Abstract
Three agr-like genes (fsrA, fsrB, and fsrC, for Enterococcus faecalis regulator) were found upstream of the previously reported gelatinase gene (gelE) and a downstream putative serine protease gene (sprE; accession number Z12296) of Enterococcus faecalis OG1RF. The deduced amino acid sequence of fsrA shows 26% identity and 38% similarity to Staphylococcus aureus AgrA (the response regulator of the accessory gene regulator system in the agr locus), FsrB shows 23% identity and 41% similarity to S. aureus AgrB, and FsrC shows 23% identity and 36% similarity to S. aureus AgrC (the sensor transducer of Agr system). Northern blot analysis suggested that gelE and sprE are cotranscribed and that fsrB and fsrC are also cotranscribed in OG1RF. Northern blot analysis of fsrA, fsrB, fsrC, gelE, and sprE insertion mutants showed that fsrB, fsrC, gelE, and sprE are not expressed in fsrA, fsrB, and fsrC mutants, while insertion in an open reading frame further upstream of fsrA did not effect the expression of these genes, suggesting that agr-like genes may be autoregulated and that they regulate gelE and sprE expression, as further confirmed by complementation of fsr gene mutations with a 6-kb fragment which contains all three fsr genes in the shuttle vector, pAT18. Testing of 95 other isolates of E. faecalis showed that 62% produced gelatinase (Gel+), while 91% (including all Gel+ strains) hybridized to a gelE probe; 71% (including all Gel+ strains) hybridized to an fsr probe, corroborating the conclusion that both gelE and fsr are necessary for gelatinase production. Testing of fsrA, fsrB, and sprE mutants in a mouse peritonitis model showed that sprE and agr-like gene mutants resulted in highly significantly prolonged survival compared to the parent strain OG1RF, a finding similar to what we had previously shown for a gelE mutant. These results suggest that sprE and agr-like genes contribute to the virulence of E. faecalis OG1RF in this model.
Enterococci are among the major causes of hospital-acquired infections, including urinary tract infections, bloodstream infections, wound infections, and endocarditis (9). Enterococcus faecalis is the most common organism recovered from enterococcal infections and accounted for 85 to 90% of clinical isolates prior to emergence of vancomycin resistance in enterococci (9). However, the mechanisms of pathogenesis of enterococci are not yet well understood. In an attempt to study the pathogenesis of enterococcal infections, we have previously generated an insertion mutation of the gelatinase gene (gelE) previously reported by Su et al. (21) in E. faecalis OG1RF and tested the mutant in a mouse peritonitis model (19). The time course of survival for animals infected with the gelE mutant was significantly prolonged compared to that seen with the parent strain OG1RF. To investigate whether the delayed mortality was caused by inactivation of gelE or by a polar effect on expression of downstream gene(s), we partially sequenced gelE flanking regions. Sequencing from OG1RF and searches in the enterococcal genome database revealed three open reading frames in the region 5′ of gelE which show homology to agrA, agrB, and agrC of Staphylococcus aureus and a previously reported gene encoding a putative serine protease (sprE, direct submission to database) 3′ of gelE.
In S. aureus, the agr/hld locus contains five genes, agrB, agrD, agrC, agrA, and hld (RNAIII), which are all required for the expression of the agr (accessory gene regulator) genes and which encode a quorum-sensing system which regulates the expression of virulence factors (5, 11, 12, 16). S. aureus agrC and agrA encode a sensor transducer and response regulator of bacterial two-component systems (11), respectively, while agrD encodes a pheromone peptide which acts as an autoinducer for sensing of cell density by the Agr system (6). The product of agrB seems to be involved in the processing of the peptide encoded by agrD (5). The Agr regulatory system in S. aureus upregulates the expression of secreted proteins such as alpha-toxin, beta-toxin, delta-toxin, enterotoxin B, toxic shock syndrome toxin 1, and serine protease and downregulates surface proteins such as protein A, coagulase, and fibronectin-binding protein (8, 16). Similar Agr systems have also been found in other staphylococcal species, including S. epidermidis (13, 22) and S. lugdunensis (5, 25).
To study the functions of these agr-like genes in E. faecalis, we generated insertion mutations of these genes, studied the expression of these genes and of gelE and sprE in these mutants, and tested the virulence of these mutants in a mouse peritonitis model. Our results suggest that E. faecalis agr-like genes, designated fsr for E. faecalis regulator, regulate the expression of gelE and sprE and are important for enterococcal virulence in the mouse peritonitis model.
MATERIALS AND METHODS
Strains and media.
