ABSTRACT
Alginate is an important polysaccharide in the ocean that supports the growth of marine microorganisms. Many widespread Vibrio species possess alginate lyases and can utilize alginate as a carbon source, but the detailed alginate degradation mechanism in Vibrio remains to be further explored. In this study, we obtained a highly efficient alginate-degrading strain, Vibrio pelagius WXL662, with 11 alginate lyases (VpAly-I to -XI) and further elucidated its molecular mechanism of alginate degradation. Three alginate utilization loci (AUL) were identified in different parts of WXL662’s genome, comprising six alginate lyases (VpAly-I, -II, -VIII, -IX, -X, and -XI) and other genes related to alginate degradation. Most of the alginate-degrading genes are strongly induced when alginate is provided as the sole carbon source. Ten alginate lyases (VpAly-I to -X) had been purified and characterized, including six from polysaccharide lyase family 7 (PL7), three from PL17, and one from PL6. These recombinant alginate lyases existing in different cellular locations were active at a wide temperature (10 to 50°C) and pH (4.0 to 9.0) range, with different substrate preferences and diverse degradation products, enabling WXL662 to efficiently utilize alginate in a changing marine environment. Importantly, outer membrane vesicles (OMVs) can act as vectors for alginate lyases (VpAly-II, -V, and -VI) in WXL662. Further investigations of public Vibrio genomes revealed that most alginate-degrading vibrios possess one AUL instead of previously reported “scattered” system. These results emphasize the specific alginate degradation strategy in Vibrio pelagius WXL662, which can be used as a model strain to study the ecological importance of effective alginate-degrading vibrios in the ocean.
IMPORTANCE Alginate is an important carbon source in the marine environment, and vibrios are major alginate utilizers. Previous studies focused only on the characteristics of individual alginate lyases in vibrios, but few of them discussed the comprehensive alginate-degrading strategy. Here, we depicted the alginate utilization mechanism and its ecological implications of a highly efficient alginate-degrading Vibrio strain, WXL662, which contained 11 alginate lyases with distinct enzymatic characteristics. Importantly, unlike other vibrios with only one alginate utilization locus (AUL) or the previously reported “scattered” system, three AUL were identified in WXL662. Additionally, the involvement of outer membrane vesicles (OMVs) in the secretion of alginate lyases is proposed for the first time.
KEYWORDS: Vibrio pelegius, alginate lyases, alginate degradation mechanism, outer membrane vesicles, OMVs
INTRODUCTION
Vibrio species are ubiquitous heterotrophic bacteria with high metabolic flexibility in the marine environment, especially in the coastal systems (1, 2). Vibrio comprises about 10% of the readily culturable bacteria, with an average abundance of 103 to 106 CFU L−1 in estuarine and coastal waters (3). Most of them could utilize various marine carbon sources, such as chitin, agar, alkane, and alginate, by encoding multiple polysaccharide lyases (PLs) to efficiently degrade these types of marine organic matter (2, 4). Thus, Vibrio spp. may exert large impacts on marine organic carbon cycling, particularly in marginal seas (2, 5).
Alginate is the main polysaccharide of brown algae (~40% of dry weight), consisting of the uronic acids β-d-mannuronopyranosyl (M) and α-l-guluronopyranosyl (G) linked by a β-1,4-glycosidic bond (6, 7). Due to the different random combinations of these two uronic acids, alginates are divided into three forms: i.e., polyM (monomer M linked by a β-1,4-glycosidic bond), polyG (monomer G linked by β-1,4-glycosidic bond), and polyMG (monomers G and M randomly arranged and linked by a β-1,4-glycosidic bond) (8). Alginate is a crucial nutrient for numerous marine heterotrophic bacteria (9, 10), and the degradation of alginate by bacterial alginate lyases is one of the important processes in the marine carbon cycle (11). Until now, numerous alginate lyases from Vibrio, Sphingomonas, Pseudoalteromonas, and Pseudomonas were identified and characterized (12). These alginate lyases degrade alginate into unsaturated oligosaccharides by cleaving glycosidic bonds via a β-elimination mechanism (13). In the Carbohydrate-Active enZYmes (CAZy) database, currently known alginate lyases are distributed in 12 PL families (PL5, -6, -7, -14, -15, -17, -18, -31, -32, -34, -36, and -39) according to their amino acid sequence composition and spatial structure characteristics (14).
To date, three different alginate degradation systems have been reported in bacteria: the polysaccharide utilization locus (PUL) system, the “scattered” system (mainly in Vibrio strains), and the “pit” transport system (only found in Sphingomonas sp. strain A1) (15). Some PULs are specific alginate utilization loci (AUL) in many alginate-utilizing bacteria, such as members within Bacteroidetes (10, 16) and Gammaproteobacteria (17). In the PUL system, extracellular alginate lyases first depolymerize the macromolecular alginate (18), and the resulting oligosaccharide products were subsequently transferred into the periplasmic space, where oligoalginate lyases (Oals) catalyze these oligosaccharides into monomers (15). The major facilitator superfamily (MFS) transporter or inner membrane transporter-like protein (ToaABC) transfers monomers into the cytoplasm (CP), and KdgF catalyzes the ketonization of unsaturated monomers into 4-deoxy-l-erythro-5-hexoseulose uronate (DEH) (15, 19). DEH is converted into the final product 2-keto-3-deoxy-6-phosphogluconate (KDPG) through multiple downstream enzymes (DehR, KdgK, and Eda) and assimilated through the Entner-Doudoroff (ED) pathway (10, 20, 21). Different from the PUL system, the scattered system mainly occurs in vibrios and comprises alginate decomposition genes scattered throughout the genome instead of in one operon (15). Besides, the scattered system employs porin KdgMN for extracellular substrate transport but not SusC and SusD (15, 22). Oligosaccharides in periplasm (PP) are transferred into cytoplasm directly via the inner membrane symporter ToaABC instead of the MFS system (15). The pit system is only found in Sphingomonas sp. strain A1, in which macromolecular alginate is incorporated into cytoplasm directly via a superchannel consisting of a “mouth-like pit” on the outer membrane (23).
In this study, a highly efficient alginate-degrading strain, Vibrio pelagius WXL662, was isolated from offshore seawater in Qingdao, China, which possessed a distinct alginate-degrading strategy instead of the previously reported scattered system in vibrios. Three AUL are distributed across different regions in the genome of V. pelagius WXL662 and cooperate to catalyze alginate. Eleven alginate lyases from strain WXL662 were identified showing diverse enzymatic properties. A novel insight into the alginate degradation strategy of vibrios is proposed and further discussed, in which outer membrane vesicles (OMVs) are detected as important vectors for alginate lyases.
RESULTS
Genes related to alginate degradation in V. pelagius WXL662.
Strain WXL662 showed strong alginate-degrading activity at 16 to 37°C (see Fig. S1 in the supplemental material). The 16S rRNA gene sequence indicated that WXL662 showed the highest similarity (99.29%) to Vibrio pelagius CECT4202T (GenBank ID AJ293802). Genomic analysis of WXL662 revealed 11 alginate lyase-encoding genes in total (named Vpaly-I to -XI) (Table 1). In addition, other genes involved in alginate degradation have also been identified, including the genes encoding three oligogalacturonate-specific porinins of KdgM (N-acetylneuraminic acid outer membrane channel protein [NanC]), three 2-dehydro-3-deoxygluconate kinases (KdgK-like proteins), three 2-dehydro-3-deoxyphosphogluconate aldolases (Eda), and multiple transporters (sodium solute symporter [SSF] and the alginate transport system, termed ToaA) (Fig. 1 and Fig. S2).
TABLE 1.
Nucleotide and amino acid sequence analysis of the alginate lyases from strain WXL662
| Protein ID | Length |
Structural domain(s) | PL | Signal peptide | Predicted MW (kDa) | pI | Most similar protein | Accession no. | Coverage (%) | Identity (%) | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Nucleic acid (bp) | Amino acid (aa) | ||||||||||
| VpAly-I | 1,752 | 583 | PL7+PL7 | PL7 | Y | 65.78 | 4.59 | Alginate lyase from Vibrio sinaloensis | P39049 | 43 | 41 |
| VpAly-II | 1,569 | 522 | CBM32+PL7_5 | PL7 | Y | 57.40 | 4.91 | Alginate lyase from Vibrio splendidus OU02 | 5ZU5_A | 95 | 69 |
| VpAly-III | 1,038 | 345 | PL7_5 | PL7 | Y | 38.16 | 5.67 | Alginate lyase from Vibrio splendidus OU02 | 5ZU5_A | 89 | 34 |
| VpAly-IV | 1,350 | 345 | CBM32+PL7_5 | PL7 | Y | 49.19 | 4.29 | Alginate lyase from Vibrio splendidus OU02 | 5ZU5_A | 91 | 48 |
| VpAly-V | 864 | 287 | PL7 | PL7 | Y | 31.82 | 7.01 | Alginate lyase (Aly C3) from Psychromonas sp. strain C-3 | 7C8G_A | 87 | 49 |
| VpAly-VI | 1,050 | 349 | PL7_5 | PL7 | Y | 39.17 | 6.06 | Alginate lyase (AlyA1) from Zobellia galactanivorans DsijT | 4BE3_A | 88 | 36 |
| VpAly-VII | 1,617 | 538 | PL6 | PL6 | Y | 59.31 | 4.74 | Alginate lyase (AlyF) from Vibrio splendidus OU02 | 5Z9T_A | 97 | 91 |
| VpAly-VIII | 2,151 | 716 | PL17_2+PL17 | PL17 | N | 80.35 | 5.05 | Oligoalginate lyase (AlyA3) from Zobellia galactanivorans DsijT | 7BJT_A | 98 | 35 |
| VpAly-IX | 2,184 | 727 | PL17_2+PL17_2 | PL17 | N | 82.22 | 5.14 | Oligoalginate lyase (AlyA3) from Zobellia galactanivorans DsijT | 7BJT_A | 97 | 34 |
| VpAly-X | 2,037 | 678 | PL17_1 | PL17 | N | 77.24 | 5.29 | Alginate lyase (Alg17c) from Saccharophagus degradans 2-40 | 4NEI_A | 52 | 27 |
| VpAly-XI | 1,059 | 352 | PL14 | PL14 | N | 40.55 | 6.64 | Alginate lyase from Bacteroides ovatus ATCC 8483 | 3NFV_A | 91 | 33 |
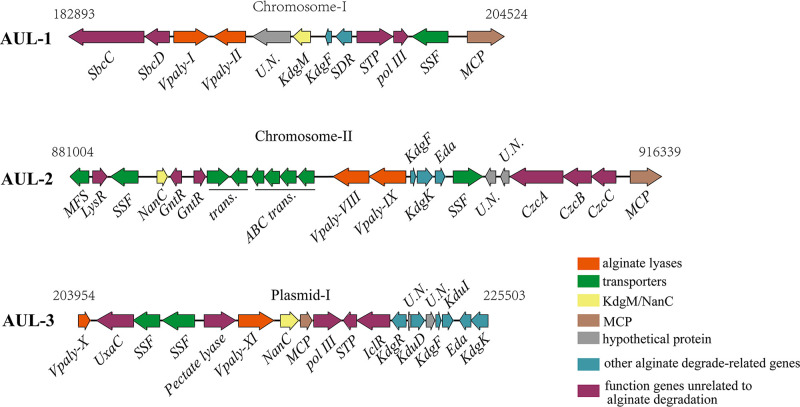
FIG 1.
