Abstract
The pro‐inflammatory factor interleukin‐8 (IL‐8) is related to poor prognosis in hepatocellular carcinoma (HCC) patients. Interleukin‐8 enhanced HCC invasion by upregulating Snail and Twist1, whether this modulation relies on microRNAs (miR) is unclear. In this study, hsa‐miR‐370‐3p was screened as candidate miRNA targeting Snail and Twist1, and its expression was downregulated by IL‐8. Luciferase assays and RNA electrophoretic mobility shift assays were used to evaluate the interaction between miR‐370‐3p and targeted mRNAs. Coimmunoprecipitation, luciferase, and ChIP assays were undertaken to investigate the mechanisms underlying IL‐8‐mediated modification of miR‐370‐3p. Gain‐ and loss‐of‐function studies, Transwell assays, and a xenograft nude mouse model were used to investigate pro‐ and antitumor activities. Interleukin‐8 and miR‐370‐3p levels were analyzed for clinical relevance in HCC patients. Our results showed that HCC patients with high levels of IL‐8 experienced more metastasis and shorter survival. Interleukin‐8 induced epithelial–mesenchymal transition and promoted liver cancer cell migration, invasion, and metastasis both in vitro and in vivo. MicroRNA‐370‐3p interacted with its cognate mRNA within the 3′‐UTR regions of Twist1 and Snail mRNA directly and specifically and attenuated IL‐8 protumoral effects on liver cancer cells. Interleukin‐8 negatively modulated miR‐370‐3p through signal transducer and activator of transcription 3 (STAT3) activation by recruiting histone deacetylase 1 (HDAC1) to miR‐370‐3p promoter. The STAT3 and HDAC antagonists inhibited liver cancer cell migration and invasion. Patients with high miR‐370‐3p and low IL‐8 levels had longer overall survival. In conclusion, our study elucidated a novel axis IL‐8/STAT3/miR‐370‐3p/Twist1 and Snail relying on HDAC1 recruitment, which showed both diagnostic and therapeutic potentials of miR‐370‐3p in HCC metastasis.
Keywords: cytokine, epithelial–mesenchymal transition, hepatocellular carcinoma, metastasis, micro RNA
Our results showed that HCC patients with high levels of IL‐8 exhibited more metastasis and shorter survival. IL‐8 induced EMT and promoted liver cancer cell migration, invasion, and metastasis both in vitro and in vivo. miR‐370‐3p interacted with its cognate mRNA within the 3'‐UTR regions of Twist1 and Snail mRNA directly and specifically and attenuated IL‐8 protumoral effects on liver cancer cells. IL‐8 negatively modulated microRNA‐370‐3p through STAT3 activation by recruiting HDAC1 to miR‐370‐3p promoter. STAT3 and HDAC antagonists inhibited liver cancer cell migration and invasion. Patients with high miR‐370‐3p and low IL‐8 levels had longer overall survival. The IL‐8/STAT3/microRNA‐370‐3p/Twist1 and Snail axis modulated HCC metastasis.
Abbreviations
- EMT
epithelial–mesenchymal transition
- EMT‐TF
epithelial–mesenchymal transition‐transcription factor
- HCC
hepatocellular carcinoma
- HDAC
histone deacetylase
- IL
interleukin
- MFE
minimum free energy
- miRNA
microRNA
- ncRNA
noncoding RNA
- OS
overall survival
- PFS
progression‐free survival
- qRT‐PCR
quantitative real‐time PCR
- STAT
signal transducer and activator of transcription
- TACE
transcatheter arterial chemoembolization
- TCGA
The Cancer Genome Atlas
- TKI
tyrosine kinase inhibitor
1. INTRODUCTION
Liver cancer, mainly HCC, is one of the most fatal cancers, especially in Asia. 1 The poor outcomes of HCC patients are largely due to metastatic spread, with 5‐year recurrence rate exceeding 70%. 2 Intrahepatic metastasis frequently manifests as intrahepatic vascular microinvasion and micrometastasis, implying the highly aggressive nature of HCC cells. 1 Uncovering the mechanisms underlying metastasis could reveal previously unappreciated features of HCC.
The complex heterogeneity of epithelial cells was described as EMT, a transient and reversible dedifferentiation to a mesenchymal‐like or mesenchymal phenotype, which partly explains the phenotype of primary HCC accompanied by portal vein tumor thrombi and intrahepatic metastasis. 3 Epithelial–mesenchymal transition is executed in response to pleiotropic signaling factors that activate specific EMT‐TFs and then trigger the dissociation of carcinoma cells from primary carcinomas. 4 Epithelial–mesenchymal transition‐TFs are pivotal regulators, as they suppress E‐cadherin, the prototypic adhesion molecule essential for mediating cell polarity, differentiation, migration, and stem cell‐like properties, which leads to the transition from well‐differentiated adenoma to invasive. 5 The most prominent EMT‐TFs, such as Snail (SNAI1), Slug (SNAI2), and ZEB, bind to E‐boxes present on the E‐cadherin promoter and suppress its transcription directly. 6 Other EMT‐TFs, such as bHLH proteins (Twist1 and Twist2), are commonly recognized as indirect repressors of E‐cadherin. 7
It is now becoming clear that chronic inflammatory stimuli are indispensable in tumorigenesis and progression. 8 Various pro‐inflammatory cytokines, including tumor necrosis factor, IL‐8, IL‐1β, IL‐6, IL‐10, IL‐18, and IL‐17, are important inflammatory mediators on neoplastic processes both in inflammation‐induced cancer and in inflammation that follows tumor development. 9 Previous studies have identified IL‐8 as a key factor involved in the progression of digestive tract tumors. 10 In liver cancer, increased IL‐8 expression is positively related to portal vein invasion and distant metastasis and negatively related to PFS and OS. 11 Hepatocellular carcinoma patients with high levels of IL‐8 also experience an impaired benefit from immunotherapy, following poor prognosis. 12 Our previous research also confirmed that IL‐8 was associated with prognostic clinical factors, including lymph node metastasis, recurrence, and alanine aminotransferase levels. Interleukin‐8 upregulates Twist1 and Snail expression and induces EMT in hepatic cancer cells, resulting in enhanced invasion. 13 However, the cellular and molecular mechanisms involved in this modulation are not completely understood.
In recent years, the expansion of knowledge of ncRNA biology has revealed critical roles in metastatic processes. 14 MicroRNAs are an emerging class of primarily ncRNAs that bind to the 3′‐UTRs of specific mRNAs, which leads to either mRNA degradation or translational pausing. 15 , 16 Functionally, miRNAs are pivotal determinants during liver cancer progression. p53 upregulates miR‐200 and miR‐192 family members, then represses ZEB1/2 expression, which normally sustains the epithelial phenotype, thus promoting EMT and HCC invasion. 17 Cross‐regulation between cytokine and miRNA pathways is beginning to be elucidated. 18 However, it is largely unknown whether miRNAs targeting EMT‐TFs involve IL‐8‐activated EMT in HCC.
Enlightened by the abovementioned progress, in this study, we combined in silico, bioinformatic, in vitro, and in vivo approaches to systematically investigate the contribution of miRNAs targeting EMT‐TFs induced by IL‐8 in liver cancer cells. The molecular mechanism underlying IL‐8‐modulated miRNA expression was also investigated.
2. MATERIALS AND METHODS
2.1. Human tissue specimens
Eighty‐seven pairs of HCC and adjacent tissues were collected immediately from patients who underwent surgical resection at the Fifth Affiliated Hospital of Sun Yat‐sen University from 2010 to 2017. A total of 100 HCC postoperatively frozen tissue and serum samples were obtained from patients who underwent surgical resection also diagnosed by professional pathologists at Sun Yat‐sen University Cancer Center from 2017 to 2018. Hepatocellular carcinoma tissues with typical macroscopic features were collected from the central part of tumor nodules, which were also examined with H&E staining. Paired adjacent nontumoral tissues without histopathologically identified tumor cells were collected at least 3 cm away from the tumor border. Informed consent was obtained from all patients and approved by the research medical ethics committee of Sun Yat‐sen University.
