ABSTRACT
Epigenetic changes in genomics provide phenotypic modification without DNA sequence alteration. This study shows that benzoic acid, a common food additive and known histone deacetylase inhibitor (HDACi), has an epigenetic effect on Saccharomyces cerevisiae. Benzoic acid stimulated formation of epigenetic histone marks H3K4Me2, H3K27Me2, H3K18ac, and H3Ser10p in S. cerevisiae and altered their phenotypic behavior, resulting in increased production of phenylethyl alcohol and ester compounds during alcoholic fermentation using wine as a representative model system. Our study demonstrates the HDACi activity of certain dietary compounds such as sodium butyrate, curcumin and anacardic acid, suggests the potential use of these dietary compounds in altering S. cerevisiae phenotypes without altering host-cell DNA. This study highlights the potential to use common dietary compounds to exploit epigenetic modifications for various fermentation and biotechnology applications as an alternative to genetic modification. These findings indicate that benzoic acid and other food additives may have potential epigenetic effects on human gut microbiota, in which several yeast species are involved.
IMPORTANCE The manuscript investigates and reports for the first time utilizing a non-GMO approach to alter the fermentation process of Pinot Noir wines. We have experimentally demonstrated that certain dietary compounds possess histone deacetylase (HDAC) inhibiting activity and can alter the wine characteristics by potentially altering yeast gene transcription, which was resulted from epigenetic effects. We have previously proposed the term “nutrifermentics” to represent this newly proposed field of research that provides insights on the effect of certain dietary compounds on microbial strains and their potential application in fermentation. This technological approach is a novel way to manipulate microorganisms for innovative food and beverage production with quality attributes catering for consumer’s needs. Using a multidisciplinary approach with an emphasis on food fermentation and biotechnology, this study will be substantially useful and of broad interest to food microbiologists and biotechnologists who seek for innovative concepts with real-world application potential.
KEYWORDS: fermentation, nutrifermentics, benzoic acid, microbial epigenetics and biotechnology
INTRODUCTION
Epigenetics is the study of phenotypic changes in organisms, which predominantly result from alterations of nucleotides and histones instead of the DNA sequence. Therefore, epigenetic modifications are considered a non-genetically modified organism (GMO) approach to manipulating organismal traits and behaviors (1, 2). Among them, DNA methylation and histone acetylation are the most common and well-studied epigenetic modifications, which are the processes of transferring a methyl group to adenine and cytosine or adding an acetyl group to lysine residues at the N terminus of histone (3). Several dietary bioactive and phytochemicals that naturally occur in fruits and vegetables can act as epigenetic modifiers and potentially can alter the target organism (4). Epigenetic modifiers can be predominantly classified as DNA methyltransferase (DNMT), DNMT inhibitors, histone deacetylase (HDAC), HDAC inhibitors, and histone acetyl (HAT) and HAT inhibitors (5, 6).
Saccharomyces cerevisiae is a well-studied model system for epigenetic regulation. Since DNA methylation systems are absent, histone modifications are the primary form of epigenetic regulation, making it a simple system for understanding the relationship between histone modifications and epigenetic states (7, 8). Recently, the fission yeast, Schizosaccharomyces pombe was subjected to higher thresholds of caffeine, resulting in epigenetic changes producing transient epimutants with phenotypic plasticity, including tolerance to caffeine and cross-resistance to antifungal agents, which was closely related to heterochromatin alterations and heterochromatin-mediated gene silencing (9).
Benzoic acid is a lipophilic weak acid that occurs naturally in many fruits, vegetables, nuts, and even in cultured dairy products as a microbial metabolite (10). Benzoic acid and its derivatives are FDA approved food additives and known histone deacetylase inhibitors (HDACi) that have been shown to stimulate a recently discovered histone mark, lysine benzoylation (11, 12). HDACi compounds play an important role in heterochromatin regulation and gene expression by affecting histone modifications (13).
Here, we investigated the possibility of developing S. cerevisiae strains with desirable characteristics for alcoholic fermentation by treating them with the epigenetic modifier, benzoic acid. Benzoic acid was selected due its known capacity to modify histone proteins, its cost-effectiveness and solubility in the aqueous system. We also demonstrate that genes responsible for aroma compounds were upregulated in epimutants compared to the original S. cerevisiae strain. The effect of benzoic acid on S. cerevisiae H3 histone marks, as benzoic acid is a known HDACi, was also investigated. The results showed that there are several other dietary compounds that could be used to epigenetically alter microbial phenotypes to produce fermented products with desirable characteristics (7, 14).
Saccharomyces cerevisiae is widely used across food and beverage industries in the production of key commercial products, including wine, beer and bread. Of these, wine is a dominant export product from many countries, including the USA and New Zealand. The wine industry is a competitive industry and developing novel wines is necessary to maintain a competitive advantage in the global market. (15). Compared to some existing approaches, such as grapevine breeding and isolation of wild yeast, epigenetic modification of wine yeast is time and cost-effective.
Our study demonstrates the possibility of using dietary epigenetic compounds to develop non-GMO microbial strains with desirable characteristics for fermented products and biotechnology applications. We use wine production as a representative model fermentation system.
RESULTS AND DISCUSSION
Project scope: the practice of altering fermentation by epigenetics.
