ABSTRACT
Members of the Campylobacter lari group are causative agents of human gastroenteritis and are frequently found in shellfish, marine waters, shorebirds, and marine mammals. Within a One Health context, we used comparative genomics to characterize isolates from a diverse range of sources and geographical locations within Europe and Australia and assess possible transmission of food, animal, and environmental isolates to the human host. A total of 158 C. lari isolates from Australia, Denmark, France, and Germany, which included 82 isolates from human stool and blood, 12 from food, 14 from domestic animal, 19 from waterbirds, and 31 from the environment were analyzed. Genome-wide analysis of the genetic diversity, virulence, and antimicrobial resistance (AMR) traits was carried-out. Most of the isolates belonged to C. lari subsp. lari (Cll; 98, 62.0%), while C. lari subsp. concheus and C. lari urease-positive thermotolerant Campylobacter (UPTC) were represented by 12 (7.6%) and 15 (9.5%) isolates, respectively. Furthermore, 33 (20.9%) isolates were not assigned a subspecies and were thus attributed to distant Campylobacter spp. clades. Whole-genome sequence-derived multilocus sequence typing (MLST) and core-genome MLST (cgMLST) analyses revealed a high genetic diversity with 97 sequence types (STs), including 60 novel STs and 14 cgMLST clusters (≤10 allele differences), respectively. The most prevalent STs were ST-21, ST-70, ST-24, and ST-58 (accounting for 13.3%, 4.4%, 3.8%, and 3.2% of isolates, respectively). A high prevalence of the 125 examined virulence-related loci (from 76.8 to 98.4% per isolate) was observed, especially in Cll isolates, suggesting a probable human pathogenicity of these strains.
IMPORTANCE Currently, relatedness between bacterial isolates impacting human health is easily monitored by molecular typing methods. These approaches rely on discrete loci or whole-genome sequence (WGS) analyses. Campylobacter lari is an emergent human pathogen isolated from diverse ecological niches, including fecal material from humans and animals, aquatic environments, and seafood. The presence of C. lari in such diverse sources underlines the importance of adopting an integrated One Health approach in studying C. lari population structure for conducting epidemiological risk assessment. This retrospective study presents a comparative genomics analysis of C. lari isolates retrieved from two different continents (Europe and Australia) and from different sources (human, domestic animals, waterbirds, food, and environment). It was designed to improve knowledge regarding C. lari ecology and pathogenicity, important for developing effective surveillance and disease prevention strategies.
KEYWORDS: Campylobacter lari group, whole-genome sequencing, virulence genes, genomic diversity, One Health
INTRODUCTION
Campylobacter is a bacterial genus belonging to the epsilon subdivision of the Proteobacteria, with some species being the leading cause of bacterial foodborne diarrheal disease worldwide. In the European Union, 246,000 human cases of campylobacteriosis are reported annually, mainly caused by thermotolerant Campylobacter such as Campylobacter jejuni and Campylobacter coli, and less frequently by Campylobacter lari (1). C. jejuni is responsible for diarrhea (sometimes bloody), abdominal pain, fever, and occasionally vomiting. C. coli causes the same disease but with a lower frequency (2). C. lari is associated with sporadic gastrointestinal infections (3), related to waterborne outbreaks (4), and to bacteremia especially in immunocompromised individuals (2, 5, 6). C. lari occasionally causes purulent pleurisy, reactive arthritis, prosthetic and urinary tract infections, and vertebral osteomyelitis (7, 8).
In the United States, an analysis of 16,549 culture-confirmed Campylobacter infections between 2010 and 2015 within the Foodborne Diseases Active Surveillance Network (FoodNet, CDC) showed that C. lari was the fourth most frequently identified species (0.6%) after C. jejuni, C. coli, and Campylobacter upsaliensis (5). C. lari infections were more prevalent in patients aged older than 40 years during the autumn and winter (5).
Campylobacter species, such as C. lari, Campylobacter concisus, Campylobacter ureolyticus, C. upsaliensis, and Campylobacter fetus are considered “emerging species” due to their inadequately understood roles in human and animal diseases (9). The clinical importance and pathogenicity of the emerging Campylobacter species have been reviewed by Costa and Iraola (10). Diagnostic laboratories may fail to detect emerging Campylobacter species owing to greater difficulties in culturing them. This is potentiated by the use of so-called syndromic PCRs that may or may not be followed by culture. These PCR tests detect C. jejuni and C. coli almost exclusively, and rarely C. upsaliensis or C. lari (11, 12). Furthermore, mass spectrometry methods like MALDI-TOF may not differentiate some of the less common species, especially those arising from environmental sources (13).
The C. lari group is composed of seven species (C. lari, Campylobacter insulaenigrae, Campylobacter volucris, Campylobacter subantarcticus, Campylobacter peloridis, Campylobacter ornithocola, and Campylobacter armoricus), two subspecies (C. lari subsp. lari [Cll] and C. lari subsp. concheus [Clc]), urease-positive thermophilic Campylobacter (UPTC), and other C. lari-like strains (14–17).
Although the C. lari group is a phylogenetically distinct clade within the genus Campylobacter, taxa within this clade are highly related (15). In fact, 70% of Cll RM2100 genes were conserved among the C. lari group species and UPTC strains (15, 18). However, the C. lari group encompasses the nalidixic acid-susceptible Campylobacter (NASC) group, nalidixic acid-resistant thermophilic Campylobacter, urease-positive thermophilic Campylobacter, and urease-producing NASC (7). C. lari UPTC isolates can be distinguished by their urease production in contrast to urease-negative strains such as Cll and Clc. The latter can be differentiated from Cll by its inability to grow on media containing 0.05% safranin (14). In addition, multiple auxotrophic phenotypes are a general feature of the C. lari group (15).
More effective detection methods and additional investigations are required to better understand how emerging Campylobacter species, including members of the C. lari group, evolve in the environment, spread through agri-food systems, and contribute to campylobacteriosis (9, 19). An integrated One Health approach in Campylobacter epidemiology and risk assessment is needed (20, 21). In contrast to C. jejuni and C. coli, enriching collections of C. lari strains from diverse sources is challenging due to the lack of selective culturing methods and their generally low prevalence. Therefore, there are limited studies on C. lari populations, particularly in different geographical regions.
Members of the C. lari group are typically isolated from similar environments, mainly coastal areas and related watersheds, shellfish, marine waters, and freshwaters and hosts including shorebirds and marine mammals (6, 15, 22). They may be also isolated from domestic animals such as poultry, dogs, cats, cattle, pigs, and sheep (6).
As with other thermotolerant Campylobacter spp., members of the C. lari group have their optimal growth at 42°C. The gastrointestinal tract of avian species whose gut temperatures are around 41 to 42°C provide optimal growth conditions (23). C. lari does not multiply in the environment outside its host. However, low environment temperatures seem to increase its survival. In France, C. lari was found to be the most frequently isolated Campylobacter species in shellfish (n = 237) from three shellfish-harvesting areas with 26.4% of the samples positive for this species versus 0.8% for C. jejuni, 2.9% for C. coli, and 1.3% for C. peloridis. Additionally, more C. lari-positive samples were observed in the autumn and winter with temperatures mainly under 15°C (22). Similarly, in Spain, C. lari was isolated in shellfish (0.07%) only in the cooler months (February and March) where water temperatures were approximately 12°C (24).
A multilocus sequence typing (MLST) scheme based on seven loci (aspA, glnA, gltA, glyA, pgm, tkt, and uncA) was used for C. jejuni and C. coli to characterize and investigate their population structure. A total of 11,899 sequence types (STs) were described for these species (https://pubmlst.org/organisms/campylobacter-jejunicoli; accessed 27 July 2022). An MLST scheme based on seven loci (adk, atpA, glnA, pgi, glyA, pgm, and tkt) was reported to both differentiate C. lari strains and identify clonal lineages (25). The non-jejuni/coli Campylobacter PubMLST database (https://pubmlst.org/bigsdb?db=pubmlst_campylobacter_nonjejuni_isolates; accessed 27 July 2022) included 429 C. lari isolates and 311 STs (including isolates from the present study). The database entries originated mainly from Europe (77.9%), especially from France (58.0%), followed by Antarctica (6.8%), South Africa (5.4%), Australia (4.2%), and Canada (3.5%). They included mainly shellfish (33.1%), animal (32.2%, especially waterbirds), human (18.2%), and environmental water (14.4%) isolates.
C. lari group pathogenicity is poorly documented. While many studies have focused on the virulence factors of C. jejuni and C. coli, there are few studies on C. lari (15, 18). However, many virulence and antibiotic resistance mechanisms present in C. jejuni were also conserved in members of the C. lari group (15, 18, 26).
Lately, the rapid development of next-generation sequencing (NGS) technologies enhanced our ability to sequence the complete genomes of members of the C. lari group and made it possible to better investigate the presence of virulence factor genes (15, 18, 26, 27).
In this study, we aimed to include a large number of C. lari isolates from different sources and geographic locations. Our objectives were to: (i) compare genomic data of a selection of C. lari isolates to evaluate their genetic diversity, (ii) identify potential reservoirs for transmission to humans, and finally (iii) assess the potential pathogenicity of these isolates by looking for the presence of virulence genes. Genomic data were found to facilitate high-level discriminatory subtyping for the identification of genetic relationships between Campylobacter isolates and potential transmission clusters (28).
RESULTS
General features of the sequenced genomes.
The genome sizes of the 158 isolates ranged from 1.344 Mb to 1.670 Mb. The G+C content ranged from 29.3% to 29.4% (see Table S1A in the supplemental material). In these isolates, 1,366 to 1,694 coding sequences (CDSs) were identified, excluding those CDSs containing prophages (Table S1A).
Taxonomic identification of isolates, using average nucleotide identity (ANI) and in silico DNA-DNA hybridization (isDDH) analyses.
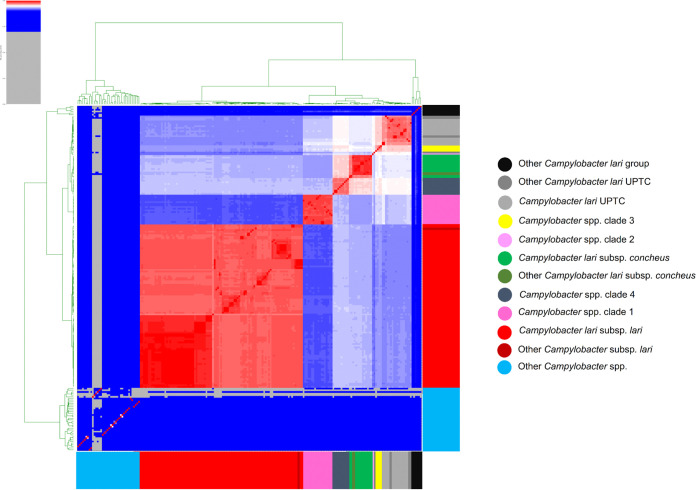
Most of the isolates belonged to C. lari subsp. lari (98 Cll; 62.0%; Fig. 1 and 2; Table S2). The other isolates were identified as C. lari subsp. concheus (12 Clc; 7.6%) and C. lari UPTC (15; 9.5%). They were found in the same groups of known and reference type strains of Cll, Clc, and C. lari UPTC, respectively.
