Abstract
A growing body of evidence suggests interplay between the gut microbiota and the pathogenesis of nonalcoholic fatty liver disease (NAFLD). However, the role of the gut microbiome in early detection of NAFLD is unclear. Prospective studies are necessary for identifying reliable, microbiome markers for early NAFLD. We evaluated 2487 individuals in a community-based cohort who were followed up 4.6 years after initial clinical examination and biospecimen sampling. Metagenomic and metabolomic characterizations using stool and serum samples taken at baseline were performed for 90 participants who progressed to NAFLD and 90 controls who remained NAFLD free at the follow-up visit. Cases and controls were matched for gender, age, body mass index (BMI) at baseline and follow-up, and 4-year BMI change. Machine learning models integrating baseline microbial signatures (14 features) correctly classified participants (auROCs of 0.72 to 0.80) based on their NAFLD status and liver fat accumulation at the 4-year follow up, outperforming other prognostic clinical models (auROCs of 0.58 to 0.60). We confirmed the biological relevance of the microbiome features by testing their diagnostic ability in four external NAFLD case-control cohorts examined by biopsy or magnetic resonance spectroscopy, from Asia, Europe, and the United States. Our findings raise the possibility of using gut microbiota for early clinical warning of NAFLD development.
INTRODUCTION
Since the 1980s, the prevalence of obesity, insulin resistance, type 2 diabetes mellitus, and obesity-associated nonalcoholic fatty liver disease (NAFLD) has grown worldwide (1–3). The occurrence of these interconnected diseases is partly driven by consumption of high-energy food and a sedentary lifestyle, and these diseases are considered critical global health and socioeconomic problems (4). Apart from associations with liver-related diseases, epidemiological studies have associated NAFLD with increased risk of developing extrahepatic chronic diseases, such as type 2 diabetes, cardiovascular disease, and chronic kidney disease (5, 6). A recent cohort study showed that overall mortality risk increases progressively with worsening NAFLD histology, and even simple steatosis increases mortality risk by 71% (7), thus simple steatosis can no longer be considered as benign as previously thought (8). Although NAFLD affects about 25% of the world’s population (9) and has a high disease burden, awareness of NAFLD is low. In a cross-sectional analysis (n = 2788) in four U.S. cities, NAFLD prevalence was 23.9%, whereas awareness of NAFLD was 2.4% in study participants with computed tomography (CT)–defined NAFLD (10). One important reason for low awareness is that most patients with NAFLD are largely asymptomatic in the disease course, where disease is mainly detected through an incidental finding of fatty liver on ultrasound or an imagining modality or routine laboratory testing (11, 12). Diagnosis by liver biopsy or imaging is reliable but difficult for large-scale screening and monitoring. Thus, the need to identify individuals who are at high risk of developing NAFLD or are at an early stage of the disease is urgent, as lifestyle interventions can reverse the disease when it is in the first stages (13). According to one study (14), weight loss and healthy diet might be sufficient to reverse simple steatosis, whereas intensified lifestyle intervention coupled with pharmacological treatment might be necessary for more advanced stages of liver diseases. Exercise programs (15), low-carbohydrate diet (16), and various types of gut microbiota–targeted treatments (17) have demonstrated their ability to prevent steatosis development and improve NAFLD outcomes in human or preclinical models. Early diagnosis and interventions to prevent NAFLD progression can also greatly reduce future health care cost, as most economic costs associated with NAFLD are incurred in advanced stages (18). Currently available methods (19–21) for early prediction of NAFLD are limited and use only a few clinical parameters or biomarkers that may not reflect the heterogeneity and complexity of NAFLD (22, 23). Thus, more convenient noninvasive alternatives are needed.
In the last 10 years, the gut microbiome has emerged as a major regulator of host energy homeostasis and substrate metabolism (24–26). The human gastrointestinal tract is colonized with 4644 bacterial species encoding 171 million genes (27). Therefore, it is not unexpected that abnormalities in gut microbiome structure and especially function might affect the brain, adipose tissue, muscle, and liver metabolism. Microbial components or metabolites such as lipopolysaccharides, secondary bile acids, dimethyl- and trimethyl-amines, and compounds derived from carbohydrate and protein fermentation appear to be strongly involved in the gut host-microbiome metabolic axis and the occurrence of metabolic diseases (28–31).
Human cross-sectional studies have delineated the role of gut bacteria in the development of NAFLD. An increased ratio of Bacteroidetes to Firmicutes phyla and a decrease in butyrate-producing Ruminococcaceae are suggested to be involved in NAFLD progression; however, the data are not always consistent (32–35). Furthermore, whether NAFLD causes taxonomic and functional changes in the microbiome or the observed dysbiosis in patients with NAFLD leads to progression of the disease is not clear. For a possible causal role in NAFLD development, gut microbiota alteration should take place long before disease is diagnosed, which would suggest prognostic value in evaluating the gut microbiome in individuals with a high risk of developing NAFLD. To assess this potential value, we conducted a 4-year prospective study in a community-based cohort of 2487 Chinese individuals. We profiled 180 matched case-control individuals who were NAFLD free at baseline using well-documented clinical information and comprehensive metagenomic and metabolomic analysis. We developed machine learning models integrating baseline microbial signatures to classify individuals based on their NAFLD status 4 years after baseline (either remaining disease free or diagnosed with the disease). We also examined whether the selected features in the model were biologically relevant to NAFLD development by exploring the diagnostic power of the model in several case-control cohorts from Asia, the United States, and Europe.
RESULTS
Characterization of the study cohort
To develop a microbiome-based prognostic model for long-term development of NAFLD, we designed a nested case-control study within a community-based prospective cohort study of Chinese adults. About 2500 participants were screened in 2014 with ultrasonography, which is recommended as the first-line diagnostic test for NAFLD (36); 1216 participants were determined as NAFLD free using criteria proposed by the Asian Pacific Association for the Study of the Liver (37). Participant enrolment is outlined in fig. S1. Stool and serum samples were obtained from participants at baseline. At the follow-up visit in 2018, after a strict exclusion process, 90 participants (38 males and 52 females) were identified as having NAFLD (NAFLD−/+) (Fig. 1). The participants in the NAFLD−/+ group were matched with 90 controls who did not have NAFLD at baseline or at the follow-up visit (NAFLD−/−). The two groups were matched in gender, age, and body mass index (BMI) at both the baseline and follow-up visits and 4-year change in BMI. There were no differences between the two groups in the prevalence of type 2 diabetes, hypertensive disease, metabolic syndrome, and medication usage at both baseline and follow-up in the cohort, apart from a significantly higher metabolic syndrome ratio in NAFLD−/+ at follow-up as expected (chi-square test, P < 0.05; table S1).
Fig. 1. Overview of the prospective study design.