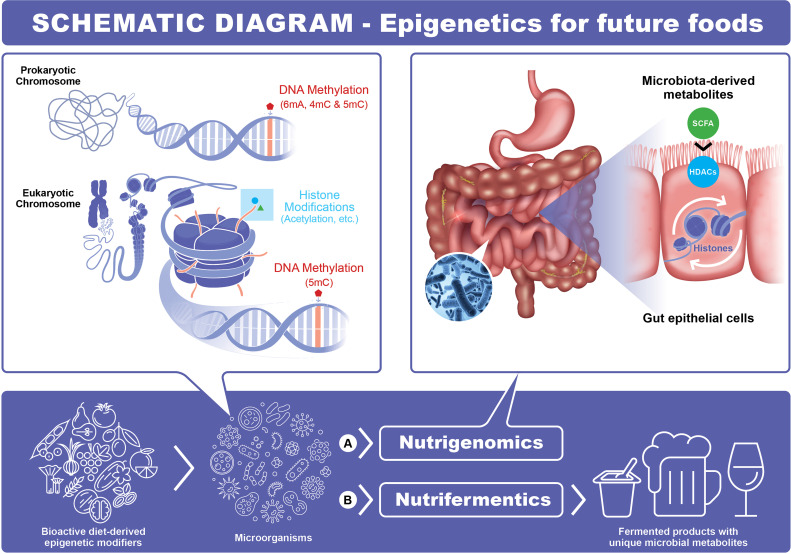
The schematic diagram put forward to fit the entire scope of the project is shown in Fig. 1 and demonstrates the proposed innovation to food fermentation by impacting gene transcription levels of microbial starters using diet-derived epigenetic modifiers. There are a range of diet-derived epigenetic modifiers, including bioactive compounds and phytochemicals, which are of health benefits to humans. For example, diet derived short-chain fatty acids are a group of HDAC inhibitors which are known to play a key role in epithelial hemostasis and repair process (14). The research investigating the effect of food and food components on gene transcription, and its role involved in the interaction between host/microbes and the nutritional environment is a well-established research field called “nutrigenomics” (Fig. 1A). In this study, we have shown that we can use these dietary epigenetic compounds such as dietary HDACi to alter the microbial phenotypes used in the fermentation process. We coined the word (16) “nutrifermentics” (Fig. 1B) to represent this new field of research. These dietary HDACi could also provide health benefits to consumers, in addition to improving starter microbial cultures in the fermentation process.
FIG 1.
Schematic diagram: An innovation to food fermentation by impacting gene transcription levels of microbial starters using diet-derived epigenetic modifiers. (A) Nutrigenomics: A well-established field of research investigating the effect of food and food components on gene transcription, and its role involved in the interaction between host/microbes and the nutritional environment. (B) Nutrifermentics: A new research direction first proposed in this study, which provides insights regarding the effect of food and food components on microbial starters and its potential application in fermentation. SCFA: short-chain fatty acids; HDACs: histone deacetylases.
Influence of benzoic acid on S. cerevisiae histone H3.
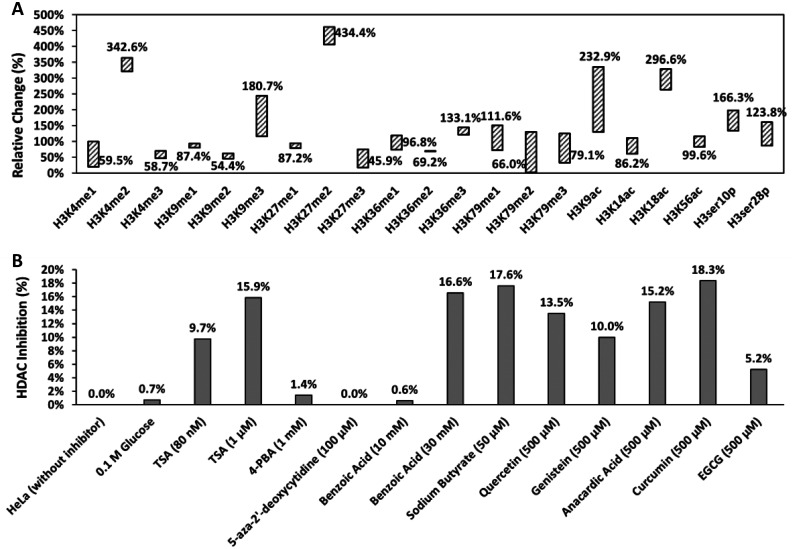
Benzoic acid and its derivatives are known HDACi (11) and recent study by Huang et al. revealed that sodium benzoate can stimulate a new histone mark, lysine benzoylation with significant physiological relevance (12). Figure 2A shows the percentage of relative changes in 21 distinct histone H3 modification patterns in S. cerevisiae, which was treated with 5 mM benzoic acid, in comparison with untreated wild-type strain. The percentages in Fig. 2A represent the relevant histone H3 modifications in 5 mM benzoic acid treated S. cerevisiae compared to that in wild type, either more modification (>100%) or less modification (<100%). The figure labels close to each floating bar indicate the mean values, and the length of each floating bar indicates the variation limits by doubling the standard deviation. Specific antibodies, including 15 for methylation, 4 for acetylation, and 2 for phosphorylation were utilized to measure the 21 patterns. Most modification patterns between treated and untreated wild-type strains were around 100% when taking the variation into consideration. However, both stimulation and inhibition in histone marks were seen with exposure to 5 mM benzoic acid. H3K4me2, H3K9me3, H3K27me2, H3K9ac, H3K18ac, and H3ser10p were stimulated more than 4-fold in treated strains, whereas few methylation patterns, including H3K4me3, H3K9me2, and H3K27me3, were about half compared to the untreated strain. Histone modifications are directly relevant to gene transcription levels in the organism.
FIG 2.
(A) Relative changes (%) of 21 histone H3 modification patterns the 5 mM benzoic acid treated S. cerevisiae using wild-type strains as the reference. (B) Histone deacetylase (HDAC) inhibition capacity of candidate epigenetic modifiers at different concentrations, in comparison with HeLa cells without inhibitors. Abbreviations: TSA: trichostatin A; 4-PBA: 4-phenylbutyric acid; EGCG: epigallocatechin gallate.
Transcription analysis by NanoString.