FIG 1.
Heatmap of pairwise average nucleotide identity (ANI) values for 158 whole-genome-sequenced C. lari group and other Campylobacter strains. The ANI was calculated using the pyani program after blastn alignment. Only regions present in all genomes were used in the ANI calculation. Values range from 0 (0%) ANI to 1 (100% ANI): Gray represents 0% ANI and clusters of highly similar isolates are highlighted in red. The dendrogram directly reflects the degree of identity between genomes. An ANI above 96% between two genomes is an indication that they belong to the same species and the colored branches represent different clusters. Other Campylobacter lari group: Campylobacter armoricus CA656T, Campylobacter peloridis LMG 23910T, Campylobacter subantarcticus LMG 24377T, Campylobacter ornithocola WBE38T, Campylobacter volucris LMG24380T, and Campylobacter insulaenigrae NCTC 12927T. Other Campylobacter lari UPTC: Campylobacter lari UPTC RM16712, Campylobacter lari UPTC RM16701, Campylobacter lari UPTC 22395, and Campylobacter lari UPTC 11845. Other Campylobacter lari subsp concheus: Campylobacter lari subsp concheus LMG 11760 and Campylobacter lari subsp concheus LMG21009T. Other Campylobacter lari subsp lari: Campylobacter lari subsp lari ATCC 35221T. Other Campylobacter spp: Campylobacter showae ATCC 51146T, Campylobacter concisus ATCC 33237T, Campylobacter hyointestinalis subsp hyointestinalis ATCC 35217T, Campylobacter helveticus ATCC 51209T, Campylobacter avium LMG 24591T, Campylobacter aviculae MIT17-670T, Campylobacter jejuni subsp. jejuni ATCC 33560T, Campylobacter jejuni subsp. doylei LMG 8843T, Campylobacter ureolyticus DSM 20703T, Campylobacter massiliensis Marseille-Q3452T, Campylobacter pinnipediorum subsp. caledonicus LMG 29473T, Campylobacter hominis ATCC BAA381T, Campylobacter corcagiensis LMG 27932T, Campylobacter fetus subsp testudinum ATCC BAA2539T, Campylobacter taeniopygiae MIT10-5678T, Campylobacter blaseri LMG 30333T, Campylobacter cuniculorum LMG 24588T, Campylobacter sputorum bv sputorum LMG7795T, Campylobacter coli ATCC 33559T, Campylobacter portucalensis FMV-PI01T, Campylobacter curvus ATCC 35224T, Campylobacter hyointestinalis subsp lawsonii LMG 14432T, Campylobacter lanienae NCTC 13004T, Campylobacter novaezeelandiae B423bT, Campylobacter rectus ATCC 33238T, Campylobacter gracilis ATCC 33236T, Campylobacter iguaniorum 1485ET, Campylobacter pinnipediorum subsp pinnipediorum LMG 29472T, Campylobacter vulpis 251-13T, Campylobacter anatolicus faydin-G140T, Campylobacter mucosalis ATCC 43264T, Campylobacter fetus subsp fetus ATCC 27374T, Campylobacter upsaliensis ATCC 43954T, Campylobacter estrildidarum MIT17-644T, Campylobacter geochelonis LMG 29375T, Campylobacter canadensis LMG24001T, and Campylobacter hepaticus NCTC 13823T.
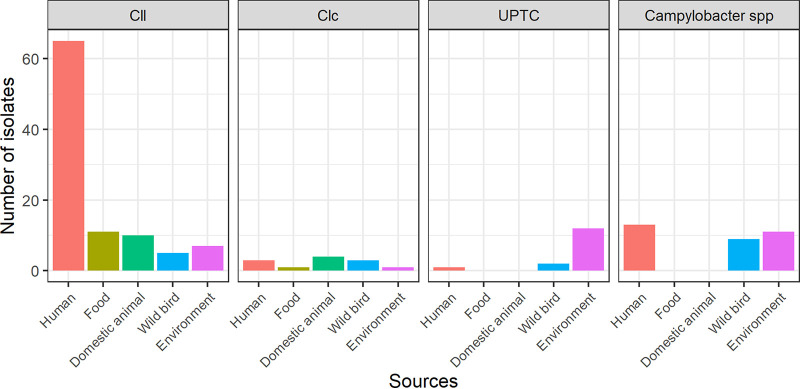
FIG 2.
Distribution of Campylobacter lari subspecies according to the sources. Cll, C. lari subsp. lari; Clc, C. lari subsp. concheus; UPTC, C. lari UPTC.
The remaining 33 isolates could not be identified to the species/subspecies level (i.e., ANI and isDDH values <96% and <70%, respectively) and were designated Campylobacter spp. (20.9%; Table S2). These isolates were confirmed to belong to the C. lari group, harboring the seven MLST genes from the C. lari scheme, with new STs that were uploaded in the non-jejuni/coli Campylobacter PubMLST database. These isolates were divided into four distinct clades probably representing novel members of the C. lari group. Three of them were closely related to the C. lari UPTC group with clade 1 formed by a homogeneous group of 18 isolates. The second clade was represented by a unique isolate (H42), while clade 3 included four isolates. Clade 4 was represented by 10 isolates close to Clc, all of human, environmental, or waterbird origin (Fig. 1; Table S1 and S2).
The source of the isolate was found to have a significant impact on the C. lari subspecies affiliation by ANI and isDDH (chi-square test, P < 0.001). However, no correlation between the affiliation to a Campylobacter subspecies and the period and country of isolation was observed (chi-square test, P = 0.014 and P = 0.0830, respectively).
Cll was the most frequent C. lari subspecies among human (79.3%), domestic animal (71.4%), and food (91.7%) isolates, whereas only 22.6% of environmental and 26.3% of waterbird isolates belonged to this subspecies (Fig. 2). Clc isolates were mainly of animal origin (four domestic animal and three waterbird isolates) and of human origin (three isolates). Only one isolate was of food origin and another from the environment (Fig. 2). Finally, C. lari UPTC isolates were mainly from the environment (13 out of the 15) and, to a lesser extent, from waterbirds (two isolates; Fig. 2).
Human isolates from France (n = 43) and Denmark (n = 15) belonged mainly to patients older than 40 years with an equivalent sex ratio and periods of isolation during the whole year regardless of the seasons (Table 1). These isolates belonged mainly to Cll (79.3%), while three isolates belonged to Clc, and one belonged to C. lari UPTC.
TABLE 1.
Description of the collection of Campylobacter lari group isolatesa
| Country | Source | Animal group | Types of samples | Period of collection | Human patients No. (%) of male patients season (sp/su/f/w)* | Median age (min; max) | No. of isolates |
|---|---|---|---|---|---|---|---|
| Australia | Human | NA | Feces | 2000–2015 | Unknown | Unknown | 24 |
| Blood | 2004 | Unknown | 1 | ||||
| Denmark | Human | NA | Feces | 2007–2019 | 8/15 (53.3%) 4/8/2/1 |
50 (0; 66) | 15 |
| Animal | Dog | Feces | 2017 | 1 | |||
| Environment | NA | Shellfish | Unknown | 5 | |||
| France | Human | NA | Feces | 2003–2016 | 20/42 (47.6%) 8/14/8/12 |
62 (9; 91) | 42 |
| Animal | Dog | Feces | 2015 | 9 | |||
| Cattle | Feces | 2016 | 1 | ||||
| Wild bird | Feces | 2017–2018 | 19 | ||||
| Environment | NA | Shellfish | 2013–2015 | 11 | |||
| NA | Freshwater | 2014 | 1 | ||||
| NA | Seawater | 2014–2015 | 4 | ||||
| NA | Sediment | 2013–2014 | 2 | ||||
| Germany | Animal | Layer/duck | Feces | 2005–2011 | 3 | ||
| Food | Duck/layer/turkey | Meat | 2002–2018 | 12 | |||
| Environment | NA | Shellfish | 2005–2017 | 7 | |||
| Soil | Unknown | 1 | |||||
| Total | 158 |
* sp, spring; su, summer; f, fall; w, winter. NA, not applicable.
Campylobacter spp. taxonomic affiliation.
To clarify the taxonomic status of the 33 Campylobacter species isolates, we selected a collection of 469 Sequence Read Archive (SRA) accessions with WGS data from Campylobacter strains from different locations and sources (Table S9). Most of the SRA accessions (n = 393) belonged to already known species/subspecies, thus we deleted them from downstream analyses. The remaining accessions were added to our 33 Campylobacter spp. genomes for an ANI analysis. Several SRA accessions matched the four clades identified in this study (Fig. S1), while new clades were observed but not included in this analysis.
Regarding the metadata obtained from the SRA database, our clade 1 isolates were closely related to 39 isolates from the USA; eight of them were collected from humans. Clade 2 isolates were also related to North American (Canada and USA) isolates, with three environmental ones. Similar geographical localization was obtained for clade 3-related isolates, most of them lacking metadata information, with one isolate from human stool, and two isolates from wild birds with no detailed information. Clade 4 included one human stool and two other isolates lacking informative details of their source or geographical location.
MLST data.
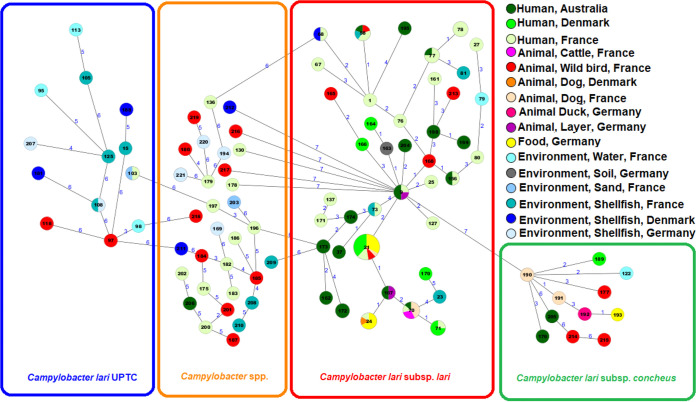
Overall, 97 STs were detected among the 158 Campylobacter isolates, suggesting a high genetic diversity within the population structure of our data set (Fig. 3; Table S2). Of these, 60 (61.8%) STs were newly described in this study (ST162 to ST221; 75 isolates, including 31 Campylobacter species isolates). The most common STs were ST-21 (21 isolates), ST-70 (seven), ST-24 (six), ST-58 (five), ST-9, ST-71, ST-77, and ST-78 (four each) belonging to the subspecies Cll (Fig. 3; Table S2).