2.2. RNA EMSA
Electrophoretic mobility shift assay was carried out according to the protocol of the LightShift Chemiluminescent RNA EMSA Kit (Thermo Scientific) and our previous studies. 19 The miRNA/mRNA and miRNA/mRNA/protein hybridizations were detected with Odyssey CLx Infrared Imaging System (LI‐COR Biosciences). Details are described in Appendix S1. The sequences of primers, siRNAs, miRNAs, and oligonucleotides for EMSA are listed in Table S1.
2.3. Chromatin immunoprecipitation assay
This procedure was carried out as described by the ChIP assay kit (Beyotime). Briefly, 10‐cm plates were seeded with cells and allowed to grow to approximately 90% confluence. A 1% final formaldehyde concentration was used to cross‐link proteins to DNA, which was then stopped by adding glycine to the suspension. The cells were collected by centrifugation, and the nuclear pellet was resuspended in ChIP buffer. The cell lysate was subjected to sonication and then incubated with 5 μg of Abs overnight, followed by incubation with protein A/G agarose beads overnight at 4°C. Bound DNA–protein complexes were eluted, and cross‐links were reversed after a series of washes. The purified DNA was resuspended in TE buffer for PCR. The sequences of primers for ChIP are listed in Table S1, and the antibodies are listed in Table S2.
2.4. Dataset information
The Cancer Genome Atlas public database (https://cancergenome.nih.gov/), which includes 40 pairs of liver cancer, was used to analyze differences in miR‐370‐3p expression between HCC and adjacent nontumor tissues. Another 424 pairs were used to analyze the correlation between IL‐8 and STAT3.
2.5. In silico analysis
The miRNAs that potentially target Snail and Twist1 transcripts were screened through three public databases: microRNA.org (http://www.microrna.org/), miRbase (https://www.mirbase.org/), and TargetScan (release 6.2, http://www.targetscan.org/vert_61/). The RNAhybrid program (http://bibiserv2.cebitec.uni‐bielefeld.de/rnahybrid/) was used to calculate the MFE of hybridization for candidate miRNAs with their cognate targets.
2.6. Statistical analysis
Statistical analysis was undertaken with SPSS software (SPSS Inc.). Values are expressed as the mean ± SD. The difference was analyzed using ANOVA or χ2‐test. Spearman's rank correlation coefficient was used to determine the correlation between two parameters. The PFS and OS curves were assessed using Kaplan–Meier plots and compared with log–rank test. p < 0.05 was considered statistically significant.
3. RESULTS
3.1. Interleukin‐8 promotes metastasis of HCC and predicts poor outcomes in patients
Increased expression of the pro‐angiogenic/pro‐inflammatory chemokine IL‐8 (CXCL8) and/or its receptors (CXCR1/CXCR2) has been characterized in various cells, such as cancer cells, endothelial cells, infiltrating neutrophils, and tumor‐associated macrophages, which suggests that IL‐8 could function as a significant regulatory factor within tumor microenvironment. 20 In a cohort of fresh‐frozen serum samples collected from primary and metastatic HCC patients (n = 76 and 24, respectively), the metastatic group showed increased IL‐8 expression (Figure 1A, p = 0.05). Moreover, the OS in the IL‐8‐high group was significantly shorter than those in the IL‐8‐low group (Figure 1B), implying that IL‐8 affects patient survival by promoting metastasis. We examined the expression of several EMT‐TFs, including Snail, Slug, Twist1, ZEB1, and ZEB2, and found that Snail and Twist1 apparently increased after IL‐8 stimulation. At the same time, the expression of epithelial marker proteins E‐cadherin and ZO‐1 decreased, and interstitial characteristic proteins N‐cadherin, fibronectin, and vimentin increased (Figure 1C). A spike‐like filopodia morphology change was also observed in IL‐8‐stimulated cells (Figure 1D). Consequently, IL‐8 promoted Huh‐7 and HepG2 migration and invasion in vitro (Figure 1E). An HCC metastasis model was established to mimic the blood spreading of cancer cells with Huh‐7 cells injected into the mouse tail vein. As shown in Figure 1F, mice treated with IL‐8 showed more severer liver metastasis than controls. Collectively, these results suggested that IL‐8 promotes cancer cell metastasis and was correlated with poor prognosis.
FIGURE 1.
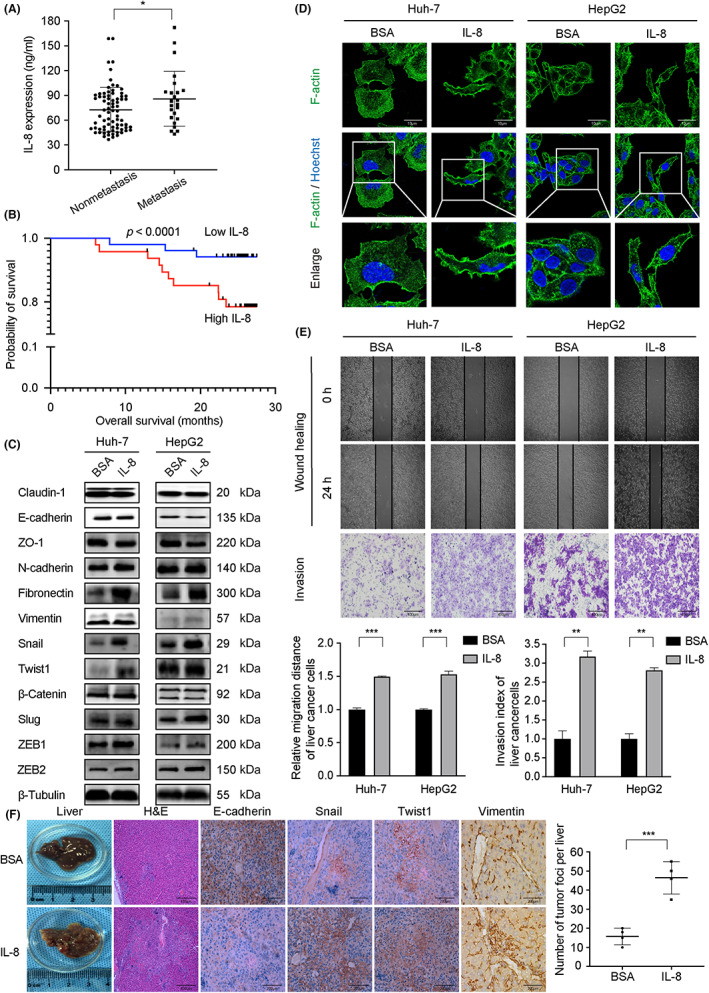
Interleukin‐8 (IL‐8) promotes metastasis of liver cancer and predicts patient outcomes. (A) Comparison of serum interleukin‐8 (IL‐8) expression levels between hepatocellular carcinoma (HCC) metastatic and nonmetastatic groups (p = 0.05). (B) HCC patients with low IL‐8 expression had better OS. (C) Expression of Snail, Twist1, N‐cadherin, fibronectin, and vimentin increased, whereas E‐cadherin and ZO‐1 decreased in liver cancer cells treated with rh‐IL‐8. (D) A spike‐like filopodia morphology change was observed in IL‐8‐stimulated cells. Cells were stained with F‐actin (green) and counterstained with Hoechst (blue). Magnification, ×400. (E) Wound healing and Transwell assays in Huh‐7 and HepG2 cells. IL‐8 stimulation enhanced liver cancer cell migration and invasion abilities. (F) Mice treated with IL‐8 showed more liver metastatic nodules and high expression of Snail, Twist1, and vimentin, whereas E‐cadherin was downregulated. Huh‐7 cells were used in the HCC metastasis model with or without IL‐8 intraperitoneal injection (100 ng per mouse, once a week), n = 6. Values are expressed as mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001.