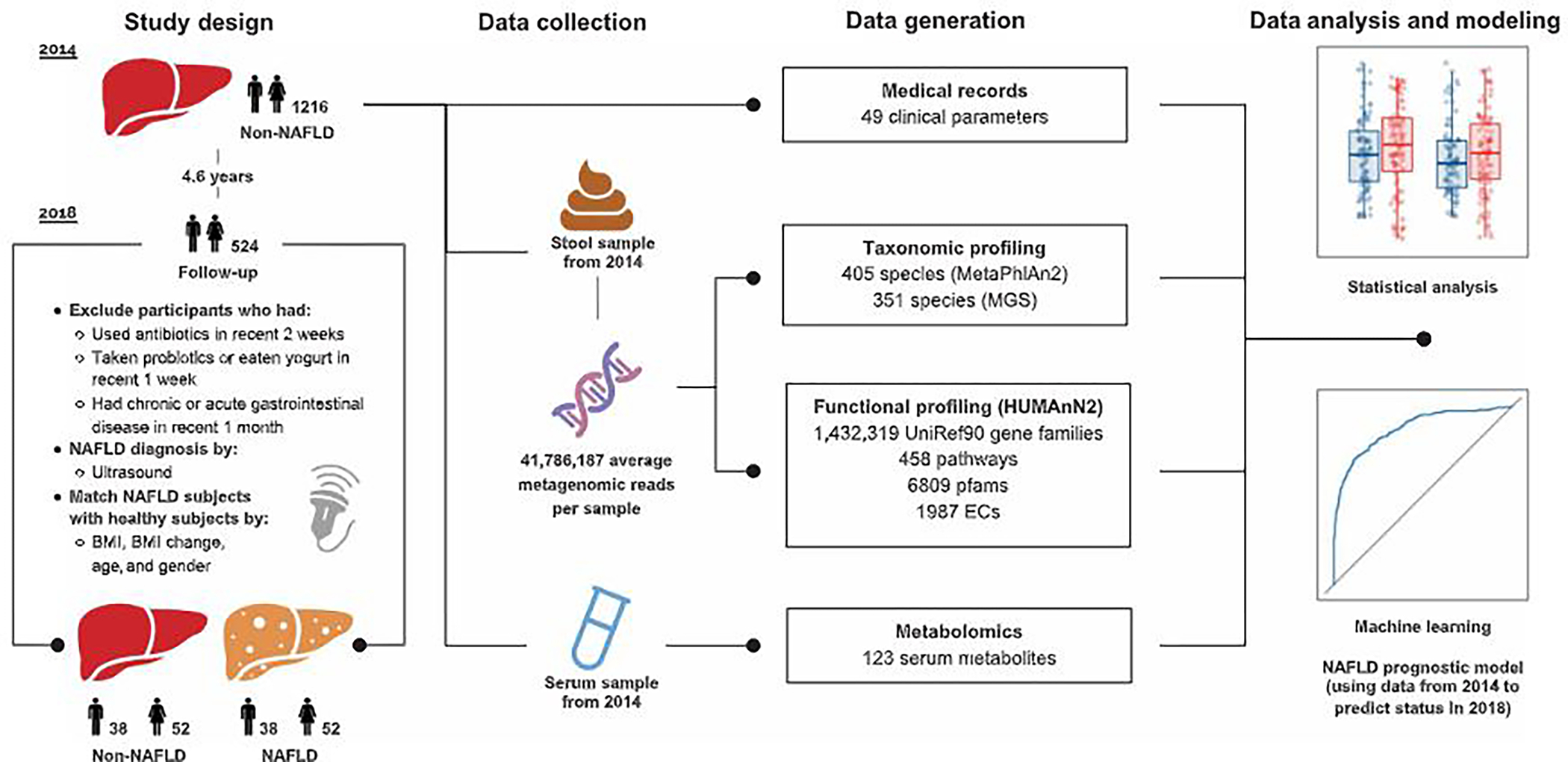
A graphical representation summarizing the study design, data collection, and the methodologies of data generation and analysis. Further details of the study design can be found in fig. S1.
Detailed baseline anthropometric parameters, glucose homeostasis parameters, serum liver enzymes and renal function, lipid profiles, and cytokines are shown in Table 1. No significant differences (t test, P > 0.05) were seen for most clinical parameters between the NAFLD−/+ and NAFLD−/− groups at baseline. Fasting insulin (FINS), homeostasis model assessment for insulin resistance (HOMA-IR), triglycerides (TGs), and high-sensitivity C-reactive protein (hs-CRP) in the NAFLD−/+ group were slightly higher than in the NAFLD−/− group (t test, P < 0.05); however, their mean or median values were within reference ranges in both groups (FINS: 5.1 to 11.2 uU/ml, HOMA-IR < 2.5, TG < 1.70 mM, and hs-CRP < 1 μg/ml) (38–41). Only TGs remained significantly different after adjusting for HOMA-IR (1.55 ± 0.90 mM versus 1.23 ± 0.61 mM; Table 1).
Table 1. Baseline characteristic of participants in INAFLD−/+ and NAFLD−/− groups.
Data are expressed as means ± SD or median (lower quartile and upper quartile) for continuous variables, and n represents percentage for categorical variables. BMI, body mass Index; SBP, systolic blood pressure; DBP, diastolic blood pressure; FBG fasting blood glucose; hs-CRP, high sensitivity C-reactive protein; TC, total cholesterol; TG, triglycerides; HDL-C high-densily lipoprotein cholesterol; FGF-21. fibroblast growth factor 21; HbA1c, hemoglobin A1C; HQMA-IR. homeostasis model assessment insulin resistance: apo, apolipoprotein; PG30, 30-min postprandial plasma glucose; PG120, 120-min postprandial plasma glucose; INS30, 30-min postprandial insulin; INS120, 120-min postprandial insulin; FINS, fasting insulin: Cr, creatinine; UAIb/Ucr, urinary albumin tocreatinine ratio; UA uric acid; FFA, free fatty acid; TBIL total bilirubin; GA, glycated albumin; AST, aspartate aminotransferase; ALT, alanine aminotransferase; GGT, gamma glutamyl transferase.
| Characteristic | Total (n = ISO) | NAFLD−/− [n=94) | NAFLD−/+ (n = 90) | P value* | P value† |
|---|---|---|---|---|---|
| Anthropometric parameters | |||||
| Sex (male) | 76 (42.22%) | 33 (42.22%) | 36 (42.22%) | - | - |
| Age (years) | 62.51 ± 3.81 | 62.03 ± 3.78 | 62.99 ± 3.81 | 0.0921 | 0.1481 |
| Weight (kg) | 62.82 ± 8.07 | 62.47 ± 7.53 | 63.16 ± 8.6 | 0.5688 | 0.3617 |
| BMI (kg/m2) | 24.55 ± 2.13 | 24.35 ± 2 | 24.75 ± 2.25 | 0.2059 | 0.0681 |
| SBP (mmHg)‡ | 130 (130, 140) | 130 (120 140) | 130 (120 140) | 0.2854 | 0.3363 |
| DBP (mmHg)‡ | 80 (80, 86) | 80 (80, 84) | 80 (80, 86) | 0.7579 | 0.7304 |
| Glucose homeostasis parameters | |||||
| FBG (mM)‡ | 5.93 (5.57, 6.36) | 5.86 (5.54, 6.33) | 6.02 (5.67, 6.39) | 0.2866 | 0.8033 |
| PG30 (mM)‡ | 10.29 (19.3, 11.45) | 10.12 (9.19, 11.16) | 10.34 (9.37, 11.76) | 0.2219 | 0.4460 |
| PG120(mM)‡ | 7.72 (16.63, 9.3) | 7.49 (6.17, 8.96) | 7.85 (7.12, 9.58) | 0.0612 | 0.2829 |
| FINS(uU/ml)‡ | 5.2 (4,03, 7.11) | 5.06 (3.99, 6.06) | 5.42 (4.11, 8.08) | 0.0266 | 0.8033 |
| INS30 (uU/ml)‡ | 38.82 (26.11, 57.25) | 36.25 (25.73, 50.64) | 40.39 (26.77, 60.31) | 0.2956 | 0.5751 |
| INSI20 (uU/ml)‡ | 37.62 (25.13, 57.42) | 34.59 (21.78, 53.46) | 41.65 (29.18, 60.03) | 0.0562 | 0.3623 |
| GA (%)‡ | 0.61 (0.57, 0.67) | 0.62 (0.57, 0.68) | 0.61 (0.57, 0.67) | 0.6013 | 0.1853 |
| HbA1c (%)‡ | 5.5 (5.2, 5.9) | 5.4 (5.2, 5.7) | 5.6 (5.3, 6) | 0.0604 | 0.2707 |
| HOHA-IR‡ | 1.35 (1.05, 1.99) | 1.33 (1.02, 1.69) | 1.5 (1.09, 2.21) | 0.0266 | - |
| HOMA-β‡ | 40.23 (31.6, 54.5) | 37.68 (31, 52.44) | 42.13 (32.75, 56.04) | 0.1038 | 0.8752 |
| Serum liver enzymes and renal lunclicn indexes | |||||
| ALT (IU/liler)‡ | 15 (12, 18) | 15 (12, 17) | 15 (13, 18) | 0.2112 | 0.2477 |
| AST (IU/liter)‡ | 21 (19, 23) | 21 (19, 23) | 21 (19, 23) | 0.9994 | 0.8621 |
| GGT (IU/liter)‡ | 18 (15, 25) | 17 (14, 22) | 20 (16, 27) | 0.2764 | 0.2087 |
| TBIL (μM)‡ | 10.7 (9, 14.4) | 10.9 (9, 14) | 10.7 (9, 14.5) | 0.9516 | 0.7712 |
| Cr (μM)‡ | 64 (56, 73) | 64 (57, 73) | 54 (55, 76) | 0.8085 | 0.4501 |
| UAIb/Ucr‡ | 6.79 (5.12, 12.59) | 6.62 (5.16, 13.03) | 6.84 (5.02, 11.91) | 0.5568 | 0.4701 |
| UA (μM)‡ | 295 (249, 341) | 269.50 (243, 334) | 302 (258, 342) | 0.1618 | 0.0913 |
| Lipid proliles | |||||
| TG (mM)‡ | 1.20 (0.86, 1.66) | 1.07 (0.80, 1.53) | 1.34 (0.90, 1,80) | 0.0051 | 0.0193 |
| TC (mM)‡ | 4.97 (4.44, 5.58) | 4.80 (4.41, 5.57) | 5.01 (4.46, 5.58) | 0.4808 | 0.7310 |
| FFA (μM)‡ | 497 (377, 666) | 497.5 (371,688) | 497 (395, 650) | 0.3526 | 0.5783 |
| HDL-C (mM)‡ | 1.35 (1.14, 1.53) | 1.40 (1.19, 1.64) | 1.30 (1.09, 1.48) | 0.0895 | 0.1451 |
| LDL-C (mM) | 3.08 ± 0.72 | 2.99 ± 0.74 | 3.17 ± 0.69 | 0.0965 | 0.1969 |
| apoA-1 (g/liter) | 1.49 ± 0.26 | 1.51 ± 0.28 | 1.46 ± 0.24 | 0.2124 | 0.3083 |
| apoB (g/liler) | 0.91 ± 0.16 | 0.9 ± 0.17 | 0.93 ± 0.16 | 0.1899 | 0.3731 |