Three AUL in the genome of strain WXL662. Numbers above the AUL are the start and end positions in the chromosome and plasmid. SSF, sodium-solute symporter (alginate transport system, termed ToaA); KdgF, pectin degradation protein KdgF; KdgK, KdgK-like protein (2-dehydro-3-deoxygluconokinase); Eda, 2-dehydro-3-deoxyphosphogluconate aldolase; SDR, short-chain dehydrogenase/reductase; KdgM, oligogalacturonate-specific porinin; NanC, N-acetylneuraminic acid outer membrane channel protein NanC; MCP, methyl-accepting chemotaxis protein, homolog 13; STP, predicted signal-transduction protein containing cAMP-binding and CBS domains; pol III, DNA polymerase III epsilon subunit; MFS, transmembrane transport MFS_1; LysR, transcriptional regulator, LysR family; GntR, transcriptional regulator, GntR family; CzcA, cation efflux system protein CusA; CzcB, cobalt/zinc/cadmium efflux RND transporter, membrane fusion protein, CzcB family; CzcC, heavy metal RND efflux outer membrane protein, CzcC family; UxaC, uronate isomerase; IclR, bacterial transcriptional regulator IclR family; KdgR, transcriptional regulator KdgR, KDG operon repressor; KduD, 2-deoxy-d-gluconate 3-dehydrogenase; KduI, predicted 4-deoxy-l-threo-5-hexosulose-uronate ketol-isomerase.
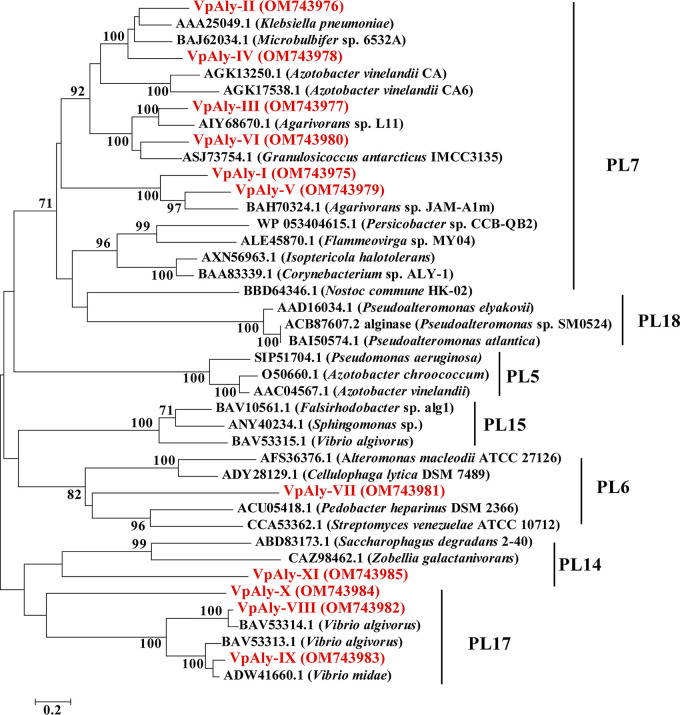
Detailed information about the 11 alginate lyases (VpAly-I to -XI) in V. pelagius WXL662 is summarized in Table 1. Six of them (VpAly-III, -VI, -VIII, -IX, -X, and -XI) showed low sequence identity with known alginate lyases (<40%). The predicted molecular weight (MW) of these 11 VpAlys varied between 31.82 to 65.78 kDa, and their pIs were between 4.29 and 7.01. These VpAlys belonged to four different PL families (Fig. 2). Six of them belonged to the PL7 family, possessing either a single catalytic domain (VpAly-III, -V, and VI), double catalytic domain (VpAly-I), or multidomain with carbohydrate-binding domain CBM32 (VpAly-II and VpAly-IV). VpAly-VII is a PL6 alginate lyase with a single catalytic domain. In the PL17 family, VpAly-VIII and VpAly-IX contained a double catalytic domain, whereas VpAly-X is a single catalytic domain alginate lyase. VpAly-XI from the PL14 family encodes one catalytic domain. Each of the VpAly genes in the PL6 and PL7 families encodes a signal peptide at its N terminus, while no signal peptides were found in four VpAlys from the PL17 and PL14 families.
FIG 2.
Neighbor-joining phylogenic tree based on amino acid sequences of 11 alginate lyases (in red) from strain WXL662 and other known alginate lyases. Bootstrap values were calculated based on 1,000 replicates. Bar, 0.2 substitution per nucleotide position. All amino acid sequences, which belonged to families PL6, PL7, PL14, and PL17, were from the CAZymes database.
The cellular localization of alginate lyases and other key enzymes in this pathway were predicted (Table S6). Three oligoalginate lyases from PL17 (VpAly-VIII, -IX, and VpAly-X) and one PL14 alginate lyase (VpAly-XI) were in cytoplasm (CP). VpAly-I to -VII seemed to be secretory alginate lyases considering the signal peptides on them. Although there is a discrepancy between different softwares when predicting the accurate cellular localization of VpAly-I to -VII, we assumed that VpAly-III was in periplasm (PP) based on the results of Cello, while the locations of other VpAlys were more likely to be in the extracellular space (EC). For other proteins involved in alginate degradation, KdgK-like protein, Eda, and phosphoglycerate kinase were in cytoplasm (CP); three alginate-degrading specific transporter proteins (KdgM and NanC) were in the outer membrane (OM).
Identification of three AUL in V. pelagius WXL662.
Although the pervious study (10, 15) showed that vibrios comprise alginate decomposition genes scattered throughout the genome instead of in one operon, we found that alginate-degrading genes in strain WXL662 existed in clusters, forming three typical alginate utilization loci (AUL1 to -3) (Fig. 1). AUL1 to -3 were located at the macrochromosome (chromosome I), microchromosome (chromosome II), and a large plasmid (plasmid I), respectively. Each AUL contains genes coding for two distinct alginate lyases, multiple transporter proteins (including KdgM/NanC in the outer membrane and ToaA in the inner membrane), and some key genes for further hydrolyses, such as the KdgF-encoding gene, which could catalyze the ketonization of unsaturated monomers into DEH. Differently, AUL1 has a gene encoding short-chain dehydrogenases/reductases (SDRs) with DEH reductase activity (24), whereas another two AUL lack this gene. In addition, AUL2 and AUL3 carry the genes that encode downstream metabolic KdgK-like protein (2-dehydro-3-deoxygluconate kinase) and Eda (2-dehydro-3-deoxyphosphogluconate aldolase), respectively. KDG (2-keto-3-deoxy-d-gluconic acid) could be catalyzed by KdgK (2-keto-3-deoxy-d-gluconic acid kinases) and Eda into KDPG, which can be directly assimilated through the Entner-Doudoroff (ED) pathway. It is worth noting that some genes, such as kduD (2-deoxy-d-gluconate 3-dehydrogenase), uxaC, and kduI (predicted 4-deoxy-l-threo-5-hexosulose-uronate ketol-isomerase) in AUL3, also play a role in pectin degradation (25), suggesting that AUL3 may be a multifunctional locus.
Transcription of alginate degradation genes with alginate as the sole carbon source.
To explore the transcription of alginate-degrading genes, transcriptome sequencing and quantitative reverse transcription-PCR (qRT-qPCR) were conducted on strain WXL662 cultured in 0.6% (wt/vol) alginate and glucose mineral medium (Tables S7 and S8 and Fig. S2). Among the alginate-degrading genes, those coding for six endo-type VpAlys in the PL7 family were obviously upregulated, which were responsible for converting alginate into AOS (degrees of polymerization [DP] = 3 to 6). Three PL17 oligoalginate lyase-encoding genes as well as one PL6 and one PL14 VpAly-encoding gene each were not differentially regulated by alginate. Additionally, three transporter protein KdgM (NanC) genes in each AUL and genes encoding some key enzymes related to downstream alginate metabolism, such as Kdgk-like protein and Eda in AUL2, were also significantly upregulated (log2 fold change [FC] of >1; P < 0.01). The KdgF- and short-chain dehydrogenase/reductase (SDR)-encoding genes in AUL1, as well as the KdgF- and DEH ketol-isomerase-encoding genes in AUL3 were upregulated (Tables S7 and S8). Moreover, transcription of many transporters located near alginate lyases was induced by the addition of alginate, such as the ABC transporter, sodium-solute symporter (SSF) and putative alginate transport system ToaA (26). The qRT-PCR confirmed the accuracy of the results mentioned above. Therefore, alginate-degrading genes distributed in three AUL can be upregulated in response to alginate, and these three AUL seems to cooperate in catabolizing alginate efficiently. Furthermore, the transcriptional levels of genes involved in the central metabolic cycle of the strain WXL662 were also elevated (log2 FC of >1; P < 0.01), suggesting that the efficient utilization of alginate boosts the energy metabolism of V. pelagius WXL662.
Alginate lyases in V. pelagius WXL662 exhibited diverse enzymatic properties.