The strains and plasmids used in this study are listed in Table 1. E. faecalis OG1RF has been described previously (10). Luria-Bertani broth and agar were used for Escherichia coli culture, and brain heart infusion (BHI; Difco Laboratories, Detroit, Mich.) was used for the growth of E. faecalis unless otherwise stated. The concentrations of antibiotics used for selection were: ampicillin (Amp), 50 μg/ml; chloramphenicol (Cm), 10 μg/ml; erythromycin (Em), 250 μg/ml; kanamycin (Kan), 50 μg/ml; tetracycline (Tet) 12.5 μg/ml for E. coli; and Cm, 10 μg/ml; Em, 10 μg/ml; and Kan, 2,000 μg/ml for E. faecalis.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant genotype or phenotype | Source or reference |
|---|---|---|
| Strain | ||
| OG1RF | Gelatinase and serine protease production positive/(Gel+, Spr+) | 10 |
| TX5128 | gelE transposon mini-γδ insertion mutant of OG1RF, Gel−, Spr−, Kanr | 19 |
| TX5240 | fsrA insertion mutant of OG1RF, Gel−, Spr−, Kanr | This study |
| TX5241 | fsrB insertion mutant of OG1RF, Gel−, Spr−, Kanr | This study |
| TX5242 | fsrC insertion mutant of OG1RF, Gel−, Spr−, Kanr | This study |
| TX5243 | sprE insertion mutant of OG1RF, Gel+, Spr−, Kanr | This study |
| TX5248 | orf1 insertion mutant of OG1RF, Gel+, Spr+, Kanr | This study |
| TX5244 | TX5240 harboring plasmid TEX5249, Gel+, Spr+, Kanr Emr | This study |
| TX5245 | TX5241 harboring plasmid TEX5249, Gel+, Spr+, Kanr Emr | This study |
| TX5246 | TX5242 harboring plasmid TEX5249, Gel+, Spr+, Kanr Emr | This study |
| TX5247 | TX5128 harboring plasmid TEX5249, Gel−, Spr−, Kanr Emr | This study |
| DH5α | E. coli host strain for routine cloning | Stratagene |
| TX5249 | DH5α(pTEX5249) | This study |
| Plasmids | ||
| pBluescript SK(−) | Cloning vector, Ampr | Stratagene |
| pTEX4577 | Suicide vector for single cross-over mutagenesis in E. faecalis, Kanr | 19 |
| pAT18 | Shuttle vector for E. coli and E. faecalis, Emr | 23 |
| pTEX5249 | pTEX5249 was constructed by cloning a 6-kb PstI/BglII fragment containing fsrA, fsrB, and fsrC into the shuttle vector pAT18, Emr | This study |
DNA techniques.
Routine isolation of plasmid DNA from E. coli was performed as previously described (3). Large-scale preparation of plasmid DNA from equilibrium centrifugation in CsCl-ethidium bromide gradient, gel electrophoresis, and Southern blot analyses was carried out according to previously described methods (17). Radioactive DNA probes were prepared by random primed DNA labeling kit (Boehringer Mannheim, Indianapolis, Ind.) according to the protocol supplied. Transformation of E. faecalis was accomplished by the method described by Li et al. (7) using a Bio-Rad Gene Pulser. Chromosomal DNA from E. faecalis was prepared according to the method described by Wilson (26). PCR amplification of DNA was performed on a DNA thermal cycler (Perkin-Elmer Corp., Norwalk, Conn.) using synthetic oligonucleotide primers from Genosys (Genosys Biotechnologies, Inc., Woodlands, Tex.) and Taq DNA polymerase from Life Technologies (Gaithersburg, Md.).
DNA sequencing and sequence analysis.
Automated sequencing was used to determine nucleotide sequence using the dideoxy-chain termination method (15, 18). PCR sequencing reactions were carried out using the Taq DyeDeoxy terminator Cycle Sequencing Kit (ABI, Foster City, Calif.), and the reactions were analyzed by an ABI Model 373A DNA sequencer. DNA sequence analysis was accomplished using the Genetics Computer Group sequence analysis package, version 7.2 (University of Wisconsin, Madison). For the DNA and protein homology search, BLAST sequence comparison programs were applied using GenEMBL and/or SWISS-PROT databases. BESTFIT or GAP was used for comparing two DNA or peptide sequences.
Mutagenesis.
Target genes in E. faecalis OG1RF were disrupted by using a suicide vector pTEX4577 (19) containing internal fragments of the genes of interest. E. faecalis OG1RF was transformed by electroporation, and transformants were selected on BHI agar plates containing kanamycin (2,000 μg/ml). Correct allelic disruption was confirmed by Southern blot or PCR analysis. The primers used to amplify internal fragments of fsrA, fsrB, fsrC, sprE, and orf1 were fsrAF1, 5′-GGG AGC TCT GAT GAT GAT TGA TTG ATG GAC; fsrAR1, 5′-GGG GTA CCA TTA CAA GTG GCA CAC CAG GAC; fsrBF1, 5′-GGG AGC TCT GGA CAA AGT ATT ATC TAA CCG; fsrBR1, 5′-TTG GTA CCC ACA CCA TCA CTG ACT TTT GC; fsrCF1, 5′-GGG AGC TCA TCG TGT GTT AGA AAA TAG C; fsrCR1, 5′-GGG GTA CCA CGA ATC ACA ACC ACT AAG TC; sprEF1 5′-TTG AGC TCC GTT CCT GCC GAA AGT CAT TC; sprER1, 5′-TTG GTA CCG ATT GGG GAA CCA GAT TGA CC; orf1F1, TTG AGC TCC TGG TTG GAA ACT AAT CGC ACG; and orf1R1, GGG GTA CCA GTA TGG CAA AGA AGT TTT AGC (linker sequences of the primers are underlined). Revertants of fsrC mutant were screened by scoring the restoration of gelatinase activity and the loss of kanamycin resistance.
Detection of gelatinase and serine protease activities.
The production of gelatinase in E. faecalis strains was scored using Todd-Hewitt (TH) (Difco Laboratories) agar plates containing 3% gelatin. After overnight incubation at 37°C, colonies that had opaque zones around them were considered gelatinase activity positive. Gelatinase and serine protease activity were detected by gelatin (0.1%) and casein (0.05%) zymogram gels (Novex, San Diego, Calif.), respectively. For electrophoresis of zymogram gels, 10 μl of 20-fold-concentrated supernatant from overnight culture was mixed with 10 μl of 2× sample buffer (0.125 M Tris-HCl [pH 6.8], 25% glycerol, 4% sodium dodecyl sulfate, 0.005% bromophenol blue) and incubated for 10 min at room temperature before loading. Renaturing and developing of zymogram gels were carried out as suggested by the supplier. S. aureus protease type XVII-B (Sigma) was used as a positive control for serine protease activity.
Northern blot analysis and RT-PCR.