| apoE (mg/dl)‡ | 3.92 (3.23, 4.67) | 3.84 (3.22, 4.56) | 4.09 (3.4, 4.87) | 0.0609 | 0.1709 |
| Lipoprotein (a) (mg/dl)‡ | 14.05 (5.81, 25.07) | 15.33 (6.12, 25.89) | 12.51 (5.78, 23.24) | 0.7654 | 0.6452 |
| Cytokines | |||||
| FGF2l (pg/ml)‡ | 302.06 (180.19, 429.52) | 268.84 (171.83, 389.11) | 332.27 (236.74, 452.81) | 0.9538 | 0.9304 |
| hs-CRP (μg/ml)‡ | 0.63 (0.35, 1.17) | 0.53 (0.28, 1.17) | 0.72 (0.43, 1.16) | 0.0351 | 0.0758 |
P value denotes differences between NAFLD−/+ and NAFLD−/− analyzed by t test without adjustment.
P value denotes differences between NAFLD−/+ and NAFLD−/− analyzed by analysis of covariance with HOMA-IR adjusted.
Log-transformed before anafysis.
Modest but distinguishable differences in baseline gut microbiome between NAFLD−/+ and NAFLD−/− individuals
We assessed the gut microbiome structure of the NAFLD−/+ and NAFLD−/− groups at baseline via shotgun metagenomic sequencing, generating 1128 gigabase pairs of high-quality reads with an average of 41,786,187 reads per sample (Fig. 1). Taxonomic profiling with MetaPhlAn2 (42) led to the identification of 405 species. Community alpha diversity measured as richness, and Shannon and Simpson indexes showed no significant differences (Wilcoxon rank-sum test, P > 0.05) at the species, genus, or family levels between the two groups (fig. S2A). Bray-Curtis, unweighted UniFrac, and weighted UniFrac distance comparisons indicated that the NAFLD−/+ and NAFLD−/− groups did not have significant community dissimilarities [permutational multivariate analysis of variance (PERMANOVA), P > 0.05; fig. S2, B and C]. The same patterns were observed when using a metagenomic species approach (43) for the taxonomic annotation (table S2).
In addition, we sequenced the baseline gut microbiota from 66 participants who were diagnosed as NAFLD in both 2014 and 2018 (NAFLD+/+) and 34 participants who were diagnosed as NAFLD in 2014 but not in 2018 (NAFLD+/−). These two groups were also matched with the other two groups described above by age, gender, BMI, and 4-year change in BMI. A thorough comparison of microbiota alpha and beta diversity among the four groups at baseline indicated that the two non-NAFLD groups were distinguishable from the two NAFLD groups (P < 0.05, Wilcoxon rank-sum test for alpha diversity comparisons and PERMANOVA for beta diversity comparisons using Bray-Curtis distances) (fig. S3). Moreover, the gut microbiota of NAFLD−/+ subjects was different from that of NAFLD+/+ and NAFLD+/− individuals. This argues that the NAFLD−/+ group was not already diseased at the baseline because they clustered with NAFLD−/− subjects at baseline. Because our focus was to identify gut microbiota signatures in disease-free individuals suggestive of NAFLD predisposition, only the NAFLD−/− and NAFLD−/+ groups were further analyzed.
A compositional analysis found that several of the 10 most abundant genera and species (Fig. 2A) were significantly associated (envfit from R package vegan, P < 0.05) with observed variation in the taxonomic profile of the study participants (fig. S2, B and C). However, their relative abundances were not significantly different (zero-inflated Gaussian mixture model, P > 0.05) between NAFLD−/+ and NAFLD−/− groups. Nevertheless, the relative abundances of 8 and 21 less-abundant genera and species, respectively, were significantly different (zero-inflated Gaussian mixture model, P < 0.05) between the two groups (fig. S2D). Methanobrevibacter [false discovery rate (FDR) = 0.01] was decreased in NAFLD−/+ compared to NAFLD−/− (a reduction in Phascolarctobacterium was insignificant at FDR = 0.2). Lower abundances of these two genera have been observed in cohort studies in obese individuals compared to lean individuals (44, 45). Slackia has been reported to be more abundant in individuals with moderate-to-severe fibrosis than in individuals with absent-to-mild fibrosis (46), and this genus was increased in the NAFLD−/+ compared to the NAFLD−/− group (FDR = 0.06). The relative abundance of Dorea formicigenerans, a species that is highly abundant in people with obesity (47), was higher in the NAFLD−/+ than the NAFLD−/− group (FDR = 0.17). Differences in the relative abundances of Methanobrevibacter, Phascolarctobacterium, Slackia, and D. formicigenerans between the two study groups remained significant even after adjusting for age, gender, BMI, and HOMA-IR (zero-inflated Gaussian mixture model, P < 0.05). Because the NAFLD−/+ and NAFLD−/− groups had no difference in BMI and in the aforementioned cohort studies, the liver status of the obese individuals was not evaluated, and our prospective design suggested that Methanobrevibacter, Phascolarctobacterium, Slackia, and D. formicigenerans could be signatures of NAFLD risk in addition to being obesity-related signatures.
Fig. 2. Global characteristics of gut microbiome and serum metabolome.