In this study, 11 potential VpAlys were expressed in E. coli BL21(DE3), and 10 of them (except VpAly-XI) were purified and showed alginate degradation activity. The crude enzyme solution of recombinant enzyme VpAly-XI was active in alginate degradation, but we failed to get purified enzyme. The detailed enzymatic property of VpAly-V was characterized by Zhang et al. (27), and those data on other nine VpAlys were obtained in this study. The molecular masses of the purified proteins were estimated by SDS-PAGE (Fig. S3), which were consistent with the predicted molecular mass (Table 1).
These 10 purified recombinant VpAlys exhibited a wide range of temperature adaptabilities, and their optimal temperatures varied between 10 and 50°C (Table 2 and Fig. S4) (27). Particularly, VpAly-VI, a potential novel member in the PL7 family, showed an optimum temperature at 10°C as a cryogenic alginate lyase, while VpAly-VII showed optimal activity at 50°C (Table 2 and Fig. S4) (27). The optimum pHs of these 10 VpAlys ranged between 6.0 and 8.0 (Table 2 and Fig. S5) (27). Among them, VpAly-I preferred an acidic environment, showing optimal activity at pH 6.0 and maintaining more than 40% enzyme activity at pH 5.0. However, VpAly-II, -III, -V, -VI, and -VII preferred an alkaline environment and showed the maximum enzymatic activity at pH 8.0. These VpAlys responded differently to metal ions and reducing agents (Table 2 and Fig. S6). For instance, 5 mM Zn2+ increased the enzyme activity of VpAly-VIII by ~2.5-fold. Al3+, Ni2+, Ca2+, Zn2+, Ba2+, and Mg2+ (at 5 mM) increased the enzyme activity of VpAly-IX by over 2.5-fold. The broad temperature and pH range and distinct responses to environmental factors shown by these alginate lyases in WXL662 could enable this strain to better utilize alginate in a changing marine environment.
TABLE 2.
Protein contents and enzymatic properties of purified alginate lyases in strain WXL662
| Protein ID | Total protein (mg) | Total enzymatic activity (U) | Sp act (U/mg) | Optimal temp (°C) | Optimal pH | Response to metal ions and reductants | Substrate preference | Degradation product(s) (DP) |
|---|---|---|---|---|---|---|---|---|
| VpAly-I | 2.13 | 415.85 | 194.83 | 40 | 6.0 | Improved: 1 mM Ca2+, Al3+, and Zn2+ | Alginate > polyG > polyM | 3–6 |
| VpAly-II | 3.12 | 391.55 | 125.50 | 42 | 8.0 | Improved: 1 mM Ca2+, Mg2+, K+, and Na+; 5 mM K+ and Na+ | Alginate > polyG > polyM | 3 |
| VpAly-III | 6.14 | 463.25 | 75.45 | 40 | 8.0 | Improved: 1 mM Ca2+, Mg2+, and K+; 5 mM Mg2+ and K+ | Alginate > polyG = polyM | 3 |
| VpAly-IV | 3.72 | 167.32 | 44.98 | 28 | 6.8 | Improved: 1 mM Ni2+and Ca2+ | Alginate > polyG > polyM | 3–4 |
| VpAly-V | 40 | 8.0 | polyM > alginate > polyG | 3 | ||||
| VpAly-VI | 3.51 | 154.98 | 44.15 | 10 | 8.0 | Improved: 1 mM urea | Alginate > polyG > polyM | 3–6 |
| VpAly-VII | 5.93 | 86.78 | 14.63 | 50 | 8.0 | Alginate > polyM > polyG | 3–6 | |
| VpAly-VIII | 9.28 | 97.26 | 10.48 | 40 | 6.8 | Improved: 5 mM Zn2+ (~2.5 times) | polyG > polyM > alginate | 1 |
| VpAly-IX | 15.33 | 152.23 | 9.93 | 40 | 7.0 | Improved: 5 mM Al3+, Ni2+, Ca2+, Zn2+, Ba2+, and Mg2+ (>2.5 times) | polyM > polyG > alginate | 1 |
| VpAly-X | 14.61 | 162.26 | 11.11 | 48 | 6.8 | Improved: 1 mM Ca2+, Sr2+; 5 mM Na+ | polyM > alginate > polyG | 1 |
| VpAly-XI | polyM > polyG > alginate |
Additionally, alginate lyases in WXL662 presented distinct substrate preference. VpAly-IX and VpAly-X preferred polyM over polyG blocks and sodium alginate as previously reported for PL17 oligoalginate lyases (28) (Table S10), while VpAly-VIII preferred polyG over polyM blocks and sodium alginate (Table 2 and Fig. S7). Considering the protein sequence identity to the most similar protein (AlyA3 from Zobellia galactanivorans DsijT) was only 35%, VpAly-VIII could be a novel PL17 oligoalginate lyase. VpAly-VIII showed distinct optimal pH and substrate preference (pH 6.8 and polyG) compared with AlyA3 (pH 8 and polyM). VpAly-V preferred to catalyze polyM, whereas the other PL7 VpAlys preferred to degrade sodium alginate (27). VpAly-VII in the PL6 family also preferred sodium alginate. Moreover, the degradation products of these alginate lyases were highly diverse, producing different types of alginate oligosaccharides (Table 2 and Fig. S8). VpAly-I to -VII (in PL6 and PL7) were all endo-type enzymes, producing different types of alginate oligosaccharides (DP = 3 to 6), while three oligoalginate lyases from PL17 (VpAly-VIII to -X) located in cytoplasm produced only monosaccharide (Table 2 and Fig. S8).
Outer membrane vesicles involved in the secretion of alginate lyases.
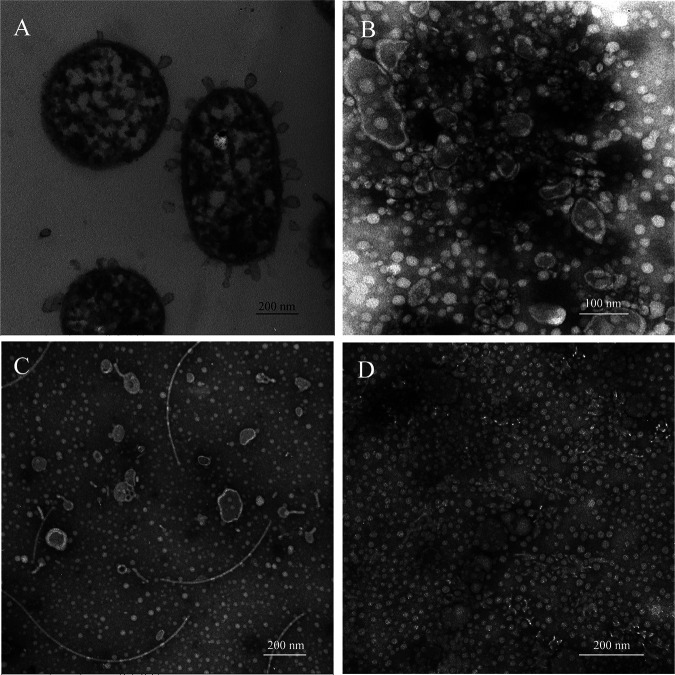
Based on the transmission electron microscopy (TEM) (Fig. S9) and AFM (Fig. S10) observation, strain WXL662 could secrete outer membrane vesicles (OMVs) when cultured in mineral medium with alginate as the sole carbon source (Fig. 3). To verify whether these OMVs function in the alginate degradation process or not, we collected and analyzed OMVs under three different cultural conditions: 0.6% (wt/vol) alginate mineral medium, 0.6% (wt/vol) glucose mineral medium, and marine ZoBell broth (MB). Crude vesicle components collected from the cultures in 0.6% (wt/vol) alginate mineral medium showed an obvious difference in the coarse bands between 25.0 and 35.0 kDa (Fig. S11). The different bands were sequenced to verify the composition of vesicles. As shown in Table S11, a total of 219 credible proteins were identified in this target band, including alginate lyases VpAly-II, VpAly-V, and VpAly-VII with relatively high content (ranking 15, 16, and 42 of 219 proteins, respectively). Therefore, these three VpAlys seemed to be secreted into the periplasm and then transported to extracellular space by OMVs. In addition, genes encoding the three most abundant proteins in OMVs located in AUL1, including one alginate lyase, VpAly-II, a putative biofilm-associated surface protein, and one oligogalacturonic acid-specific porin, KdgM. Thus, it is hypothesized that AUL1 might be the key locus responsible for the extracellular degradation of alginate through OMVs.
FIG 3.
Morphology of purified vesicle samples under different culture conditions. (A) Morphology and internal structure of strain WXL662 cultured with alginate as the sole carbon source; (B) morphology of pure OMVs cultured with alginate as the sole carbon source (bar, 100 nm); (C) morphology of pure OMVs cultured in MB medium (bar, 200 nm); (D) morphology of WXL662 with glucose as the sole carbon source, which produced no OMVs (bar, 200 nm).
AUL are commonly found in the genome of the alginate-degrading Vibrio strains.