Total RNA from E. faecalis was isolated from cells in exponential phase using RNeasy Mini Kit (Qiagen, Santa Clarita, Calif.) according to the protocol of the supplier with slight modifications. A 6-mg/ml lysozyme solution was used instead of a concentration of 3 mg/ml for the initial lysis step. Total RNA (20 to 40 μg) was treated with 5 U of RQ1 DNase (Promega, Madison, Wis.) for 20 min at 37°C and extracted once with phenol and twice with chloroform-phenol-isoamyl alcohol (25:24:1). Gel electrophoresis, Northern blotting, and hybridization were performed as previously described (17). For reverse transcription PCR (RT-PCR), 50 ng of total RNA was mixed with primers (0.1 μg each), and RT was performed at 37°C for 1 h using Superscript reverse transcriptase (Life Technologies) after heating at 80°C for 5 min. After heat inactivation of the RT mixture at 94°C for 5 min, PCR was performed to amplify cDNA using Taq DNA polymerase (Life Technologies). Reactions without reverse transcriptase were used as controls to detect DNA contamination in total RNA. The internal fragments used for mutagenesis of fsrA, fsrB, fsrC, and sprE were also used as probes for Northern blot analysis. gelE probe for Northern blot analysis was amplified by PCR using gelEF1 (5′-TGG TTG TGA TTC GTT TGT TGG-3′) and gelER1 (5′-TGA ATA AAC TTG TTC TTC TGC G-3′) primers. Two E. faecalis autolysin (2) primers, lytF1 (5′-ACA CCA ACC ACA GAA ACT AC-3′) and lytR1 (5′-GGC AAT AAA TTC TGA AGG AC-3′), were used to amplify a 330-bp internal fragment of autolysin as a probe in the control of Northern blot. These two autolysin primers were also used as a positive control in RT-PCR.
Complementation of mutants of agr-like genes.
To complement the agr-like genes in trans, a 6-kb BglII/PstI fragment containing the three agr-like genes but only the first 395 bp of the gelE gene was cloned into a shuttle vector, pAT18 (Emr) (23), and transformed into fsrA, fsrB, fsrC, and gelE mutants. The Emr transformants were plated into gelatin containing TH agar plates to determine the complementation of the production of gelatinase activity. The expression of fsr and sprE genes in the complemented mutants was determined by Northern blot analysis.
Peritonitis model.
E. faecalis OG1RF and mutants were grown overnight in BHI broth. The cells were harvested by centrifugation, washed once with 0.9% saline, then resuspended in saline to an optical density at 600 nm of 2.2 to 2.8; the CFU of the cell suspensions were determined by plating serial dilutions onto BHI agar plates. Serial dilutions were made in saline and were mixed (1:10) with 50% of sterile rat fecal extract (SRFE) to the desired inocula (19). Groups of 4- to 6-week-old (22 to 25 g), outbred (ICR) female mice were challenged intraperitoneally with different dilutions. A control group of mice was injected with 50% SRFE only. Survival was monitored every 3 to 6 h. Kaplan Meier survival curves and log rank analysis were performed as described previously (19). Results from same-day experiments, when equal inocula or when OG1RF was used at a lower inoculum than a mutant, were compared.
Growth curves.
To determine the growth rates of OG1RF and mutants, overnight cultures were diluted 1:20 in BHI and grown at 37°C with shaking. The turbidity was measured at different time points using a Manostat (Manostat, New York, N.Y.). CFU were determined by serial dilution of the cultures at different time points in saline and plating onto BHI agar plates in duplicate.
Colony hybridization.
Colony hybridization was carried out as previously described (4) to determine the presence of the genes of interest in clinical isolates. To study the presence of fsr genes in clinical isolates, an intragenic fsrB fragment was amplified using fsrBF1 and fsrBR1 primers by PCR and used as a probe for colony hybridization. Probes for gelE and sprE were amplified by PCR using two pairs of primers, primers gelEF1 and gelER1 and primers sprEF1 and sprER1, respectively.
RESULTS
Sequencing and database search of gelE flanking regions.
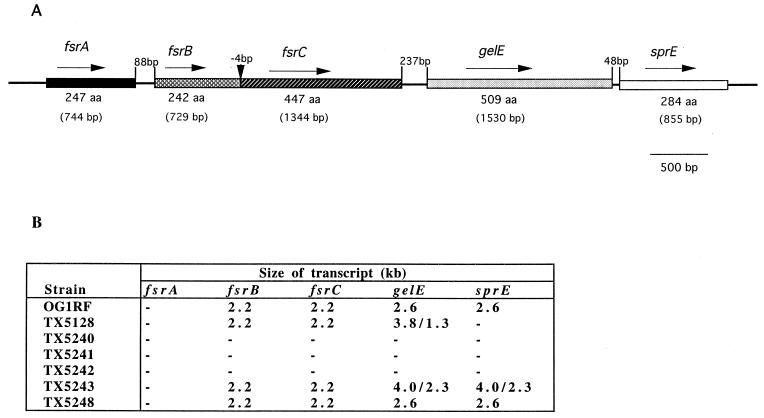
Sequencing of the region 3′ of gelE of OG1RF revealed an open reading frame of 855 bp (sprE) which has been previously deposited in GenBank (accession number Z12296). It encodes a 284-amino-acid peptide which shows homology to bacterial serine proteases. Sequencing and analysis of 2-kb sequence 5′ to gelE revealed two open reading frames (Fig. 1A), both oriented in the same direction as gelE. The peptide deduced from the first open reading frame (447 amino acids) shows significant homology to histidine kinases of bacterial two-component systems, including AgrC of staphylococci (23% identity and 36% similarity to AgrC of the Agr system in S. aureus [accession number AF001783]) (5), and contains residues commonly conserved among these enzymes (Fig. 2). The deduced peptide from the second open reading frame shows 23% identity and 41% similarity to AgrB (accession number AF001782) (5) (Fig. 2), which is unique for the Agr system of staphylococci; thus, we designated them as fsrC and fsrB (E. faecalis regulator; GenBank accession number AF108141). A BLAST search in the E. faecalis genome database of The Institute for Genomic Research using this 2-kb sequence 5′ of gelE gave a perfect match to a contig and the analysis of the sequence further upstream of this 2-kb region showed one open reading frame (designated fsrA) encoding a peptide of 247 amino acids with homology to response regulators of bacterial two-component systems such as AgrA of S. aureus (5), including highly conserved N-terminal residues of Asp and Lys (Fig. 1A and 2). Another open reading frame encoding 652 amino acids was found further upstream of fsrA, termed orf1, which shows homology to bacterial N-acetylmuramidases. Putative promoter sequences were found upstream of fsrA and fsrB but not upstream of fsrC or sprE. In fact, fsrB and fsrC overlap by 4 bp. The observation that the open reading frames in the gelE 5′ region show homology to all of the agr genes except agrD of the S. aureus agr locus, which functions as a virulence regulator, suggested that this locus in E. faecalis may have a similar regulatory function.