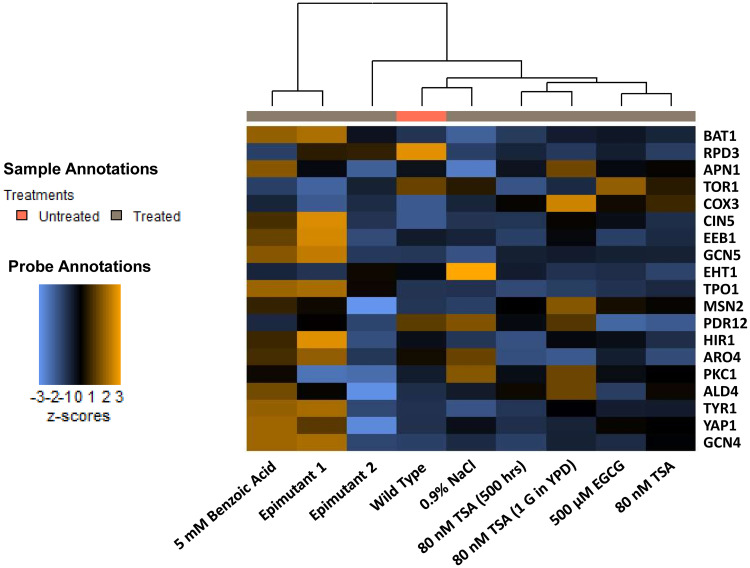
NanoString transcription analysis revealed the transcript abundance of 19 protein coding genes (excluding five housekeeping genes used as assay controls) on 9 refined samples. Fig. 3 shows the gene transcription levels of S. cerevisiae under different treatments, TSA treatments, including first-time exposure, 500 h treatment (20 h/subculture) and 1 generation w/o treatment after 500 h exposure were included as the HDACi controls, which were in accordance with benzoic acid treatments (first-time exposure to 5 mM benzoic acid, 500 h treatment [20 h/subculture] at 10 mM benzoic acid/epimutant 1 and 1 generation w/o treatment after 500h exposure/epimutant 2). The untreated wild-type strain was included as a negative-control along with 0.9% sodium chloride and with a dietary polyphenol epigallocatechin gallate (EGCG) that has been reported to inhibit DNMTs (4).
FIG 3.
Heat map showing unsupervised hierarchical clustering of 9 S. cerevisiae samples under different treatments, based on their transcription levels of 19 selected protein coding genes. TSA: trichostatin A; EGCG: epigallocatechin gallate.
Results are presented as a heat map graph after Z-score transformation, ranging from −3 to 3, blue (downregulation) to orange (upregulation), Fig. 3. As shown in Fig. 3, RNA samples with different treatments were clustered after data normalization, in which benzoic acid supplementation became a distinct influencing factor.
Genes responsible for overproducing phenylethyl alcohol (ARO4 and TYR1) (17), fusel alcohol, and ester synthesis (EEB1) (18), biosynthesis of higher alcohols (BAT1) (19) were all upregulated in 5 mM benzoic acid treated S. cerevisiae as well as epimutant 1. There was no significant change in the transcription levels of another ester synthase gene (EHT1) (18) between samples, particularly between benzoic acid treated group and others. Several genes responsible for stress tolerance and cell cycle were analyzed finding the histone deacetylase gene (RPD3) (20) transcription was downregulated while the histone acetyltransferase gene (GCN5) (21) was upregulated in both 5 mM benzoic acid-treated strain and epimutant 1, clearly supporting the role of benzoic acid as an HDACi. Kurat et al. (21) previously reported that upregulation of the GCN5 expression led to histone acetylation and global transcriptional activation. The variation observed could potentially indicate alternative acetylation mechanisms in S. cerevisiae resulted from different concentrations and exposure time of certain epigenetic compounds such as benzoic acid. With respect to the clusters, RNA from S. cerevisiae epimutant 1 (500 h growth in YPD broth containing 10 mM benzoic acid, 20 h/subculture) showed quite similar transcription patterns to 5 mM benzoic acid treatment (first time exposure). Moreover, the epimutant 2 (500 h growth in YPD broth containing 10 mM benzoic acid, followed by 20 h growth in regular YPD broth w/o stress, 20 h/subculture) exhibited significantly different RNA transcription patterns compared with the benzoic acid treatment group. The epimutant 2 tended to be more relevant to wild type and other S. cerevisiae treatments. This observation suggests that the alteration of gene transcription caused by dietary epigenetic compounds, which is revealed by direct counts of RNA transcripts, is transient and tends to ease out once the stress inducer is eliminated from the environment.
Influence of benzoic acid on S. cerevisiae nucleus.
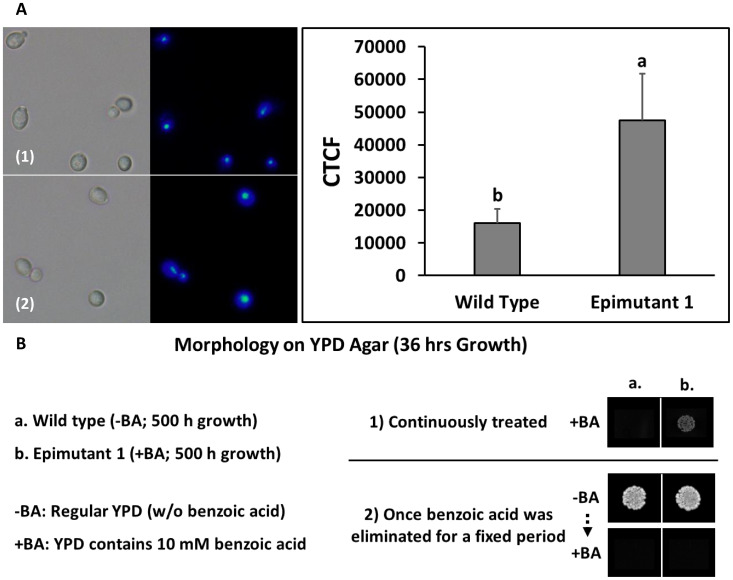
As shown in Fig. 4A, DAPI (4’, 6-diamidino-2-phenylindole) staining was applied to wild type and epimutant 1 to visualize their nuclei in terms of any size changes (expansion) that may have resulted from benzoic acid treatment. As DAPI stoichiometrically binds to DNA, which enables the detection and comparison of DNA content variation by fluorescence microscopy (22). The corrected total cell fluorescence (CTCF) was calculated based on the integrated density in the nucleus region. The mean comparison results indicate that there is a significant difference between two samples (P < 0.05), which suggests an expansion to the nucleus region in S. cerevisiae when they are exposed to 10 mM benzoic acid. Walters et al. (23) suggested that the expansion of nuclear envelope in budding yeast is independent from cell growth, but potentially related to nucleoplasmic factors, such as one or more nucleoplasmic proteins that are synthesized or imported into the nucleus. As HDAC and HDACi have been well researched being involved in multiple cell processes, such as cytokinesis and apoptosis (24, 25). Therefore, benzoic acid induced, HDACi related modifications could have occurred in nucleoplasm, resulting in expanded nucleus or relaxed genome state, which led to more fluorescence in this study.
