Abstract
Lipid nanoparticles (LNPs) have been commonly used as a vehicle for nucleic acids, such as small interfering RNA (siRNA); the surface modification of LNPs is one of the determinants of their delivery efficiency especially in systemic administration. However, the applications of siRNA‐encapsulated LNPs are limited due to a lack effective systems to deliver to solid tumors. Here, we report a smart surface modification using a charge‐switchable ethylenediamine‐based polycarboxybetaine for enhancing tumor accumulation via interaction with anionic tumorous tissue constituents due to selective switching to cationic charge in response to cancerous acidic pH. Our polycarboxybetaine‐modified LNP could enhance cellular uptake in cancerous pH, resulting in facilitated endosomal escape and gene knockdown efficiency. After systemic administration, the polycarboxybetaine‐modified LNP accomplished high tumor accumulation in SKOV3‐luc and CT 26 subcutaneous tumor models. The siPLK‐1‐encapsulated LNP thereby accomplished significant tumor growth inhibition. This study demonstrates a promising potential of the pH‐responsive polycarboxybetaine as a material for modifying the surface of LNPs for efficient nucleic acid delivery.
Keywords: drug delivery systems, lipid nanoparticle, pH responsiveness, polyzwitterion, siRNA
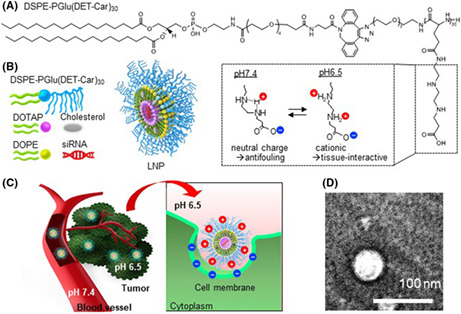
Schematic illustration of DSPE‐PGlu(DET‐Car)30 and siRNA‐encapsulated PGlu(DET‐Car)30 LNPs
1. INTRODUCTION
Small interfering RNA (siRNA) can be applied for the treatment of countless diseases, including cancer, by downregulating gene expression. 1 , 2 , 3 However, the innate physiochemical properties of siRNA molecules, such as their susceptibility to degradation by serum nucleases, creates an imposing barrier for systemic delivery. 4 Among the various siRNA carriers developed in the past decade, lipid nanoparticles (LNPs) continue to garner significant attention for their biocompatibility, low toxicity, and ability to encapsulate siRNA at high efficiencies. 3 A prominent formulation strategy among lipid nanoparticle‐based siRNA carriers consists of a pH‐responsive (ionizable tertiary amine) lipid together with polyethylene glycol (PEG) lipid for enhanced nucleic acid delivery. For instance, Patisiran (Onpattro®), whose United States Food and Drug Administration (US FDA) approval in 2018 represents a milestone in siRNA therapeutics, was the first FDA‐approved siRNA‐LNP system. 1 , 2 So far it is the only siRNA‐based medicine to receive FDA approval, despite intense efforts by researchers for over two decades, Patisiran has ignited optimism regarding the potential of LNPs as siRNA delivery conduits, forming a blueprint for the future design of successful siRNA medicines.
siRNA is a promising technology for cancer therapy. However, the applications of siRNA are limited due to a lack of effective systems to deliver to target organs and tissues other than the liver. The ability to pinpoint tumor targeting, together with efficient nucleic acid transfection, is a prerequisite for a successful LNP system. Tumor‐specific conditions can be used to enhance tumor targeting. The extracellular acidic pH of the tumor microenvironment is a prevalent characteristic that results from hypoxia, as well as rapid glycolysis and lactate production. 5 , 6 , 7 , 8 One common strategy to facilitate passive tumor targeting via the enhanced permeability and retention (EPR) effect is the use of pH‐sensitive ionizable lipids that can be designed to exploit the differences in pH observed between physiological and tumorous pH3. Similarly, ionizable lipids with a pK a <6.5 such as DLin‐MC3‐DMA are regularly utilized to improve endosomal escape via head group protonation within the acidic environment of endosomes. 3 , 9
Envisaging a molecule simultaneously possessing antifouling and pH‐responsive tumor targeting functionality and that could be ornamented upon various nanocarriers, we previously developed an ethylenediamine‐based polycarboxybetaine zwitterion [PGlu(DET‐Car)] that demonstrated antifouling behavior, as well as significantly enhanced tumor accumulation by means of its fine‐tuned pH‐regulated switch to net cationic, occurring selectively within the acidic tumor tissue. 10 We confirmed the two distinct pK a values of the ethylenediamine group in the PGlu(DET‐Car) side chains to be ~6.3 and 9.0. 10 The gauche ring system adopted by monoprotonated ethylenediamine and the, consequently, high thermodynamic cost was necessary to achieve the favored anticonformer of bisprotonated ethylenediamine. This accounts for the two orders of magnitude separation in pK a observed for the group, a protonation pattern atypical of alkyl diamines with two close pK a values. 11 , 12 , 13
Here, we corroborated these pK a properties in LNP, in which they were determined to be near neutral at physiological pH and switch to cationic at cancerous pH (6.5). We sought to adapt this system to create a cancerous pH‐responsive stealth LNP device by means of a head group polymer modification of 1,2‐distearoyl‐sn‐glycero‐3‐phosphorylethanolamine (DSPE). The LNP surface charge was designed to be near net neutral in physiological pH, and switch to net cationic via bisprotonation of the polycarboxybetaine ethylenediamine moiety in the reduced pH microenvironment commonly found among solid tumors. This is, to our knowledge, the first instance of an antifouling ethylenediamine‐based polycarboxybetaine‐modified LNP possessing cancerous pH responsivity, demonstrating successful nucleic acid delivery in vitro and in vivo. This system constitutes a promising LNP surface modification strategy for use in tumor targeting and nucleic acid delivery.
2. MATERIALS AND METHODS
2.1. Materials
Information regarding materials, cell lines (human ovarian carcinoma cells expressing the firefly luciferase gene [SKOV3‐luc], murine colon cancer cells [CT26]) and animals is described in Appendix S1. All animal experiments were approved by the Animal Care and Use Committee and performed in accordance with the Guidelines for the Care and Use of Laboratory Animals set forth by the Tokyo Institute of Technology.
2.2. Preparation of DSPE‐PGlu(DET‐Car)30
PGlu(DET‐Car)30 was synthesized according to our previous report. 10 Detailed procedures for DSPE‐PGlu(DET‐Car)30 synthesis and characterization are described in Appendix S1.