FIG 3.
Minimum spanning tree of C. lari ST sequences (n = 158 isolates). Colors correspond to the origin of the samples. Allelic distances between isolates are indicated. The numbers given in the circles correspond to the sequence type numbers. The sequence type numbers >162 are new STs.
The new STs represented 17 of the 41 (41.5%) STs of Cll, 10 of the 11 (90.9%) STs of Clc, three of the 13 STs (23.1%) of C. lari UPTC isolates, and 30 of the 32 STs (93.7%) of Campylobacter species isolates (Table S2).
Three of the 60 new STs had new allele sequences detected in all seven loci (one waterbird and two shellfish isolates), 47 STs resulted from a combination of already described alleles and new alleles, while 10 STs resulted from new combinations of already described alleles.
Most of the new STs (81.7%) were represented by a single isolate and most found among waterbird isolates (15/19; 79.0%), followed by isolates from the environment (15/31; 48.4%) and human isolates (33/82; 40.2%). Only one new ST was obtained among food isolates (1/11; 9.1%). New STs were most common among Australian isolates (18/25; 72%), followed by German (10/23; 43.5%), Danish (8/21; 38.1%), and French isolates (39/89; 43.8%).
Twenty of the 97 STs were also detected in 150 of the 469 SRA accessions and three of the new STs (i.e., ST-163, ST-164, and ST-182) were detected in eight SRA accessions. ST-21 (66 strains), ST-24 (nine), and ST-70 (seven) were also common STs of this collection.
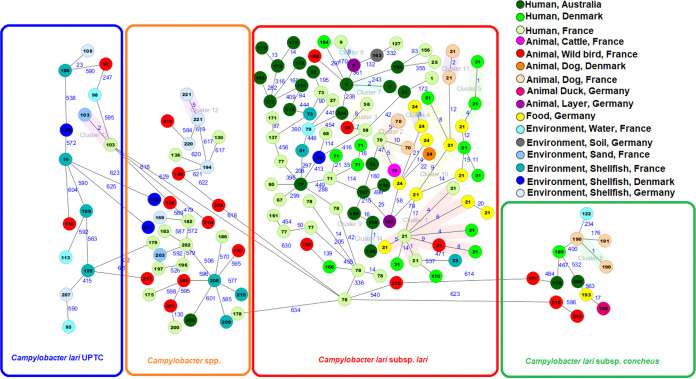
Core-genome cgMLST data.
The data were analyzed with an ad hoc cgMLST scheme created based on our collection. The resulting minimum spanning tree confirmed the presence of different branches corresponding to the different subspecies identified by ANI and isDDH analysis (i.e., Cll, Clc, C. lari UPTC, and Campylobacter spp.; Fig. 4). To identify closely related strains and potential transmission events between the four different compartments, i.e., human, animal (waterbirds and domestic animals), food, and environmental, we chose a pairwise allelic distance (AD) threshold of 10 alleles. In total, 14 clusters were identified, seven comprised of isolates from different compartments (Fig. 4 and Table S3). The largest cluster with eight isolates of Cll (ST-21) contained one isolate of animal origin, five isolates of human origin and two isolates of food origin, indicating probable transmission between these compartments. Moreover, there were smaller clusters consisting of human isolates and animal or food isolates. There was only one cluster (ST-103; two C. lari UPTC isolates) with related genomes from samples from human and environmental origin. The other isolates with a common ST (ST-73) found in both humans and the environment (i.e., shellfish) present an AD greater than 50 alleles.
FIG 4.
Minimum spanning tree of core genome multilocus sequence typing (cgMLST) with 662 targets of 155 Campylobacter species isolates. Allelic distances between isolates are indicated, clusters with allele difference <10 are indicated by shaded colors (genetically closely related isolates). The numbers given in the circles correspond to the sequence type numbers. Nodes are colored according to the isolation source.
The comparison of the 21 C. lari genomes to 66 SRA accessions, both belonging to ST-21 showed the presence of 10 clusters with an AD threshold of 10, three of which included isolates from this study. In fact, one waterbird (A23) and two human isolates (the Danish H74 and the French H39) were closely related to three isolates from the USA, the first two from chicken and the last one from a source that was not specified. (Fig. S2).
The human isolates were highly diverse (51 STs among 82 isolates, including 27 new STs) with ST21 as the most common (11 isolates). Furthermore, few STs were present in isolates from several countries and a lower allele distance between inter-Europe isolates was observed (between Danish and French isolates, four and 21 allele differences for ST21 and ST71, respectively) than intercontinental isolates (i.e., between Australian and French isolates; 42, 47, 93, 208, and 370 allele differences for ST58, ST70, ST156, ST77, and ST9, respectively). A cgMLST cluster, including human isolates from two different countries (ST-21, cluster 1; Fig. 4) was identified.
Whole-genome single nucleotide polymorphism (SNP) analysis.
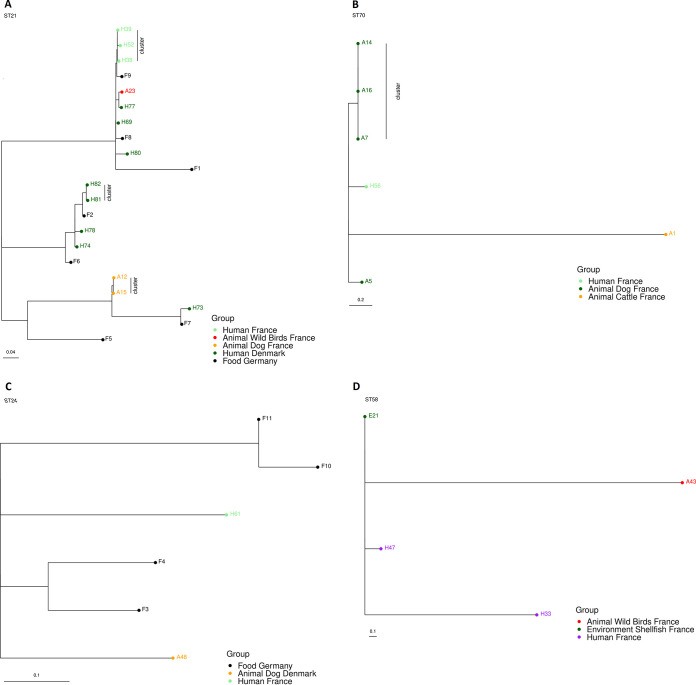
Isolates belonging to the four predominant sequence types, ST-21, ST-70, ST-24, and ST-58 (accounting for 13.3%, 4.4%, 3.8%, and 3.2% of the isolates, respectively) were further evaluated by SNP analysis, and the retrieved maximum likelihood phylogenetic SNP trees are presented in Fig. 5A to D.
FIG 5.
Maximum likelihood phylogenetic SNP trees of the four most common sequence types. (A) ST-21; (B) ST-70; (C) ST-24; (D) ST-58. The source group is shown with colored circles.
ST-21 was found in 21 isolates, specifically human isolates from Denmark (eight) and France (three), in animal isolates from France (two from dogs and one from gull feces), and in food isolates from Germany (from layer, duck, or turkey meat samples). SNP analysis provided three distinct SNP clusters, while four cgMLST (with 10 AD) clusters were obtained. Specifically, one cluster included two dog isolates from France (1 SNP), another two human isolates from Denmark (5 SNPs), and two human isolates from France (4 SNPs; Table S8).
ST-70 was found in seven isolates, specifically human isolates from both France (one) and Australia (one), and animal isolates from France (four from dogs and one from cattle). The final SNP analysis included only six of the ST-70 isolates since the human H10 isolate was excluded, as it was very distant (>1,000 SNPs) from the remaining isolates. While the cgMLST grouped four of the seven isolates, the SNP analysis detected only one cluster among three French dog isolates (0 SNPs; Table S8).
ST-24 was found in six isolates: one French human isolate, one Danish dog isolate and four isolates from food in Germany. No SNP cluster was detected. The SNP differences among isolates ranged from 10 to 62 SNPs (Table S8).
Finally, ST-58 was found in five isolates: human isolates from France (two) and Australia (one), one waterbird isolate, and one environmental isolate, both from France. Except for the Australian human isolate (H6), which was very distant (>1,000 SNPs) from the other isolates and thus not included in the final tree, the isolates within this ST were genetically close (all within 24 SNPs). Although no SNP clusters were observed among the ST58-isolates, two French isolates from shellfish and human were only six SNPs apart (Table S8).
Virulence genes.
The potential virulence and survival factors (n = 125) we investigated can be categorized into motility (flagella and chemotaxis; n = 50), adhesion (seven), invasion factors (n = 10), toxin production (n = 3), carbohydrate structures (n = 40), iron uptake system (n = 2), stress response (n = 10), and other virulence factors (n = 3) (Table S4). In silico screening results of the presence/absence of these genes are presented in Table S1. The 158 isolates harbored between 96 (76.8%) and 121 (96.8%) genes of the 125 screened genes (mean 112.6 ± 4.3).
The most prevalent genes were those coding for the cytolethal distending toxin (CDT) (n = 3; 100%; genes coding for the three subunits: CdtA, CdtB, and CdtC), iron uptake system (fur and cfrA genes; 99.7% ± 4.0), stress response (n = 10; 98.7% ± 3.9), invasion factors (n = 10; 95.6% ± 5.5), and, to a lesser extent, motility (n = 50; 91.7% ± 2.8), carbohydrate structures (n = 40; 89.1% ± 8.4), other virulence factors (mviN, encoding a virulence factor protein, and eptC and fcl genes; 78.1% ± 19.5), and adhesion (n = 7; 61.7% ± 11.4).