FIG. 1.
Open reading frames and their transcripts in gelE flanking regions. (A) Open reading frames. The solid line represents the chromosome, and the genes and open reading frames are indicated by boxes in different shades. The orientation of the genes and open reading frames are indicated by arrows. (B) Summary of the Northern blot results using RNA from different strains and different gene probes. −, No signal in Northern blot analysis.
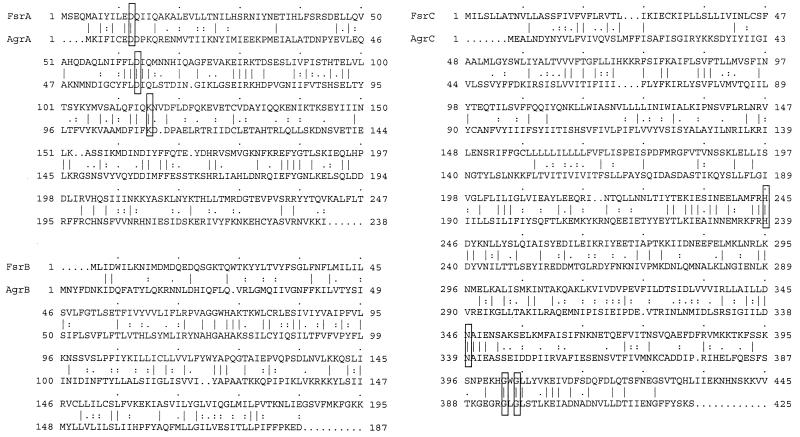
FIG. 2.
Sequence alignment of FsrA, FsrB, and FsrC with AgrA, AgrB, and AgrC from S. aureus. Identical amino acids are indicated by a symbol as “|”, and similar amino acids are indicated by a symbol as “:” or “.” between them. Conserved residues in H, N, and G2 blocks of histidine protein kinases and conserved Asp and Lys residues in response regulators of bacterial two-component systems (20) are boxed. The sequences from S. aureus were as follows: AgrA, accession number M21854 (14); AgrB, accession number AF001782 (5); and AgrC, accession number AF001783 (5).
Disruption of fsr genes and sprE and detection of gelatinase and serine protease activities.
E. faecalis serine protease and agr-like genes were disrupted by transforming OG1RF with internal fragments of the target genes in suicide vector pTEX4577 using selection for Kan resistance. All the Kanr transformants analyzed by Southern blot analysis were found to have the correct insertions in the target genes (data not shown). The resulting mutants are listed in Table 1.
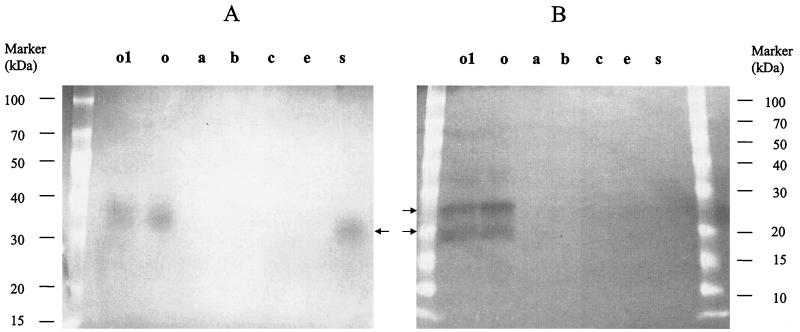
Gelatinase activity was evidenced on TH agar plates containing 3% gelatin and on a gelatin zymogram gel for the parent strain OG1RF and the sprE and orf1 mutants (TX5243 and TX5248) (Fig. 3A), indicating that the disruption of sprE and orf1 did not effect gelE expression. However, fsrA, fsrB, and fsrC mutants (TX5240, TX5241, and TX5242) did not show detectable gelatinase activity either on gelatin containing TH agar plates or on a gelatin zymogram gel (Fig. 3A). Similarly, serine protease activity was not detected for agr-like gene mutants (Fig. 3B), while OG1RF and the orf1 mutant had two lysis bands (26 and 20 kDa) on a casein zymogram gel, suggesting a possible regulatory effect or polar effect of fsrA, fsrB, and fsrC on the expression of gelE and sprE. Neither the sprE (TX4243) mutant nor the previously described gelE mutant (19) showed serine protease activity on a casein zymogram gel (Fig. 3B), suggesting that the insertion in gelE has a polar effect on sprE expression. The molecular mass of 26 kDa of the higher band shown on the casein gel for OG1RF matches the predicted size of the product of sprE. The lower band may be the mature or degraded form of the protease.
FIG. 3.
