal. Mach. Intell. 2019;43(2):712–728. doi: 10.1109/TPAMI.2019.2932062. [DOI] [PubMed] [Google Scholar]
- 24.Ma F.Z., Sun T., Liu L.Y., Jing H.Y. Detection and diagnosis of chronic kidney disease using deep learning-based heterogeneous modified artificial neural network. Futur. Gener. Comput. Syst. 2020;111:17–26. [Google Scholar]
- 25.J. Ma, Y.X. Wang, X. An, C. Ge, Z. Yu, J. Chen, Q. Zhu, G. Dong, J. He, Z. He, Towards Efficient COVID-19 CT Annotation: A Benchmark for Lung and Infection Segmentation, arXiv preprint, 2020, Available: http://arxiv.org/abs/2004.12537.
- 26.Mahmud T., Alam M.J., Chowdhury S., Ali S.N., Rahman M.M., Fattah S.A., Saquib M. CovTANet: a hybrid tri-level attention-based network for lesion segmentation, diagnosis, and severity prediction of COVID-19 chest CT scans. IEEE Trans. Ind. Inf. 2020;17(9):6489–6498. doi: 10.1109/TII.2020.3048391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mansour R.F., Escorcia-Gutierrez J., Gamarra M., Gupta D., Castillo O., Kumar S. Unsupervised deep learning based variational autoencoder model for COVID-19 diagnosis and classification. Pattern Recogn. Lett. 2021;151:267–274. doi: 10.1016/j.patrec.2021.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.“MedSeg” [Online]. Available: https://medicalsegmentation.com/covid19/.
- 29.S. Mehta, M. Rastegari, Separable Self-attention for Mobile Vision Transformers, arXiv preprint, 2022, Available: http://arxiv.org/abs/2206.02680.
- 30.Morozov S.P., Andreychenko A.E., Blokhin I.A., Gelezhe P.B., Gonchar A.P., Nikolaev A.E., Pavlov N.A., Chernina V.Y., Gombolevskiy V.A. MosMedData: data set of 1110 chest CT scans performed during the COVID-19 epidemic. Digital Diagnostics. 2020;1(1):49–59. [Google Scholar]
- 31.Nguyen D.C., Pham Q.V., Pathirana P.N., Ding M., Seneviratne A., Lin Z.H., Dobre Q., Hwang W. Federated learning for smart healthcare: A survey. ACM Comput. Surveys (CSUR) 2022;55(3):1–37. [Google Scholar]
- 32.Nishio T., Yonetani R. 2019 IEEE International Conference on Communications (ICC) 2019. Client selection for federated learning with heterogeneous resources in mobile edge; pp. 1–7. [Google Scholar]
- 33.Pathak N., Deb P.K., Mukherjee A., Misra S. IoT-to-the-rescue: A survey of IoT solutions for COVID-19-like pandemics. IEEE Internet Things J. 2021;8(17):13145–13164. [Google Scholar]
- 34.S.Y. Qiao, H.Y. Wang, C.X. Liu, W. Shen, A. Yuille, Micro-Batch Training with Batch-Channel Normalization and Weight Standardization, arXiv preprint, 2019, Available: http://arxiv.org/abs/1903.10520.
- 35.Ronneberger O., Fischer P., Brox T. International Conference on Medical Image Computing and Computer-assisted Intervention. Springer; Cham: 2015. U-net: Convolutional networks for biomedical image segmentation; pp. 234–241. [Google Scholar]
- 36.Roy A.G., Navab N., Wachinger C. Recalibrating Fully Convolutional Networks With Spatial and Channel ‘Squeeze and Excitation’ Blocks. IEEE Trans. Med. Imaging. 2018;38(2):540–549. doi: 10.1109/TMI.2018.2867261. [DOI] [PubMed] [Google Scholar]
- 37.Valanarasu J.M.J., Yasarla R., Wang P.Y., Hacihaliloglu I., Patel V.M. Learning to Segment Brain Anatomy from 2D Ultrasound with Less Data. IEEE J. Sel. Top. Signal Process. 2020;14(6):1221–1234. [Google Scholar]
- 38.Hasselt H.V., Guez A., Silver D. Proceedings of the AAAI Conference on Artificial Intelligence. 2016. Deep reinforcement learning with double q-learning; pp. 2094–2100. [Google Scholar]
- 39.Varela-Santos S., Melin P. A new approach for classifying coronavirus COVID-19 based on its manifestation on chest X-rays using texture features and neural networks. Inf. Sci. 2021;545:403–414. doi: 10.1016/j.ins.2020.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vaswani A., Shazeer N., Parmar N., Uszkoreit J., Jones L., Gomez A.N., Kaiser L., Polosukhin I. Attention is all you need. Adv. Neural Inf. Proces. Syst. 2017:5998–6008. [Google Scholar]
- 41.Wang G.T., Liu X.L., Li C.P., Xu Z.Y., Ruan J.G., Zhu H.F., Meng T., Li K., Huang N., Zhang S.T. A noise-robust framework for automatic segmentation of COVID-19 pneumonia lesions from CT images. IEEE Trans. Med. Imaging. 2020;39(8):2653–2663. doi: 10.1109/TMI.2020.3000314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang R.Z., Zheng G.Y. CyCMIS: Cycle-consistent Cross-domain Medical Image Segmentation via diverse image augmentation. Med. Image Anal. 2022;76 doi: 10.1016/j.media.2021.102328. [DOI] [PubMed] [Google Scholar]
- 43.Wang X.F., Jiang L., Li L., Xu M., Deng X., Dai L.S., Xu X.Y., Li T.Y., Guo Y.C., Wang Z.L., Dragotti P.L. Joint learning of 3D lesion segmentation and classification for explainable COVID-19 diagnosis. IEEE Trans. Med. Imaging. 2021;40(9):2463–2476. doi: 10.1109/TMI.2021.3079709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang X.Y., Yuan Y.W., Guo D.Y., Huang X.J., Cui Y., Xia M., Wang Z.H., Bai C., Chen S.Y. SSA-Net: Spatial self-attention network for COVID-19 pneumonia infection segmentation with semi-supervised few-shot learning. Med. Image Anal. 2022;79 doi: 10.1016/j.media.2022.102459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Z., Liu Q.D., Dou Q. Contrastive Cross-Site Learning with Redesigned Net for COVID-19 CT Classification. IEEE J. Biomed. Health Inform. 2020;24(10):2806–2813. doi: 10.1109/JBHI.2020.3023246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yao C.H., Wu S.D., Liu Z., Li P. A deep learning model for predicting chemical composition of gallstones with big data in medical Internet of Things. Futur. Gener. Comput. Syst. 2019;94:140–147. [Google Scholar]
- 47.Zhang W.S., Zhou T., Lu Q.H., Wang X., Zhu C.S., Sun H.Y., Wang Z.P., Lo S.K., Wang F.Y. Dynamic-fusion-based federated learning for COVID-19 detection. IEEE Internet Things J. 2021;8(21):15884–15891. doi: 10.1109/JIOT.2021.3056185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou L.X., Li Z.X., Zhou J.X., Li H.Y., Chen Y.P., Huang Y.X., Xie D.X., Zhao L.T., Fan M., Hashmi S., Abdelkareem F., Eiada R., Xiao X.G., Li L.H., Qiu Z.W., Gao X. A rapid, accurate and machine-agnostic segmentation and quantification method for CT-based COVID-19 diagnosis. IEEE Trans. Med. Imaging. 2020;39(8):2638–2652. doi: 10.1109/TMI.2020.3001810. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.