FIG 4.
(A) DAPI staining (bright-field/fluorescent, at 100×) and corrected total cell fluorescence levels of benzoic acid-treated S. cerevisiae in comparison with wild type. (B) Phenotypic plasticity of benzoic acid-treated S. cerevisiae epimutant. (16): Wild Type; (1): Epimutant 1; CTCF: corrected total cell fluorescence; a–b: different letters indicate significant difference based on Tukey pairwise mean comparison results (P < 0.05).
Yeast morphology in relation to epigenetic alteration.
The cellular morphology of benzoic acid-treated S. cerevisiae was recorded to depict phenotypic differences compared to the wild type (Fig. 4B). Epimutant 1 was more tolerant to benzoic acid treatment and showed visible alteration in colony morphology. Observed tolerance and adaptation to benzoic acid faded soon after the stress was eliminated from the environment, demonstrating the transient nature of the treatment and possibly epigenetic change. The observation is supported by S. Torres-Garcia et al. (9); phenotypic plasticity can be promoted by epigenetic processes that let the wild-type cells adapt to certain unfavorable environments without altering genetic information, although these alterations are generally unstable and will be gradually lost without the stress. This observation is in line with NanoString assay, where S. cerevisiae epimutant 1 showed very similar transcription patterns to 5 mM benzoic acid-treated strain (first time exposure). However, epimutant 2 exhibited significantly different gene transcription patterns compared within the benzoic acid treatment group. Epimutant 2 gene transcription patterns were more similar to wild type than other treatments. This suggests that the alteration of gene transcript abundance caused by dietary epigenetic compounds is transient and tends to fade once the compound is eliminated from the environment. Overall, the robustness of S. cerevisiae epimutants and their adaption to stressed environment were improved by continuously treating the strain with the threshold levels of benzoic acid. However, epigenetic plasticity could be an issue in retaining the robust characteristics for future generations once the epigenetic modifiers is removed from the environment. However, commercial yeast starter culture producer could potentially prefer single/double use strains similar to commercial seed companies.
Wine characteristics changes due to epigenetic alteration.
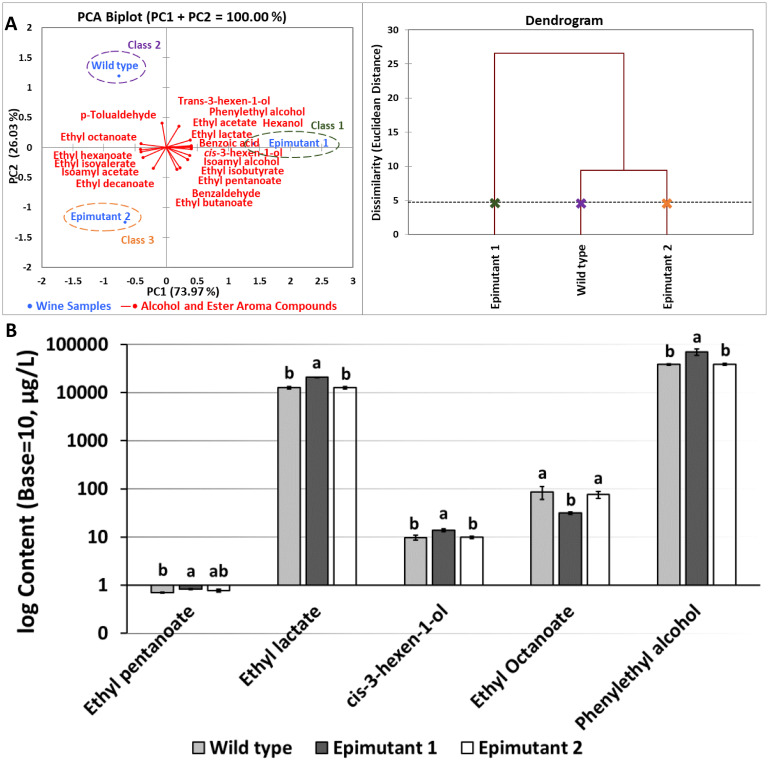
To test the impact of benzoic acid-stimulated epigenetic changes on fermentation characteristics of S. cerevisiae, treated cultures were used to ferment wine samples. Wines were fermented using three S. cerevisiae starters, including wild type, epimutant 1 (500 h growth in YPD broth containing 10 mM benzoic acid, 20 h/subculture), and epimutant 2 (epimutant 1 followed by 20 h growth in regular YPD broth without stress). Principal-component analysis (PCA) of aromatic attributes of wine samples and agglomerative hierarchical clustering (AHC) were utilized for classification of fermented wine samples (Fig. 5A). In addition, GC-MS analysis was carried out on wine samples, distinct results are shown in Fig. 5B, with full analysis of 18 compounds listed in Table 1. The three starters resulted in three wine categories, each with distinct aromatic profiles (Fig. 5A and 5B). The positive correlation between epimutant 1 and cis-3-hexen-1-ol may indicate a kiwifruit and leaf-like aroma is potentially associated with wine produced by epimutant 1 (26). Since cis-3-hexen-1-ol is an important aroma compound in many white wines, it might confer a complex aromatic profile on the altered wine, by adding partial aromatic features of white wine. As shown in Fig. 5B, the content of five ester and higher alcohol compounds is listed as potential indication of wine aroma alterations resulting from epimutation of the starters. Wine fermented by epimutant 1 possessed significantly increased phenylethyl alcohol (rose scent), ethyl lactate (butter aroma), cis-3-hexen-1-ol (leaf alcohol confers grassy-green odor) and ethyl pentanoate (fruity aroma), whereas the content of ethyl octanoate was reduced (soapy, floral aroma) (P < 0.05) (27). This GC-MS analysis clearly supports the gene transcription levels observed using NanoString assay. This has potential positive applications in commercial fermentation, considering that the treatments lead to overtranscription of genes associated with favorable aromas such as Phenyl ethyl alcohol and overproduction of these compounds is confirmed by GC-MS analysis.