Gelatinase and serine protease activities in fsr and sprE mutants. These pictures are the negative images of gelatin and casein zymogram gels. (A) Gelatinase activity of mutants on gelatin zymogram gel. (B) Serine protease activity of mutants on casein zymogram gel. Lanes: o1, mutant with disruption of orf1; o, OG1RF; a, mutant with disruption of fsrA; b, mutant with disruption of fsrB; c, mutant with disruption of fsrC; e, mutant with disruption of gelE; s, mutant with disruption of sprE.
Two revertants from the fsrC mutant detected by scoring for restoration of gelatinase activity were found to be sensitive to kanamycin and to have restored the gelatinase activity. One of them was subsequently used in the animal experiment.
Northern blot analysis and RT-PCR.
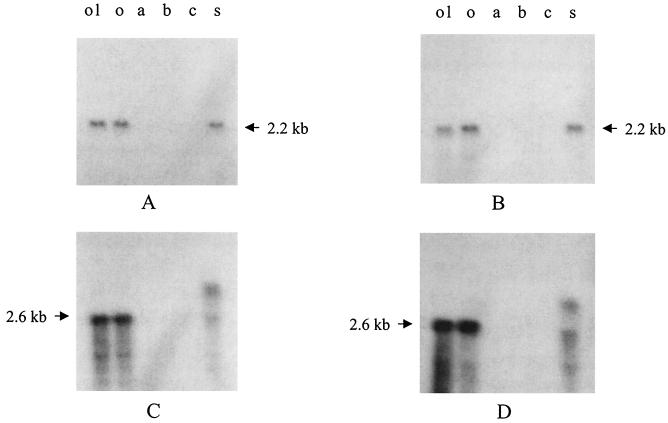
Northern blot analysis was conducted to investigate whether the loss of gelatinase activity in the fsr gene mutants was caused by polar effects. Results of blots of total RNA from TX5240, TX5241, TX5242, TX5243, TX5248, TX5128, and OG1RF hybridized with internal fragments of fsrA, fsrB, fsrC, gelE, sprE, and orf1 are summarized in Fig. 1B. Hybridization using fsrB and fsrC probes gave a 2.2-kb band for wild-type OG1RF, gelE, sprE, and orf1 mutants (Fig. 4, data not shown for the gelE mutant), suggesting that fsrB (729 bp) and fsrC (1,344 bp) are cotranscribed and that disruption of orf1, gelE, and sprE does not effect the expression of fsrB and fsrC. Interestingly, expression of fsrB and fsrC was not detectable in fsrA, fsrB, or fsrC mutants (Fig. 4), suggesting autoregulation of fsr genes. A Northern blot was not able to detect expression of fsrA or orf1 in either OG1RF or fsr gene mutants. However, RT-PCR showed that fsrA was expressed in OG1RF (data not shown), suggesting that fsrA is expressed at a low level, which is different from fsrB and fsrC.
FIG. 4.
Northern blot analysis of fsr and sprE mutants. (A, B, C, and D) Northern blots of mutants using fsrB, fsrC, gelE, and sprE probes, respectively. Lanes: o1, mutant with disruption of orf1; o, OG1RF; a, mutant with disruption of fsrA; b, mutant with disruption of fsrB; c, mutant with disruption of fsrC; s, mutant with disruption of sprE.
Northern blot analysis using gelE and sprE probes gave a single 2.6-kb band for OG1RF (Fig. 4), which is approximately 200 bp greater than the sum of sizes of gelE (1531 bp) and sprE (855 bp), suggesting that gelE and sprE are cotranscribed. The gelE and sprE mRNA was not detectable in fsrA, fsrB, and fsrC mutants (Fig. 4), suggesting that these genes may regulate the expression of gelE and/or sprE. The gelE probe hybridized with two bands in the gelE and sprE mutants (1.3 and 3.8 kb and 2.3 and 4.0 kb, respectively) (Fig. 4, data not shown for the gelE mutant). The lower bands were the truncated form of the gelE transcript since it only hybridized to the gelE probe but not to the kanamycin resistance gene probe from the suicide vector (data not shown), while the higher band was the fusion of the truncated transcript and the readthrough to the insertion sequence since it also hybridized with the kanamycin resistance gene (data not shown). Similarly, the sprE probe hybridized with two bands (2.3 and 4.0 kb) in the sprE mutant (Fig. 4). No sprE mRNA was detected in the gelE mutant (data not shown), indicating a polar effect on sprE by insertion in gelE.
Complementation of agr-like genes.
To further confirm that abolishing gelE and sprE expression in fsr gene mutants was due to a regulatory effect, not a polar effect, complementation was carried out using a 6-kb PstI/BglII fragment in shuttle vector pAT18 (pTEX5249) containing fsrA, fsrB, and fsrC genes. fsrA, fsrB, and fsrC mutants which harbored plasmid pTEX5249 (TX5244, TX5245, and TX5246) all produced gelatinase activity as detected by TH agar containing 3% gelatin (data not shown), indicating that fsr gene function can be provided in trans. Northern blot analysis also showed that fsrB and sprE were expressed in complemented fsr gene mutants (data not shown). No gelatinase activity was detected in the gelE mutant harboring pTEX5249 (TX5247). These results further confirm that fsr genes positively regulate the expression of the gelE and sprE genes.
Animal model and growth curves.