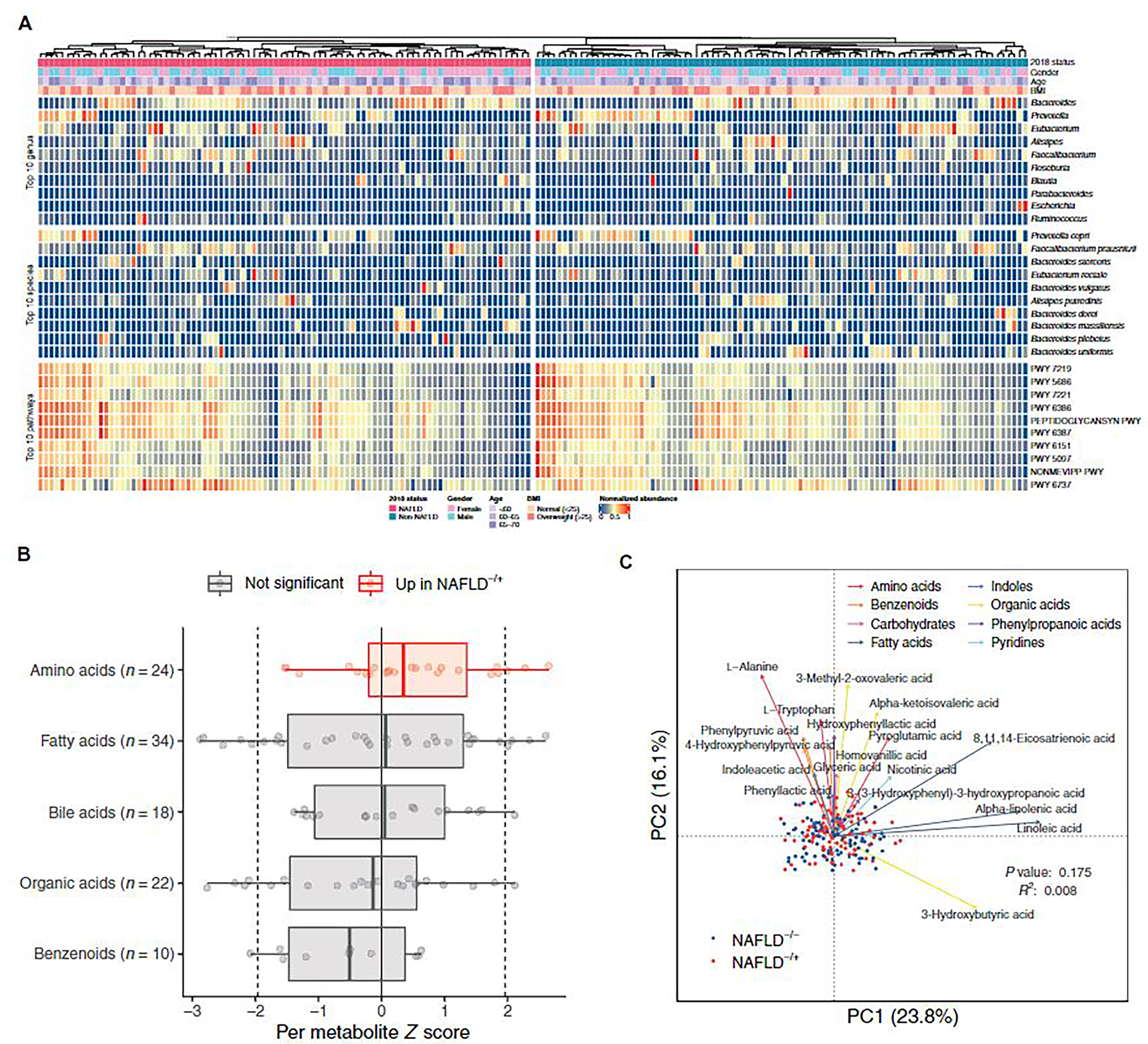
(A) Relative abundance of the 10 most abundant genera, species, and pathways for the 180 participants at baseline, grouped by NAFLD status at the follow-up visit. Anthropometric characteristics of the participants at baseline are also shown. Abundance values are normalized to the range of 0 and 1. (B) Changes of metabolites (μM) in metabolite classes containing at least 10 metabolites. Each point represents a metabolite and its z score from Wilcoxon rank-sum test comparing the two groups (negative indicates higher abundance in NAFLD−/−; positive indicates higher abundance in NAFLD−/+). Dotted lines at −1.96 and 1.96 denote the significance threshold. Colors indicate comparisons between z scores of metabolites in a metabolite class against the z scores of metabolites in all other classes. Box plots show median, lower/upper quartiles, and whiskers (the last data points 1.5 times interquartile range from the lower or upper quartiles). (C) Principal coordinates analysis for 180 participants based on Bray-Curtis distances using baseline serum concentrations of 123 metabolites. For each metabolite class, the top 3 (or fewer) metabolites that were significantly associated with the metabolome variation in the study cohort are shown. PC, principal coordinates.
We used HUMAnN2 (48) for functional profiling of the microbial communities and identified 458 pathways. Likewise, the taxonomic profile and the microbiota functional potential could not differentiate between NAFLD−/+ and NAFLD−/− groups by alpha and beta diversity (fig. S4, A and B). Four of the most abundant pathways detected, uridine monophosphate biosynthesis I, uridine diphosphate–N–acetylmuramoyl-pentapeptide biosynthesis I and II, and peptidoglycan biosynthesis I (Fig. 2A), were significantly associated with observed variation in the functional profiles of study participants (envfit from R package vegan, P < 0.05; fig. S4B). These pathways were proposed to be discriminatory for NAFLD cirrhosis against control groups in a recent U.S. cohort study (49); however, their relative abundances were not significantly different (zero-inflated Gaussian mixture model, P > 0.05) between the NAFLD−/+ and NAFLD−/− groups in our prospective study. Nevertheless, we found 19 biosynthetic pathways significantly different in relative abundance between the two groups (zero-inflated Gaussian mixture model, P < 0.05) (fig. S4C). We observed a significantly higher relative abundance of geranylgeranyl diphosphate biosynthesis and the mevalonate pathway in the NAFLD−/− group. These pathways are dysregulated in mice and humans with nonalcoholic steatohepatitis (NASH) (50). Two genes encoding enzymes involved in these pathways, hydroxymethyglutaryl–coenzyme A (CoA) reductase (EC 1.1.1.34) and mevalonate kinase (EC 2.7.1.36), were significantly enriched in the NAFLD−/− group (zero-inflated Gaussian mixture model, P < 0.05; table S3). Methanobrevibacter smithii was the major contributor of gene expression abundance of hydroxymethyglutaryl-CoA reductase (95%) and mevalonate kinase (40%). In contrast, the NAFLD−/+ group had a higher relative abundance of phosphatidate metabolism and cholic acid degradation. Cholic acid is a primary bile acid that decreases substantially in rats on a Western diet and is proposed as an early marker of NAFLD development (51). Genes encoding phospholipase D (EC 3.1.4.4) and bile-acid-7-alpha-dehydratase (EC 4.2.1.106) were also significantly enriched in the NAFLD−/+ group (zero-inflated Gaussian mixture model, P < 0.05, table S3). The four significant pathways above (geranylgeranyl diphosphate biosynthesis, mevalonate pathway, phosphatidate metabolism, and cholic acid degradation) remained significantly different (zero-inflated Gaussian mixture model, P < 0.05) between the two groups after adjusting for age, gender, BMI, and HOMA-IR, except for cholic acid degradation that was marginally significant (P = 0.050).
Metabolite enrichment and metabolic shifts in NAFLD−/+ versus NAFLD−/− groups
We next performed targeted metabolomic analysis of serum samples collected at baseline to interrogate whether differences in species and pathway abundance of gut microbiota led to distinct profiles of microbial metabolites in the NAFLD−/+ and NAFLD−/− groups. We detected 123 metabolites grouped into nine metabolite classes (Fig. 2B). We performed enrichment analysis to identify metabolite classes that were significantly overabundant or underabundant in the NAFLD−/+ or the NAFLD−/− group, and found amino acids were significantly elevated in the NAFLD−/+ group (Wilcoxon rank-sum test, P < 0.05; table S4). We further analyzed the untargeted metabolomic data of a European case-control cohort (MICROBARIA) involving 52 obese women including 26 with biopsy-confirmed NAFLD and 26 non-NAFLD (52). Two amino acids positively correlated with NAFLD-related liver enzymes, including the branched-chain amino acid valine with alanine transaminase (ALT) (P < 0.05, Spearman correlation) and the aromatic amino acid tyrosine with aspartate transaminase (AST) (P < 0.05, Spearman correlation). Our findings in the Asian prospective and European cohort–based datasets are further supported by recent metabolomic-based studies, suggesting that perturbations in amino acid metabolism are involved in NAFLD and NASH pathogenesis (53–55).