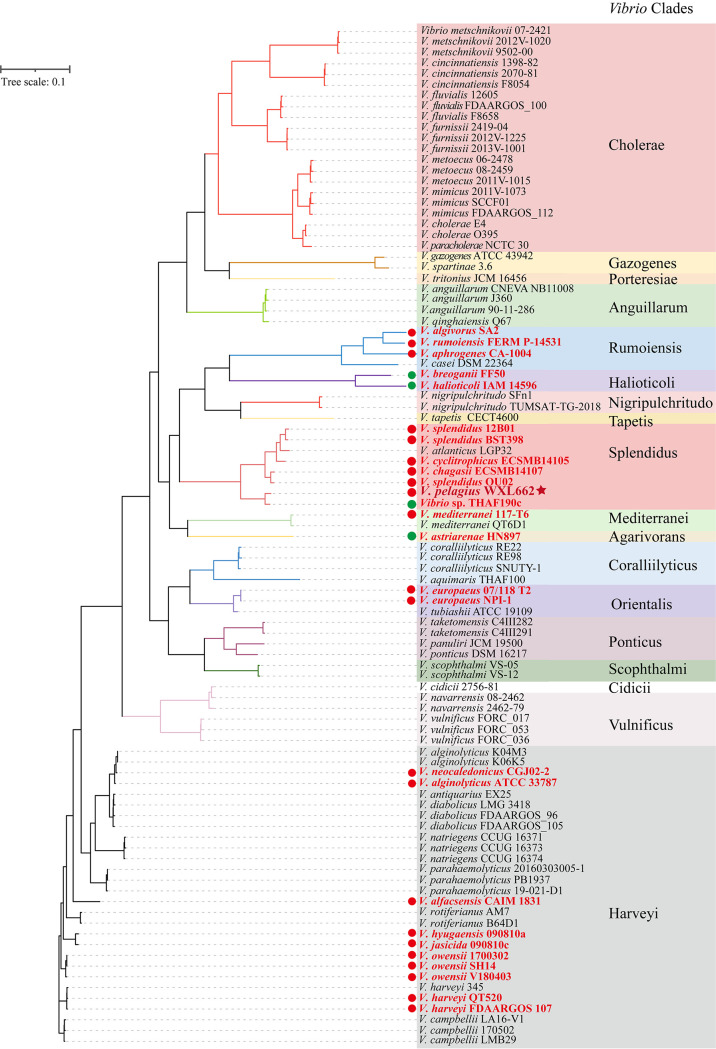
To further determined the alginate-degrading species in the genus Vibrio, a total of 94 nearly complete Vibrio genomes were downloaded from NCBI database and annotated in this study (Table S2). Maximum likelihood (ML) tree of these Vibrio genomes from 18 Vibrio clades was constructed (Fig. 4). The alginate-degrading genes were identified in a total of 25 (~26%) Vibrio strains from seven Vibrio clades (Rumoiensis, Halioticoli, Splendidus, Mediterranei, Agarivorans, Orientalis, and Harveyi) (Fig. 4) (29–31). The alginate-degrading genes in 21 alginate-degrading Vibrio strains existed in clusters, with only one complete AUL in their genomes (red circle labeled in Fig. 4; Fig. S12 and Table S9). Except for the AUL in the genomes of Vibrio owensii 1700302 and V. owensii SH14, which lack genes encoding KdgM and NanC (the alginate-degrading specific transporter proteins), other AUL contain all the genes encoding the key proteins in the alginate degradation pathway, including multiple alginate lyases and polyM lyases (n = 2 to 10), specific transporter KdgM and/or NanC, three types of DEH reductase, KdgF, KdgK-like protein, and Eda. For three AUL in strain WXL662, the gene organizations of AUL2 and AUL3 are similar to those of the AUL in other vibrios, while AUL1 lacks the genes coding for KdgK-like protein and Eda (Fig. 1 and Fig. S12). However, the alginate-degrading genes in four alginate-degrading Vibrio strains, including V. breoganii FF50, V. astriarenae HN897, Vibrio sp. strain THAF190c, and V. halioticoli IAM 14596T, were scattered in the genomes. These results indicated that the AUL is more commonly found in Vibrio species than the “scattered” system of alginate-degrading genes. Most Vibrio strains possess only one AUL, except V. pelagius WXL662, in which three AUL have been identified.
FIG 4.
Maximum likelihood (ML) phylogenetic tree of 95 Vibrio genomes from 18 Vibrio clades. Genomes with alginate degradation genes are marked with solid circles (red in the form of AUL and green for the scattered system). The genome of V. pelagius WXL662 is marked with a star.
DISCUSSION
Many widespread Vibrio species could utilize alginate as a carbon source (22, 32, 33). Alginate-utilizing Vibrio strains encode multiple alginate lyases and own a specific strategy (i.e., the scattered system) to degrade alginate (15, 22, 34). In the present study, we described an efficient alginate degradation mechanism in V. pelagius WXL662 with identification of multiple AUL and alginate lyase-containing OMVs. Alginate lyases with diverse enzymatic properties have been characterized, emphasizing the strong alginate utilization ability of V. pelagius WXL662 under different conditions and the potential ecological significance of such alginate-degrading vibrios.
Specific AUL in the genome of V. pelagius WXL662.
AUL consisting of the alginate-degrading genes are commonly found in the genome of the alginate-degrading bacteria. For instance, the AUL in Sphingomonas sp. strain A1 consists of the genes encoding ABC transporters, alginate binding proteins, exoalginate lyase (Aly-IV) and three types of endoalginate lyase (Aly-I, -II, and -III) (35–39). Flavobacterium sp. strain UMi-01 also possessed only one AUL (the length of this AUL is about 15.6 kb)-encoded transporter SusC/SusD-like protein, two alginate lyase genes, one SDR-like enzyme gene, and other genes necessary to degrade alginate downstream (coding for KdgF-like protein, KdgK, and Eda) (18). However, a previous study reported that alginate decomposition genes in Vibrio species are not colocalized in the same operon but distributed across different regions of the whole genome, as described in V. splendidus 12B01 (40), which was named as the “scattered” alginate degradation system (15). However, we actually identified an AUL in the genome of V. splendidus 12B01 (see Fig. S12 in the supplemental material): even though there is an alginate lyase not present in AUL, the distribution pattern of alginate-degrading genes in the genome was still defined as an “AUL system” in our study. We did identify the scattered system in 4 out of 25 alginate-degrading Vibrio strains, confirming that this scattered system is not the mainstream form for alginate degradation in vibrios.
Our results found that although the AUL are widely present in Vibrio genomes, most vibrios contained only one AUL. In contrast, three alginate-degrading AUL located at different positions of WXL662’s genome (AUL1 in chromosome I, AUL2 in chromosome II, and AUL3 in plasmid I, respectively) (Fig. 1). All three AUL are capable of degrading alginate into DEH and further generating KDG through DEH reductase, which can be incorporated into the central metabolic cycle. The transcriptome results with alginate as the sole carbon source showed that all genes in AUL1 were significantly upregulated, and part of the alginate-degrading genes in AUL2 and AUL3 were also upregulated, indicating all three AUL cooperated in the utilization of alginate by WXL662. Multiple AUL may help Vibrio species to rapidly transcribe alginate-degrading genes in response to alginate in the marine environment and confer higher flexibility in the regulation of alginate degradation process.
Cooperation of diverse alginate lyases in V. pelagius WXL662.
In addition to the specific AUL in Vibrio sp. strain WXL662, this strain is also characterized by considerable numbers of alginate lyases (VpAly-I to -XI). Many alginate-degrading bacteria encode multiple alginate lyases that collaborate to degrade alginate (22, 41–44). For instance, 12 potential alginate lyase genes that belong to the PL6, -7, -17, and -38 families were identified in the whole genome of marine bacterium Agarivorans sp. strain B2Z047 (42). Strain Cellulophaga algicola DSM 14237 encodes five alginate lyases, with three endolytic enzymes and two exolytic enzymes. For vibrios, seven alginate lyases from V. splendidus 12B01 were characterized (22, 44), showing optimal activity between pH 7.5 and 8.5 and at 16 to 35°C. In comparation, more alginate lyases were identified in WXL662, and these lyases showed a broader optimal temperature range (10 to 50°C), indicating this strain can efficiently degrade alginate under extremely temperature fluctuations. The wide pH and temperature adaptations, distinct substrate preference, and cell locations of these alginate lyases in WXL662 make it a powerful alginate utilizer in marine environments.
Previously reported alginate lyases in vibrios have been found in PL6, -7, and -17, while PL14 alginate lyase (VpAly-XI) in vibrios was reported for the first time in this study (Table 2 and Table S10). Some of these alginate lyases in WXL662 shared high protein sequence identity with other alginate lyases in vibrios but varied in enzymatic activity. For instance, VpAly-VII in the PL6 family showed 90.2% identity with AlyF in Vibrio splendidus OU02 (45), but the degradation products of VpAly-VII were oligosaccharides (DP = 3 to 6), while those of AlyF were trisaccharide. The enzymatic characteristics of 91 previously studied alginate lyases from different species and protein families (PL6, -7, -14, -17) are summarized in Table S10. Six of alginate lyases in WXL662 shared low protein sequence identity with known alginate lyases (<40%) (Table 1), and some of them exhibited distinct enzymatic characteristics compared with their most similar enzyme or enzymes in the same protein family. For example, VpAly-VIII in the PL17 family shared only 35% protein sequence identity with known alginate lyases and exhibits a preference for polyG blocks, while other previously reported PL17 oligoalginate lyases preferred polyM or alginate (28) (Table S10). Moreover, currently reported alginate lyases of the PL7 family exhibit an optimum temperature from 16°C to 70°C (Table S10), while VpAly-VI showed the highest enzymatic activity at 10°C and expanded the range of optimum temperatures of PL7 alginate lyases. Therefore, VpAly-VIII and -VI might represent novel alginate lyases in vibrios.
OMVs assist in the alginate degradation process of strain WXL662.
Vesicles produced by bacteria have a variety of biological functions, including virulence factor transport, DNA transfer, phage interception, antibiotic and eukaryotic host defense, cell detoxification, and communication between bacteria (46–48). Different bacteria, including Vibrio species, could secrete OMVs in various shapes and functions. For instance, V. vulnificus could form segmented tubular OMVs functioning as virulence factors in the logarithmic phase, while OMVs in the plateau phase had low production and irregular shape (49). V. shilonii with chitin degradation ability could release two types of OMVs, with a single membrane or two membranes, which can release signaling molecules and active enzymes such as chitinase to its environment (50).
In this study, we have proposed the roles of OMVs in the alginate degradation process for the first time. The OMVs secreted by strain WXL662 have two forms: single vesicle and the segmented tubular (Fig. S9C and D). The formation of OMVs in strain WXL662 may also related to the growth state of the cell. The new generation of WXL662 cells that have just completed division cannot secrete OMVs immediately. After a period of growth, the cytoplasm begins to spread to the edge of the cell and the OMVs begin to form, subsequently releasing alginate lyases into the environment. Like V. shilonii, which with its chitin degradation ability could secret OMVs that function as the conduits of chitinase, the OMVs secreted by V. pelagius WXL662 could be a vector for several alginate lyases (VpAlyII, -V, and -VII) and other components. By releasing OMVs, vibrios can utilize alginate in the surrounding environment more efficiently. Whether OMVs are commonly involved in the alginate metabolism of vibrios is yet to be confirmed.
Novel insights into the alginate-degrading process of Vibrio species and its ecological significance.