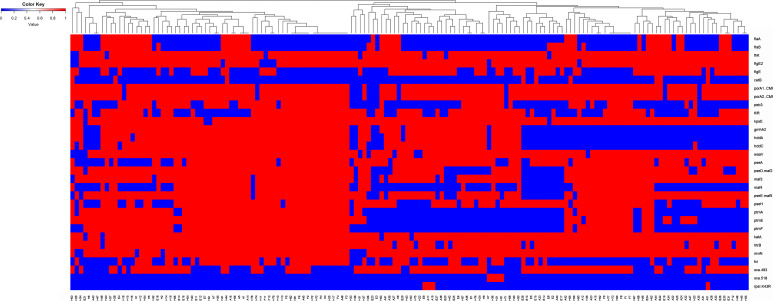
Ninety-four (75.2%) selected genes were present in more than 98% of the isolates (>155 isolates), while three genes (flaJ, capA, and jlpA) were absent in all isolates. The presence or absence of the 28 other virulence genes in the 158 isolates is presented in a heatmap (Fig. 6). Twenty (16.0%) genes were present in 50% to 98% of the isolates (two motility genes, fliK and flgE2; two adhesion genes, porA1 and porA2; one invasion gene, fliR; 12 carbohydrate structure genes; two stress response genes, katA and htrB; and the mviN gene). Eight (6.4%) genes were present in less than 50% of the isolates (from 9 to 78 isolates; cetB [9, motility], flaA [43, motility], flaB [50, motility], fcl [65, other virulence factor], maf4 [70, carbohydrate structure], ptmF [78, carbohydrate structure], flgE [75, motility], and peb3 [75, adhesion]).
FIG 6.
Heatmap of 31 virulence and resistance genes in the 158 isolates. The genes present in >98% of isolates and those present in <2% the isolates were not included in this heatmap.
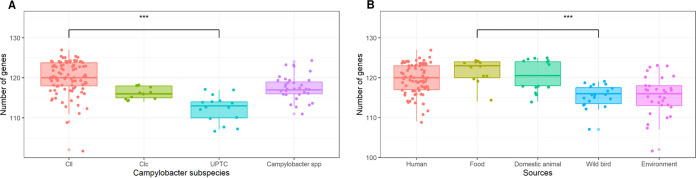
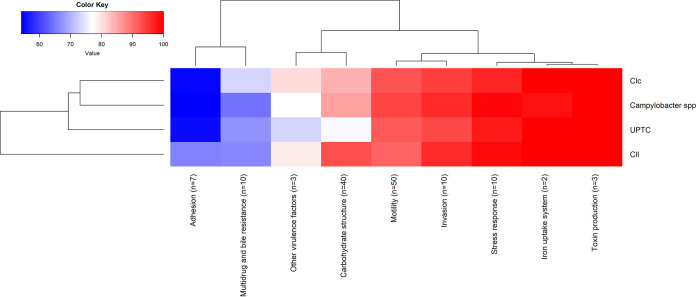
A significant correlation was found between the taxonomic affiliation and the number of detected virulence genes per isolate (F-test, P < 0.001), with Cll isolates having more virulence genes (mean 114.2 ± 4.2) than the other subspecies (C. lari UPTC, mean 106.7 ± 3.3; Clc, mean 110 ± 1.6; Campylobacter spp., mean 111.7 ± 3.1; Fig. 7A). The number of virulence genes in Cll and C. lari UPTC were significantly different (t test, P < 0.001). Compared to C. lari UPTC, the Cll isolates had a higher number of genes encoding adhesion factors (mean 4.6 versus 3.8 to 3.9) and carbohydrate structures (mean 37.2 versus 30.5 to 34.3), including lipooligosaccharide biosynthesis (LOS; mean 10.1 versus 8.6 to 9.3), and O-linked flagellar glycosylation (mean 13.2 versus 8.2 to 11.2) (Fig. 7 and 8). Furthermore, the peb3 gene (adhesion) was only present in Cll isolates (76.5% of Cll isolates) and the maf4, ptmA, ptmE, ptmF (carbohydrate structure; O-linked flagellar glycosylation) genes were more prevalent in Cll isolates than in other C. lari subspecies (62.2%, 75.5%, 82.7%, and 71.4% versus <27.3%, <24.2%, <33.3%, and <24.2%, respectively). In contrast, the flgE gene was weakly present in Cll isolates (only 24.5% versus >73.3% of the other subspecies isolates).
FIG 7.
Box plots showing the distribution of virulence, and AMR- and bile-resistant genes present in the isolates according to the C. lari subspecies (A) and sources (B). Cll isolates had a significantly higher number of virulence genes compared to the other Campylobacter subspecies. Domestic animal isolates had a significantly higher number of virulence genes compared to human, food, wild bird, and environmental isolates.
FIG 8.
Heatmap of virulence and resistance genes classified in different categories of factors according to the subspecies.
Isolate source was also found to have a significant impact on the number of virulence genes (F-test, P < 0.001). Domestic animal (114.6 ± 4.0), food (115.6 ± 3.1), and human (113.7 ± 3.3) isolates had more virulence genes than isolates from the environment (109.5 ± 4.9) and waterbird (109.5 ± 2.5) isolates (Fig. 7B). In addition, the number of virulence genes in isolates from food and those from waterbirds and the environment were significantly different (t test, P < 0.001).
AMR and bile resistance.
The cmeA, cmeB, and cmeC genes constituting the cmeABC efflux complex genes (29) were present in 155 (98.1%), 157 (99.4%), and 157 (99.4%) isolates, respectively. The cmeR regulatory gene was present in all the isolates. Finally, the macA and macB genes constituting the macAB efflux locus (multicomponent efflux complex which confers macrolide resistance) were present in 157 (99.4%) and all the isolates, respectively. Out of the genomes sequenced in this study, 67.1% of the isolates contained acquired genes associated with resistance to β-lactam antibiotics, i.e., class D oxacillinase (OXA)-type β-lactamase genes. Specifically, 63.9% (101 isolates) contained the blaOXA-493 resistance determinant, whereas 2.5% (four isolates; human Clc isolates) contained the blaOXA-518 resistance determinant (Fig. 6).
The tetO locus conferring resistance to tetracycline was only found in one German food isolate, whereas the rpsL K43R mutation conferring potential resistance to streptomycin (30) was found in three dog isolates and one water isolate, all from France and belonging to Clc. The latter isolates also harbored the blaOXA-493 resistance determinant (Table S1 and S5).
Pangenome.
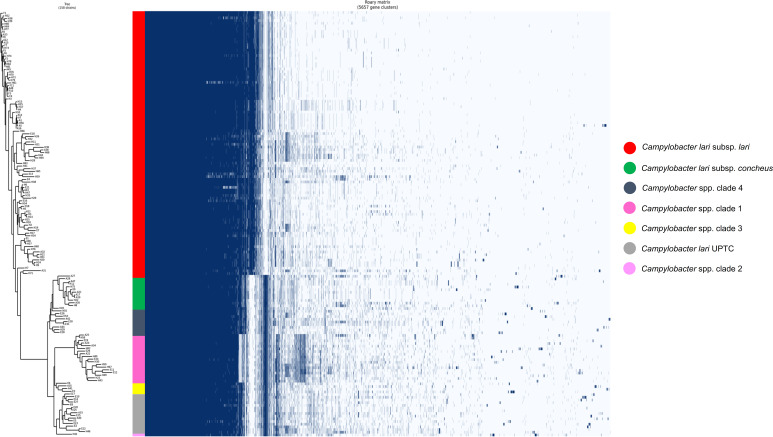
The comparative genomic analysis was performed to determine whether the functional genome content differed between the 158 isolates. The Roary pangenome analysis revealed 5,657 genes clusters (GC), of which only 951 were core genes (Fig. 9), 661 were shell genes (identified in 23 to 150 isolates), and 3,880 were cloud genes (23 isolates). Approximately 68.8% of all genes were cloud genes, indicating that each isolate tended to contribute an average of 24 unique genes to the pangenome. Cluster Orthologous Group (COG) functional category analysis of these GC according to each Campylobacter clade did not indicate functions related to pathogenesis (data not shown). According to the presence or absence of particular genes or groups of genes, isolates were split into six main clades: the Cll isolates (n = 98), the clc clade with its related Campylobacter spp. Clade 4 (n = 10), the C. lari UPTC and its related Campylobacter spp. Clades 2 and 3 (one and four isolates, respectively), and the Campylobacter spp. Clade 1 (including a cluster of 18 isolates and one isolate). Whereas the Cll group seems to be quite homogeneous, isolates belonging to the other subspecies exhibit greater variations. Clc, C. lari UPTC, and their related Campylobacter species isolates presented quite conserved profiles, while the Campylobacter spp. clade 1 presented the highest genetic distance.
FIG 9.
Pangenome analysis of Campylobacter genomes using Roary (71). Gene clusters (n = 5,657) were ordered according to the hierarchical clustering of their presence/absence (dendrogram). Presence, blue; absence, white. Genomes were ordered based on bacterial sp. or Campylobacter spp. groups.
DISCUSSION
Based on a wide geographic distribution and diverse sample sources, this multicountry retrospective study provides a comprehensive genomic comparison of C. lari isolates (n = 158) from humans, environment, domestic animals, waterbirds, and food. Very few studies have described C. lari isolates from several sources (6, 15, 24, 31).
This study resulted in the discovery of 33 potentially new species or subspecies of the C. lari group and the identification of 60 new STs, contributing to the non-jejuni/coli Campylobacter PubMLST database.
A major difficulty we encountered during this study was compiling a large collection of C. lari group strains from different countries/continents. In fact, they are much less frequently recovered from human, animal, environmental, or food sources than C. jejuni and C. coli: e.g., C. lari represented only 2.5% to 6.7% of Campylobacter species isolated from domestic animals (poultry, dogs, and cattle) in Sweden, Italy, and Lithuania (32–34) and 0.2% and 1.9% of human clinical Campylobacter species isolates in France and Ireland, respectively (35, 36). However, C. lari may be frequently isolated from seagulls, the coastal environment, and from shellfish (6, 22, 24). Although C. lari group members are relatively rare in human clinical samples, it is still important to understand their population structure and epidemiology, and to decipher host and sources of this emerging group of potential pathogens.
We sought to understand the clades and subspecies of the C. lari group isolates and their STs; data that are seldom available in studies on the C. lari group. In fact, few studies (15) described C. lari isolates at the subspecies level; most often the subspecies were not given, or the isolates were only separated into UPTC and urease-negative (UN) isolates (6, 37). In addition, while many studies have described the STs of C. jejuni and C. coli isolates (38, 39), very few studies described the STs of C. lari isolates and their related population structure (i.e., epidemic clones and/or cross-source connections) (24).