Of the 15 significantly different metabolites between the NAFLD−/+ and NAFLD−/− groups at baseline (generalized linear model, P < 0.05; fig. S5A) and the metabolites that were significantly associated with the observed metabolomic variation (envfit from R package vegan, P < 0.05; Fig. 2C), several are reported to be involved in NAFLD in case-control human or animal studies. For example, 3-chlorotyrosine, arachidonic acid, and oxoglutaric acid are markers, respectively, of liver damage and NAFLD development in mouse (56) and rat (57) models and a human NAFLD study (58). Tryptophan was also significantly associated with the metabolome variation (envfit from R package vegan, P < 0.05; Fig. 2C) in our cohort, and aromatic amino acids have been associated with NAFLD (54). These metabolites were higher in the NAFLD−/+ group than the NAFLD−/− group. Concentration of a gut microbiota–regulated fatty acid, 8,11,14-eicosatrienoic acid, linked to obesity and insulin resistance (59, 60), was also significantly higher (generalized linear model, P < 0.05; fig. S5A) in the NAFLD−/+ group in our prospective study. Phenyllactic acid, produced by lactic acid bacteria and suggested to reduce reactive oxygen species production in rodents (61), was significantly higher in the NAFLD−/− group (generalized linear model, P < 0.05; fig. S5A). On the contrary, the direction of concentration differences in the two study groups for isovaleric and docosahexaenoic acids (both higher in NAFLD−/+) (generalized linear model, P < 0.05; fig. S5A) was inconsistent with proposals in the literature from case-control NAFLD studies about the possible roles of these compounds (62–64). These agreements and discrepancies in metabolite abundances in our prospective study with case-control cohort and mouse studies in the literature should help to narrow the metabolic marker possibilities for NAFLD progression. The concentrations of additional fatty acids were significantly different between the NAFLD−/+ and NAFLD−/− groups (fig. S5A), but the functional significance of these metabolites in NAFLD is relatively unknown. Last, the concentrations of measured serum metabolites such as 3-chlorotyrosine and phenyllactic acid were significantly associated with gut microbiota species composition (Mantel test, P < 0.05; fig. S5B and table S5).
A machine learning prospective model to detect early signatures of NAFLD
We built a noninvasive risk assessment model (random forest algorithm) to classify healthy subjects based on their NAFLD status after 4.6 years, using a combination of baseline metagenomic and metabolomic features. A leave-one-out iterative approach was applied to build and evaluate our model due to the relatively small cohort size (n = 180). We built a prospective model using 14 taxonomic, functional, and metabolomic features of the study participants at baseline that enabled classification based on their NAFLD status 4.6 years later with an area under the receiver operating characteristic curve (auROC) of 0.72 (Fig. 3A). The performance of the model was significantly improved to an auROC of 0.79 (DeLong test, P value for difference < 0.05) with the addition of only two more noninvasive clinical features (Fig. 3B). We then slightly improved our model by also including the most accessible anthropometric parameters, BMI and age, to obtain our final model (auROC, 0.80; Fig. 3C and fig. S6).
Fig. 3. Predictive performance of machine learning models in the study cohort and diagnostic performance of the final model in external cohorts.
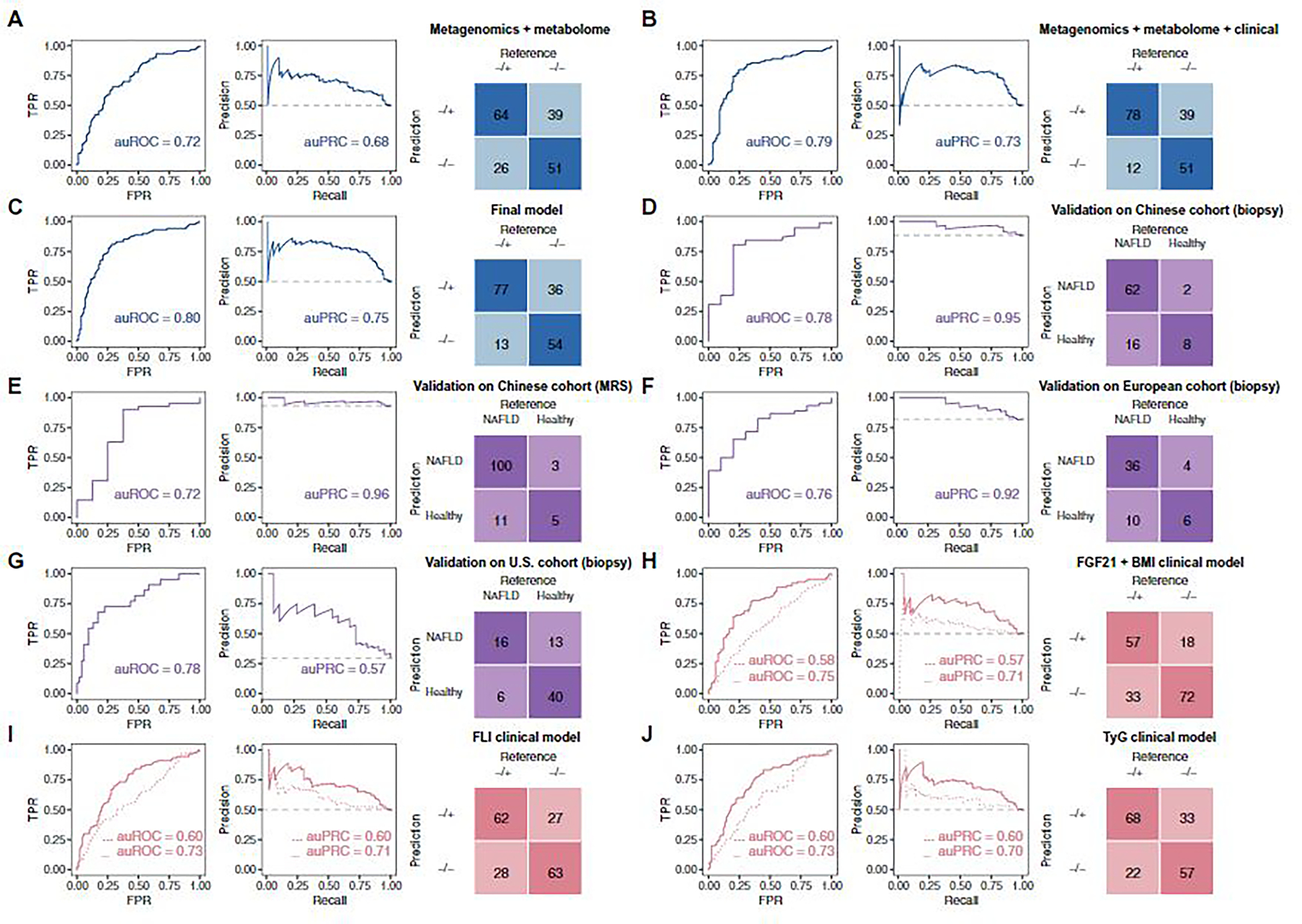
(A to C, in blue) Performance of leave-one-out iterative machine learning models discriminating between NAFLD−/+ and NAFLD−/− groups using features of the following: (A) metagenomics + metabolome, (B) metagenomics + metabolome + 2 clinical parameters (HDL and fasting insulin), and (C) metagenomics + metabolome + 2 clinical parameters (HDL and fasting insulin) + anthropometrics (BMI and age). (D to G, in purple) Diagnostic performances of a model built based on subsets of the selected features to discriminate between participants who were healthy or had NAFLD in four external cohorts: (D) a Chinese cohort in which NAFLD diagnosis was determined with biopsy, (E) a Chinese cohort in which NAFLD diagnosis was based on MRS, (F) a biopsy-diagnosed European NAFLD cohort, and (G) a biopsy-diagnosed U.S. cirrhosis cohort. (H to J, in peach) Leave-one-out iterative machine learning performance to discriminate between NAFLD−/+ and NAFLD−/− groups in models of: (H) FGF21 + BMI clinical model, with and without metagenomics + metabolome features; (I) FLI clinical model, with and without metagenomics + metabolome features; and (J) TyG clinical model, with and without metagenomics + metabolome features. (H to J) Models without metagenomics and metabolome features were trained by logistic regression (dotted lines); models including metagenomics and metabolome features were trained by random forest (solid lines). Confusion matrices in (F) to (H) are from models with metagenomics and metabolome features. The figure colors represent the purpose of the model: blue, model construction; purple, external validation in cohorts of different ethnicity; peach, testing performance of previous clinical models in our cohort. Further details of the overall machine learning analysis framework can be found in fig. S6. auROC, area under the receiver operating characteristics curve; auPRC, area under the precision-recall curve; TPR, true-positive rate; FPR, false-positive rate.