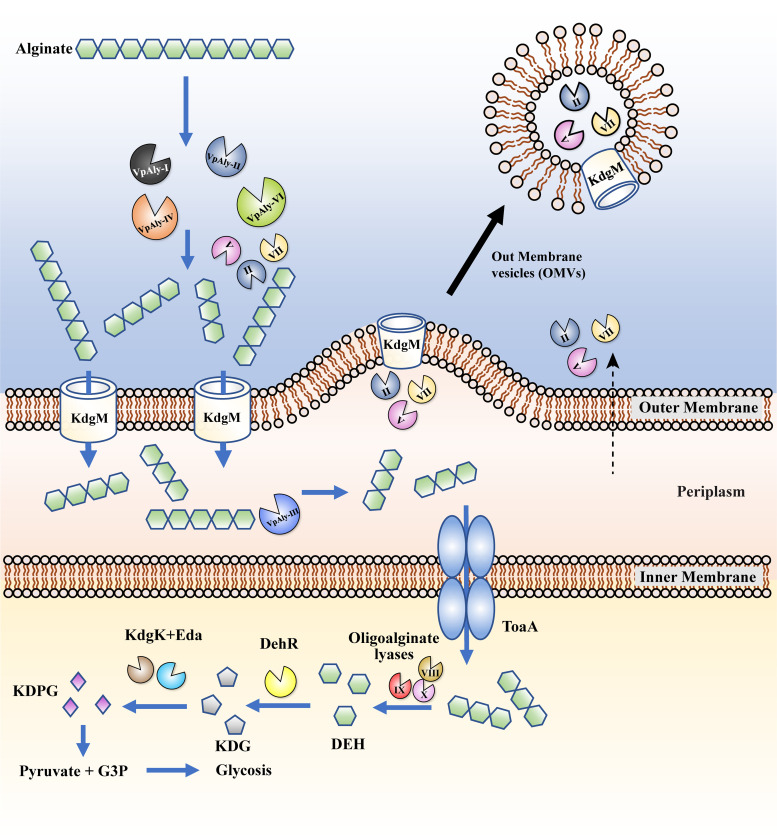
Based on our results, we elucidated the alginate degradation process of strain WXL662 as illustrated in Fig. 5. Briefly, the process occurs as follows. (i) The extracellular enzymes (VpAly-I, -II, -V, -IV, -VI, and -VII) are transported across the membrane and degrade macromolecules of alginate into oligosaccharides with 3 to 6 degrees of polymerization (DP), completing the first step of alginate metabolism. OMVs are involved in the secretion of VpAly-II, -V, and -VII. (ii) Some transporters such as KdgM, which is located on the outer membrane, could transport alginate oligomers into the periplasm. The alginate lyases located in the periplasm (VpAly-III) further degrade oligosaccharides (DP = 3 to 6) into trisaccharides. (iii) Trisaccharide is transferred into the cytoplasm by inner membrane transporter-like ToaABC (15). The oligoalginate lyases (VpAly-VIII, -IX, and -X) reduce trisaccharide into monosaccharides, forming 4-deoxy-l-erythro-5-hexoseulose uronic acid (DEH). DEH is converted into the final product 2-keto-3-deoxy-6-phosphogluconate (KDPG) through multiple downstream enzymes (DehR, KdgK-like protein, and Eda) and assimilated through the central metabolic cycle.
FIG 5.
Alginate degradation strategy of V. pelagius WXL662. The extracellular alginate lyases (VpAly-I, -II, -V, -IV, -VI, and -VII) degrade macromolecule alginate into oligosaccharides with 3 to 6 degrees of polymerization (DP), completing the first step of alginate metabolism. Three alginate lyases (VpAly-II, -V, and -VII) might be transferred into the environment by outer membrane vesicles (OMVs). The specific outer membrane transporter KdgM transfers oligosaccharides (DP = 3 to 6) into the periplasm, where VpAly-III catalyze oligosaccharides into trisaccharides. Furthermore, abundant trisaccharides are transferred into the cytoplasm by inner membrane transporter ToaA, and the oligoalginate lyases (VpAly-IX, -X, XI) in the cytoplasm reduced trisaccharides into monosaccharides. The monosaccharide 4-deoxy-l-erythro-5-hexoseulose uronic acid (DEH) is converted into the final product 2-keto-3-deoxy-6-phosphogluconate (KDPG) through multiple downstream enzymes (DehR, KdgK, and Eda) and assimilated through the central metabolic cycle.
Vibrios are ubiquitous in marine environments, especially in coastal areas, and comprise up to 10% of the readily culturable marine bacteria in culture-based studies and generally ~1% of the total bacterioplankton with culture-independent methods (2). Vibrio strains encoding various alginate lyases are efficient utilizers of alginate in the marine environment and promote the organic carbon cycle in the ocean. In the current study, we found the strong alginate-degrading activity of V. pelagius WXL662 was due to multiple alginate lyases with diverse enzymatic properties. These lyases are organized into three AUL and cooperate during the alginate degradation process. The involvement of OMVs can also further enhance the degrading efficiency of extracellular alginate. These results emphasized the specific alginate utilization strategy in V. pelagius WXL662, which may represent an ideal model to study highly efficient alginate-utilizing Vibrio species.
Conclusion.
In this study, we depicted the alginate utilization mechanism of a highly efficient alginate-degrading strain of V. pelagius, WXL662. The genome of WXL662 encodes 11 alginate lyases and contains other genes involved in the complete degradation of alginates. Ten of these alginate lyases (VpAly-I to -X) were purified and showed activity at a wide range of temperatures and pHs with distinct substrate preference and degradation products. Some of these alginate lyases and other alginate degradation-related genes are organized into three AUL and can be induced with alginate as the sole carbon source. Furthermore, OMVs were involved in the transportation of alginate lyases. These results help us to better understand the alginate utilization process in vibrios and emphasize the ecological importance of Vibrio species in the global carbon cycle.
MATERIALS AND METHODS
Strains, media, and growth conditions.
V. pelagius WXL662 was isolated from the offshore seawater (0.3 m) in Qingdao, China (120.3183°E, 36.0618°N), in July 2015 and was routinely cultured on marine agar 2216E (MA) (Hopebio, Qingdao, China). To detect the alginate-degrading ability at different temperatures, strain WXL662 were cultured on 0.6% sodium alginate agar plates [per L: 30 g NaCl, 7 g K2HPO4, 3 g K2HPO4, 2 g (NH4)2SO4, 0.05 g FeSO4·7H2O, 0.1 g MgSO4·7H2O, 6 g sodium alginate] at 0, 4, 16, 28, 37, and 45°C for 7 days, and then 10% CaCl2 was poured onto the plates and the colonies were soaked for 30 min to observe whether transparent circles appeared around the colonies. Escherichia coli BL21(DE3) was cultured on Luria-Bertani (LB) agar at 37°C, used as a host for plasmid pET28a(+) (Novagen, Beijing, China). The alginate used in this study was purchased from Shanghai Yuanye Bio-Technology Co., Ltd. The substrates polyM and polyG were kindly provided by Xia Zhao at the Ocean University of China.
Morphological characteristics of strain WXL662.
The morphological characteristics of strain WXL662 were observed using transmission electron microscopy (TEM) and atomic force microscopy (AFM). Cells were prepared by growing at 28°C to the mid-logarithmic growth phase (optical density at 600 nm [OD600] = 0.6) in synthetic marine ZoBell broth (MB) (per L: 5 g peptone, 1 g yeast extract, 0.1 g FePO4) and mineral medium [per L: 30 g NaCl, 7 g K2HPO4, 3 g K2HPO4, 2 g (NH4)2SO4, 0.05 g FeSO4·7H2O, 0.1 g MgSO4·7H2O] containing 0.6% (wt/vol) alginate and glucose as the sole carbon sources, respectively. The cell samples were prepared as follows: 2-mL cultures were collected into a 2.5-mL Eppendorf tube and centrifuged at 2,000 rpm for 5 min. The cells were washed with 0.85% (wt/vol) normal saline solution three times and fixed by 10% (wt/vol) glutaraldehyde solution to avoid bacteriolysis. After negative staining with phosphotungstic acid, the morphology of bacteria was observed by TEM (51, 52). Bacterial cultures were applied to the dissociated mica and washed with deionized water to remove salt ions. After drying, the samples were placed under an AFM microscope to observe the bacteria under vacuum (53).
Genome sequencing and annotation of strain WXL662.
The genome sequencing and annotation of strain WXL662 were performed according to Lin et al. (54). The extraction of total genomic DNA and whole-genome shotgun sequencing of strain WXL662 was carried out with paired-end sequencing by HiSeq 4000 (Illumina) and long sequencing by PacBio RS II Sequencer (PacificBiosciences, Menlo Park, CA). The long sequence was assembled using Canu v.1.1 (55), and Pilon v.1.16 (56) was used to polish PacBio assemblies according to Illumina data. Misassemblies were identified and corrected using REAPR (57) and manual inspection. Open reading frame (ORF) prediction and annotation were carried out using prokka 1.14-dev (58) and the RASTtk pipeline (59) with the default setting. Gene functional annotations were performed using Cluster of Orthologous Groups of proteins (COG) (60), GO (Gene Ontology Consortium, 2017), KEGG (61), Pfam (62), and dbCAN (63). Detailed genomic information of V. pelagius WXL662 is listed in Table S1.
The amino acid sequences of alginate lyases are analyzed by blastp against the Protein Data Bank (PDB) database and SwissProt database (https://blast.ncbi.nlm.nih.gov/); the protein with the highest similarity in either PDB or SwissProt was chosen for further analysis. The available domains and protein annotation of alginate lyases were predicted by the SMART database (http://smart.embl-heidelberg.de/), and the amino acid residues encoding a putative signal peptide were predicted by the SignaIP 5.0 server (http://www.cbs.dtu.dk/services/SignalP/). Molecular weight (MW) and theoretical isoelectric point (pI) were predicted by the ExPASy database (https://web.expasy.org/compute_pi/). The reference alginate lyase genes using to construct the neighbor-joining phylogenic tree were randomly selected from the CAZy database. The ClustalW software was used for multisequence comparison, and the phylogenetic tree was constructed by MEGA 7.0 (64). The subcellular localization of proteins was predicted by several different computational tools as previously described by Romine (65) and Kabisch (16), using Cello (66), PsortB (67), Phobius TM and Phobius SP (68), TMHMM (69), Bomp (70), SMART (71), Lipop (72) and Bomp category (70).