3.2. Identification of EMT‐TFs targeting miRNA
We wanted to explore whether a factor regulates Snail and Twist1 simultaneously, perhaps a key link between IL‐8 and EMT. Certain miRNAs target multiple genes simultaneously and exert synergistic effects on cellular biofunctions. 21
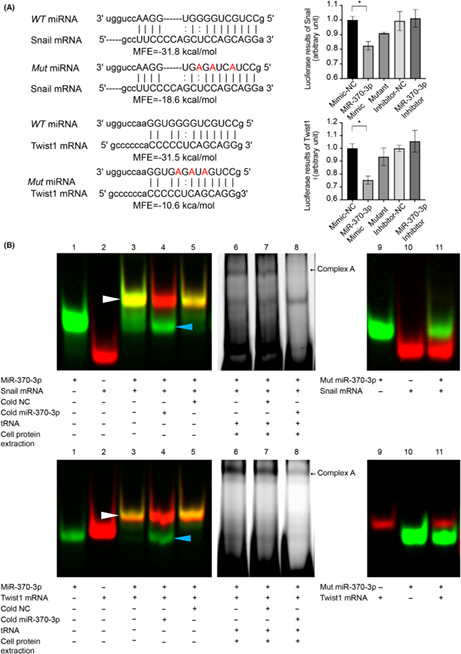
We undertook an in silico analysis using three algorithms (TargetScan, miRbase, and microRNA.org) that predict targeting transcripts by miRNAs based on the 3′‐UTR of Snail and Twist1. Minimum free energy represents the hybridization ability between miRNAs and targeting sequences, and we have previously reported that MFE < −20 kcal/mol is a key parameter for efficient binding. 19 As shown in Figure S1, there are 11 miRNAs that potentially target Snail and Twist1; miR‐370‐3p is unique in that its MFE is lower than −20 kcal/mol. Mutated miR‐370‐3p was constructed as three nucleotides in the WT sequence, and MFE between miRNAs and mRNAs was significantly reduced to −18.6 kcal/mol and − 10.6 kcal/mol, respectively (Figure 2A). We then explored the differentially expressed miRNAs between HCC and normal samples in TCGA. A Venn diagram of the screened differential genes and 11 candidate miRNAs was constructed, which identified miR‐370 and miR‐381 at the intersection. Our review of other studies found that the regulation of miR‐370 on EMT‐TFs has not been reported yet. MicroRNA‐381 was reported to influence IL‐8 expression, indicating that miR‐381 might be an upstream factor of IL‐8. 22 Based on the above results, we chose miR‐370‐3p as the candidate gene for further study (Figure S2, S5).
FIGURE 2.
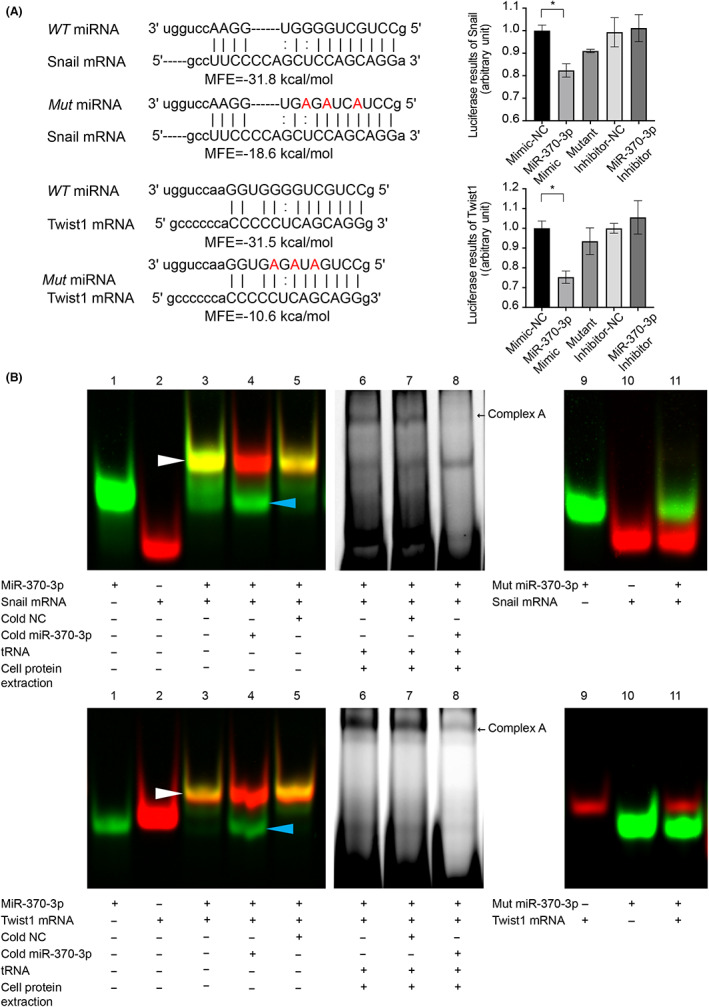
Identification of Snail and Twist1 as direct effectors of microRNA (miR)‐370‐3p. (A) Predicted hybrid complexes formed by miR‐370‐3p and sequences within the 3′‐UTR of WT Snail and Twist1 mRNA. The free energy of hybridization between miR‐370‐3p and Snail 3′‐UTR was −31.8 kcal/mol, and Twist1 3′‐UTR was −31.5 kcal/mol. When three nucleotides in miR‐370‐3p sequence were mutated, the free energy of hybridization was reduced to −18.6 and − 10.6 kcal/mol, respectively. (B) RNA EMSAs showed that miR‐370‐3p oligonucleotides interacted with Snail and Twist1 mRNA oligonucleotides directly. Lane 1, free miRNA; lane 2, free mRNA; lane 3, miRNA + mRNA; lane 4, miRNA + mRNA + cold hsa‐miR‐370‐3p; lane 5, miRNA + mRNA + cold negative control (NC, cold NC does not affect hybridization); lanes 6–8, RNA : protein interactions. After adding cell cytoplasmic extract to reaction mixtures, miRNA/mRNA/protein complex formation occurred (complex A). Complex A was competitively inhibited by 50‐fold molar excess of cold miR‐370‐3p (lane 8). Mutated miR‐370‐3p failed to form miRNA/mRNA hybridization (lanes 9–11). (C) Luciferase activity of the luciferase reporter constructs containing either WT or mutated 3′‐UTRs of Snail and Twist1 after miR‐370‐3p mimic treatment. *p < 0.05.
RNA EMSA confirmed the direct interaction between miR‐370‐3p and the 3′‐UTR of Snail and Twist1 mRNA. As shown in Figure 2B, both target sequences from either Snail or Twist1 3′‐UTR compound with miR‐370‐3p (white arrowhead), as well as the miRNA/mRNA/protein complex (black arrow, complex A). The complexes were inhibited by 50‐fold molar excess of cold miR‐370‐3p (lane 4 and lane 8) but not inhibited by negative control (lane 5 and lane 7), indicating the binding specification between miRNA and cognate targets. In addition, mutant miR‐370‐3p could not bind to its targets (lanes 9–11).
To further confirm the direct regulation of miR‐370‐3p on 3′‐UTR of Snail and Twist1 mRNA, HEK293T cells were transfected with luciferase reporter vectors containing WT 3′‐UTR of Snail or Twist1. As shown in Figure 2C, miR‐370‐3p mimic suppressed the luciferase activity of Snail 3′‐UTR by approximately 20% and Twist1 3′‐UTR by approximately 25%, whereas mutant miR‐370‐3p did not show any effect. We concluded that miR‐370‐3p targets Snail and Twist1 directly and specifically.
3.3. Hsa‐miR‐370‐3p inhibited hepatic cancer cell EMT and metastasis
The function of miR‐370‐3p in EMT is less clear, nor has it been studied in the context of HCC metastasis. We first transfected Huh‐7 and HepG2 cells with synthetic miR‐370‐3p molecules (mimic, inhibitor, and corresponding negative controls) and verified that miR‐370‐3p downregulated Snail and Twist1 expression at both mRNA and protein levels (Figure 3A,B). MicroRNA‐370‐3p mimic group showed increased E‐cadherin and ZO‐1, but decreased N‐cadherin, vimentin, and fibronectin expression, and miR‐370‐3p inhibitor exerted the opposite effects (Figure 3B). Consistently, miR‐370‐3p mimics inhibited Huh‐7 and HepG2 cell migration and invasion in vitro (Figure 3C). The effects of miR‐370‐3p mimics were similar to those of siRNA Snail and siRNA Twist1 (Figure S2 and S3), mice bearing Huh‐7miR‐370‐3p showed much less liver metastasis than control. As we did not detect apparent changes in Huh‐7 or HepG2 cell proliferation after miR‐370‐3p overexpression (Figure S4A), we believed that the diminished invasion and metastasis were due to the anti‐EMT effect of miR‐370‐3p.