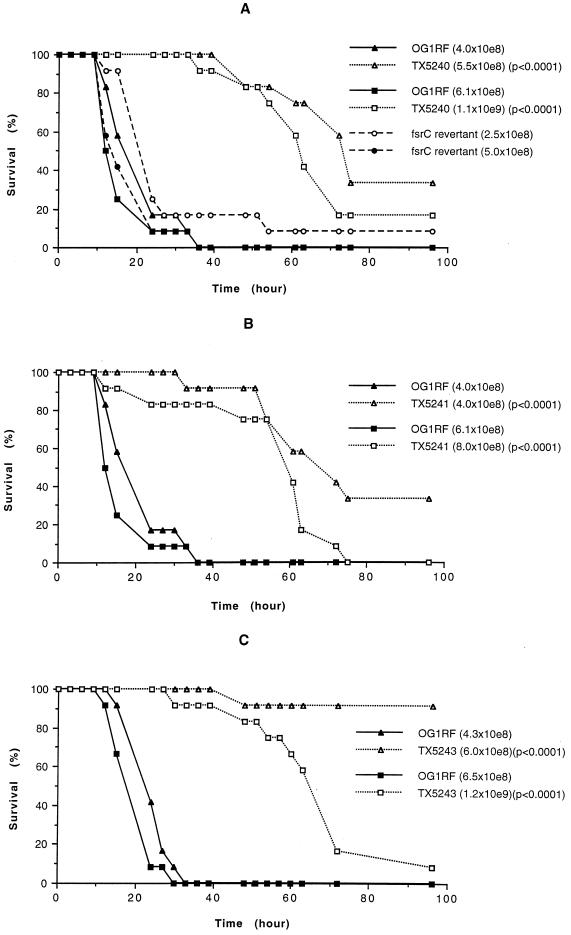
Using a mouse peritonitis model, we studied the virulence of fsr and sprE mutants in comparison to OG1RF. In general, the mutants had a similar 50% lethal dose to the parent strain OG1RF. The time course of survival for TX5248 was also similar to that of OG1RF (data not shown), suggesting that orf1 does not play an important role in enterococcal infection in this animal model. However, the time courses of survival for TX5240 (fsrA mutant), TX5241 (fsrB mutant), and TX5243 (sprE mutant) were significantly prolonged compared to the parent strain OG1RF (e.g., P < 0.0001 by log rank for survival at the inocula of 5.5 × 108 CFU and 4.0 × 108 of TX5240 and TX5241 versus 4.0 × 108 CFU of OG1RF [n = 12]; P < 0.0001 at the inocula of 1.1 × 109 and 8.0 × 108 of TX5240, TX5241 versus 6.1 × 108 of OG1RF [n = 12]; P < 0.0001 at the inocula of 6.0 × 108 and 1.2 × 109 CFU of TX5243 versus 4.3 × 108 and 6.5 × 108 of OG1RF [n = 12]) (Fig. 5). The same highly significant difference was also noted for the fsrC revertant compared to the mutants. The delays in lethality were also seen when groups of six animals were used in another trial (data not shown). In separate experiments testing TX5242 (fsrC mutant), there was also a trend to delayed death with groups of six mice (P = 0.0787, 0.0569, and 0.0835 at inocula of 6.2 × 108, 1.2 × 109, and 2.5 × 109 of TX5242 versus 5.7 × 108, 1.1 × 109, and 2.3 × 109 of OG1RF, respectively); when a single inoculum was tested with 20 mice, the delay was significant (P = 0.0040), but the inoculum of OG1RF was slightly higher than TX5242 (4.2 × 109 of TX5242 versus 4.8 × 109 of OG1RF) (data not shown). Because all results for the fsrC mutant showed a trend similar to fsrA and fsrB mutants, these results were not repeated so that additional animals would not be subjected to this lethality assay. The revertant from TX5242 (fsrC mutant) whose gelatinase activity was restored was also used as a control in the experiments and the time course to death for the revertant was the same as OG1RF (Fig. 5), indicating that the prolonged survival was not caused by some other mutation that may have occurred during the generation of the mutants.
FIG. 5.
Kaplan-Meier survival plots of wild-type OG1RF and fsr and sprE mutants in the mouse peritonitis model. (A) Kaplan-Meier survival plots of OG1RF, the fsrA mutant (TX5240) and an fsrC revertant. (B) OG1RF and the fsrB mutant (TX5241). (C) OG1RF and the sprE mutant (TX5243). Twelve mice were tested with each inoculum of each of the strains shown. The P value refers to mutant versus OG1RF at the same or a smaller inoculum (▴ versus ▵ or ■ versus □).
To make certain that the delay in death in the animal model was not caused by a growth defect of the mutants, we determined the growth rates of fsr and sprE mutants in comparison to that of OG1RF. The growth rates of the mutants were the same as that of OG1RF (data not shown), which further suggests that agr-like genes and sprE are virulence factors.
Comparison of colony hybridization results with gelatinase phenotype.
Lysates of 152 E. faecalis isolates were hybridized with fsrB, gelE, and sprE probes. A total of 105 isolates (∼69%) were positive with the fsrB probe, while 141 (∼93%) isolates were positive with gelE and sprE probes, respectively. All isolates that hybridized with the fsrB probe were also gelE and sprE probe positive, and gelE and sprE were both found in the same isolates. Based on these and previous results (4) and retesting of 95 isolates for gelatinase activity and 32 isolates (from the 95 isolates) for the presence of fsrB and gelE by PCR, 62% (59 of 95), 71% (67 of 95), and 91% (86 of 95) were gelatinase, fsrB, and gelE positive, respectively. No isolates lacking gelE or fsrB produced gelatinase, but eight isolates which hybridized to both gelE and fsrB probes were gelatinase negative.
DISCUSSION
The agr loci in S. aureus (11, 12, 14), S. epidermidis (13, 24), and S. ludgunensis (25) contain five genes (agrA, agrB, agrD, agrC, and hld), whose sequences and organization are similar to each other. The three agr-like genes (fsrA, fsrB, and fsrC) that we found in E. faecalis OG1RF show significant homology to three agr genes (agrA, agrB, and agrC) in staphylococci, although considerably less than is seen among the staphylococcal homologs. FsrA and FsrC resemble response regulators and sensor transducers of bacterial two-component systems, respectively. FsrC was found to have the greatest similarity to AgrC of staphylococci, although FsrA showed greater similarity to some other response regulators than to AgrA. The organization of the fsr genes in E. faecalis is also different. That is, fsrA (the agrA analog) is upstream of fsrB and fsrC (the agrB and agrC analogs) in E. faecalis instead of downstream of these genes as in staphylococci.