2.3. Preparation of siRNA‐encapsulated PGlu(DET‐Car)30 LNPs
PGlu(DET‐Car)30 LNPs were prepared according to the t‐BuOH dilution procedure used by Sato et al. 14 Briefly, DOPE (5 mM)/cholesterol (5 mM)/1,2‐dioleoyl‐3‐trimethylammonium‐propane (DOTAP) (5 mM) in varying compositions were dissolved in 200 μl of 90% t‐BuOH solution. Separately, aqueous solutions of DSPE‐PGlu(DET‐Car)30 were mixed with 20 μl 1 mg/ml siRNA buffered in pH 7.4 4‐(2‐hydroxyethyl)‐1‐piperazineethanesulfonic acid (HEPES), and the contents thoroughly mixed. The mixtures were then added into 2 ml pH 7.4 HEPES buffer under vigorous stirring to facilitate the formation of LNPs. Residual t‐BuOH was removed by consecutive dilution with D‐PBS (−) followed by ultrafiltration (Amicon® Ultra‐15 Centrifugal Filter Unit‐50 K, Merck Millipore, Burlington, MA, USA). Control LNP were prepared following the methods outlined here using DSPE‐PEG5k (20 μl, 1 mM) instead of DSPE‐PGlu(DET‐Car)30 and having the similar particle size without pH‐responsive ability.
2.4. Characterization of LNPs
Hydrodynamic size, polydispersity index (PDI), and ζ‐potential of LNPs were determined using Zetasizer Nano‐ZS90 (Malvern, UK). ζ‐Potential of LNPs was measured in 2‐(N‐morpholino)ethanesulfonic acid (MES) buffer (pH 6.5), and HEPES buffer (pH 7.4). Stability of LNPs in serum was evaluated with 50% FBS and incubation for 24 or 48 h at 37°C (5% CO2, 95% humidity). The morphology of LNP was observed using a transmission electron microscope (TEM; JEM‐1400, JEOL, Tokyo, Japan) at the accelerated voltage of 100 V. For TEM sample preparation, LNPs were placed on a copper grid, and the samples were stained with gadolinium acetate and then washed with deionized (DI) water.
2.5. Encapsulation efficiency (EE%)
siRNA EE was determined using the RiboGreen assay according to the manufacturer's instructions. For calculation of total siRNA, Quant‐iT™ RiboGreen® RNA reagent (Thermo Fisher Scientific, Waltham, MA, USA) was diluted in HEPES buffer (pH 7.4, 10 mM) containing 0.4% (v/v) Triton X‐100 and 80 μg/ml dextran sulfate (Merck Millipore, Burlington, MA, USA), and subsequently mixed with the LNP solution. For free siRNA, Quant‐iT™ RiboGreen® RNA reagent was diluted with HEPES buffer (pH 7.4, 10 mM), and mixed with the LNP solution. The fluorescence of total and free siRNA was measured using a microplate reader (SPARK TKS01, TECAN, Zürich, Switzerland) (λex/λem = 480/525 nm). Separate calibration curves were prepared for each solution to consider the effects of Triton X‐100 and dextran sulfate on fluorescence intensities. EE % was determined using the following equation:
2.6. Cellular uptake
CT26 and SKOV3‐luc cells were seeded into a 24‐well plate at a density of 5 × 104 cells/well. DMEM containing 10% FBS and 1% penicillin at pH 6.5 was prepared by the addition of diluted HCl, followed by filtration through a Millex‐GV Filter, 0.22 μM (Merck Millipore, Burlington, MA, USA). After 24 h incubation, the culture medium was replaced with fresh medium (pH 7.4 and 6.5) containing Alexa647‐siGL3‐encapsulated LNPs at a final siRNA concentration of 100 nM, followed by additional 2 and 6 h incubations. Subsequently, cells were washed twice using D‐PBS(−), detached with 150 μl of trypsin/EDTA (Merck Millipore, Burlington, MA, USA) and suspended in 350 μl of the medium. The fluorescence intensity of cells was measured by flow cytometry (FCM) (Guava® easyCyte™, Malvern, UK) (λex/λem = 642/661 nm). In total, 3000 cells were acquired for each sample.
2.7. Cytotoxicity
SKOV3‐luc cells were seeded into 96‐well plates at a density of 2500 cells/well with DMEM containing 10% FBS and 1% penicillin. After 24 h incubation, cells were treated with fresh medium (pH 6.5 or 7.4) containing siGL3‐loaded LNPs at a final siRNA concentration of 100 nM. After 24 and 48 h incubations, cell viability was evaluated using the Cell Counting Kit‐8 (Dojindo, Kumamoto, Japan). Absorbance at 450 nm was measured using an iMark™ Microplate Absorbance Reader (Bio‐Rad Laboratories, Inc., Hercules, CA, USA). Cell viability was determined using the following equation:
where [A]test, [A]control, and [A]blank are the absorbance values of the wells belonging to treated cells, nontreated cells (pH 7.4), and empty medium, respectively.
2.8. Endosomal escape assessment via subcellular distribution
CT26 cells at a density of 1 × 105 cells/dish and SKOV3‐luc cells at a density of 5 × 105 cells/dish were seeded into 35‐mm glass dishes with DMEM containing 10% FBS and 1% penicillin. After a 24 h incubation, the cells were treated with fresh medium (pH 6.5) containing Alexa647‐siGL3‐encapsulated LNPs at a final siRNA concentration of 100 nM, followed by additional 1 and 8 h incubations. The cells were subsequently washed twice using D‐PBS(−). Cells were stained with Lysotracker Red DND‐99 (50 nM) for 30 min and with Hoechst33342 (10 μg/ml) for 5 min. Cells were washed with D‐PBS(−) twice after each staining step, and were then observed using a confocal laser scanning microscope (CLSM; LSM710, Carl Zeiss AG, Oberkochen, Germany) at excitation wavelengths 405 nm for Hoechst 33342, 561 nm for Lysotracker Red DND‐99, and 633 nm for Alexa‐647 siRNA‐loaded LNPs. Quantification analysis of the co‐localization ratio was performed using zeiss zen software (ver. 3.3) for LSM710 with 15 cells for SKOV3‐luc and 23 cells for CT26.
2.9. Endosomal escape assessment via calecin release
SKOV3‐luc and CT26 cells were seeded overnight at a density of 5000 cells/well in DMEM containing 10% FBS and 1% penicillin in Lab‐Tek® Chambered #1.0 Borosilicate Coverglass wells (Nunc, Rochester, NY). Cells were washed with D‐PBS(−), and medium was replaced with fresh medium (pH 6.5) containing 250 μM calcein (Dojindo, Kumamoto, Japan) and siGL3‐encapsulated LNPs at an siRNA dose of 100 nM, followed by a 2 h incubation. Endosomal escape of LNPs was observed in DMEM using a CLSM (LSM710, Carl Zeiss AG, Oberkochen, Germany) at excitation wavelengths 405 nm for Hoechst 33342 and 488 nm for calcein.