As observed by Miller et al. (15), molecular typing shows a high genetic diversity within the C. lari group, with a greater diversity of isolates of C. lari UPTC and Campylobacter spp. than Cll. Our isolates are distributed in several subspecies or groups of members, i.e., Cll, Clc, C. lari UPTC, and four main Campylobacter spp. clades. For these latter groups, the species and the subspecies are yet to be identified, suggesting a new clade within the C. lari group, or even a new species (i.e., the high genetic distance between the Campylobacter spp. clade 1 and the other groups). A significantly different distribution of isolates to subspecies level within the sources was highlighted in this study (Chi-Square test, P < 0.001). In fact, human, domestic animal, and food isolates belong mainly to Cll (>70% of the isolates), whereas waterbird and environmental isolates belong mainly to Campylobacter spp. (47.4%) and Cll (26.3%), and to Campylobacter spp. (35.5%) and C. lari UPTC (38.7%), respectively. These findings agree with those presented by Matsuda and Moore (6), in a review which described (i) the presence of urease-negative (UN) Campylobacter (i.e., Cll and Clc) and C. lari UPTC in waterbirds and in the environment (water and shellfish) and (ii) the presence of UN Campylobacter in domestic animals, food, and humans. Few C. lari UPTC isolates have been identified from humans (only one) and any association between these isolates and human disease remains unclear (6). However, we are aware that a C. lari UPTC strain was recently isolated from the urine of a patient with a urinary tract infection (40).
Although C. lari isolates are characterized by high diversity, ST-21 is prevalent in our collection. It includes 21 isolates, of which 18 isolates are included in four distinct cgMLST clusters (AD < 10) and five isolates are included in three distinct SNP clusters (<5 SNPs). ST-21 was identified in humans in both France and Denmark, in domestic animals and a waterbird in France, and in poultry meat in Germany. In agreement with our results, this ST was also found in the USA in human clinical isolates (9/13 C. lari isolates) (31), in domestic animals (i.e., chicken, turkey, cattle, and swine), and in crows (41), confirming its generalist status (66 genomes of 469 SRA accessions).
This study provides insights into potential sources of human infections by C. lari group members through the identification of cgMLST clusters, comprising both isolates of human and other origins or more generally of common STs. Thus, of the 14 cgMLST clusters (AD 10), seven clusters included both human isolates and isolates from (i) food (ST-21), (ii) domestic animals (ST-70, ST-21, and ST-9), (iii) waterbirds (ST-21 and ST-58), and (iv) the environment (ST-103). This suggests that transmission to humans can occur from multiple sources, even from waterbirds and the environment. However, there are very few point source outbreaks of C. lari reported in the literature. Ideally, public health investigators would conduct epidemiological studies of C. lari infections in humans to examine risk factors and sources of infections.
Our findings agree with other studies investigating the C. lari group, which show that although domestic animals and food isolates are more likely sources of human infections (e.g., of Cll subspecies, with a higher number of virulence genes), waterbirds and the environment may constitute reservoirs for the C. lari group that can infect humans, e.g., via raw shellfish (22, 24). In addition, the presence of shared STs between human and waterbird isolates is confirmed by human isolates from this study sharing the same STs found among waterbird isolates in the pubMLST database (https://pubmlst.org/organisms/campylobacter-non-jejunicoli).
Few studies have targeted the pathogenicity of the C. lari group (15, 18, 26). This study identified several genes involved in the carbohydrate structures that were more frequently present in human isolates than in nonhuman isolates, especially from the environment. These included the following genes: pseD/maf2, maf3, maf4, pse/maf5, ptmA, ptmE, ptmF (carbohydrate structure; O-linked flagellar glycosylation) and gmhA2, hddA, and hddC (lipooligosaccharide biosynthesis; LOS). In the same way, the fliR (encoding a flagellar biosynthetic protein; invasion) and peb3 (an adhesion factor) genes were also more frequent in human isolates. These differences, according to the sources, correlated with the assignment of isolates to species or subspecies in humans and the environment. Thus, Cll represents more than 70% of the human, domestic animal, and food strains, while Cll constitutes only 22.6% of the isolates from the environment. The pseD/maf2, maf3, maf4, pse/maf5, ptmA, ptmE, ptmF, gmA2, hddA, hddC, peB3, fliR flaA, and fcl genes were found more frequently in Cll than in C. lari UPTC isolates. Interestingly, peb3 was found only in Cll strains. In contrast, the flgE gene encoding a flagellar hook protein was more prevalent in environmental (71%) than in human (41.5%) isolates, and in the UPTC isolates (73.3%), Campylobacter spp., and Clc (91.7%) than in Cll (24.5%).
Some genes (i.e., flaJ, capA, and jlpA) were absent. The absence of the jlpA lipoprotein coding gene is in line with the absence of this lipoprotein in other members of the C. lari group (e.g., the Cll RM2100 and LMG11760 strains, the hyperaerotolerant C. lari SCHS02 strain, and C. armoricus isolates [17, 18, 26]). These C. lari isolates contained a periplasmic Cu/Zn superoxide dismutase (SodC), also previously reported in some C. lari isolates (18, 26) and the cytoplasmic Fe superoxide dismutase (SodB), the only one harbored by C. jejuni.
Depending on source and subspecies, 75.5% to 94.8% (n = 135) of the investigated virulence and AMR genes were present in each isolate.
The frequent presence of these virulence genes could potentially explain the ability to cause disease in humans. All human and other source isolate genomes contained genes associated with flagella-mediated motility, adhesion, toxin production, carbohydrate structure, stress response, and iron uptake system. Furthermore, resistance to antibiotics was also observed in some of these C. lari isolates. Out of the total genomes sequenced, 67.1% of the isolates contained class D β-lactamase genes. Specifically, 63.9% (101 isolates) contained blaOXA-493, whereas 2.5% contained blaOXA-518, the latter observed only in Clc. This finding agrees with results from Rivera-Mendoza et al. (42) who recently reported the blaOXA-493 gene detection exclusively within members of the C. lari group. The blaOXA-493 gene was also found in C. armoricus (17), another member of this group. The tetO locus conferring resistance to tetracycline was only found in one German food Cll isolate (probably harbored on a plasmid), whereas the rpsL K43R mutation conferring a potential resistance to streptomycin was found in three isolates from dogs and one from water, all originating in France and belonging to Clc. The prevalence of AMR genes in C. lari isolates from a wide host range may pose a public health risk. However, the expression of these AMR genes should be confirmed by in vitro tests. Our results show that several genomes from all the investigated sources in this study display high similarity to sequences of isolates implicated in human diseases, suggesting that these isolates are potential pathogens of public health importance and that a zoonotic transfer may occur. Whether those virulence genes are expressed still needs to be determined.
From a pangenome perspective, and in agreement with MLST and cgMLST analyses, this study identified the presence of novel clades within the known members of the C. lari group. The pangenome analysis indicated the presence/absence of several clusters of genes which will be investigated soon, with the aim of detecting genetic determinants (i.e., metabolic potential, stress response, adhesion, etc.) that may explain the success of some STs or clones in colonizing hosts, or tropism for specific ecosystems (e.g., coastal areas).
Conclusion.
WGS analyses of isolates of the C. lari group from various sources and countries highlighted great genetic diversity within this group of thermotolerant Campylobacter. The identification of many novel STs and the lack of subspecies identification of some isolates confirm the need for further investigation to explore the overall diversity within this group.
C. lari group members harbor many of the virulence-related genes previously identified in C. jejuni and C. coli (76.8% to 96.8% per isolate), suggesting that members of this group might be pathogenic for humans. In addition, they contain multidrug-resistant genes such as the genes encoding the CmeABC efflux complex and class D β-lactamase genes, further boosting their pathogenic profile.
Finally, these data corroborate the importance of the One Health concept by providing essential data on the presence of potentially pathogenic bacteria such as C. lari in the environment, animals, food, and humans and their transmission between the different ecosystems and hosts. This study could be a first step in setting up a prospective multicountry project to investigate the prevalence and characteristics of C. lari in the main sources we have identified, according to a standardized protocol, and to compare isolates based on genomic data similar to this study.
MATERIALS AND METHODS
Collection of isolates.
A set of 158 isolates of the C. lari group from human, animal, food, and environmental sources were collected from three European countries (Denmark, France, and Germany; n = 133) and from Australia (n = 25; Table 1 and S1) as described below.
(i) Data sampling in Denmark. Clinical cases of Campylobacter spp. in humans in Denmark (DK) are notifiable through the laboratory surveillance systems at the Statens Serum Institut (SSI). Campylobacter species isolates from patients diagnosed with campylobacteriosis were referred to the SSI for further characterization. The 15 isolates from human stool samples were collected from 2007 to 2019 (Table 1 and S1). The five Danish shellfish isolates came from a collection of isolates from shellfish built by Freie Universität Berlin (FU) and were collected at retail from 2010 to 2013. One dog isolate was collected in 2017 as part of a former WGS project (28).
(ii) Data sampling in France. The clinical isolates (n = 42; isolated between 2003 and 2016; Table 1 and S1) were received from French laboratories and hospitals as pure cultures by the French National Reference Center for Campylobacters and Helicobacters (CNRCH).
The 10 domestic animal isolates were collected by ANSES (French Agency for Food Environmental and Occupational Health & Safety, Ploufragan, France) and came from cattle (n = 1; 2016) and dog feces (n = 9; 2015; Table 1 and S1). The 19 wild bird isolates were collected by IFREMER from gulls (n = 7), Eurasian curlews (n = 4), common shelducks (n = 3), Eurasian oystercatchers (n = 4), and Brent goose (n = 1) fresh droppings in Brittany during another research project (Campyshell).
Finally, the environmental isolates (shellfish [n = 11], freshwater [n = 1], seawater [n = 4] and sediment [n = 2]) were collected by IFREMER and originated from another research project (22).
All French isolates were identified as C. lari using a MALDI-TOF mass spectrometer (MALDI-TOF Brucker Microflex) (13).
(iii) Data sampling in Germany. The collection of German (DE) isolates was comprised of three animal isolates (layer and duck) from 2005 to 2008, 12 food isolates (duck, layer, and turkey meat) from 2002 to 2018, and eight environmental isolates (soil [n = 1] and shellfish [n = 7] from 2005 to 2017; Table 1 and S1).
(iv) Data sampling in Australia. The collection of Australian (AUS) isolates was comprised of 25 human clinical isolates from feces (n = 24) and blood (n = 1) obtained between 2000 and 2015 (Table 1 and S1).
Bacterial culture and genomic DNA extraction.
French isolates, stored at −80°C in homemade brucella broth with 10% glycerol, were subcultured twice onto Karmali (Oxoid) plates incubated at 41.5°C for 48 h under microaerobic conditions (Oxoid CampyGen, Dardilly, France). Genomic DNA (gDNA) was extracted from fresh colonies using the GenElute bacterial genomic DNA kit (Sigma).
Danish clinical isolates were cultured (and subcultured) on blood agar plates and incubated under microaerobic conditions (85% N2, 10% CO2, 5% O2) at 41°C for 48 h. Subsequently, gDNA was extracted using the DNeasy blood and tissue kit (Qiagen).