FIGURE 3.
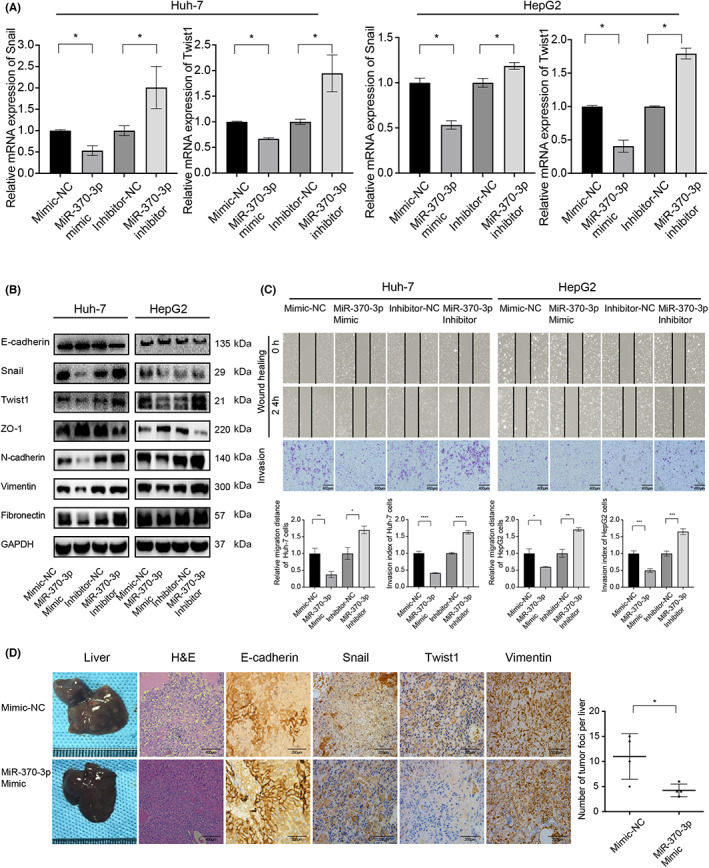
MicroRNA (miR)‐370‐3p inhibits the migration and invasion of liver cancer cells in vitro and in vivo. (A) Real‐time PCR quantification of Snail and Twist1 expression in Huh‐7 and HepG2 cells transfected with miR‐370‐3p mimic or inhibitor for 48 h. mRNA expression was normalized to β‐actin. (B) Transfection of miR‐370‐3p mimic inhibited Snail and Twist1 at the protein level in Huh‐7 and HepG2 cells. At the same time, E‐cadherin and ZO‐1 increased, whereas the expression of N‐cadherin, fibronectin, and vimentin decreased. However, miR‐370‐3p inhibitor exerted opposite effects. β‐Tubulin was used as a loading control. (C) miR‐370‐3p inhibited Huh‐7 and HepG2 cell migration and invasion. (D) Mice bearing Huh‐7miR‐370‐3pmimic showed much less liver metastasis than control with two‐tailed Student's t‐test, n = 6. Experiments were performed in triplicate, and data are shown as mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
3.4. MicroRNA‐370‐3p attenuated IL‐8‐induced HCC metastasis
We note that miR‐370‐3p significantly impedes liver cancer cell migration and invasion through targeting Snail and Twist1, properties reminiscent of IL‐8 activation. Thus, we proposed that miR‐370‐3p antagonizes IL‐8‐mediated tumor progression. Intriguingly, IL‐8‐induced expression of Snail and Twist1 was attenuated by miR‐370‐3p mimic transfection prior to IL‐8 treatment (Figure 4A). Consistent with these observations, overexpression of miR‐370‐3p in liver cancer cells was sufficient to rescue the promotion of migration and invasion induced by IL‐8 (Figure 4B). Accordingly, miR‐370‐3p‐treated mice showed fewer liver metastatic nodules after intraperitoneal injection of IL‐8 suspension (Figure 4C). These findings indicated that miR‐370‐3p was involved in the IL‐8/Snail and Twist1 axis in HCC.
FIGURE 4.
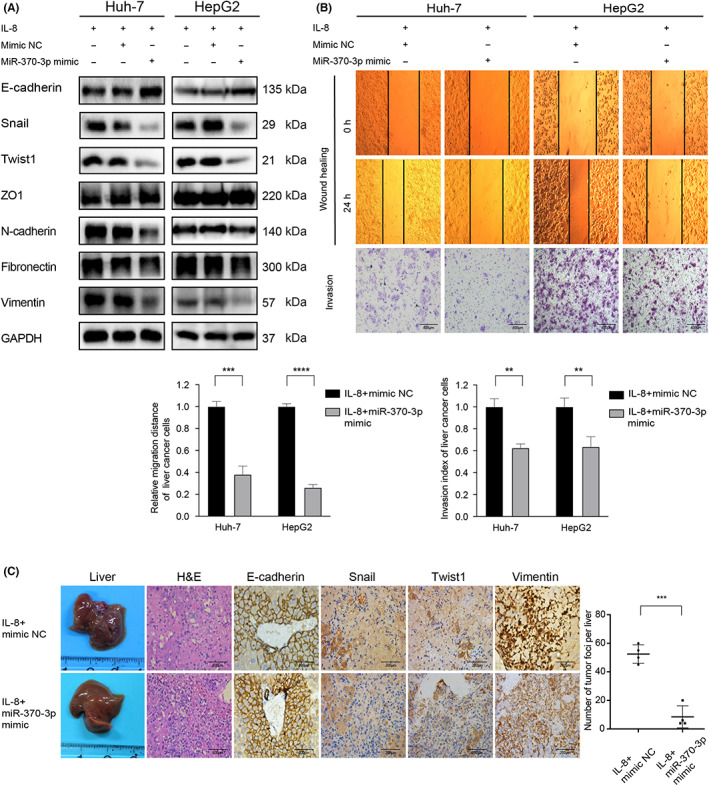
MicroRNA (miR)‐370‐3p attenuated interleukin‐8 (IL‐8)‐induced liver cancer cell metastasis. (A) IL‐8‐induced expression of Snail and Twist1 was attenuated when miR‐370‐3p mimic molecules were transfected into cells prior to IL‐8 treatment. (B) Overexpression of miR‐370‐3p in Huh‐7 and HepG2 cells rescued the promotion of migration and invasion induced by IL‐8. (C) miR‐370‐3p‐treated mice showed fewer liver metastatic nodules after IL‐8 intraperitoneal injection. Immunohistochemical analysis of tumor tissue sections showed that expression of E‐cadherin increased, whereas Snail, Twist1, and vimentin decreased in Huh‐7miR‐370‐3pmimic mice. **p < 0.01, ***p < 0.001, ****p < 0.0001.
3.5. Interleukin‐8 suppressed miR‐370‐3p expression by STAT3 occupying the promoter
A previous study showed that miR‐370 expression was influenced by IL‐6, 23 which prompted us to further explore whether IL‐8 regulates the expression of miR‐370‐3p. Interestingly, miR‐370‐3p expression was dramatically decreased after IL‐8 treatment (Figure 5A). We previously detected apparent phosphorylation of the Tyr705 residue of STAT3 in IL‐8‐treated liver cancer cells. 13 Using HCC data from TCGA, we found that the expression of STAT3 was positively correlated with IL‐8 expression (Figure 5B). We also found that IL‐8‐downregulated miR‐370‐3p expression was significantly reversed by STAT3 phosphorylation inhibitor Stattic (Figure 5C). Consistently, Stattic attenuated the promoting effects exerted by IL‐8 on liver cancer cell migration and invasion (Figure 5D).