2.10. In vitro gene silencing via luciferase assay
SKOV3‐luc cells were seeded into a 24‐well plate at density of 5 × 104 cells/well in DMEM containing 10% FBS and 1% penicillin followed by a 24 h incubation. After washing twice with D‐PBS(−), the cells were treated with pH 6.5 medium containing siGL3‐encapsulated LNPs at a final siRNA concentration of 100 nM followed by a 6 h incubation. Subsequently, the medium was replaced by fresh DMEM containing 10% FBS and 1% penicillin, and the cells were incubated for 24 h. The cells were then washed with D‐PBS(−) and lysed in 200 μl/well of Passive Lysis Buffer (Promega Corporation, Madison, WI, USA) for 30 min, after which the supernatant (20 μl) was transferred to white plates (Sumitomo Bakelite Co., LTD), and finally subjected to treatment with luciferin (70 μl) (Luciferase Assay System, Promega Corporation, Madison, WI, USA). Relative luminescence units (RLU) were measured using a GloMax® 96 Microplate Luminometer (Promega Corporation). Gene silencing efficiency was calculated based on the reduction in RLU compared with nontreated (NT) cells.
2.11. Biodistribution study
Subcutaneous CT26 tumor models were prepared by subcutaneous inoculation of CT26 cells (5 × 105 cells/mouse) into the back or the right flank of BALB/c mice. When the tumor size reached ~200 mm3, Alexa‐647‐labeled siGL3‐encapsulated LNPs were intravenously injected into mice at a siRNA dose of 0.5 mg siRNA/kg. At 1, 6, and 24 h post‐administration, blood was obtained from the inferior vena cava and heparinized. The organs and tumors were collected and subsequently homogenized in Passive Lysis Buffer, followed by centrifugation. After demulsification by mixing the supernatant with 3% SDS in 60% t‐BuOH solution, the fluorescence intensity of Alexa‐647 in the samples was measured using a microplate reader (SPARK TKS01) (λex/λem = 630/690 nm). The tumor accumulation of Alexa‐647‐labeled siGL3‐encapsulated LNPs was also studied using subcutaneous SKOV3‐luc tumor models that were prepared by subcutaneous inoculation of SKOV3‐luc cells (5 × 106 cells per mouse) into BALB/c nude mice. When the tumor size reached ~200 mm3, LNPs were intravenously injected and the tumor accumulation of siRNA at 6 and 24 h post‐administration was evaluated in a similar manner as described previously.
2.12. In vivo gene silencing via luciferase activity
Subcutaneous SKOV3‐luc models were prepared in the same way with biodistribution. When tumor size reached ~100 mm3, the mice received an intraperitoneal (i.p.) injection of 6 mg VivoGlo™ Luciferin (Promega Corporation, Madison, WI, USA), and 13 min later, imaged under an in vivo imaging system (IVIS) (PerkinElmer, Waltham, MA, US) as the 0 h time point. After 24 h post‐i.v. injection of siGL3‐encapsulated LNP at siRNA dose of 0.6 mg/kg or D‐PBS(−), mice were i.p. injected with same dose of luciferin 13 min before imaging. Gene knockdown efficacy as determined as inhibition of radiant efficiency of each group in 24 h was compared with radiant efficiency in 0 h. Radiant efficiency measurement of the tumor luminescence was automatically assigned by the Live Image software used for image processing.
2.13. In vivo gene silencing via RT‐PCR
Subcutaneous SKOV3‐luc bearing mice (100 mm3 of tumor size) received an i.v. injection of siGL3‐encapsulated LNP at the siRNA dose of 0.5 mg/kg or D‐PBS(−). The sample‐treated mice were sacrificed 24 h post‐injection, and the tumor were taken and washed with D‐PBS(−). Next 30 mg of tumor tissue was homogenized and the RNA in the homogenization solution was isolated using the RNeasy Protect Mini Kit (Qiagen, Hilden, Germany), according to the manufacturer's instructions. DNase I (TaKaRa Bio Inc., Shiga, Japan) was used to remove DNA contamination, followed by reverse transcription (RT) of the total RNA (5 μg for each sample) using the PrimeScript RT reagent Kit (TaKaRa Bio Inc. Shiga, Japan) with an oligo(dT) primer. PCR was performed using the PIKOREAL 96 Real‐Time PCR System (Thermo Scientific™, Waltham, MA, USA). The reaction mixtures contained 2 μg of cDNA with appropriate primer pairs and the FastStart Universal SYBR Green Master (Merck Millipore, Burlington, MA, USA). GL3 levels were calculated using the comparative Ct method with GAPDH as the endogenous housekeeping gene. Gene expression was normalized to the value for nontreated cells. The following primer pairs were used: GL3: 5′‐TGAGTACTTCGAAATGTCCGTTC‐3′ (forward); 5′‐GTATTCAGCCCATATCGTTTCAT‐3′ (reverse); GAPDH: 5′‐CCTCTGACTTCAACAGCGAC‐3′ (forward); 5′‐GCCACATACCAGGAAATGAG‐3′ (reverse).
2.14. Tumor growth suppression effect
Tumor growth suppression effect was evaluated using a SKOV3‐luc tumor‐bearing model, prepared as described above. When tumors reached 25 mm3, mice were intravenously injected with siPLK1‐encapsulated LNPs at an siRNA dose of 2.5 mg/kg every other day. The tumor volume (V) was calculated using the following equation:
where a and b are the major and minor axes of the tumor, respectively.
2.15. Statistical analysis
Statistical analysis was conducted using Student's t‐test, Tukey's multiple comparisons test, and Sidak's multiple comparisons. A p‐value <0.05 was considered statistically significant.
3. RESULTS
3.1. Preparation and characterization of PGlu(DET‐Car)30 LNPs
To synthesize a polyzwitterion‐conjugated lipid composed of a hydrophobic DSPE moiety and hydrophilic PGlu(DET‐Car)30 segment “DSPE‐PGlu(DET‐Car)30” (Figure 1A and Figure S1), N3‐PGlu(DET‐Car)30 was reacted with DSPE‐PEG4‐DBCO via click chemistry. DSPE‐PEG4‐DBCO‐PGlu(DET‐Car)30 was used for the preparation of siRNA‐encapsulated PGlu(DET‐Car)30 LNPs (Figure 1B) at different lipid compositions (Table 1) using the t‐BuOH dilution procedure. 14 Characterization of LNPs was performed using dynamic laser scattering (DLS) (Tables 1 and 2). The size of PGlu(DET‐Car)30 LNPs decreased with increasing ratios of DSPE‐PGlu(DET‐Car)30, indicating that the ratio of PGlu(DET‐Car)30 may influence the spatial structure of the LNPs (Table 1). The optimized PGlu(DET‐Car)30 LNP containing 7% DSPE‐PGlu(DET‐Car)30 and siRNA had a size of 130 nm (Table 2). The particle size was suitable for passive accumulation in tumor tissues via the EPR effect while avoiding quick clearance by the mononuclear phagocyte system (MPS) cells of the spleen and liver for prolonged blood circulation. 15 , 16 The optimized DSPE‐PGlu(DET‐Car)30‐modified LNP showed a unimodal size distribution and high siRNA EE (Table 2). The morphology of LNP was carried out using TEM (Figure 1D). The LNP had spherical shape, and the size of the LNP observed in the TEM image was slightly smaller than that measured by DLS, due to the dehydration process in sample preparation. The high siRNA EE resulted from sufficient DOTAP with an N/P ratio of 4. In addition, the LNP was resistant to changes in size with time in the presence of serum (Figure S3) and at different pH (Figure S4), suggesting its high stability under biological conditions. The size of LNP in serum was slightly smaller than in D‐PBS(−) due to osmotic pressure. Importantly, the ζ‐potential of PGlu(DET‐Car)30 LNP switched from nearly neutral at pH 7.4 to an appreciably positive charge at pH 6.5 (Table 2 and Figure 1C). This increased ζ‐potential of LNP should be due to bisprotonation of the ethylenediamine moieties of PGlu(DET‐Car), as illustrated in Figure 1. The size of LNPs was controllable by changing the mixing ratio of DSPE‐PGlu(DET‐Car)30 (Table 1), suggesting the facile preparation of DSPE‐PGlu(DET‐Car)30 LNPs similar to conventional PEGylated LNPs. 17 Given that the pH of the tumor microenvironment was a pH of ~6.5, 18 PGlu(DET‐Car)30 LNP was expected to exert facilitated cellular uptake in response to cancerous pH.