The German C. lari strains from BfR and the soil sample as well as the five Danish shellfish strains from the FU collection were isolated according to ISO 10272-1:2005 and ISO 10272-1:2017. The species was further verified by real-time PCR as described by Mayr et al. (43) (BfR collection) or by mPCR as described by Wang et al. (44) (FU collection).
C. lari isolates, stored at −80°C in cryotubes (MAST Diagnostica, Reinfeld, Germany), were reisolated on Mueller-Hinton agar (Oxoid, Wesel, Germany) supplemented with 5% sheep blood (Oxoid) and incubated for 48 h at 37°C in microaerobic atmosphere (6% O2, 10% CO2, 84% N2) generated by Anoxomat (Mart Microbiology, Drachten, the Netherlands). Subsequently, gDNA was extracted with the MasterPure DNA purification kit for blood v.2 according to the manufacturer’s instructions (Biozym, Oldendorf, Germany). The gDNA of other German isolates was extracted with the GenElute bacterial genomic DNA kit (Sigma).
Australian isolates, stored at −70°C in 20% (vol/vol) glycerol in nutrient broth 2 (Oxoid, UK), were subcultured twice onto horse blood agar (Oxoid) plates incubated at 37°C for 48 h under microaerobic conditions (CampyGen, Oxoid,). gDNA from the Australian isolates was extracted using previously described methods (45).
Whole-genome sequencing (WGS).
All isolates were sequenced using Illumina MiSeq (France, 2 × 150, 2 × 250, or 2 × 300 bp; Germany and Denmark, 2 × 300 bp) and NextSeq (Denmark, Australia and Germany, 2 ×150 bp) sequencing platforms (Table S1).
Raw data quality control, genome assemblies, and annotations.
Data were analyzed using the CELIA v1.0 workflow (https://github.com/ifremer-bioinformatics/celia) developed by the SeBiMER (Ifremer’s Bioinformatics Core Facility) as an open-source modular workflow to assemble and annotate prokaryotic genomes. CELIA was developed using the NextFlow workflow manager (46) and tools were containerized using Docker. Data were processed as follows: genomic data quality was assessed using FastQC v.0.11.8 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc) and MultiQC v.1.8 (47). De novo assembly of genomes from raw reads was performed using Unicyler (v.0.4.8) with minimal output contig length (–min_fasta_length) set to 500 bp (48). To remove potential contaminants from the genome assemblies, contigs were screened for similarities against the UniVec database (v.03–20-2017) using BLASTN (49) according to UniVec documentation. Sequencing coverage was estimated by mapping paired-end reads to the corresponding assembled genome with Bowtie2 (v.2.3.5) (50) in “sensitive” mode and computed with Mosdepth (v.0.2.7) (51). The completeness of the genome assembly was assessed by searching for similarities against highly conserved genes among Campylobacterales. For this purpose, we ran BUSCO (v.4.0.0) (52) in “genome” mode specifying the Campylobacterales profile library containing 628 core proteins (released April 2019).
The obtained draft genomes were checked for consistency, e.g., the number of contigs, DNA G+C content and total size of assembly, N50 values, and percentage of coverage using Quast v.5.0.0 (53).
Automatic gene prediction was executed using the Prokka pipeline v.1.14.5 with the following parameters: -force, -addgenes, -compliant, -genus Campylobacter –usegenus –rfam (54). Transfer RNAs (tRNAs) and transfer-mRNA (tmRNA) were predicted with the ARAGORN program (55) implemented in Prokka. Ribosomal RNA (rRNA) loci and prophages were predicted through RAST subsystem database (https://rast.nmpdr.org/; accessed 20 November 2020 [56]).
Details of the assembly metrics, genome annotations and GenBank accession numbers are provided in Table S1.
Average nucleotide identity (ANI) and in silico DNA-DNA hybridization (isDDH).
Whole-genome sequence similarities, based on thresholds below 96 and 70% for the ANI and isDDH, respectively, were used for the delimitation of closely related species (57, 58). ANI values were calculated on our assembled genomes compared to the 46 Campylobacter type strains (as described in https://www.ezbiocloud.net/; accessed 12 February 2022) and 469 Campylobacter spp. retrieved from NCBI SRA (assembled as described previously) to expand our analysis of C. lari population genetics, especially to Campylobacter species isolates from our data set (Table S6) using fastANI (v.1.3) (59) and PYANI with blast option (v.0.2.7) (60, 61) to estimate their intergenomic similarities, and thus validate their clustering within the C. lari group. In silico DNA-DNA hybridization values were calculated using the Genome-to-Genome Distance Calculator (GGDC; https://ggdc.dsmz.de/ggdc.php) (62). The isDDH model “formula2” was used as recommended for draft genomes. Wolinella succinogenes ATCC29543T was used as an outliner to root the trees.
Molecular typing: MLST, cgMLST, and SNP analyses.
(i) New alleles and STs submission. A total of 144 new alleles and 60 new STs were submitted to the pubMLST Campylobacter non-jejuni/coli database (https://pubmlst.org/bigsdb?db=pubmlst_campylobacter_nonjejuni_isolates).
(ii) WGS-derived MLST and ad hoc cgMLST. The MLST profile of each isolate was determined after trimming and assembly using SeqSphere+ v.6.0 (Ridom, Munster, Germany [63]) from de novo assembled contig sequences using the software package MLST v.2.15.1 (64) based on the Campylobacter PubMLST database (http://pubmlst.org/campylobacter). The typing consists of assigning a “Sequence Type” to each of the strains based on the concatenated sequences of seven housekeeping genes (adk, pgi, glnA, glyA, pgm, tkt, and atpA) located on the chromosome. A minimum spanning tree (MST) showing the MLST profiles of the 158 Campylobacter species isolates with the host species of origin was drawn.
A core genome MLST (cgMLST) scheme defining a comprehensive set of those loci present in most members of C. lari group was also performed using SeqSphere+ software. The annotated reference strain C. lari FDAARGOS (NZ_CP068172.1; C. lari RM2100) was used to seed the database in addition to 27 Campylobacter strains of the C. lari group (details in Table S6). The cgMLST was conducted also using the generated cgMLST scheme of 662 target loci. The presence of these loci in each draft genome was compared using BLASTN to identify genes with 100% overlap and ≥95% sequence similarity. Only isolates, which possessed at least 95% of the loci (“percent good targets”) were included in the analysis. Three isolates (H61, E21, and E5) were withdrawn from the cgMLST analysis because they had less than 95% good targets.
The SeqSphere+ tool was used to map the reads against the reference genome (C. lari FDAARGOS) using BWA v 0.6.2 software (parameters setting: minimum coverage of five and Phred value >30) and to determine the cgMLST gene alleles. The combination of all these alleles in each strain formed an allelic profile that was used to generate a minimum spanning tree (MST) using SeqSphere+ with the “pairwise ignore missing values” parameter. A threshold of ≤10 allelic differences was used to define clusters.
The same method was used for a more specific cgMLST of C. lari ST-21 strains. To the 21 genomes of ST-21 isolates in our study, the 66 C. lari ST-21 genomes of the 469 SRA genomes extracted from the NCBI SRA were added.
(iii) SNP analysis. SNPs analysis was performed using NASP (65) with BWA mapping, variant calling by GATK with UnifiedGenotyper, minimum 10× genome coverage, and a proportion of 0.9 of reads matching the call. A subsequent step for removal of recombination was performed with cleanrecomb (https://www.biorxiv.org/search/cleanrecomb) (65). Due to the high diversity of strains of the C. lari group, SNP analysis was performed separately for each 7-locus MLST, and only the four major STs were further included:ST-21 (21 isolates), ST-24 (six), ST-58 (five), ST-70 (seven). For each SNP analysis, a genome of the specific ST was employed as reference, specifically H77 (ST-21), H61 (ST-24), E21 (ST-58), and A1 (ST-70). Maximum likelihood (ML) trees were constructed using RAxML-NG with HKY as tree model (modeltest-ng to find the best model) and option all (66). Trees were plotted with ggtree (v.3.2.1) in R (v.4.1.2). Isolates with ≤5 SNP differences were considered part of SNP clusters.
Virulence genes and antimicrobial resistance.
Three additional approaches were used to identify virulence as well as multidrug- and bile-resistant encoding genes in the genomes. Their presence was determined in silico with ABRicate software (v.0.9.8) (67) first using the Virulence Factor database (VFDB) dedicated to C. jejuni and C. coli species (68) (query date, March 2020). The second approach involved the use of an in-house created virulence and AMR gene database (n = 74) screened within ABRicate with a minimum sequence identity set at 65% and a minimum length coverage of 80%. It contained gene sequences associated with motility, invasion, chemotaxis, adhesion, toxin, capsule, multidrug and stress response in the C. lari RM2100 strain (accession no. CP000932.1). The third approach corresponded to a manual search from annotation outputs, followed by a comparison of the sequences of each candidate marker, against the information available in NCBI databases, using BLAST algorithm. A list of the virulence and antibiotic resistance genes and their accession numbers is available in Table S4.
Thus, a total of 125 virulence-related genes were investigated (Table S4). In addition, six multidrug- and bile-resistant genes coding for efflux systems (cmeA, cmeB, cmeC, cmeR, macA, and macB) were also investigated.
Furthermore, chromosomal point mutations and genes associated with resistance to antibiotics such as streptomycin and β-lactams, respectively, were determined using staramr v.0.7.2 (https://github.com/phac-nml/staramr) (69, 70).
Pangenome.
The core and accessory genome of the 158 isolates were determined at 90% identity using Roary v.3.13.0 (71) with the following flags: -e (create a multiFASTA alignment of core genes using PRANK); -n (fast core gene alignment with MAFFT); -v (verbose output to STDOUT); -i 90 (minimum percentage identity for blastp; 90%). The Roary analysis was repeated at the 95% and 85% identity cutoffs to check for any major variations in the core and accessory genomes. The number of core, soft-core, shell, and cloud genes as well as the overall core and accessory genome determined by the Roary analysis were visualized using the roary_plots.py script retrieved from https://github.com/sanger-pathogens/Roary/tree/master/contrib/roary_plots. The obtained pangenome reference coding genes were translated to protein sequences and functionally annotated using eggNOG-mapper v2.1.6 (72).
Statistical analysis.
Statistical analyses were conducted in R (v.4.1.1) implemented in Rstudio (v.2021.09.0). Chi-square test, F test and t test were performed. A P value of <0.05 was considered statistically significant.
Data availability.