We evaluated the biological relevance of the selected features by testing the diagnostic ability of components of our model to distinguish between healthy individuals and patients with NAFLD in two publicly available independent external Asian case-control cohorts; one cohort was diagnosed by biopsy and the other was diagnosed by magnetic resonance spectroscopy (MRS). This allowed us to further explore whether the patient diagnosis method had an impact on the model performance. We built a new prospective model using only nine features from the final model that were available in these external cohorts (fig. S6). The new model derived based on our study cohort discriminated healthy and NAFLD groups in the two external cohorts with auROCs of 0.78 and 0.72 (Fig. 3, D and E), showcasing that the features we identified were closely related to NAFLD development or pathophysiology. Besides the Asian cohorts, we further validated our prospective model in other case-control cohorts of different ethnicity. In the European cohort FLORINASH (54), the model (with the same nine features as in the Asian cohorts) reached an auROC of 0.76 (Fig. 3F), whereas in a U.S. cohort (49), the validation auROC (with seven available features) was 0.78 (Fig. 3G). Taking into consideration that only no more than half of features in our original prospective model were available in the external cohorts, we expect that the true accuracy may be higher.
Previous clinical prospective NAFLD studies demonstrated that fibroblast growth factor 21 and BMI (FGF21 + BMI), fatty liver index (FLI), and TG and glucose index (TyG) predict NAFLD development from 3 up to 9 years before diagnosis (auROCs of 0.71 to 0.82) (19–21). We compared the performance of our prospective model with FGF21 + BMI, FLI, and TyG to predict NAFLD occurrence in our cohort with matched baseline characteristics. The performance of our final model (auROC, 0.80) was significantly better than all three clinical models (auROCs of 0.58 to 0.60, P values for difference < 0.01; Fig. 3, H to J). To confirm the importance of metagenomic and metabolomic information in prospective NAFLD prediction, we added metagenomic and metabolomic features from our final prospective model to the clinical models (fig. S6) and observed significant improvements in all (auROCs of 0.73 to 0.75, P values for difference < 0.05; Fig. 3, H to J); however, none of the models reached the auROCs of our final model.
In total, 18 features were used in the final model: two genera, three pathways, nine metabolites, and four anthropometric and clinical parameters (Fig. 4, A and B). Our analysis revealed that the most important feature of our risk assessment model was phenyllactic acid (Fig. 4A). By analyzing the untargeted metabolomic data from the European MICROBARIA cohort (52), we found that phenyllactic acid negatively correlated with ALT, AST, and gamma-glutamyl transferase (correlation coefficients = −0.35, −0.45, and −0.45; P = 0.004, 0.14, and 0.053; Pearson’s correlation adjusted for age, BMI, fasting glucose, and insulin).
Fig. 4. SHAP-based model interpretation.
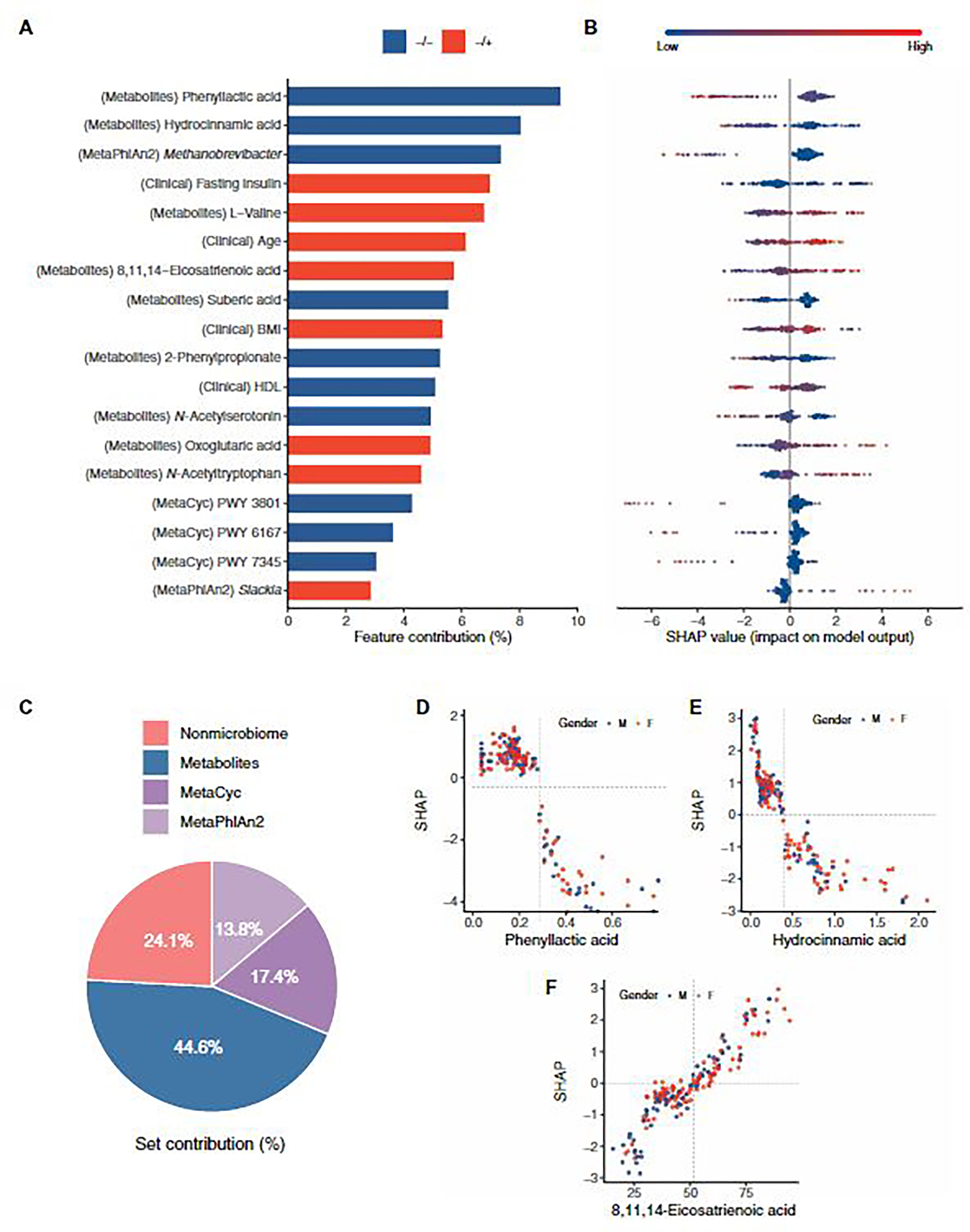
(A) Bar plot of selected features and their contribution in the NAFLD prediction model. Features are in descending order by contribution (also known as importance) in the model. Blue bar, higher value of the feature for association with NAFLD−/−; red bar, higher value of the feature for association with NAFLD−/+. Details of associations are shown in (B) a bee swarm plot in which each point represents a participant (n = 180). Color indicates the value of the feature, with red higher and blue lower. Negative SHAP value indicates the feature attribution for prediction of NAFLD−/−; Positive SHAP value indicates the feature attribution for prediction of NAFLD−/+. (C) Feature category contribution calculated by summing the SHAP values per set. (D to F) Examples of SHAP dependence plots, showing the effect the feature has on model prediction. Each point represents a participant (n = 180). Color indicates sex with blue for male and red for female. X axis is the feature value, and y axis is the SHAP value for the feature. The optimal thresholds for features are indicated by the vertical dotted lines.
SHapley Additive exPlanations (SHAP) (65) analysis also revealed that Methanobrevibacter was associated with NAFLD−/−, and Slackia was associated with NAFLD−/+ (Fig. 4, A and B). These genera were differentially abundant in our two study groups (fig. S2D). Furthermore, 8,11,14-eicosatrienoic acid, hydrocinnamic acid, and oxoglutaric acid are associated with type 2 diabetes, obesity, insulin resistance, and NAFLD (58, 60, 61, 66), and our model revealed similar trends (Fig. 4, A and B).
The feature set contribution was also computed by summing the SHAP values per category. Metabolites were the most important in the model, contributing 44.6% to model performance, followed by the microbiome and nonmicrobiome features, with contributions of 31.2 and 24.1%, respectively (Fig. 4C).