FIGURE 1.
(A) Structure of DSPE‐PGlu(DET‐Car)30. (B) Schematic illustration of siRNA‐encapsulated PGlu(DET‐Car)30 LNPs. (C) How PGlu(DET‐Car)30 LNPs facilitate tumor accumulation through pH switchable properties. (D) TEM image of LNP. LNP would switch to a positive charge in response to a tumor acidic environment via protonated amino groups of polymers to enhance tumor accumulation. Cationic LNP would improve the interaction with anionic cell membrane for effective siRNA delivery
TABLE 1.
Characteristic of PGlu(DET‐Car)30 LNPs through different lipid composition with various molar ratios using DLS (n = 3)
| Lipid composition (%) | Diameter a | PDI a | |||
|---|---|---|---|---|---|
| DOTAP | DOPE | Cholesterol | PGlu(DET‐Car)30 | ||
| 30 | 40 | 30 | 1 | 290 ± 1.3 | 0.199 ± 0.037 |
| 40 | 30 | 30 | 1 | 270 ± 0.2 | 0.180 ± 0.001 |
| 35 | 35 | 30 | 1 | 220 ± 2.0 | 0.144 ± 0.036 |
| 34 | 34 | 30 | 2 | 160 ± 1.4 | 0.165 ± 0.016 |
| 34 | 34 | 29 | 3 | 140 ± 0.1 | 0.195 ± 0.003 |
| 32 | 32 | 28 | 7 | 130 ± 0.7 | 0.275 ± 0.004 |
LNP was suspended in D‐PBS(−) and the hydrodynamic diameter and polydispersity index (PDI) were determined using dynamic light scattering.
TABLE 2.
Characterization of siRNA‐encapsulated PGlu(DET‐Car)30 LNP. The results are shown as the mean ± SD (n = 10)
| Diameter (nm) | PDI | EE (%) | ζ‐potential (mV) |
|---|---|---|---|
| 130 ± 3.8 | 0.269 ± 0.019 | 95 ± 3 |
17.6 ± 1.0 (pH 6.5) 3.6 ± 2.9 (pH 7.4) |
Note: LNP was suspended in PBS(−) and the hydrodynamic diameter and polydispersity index (PDI) were determined using dynamic light scattering. For ζ‐potential measurement, LNP was suspended in MES and HEPES buffer (pH 6.5 and 7.4) and the ζ‐potential were measured using a Zetasizer Nano‐ZS90 system. Encapsulation efficiency (EE) was determined by RiboGreen assay.
3.2. Cellular uptake
The cellular uptake of Alexa647‐siGL3‐encapsulated LNP was evaluated in the SKOV3‐luc and CT26 cell lines using FCM (Figure 2). PGlu(DET‐Car)30 LNP exhibited time‐dependent cellular uptake in both cell lines (Figure 2A–D). Moreover, PGlu(DET‐Car)30 LNP showed significantly higher cellular uptake at pH 6.5 compared with pH 7.4 (Figure 2A–F), suggesting that the increased surface cationic charge at cancerous pH 6.5 should contribute to the enhanced cellular uptake. Considering that the acidic environment and cationic particle might cause cytotoxicity, we confirmed that there was no significant cytotoxicity during the 24 h co‐incubation with PGlu(DET‐Car)30 LNPs at pH 6.5 (Figure S5).
FIGURE 2.
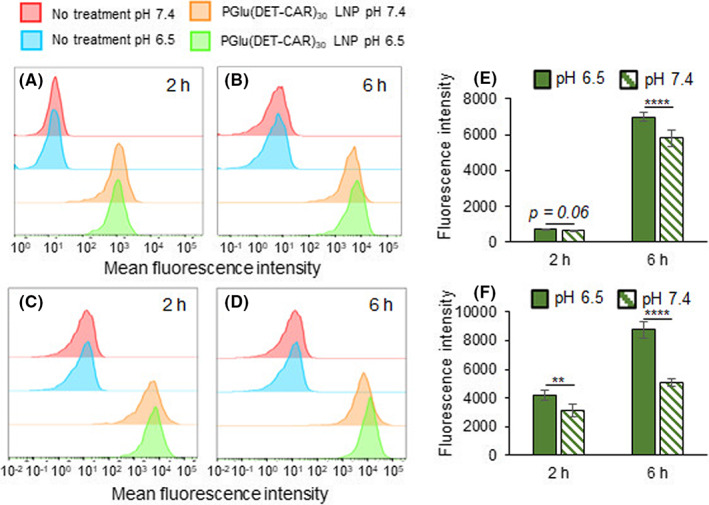
Flow cytometric analysis of cellular uptake. Population of Alexa647‐labeled siRNA‐encapsulated PGlu(DET‐CAR)30 LNP in SKOV3‐luc cells with (A) a 2 h incubation and (B) a 6 h incubation. Population of Alexa647‐labeled siRNA‐encapsulated PGlu(DET‐CAR)30 LNP in CT 26 cells with (C) a 2 h incubation and (D) a 6 h incubation. Quantification analysis of cellular uptake in (E) SKOV3‐luc cells and (F) CT26 cells. In total, 3000 events were acquired for each sample. The results are shown as the mean ± SD (n = 3), **** p < 0.0001 (Student's t‐test)
3.3. Subcellular distribution and endosomal escape
After confirming the cellular uptake, we investigated the endosomal escape by subcellular distribution using CLSM imaging with 1 and 8 h treatments of LNP at pH 6.5 (Figure 3A,B). PGlu(DET‐Car)30 LNP showed a significantly decreased co‐localization ratio of Alexa647‐siRNA with Lyso Tracker Red‐stained organelles were subjected to 1–8 h incubations in both SKOV3‐luc and CT26 (Figure 3B), and indicated that the release of siRNA from the endosome was between 1 and 8 h. We further examined endosomal escape through co‐incubation of LNP with calcein for 2 h. Calcein is a small membrane‐impermeable dye that is taken up by cells through endocytosis. Therefore, diffusion of calcein into the cytosol indicated the disruption of the endosomal membrane. As shown in Figure 3C, PGlu(DET‐Car)30 LNPs at pH 6.5 exhibited distinct diffusion of calcein to the cytosol in both CT26 and SKOV3‐luc cells, suggesting endosomal escape. As the ζ‐potential measurement indicated an appreciable positive charge of PGlu(DET‐Car)30 LNP in an acidic environment (Table 2), the internalized cationic LNPs might induce endosomal escape through enhanced electrostatic interaction between the cationic LNP surface and the anionic endosomal membrane due to the more acidic endosomal environment. 19
FIGURE 3.