FIG 5.
(A) Left: Principal-component analysis (PCA) bi-plot illustrating the relationship between wine samples fermented under different conditions and the variance of alcohol and ester aroma compounds; Right: Wine samples grouped using agglomerative hierarchical clustering (HCA) according to dissimilarity levels based on GC-MS analysis. (B) The content of ester and higher alcohol compounds potentially contributing to distinct aromatic profiles of wine samples fermented by epimutated S. cerevisiae. a–b: different letters indicate significant difference based on Tukey pairwise mean comparison results (P < 0.05).
TABLE 1.
Analysis of alcohol and ester aroma compounds detected in Pinot Noir winea
| Wine samples | Ethyl acetate (mg/L) | Ethyl isobutyrate (μg/L) | Ethyl butanoate (μg/L) | Ethyl isovalerate (μg/L) | Isoamyl acetate (μg/L) | Ethyl pentanoate (μg/L) | Isoamyl alcohol (mg/L) | Ethyl hexanoate (μg/L) | Ethyl lactate (mg/L) |
|---|---|---|---|---|---|---|---|---|---|
| Wild type | 27.5 ± 0.5ab | 43.8 ± 4.4a | 64.5 ± 9.0a | 4.1 ± 0.5a | 165.7 ± 24.9a | 0.7 ± 0.0b | 143.7 ± 6.1a | 75.0 ± 16.4a | 12.6 ± 0.6b |
| Epimutant 1 | 30.9 ± 1.9a | 51.6 ± 4.0a | 66.9 ± 4.9a | 3.3 ± 0.7a | 151.3 ± 20.7a | 0.8 ± 0.0a | 175.2 ± 23.7a | 54.0 ± 9.5a | 20.8 ± 0.3a |
| Epimutant 2 | 26.2 ±2.1b | 46.8 ± 2.5a | 67.3 ± 5.6a | 4.3 ± 0.3a | 174.5 ± 19.1a | 0.8 ± 0.1ab | 147.6 ± 4.4a | 76.2 ± 2.5a | 12.6 ± 0.7b |
| Wine samples | Hexanol (μg/L) | Trans-3-hexen-1-ol (μg/L) | Cis-3-hexen-1-ol (μg/L) | Ethyl octanoate (μg/L) | Benzaldehyde (μg/L) | Ethyl decanoate (μg/L) | Phenylethyl alcohol (mg/L) | p-Tolualdehyde (μg/L) | Benzoic acid (mg/L) |
| Wild type | 605.7 ± 14.1a | 54.4 ± 10.4a | 9.7 ± 1.2b | 86.1 ± 25.6a | 35.5 ± 11.3a | 20.2 ± 4.0a | 38.4 ± 1.4b | 159.6 ± 21.1a | 2.4 ± 0.1b |
| Epimutant 1 | 694.1 ± 24.8a | 56.4 ± 7.8a | 13.8 ± 0.9a | 31.6 ± 1.7b | 45.3 ± 11.7a | 20.0 ± 2.2a | 69.9 ± 10.6a | 146.1 ± 13.6a | 3.3 ± 0.1a |
| Epimutant 2 | 608.1 ± 65.1a | 21.3 ± 11.0b | 9.9 ± 0.6b | 75.4 ± 12.1a | 45.2 ± 12.2a | 24.4 ± 6.0a | 38.8 ± 1.9b | 137.1 ± 13.0a | 2.3 ± 0.1b |
aResults are presented as mean ± SD (standard deviation, N = 3). Results with different superscripts within the same column indicate significant differences between three types of wines (Tukey’s HSD test, p < 0.05). Wine samples were fermented using three types of the starters: Wild Type: 500 h growth in regular YPD broth, 20 h/subculture; Epimutant 1: 500 h growth in YPD broth containing 10 mM benzoic acid, 20 h/subculture; Epimutant 2: 500 h growth in YPD broth containing 10 mM benzoic acid, followed by 20 h growth in regular YPD broth w/o stress, 20 h/subculture.
In addition to aroma alterations, major chemical composition changes due to epigenetic alterations were also investigated, including residual sugars (glucose and fructose), glycerol, and ethanol (Data set S1). Generally, epimutant 1 tended to increase the fructose content in wine (P < 0.05), whereas not significantly affect the content of other chemicals analyzed (P > 0.05). The study detected a few more distinct aromatic compounds by qualitative GC-MS in these wine samples that are listed in Data set S2.
HDAC inhibition capacity.
As in the previous NanoString assay, significantly different patterns of transcribed genes by other treatments were compared to benzoic acid group, suggesting that HDACi and compounds capable of modifying epigenetic states have different effects on histone proteins and gene transcription patterns, potentially indicating wide range of application for these HDACi (Fig. 3). This led us to consider the HDACi activity of dietary epigenetic compounds. To investigate this, we applied a well-established HDAC assay using HeLa cell lines (6, 14). The dietary compounds tested were sodium butyrate, quercetin, genistein, anacardic acid, curcumin, and EGCG (Fig. 2B). Untreated HeLa nuclear extracts were used as the negative control. 5-Aza-2’-deoxycytidine, which is a well-recognized DNMT inhibitor but not HDAC inhibitor (28), and glucose were also included as negative controls for assay calibration. A well-known HDACi, namely, TSA, was used as a generic inhibitor of histone acetylation to test the impact on gene transcription (29). Relevant half-maximal inhibitory concentrations (IC50) were referred for determining the testing concentrations of candidate chemicals, except for benzoic acid since an IC50 was not determined. In general, most of the tested dietary compounds exhibited equivalent or better HDACi capacity compared to TSA, the positive control, suggesting their potential application in the food industry, particularly the food fermentation field.
Conclusions.