In S. aureus, agrB, agrD, agrC, and agrA, which are autoregulated, are under the control of a promoter designated P2 and agrA is also transcribed individually from a different promoter, P1 (11, 12, 14, 24). Our Northern blot and RT-PCR analyses suggest that fsrB and fsrC are cotranscribed and that fsrA may be transcribed from a promoter different from that of fsrB and fsrC, since its expression level was much lower than that of fsrB and fsrC. Northern blot analysis of fsrA, fsrB, and fsrC mutants suggests that the fsr genes in E. faecalis may also be autoregulated since insertion in fsrC abolished the expression of fsrB, which is upstream of fsrC, and disruption of fsrA also terminated the expression of fsrB and fsrC, although the latter two are on a transcript that does not contain fsrA. A transcript, RNAIII, which is transcribed from hld upstream of agrB and encodes delta-hemolysin in the agr/hld locus in S. aureus has been demonstrated to be the effector for the Agr response (12). Insertion mutagenesis of gelE, sprE, and orf1 in flanking regions of the fsr locus and Northern blot analysis of the mutants indicate that the transcripts of fsr flanking regions are not responsible for the autoregulated function of the locus since fsrB and fsrC were both expressed in these mutants as detected by Northern blot. In addition, no agrD or hld homolog was found by search in the E. faecalis genome database. In different groups of S. aureus, which can be divided based on their cross-activation and cross-inhibition of the Agr response (5), the agrD homologs (which encode the precursor of an autoinducing peptide of these different groups) show very limited sequence resemblance, suggesting that the peptide inducer of E. faecalis would also be different from those of S. aureus. Sequence analysis of the putative FsrB of E. faecalis and AgrB of S. aureus reveals that FsrB is about 50 amino acids longer than AgrB and that ca. 50 amino acids at the C terminus of FsrB do not show sequence homology to AgrB. This raises the possibility that this portion of FsrB, which is approximately the size of the autoinducing peptide precursor encoded by agrD in staphylococci, could be the precursor of an autoinducing peptide in E. faecalis OG1RF if, indeed, there is a inducer in E. faecalis.
We have demonstrated here that the fsr locus in E. faecalis OG1RF positively regulates the expression of gelE and sprE, since the expression of both gelE and sprE was abolished in fsrA, fsrB, and fsrC mutants as detected by Northern blot analysis. Gelatinase and serine protease activities were also undetectable in the fsr mutants. Complementation experiments indicate that the elimination of gelE and sprE expression was not due to a polar effect of the insertions in fsr mutants since the expression of gelE and sprE could be restored by providing the fsr genes in trans. These results suggest that the fsr locus in E. faecalis may have a regulator function similar to that of the agr locus in S. aureus, which positively regulates the expression of secreted proteins. We have not yet tested the effect of the insertions in the fsr genes on other surface or secreted proteins in E. faecalis.
In a previous study, we had shown that an insertion mutant of gelE had a significantly delayed time to death in a mouse peritonitis model (19), which suggests that gelE or gene(s) downstream of gelE are important for this aspect of the infection. The fact that expression of sprE was not detected in the gelE mutant and that the gelatinase-positive mutant with an insertion in sprE also showed delayed killing indicates that sprE is important in the infection in this animal model. However, we still do not know if gelE independently influences the outcome of this enterococcal infection. Testing of a gelE deletion mutant may provide further information to address this question. In S. aureus, an agr mutant has been shown to have a significant effect on infection compared to the wild-type strain as shown by a murine arthritis model (1). We found that insertions in the fsr genes also caused significant delays in killing in the mouse peritonitis model and that the pattern of delay was similar to that of gelE and sprE mutants. Since the agr locus in S. aureus controls the expression of several virulence properties, some of which are activated while some are inhibited (8), the effects of agr mutants in animal models may not be attributable to a simple activation or inhibition effect. Further study is needed to address whether the fsr genes in E. faecalis also regulate other (virulence) factors besides gelE and sprE and if regulation occurs in a fashion similar to that of S. aureus agr system.
It was interesting to note that the presence of fsrB was lower than that of gelE and sprE in our survey of 152 clinical isolates of E. faecalis (69% versus 93 and 93%). Of 95 isolates retested for gelatinase in this study, 59 (62%) were gelatinase production positive and all 59 were fsrB positive, a finding consistent with the genetic data obtained with strain OG1RF, indicating that fsr genes are also required for the expression of gelE gene in E. faecalis clinical isolates. No isolates lacking gelE produced detectable gelatinase. Whether expression of gelE can be elicited in fsr lacking strains under other conditions has yet to be determined.
In conclusion, E. faecalis fsr genes positively regulate the expression of gelE and sprE which are important for enterococcal infection in the mouse peritonitis model and these fsr genes appear to be autoregulated.
ACKNOWLEDGMENT
This work was supported National Institutes of Health grant AI33516 from the Division of Microbiology and Infectious Diseases, National Institute of Allergy and Infectious Diseases, to B. E. Murray.