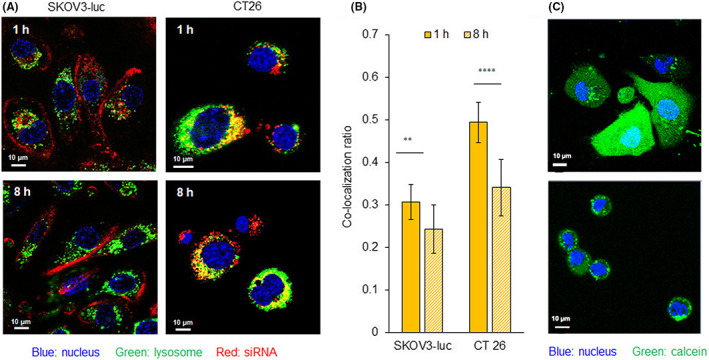
Assessment of endosomal escape. (A) CLSM image of 1 and 8 h treatments of Alexa647‐labeled siRNA‐encapsulated PGlu(DET‐Car)30 LNP in SKOV3‐luc and CT 26 cells. (B) Co‐localization ratio of Alexa647‐siRNA‐ and Lyso Tracker Red‐labeled organelles in SKOV3‐luc and CT 26 cells. The cells were incubated at pH 6.5 with PGlu(DET‐Car)30 LNP (100 nM Alexa647‐siGL3) for 1 and 8 h. All pictures show merged images, which include the nuclei (blue) and LNP (red). Scale bars represent 10 μm at all images. (C) Cytosolic localization of calcein (green) in SKOV3‐luc and CT 26 cells. The cells were incubated at pH 6.5 with PGlu(DET‐Car)30 LNP (100 nM siGL3) for 2 h. Nuclei were stained using Hoechst 33342 (blue). Scale bar (white) = 10 μm. The results are shown as the mean ± SD (n = 15 for SKOV3‐luc, n = 23 for CT 26), ** p < 0.01, **** p < 0.0001 (Student's t‐test)
3.4. In vitro gene silencing
The in vitro gene silencing efficiency of siGL3‐encapsulated PGlu(DET‐Car)30 LNPs was investigated in SKOV3‐luc cells using a luciferase assay (Figure 4). The siRNA delivery capabilities of PGlu(DET‐Car)30 LNP were compared with those of the control LNP, which was designed to be analogous in size and EE (Table S1). Compared with the control LNP, the PGlu(DET‐Car)30 LNPs showed a significantly enhanced gene silencing activity due to its charge‐switchable properties, which might facilitate cellular uptake and endosomal escape.
FIGURE 4.
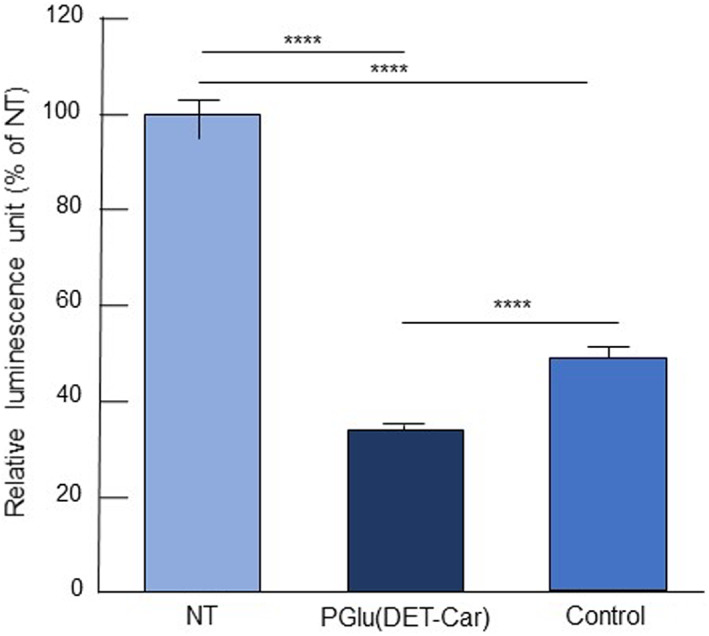
In vitro gene silencing efficiency of LNPs using a luciferase assay. SKOV3‐luc cells were treated with siGL3‐encapsulated PGlu(DET‐Car)30 LNP and non‐pH‐responsive control LNP at pH 6.5 for 6 h followed by incubation in fresh medium for another 24 h. The siRNA dose was 100 nM. The results are shown as the mean ± SD (n = 7), **** p < 0.0001 (Tukey's multiple comparisons test)
3.5. Biodistribution study
The in vivo biodistribution of Alexa647‐siGL3‐encapsulated PGlu(DET‐Car)30 LNP and control LNP was evaluated using a CT26 tumor‐bearing mouse model (Figure 5). PGlu(DET‐Car)30 LNP exhibited prolonged blood circulation and greatly higher tumor accumulation compared with the control LNP (Figure 5A,B). It is worth mentioning that PGlu(DET‐Car)30 LNP maintained a significantly higher tumor accumulation level for up to 24 h, whereas the control LNP showed a sharp decline after 6 h. The similarly higher tumor accumulation of PGlu(DET‐Car)30 LNP was also observed in SKOV3‐luc tumor‐bearing mice (Figure S6). In addition, both LNPs showed considerable accumulation within the liver and spleen, suggesting that their mechanism of clearance may be mediated by the MPS (Figure 5C,D). PGlu(DET‐Car)30 LNP showed a lower accumulation in spleen compared with the control (Figure 5C,D). Although the mechanism of relatively low splenic accumulation of PGlu(DET‐Car)30 LNP is still under investigation, the surface modification of PGlu(DET‐Car)30 onto LNP might cause a distinct accumulation behavior in spleen, probably due to the stealth ability difference of the surface material. Therefore, the sustained tumor accumulation behavior of PGlu(DET‐Car)30 LNP was assumed to be due to the cationic charge of PGlu(DET‐Car)30 LNP within the cancerous pH, as well as a prolonged blood circulation. That is, the strong electrostatic interaction with the cell membranes of the cancer cells led to prolonged intratumoral retention. This process was not readily available to non‐pH‐responsive control LNP, which consequently experienced only a transient sojourn in cancer tissue.
FIGURE 5.