Heterologous expression, purified and characterized, of recombinant alginate lyases.
Putative alginate lyase-encoding genes were searched from the whole-genome sequence of WXL662. The expression and purification of recombinant alginate lyases (without signal peptides) in E. coli BL21(DE3) were performed according to Tang et al. (73) with minor modifications. Briefly, the putative gene was amplified with corresponding primer pairs (Table S3) and cloned into pET28(+). The resulting vectors were transformed into E. coli BL21(DE3). The recombinant cells were grown in LB medium supplemented with kanamycin (50 μg/mL) at 37°C until they reached the logarithmic phase (OD600 = 0.4 to 0.6). The recombinant protein was induced by the addition of 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at 16°C for 12 h. Cells were harvested by centrifugation at 8,000 × g for 10 min and then washed with Tris-HCl buffer (20 mM Tris, 50 mM NaCl [pH 8.0]). The suspensions were sonicated on ice, centrifuged at 12,000 rpm for 30 min, and filtered through a 0.22-μm-pore-size filter to remove intact cells and debris. The sterile supernatant was loaded onto nitrilotriacetic acid (NTA)-Ni (Qiagen). Proteins were purified according to the manufacturer’s recommendations. The purified recombinant alginate lyases were assessed by 12% sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) according to the method of Laemmli et al. (74).
The enzymatic activity of the recombinant alginate lyases was determined according to Thomas et al. (8) with minor modifications. Briefly, enzyme (20 μL) was added to Tris-HCl buffer (50 mM [pH 8.0]) with 200 mM NaCl and 0.3% (wt/vol) alginate (180 μL), and then the resulting mixture was incubated at 30°C for 10 min. One unit of enzyme activity was defined as an increase of 1 OD235 unit per min. Unless otherwise stated, 20 μL purified enzyme was added in 180 μL substrate under different conditions and reacted for 10 min; 20 μL reaction mixture was then added to 180 μL double-distilled water (ddH2O) in 96-well plates (Corning Life Sciences, Acton, MA), and the increase of absorbance value of the reaction system was measured within 10 min.
To determine the optimum catalysis temperature, the alginate lyase activity was measured at 10 to 60°C at different intervals. The optimal pH values of the purified alginate lyases were measured between pH 3.0 and 10.6 using three kinds of buffer systems: 0.05 M citric acid-sodium dihydrogen phosphate buffer (pH 3.0, 4.0, 5.0, 6.0, 6.2, 6.8, 7.0, 7.2, 7.8, and 8.0), 0.05 M Tris-HCl (pH 8.2 and 8.6), and 0.05 M glycine-NaOH (pH 9.0, 10.0, and 10.6). To analyze the effects of metal ions on the activity of eight recombinant alginate lyases (except Aly-VII, a polyM preference enzyme), different chemicals (Na+, K+, Ca2+, Fe3+, Mn2+, Al3+, Co2+, Ni2+, Fe2+, Cu2+, Mg2+, Zn2+, EDTA, SDS, and urea) were added to the reaction mixture at a final concentration of 1 or 10 mM. Substrate specificity of alginate lyases was tested by measuring their activities toward polyM, polyG, and sodium alginate. To determine the oligosaccharide compositions of the final digests, oligomers released from polyM, polyG, and sodium alginate by alginate lyases were analyzed by thin-layer chromatography (TLC) (Merck, Darmstadt, Germany), and the solvent system was 1-butanol–acetic acid–water (4:6:1 [vol/vol]). The products were visualized by heating TLC plates at 100°C for 15 min after spraying them with 10% (vol/vol) sulfuric acid in ethanol.
Transcriptomic analysis and qRT-PCR.
Strain WXL662 was grown in a mineral medium containing 0.6% (wt/vol) alginate or glucose as the sole carbon source at 28°C and 170 rpm in triplicates. The cells were collected when they reached the mid-log phase (~8 h) (Fig. S2) and frozen in liquid nitrogen immediately. RNA extraction and purification, reverse transcription, library construction, and sequencing were performed at Shanghai Majorbio Bio-pharm Biotechnology Co., Ltd. (Shanghai, China), according to the manufacturer’s instructions (Illumina, San Diego, CA, USA) as follows. Briefly, total RNA was extracted using TRIzol reagent and the genomic DNA was removed using DNase I (TaKara). The RNA-seq transcriptome library was prepared following the TruSeq RNA sample preparation kit (Illumina, San Diego, CA) using 2 μg of high-quality total RNA. The paired-end RNA-seq sequencing library was sequenced with the Illumina HiSeq×TEN (2 × 150-bp read length). The processing of original images to sequences, base calling, and quality value calculations were performed using the Illumina GA Pipeline (version 1.6), in which 150-bp paired-end reads were obtained. A Perl program was written to select clean reads by removing low-quality sequences, reads with more than 5% N bases (unknown bases), and reads containing adaptor sequences. High-quality reads in each sample were mapped to the genome of strain WXL662 using Bowtie2 (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml). Differentially expressed genes (DEGs) were obtained based on the normalized FPKM (fragments per kilobase of transcript per million mapped reads) values in alginate- and glucose-treated samples. A false-discovery rate (FDR) control (75) was used to correct for the P value. Genes with an FDR value of ≤0.05 and |log2 FC| of ≥1 were assigned as differentially expressed. Differences in gene transcription were also analyzed by the R package EdgeR (76), which was used for differential transcript analysis and identification of DEGs.
Variations in transcription levels of 20 target genes (including housekeeping genes and alginate-degrading genes) (Table S4) between cultures grown with 0.6% alginate mineral medium relative to those grown with 0.6% glucose mineral medium were evaluated by quantitative RT-PCR (qRT-PCR). The total RNA from cells was extracted as described above. The PrimeScript RT reagent kit with gDNA Eraser (TaKaRa) was used to reverse transcribe the RNA into cDNA. qRT-PCR was performed with ABI 7500 real-time PCR (Applied Biosystems, Foster City, CA, USA) using SYBR Premix Ex Taq (TaKaRa) and gene-specific primers listed in Table S4. The amplification efficiencies of all primer pairs were tested using standard dilution procedures. The cycle threshold (CT) value was determined by the StepOnePlus real-time PCR system (Thermo Fisher Scientific, Waltham, MA, USA). The fold change of gene transcription (alginate versus glucose) was normalized using the housekeeping gene ftsZ as a reference.
Preparation and detection of OMVs from WXL662.
The OMVs were prepared from strain WXL662 cultured in a medium containing 0.6% alginate or glucose mineral medium and MB at 28°C to the mid-exponential growth phase. OMVs were collected according to Li et al. (50) with minor modifications. Liquid cultures of strain WXL662 (4 L) were collected, and the cells were pelleted by centrifugation at 8,000 rpm for 10 min at 4°C (Eppendorf F34-6-38 rotor). The supernatant was collected and filtered through 0.22-μm-pore-size membrane filter (GVWP; Millipore) to remove the remaining bacterial cells. The filtrate was concentrated using a Amicon Ultra15 centrifugal filter (Merck; 100-kDa cutoff), and the culture volume was reduced to about 250 mL. This concentrated supernatant was pelleted by ultracentrifugation at 100,000 × g for 2 h at 4°C (Beckman Coulter SW-41 Ti rotor). The OMVs were further purified using an Optiprep (iodixanol; Sigma-Aldrich) density gradient centrifugation. The crude vesicle sample was adjusted to 45% OptiPrep in a buffer containing 3% (wt/vol) NaCl and 10 mM HEPES (pH 8.0). Equal volumes of 40, 35, 30, 25, 20, 15, 10, and 0% Optiprep (in the same buffer) were layered over the 1.3-mL crude vesicle sample and centrifuged at 30,000 rpm for 6 h at 4°C (Beckman Coulter, optima XPN-100 ultracentrifuge, SW41 Ti rotor), and fractions of equal volume were removed sequentially from the top. The material in each fraction was diluted 10-fold with buffer (3% NaCl with 10 mM HEPES) and pelleted in an ultracentrifuge (100,000 × g, 2 h, 4°C, 40,000 rpm; Hitachi P90AT rotor). The resulting pellet was resuspended in fresh buffer and frozen at −80°C.
The presence and purity of OMVs were visualized using negative staining followed by TEM (Mic JEM-1200EX; Japan). According to the method of Laemmli et al. (74), the purified OMVs were assessed by 12% sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Compared with the purified OMV samples under the conditions of MB and 0.6% glucose mineral cultures, the different protein bands under the condition of 0.6% alginate mineral culture were collected and sent to Shanghai Bioengineering Co., Ltd., for liquid chromatography-tandem mass spectrometry (LC-MS/MS) 30-min sequencing.
Analysis of alginate-degrading genes in other Vibrio genomes.
A total of 94 nearly complete Vibrio genomes were downloaded from NCBI database (Table S5). Open reading frame (ORF) prediction and gene annotation for these genomes were carried out using prokka v.1.14 (58) and the RASTtk online server (59) with the default setting. The hypothetical proteins in AUL were further annotated using SMART database (http://smart.embl-heidelberg.de/).
A maximum likelihood (ML) tree of 95 Vibrio genomes was constructed by Fasttree (77) based on single-copy core genes identified by OrthoFinder v.2.5.4 (78). Specifically, sequences of all the single-copy core genes were aligned by Mafft (79) and trimmed by Trimal (80). The trimmed sequences for each single-copy core genes were concatenated by a customized script for tree construction.
Data availability.
The GenBank accession numbers for alginate lyase genes Vpaly-I to Vpaly-XI are OM743975 to OM743985, respectively. The complete genome sequences of V. pelagius WXL662 were deposited in the NCBI GenBank server under accession no. PRJNA793771. The transcriptome shotgun assembly project has been deposited at GenBank under accession no. PRJNA832561.
ACKNOWLEDGMENTS
This work was supported the National Natural Science Foundation of China (41730530 and 92251303), the National Key Research and Development Program of China (2018YFE0124100), and the Fundamental Research Funds for the Central Universities (no. 202172002).