WGS data sets used in this study were deposited at DDBJ/EMBL/GenBank. The sequences were published under BioProject no. PRJNA798893 and PRJNA818070, BioSample no. SAMN25132837 to SAMN25132984 and SAMN26815012 to SAMN26815036, Genome no. JAKMUQ000000000 to JAKMPA000000000, and SRA accession no. SRR17832004 to SRR17831927 and SRR18392311 to SRR18392294 at the NCBI sequence read archive (SRA). Accession numbers are listed in Table S1B.
ACKNOWLEDGMENTS
We thank Katell Rivoal (ANSES, Ploufragan, France) and Kerstin Stingl (BfR, Germany) for providing French dog and cattle isolates, and German duck, poultry meat, and shellfish isolates, respectively. We also thank Christian Penny and Cécile Walczak (LIST, Luxembourg) for discussions during the implementation of the study and for sequencing a part of the French strains. Finally, we also thank Lindsay Mégraud for proofreading the manuscript.
This work was supported by the project “COllaborative Management Platform for detection and Analyses of (Re-)emerging and foodborne outbreaks in Europe” (COMPARE), which received funding from the European Union’s Horizon 2020 Research and Innovation Program under grant agreement no. 643476. The Australian National Health and Medical Research Council (NHMRC) provided funding for D.J.I. and M.D.K. to participate in the COMPARE study (GNT1103694). D.J.I. is supported by an NHMRC Investigator Grant (GNT1195210) and M.D.K. is supported by an NHMRC Career Development Fellowship (GNT1145997).
M.G. conceived the study and E.M.N. and K.G.J. assisted in designing and planning the study. M.G., J.S., F.M., and P.L. collected and selected the French isolates; K.G.J., S.B., and D.J.I. provided the Danish, German, and Australian isolates, respectively. J.S. carried out the wet lab work for the French isolates. A.C. did the assemblies of the dataset genomes of this study and the description of the general features. N.N. did the genome annotations and the identification of subspecies using ANI and DDH. N.N., S.B., and K.G.J. performed the MLST, cgMLST, and SNP analyses, respectively. N.N. performed the virulence gene analysis. A.M.B. retrieved Campylobacter type strain genomes, downloaded and assembled SRA dataset, performed the pangenome and pairwise ANI analysis and the screening for antibiotic resistance and point-mutation genes. M.G. drafted the manuscript with help from the other authors. All authors approved of the final manuscript.
Footnotes
Supplemental material is available online only.
Contributor Information
Michele Gourmelon, Email: Michele.Gourmelon@ifremer.fr.
Knut Rudi, Norwegian University of Life Sciences.
REFERENCES
- 1.The European Union One Health. 2019. 2018 Zoonoses Report. EFSA J - Wiley Online Library. https://efsa.onlinelibrary.wiley.com/doi/10.2903/j.efsa.2019.5926. [Google Scholar]
- 2.Martinot M, Jaulhac B, Moog R, Martino SD, Kehrli P, Monteil H, Piemont Y. 2001. Campylobacter lari bacteremia. Clin Microbiol Infect 7:96–97. 10.1046/j.1469-0691.2001.00212.x. [DOI] [PubMed] [Google Scholar]
- 3.Tauxe RV, Patton CM, Edmonds P, Barrett TJ, Brenner J, Blake PA. 1985. Illness associated with Campylobacter laridis, a newly recognized Campylobacter species. J Clin Microbiol 21:222–225. 10.1128/jcm.21.2.222-225.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Broczyk A, Thompson S, Smith D, Lior H. 1987. Water-borne outbreak of Campylobacter laridis-associated gastroenteritis. Lancet 1:164–165. 10.1016/S0140-6736(87)92003-4. [DOI] [PubMed] [Google Scholar]
- 5.Patrick ME, Henao OL, Robinson T, Geissler AL, Cronquist A, Hanna S, Hurd S, Medalla F, Pruckler J, Mahon BE. 2018. Features of illnesses caused by five species of Campylobacter, Foodborne Diseases Active Surveillance Network (FoodNet) - 2010–2015. Epidemiol Infect 146:1–10. 10.1017/S0950268817002370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsuda M, Moore JE. 2011. The epidemiology and zoonotic transmission of thermophilic Campylobacter lari. BMRJ 1:104–121. 10.9734/BMRJ/2011/517. [DOI] [Google Scholar]
- 7.Igwaran A, Okoh AI. 2019. Human campylobacteriosis: a public health concern of global importance. Heliyon 5:e02814. 10.1016/j.heliyon.2019.e02814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mori E, Hashimoto T, Yahiro T, Miura M, Ishihara T, Miyazaki M, Komiya K, Takahashi N, Nishizono A, Hiramatsu K. 2022. Campylobacter lari vertebral osteomyelitis. Jpn J Infect Dis 75:322–324. 10.7883/yoken.JJID.2021.532. [DOI] [PubMed] [Google Scholar]
- 9.Kaakoush NO, Castaño-Rodríguez N, Mitchell HM, Man SM. 2015. Global epidemiology of Campylobacter infection. Clin Microbiol Rev 28:687–720. 10.1128/CMR.00006-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costa D, Iraola G. 2019. Pathogenomics of emerging Campylobacter Species. Clin Microbiol Rev 32:e00072-18. 10.1128/CMR.00072-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berenger BM, Chui L, Ferrato C, Lloyd T, Li V, Pillai DR. 2022. Performance of four commercial real-time PCR assays for the detection of bacterial enteric pathogens in clinical samples. Int J Infect Dis 114:195–201. 10.1016/j.ijid.2021.10.035. [DOI] [PubMed] [Google Scholar]
- 12.Roy C, Robert D, Bénéjat L, Buissonnière A, Ducournau A, Mégraud F, Bessède E, Boraud D, Lehours P. 2020. Performance evaluation of the Novodiag bacterial GE+ multiplex PCR assay. J Clin Microbiol 58:e01033-20. 10.1128/JCM.01033-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bessède E, Solecki O, Sifré E, Labadi L, Mégraud F. 2011. Identification of Campylobacter species and related organisms by matrix assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry. Clin Microbiol Infect 17:1735–1739. 10.1111/j.1469-0691.2011.03468.x. [DOI] [PubMed] [Google Scholar]
- 14.Debruyne L, On SLW, De Brandt E, Vandamme P. 2009. Novel Campylobacter lari-like bacteria from humans and molluscs: description of Campylobacter peloridis sp. nov., Campylobacter lari subsp. concheus subsp. nov. and Campylobacter lari subsp. lari subsp. nov. Int J Syst Evol Microbiol 59:1126–1132. 10.1099/ijs.0.000851-0. [DOI] [PubMed] [Google Scholar]
- 15.Miller WG, Yee E, Chapman MH, Smith TPL, Bono JL, Huynh S, Parker CT, Vandamme P, Luong K, Korlach J. 2014. Comparative genomics of the Campylobacter lari group. Genome Biol Evol 6:3252–3266. 10.1093/gbe/evu249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caceres A, Munoz I, Iraola G, Diaz-Viraque F, Collado L. 2017. Campylobacter ornithocola sp. nov., a novel member of the Campylobacter lari group isolated from wild bird faecal samples. Int J Syst Evol 67:1643–1649. 10.1099/ijsem.0.001822. [DOI] [PubMed] [Google Scholar]
- 17.Boukerb AM, Penny C, Serghine J, Walczak C, Cauchie H-M, Miller WG, Losch S, Ragimbeau C, Mossong J, Megraud F, Lehours P, Benejat L, Gourmelon M. 2019. Campylobacter armoricus sp. nov., a novel member of the Campylobacter lari group isolated from surface water and stools from humans with enteric infection. Int J Syst Evol Microbiol 69:3969–3979. 10.1099/ijsem.0.003836. [DOI] [PubMed] [Google Scholar]
- 18.Miller WG, Wang G, Binnewies TT, Parker CT. 2008. The complete genome sequence and analysis of the human pathogen Campylobacter lari. Foodborne Pathog Dis 5:371–386. 10.1089/fpd.2008.0101. [DOI] [PubMed] [Google Scholar]
- 19.Hakeem MJ, Lu X. 2020. Survival and control of Campylobacter in poultry production environment. Front Cell Infect Microbiol 10:615049. 10.3389/fcimb.2020.615049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pitkänen T, Hänninen M-L. 2017. Members of the family Campylobacteraceae: Campylobacter jejuni, Campylobacter coli. Global Water Pathogens Project; Part 3 Specific excreted pathogens: environmental and epidemiology aspects. http://www.waterpathogens.org/book/campylobacter. [Google Scholar]
- 21.Gölz G, Rosner B, Hofreuter D, Josenhans C, Kreienbrock L, Löwenstein A, Schielke A, Stark K, Suerbaum S, Wieler LH, Alter T. 2014. Relevance of Campylobacter to public health-The need for a One Health approach. Int J Med Microbiol 304:817–823. 10.1016/j.ijmm.2014.08.015. [DOI] [PubMed] [Google Scholar]
- 22.Rincé A, Balière C, Hervio-Heath D, Cozien J, Lozach S, Parnaudeau S, Le Guyader FS, Le Hello S, Giard J-C, Sauvageot N, Benachour A, Strubbia S, Gourmelon M. 2018. Occurrence of bacterial pathogens and human noroviruses in shellfish-harvesting areas and their catchments in France. Front Microbiol 9:2443. 10.3389/fmicb.2018.02443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jeon B, Saisom T, Sasipreeyajan J, Luangtongkum T. 2022. Live-attenuated oral vaccines to reduce Campylobacter colonization in poultry. Vaccines 10:685. 10.3390/vaccines10050685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lozano-León A, Rodríguez-Souto RR, González-Escalona N, Llovo-Taboada J, Iglesias-Canle J, Álvarez-Castro A, Garrido-Maestu A. 2021. Detection, molecular characterization, and antimicrobial susceptibility, of Campylobacter spp. isolated from shellfish. Microb Risk Anal 18:100176. 10.1016/j.mran.2021.100176. [DOI] [Google Scholar]
- 25.Miller WG, On SLW, Wang G, Fontanoz S, Lastovica AJ, Mandrell RE. 2005. Extended multilocus sequence typing system for Campylobacter coli, C. lari, C. upsaliensis, and C. helveticus. J Clin Microbiol 43:2315–2329. 10.1128/JCM.43.5.2315-2329.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song H, Kim J, Guk J-H, An J-U, Lee S, Cho S. 2020. Complete genome sequence and comparative genomic analysis of hyper-aerotolerant Campylobacter lari strain SCHS02 isolated from duck for its potential pathogenicity. Microb Pathog 142:104110. 10.1016/j.micpath.2020.104110. [DOI] [PubMed] [Google Scholar]