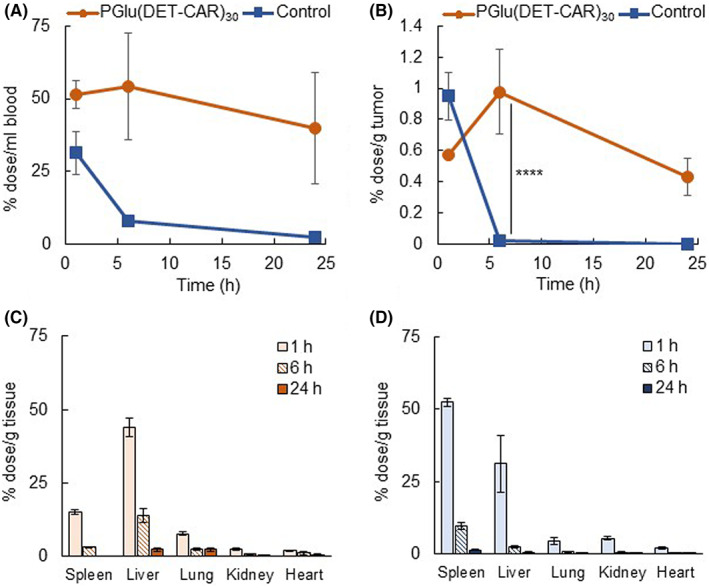
Biodistribution after i.v. administration of Alexa647‐siGL3‐encapsulated PGlu(DET‐Car)30 LNP and control LNP. (A) Blood circulation. (B) Tumor accumulation. Accumulation of (C) PGlu(DET‐Car)30 LNP and (D) control LNP in other organs. Each result is expressed as the mean ± SEM (n = 3), **** p < 0.0001 (two‐way anova with Sidak's multiple comparisons test)
3.6. In vivo gene silencing and tumor suppression effect
The in vivo gene silencing ability and the application to tumor suppression of PGlu(DET‐Car)30 LNP were investigated (Figures 6 and 7). The in vivo gene knockdown ability of PGlu(DET‐Car)30 LNP was confirmed via the reduction of luciferase activity and mRNA expression compared with D‐PBS(−) treatment (Figure 6A,B). After the confirming tumor accumulation and in vivo gene silencing activity of PGlu(DET‐Car)30 LNP, the tumor suppression effect was examined using siPLK1‐encapsulated LNPs in SKOV3‐luc tumor‐bearing mice. siPLK1 has been shown to interfere with mitosis and suppress the proliferation of human cancer cells. 20 siPLK1‐encapsulated LNPs at a dose of 2.5 mg/kg siPLK1 were administrated through periodic i.v. administration, as shown in Figure 7A. Compared with both siPLK1‐encapsulated control LNP and NT groups, siPLK1‐encapsulated PGlu(DET‐Car)30 LNP significantly inhibited tumor growth without the loss of body weight (Figure 7B–D). The pH‐responsive nature and slower clearance of PGlu(DET‐Car)30 LNP (Figure 5A,B) led to enhanced tumor accumulation compared with the control LNP, and therefore led to siRNA delivery at a level adequate for tumor growth suppression.
FIGURE 6.
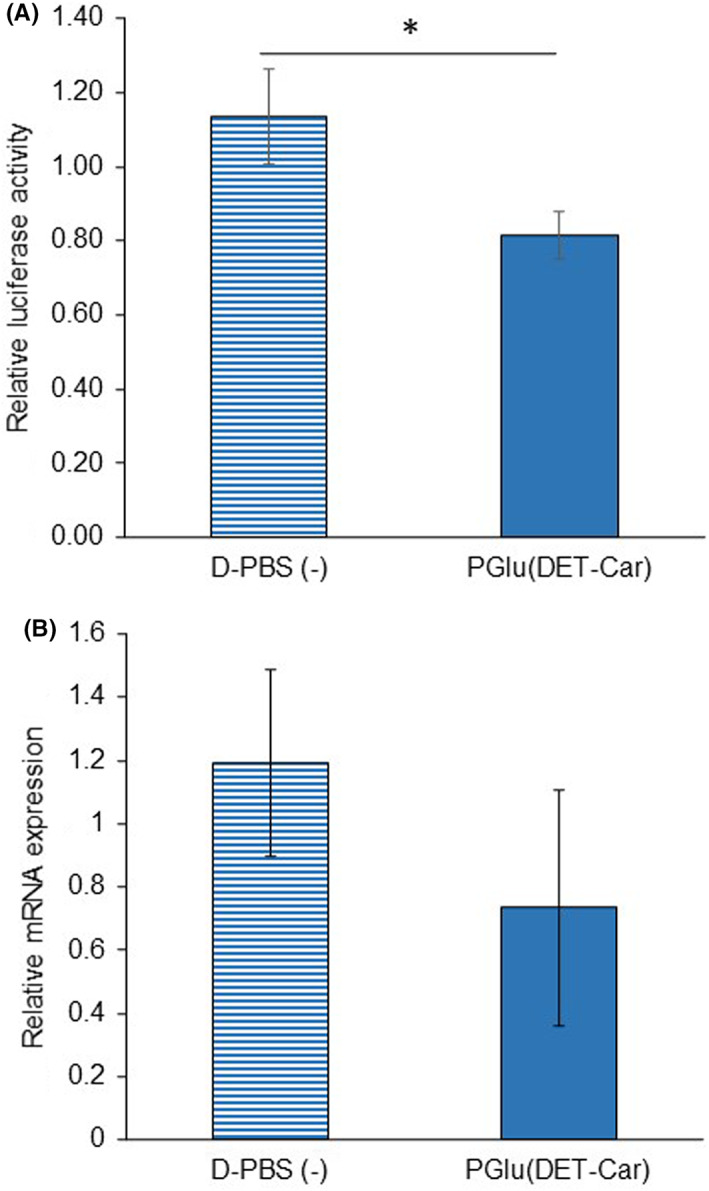
In vivo gene silencing assessment of PGlu(DET‐Car)30 LNP. Gene knockdown ability was examined by (A) bioluminescence (n = 5) and (B) RT‐PCR (n = 3) after 24 h post‐i.v. injection of siGL3‐encapsulated PGlu(DET‐Car)30 LNP. Each result is expressed as the mean ± SEM, * p < 0.05 (Student's t‐test)
FIGURE 7.