We thank all of the scientists and crews for their assistance with sampling during the cruise conducted by Dongfanghong 2 of Ocean University of China. Sincere thanks goes to Jiwen Liu and Min Yu from Ocean University of China for technical assistance.
We confirm that the content of the manuscript is original and the manuscript has neither been published previously nor is being considered for publication elsewhere.
We declare no conflict of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Xiao-Hua Zhang, Email: xhzhang@ouc.edu.cn.
Haruyuki Atomi, Kyoto University.
REFERENCES
- 1.Wang X, Liu J, Li B, Liang J, Sun H, Zhou S, Zhang X-H. 2019. Spatial heterogeneity of Vibrio spp. in sediments of Chinese marginal seas. Appl Environ Microbiol 85:e03064-18. 10.1128/AEM.03064-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang X, Lin H, Wang X, Austin B. 2018. Significance of Vibrio species in the marine organic carbon cycle—a review. Sci China Earth Sci 61:1357–1368. 10.1007/s11430-017-9229-x. [DOI] [Google Scholar]
- 3.Eilers H, Pernthaler J, Glöckner FO, Amann R. 2000. Culturability and in situ abundance of pelagic bacteria from the North Sea. Appl Environ Microbiol 66:3044–3051. 10.1128/AEM.66.7.3044-3051.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He X, Yu M, Wu Y, Ran L, Liu W, Zhang X-H. 2020. Two highly similar chitinases from marine Vibrio species have different enzymatic properties. Mar Drugs 18:139. 10.3390/md18030139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liang J, Liu J, Wang X, Lin H, Liu J, Zhou S, Sun H, Zhang X-H. 2019. Spatiotemporal dynamics of free-living and particle-associated Vibrio communities in the northern Chinese marginal seas. Appl Environ Microbiol 85:e00217-19. 10.1128/AEM.00217-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haug A, Larsen B, Smidsrød O, Smidsrød O, Eriksson G, Blinc R, Paušak S, Ehrenberg L, Dumanović J. 1967. Studies on the sequence of uronic acid residues in alginic acid. Acta Chem Scand 21:691–704. 10.3891/acta.chem.scand.21-0691. [DOI] [Google Scholar]
- 7.Mabeau S, Kloareg B. 1987. Isolation and analysis of the cell walls of brown algae: Fucus spiralis, F. ceranoides, F. vesiculosus, F. serratus, Bifurcaria bifurcata and Laminaria digitata. J Exp Bot 38:1573–1580. 10.1093/jxb/38.9.1573. [DOI] [Google Scholar]
- 8.Thomas F, Lundqvist LC, Jam M, Jeudy A, Barbeyron T, Sandström C, Michel G, Czjzek M. 2013. Comparative characterization of two marine alginate lyases from Zobellia galactanivorans reveals distinct modes of action and exquisite adaptation to their natural substrate. J Biol Chem 288:23021–23037. 10.1074/jbc.M113.467217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stokstad E. 2012. Biofuels. Engineered superbugs boost hopes of turning seaweed into fuel. Science 335:273. 10.1126/science.335.6066.273. [DOI] [PubMed] [Google Scholar]
- 10.Thomas F, Barbeyron T, Tonon T, Génicot S, Czjzek M, Michel G. 2012. Characterization of the first alginolytic operons in a marine bacterium: from their emergence in marine Flavobacteria to their independent transfers to marine Proteobacteria and human gut Bacteroides. Environ Microbiol 14:2379–2394. 10.1111/j.1462-2920.2012.02751.x. [DOI] [PubMed] [Google Scholar]
- 11.Chen X-L, Dong S, Xu F, Dong F, Li P-Y, Zhang X-Y, Zhou B-C, Zhang Y-Z, Xie B-B. 2016. Characterization of a new cold-adapted and salt-activated polysaccharide lyase family 7 alginate lyase from Pseudoalteromonas sp. SM0524. Front Microbiol 7:1120. 10.3389/fmicb.2016.01120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsushima R, Danno H, Uchida M, Ishihara K, Suzuki T, Kaneniwa M, Ohtsubo Y, Nagata Y, Tsuda M. 2010. Analysis of extracellular alginate lyase and its gene from a marine bacterial strain, Pseudoalteromonas atlantica AR06. Appl Microbiol Biotechnol 86:567–576. 10.1007/s00253-009-2278-z. [DOI] [PubMed] [Google Scholar]
- 13.Ertesvåg H. 2015. Alginate-modifying enzymes: biological roles and biotechnological uses. Front Microbiol 6:523. 10.3389/fmicb.2015.00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cha Q-Q, Wang X-J, Ren X-B, Li D, Wang P, Li P -Y, Fu H-H, Zhang X-Y, Chen X-L, Zhang Y-Z, Xu F, Qin Q-L. 2021. Comparison of alginate utilization pathways in culturable bacteria isolated from Arctic and Antarctic marine environments. Front Microbiol 12:609393. 10.3389/fmicb.2021.609393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang L, Li X, Zhang X, Li Y, Wang L. 2021. Bacterial alginate metabolism: an important pathway for bioconversion of brown algae. Biotechnol Biofuels 14:158. 10.1186/s13068-021-02007-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kabisch A, Otto A, König S, Becher D, Albrecht D, Schüler M, Teeling H, Amann RI, Schweder T. 2014. Functional characterization of polysaccharide utilization loci in the marine Bacteroidetes ‘Gramella forsetii’ KT0803. ISME J 8:1492–1502. 10.1038/ismej.2014.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neumann AM, Balmonte JP, Berger M, Giebel HA, Arnosti C, Voget S, Simon M, Brinkhoff T, Wietz M. 2015. Different utilization of alginate and other algal polysaccharides by marine Alteromonas macleodii ecotypes. Environ Microbiol 17:3857–3868. 10.1111/1462-2920.12862. [DOI] [PubMed] [Google Scholar]
- 18.Inoue A, Nishiyama R, Ojima T. 2016. The alginate lyases FlAlyA, FlAlyB, FlAlyC, and FlAlex from Flavobacterium sp. UMI-01 have distinct roles in the complete degradation of alginate. Algal Res 19:355–362. 10.1016/j.algal.2016.03.008. [DOI] [Google Scholar]
- 19.Hobbs JK, Lee SM, Robb M, Hof F, Barr C, Abe KT, Hehemann J-H, McLean R, Abbott DW, Boraston AB. 2016. KdgF, the missing link in the microbial metabolism of uronate sugars from pectin and alginate. Proc Natl Acad Sci USA 113:6188–6193. 10.1073/pnas.1524214113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Preiss J, Ashwell G. 1962. Alginic acid metabolism in bacteria. I. Enzymatic formation of unsaturated oligosaccharides and 4-deoxy-l-erythro-5-hexoseulose uronic acid. J Biol Chem 237:309–316. 10.1016/S0021-9258(18)93920-7. [DOI] [PubMed] [Google Scholar]
- 21.Preiss J, Ashwell G. 1962. Alginic acid metabolism in bacteria. II. The enzymatic reduction of 4-deoxy-l-erythro-5-hexoseulose uronic acid to 2-keto-3-deoxy-d-gluconic acid. J Biol Chem 237:317–321. 10.1016/S0021-9258(18)93921-9. [DOI] [PubMed] [Google Scholar]
- 22.Jagtap SS, Hehemann J-H, Polz MF, Lee J-K, Zhao H. 2014. Comparative biochemical characterization of three exolytic oligoalginate lyases from Vibrio splendidus reveals complementary substrate scope, temperature, and pH adaptations. Appl Environ Microbiol 80:4207–4214. 10.1128/AEM.01285-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hashimoto W, Kawai S, Murata K. 2010. Bacterial supersystem for alginate import/metabolism and its environmental and bioenergy applications. Bioeng Bugs 1:97–109. 10.4161/bbug.1.2.10322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takase R, Ochiai A, Mikami B, Hashimoto W, Murata K. 2010. Molecular identification of unsaturated uronate reductase prerequisite for alginate metabolism in Sphingomonas sp. A1. Biochim Biophys Acta 1804:1925–1936. 10.1016/j.bbapap.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 25.Tang K, Lin Y, Han Y, Jiao N. 2017. Characterization of potential polysaccharide utilization systems in the marine Bacteroidetes Gramella Flava JLT2011 using a multi-omics approach. Front Microbiol 8:220. 10.3389/fmicb.2017.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee OK, Lee EY. 2016. Sustainable production of bioethanol from renewable brown algae biomass. Biomass Bioenergy 92:70–75. 10.1016/j.biombioe.2016.03.038. [DOI] [Google Scholar]
- 27.Zhang K, Li Z, Zhu Q, Cao H, He X, Zhang X-H, Liu W, Lyu Q. 2022. Determination of oligosaccharide product distributions of PL7 alginate lyases by their structural elements. Commun Biol 5:782. 10.1038/s42003-022-03721-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li S, Wang L, Jung S, Lee BS, He N, Lee M-S. 2020. Biochemical characterization of a new oligoalginate lyase and its biotechnological application in Laminaria japonica degradation. Front Microbiol 11:316. 10.3389/fmicb.2020.00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sawabe T, Ogura Y, Matsumura Y, Feng G, Amin AR, Mino S, Nakagawa S, Sawabe T, Kumar R, Fukui Y, Satomi M, Matsushima R, Thompson FL, Gomez-Gil B, Christen R, Maruyama F, Kurokawa K, Hayashi T. 2013. Updating the Vibrio clades defined by multilocus sequence phylogeny: proposal of eight new clades, and the description of Vibrio tritonius sp. nov. Front Microbiol 4:414. 10.3389/fmicb.2013.00414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goudenège D, Labreuche Y, Krin E, Ansquer D, Mangenot S, Calteau A, Médigue C, Mazel D, Polz MF, Le Roux F. 2013. Comparative genomics of pathogenic lineages of Vibrio nigripulchritudo identifies virulence-associated traits. ISME J 7:1985–1996. 10.1038/ismej.2013.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pollock FJ, Wilson B, Johnson WR, Morris PJ, Willis BL, Bourne DG. 2010. Phylogeny of the coral pathogen Vibrio coralliilyticus. Environ Microbiol Rep 2:172–178. 10.1111/j.1758-2229.2009.00131.x. [DOI] [PubMed] [Google Scholar]
- 32.Sugimura I, Sawabe T, Ezura Y. 2000. Cloning and sequence analysis of Vibrio halioticoli genes encoding three types of polyguluronate lyase. Mar Biotechnol (NY) 2:65–73. 10.1007/s101269900010. [DOI] [PubMed] [Google Scholar]