- 27.Boukerb AM, Schaeffer J, Serghine J, Carrier G, Le Guyader FS, Gourmelon M. 2020. Complete genome sequence of Campylobacter armoricus CA639, which carries two plasmids, compiled using Oxford Nanopore and Illumina sequencing technologies. Microbiol Resour Ann 9:e00045-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joensen KG, Kiil K, Gantzhorn MR, Nauerby B, Engberg J, Holt HM, Nielsen HL, Petersen AM, Kuhn KG, Sando G, Ethelberg S, Nielsen EM. 2020. Whole-genome sequencing to detect numerous Campylobacter jejuni outbreaks and match patient isolates to Sources, Denmark, 2015–2017. Emerg Infect Dis 26:523–532. 10.3201/eid2603.190947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yao H, Zhao W, Jiao D, Schwarz S, Zhang R, Li X-S, Du X-D. 2021. Global distribution, dissemination and overexpression of potent multidrug efflux pump RE-CmeABC in Campylobacter jejuni. J Antimicrob Chemother 76:596–600. 10.1093/jac/dkaa483. [DOI] [PubMed] [Google Scholar]
- 30.Olkkola S, Juntunen P, Heiska H, Hyytiäinen H, Hänninen M-L. 2010. Mutations in the rpsL gene are involved in streptomycin resistance in Campylobacter coli. Microb Drug Resist 16:105–110. 10.1089/mdr.2009.0128. [DOI] [PubMed] [Google Scholar]
- 31.Hudson LK, Andershock WE, Yan R, Golwalkar M, M’ikanatha NM, Nachamkin I, Thomas LS, Moore C, Qian X, Steece R, Garman KN, Dunn JR, Kovac J, Denes TG. 2021. Phylogenetic Analysis Reveals Source Attribution Patterns for Campylobacter spp. in Tennessee and Pennsylvania. Microorganisms 9:2300. 10.3390/microorganisms9112300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giacomelli M, Follador N, Coppola LM, Martini M, Piccirillo A. 2015. Survey of Campylobacter spp. in owned and unowned dogs and cats in Northern Italy. Vet J 204:333–337. 10.1016/j.tvjl.2015.03.017. [DOI] [PubMed] [Google Scholar]
- 33.Ramonaitė S, Rokaitytė A, Tamulevičienė E, Malakauskas A, Alter T, Malakauskas M. 2013. Prevalence, quantitative load and genetic diversity of Campylobacter spp. in dairy cattle herds in Lithuania. Acta Vet Scand 55:87. 10.1186/1751-0147-55-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frosth S, Karlsson-Lindsjö O, Niazi A, Fernström L-L, Hansson I. 2020. Identification of transmission routes of Campylobacter and on-farm measures to reduce Campylobacter in Chicken. Pathogens 9:363. 10.3390/pathogens9050363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chereau F, Bessède E, De Valk H, Lehours P. 2020. Bilan de la surveillance des infections à Campylobacter en France en. 2019. 1:7. [Google Scholar]
- 36.Brehony C, Lanigan D, Carroll A, McNamara E. 2021. Establishment of sentinel surveillance of human clinical campylobacteriosis in Ireland. Zoonoses Public Health 68:121–130. 10.1111/zph.12802. [DOI] [PubMed] [Google Scholar]
- 37.Hirayama J, Sekizuka T, Tazumi A, Taneike I, Moore JE, Millar BC, Matsuda M. 2009. Structural analysis of the full-length gene encoding a fibronectin-binding-like protein (CadF) and its adjacent genetic loci within Campylobacter lari. BMC Microbiol 9:192. 10.1186/1471-2180-9-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Haan CPA, Lampén K, Corander J, Hänninen M-L. 2013. Multilocus sequence types of environmental Campylobacter jejuni isolates and their similarities to those of human, poultry and bovine C. jejuni isolates. Zoonoses Public Health 60:125–133. 10.1111/j.1863-2378.2012.01525.x. [DOI] [PubMed] [Google Scholar]
- 39.Mulder AC, Franz E, de Rijk S, Versluis MAJ, Coipan C, Buij R, Müskens G, Koene M, Pijnacker R, Duim B, Bloois L van der G, Veldman K, Wagenaar JA, Zomer AL, Schets FM, Blaak H, Mughini-Gras L. 2020. Tracing the animal sources of surface water contamination with Campylobacter jejuni and Campylobacter coli. Water Res 187:116421. 10.1016/j.watres.2020.116421. [DOI] [PubMed] [Google Scholar]
- 40.Bézian MC, Ribou G, Barberis-Giletti C, Mégraud F. 1990. Isolation of a urease positive thermophilic variant of Campylobacter lari from a patient with urinary tract infection. Eur J Clin Microbiol Infect Dis 9:895–897. 10.1007/BF01967506. [DOI] [PubMed] [Google Scholar]
- 41.Weis AM, Storey DB, Taff CC, Townsend AK, Huang BC, Kong NT, Clothier KA, Spinner A, Byrne BA, Weimer BC. 2016. Genomic comparison of Campylobacter spp. and their potential for zoonotic transmission between birds, primates, and livestock. Appl Environ Microbiol 82:7165–7175. 10.1128/AEM.01746-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rivera-Mendoza D, Martínez-Flores I, Santamaría RI, Lozano L, Bustamante VH, Pérez-Morales D. 2020. Genomic analysis reveals the genetic determinants associated with antibiotic resistance in the zoonotic pathogen Campylobacter spp. distributed globally. Front Microbiol 11:513070. 10.3389/fmicb.2020.513070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mayr AM, Lick S, Bauer J, Thärigen D, Busch U, Huber I. 2010. Rapid detection and differentiation of Campylobacter jejuni, Campylobacter coli, and Campylobacter lari in Food, using multiplex Real-Time PCR. J Food Prot 73:241–250. 10.4315/0362-028x-73.2.241. [DOI] [PubMed] [Google Scholar]
- 44.Wang G, Clark CG, Taylor TM, Pucknell C, Barton C, Price L, Woodward DL, Rodgers FG. 2002. Colony multiplex PCR assay for identification and differentiation of Campylobacter jejuni, C. coli, C. lari, C. upsaliensis, and C. fetus subsp. fetus. J Clin Microbiol 40:4744–4747. 10.1128/JCM.40.12.4744-4747.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ingle DJ, Gonçalves da Silva A, Valcanis M, Ballard SA, Seemann T, Jennison AV, Bastian I, Wise R, Kirk MD, Howden BP, Williamson DA. 2019. Emergence and divergence of major lineages of Shiga-toxin-producing Escherichia coli in Australia. Microb Genom 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Di Tommaso P, Chatzou M, Floden EW, Barja PP, Palumbo E, Notredame C. 2017. Nextflow enables reproducible computational workflows. Nat Biotechnol 35:316–319. 10.1038/nbt.3820. [DOI] [PubMed] [Google Scholar]
- 47.Ewels P, Magnusson M, Lundin S, Käller M. 2016. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32:3047–3048. 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 50.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pedersen BS, Quinlan AR. 2018. Mosdepth: quick coverage calculation for genomes and exomes. Bioinformatics 34:867–868. 10.1093/bioinformatics/btx699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31:3210–3212. 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 53.Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 55.Laslett D, Canback B. 2004. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res 32:11–16. 10.1093/nar/gkh152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Auch AF, von Jan M, Klenk H-P, Göker M. 2010. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand Genomic Sci 2:117–134. 10.4056/sigs.531120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Konstantinidis KT, Tiedje JM. 2005. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci USA 102:2567–2572. 10.1073/pnas.0409727102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. 2018. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun 9:5114. 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pritchard L. PYANI: Python module for average nucleotide identity analyses. https://github.com/widdowquinn/pyani/releases/tag/v0.2.7.
- 61.Pritchard L, Glover RH, Humphris S, Elphinstone JG, Toth IK. 2016. Genomics and taxonomy in diagnostics for food security: soft-rotting enterobacterial plant pathogens. Anal Methods 8:12–24. 10.1039/C5AY02550H. [DOI] [Google Scholar]
- 62.Meier-Kolthoff JP, Auch AF, Klenk H-P, Göker M. 2013. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60. 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jünemann S, Sedlazeck FJ, Prior K, Albersmeier A, John U, Kalinowski J, Mellmann A, Goesmann A, von Haeseler A, Stoye J, Harmsen D. 2013. Updating benchtop sequencing performance comparison. Nat Biotechnol 31:294–296. 10.1038/nbt.2522. [DOI] [PubMed] [Google Scholar]
- 64.Seemann T. 2016. Scan contig files against PubMLST typing schemes. https://github.com/tseemann/mlst.
- 65.Sahl JW, Lemmer D, Travis J, Schupp JM, Gillece JD, Aziz M, Driebe EM, Drees KP, Hicks ND, Williamson CHD, Hepp CM, Smith DE, Roe C, Engelthaler DM, Wagner DM, Keim P. 2016. NASP: an accurate, rapid method for the identification of SNPs in WGS datasets that supports flexible input and output formats. Microb Genom 2:e000074. 10.1099/mgen.0.000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. 2019. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35:4453–4455. 10.1093/bioinformatics/btz305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Seeman T. 2018. Abricate, Github. https://github.com/tseemann/abricate. [Google Scholar]
- 68.Chen L, Yang J, Yu J, Yao Z, Sun L, Shen Y, Jin Q. 2005. VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res 33:D325–328. 10.1093/nar/gki008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zankari E, Allesøe R, Joensen KG, Cavaco LM, Lund O, Aarestrup FM. 2017. PointFinder: a novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J Antimicrob Chemother 72:2764–2768. 10.1093/jac/dkx217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, Fookes M, Falush D, Keane JA, Parkhill J. 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31:3691–3693. 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cantalapiedra CP, Hernández-Plaza A, Letunic I, Bork P, Huerta-Cepas J. 2021. eggNOG-mapper v2: functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol Biol Evol 38:5825–5829. 10.1093/molbev/msab293. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 and S2. Download aem.01368-22-s0001.pdf, PDF file, 0.8 MB (786.6KB, pdf)
Tables S1 to S9. Download aem.01368-22-s0002.xlsx, XLSX file, 3.6 MB (3.6MB, xlsx)
Data Availability Statement
WGS data sets used in this study were deposited at DDBJ/EMBL/GenBank. The sequences were published under BioProject no. PRJNA798893 and PRJNA818070, BioSample no. SAMN25132837 to SAMN25132984 and SAMN26815012 to SAMN26815036, Genome no. JAKMUQ000000000 to JAKMPA000000000, and SRA accession no. SRR17832004 to SRR17831927 and SRR18392311 to SRR18392294 at the NCBI sequence read archive (SRA). Accession numbers are listed in Table S1B.