This study showed the potential applications of dietary epigenetic compounds in food research. As a proof of concept, it has been shown that epigenetic changes in yeast S. cerevisiae can be induced using dietary compounds resulting in different aromatic profiles in alcoholic fermentation. This opens the exciting possibility of using a non-GMO approach to obtain microbial strains with desirable characteristics for fermented food products. Interestingly, it was observed that the downregulation of H3K27me3 histone mark and upregulation of GCN4 gene, which are associated with life span extension in C.elgans and S. cerevisiae, respectively (30–32). Understanding the role of these dietary epigenetic compounds on cell aging is also potentially an interesting direction for future research.
MATERIALS AND METHODS
S. cerevisiae starter preparation for wine fermentation.
A commercial wine yeast S. cerevisiae EC-1118 was used as fermentation starters in this study. Three types of starters were involved and applied, including wild type (500 h growth in regular YPD broth, 20 h/subculture), epimutant 1 (500 h growth in YPD broth containing 10 mM benzoic acid, 20 h/subculture), and epimutant 2 (500 h growth in YPD broth containing 10 mM benzoic acid, followed by 20 h growth in regular YPD broth w/o stress, 20 h/subculture). For each subculture, 0.5 mL of cultured broth was transferred to 25 mL of relevant YPD broth with or without stress, as an inoculum at an OD600 of 1.0 ± 0.2.
Three starters, namely, wild type, epimutant 1, and epimutant 2, were then inoculated at 107 CFU/mL to 400 mL of Pinot Noir grape juice (Lincoln University Farm, harvested in 2020) contained in a 500 mL Schott bottle for fermentation at 32°C, in triplicate. Brix values were monitored every 2 days as an indicator of the fermentation progress. Wine samples were collected and stored at 4°C for downstream analysis upon completion of fermentation.
Histone H3 modification multiplex assay.
The 21 histone H3 modification patterns of 5 mM benzoic acid treated S. cerevisiae compared to untreated wild-type strain were measured using EpiQuik Histone H3 modification multiplex assay kit (Colorimetric; EpiGentek, NY, USA) following manufacturer’s instructions. Absorbance was measured using FLUOstar Omega microplate reader (BMG LABTECH, Ortenberg, Germany) at 450 nm with a reference wavelength of 655 nm.
RNA purification and transcription analysis.
S. cerevisiae under different treatments were harvested after 12 h growth to reach a sample size of 1 × 108 cells for RNA purification. The treatments include wild type, benzoic acid epimutant 1 & 2 as previously described, and alternative epimutants, which were continuously treated with 80 nM Trichostatin A (TSA) as positive controls. Strains which were first-time exposed to 5 mM benzoic acid, 80 nM TSA, 500 μM EGCG were also included, as well as 0.9% NaCl as the negative control. Total RNA from S. cerevisiae was isolated using RiboPure RNA purification kit (Invitrogen, MA, USA), following manufacturer’s instructions. RNA purity was measured by DeNovix DS-11 Spectrophotometer (DeNovix Inc., DE, USA). The RNA transcription analysis was measured using nCounter technology (NanoString Technologies, Inc., WA, USA). RNA samples were posted to The University of Auckland, where all the preparation and measurement were completed. Assay was carried out on 12 samples/24 genes (including 5 housekeeping genes, PGK1, TDH2, TFC1, TPI1, and ACT1, please refer to SD 4 Gene List for detailed information), the RNA input amount was 300 ng for each sample. Transcript counts were normalized and analyzed using the nSolver 4.0 software (NanoString Technologies, Inc., WA, USA).
DAPI staining.
S. cerevisiae strains were cultured overnight to an OD600 = 1.0 ± 0.2, followed by being treated with 2 volumes of 100% Ethanol for 45 min at room temperature. The mixture was centrifuged at 2500 rpm for 1 min, 1 mL 1 × PBS was used to wash the cells, followed by another centrifugation at 2500 rpm for 1 min. The pellet was resuspended in 200 μL of 1 × PBS/1:2000 dilution DAPI mixture, and was observed under Nikon Eclipse 50i fluorescence microscope (Nikon, Tokyo, Japan) after 45 min.
Yeast morphology.
S. cerevisiae starters were transferred from YPD broth onto corresponding YPD agar plates, either regular YPD or YPD containing 10 mM benzoic acid. Cultured media were serially diluted to OD600 = 0.1, 5 μL strain solution was spotted onto corresponding YPD agar plates after an additional 10 times dilution being applied, 1 × PBS was used for dilution. The growth temperature was set at 32°C.
GC-MS and chemical analysis of wine samples.
The alcohol and ester aroma compounds analysis was conducted using headspace-solid-phase microextraction (HS-SPME) and Shimadzu QP-2010 GC-MS (Shimadzu, Kyoto, Japan). The methodology was adopted from previous published articles, with slight modification regarding the diluent and sample matrix used with the standards (33, 34). Detailly, 0.9 mL of sample was pipetted into a 20 mL amber SPME vial and diluted with 8.06 mL of 5 g/L tartaric acid buffer (pH 3.5), 40 μL of composite internal standard was added followed by 4.5 g of sodium chloride before the vial was immediately capped.
For the preparation of the highest standard of the calibration curve, the composite standard was diluted in 136 mL sample matrix which was rotary evaporated at 36°C for 40 min to remove volatile background and reconstituted with 14.2% Ethanol as well as 40 μL of 5 M sodium hydroxide which returned the pH back to 3.15. It was then serially diluted in the provided matrix to ensure each vial had a maximum volume of 0.9 mL of matrix present. Each vial was then diluted further with 8.06 mL of tartaric acid buffer as in the samples with 40 μL of composite internal standard being added, followed by 4.5g of sodium chloride before the vials were immediately capped.
Ethanol content was analyzed by GC-FID, which was carried out on a Shimadzu GC-2010 gas chromatograph-flame ionization detector equipped with an AOC-20i autoinjector and AOC-20s autosampler. The chromatography was performed using an 19091N-133 HP-Innowax GC column (Polyethylene Glycol-Agilent Technologies, CA, USA). Residual sugars, including glucose and fructose were measured using Vintessential enzymatic test kit (Vintessential Laboratories – Tasmania, TAS, Australia), and glycerol content was measured using Megazyme glycerol assay kit (Megazyme, Wicklow, Ireland).
HDAC inhibition assay.