FIGURE 5.
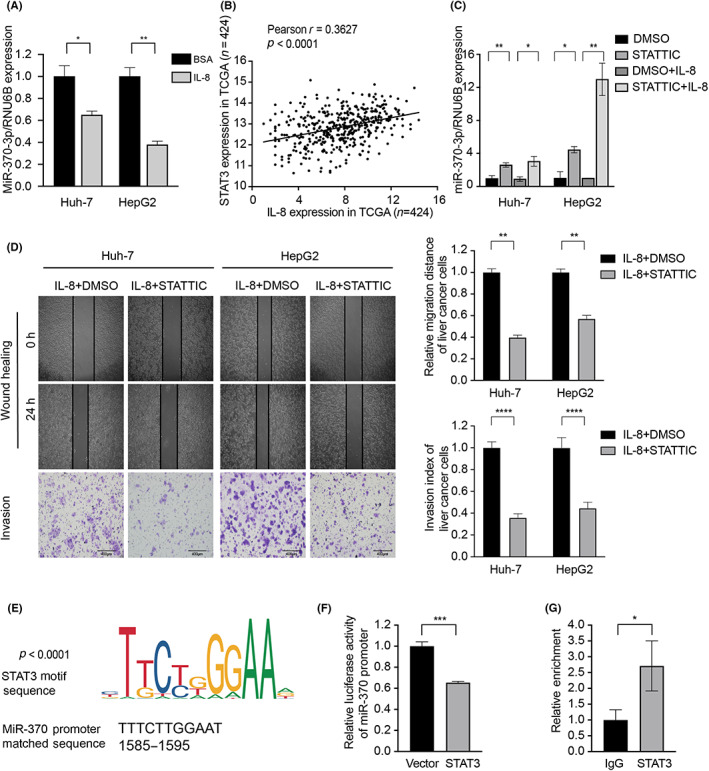
Interleukin‐8 (IL‐8) suppressed microRNA (miR)‐370‐3p expression by signal transducer and activator of transcription 3 (STAT3) occupying the miR‐370‐3p promoter. (A) Expression of miR‐370‐3p was dramatically decreased after IL‐8 treatment. (B) Spearman's correlation analysis indicated that the STAT3 level was positively correlated with IL‐8 level in The Cancer Genome Atlas database. (D) miR‐370‐3p levels were rescued by Stattic after IL‐8 treatment. (D) Stattic attenuated the promoting effects exerted by IL‐8 on Huh‐7 and HepG2 cell migration and invasion. (E) Schematic illustration of miR‐370‐3p promoter with the indicated STAT3‐binding capability. (F) HEK293T cells were cotransfected with miR‐370‐3p‐luc reporter and STAT3 for 48 h and then subjected to luciferase activity assay. STAT3 suppressed miR‐370‐3p promoter activity. (G) Direct interaction between STAT3 and miR‐370‐3p promoter was confirmed by ChIP assays. *p < 0.05, **p < 0.01, ***p < 0.001,****p < 0.0001.
As a transcription factor, STAT3 frequently binds to the promoter of genes and mediates their epigenetic modification. 24 Some miRNAs interact with the JAK–STAT3 pathway to regulate inflammation and oncogenesis, which could be potentially applied as clinical inhibitors. 25 We hypothesized that STAT3 might display transcription‐modulatory properties between IL‐8 and miR‐370‐3p through chromatin topological modulation. We confirmed this hypothesis by investigating UCSC and MEME websites, which identified the potential STAT3 binding motif on miR‐370‐3p promoter (Figure 5E). A dual‐luciferase reporter assay verified that miR‐370‐3p promoter activity was significantly decreased upon STAT3 overexpression (Figure 5F). The direct interaction between STAT3 and miR‐370‐3p promoter was further confirmed by ChIP assays (Figure 5G). Taken together, these results showed that IL‐8 regulates miR‐370‐3p expression through STAT3 directly binding to miR‐370‐3p promoter.
3.6. Signal transducer and activator of transcription 3 recruits HDAC1 and inhibits miR‐370‐3p promoter activity
A previous study reported that IL‐6 reduced miRNA‐370 through DNA methylation manipulations in cholangiocarcinoma cells. 23 In the present study, 5‐aza‐2′‐deoxycytidine treatment did not alter miR‐370‐3p levels in liver cancer cells after IL‐8 incubation, indicating that DNA methylation was not attributable to IL‐8‐decreased miR‐370‐3p expression (Figure S4B). However, HDACs (e.g., HDAC7 and p300/HDAC3) repress STAT3 activation by interfering with STAT3 dimerization, DNA binding, and transcriptional activity. 26 We attempted to identify HDACs that might be recruited to miR‐370‐3p promoter by STAT3. Interestingly, among the HDAC family (HDAC1–HDAC11), STAT3 strongly bound with HDAC1, HDAC2, HDAC3, HDAC4, HDAC5, HDAC6, HDAC7, and HDAC11 (Figure 6A and S3). A complementary function assay showed that miR‐370‐3p expression was tightly related to HDAC1 and HDAC11 (Figure 6B). siHDAC1 significantly increased miR‐370‐3p promoter luciferase activity (Figure 6C). As observed with these results, nonspecific HDAC antagonists SAHA and chidamide resulted in upregulation of miR‐370‐3p expression (Figure 6D). Consistently, SAHA attenuated the promoting effects exerted by IL‐8 on migration and invasion (Figure 6E), indicating that HDAC antagonists could be a promising treatment for liver cancer. Collectively, these data showed that STAT3 interacts with HDAC1 to inhibit miR‐370‐3p promoter activity and repress miR‐370‐3p expression.
FIGURE 6.
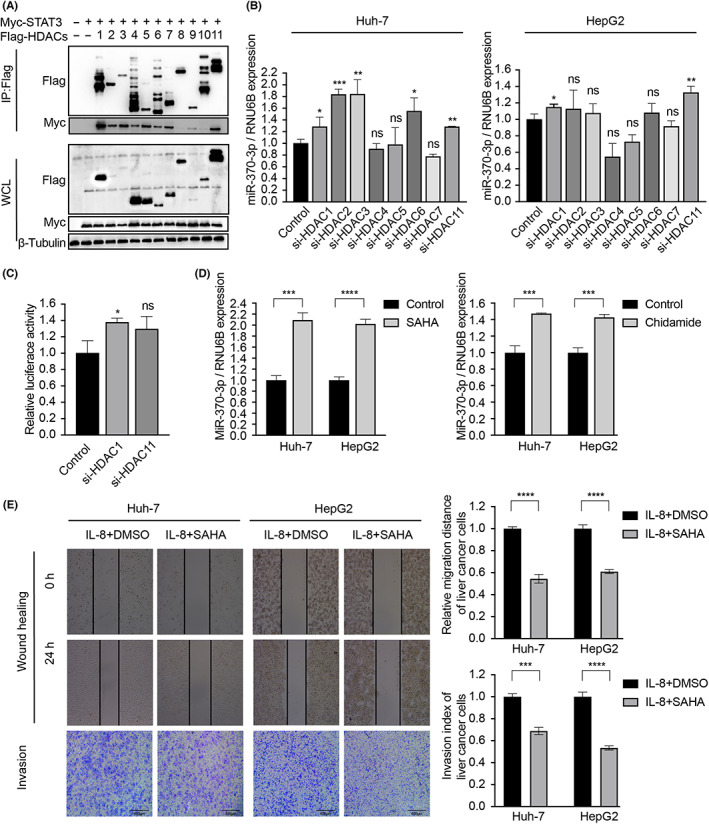
Signal transducer and activator of transcription 3 (STAT3) recruits histone deacetylase 1 (HDAC1) and inhibits microRNA (miR)‐370‐3p promoter activity (A) HEK293T cells were cotransfected with Myc‐STAT3 and Flag‐HDACs for 48 h, then lysed and analyzed by immunoprecipitation (IP). (B) After transfection of siHDACs, the expression of miR‐370‐3p were examined by quantitative RT‐PCR. (C) HEK293T cells were cotransfected with miR‐370‐3p‐Luc reporter and siHDACs for 48 h, and the luciferase activity assay showed that siHDAC1 significantly increased miR‐370‐3p promoter luciferase activity. (D) HDAC inhibitors (SAHA and Chidamide) increased miR‐370‐3p expression. (E) SAHA inhibited Huh‐7 and HepG2 cell migration and invasion. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. ns, not significant.