Dependence plots were built to reveal the nonlinear correlations of features and risk of NAFLD. The optimal thresholds of each feature were identified (fig. S7). We found that high-density lipoprotein (HDL) was associated with NAFLD occurrence after 4.6 years when <1.39 mM, which is close to the diagnostic criteria for metabolic syndrome when HDL was <1.0 mM (male) or <1.3 mM (female) (67). We also examined the dependence plots of microbial metabolite phenyllactic acid, hydrocinnamic acid, and 8,11,14-eicostrienoic acid (Fig. 4, D to F). Phenyllactic acid was associated with protection against NAFLD at a concentration of >0.25 μM. The concentration of 8,11,14-eicosatrienoic acid increased the risk of NAFLD at >51.5 μM, and hydrocinnamic acid was associated with NAFLD at a concentration of <0.39 μM. Visual inspection of the dependence plots did not indicate any differences by sex. We converted the features into binary variables (≥ or < thresholds) according to their optimal threshold and found that 12 of 18 features showed significant association with NAFLD progression (chi-square test, P < 0.05; table S6). These results demonstrated the importance of including an interpretable machine learning framework, such as SHAP, to provide insights when analyzing microbiome data.
We further examined whether the features of our risk assessment model could be used to classify subjects of the NAFLD−/+ group based on different degrees of steatosis. We initially divided the NAFLD−/+ group based on their liver fat percentage at the time of diagnosis (4.6 years after enrolment). Subsequently, using the values of the 18 features at baseline, we built a model classifying mild and severe steatosis cases. This new random forest model had an auROC of 0.78 (fig. S8A). Similarly as above, we attempted to confirm the biological relevance of the selected features by testing the diagnostic power of our prospective model in an independent external case-control cohort from the United States (49). Despite the lack of absolute quantification of metabolomic data, our model showed an accuracy of 71.4% to correctly identify severe steatosis cases with only gut microbial and clinical features.
Previous work has demonstrated the value of gut microbiome-based diagnostic tests for advanced fibrosis (68). The participants in our cohort were unlikely to develop advanced fibrosis after 4 years, starting as NAFLD free at baseline. Nevertheless, the prospective design of our study enabled us to explore whether the baseline microbiota is associated with the change or deterioration of fibrosis. Grouping our NAFLD−/+ participants by the change of fibrosis 4 (FIB-4) index from 2014 to 2018, we built a new risk assessment model using five gut microbiota functional pathways, classifying subjects by the fibrosis deterioration with an auROC of 0.72 (fig. S8B). In a U.S. case-control cohort (49), the pathway with the highest importance in our model, phosphopantothenate biosynthesis, was significantly higher (zero-inflated Gaussian mixture model, P < 0.05) in the cirrhosis group than in non-NAFLD controls. Methanobrevibacter, which was the top taxonomic feature in the prospective model, was also significantly lower (zero-inflated Gaussian mixture model, P < 0.05) in patients with cirrhosis.
DISCUSSION
NAFLD prevalence has rapidly increased over a short time, especially in China (69). China is projected to have the largest number of liver-related deaths among the most economically developed countries by 2030 (70). Accumulating evidence suggests that the gut microbiome may emerge as an active player in NAFLD development (71). Human studies demonstrated different gut microbiota profiles among individuals with NAFLD and those without, as well as in individuals at different stages of NAFLD (68, 72). In the recently proposed concept of metabolic-associated fatty liver disease (MAFLD) that extends beyond NAFLD (23), gut microbiota is suggested to be a major factor related to the heterogeneous phenotype of MAFLD. In both NAFLD and MAFLD, the disease complexity and heterogeneity may be better resolved by the inclusion of omics technologies that integrate patient clinical phenotypes and molecular phenomics and gut microbial features. This approach has shown its potential in the classification of hepatic (73) and, more recently, extrahepatic diseases including ischemic heart disease (74) and coronary artery disease (75). Both studies of cardiovascular diseases suggested that major alterations of the gut microbiome and metabolome might occur earlier than clinical onset of disease, suggesting the utility of gut microbiota–based risk assessment. A recent prospective study extended cross-sectional evidence and demonstrated that gut microbiota composition is predictive of incident type 2 diabetes after 15.8 years (76). Our study comprehensively characterized the gut microbiome of Chinese participants using stool samples taken 4.6 years before the NAFLD diagnosis and matched controls. We assessed the ability of metagenomic and metabolomic features as a risk assessment tool of NAFLD occurrence within 4.6 years and developed a random forest machine learning model that distinguished individuals at risk for NAFLD from controls with a performance of 0.80 auROC. The final model consisted of 18 features of mainly bacterial genera, pathways, and metabolites, with two clinical and two anthropometric parameters. Using subsets of those features available in external case-control cohorts also showed good ability (auROC of 0.73 to 0.78) to classify individuals with and without NAFLD, including in cohorts with the biopsy-confirmed present/absence of NAFLD and of different ethnicities, supporting the biological relevance and generalizability of our prospective model.
Diagnosis of NAFLD requires evidence of hepatic steatosis, either by histology or imaging. Liver biopsies have a risk of severe complications, and the sampling procedure may leave some people with NAFLD undiagnosed if they have unevenly distributed histological lesions (13). Steatosis evaluation based on imaging such as MRS, CT, or ultrasonography has limitations in clinical practice such as high price, radiation exposure, and limited sensitivity. Numerous research efforts have searched for other reliable, cost-effective, non-invasive diagnostic approaches, including using features that are clinical (age, gender, diabetes, and BMI), biochemical (aminotransferases, bilirubin, and ferritin), metabolic (glycated hemoglobin, insulin, and HOMA-IR), or lipid (TG and cholesterol) parameters or other markers such as FGF21 and adiponectin (77–79). A few prospective studies have also attempted to predict the development of NAFLD over the long term (19–21). However, the predictive power of these models was evaluated in study groups with unmatched baseline characteristics, which may have led to overestimation of model performance. In our community-based prospective study, these models showed limited performance (auROC in the range of 0.58 to 0.60) when our nested case-control design included matching for gender, age, BMI, and 4-year BMI change. This matching is particularly important for removing confounding effects and to uncover microbiome-related risk factors for NAFLD development, given that obesity is a major risk factor for NAFLD. Our microbiome-based model demonstrated a good performance (auROC of 0.72) for predicting the NAFLD status of NAFLD-free individuals after 4.6 years.
Our study has limitations. The classification of patients into two groups was not based on liver biopsy, which remains the gold standard for NAFLD diagnosis. However, this method is impractical in a community study with thousands of participants, as in our study, and is unethical for participants who do not show any sign of the disease (matched controls). Moreover, according to guidelines from the European Association for the Study of the Liver, European Association for the Study of Diabetes, and European Association for the Study of Obesity, ultrasound is the first-line diagnostic test for NAFLD (36), especially for large-scale screening studies. This diagnostic criterion has been extensively used in previous studies, such as the Rotterdam cohort (80), the Golestan cohort (81), and the Kangbuk Samsung Health Study (82). We note that our cohort is of high quality, with relatively comprehensive indexes acquired in a large population. For example, in the measurement of glucose metabolism, oral glucose tolerance tests were conducted for all participants. This test is usually replaced by fasting glucose or FINS tests in many population-based studies. Second, we could not predict the development of more severe outcomes such as fibrosis because of their low incidence. This was mainly due to the nature of our community-based epidemiological investigation. However, using baseline microbiota, we were able to classify subjects by fibrosis deterioration with an auROC of 0.72.
Furthermore, serum ferritin was not measured in our study, although several studies have indicated its relevance in NAFLD (83–85). Therefore adding ferritin in our prospective model could potentially enhance performance. The predictive power of our prospective model (auROC of 0.80) was an advance compared to existing clinical models (auROCs of 0.58 to 0.60). However, further improvements, for example, integrating additional biochemical parameters, will be necessary for clinical applications. Our metabolic signatures and their taxonomic drivers revealed by our prospective model imply but do not prove causality; thus, additional studies are required to clarify the molecular mechanisms involved in NAFLD development.