REFERENCES
- 1.Abdelnour A, Arvidson S, Bremell T, Ryden C, Tarkowski A. The accessory gene regulator (agr) controls Staphylococcus aureus virulence in a murine arthritis model. Infect Immun. 1993;61:3879–3885. doi: 10.1128/iai.61.9.3879-3885.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beliveau C, Potvin C, Trudel J, Asselin A, Bellemare G. Cloning, sequencing, and expression in Escherichia coli of a Streptococcus faecalis autolysin. J Bacteriol. 1991;173:5619–5623. doi: 10.1128/jb.173.18.5619-5623.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coque T M, Patterson J E, Steckelberg J M, Murray B E. Incidence of hemolysin, gelatinase and aggregative substance among enterococci isolated from patients with endocarditis and other infections and from feces of hospitalized and community-based persons. J Infect Dis. 1995;171:1223–1229. doi: 10.1093/infdis/171.5.1223. [DOI] [PubMed] [Google Scholar]
- 5.Ji G, Beavis R, Novick R P. Bacterial interference caused by autoinducing variants. Science. 1997;276:2027–2030. doi: 10.1126/science.276.5321.2027. [DOI] [PubMed] [Google Scholar]
- 6.Ji G, Beavis R C, Novick R P. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc Natl Acad Sci USA. 1995;92:12055–12059. doi: 10.1073/pnas.92.26.12055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li X, Weinstock G M, Murray B E. Generation of auxotrophic mutants of Enterococcus faecalis. J Bacteriol. 1995;177:6866–6873. doi: 10.1128/jb.177.23.6866-6873.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lindberg M, Jonsson K, Muller H, Jonsson H, Signas C, Hook M, Raja R, Raucci G, Anantharamaiah G M. Fibronectin-binding proteins in Staphylococcus aureus. In: Novick R P, editor. Molecular biology of the staphylococci. New York, N.Y: VCH; 1990. pp. 343–356. [Google Scholar]
- 9.Murray B E. The life and times of the enterococcus. Clin Microbiol Rev. 1990;3:46–65. doi: 10.1128/cmr.3.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murray B E, Singh K V, Ross R P, Heath J D, Dunny G M, Weinstock G M. Generation of restriction map of Enterococcus faecalis strain OG1 and investigation of growth requirements and regions encoding biosynthetic function. J Bacteriol. 1993;175:5216–5223. doi: 10.1128/jb.175.16.5216-5223.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Novick R P, Projan S J, Kornblum J, Ross H F, Ji G, Kreiswirth B, Vandenesch F, Moghazeh S. The agr P2 operon: an autocatalytic sensory transduction system in Staphylococcus aureus. Mol Gen Genet. 1995;248:446–458. doi: 10.1007/BF02191645. [DOI] [PubMed] [Google Scholar]
- 12.Novick R P, Ross H F, Projan S J, Kornblum J, Kreiswirth B, Moghazeh S. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 1993;12:3967–3975. doi: 10.1002/j.1460-2075.1993.tb06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Otto M, SuBmuth R, Jung G, Gotz F. Structure of the pheromone peptide of the Staphylococcus epidermidis agr system. FEBS Lett. 1998;424:89–94. doi: 10.1016/s0014-5793(98)00145-8. [DOI] [PubMed] [Google Scholar]
- 14.Peng H-L, Novick R P, Kreiswirth B, Kornblum J, Schlievert P. Cloning, characterization, and sequencing of an accessory gene regulator (agr) in Staphylococcus aureus. J Bacteriol. 1988;170:4365–4372. doi: 10.1128/jb.170.9.4365-4372.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prober J M, Trainor G L, Dam R J, Hobbs F W, Robertson C W, Zagursky R J, Cocuzza A J, Jensen M A, Baumeister K. A system for rapid DNA sequencing with fluorescent chain-terminating dideoxynucleotides. Science. 1987;238:336–341. doi: 10.1126/science.2443975. [DOI] [PubMed] [Google Scholar]
- 16.Recsei P, Kreiswirth B, O'Reilly M, Schlievert P, Gruss A, Novick R P. Regulation of exoprotein gene expression by agr. Mol Gen Genet. 1985;202:58–61. doi: 10.1007/BF00330517. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 18.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh K V, Qin X, Weinstock G M, Murray B E. Generation and testing of mutants of Enterococcus faecalis in a mouse peritonitis model. J Infect Dis. 1998;178:1416–1420. doi: 10.1086/314453. [DOI] [PubMed] [Google Scholar]
- 20.Stock J B, Ninfa A J, Stock A M. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol Rev. 1989;53:450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Su Y A, Sulavik M C, He P, Makinen K K, Makinen P-L, Fiedler S, Wirth R, Clewell D B. Nucleotide sequence of the gelatinase gene (gelE) from Enterococcus faecalis subsp. liquefaciens. Infect Immun. 1991;59:415–420. doi: 10.1128/iai.59.1.415-420.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tegmark K, Morfeldt E, Arvidson S. Regulation of agr-dependent virulence genes in Staphylococcus aureus by RNAIII from coagulase-negative staphylococci. J Bacteriol. 1998;180:3181–3186. doi: 10.1128/jb.180.12.3181-3186.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trieu-Cuot P, Carlier C, Poyart-Salmeron C, Courvalin P. Shuttle vectors containing a multiple cloning site and a lacZα gene for conjugal transfer of DNA from Escherichia coli to gram-positive bacteria. Gene. 1991;102:99–104. doi: 10.1016/0378-1119(91)90546-n. [DOI] [PubMed] [Google Scholar]
- 24.Van Wamel W J, Van Rossum G, Verhoef J, Vandenbroucke-Grauls C M, Fluit A C. Cloning and characterization of an accessory gene regulator (agr)-like locus from Staphylococcus epidermidis. FEMS Microbiol Lett. 1998;163:1–9. doi: 10.1111/j.1574-6968.1998.tb13018.x. [DOI] [PubMed] [Google Scholar]
- 25.Vandenesch F, Projan S, Kreiswirth B, Etienne J, Novick R P. agr-related sequences in Staphylococcus ludunensis. FEMS Microbiol Lett. 1993;111:115–122. doi: 10.1111/j.1574-6968.1993.tb06370.x. [DOI] [PubMed] [Google Scholar]
- 26.Wilson K. Current protocols in molecular biology. Brooklyn, N.Y: Green Publishing Associates; 1990. pp. 2.4.1–2.4.2. [Google Scholar]