The HDAC inhibition capacity of candidate epigenetic modifiers was measured by a fluorometric HDAC assay kit (Active Motif, Inc., CA, USA), following the manufacturer’s instructions with slight modification to suit the objectives of this research. HeLa nuclear extract was used as the HDAC source, with an input volume of 5 μL. Candidate epigenetic modifiers/HDAC inhibitors, including the positive-control TSA, were added at the volume of 10 μL. The volume of HDAC assay buffer was adjusted to reach a total volume of 50 μL in each well. Fluorescence was measured using FLUOstar Omega microplate reader (BMG LABTECH, Ortenberg, Germany) with excitation wavelength at 360 nm and emission wavelength at 460 nm.
Statistical analysis.
Results were gathered from three independent biological replicates unless otherwise stated. Data were analyzed using analysis of variance (ANOVA) with a generalized linear model, followed by post hoc Tukey’s mean comparison test, using Minitab 20 (Minitab, LLC, PA, USA). PCA and AHC were analyzed using XLSTAT Statistical Software 2016 (Addinsoft, Paris, France). A confidence level of 95% was applied to the statistical analysis and data are presented as mean ± SD.
ACKNOWLEDGMENTS
The work described is in relation to Australian Provisional Patent Number 2022901674. We thank Shibani Suresh for her contributions to the initial part of the research, Jason Breitmeyer for his technical and experimental support toward GC-MS analysis, and Lincoln University Marketing and Customer Engagement Team for designing the schematic diagram.
Yanzhuo Kong: investigation, validation, formal analysis, writing–original draft, and writing–review and editing; Kenneth J. Olejar: investigation, validation, and writing–review and editing; Stephen L.W. On: validation and writing–review and editing; Christopher Winefield: validation and writing–review and editing; Philip A. Wescombe: validation and writing–review and editing; Charles S. Brennan: writing–review and editing; Richard N. Hider: investigation and validation; Venkata Chelikani: conceptualization, writing–review and editing, resources, supervision, and funding acquisition.
We declare that we have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
This work was supported by Lincoln University Startup grant (Grant INT4665) and KiwiNet Emerging Innovator award (Grant No: 46465).
Footnotes
Supplemental material is available online only.
Contributor Information
Venkata Chelikani, Email: Venkata.Chelikani@lincoln.ac.nz.
Johanna Björkroth, University of Helsinki.
REFERENCES
- 1.Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. 2009. An operational definition of epigenetics. Genes Dev 23:781–783. 10.1101/gad.1787609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morovic W, Budinoff CR. 2020. Epigenetics: a new frontier in probiotic research. Trends Microbiol 29:117–126. 10.1016/j.tim.2020.04.008. [DOI] [PubMed] [Google Scholar]
- 3.Damodaran TV. 2011. Genotoxicities and infertility. Reproductive and Developmental Toxicology doi:10.1016/B978-0–12-382032–7.10071–2.:923–947. 10.1016/B978-0-12-382032-7.10071-2. [DOI] [Google Scholar]
- 4.Hardy TM, Tollefsbol TO. 2011. Epigenetic diet: impact on the epigenome and cancer. Epigenomics 3:503–518. 10.2217/epi.11.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Link A, Balaguer F, Goel A. 2010. Cancer chemoprevention by dietary polyphenols: promising role for epigenetics. Biochem Pharmacol 80:1771–1792. 10.1016/j.bcp.2010.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malcomson FC, Mathers JC. 2017. Nutrition, epigenetics and health through life. Nutr Bull 42:254–265. 10.1111/nbu.12281. [DOI] [Google Scholar]
- 7.Zemach A, McDaniel IE, Silva P, Zilberman D. 2010. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 328:916–919. 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- 8.O’Kane CJ, Hyland EM. 2019. Yeast epigenetics: the inheritance of histone modification states. Bioscience Rep 39:1–3. 10.1042/BSR20182006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Torres-Garcia S, Yaseen I, Shukla M, Audergon PNCB, White SA, Pidoux AL, Allshire RC. 2020. Epigenetic gene silencing by heterochromatin primes fungal resistance. Nature 585:453–458. 10.1038/s41586-020-2706-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park SY, Yoo MY, Paik HD, Lim SD. 2017. Production of benzoic acid as a natural compound in fermented skim milk using commercial cheese starter. J Dairy Sci 100:4269–4275. 10.3168/jds.2016-12399. [DOI] [PubMed] [Google Scholar]
- 11.Anantharaju PG, Reddy BD, Padukudru MA, Kumari Chitturi CHM, Vimalambike MG, Madhunapantula SV. 2017. Naturally occurring benzoic acid derivatives retard cancer cell growth by inhibiting histone deacetylases (HDAC). Cancer Biol Ther 18:492–504. 10.1080/15384047.2017.1324374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang H, Zhang D, Wang Y, Perez-Neut M, Han Z, Zheng YG, Hao Q, Zhao Y. 2018. Lysine benzoylation is a histone mark regulated by SIRT2. Nat Commun 9:1–11. 10.1038/s41467-018-05567-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schnekenburger M, Diederich M. 2015. Nutritional epigenetic regulators in the field of cancer: new avenues for chemopreventive approaches, p 393–425, Epigenetic cancer therapy. Elsevier. [Google Scholar]
- 14.Wu SE, Hashimoto-Hill S, Woo V, Eshleman EM, Whitt J, Engleman L, Karns R, Denson LA, Haslam DB, Alenghat T. 2020. Microbiota-derived metabolite promotes HDAC3 activity in the gut. Nature 586:108–112. 10.1038/s41586-020-2604-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anonymous. Wine market size, share & trends analysis report by product (table wine, dessert wine, sparkling wine), by distribution channel (on-trade, off-trade), by region, and segment forecasts, 2021–2028. https://www.grandviewresearch.com/industry-analysis/wine-market.