3.7. Interleukin‐8 and miR‐370‐3p levels correlate with prognosis of HCC patients
All the above results in vitro and in vivo verified that miR‐370‐3p plays critical roles in the IL‐8‐dependent EMT regulatory circuit, and we consequently explored the clinical value in patients. Transcatheter arterial chemoembolization, chemotherapy, and TKIs are the main treatments for liver cancer, but TACE trauma aggravates hypoxia and inflammatory reactions. It has been reported that chemotherapy and radiotherapy also increase inflammation. 27 We measured serum IL‐8 levels in 17 untreated HCC patients, 9 of whom subsequently underwent TACE. We found that IL‐8 levels increased after TACE treatment (Figure 7A). The level of IL‐8 also increased in the culture supernatant of HepG2 treated with several common chemotherapeutic drugs, such as Doxorubicin and Oxaliplatin, but TKI Sorafenib had no significant effect (Figure 7B).
FIGURE 7.
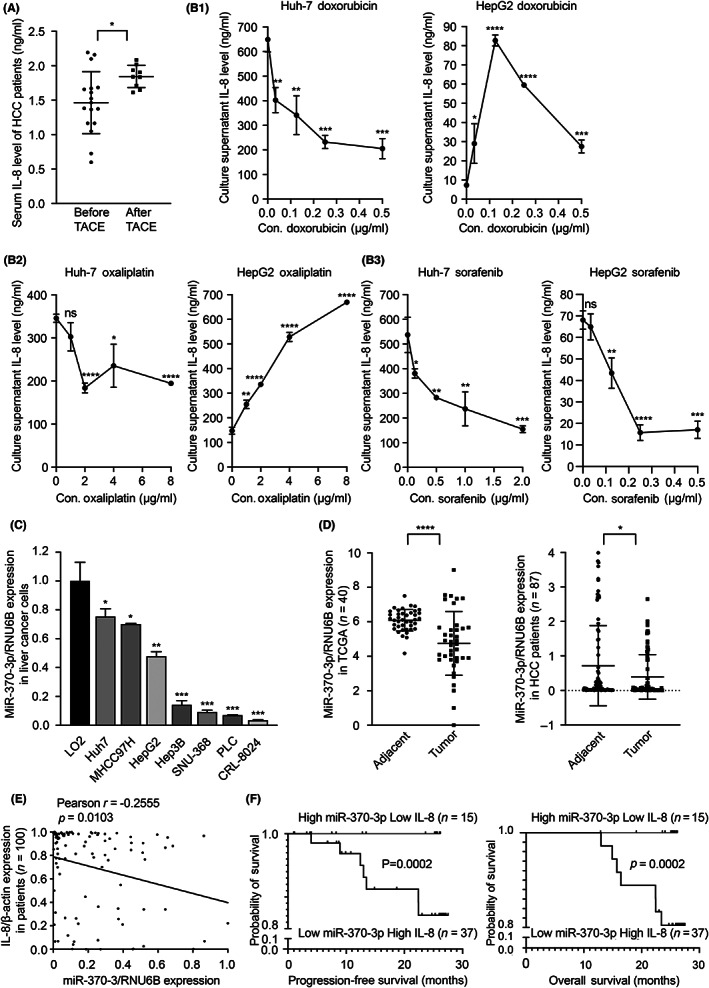
Interleukin‐8 (IL‐8) and microRNA (miR)‐370‐3p levels correlate with hepatocellular carcinoma (HCC) patient prognosis. (A) Serum IL‐8 levels of HCC patients before and after transcatheter arterial chemoembolization (TACE) were tested by ELISA. (B) IL‐8 concentration in the culture supernatant of HCC cells treated with doxorubicin, oxaliplatin, and sorafenib. (C) miR‐370‐3p levels in seven different genetic HCC cell lines were significantly lower than nontransformed hepatic cell line LO2. (D) miR‐370‐3p levels were lower in cancer in both The Cancer Genome Atlas (TCGA) database and clinical samples. (E) Spearman's correlation analysis indicated that miR‐370‐3p expression was inversely correlated with IL‐8 mRNA levels in HCC patients. (F) Comparison of progression‐free survival and overall survival between different expression levels of miR‐370‐3p and IL‐8 in HCC patients. Actuarial probabilities were calculated using the Kaplan–Meier method and compared using the log–rank test. HCC patients with miR‐370‐3phigh and IL‐8low showed much better PFS and OS. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. ns, not significant.
As miR‐370‐3p targets Snail and Twist1, it is rational to suppose that miR‐370‐3p plays an important modulatory effect on multiple signaling pathways during EMT. In fact, miR‐370‐3p was significantly downregulated in liver cancer cell lines Huh‐7, MHCC97H, HepG2, Hep3B, SNU‐368, PLC, and CRL‐8024 relative to the nontransformed hepatic LO2 cell line (Figure 7C). Similarly, the expression of miR‐370‐3p in HCC tumor tissues was decreased compared to adjacent nontumor tissues in both TCGA and clinical samples (Figure 7D). In addition, patients' miR‐370‐3p levels were inversely correlated with IL‐8 mRNA levels (Pearson r = −0.2555, p = 0.0103; Figure 7E). Hepatocellular carcinoma patients with high miR‐370‐3p and low IL‐8 showed much better PFS and OS than those with low miR‐370‐3p and high IL‐8 (PFS: 23.63 months vs. 1.1 months, respectively, p = 0.0002; OS: 25.45 months vs. 12.9 months, respectively, p = 0.0002) (Figure 7F), indicating that miR‐370‐3p and IL‐8 are valuable prognostic predictive factors.
4. DISCUSSION
Tumor cells undergoing EMT have increased migratory and invasive capabilities that induce HCC metastasis and poor prognosis. Epithelial–mesenchymal transition requires the reprogramming of the epithelial phenotype, which predominantly involves the activation of EMT‐TFs. 3 The expression of E‐cadherin is mainly controlled by three families of transcription factors: Snail and Slug, ZEB1 and ZEB2, and Twist1 and Twist2. 28 Inflammatory mediators induce EMT and promote metastasis through upregulating EMT‐TF expression. 8 Interleukin‐8, a pro‐inflammatory cytokine, is upregulated in cancerous tissues and is associated with tumor differentiation, proliferation, migration, invasion, and metastasis. 10 Continuously high expression of IL‐8 stimulates downstream signaling through the PI3K/Akt, 29 Erk–MAPK, 30 p38 MAPK, 31 and PKC 32 pathways and consequently promotes tumor progression and dissemination. We have previously shown that IL‐8 induces EMT through upregulation of Snail and Twist1. 13 We would like to further explore which endogenous factor is involved in IL‐8‐induced EMT and implicates clinical treatment.
Accumulating evidence indicated that miRNAs are important components involved in posttranscriptional regulation of EMT. Members of the miR‐200 family (miR‐200a, miR‐200b, miR‐200c, miR‐141, and miR‐429) have emerged as important regulators of EMT, in part by targeting ZEB1 and ZEB2. 28 The miR‐7/IGF1R/Snail axis increases E‐cadherin expression and partially reverses EMT in gastric cancer. 33 However, it has not been reported whether miRNAs regulate multiple EMT‐TFs simultaneously.