- 33.Zhuang J, Zhang K, Liu X, Liu W, Lyu Q, Ji A. 2018. Characterization of a novel polyM-preferred alginate lyase from marine Vibrio splendidus OU02. Mar Drugs 16:295. 10.3390/md16090295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kawamoto H, Horibe A, Miki Y, Kimura T, Tanaka K, Nakagawa T, Kawamukai M, Matsuda H. 2006. Cloning and sequencing analysis of alginate lyase genes from the marine bacterium Vibrio sp. O2. Mar Biotechnol (NY) 8:481–490. 10.1007/s10126-005-6157-z. [DOI] [PubMed] [Google Scholar]
- 35.Hashimoto W, Miyake O, Momma K, Kawai S, Murata K. 2000. Molecular identification of oligoalginate lyase of Sphingomonas sp. strain A1 as one of the enzymes required for complete depolymerization of alginate. J Bacteriol 182:4572–4577. 10.1128/JB.182.16.4572-4577.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Momma K, Mishima Y, Hashimoto W, Mikami B, Murata K. 2005. Direct evidence for Sphingomonas sp. A1 periplasmic proteins as macromolecule-binding proteins associated with the ABC transporter: molecular insights into alginate transport in the periplasm. Biochemistry 44:5053–5064. 10.1021/bi047781r. [DOI] [PubMed] [Google Scholar]
- 37.Murata K, Kawai S, Mikami B, Hashimoto W. 2008. Superchannel of bacteria: biological significance and new horizons. Biosci Biotechnol Biochem 72:265–277. 10.1271/bbb.70635. [DOI] [PubMed] [Google Scholar]
- 38.Miyake O, Ochiai A, Hashimoto W, Murata K. 2004. Origin and diversity of alginate lyases of families PL-5 and -7 in Sphingomonas sp. strain A1. J Bacteriol 186:2891–2896. 10.1128/JB.186.9.2891-2896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hayashi C, Takase R, Momma K, Maruyama Y, Murata K, Hashimoto W. 2014. Alginate-dependent gene expression mechanism in Sphingomonas sp. strain A1. J Bacteriol 196:2691–2700. 10.1128/JB.01666-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wargacki AJ, Leonard E, Win MN, Regitsky DD, Santos CNS, Kim PB, Cooper SR, Raisner RM, Herman A, Sivitz AB, Lakshmanaswamy A, Kashiyama Y, Baker D, Yoshikuni Y. 2012. An engineered microbial platform for direct biofuel production from brown macroalgae. Science 335:308–313. 10.1126/science.1214547. [DOI] [PubMed] [Google Scholar]
- 41.Xu F, Cha Q-Q, Zhang Y-Z, Chen X-L. 2021. Degradation and utilization of alginate by marine Pseudoalteromonas: a review. Appl Environ Microbiol 87:e00368-21. 10.1128/AEM.00368-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun X-K, Gong Y, Shang D-D, Liu B-T, Du Z-J, Chen G-J. 2022. Degradation of alginate by a newly isolated marine bacterium Agarivorans sp. B2Z047. Mar Drugs 20:254. 10.3390/md20040254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fischer A, Wefers D. 2019. Chromatographic analysis of alginate degradation by five recombinant alginate lyases from Cellulophaga algicola DSM 14237. Food Chem 299:125142. 10.1016/j.foodchem.2019.125142. [DOI] [PubMed] [Google Scholar]
- 44.Badur AH, Jagtap SS, Yalamanchili G, Lee J-K, Zhao H, Rao CV. 2015. Alginate lyases from alginate-degrading Vibrio splendidus 12B01 are endolytic. Appl Environ Microbiol 81:1865–1873. 10.1128/AEM.03460-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lyu Q, Zhang K, Shi Y, Li W, Diao X, Liu W. 2019. Structural insights into a novel Ca2+-independent PL-6 alginate lyase from Vibrio OU02 identify the possible subsites responsible for product distribution. Biochim Biophys Acta Gen Subj 1863:1167–1176. 10.1016/j.bbagen.2019.04.013. [DOI] [PubMed] [Google Scholar]
- 46.Cho J-C, Giovannoni SJ. 2004. Oceanicola granulosus gen. nov., sp. nov. and Oceanicola batsensis sp. nov., poly-β-hydroxybutyrate-producing marine bacteria in the order ‘Rhodobacterales’. Int J Syst Evol Microbiol 54:1129–1136. 10.1099/ijs.0.03015-0. [DOI] [PubMed] [Google Scholar]
- 47.Orench Rivera N, Kuehn MJ. 2016. Environmentally controlled bacterial vesicle-mediated export. Cell Microbiol 18:1525–1536. 10.1111/cmi.12676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schwechheimer C, Kuehn MJ. 2015. Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat Rev Microbiol 13:605–619. 10.1038/nrmicro3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hampton CM, Guerrero-Ferreira RC, Storms RE, Taylor JV, Yi H, Gulig PA, Wright ER. 2017. The opportunistic pathogen Vibrio vulnificus produces outer membrane vesicles in a spatially distinct manner related to capsular polysaccharide. Front Microbiol 8:2177. 10.3389/fmicb.2017.02177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li J, Azam F, Zhang S. 2016. Outer membrane vesicles containing signalling molecules and active hydrolytic enzymes released by a coral pathogen Vibrio shilonii AK1. Environ Microbiol 18:3850–3866. 10.1111/1462-2920.13344. [DOI] [PubMed] [Google Scholar]
- 51.Kohler T, Curty LK, Barja F, Van Delden C, Pechère J-C. 2000. Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J Bacteriol 182:5990–5996. 10.1128/JB.182.21.5990-5996.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rashid MH, Kornberg A. 2000. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc Natl Acad Sci USA 97:4885–4890. 10.1073/pnas.060030097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Braga PC, Ricci D. 1998. Atomic force microscopy: application to investigation of Escherichia coli morphology before and after exposure to cefodizime. Antimicrob Agents Chemother 42:18–22. 10.1128/AAC.42.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin H, Yu M, Wang X, Zhang X-H. 2018. Comparative genomic analysis reveals the evolution and environmental adaptation strategies of vibrios. BMC Genomics 19:135. 10.1186/s12864-018-4531-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res 27:722–736. 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, Earl AM. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hunt M, Kikuchi T, Sanders M, Newbold C, Berriman M, Otto TD. 2013. REAPR: a universal tool for genome assembly evaluation. Genome Biol 14:R47. 10.1186/gb-2013-14-5-r47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 59.Brettin T, Davis JJ, Disz T, Edwards RA, Gerdes S, Olsen GJ, Olson R, Overbeek R, Parrello B, Pusch GD, Shukla M, Thomason JA, Stevens R, Vonstein V, Wattam AR, Xia F. 2015. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep 5:8365–8366. 10.1038/srep08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tatusov RL, Galperin MY, Natale DA, Koonin EV. 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res 28:33–36. 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kanehisa M, Goto S. 2000. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28:27–30. 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, Salazar GA, Tate J, Bateman A. 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44:D279–D285. 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yin Y, Mao X, Yang J, Chen X, Mao F, Xu Y. 2012. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res 40:W445–W451. 10.1093/nar/gks479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Romine MF. 2011. Genome-wide protein localization prediction strategies for gram negative bacteria. BMC Genomics 12(Suppl 1):S1. 10.1186/1471-2164-12-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu C-S, Cheng C-W, Su W-C, Chang K-C, Huang S-W, Hwang J-K, Lu C-H. 2014. CELLO2GO: a web server for protein subCELlular LOcalization prediction with functional gene ontology annotation. PLoS One 9:e99368. 10.1371/journal.pone.0099368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Sahinalp SC, Ester M, Foster LJ, Brinkman FSL. 2010. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26:1608–1615. 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Käll L, Krogh A, Sonnhammer EL. 2007. Advantages of combined transmembrane topology and signal peptide prediction-the Phobius web server. Nucleic Acids Res 35:W429–W432. 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen Y, Yu P, Luo J, Jiang Y. 2003. Secreted protein prediction system combining CJ-SPHMM, TMHMM, and PSORT. Mamm Genome 14:859–865. 10.1007/s00335-003-2296-6. [DOI] [PubMed] [Google Scholar]
- 70.Berven FS, Flikka K, Jensen HB, Eidhammer I. 2004. BOMP: a program to predict integral β-barrel outer membrane proteins encoded within genomes of Gram-negative bacteria. Nucleic Acids Res 32:W394–W399. 10.1093/nar/gkh351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Letunic I, Doerks T, Bork P. 2012. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res 40:D302–D305. 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Juncker AS, Willenbrock H, Von Heijne G, Brunak S, Nielsen H, Krogh A. 2003. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci 12:1652–1662. 10.1110/ps.0303703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tang K, Su Y, Brackman G, Cui F, Zhang Y, Shi X, Coenye T, Zhang X-H. 2015. MomL, a novel marine-derived N-acyl homoserine lactonase from Muricauda olearia. Appl Environ Microbiol 81:774–782. 10.1128/AEM.02805-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 75.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, Van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Price MN, Dehal PS, Arkin AP. 2009. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26:1641–1650. 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Emms DM, Kelly S. 2019. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol 20:238. 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S12 and Tables S1 to S9. Download aem.01389-22-s0001.pdf, PDF file, 1.6 MB (1.6MB, pdf)
Table S10. Download aem.01389-22-s0002.xlsx, XLSX file, 0.04 MB (41.1KB, xlsx)
Table S11. Download aem.01389-22-s0003.xlsx, XLSX file, 0.03 MB (29KB, xlsx)
Data Availability Statement
The GenBank accession numbers for alginate lyase genes Vpaly-I to Vpaly-XI are OM743975 to OM743985, respectively. The complete genome sequences of V. pelagius WXL662 were deposited in the NCBI GenBank server under accession no. PRJNA793771. The transcriptome shotgun assembly project has been deposited at GenBank under accession no. PRJNA832561.