- 16.Suresh S. 2021. Role of DNA methylation in wine yeast: a dissertation submitted in partial fulfilment of the requirements for the degree of master of science in food innovation at Lincoln University. Lincoln University. [Google Scholar]
- 17.Cordente AG, Solomon M, Schulkin A, Leigh Francis I, Barker A, Borneman AR, Curtin CD. 2018. Novel wine yeast with ARO4 and TYR1 mutations that overproduce ‘floral’aroma compounds 2-phenylethanol and 2-phenylethyl acetate. Appl Microbiol Biotechnol 102:5977–5988. 10.1007/s00253-018-9054-x. [DOI] [PubMed] [Google Scholar]
- 18.Saerens SM, Verstrepen KJ, Van Laere SD, Voet AR, Van Dijck P, Delvaux FR, Thevelein JM. 2006. The Saccharomyces cerevisiae EHT1 and EEB1 genes encode novel enzymes with medium-chain fatty acid ethyl ester synthesis and hydrolysis capacity. J Biol Chem 281:4446–4456. 10.1074/jbc.M512028200. [DOI] [PubMed] [Google Scholar]
- 19.Lilly M, Bauer FF, Styger G, Lambrechts MG, Pretorius IS. 2006. The effect of increased branched-chain amino acid transaminase activity in yeast on the production of higher alcohols and on the flavour profiles of wine and distillates. FEMS Yeast Res 6:726–743. 10.1111/j.1567-1364.2006.00057.x. [DOI] [PubMed] [Google Scholar]
- 20.Rundlett SE, Carmen AA, Kobayashi R, Bavykin S, Turner BM, Grunstein M. 1996. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc Natl Acad Sci USA 93:14503–14508. 10.1073/pnas.93.25.14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kurat CF, Lambert J-P, Petschnigg J, Friesen H, Pawson T, Rosebrock A, Gingras A-C, Fillingham J, Andrews B. 2014. Cell cycle-regulated oscillator coordinates core histone gene transcription through histone acetylation. Proc Natl Acad Sci USA 111:14124–14129. 10.1073/pnas.1414024111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferro A, Mestre T, Carneiro P, Sahumbaiev I, Seruca R, Sanches JM. 2017. Blue intensity matters for cell cycle profiling in fluorescence DAPI-stained images. Lab Invest 97:615–625. 10.1038/labinvest.2017.13. [DOI] [PubMed] [Google Scholar]
- 23.Walters AD, Amoateng K, Wang R, Chen J-H, McDermott G, Larabell CA, Gadal O, Cohen-Fix O. 2019. Nuclear envelope expansion in budding yeast is independent of cell growth and does not determine nuclear volume. Mol Biol Cell 30:131–145. 10.1091/mbc.E18-04-0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richa S, Dey P, Park C, Yang J, Son JY, Park JH, Lee SH, Ahn M-Y, Kim IS, Moon HR, Kim HS. 2020. A new histone deacetylase inhibitor, MHY4381, induces apoptosis via generation of reactive oxygen species in human prostate cancer cells. Biomol Ther (Seoul) 28:184–194. 10.4062/biomolther.2019.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grewal C, Hickmott J, Rentas S, Karagiannis J. 2012. A conserved histone deacetylase with a role in the regulation of cytokinesis in Schizosaccharomyces pombe. Cell Div 7:13–14. 10.1186/1747-1028-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Newcomb R, McRae J, Ingram J, Elborough K, Jaeger SR. 2010. Genetic variation in taste and odour perception: an emerging science to guide new product development. Consumer-driven innovation in food and personal care products, p 570–596. 10.1533/9781845699970.5.570. [DOI] [Google Scholar]
- 27.Anonymous. Diagnostic key classes of off-character compounds esters. https://wineserver.ucdavis.edu/industry-info/enology/fermentation-management-guides/key-diagnosing-problem-fermentations/diagnostic-key-classes-character-compounds-esters.
- 28.Christman JK. 2002. 5-Azacytidine and 5-aza-2′-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene 21:5483–5495. 10.1038/sj.onc.1205699. [DOI] [PubMed] [Google Scholar]
- 29.Yamashita YI, Shimada M, Harimoto N, Rikimaru T, Shirabe K, Tanaka S, Sugimachi K. 2003. Histone deacetylase inhibitor trichostatin a induces cell-cycle arrest/apoptosis and hepatocyte differentiation in human hepatoma cells. Int J Cancer 103:572–576. 10.1002/ijc.10699. [DOI] [PubMed] [Google Scholar]
- 30.Merkwirth C, Jovaisaite V, Durieux J, Matilainen O, Jordan SD, Quiros PM, Steffen KK, Williams EG, Mouchiroud L, Tronnes SU, Murillo V, Wolff SC, Shaw RJ, Auwerx J, Dillin A. 2016. Two conserved histone demethylases regulate mitochondrial stress-induced longevity. Cell 165:1209–1223. 10.1016/j.cell.2016.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steffen KK, MacKay VL, Kerr EO, Tsuchiya M, Hu D, Fox LA, Dang N, Johnston ED, Oakes JA, Tchao BN, Pak DN, Fields S, Kennedy BK, Kaeberlein M. 2008. Yeast life span extension by depletion of 60s ribosomal subunits is mediated by Gcn4. Cell 133:292–302. 10.1016/j.cell.2008.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mittal N, Guimaraes JC, Gross T, Schmidt A, Vina-Vilaseca A, Nedialkova DD, Aeschimann F, Leidel SA, Spang A, Zavolan M. 2017. The Gcn4 transcription factor reduces protein synthesis capacity and extends yeast lifespan. Nat Commun 8:1–12. 10.1038/s41467-017-00539-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tomasino E, Harrison R, Breitmeyer J, Sedcole R, Sherlock R, Frost A. 2015. Aroma composition of 2-year-old N ew Z ealand P inot N oir wine and its relationship to sensory characteristics using canonical correlation analysis and addition/omission tests. Australian J Grape and Wine Res 21:376–388. 10.1111/ajgw.12149. [DOI] [Google Scholar]
- 34.Green J, Parr W, Breitmeyer J, Valentin D, Sherlock R. 2011. Sensory and chemical characterisation of Sauvignon blanc wine: influence of source of origin. Food Res Int 44:2788–2797. 10.1016/j.foodres.2011.06.005. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download aem.01528-22-s0001.xlsx, XLSX file, 0.02 MB (25.2KB, xlsx)