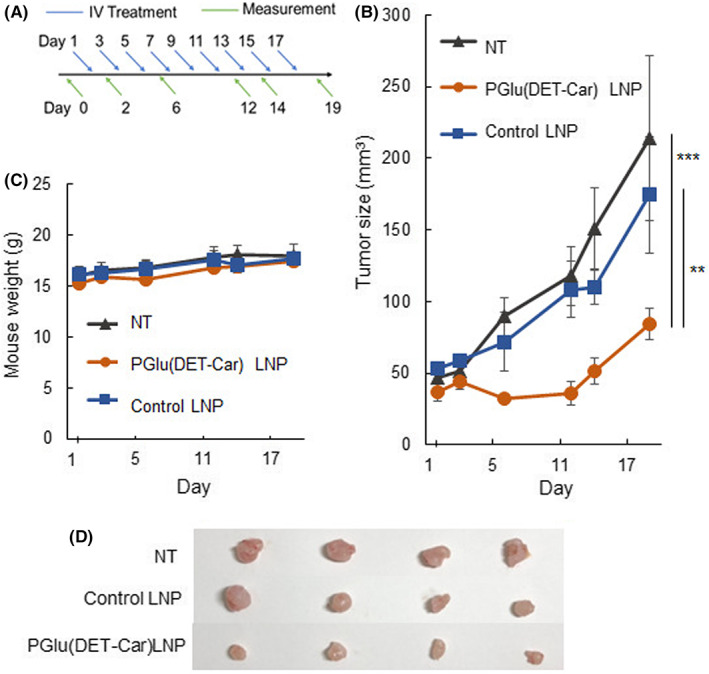
Tumor suppression effect. (A) Scheme of the therapeutic regimen. (B) Tumor suppression effect and (C) mouse body weight change during periodic i.v. administration of siPLK1‐loaded LNPs. Each point is expressed as the mean ± SEM (n = 4), ** p < 0.01, and *** p < 0.001 (two‐way anova with Tukey's multiple comparisons test). (D) Snapshots of the tumor in the SKOV3‐luc subcutaneous model on the final day
4. DISCUSSION
In siRNA delivery targeting solid tumors, nanocarriers should display efficient cellular uptake by cancer cells at the tumor site, while showing prolonged blood circulation, which is a prerequisite for tumor accumulation based on the EPR effect. To achieve this paradoxical objective, in this study, we designed a cancerous pH‐responsive polycarboxybetain [PGlu(DET‐Car)], which exhibits switchable net charge when responding to the surrounding pH, as a smart shell material for LNPs. That is, the PGlu(DET‐Car) displayed a neutral charge at physiological pH 7.4, therefore exhibiting stealth properties during circulation; however, it displayed a cationic property in acidic tumor microenvironments, facilitating the siRNA delivery to cancer cells.
Surface charge plays a critical role in the cellular uptake and transfection efficiency of siRNA. Under acidic conditions mimicking the tumor microenvironment, PGlu(DET‐Car)30 molecules are in an overall cationic state for electrostatic interactions between the LNP surface and the negatively charged cell membranes of cancer cells (Figure 1C), leading to acidic pH‐responsive enhancement in cellular uptake (Figure 2). Furthermore, the cationic surface on PGlu(DET‐Car)30 LNP in the endosomal compartment enables the electrostatic interaction with the endosomal membrane, allowing facilitated endosomal escape of siRNA through membrane fusion (Figure 3). In addition, the pH‐dependent protonation of the amino groups within the endosomal lumen may result in osmotic swelling and subsequent endosome disruption via the proton sponge effect. 21 Therefore, the PGlu(DET‐Car)30 LNP achieved more efficient gene knockdown in cancer cells compared with the control LNP (Figure 4).
PGlu(DET‐Car)30 LNP exhibited extended blood circulation (Figure 5A), as polycarboxybetaine‐coated liposomes demonstrated the longevity in the bloodstraem. 22 The antifouling properties of polyzwitterions are attributed to the formation of hydrating layers with water molecules based in ion–dipole interactions. 23 , 24 The polyzwitterion structure of PGlu(DET‐Car)30 LNP might influence its affinity for serum proteins in blood circulation, for which a further understanding of the relationship between the physicochemical characteristics of PGlu(DET‐Car) LNPs and their in vivo behavior will be required.
The PGlu(DET‐Car)30 LNP demonstrated an enhanced tumor accumulation and retention within two distinct tumor models, (Figure 5B and Figure S6), due to the cationic surface charge of PGlu(DET‐Car)30 LNP, endowing tissue‐interactive properties under cancerous pH conditions. Given that pH of tumor microenvironment has been observed to be persistently acidic, 25 the pH‐responsive PGlu(DET‐Car)30 LNP might possess a great potential for siRNA delivery to various solid tumors. Conversely, the penetration ability of LNPs was also an important feature for use as nanomedicines. Therefore, the current limitation in this study might be the practicality of this system against heterogenic and stromal‐rich tumors, such as human pancreatic cancer, because we used subcutaneous tumor models with CT26 cells and SKOV3 cells. Although further optimization of the PGlu(DET‐Car) LNPs, including the use of pH‐responsive ionizable lipids such as 1,2‐dioleoyl‐3‐dimethylammonium propane (DODAP), is desirable to further bolster tumor accumulation, penetration, and siRNA transfection activity, the tumor targeting ability of PGlu(DET‐Car)30 LNP makes it an attractive choice as a vehicle for siRNA‐based cancer therapy. In addition, PGlu(DET‐Car) LNPs might be useful for siRNA‐based treatment of various intractable diseases, because the acidic environment is universally recognized at the site of inflammation and within the endosomes/lysosomes of the cells.
In summary, we demonstrated that PGlu(DET‐Car)30 LNP shows cancerous pH‐responsive enhancement in cellular uptake, endosomal escape, and siRNA transfection in vitro and enables successful delivery of therapeutic siRNA in subcutaneous tumor models. These results strongly signify the utility of PGlu(DET‐Car)30 as a smart shell material for surface modification of LNPs for systemic delivery of siRNA payloads to malignant tumors.
AUTHOR CONTRIBUTIONS
Y.S. performed all experiments and wrote the manuscript. H.G., A.G., X.S., W.C., M.K., M. T., M. M., Y. H., T. N., and H. T. assisted in vitro/in vivo experiments and manuscript preparation. X.S., K.O., J.M., T.O. and H. T. assisted synthesis and characterization of DSPE‐PGlu(DET‐Car)30. Y.M. and N.N. supervised the whole project and conceived the concepts of this study.
FUNDING INFORMATION
This work was supported by the Center of Innovation (COI) program (JPMJCE1305) from the Japan Science and Technology Agency (JST), Science and Technology Platform Program for Advanced Biological Medicine (JP21am0401018) from Japan Agency for Medical Research and Development (AMED), the Project for Cancer Research And Therapeutic Evolution (P‐CREATE) (JP19cm0106202) from AMED, JSPS KAKENHI (JP18H04163, JP21K18322, 21 K20513), and Five‐star Alliance from the Ministry of Education, Culture, Sports, Science and Technology (MEXT).
DISCLOSURE
N.N. is an editorial board member. K.O., J.M., T.O. are employees of NOF CORPORATION. Other authors do not have any conflict.
ETHICS STATEMENT
Approval of the research protocol by an Institutional Reviewer Board: N/A. Informed Consent: N/A. Registry and the Registration No. of the study/trial: N/A. Animal Studies: All animal experiments were approved by the Animal Care and Use Committee and performed in accordance with the Guidelines for the Care and Use of Laboratory Animals set forth by Tokyo Institute of Technology.
Supporting information
Appendix S1
ACKNOWLEDGMENTS
We thank Materials Analysis Suzukake‐dai, Technical Department, Tokyo Institute of Technology, for NMR measurement and TEM observation.