According to the library screening results, we undertook comprehensive studies to verify that miR‐370‐3p modulates Snail as well as Twist1 directly and specifically. Although miR‐370‐3p suppressed the expression of Snail and Twist1 by approximately 20% and 25%, respectively, they significantly reduced liver cancer cell invasion and metastasis by approximately 60%, due to the synergistic inhibition effects on two targets. Epithelial–mesenchymal transition‐TFs play pleiotropic roles in cancer progression and are involved in cellular processes both in classical EMT (the loss of cell‐to‐cell adhesion) and other cellular processes (cell apoptosis, cell proliferation, stemness, and immunosuppression). 1 However, the synergistic effect of other target genes of miR‐370‐3p might also contribute to inhibition of invasion. In fact, miR‐370‐3p acts as a tumor suppressor in different types of cancer by targeting multiple protumorigenic genes. 34 , 35 For example, miRNA‐370 suppresses the progression and proliferation of human astrocytoma and glioblastoma by negatively regulating β‐catenin and causing activation of FOXO3a. 36 Upregulation of miRNA‐370 promotes cell apoptosis and inhibits proliferation by targeting PTEN in human gastric cancer. 37 Other indirect molecular mechanisms underlying miR‐370‐3p antitumor activities have also been reported, such as regulating phosphatidylinositol, MAPK, and B‐cell receptor signaling. 38 MicroRNA‐370‐3p alleviated ulcerative colitis‐associated colorectal cancer by inhibiting the inflammatory response and EMT. 39 The androgen receptor‐mediated ADAR2/circFNTA/miR‐370‐3p/FNTA pathway activates KRAS signaling to alter bladder cancer cell invasion and chemosensitivity. 40
In addition, several interactions among miRNAs, EMT inducers, and EMT‐TFs facilitate cancer progression. Tight interconnected and complex regulatory loops between miRNAs and EMT‐TFs control epithelial cell plasticity, providing self‐enhancing signals and robustness to maintain epithelial or mesenchymal status. 41 The ZEB/miR‐200 and Snail/miR‐34 networks are significant feedback loops during EMT. 42 , 43 Twist1 induces the production of IL‐8, which activates MMPs and promotes invasion of breast epithelial and cancer cells. 44 Additionally, miRNAs modulate genes related to inflammation; for example, the CXCL12/miR‐370‐3p/HMGA2 pathway is involved in tumor growth and invasiveness of nonfunctional pituitary adenomas. 45 Reduced Let‐7 directly activates the IL‐6 cascade, which involves STAT3, increasing nuclear factor‐κB transcriptional activity and transformation from normal to cancer. 46
A previous study showed that hypermethylation of CpG was involved in IL‐6‐reduced miR‐370 suppression. 23 In the present study, we reported a novel mechanism by which IL‐8 downregulated miR‐370‐3p expression through the STAT3 pathway and HDAC recruitment. Many HDAC inhibitors have already shown remarkable antitumor activities in the preclinical setting. 47 Our results confirmed that SAHA and chidamide significantly reduced the migration and invasion of cancer cells, which implicates potential therapy for HCC. We noticed that many ongoing clinical trials are exploring the therapeutic value of HDAC inhibitors in HCC (NCT01075113 and NCT00321594). From a new perspective, our study showed that miR‐370‐3p inhibits metastasis by regulating EMT‐TFs both Snail and Twist1; we also elucidated that the IL‐8/STAT3/miR‐370‐3p pathway relies on HDAC1 recruitment, which characterizes a crucial mechanism underlying the metastasis of HCC. This has opened up a new avenue for further pharmacological research and suggests that HDAC inhibitors might be an adjuvant therapy to prevent HCC metastasis.
Nevertheless, our study is limited and cannot explain the full mechanism by which IL‐8 promotes or miRNA‐370‐3p inhibits HCC invasion and metastasis. First, other EMT‐TFs might also be involved in the process of IL‐8‐induced HCC invasion. Second, IL‐8 can active many signal pathways. such as the PI3K/Akt pathway, to exert protumor effects. In addition to inducing EMT, IL‐8 promotes tumor metastasis as well as inducing angiogenesis or recruiting larger numbers of immunosuppressive cells into the tumor microenvironment. Third, miR‐370‐3p could modulate other targeting genes to attenuate the protumor functions of IL‐8. Finally, other miRNAs that regulate multiple EMT‐TFs also possibly play important roles underlying IL‐8‐induced HCC invasion and metastasis. What we have reported so far is only one of the potential mechanisms by which IL‐8 promotes the metastasis of HCC and miRNA‐370‐3p interference, and more possible mechanisms need to be explored deeply.
FUNDING INFORMATION
This work was supported by National Natural Science Foundation of China (grant no. 81872348, 81,971,773, 82,073,209), Guangdong Basic and Applied Basic Research Foundation (grant no. 2020A1515010808, 2019A1515011356, 2019A030317023). The Fundamental Research Funds for the Central Universities, Sun Yat‐sen University (grant no. 19ykzd08).
DISCLOSURE
The authors have no conflicts of interest to declare.
ETHICS STATEMENT
Approval of the research protocol: The study was approved by the research medical ethics committee of Sun Yat‐sen University.
Informed consent: Informed consent was obtained from all patients.
Registry and registration no. of the study/trial: N/A.
Animal studies: All experimental procedures involving animals were approved by the Institutional Animal Care and Use Committee (IACUC) of the Fifth Affiliated Hospital of Sun Yat‐sen University.
Supporting information
Figure S1
Figure S2
Figure S3
Figure S4
Figure S5
Appendix S1
Table S1
Table S2
ACKNOWLEDGMENTS
The Flag‐HDAC constructs were generously provided by Professor Tiebang Kang (Sun Yat‐sen University Cancer Center). We thank Professor Rongping Guo (Sun Yat‐sen University Cancer Center) for providing MHCC97H, Hep3B, SNU‐368, PLC and CRL‐8024 cell lines.
Peng S, Chen Y, Li T, et al. Hsa‐microRNA‐370‐3p targeting Snail and Twist1 suppresses IL‐8/STAT3‐driven hepatocellular carcinoma metastasis. Cancer Sci. 2022;113:4120‐4134. doi: 10.1111/cas.15571
Siqi Peng, Yutong Chen, and Ting Li contributed equally to this work.
Contributor Information
Jian Li, Email: lijian5@mail.sysu.edu.cn.
Linjuan Zeng, Email: zenglinj@mail.sysu.edu.cn.