Sung Y‐J, Guo H, Ghasemizadeh A, et al. Cancerous pH‐responsive polycarboxybetaine‐coated lipid nanoparticle for smart delivery of siRNA against subcutaneous tumor model in mice. Cancer Sci. 2022;113:4339‐4349. doi: 10.1111/cas.15554
Contributor Information
Yutaka Miura, Email: miura.y.ai@m.titech.ac.jp.
Nobuhiro Nishiyama, Email: nishiyama.n.ad@m.titech.ac.jp.
REFERENCES
- 1. Hu B, Weng Y, Xia XH, Liang XJ, Huang Y. Clinical advances of siRNA therapeutics. J Gene Med. 2019;21:e3097. doi: 10.1002/jgm.3097 [DOI] [PubMed] [Google Scholar]
- 2. Weng Y, Xiao H, Zhang J, Liang XJ, Huang Y. RNAi therapeutic and its innovative biotechnological evolution. Biotechnol Adv. 2019;37:801‐825. doi: 10.1016/j.biotechadv.2019.04.012 [DOI] [PubMed] [Google Scholar]
- 3. Lu Z‐R, Laney VEA, Hall R, Ayat N. Environment‐responsive Lipid/siRNA nanoparticles for cancer therapy. Adv Healthc Mater. 2021;10:2001294. doi: 10.1002/adhm.202001294 [DOI] [PubMed] [Google Scholar]
- 4. Wang J, Lu Z, Wientjes MG, Au JL. Delivery of siRNA therapeutics: barriers and carriers. AAPS J. 2010;12:492‐503. doi: 10.1208/s12248-010-9210-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liberti MV, Locasale JW. The Warburg effect: how does it benefit cancer cells? Trends Biochem Sci. 2016;41:211‐218. doi: 10.1016/j.tibs.2015.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4:891‐899. doi: 10.1038/nrc1478 [DOI] [PubMed] [Google Scholar]
- 7. Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029‐1033. doi: 10.1126/science.1160809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Helmlinger G, Yuan F, Dellian M, Jain RK. Interstitial pH and pO2 gradients in solid tumors in vivo: high‐resolution measurements reveal a lack of correlation. Nat Med. 1997;3:177‐182. doi: 10.1038/nm0297-177 [DOI] [PubMed] [Google Scholar]
- 9. Schlich M, Palomba R, Costabile G, et al. Cytosolic delivery of nucleic acids: the case of ionizable lipid nanoparticles. Bioeng Transl Med. 2021;6:e10213. doi: 10.1002/btm2.10213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ranneh AH, Takemoto H, Sakuma S, et al. An ethylenediamine‐based switch to render the polyzwitterion cationic at tumorous pH for effective tumor accumulation of coated nanomaterials. Angew Chem Int Ed Engl. 2018;57:5057‐5061. doi: 10.1002/anie.201801641 [DOI] [PubMed] [Google Scholar]
- 11. Takemoto H, Nishiyama N. Construction of nanomaterials based on pH‐responsive polymers for effective tumor delivery. Polym J. 2021;53:1353‐1360. doi: 10.1038/s41428-021-00542-7 [DOI] [Google Scholar]
- 12. De Corte D, Schläpfer C‐W, Daul C. A density functional theory study of the conformational properties of 1,2‐ethanediamine: protonation and solvent effects. Theor Chem Accounts. 2000;105:39‐45. doi: 10.1007/s002140000177 [DOI] [Google Scholar]
- 13. Miyata K, Oba M, Nakanishi M, et al. Polyplexes from poly (aspartamide) bearing 1,2‐diaminoethane side chains induce pH‐selective, endosomal membrane destabilization with amplified transfection and negligible cytotoxicity. J Am Chem Soc. 2008;130:16287‐16294. doi: 10.1021/ja804561g [DOI] [PubMed] [Google Scholar]
- 14. Sato Y, Hatakeyama H, Sakurai Y, Hyodo M, Akita H, Harashima H. A pH‐sensitive cationic lipid facilitates the delivery of liposomal siRNA and gene silencing activity in vitro and in vivo. J Control Release. 2012;163:267‐276. doi: 10.1016/j.jconrel.2012.09.009 [DOI] [PubMed] [Google Scholar]
- 15. Yoo JW, Doshi N, Mitragotri S. Adaptive micro and nanoparticles: temporal control over carrier properties to facilitate drug delivery. Adv Drug Deliv Rev. 2011;63:1247‐1256. doi: 10.1016/j.addr.2011.05.004 [DOI] [PubMed] [Google Scholar]
- 16. Blanco E, Shen H, Ferrari M. Principles of nanoparticle design for overcoming biological barriers to drug delivery. Nat Biotechnol. 2015;33:941‐951. doi: 10.1038/nbt.3330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bao Y, Jin Y, Chivukula P, et al. Effect of PEGylation on biodistribution and gene silencing of siRNA/lipid nanoparticle complexes. Pharm Res. 2013;30:342‐351. doi: 10.1007/s11095-012-0874-6 [DOI] [PubMed] [Google Scholar]
- 18. Neri D, Supuran CT. Interfering with pH regulation in tumours as a therapeutic strategy. Nat Rev Drug Discov. 2011;10:767‐777. doi: 10.1038/nrd3554 [DOI] [PubMed] [Google Scholar]
- 19. Geisow MJ, Evans WH. pH in the endosome. Measurements during pinocytosis and receptor‐mediated endocytosis. Exp Cell Res. 1984;150:36‐46. doi: 10.1016/0014-4827(84)90699-2 [DOI] [PubMed] [Google Scholar]
- 20. Spänkuch‐Schmitt B, Bereiter‐Hahn J, Kaufmann M, Strebhardt K. Effect of RNA silencing of polo‐like kinase‐1 (PLK1) on apoptosis and spindle formation in human cancer cells. J Natl Cancer Inst. 2002;94:1863‐1877. doi: 10.1093/jnci/94.24.1863 [DOI] [PubMed] [Google Scholar]
- 21. Bus T, Traeger A, Schubert US. The great escape: how cationic polyplexes overcome the endosomal barrier. J Mater Chem B. 2018;6:6904‐6918. doi: 10.1039/C8TB00967H [DOI] [PubMed] [Google Scholar]
- 22. Cao Z, Zhang L, Jiang S. Superhydrophilic zwitterionic polymers stabilize liposomes. Langmuir. 2012;28:11625‐11632. doi: 10.1021/la302433a [DOI] [PubMed] [Google Scholar]
- 23. Erfani A, Seaberg J, Aichele CP, Ramsey JD. Interactions between biomolecules and zwitterionic moieties: a review. Biomacromolecules. 2020;21:2557‐2573. doi: 10.1021/acs.biomac.0c00497 [DOI] [PubMed] [Google Scholar]
- 24. Keefe AJ, Jiang S. Poly (zwitterionic) protein conjugates offer increased stability without sacrificing binding affinity or bioactivity. Nat Chem. 2011;4:59‐63. doi: 10.1038/nchem.1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kato Y, Ozawa S, Miyamoto C, et al. Acidic extracellular microenvironment and cancer. Cancer Cell Int. 2013;13:89. doi: 10.1186/1475-2867-13-89 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1