REFERENCES
- 1. Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391:1301‐1314. [DOI] [PubMed] [Google Scholar]
- 2. Yang JD, Hainaut P, Gores GJ, Amadou A, Plymoth A, Roberts LR. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16:589‐604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Giannelli G, Koudelkova P, Dituri F, Mikulits W. Role of epithelial to mesenchymal transition in hepatocellular carcinoma. J Hepatol. 2016;65:798‐808. [DOI] [PubMed] [Google Scholar]
- 4. Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial‐mesenchymal transitions in development and disease. Cell. 2009;139:871‐890. [DOI] [PubMed] [Google Scholar]
- 5. Perl AK, Wilgenbus P, Dahl U, Semb H, Christofori G. A causal role for E‐cadherin in the transition from adenoma to carcinoma. Nature. 1998;392:190‐193. [DOI] [PubMed] [Google Scholar]
- 6. Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415‐428. [DOI] [PubMed] [Google Scholar]
- 7. Yang J, Weinberg RA. Epithelial‐mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell. 2008;14:818‐829. [DOI] [PubMed] [Google Scholar]
- 8. Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860‐867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sun B, Karin M. Obesity, inflammation, and liver cancer. J Hepatol. 2012;56:704‐713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Itoh Y, Joh T, Tanida S, et al. IL‐8 promotes cell proliferation and migration through metalloproteinase‐cleavage proHB‐EGF in human colon carcinoma cells. Cytokine. 2005;29:275‐282. [DOI] [PubMed] [Google Scholar]
- 11. Liu Y, Zhang Y, Wang S, et al. Prospero‐related homeobox 1 drives angiogenesis of hepatocellular carcinoma through selectively activating interleukin‐8 expression. Hepatology. 2017;66:1894‐1909. [DOI] [PubMed] [Google Scholar]
- 12. Schalper KA, Carleton M, Zhou M, et al. Elevated serum interleukin‐8 is associated with enhanced intratumor neutrophils and reduced clinical benefit of immune‐checkpoint inhibitors. Nat Med. 2020;26:688‐692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Peng S, Chen Y, Gong Y, et al. Predictive value of intratumour inflammatory cytokine mRNA levels of hepatocellular carcinoma patients and activation of two distinct pathways govern IL‐8 induced epithelial‐mesenchymal transition in human hepatic cancer cell lines. Cytokine. 2019;119:81‐89. [DOI] [PubMed] [Google Scholar]
- 14. Gupta RA, Shah N, Wang KC, et al. Long non‐coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071‐1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post‐transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102‐114. [DOI] [PubMed] [Google Scholar]
- 16. Chen CZ. MicroRNAs as oncogenes and tumor suppressors. N Engl J Med. 2005;353:1768‐1771. [DOI] [PubMed] [Google Scholar]
- 17. Kim T, Veronese A, Pichiorri F, et al. p53 regulates epithelial‐mesenchymal transition through microRNAs targeting ZEB1 and ZEB2. J Exp Med. 2011;208:875‐883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Amado T, Schmolka N, Metwally H, Silva‐Santos B, Gomes AQ. Cross‐regulation between cytokine and microRNA pathways in T cells. Eur J Immunol. 2015;45:1584‐1595. [DOI] [PubMed] [Google Scholar]
- 19. Zeng L, Chen Y, Wang Y, et al. MicroRNA hsa‐miR‐370‐3p suppresses the expression and induction of CYP2D6 by facilitating mRNA degradation. Biochem Pharmacol. 2017;140:139‐149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Waugh DJ, Wilson C. The interleukin‐8 pathway in cancer. Clin Cancer Res. 2008;14:6735‐6741. [DOI] [PubMed] [Google Scholar]
- 21. Hydbring P, Wang Y, Fassl A, et al. Cell‐cycle‐targeting microRNAs as therapeutic tools against refractory cancers. Cancer Cell. 2017;31(4):576‐590. e578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lin CC, Law BF, Hettick JM. MicroRNA‐mediated calcineurin signaling activation induces CCL2, CCL3, CCL5, IL8, and chemotactic activities in 4,4′‐methylene diphenyl diisocyanate exposed macrophages. Xenobiotica. 2021;51:1436‐1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Meng F, Wehbe‐Janek H, Henson R, Smith H, Patel T. Epigenetic regulation of microRNA‐370 by interleukin‐6 in malignant human cholangiocytes. Oncogene. 2008;27:378‐386. [DOI] [PubMed] [Google Scholar]
- 24. Aaronson DS, Horvath CM. A road map for those who don't know JAK‐STAT. Science. 2002;296:1653‐1655. [DOI] [PubMed] [Google Scholar]
- 25. Yu H, Lee H, Herrmann A, Buettner R, Jove R. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat Rev Cancer. 2014;14:736‐746. [DOI] [PubMed] [Google Scholar]
- 26. Yuan ZL, Guan YJ, Chatterjee D, Chin YE. Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science. 2005;307:269‐273. [DOI] [PubMed] [Google Scholar]
- 27. Dillon MT, Bergerhoff KF, Pedersen M, et al. ATR Inhibition Potentiates the Radiation‐induced Inflammatory Tumor Microenvironment. Clin Cancer Res. 2019;25:3392‐3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Guo F, Parker Kerrigan BC, Yang D, et al. Post‐transcriptional regulatory network of epithelial‐to‐mesenchymal and mesenchymal‐to‐epithelial transitions. J Hematol Oncol. 2014;7:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shao N, Lu Z, Zhang Y, et al. Interleukin‐8 upregulates integrin beta3 expression and promotes estrogen receptor‐negative breast cancer cell invasion by activating the PI3K/Akt/NF‐kappaB pathway. Cancer Lett. 2015;364:165‐172. [DOI] [PubMed] [Google Scholar]
- 30. Venkatakrishnan G, Salgia R, Groopman JE. Chemokine receptors CXCR‐1/2 activate mitogen‐activated protein kinase via the epidermal growth factor receptor in ovarian cancer cells. J Biol Chem. 2000;275:6868‐6875. [DOI] [PubMed] [Google Scholar]
- 31. Murphy C, McGurk M, Pettigrew J, et al. Nonapical and cytoplasmic expression of interleukin‐8, CXCR1, and CXCR2 correlates with cell proliferation and microvessel density in prostate cancer. Clin Cancer Res. 2005;11:4117‐4127. [DOI] [PubMed] [Google Scholar]
- 32. Lang K, Niggemann B, Zanker KS, Entschladen F. Signal processing in migrating T24 human bladder carcinoma cells: role of the autocrine interleukin‐8 loop. Int J Cancer. 2002;99:673‐680. [DOI] [PubMed] [Google Scholar]
- 33. Zhao X, Dou W, He L, et al. MicroRNA‐7 functions as an anti‐metastatic microRNA in gastric cancer by targeting insulin‐like growth factor‐1 receptor. Oncogene. 2013;32:1363‐1372. [DOI] [PubMed] [Google Scholar]
- 34. Chen J, Liu G, Wu Y, et al. CircMYO10 promotes osteosarcoma progression by regulating miR‐370‐3p/RUVBL1 axis to enhance the transcriptional activity of beta‐catenin/LEF1 complex via effects on chromatin remodeling. Mol Cancer. 2019;18:150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wei CY, Zhu MX, Lu NH, et al. Circular RNA circ_0020710 drives tumor progression and immune evasion by regulating the miR‐370‐3p/CXCL12 axis in melanoma. Mol Cancer. 2020;19:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lu M, Wang Y, Zhou S, et al. MicroRNA‐370 suppresses the progression and proliferation of human astrocytoma and glioblastoma by negatively regulating β‐catenin and causing activation of FOXO3a. Exp Ther Med. 2018;15:1093‐1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zeng Y, Fu M, Wu GW, et al. Upregulation of microRNA‐370 promotes cell apoptosis and inhibits proliferation by targeting PTEN in human gastric cancer. Int J Oncol. 2016;49:1589‐1599. [DOI] [PubMed] [Google Scholar]
- 38. Leivonen SK, Icay K, Jantti K, et al. MicroRNAs regulate key cell survival pathways and mediate chemosensitivity during progression of diffuse large B‐cell lymphoma. Blood Cancer J. 2017;7:654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lin L, Wang D, Qu S, Zhao H, Lin Y. miR‐370‐3p alleviates ulcerative colitis‐related colorectal cancer in mice through inhibiting the inflammatory response and epithelial‐mesenchymal transition. Drug Des Devel Ther. 2020;14:1127‐1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chen J, Sun Y, Ou Z, et al. Androgen receptor‐regulated circFNTA activates KRAS signaling to promote bladder cancer invasion. EMBO Rep. 2020;21:e48467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Diaz‐Lopez A, Moreno‐Bueno G, Cano A. Role of microRNA in epithelial to mesenchymal transition and metastasis and clinical perspectives. Cancer Manag Res. 2014;6:205‐216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Park SM, Gaur AB, Lengyel E, Peter ME. The miR‐200 family determines the epithelial phenotype of cancer cells by targeting the E‐cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894‐907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Siemens H, Jackstadt R, Hunten S, et al. miR‐34 and SNAIL form a double‐negative feedback loop to regulate epithelial‐mesenchymal transitions. Cell Cycle. 2011;10:4256‐4271. [DOI] [PubMed] [Google Scholar]
- 44. Li S, Kendall SE, Raices R, et al. TWIST1 associates with NF‐kappaB subunit RELA via carboxyl‐terminal WR domain to promote cell autonomous invasion through IL8 production. BMC Biol. 2012;10:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cai F, Dai C, Chen S, et al. CXCL12‐regulated miR‐370‐3p functions as a tumor suppressor gene by targeting HMGA2 in nonfunctional pituitary adenomas. Mol Cell Endocrinol. 2019;488:25‐35. [DOI] [PubMed] [Google Scholar]
- 46. Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF‐kappaB, Lin28, Let‐7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139:693‐706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liva SG, Tseng YC, Dauki AM, et al. Overcoming resistance to anabolic SARM therapy in experimental cancer cachexia with an HDAC inhibitor. EMBO Mol Med. 2020;12:e9910. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Figure S2
Figure S3
Figure S4
Figure S5
Appendix S1
Table S1
Table S2