Integrating bacterial species and functions in machine learning models for predicting host response to treatment or lifestyle interventions and disease progression has shown great potential (86–89). For NAFLD and its complications, gut microbiota changes can independently predict the risk of short-term hospitalizations (90 days) in patients with cirrhosis with an auROC of 0.83 (90). Elucidating the importance of the gut microbiome as a long-term risk assessment tool in NAFLD is important because of the current limited therapeutic landscape for NAFLD and findings that early detection can substantially improve outcomes for patients with NAFLD (91, 92). Our proof-of-concept study identified a microbiome signature in participants at risk of developing NAFLD in the next 4 years and points to the potential of noninvasive diagnostic tests to complement existing clinical screening tools for NAFLD. Moreover, identifying microbiome signatures also opens a window of opportunities for microbiome-based prophylactic and therapeutic interventions such as the utility of propionic acid as a potent immunomodulatory supplement to multiple sclerosis drugs (93), which is not offered by a clinical predictive model built upon only a few clinical parameters or other features. Evaluation and further improvement of our NAFLD risk assessment model using larger prospective studies that are heterogeneous for ethnicity and lifestyle patterns will increase the model’s generalizability and obtain more refined estimations of its accuracy.
MATERIALS AND METHODS
Study design
The aim of this study was to identify potential predictive signatures for early clinical warning of NAFLD and to develop a prognostic risk assessment model for long-term NAFLD development. For this purpose, we conducted a nested case-control study within a 4.6-year prospective study in 2487 Chinese individuals, and we profiled 180 individuals from 1216 NAFLD-free participants at baseline, including 90 that were diagnosed with NAFLD in the follow-up visit (NAFLD−/+), which were matched with 90 controls without NAFLD (NAFLD−/−) by gender, age, BMI, and 4.6-year BMI change. We performed comprehensive metagenomic and metabolomic analyses using stool and serum samples taken at baseline, including taxonomic diversity and profiles at family, genus and species levels, microbial enzymes, metabolic pathways, and metabolites. An interpretable machine learning model integrating baseline microbial signatures was built to predict NAFLD development after 4 years. The biological relevance of selected features in the model to NAFLD development was further validated in external cohorts, including three cohorts with the biopsy-confirmed presence/absence of NAFLD. New models were built for validation, given that some features were not available in the external cohorts. All validation models were trained on our cohort and tested in the external cohorts. Further materials and methods details are available in the Supplementary Materials.
Study participants
All participants were from the Nicheng Diabetes Screening Project (also called the Shanghai Nicheng Cohort Study) previously described (94, 95). This population-based, prospective study was designed to assess the prevalence, incidence, and factors related to cardiometabolic diseases among adults in Nicheng County, a suburb of Shanghai, China. On the basis of the project, we designed a nested case-control study to explore the potential causal role of the gut microbiome in NAFLD in three randomly selected Nicheng communities (involving 2487 participants). Figure S1 outlines study enrolment. Of 2487 participants, 1216 were identified as not having NAFLD at baseline; among them, 524 completed a follow-up visit 4.6 years after baseline and were screened by ultrasonography. Incident cases of NAFLD (n = 146) were identified at the 4.6-year follow-up visit, of which 90 participants were eligible for this study involving gut microbiota, according to the following criteria to exclude participants: existed fatty liver, acute infectious disease, biliary obstructive diseases, alcohol abuse (more than 140 g of ethanol/week for men or 70 g of ethanol/week for women), acute or chronic cholecystitis, acute or chronic viral hepatitis, cirrhosis, diarrhea, known hyperthyroidism or hypothyroidism, chronic renal insufficiency, heart failure, presence of cancer, pregnancy, stroke in acute phase, receipt of any antibiotic treatment within 2 weeks or receipt of any probiotic or prebiotic within 1 week before sample collection, and suffering from chronic or acute gastrointestinal diseases (including diarrhea, gastrointestinal infection, and inflammatory bowel disease) in recent 1 month before sample collection. Controls (n = 90 for a case-control ratio of 1:1) were chosen from the remaining participants who did not develop NAFLD by the follow-up visit. To control for the risk profiles in patients who developed NAFLD and those who did not, controls were matched for age (±3 years), sex (male and female), BMI (±3 kg/m2) at both baseline and follow-up, and BMI change (±0.5 kg/m2). The study was approved by the ethics committee of the Shanghai Sixth People’s Hospital (approval no: 2014–27), following the principles of the Declaration of Helsinki. Written informed consent was obtained from all participants.
Evaluating the diagnostic ability of the model in external cohorts
To our knowledge, no similar studies have conducted long-term follow-up of NAFLD development in healthy individuals using a combination of gut metagenome, metabolome, and clinical features as a risk assessment tool. Thus, we were unable to test our prospective model directly in an external cohort. Instead, we used external case-control cohorts to examine the ability of our final prognostic model to classify correctly NAFLD and healthy participants. Four cohorts were used, including two cohorts of Chinese: (i) 78 patients with NAFLD and 10 controls without NAFLD, as diagnosed with biopsy (BioProject ID: PRJNA732131), and (ii) 111 MRS-diagnosed NAFLD patients and 8 controls (BioProject IDs: PRJNA703757 and PRJNA414688); and two biopsy-diagnosed cohorts of other ethnicity: (iii) a European cohort of 46 patients with NAFLD and 10 controls (54) and (iv) a U.S. cohort of 26 cirrhosis patients and 54 controls (49). For further additional data (e.g., anthropometric and/or available clinical data) for the two Chinese validation cohorts besides grouping information, please contact the corresponding author.
Because some selected features included in the final model were not available in the external cohorts, we were unable to test our model directly. Instead, we built a new prognostic model based on the NAFLD−/+ and NAFLD−/− groups using a subset of the 18 selected features that were available in the external cohorts. In the model for the two Chinese cohorts and the European cohort, 9 of the 18 selected features were used: two genera, three pathways, two anthropometric parameters, and two noninvasive clinical metadata; whereas 7 of the 18 selected features were used in the model for the U.S. cohort: two genera, one pathway, two anthropometric parameters, and two noninvasive clinical metadata. Performances of models, including ROC curves, precision-recall curves, and confusion matrices (generated with the optimal probability cutoff of the ROC curve), were produced by applying the model to the unseen external cohort data.
Statistical analysis
Statistical analyses of clinical data were performed with SAS version 9.4 (SAS Institute Inc.). Normally distributed data were expressed as means ± SD. Data that were not normally distributed, as determined using the Kolmogorov-Smirnov test, were logarithmically transformed before analysis and expressed as median with lower and upper quartiles. Student’s t test and chi-square tests were used to assess differences between two groups for continuous and categorical variables, respectively. In addition, analysis of covariance was used for continuous variables to assess the difference between the two groups after adjusting for HOMA-IR.
Metagenomic data, including taxonomy and functional data, and metabolomic data were analyzed in R software version 3.6.3. Metagenomic data were analyzed with the zero-inflated Gaussian mixture model, using the function fitZig from R package metagenomeSeq (96) with the default settings; metabolomic data were analyzed using the generalized linear model with inverse gamma distribution. Wilcoxon rank-sum tests were used to test for significant differences in alpha diversity. PERMANOVA was used to analyze beta diversity with adonis function from R package vegan. A Mantel test, implemented in mantel from R package vegan, using Spearman’s correlation coefficient was used to analyze the associations between microbiome and metabolites. Bray-Curtis dissimilarity matrices based on taxonomic relative abundance and Euclidean dissimilarity matrix for each metabolite were computed to perform this test. The auROCs of different models were compared with the DeLong test, using the roc. test function from R package pROC (97). Data were considered statistically significant at P value < 0.05. The Benjamini-Hochberg procedure was applied to calculate the FDR to adjust P values for multiple hypothesis testing.
Supplementary Material
Acknowledgments:
We thank K. Zych (Clinical Microbiomics) for reviewing the machine learning analysis.
Funding:
This work was supported by Marie Sklodowska-Curie Actions (MSCA) and Innovative Training Networks, H2020-MSCA-ITN-2018 813781 “BestTreat” (G.P., H. Leung, S.L.S., D.P., E.N., and H.B.N.); the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy, EXC 2051, Project ID 390713860 (G.P.); the Hong Kong Research Grants Council/Area of Excellence (AoE/M/707-18) (A.X.); the EU-HK Co-funding Mechanism (E/HKU703/20) (A.X.); the National Natural Science Foundation of China (NSFC)–NHMRC joint research grant (81561128016) (W.J.); the Shanghai Municipal Key Clinical Specialty (W.J.); the National Key Research and Development Program of China (2018YFA0800402) (W.J.); the General Program of NSFC (81870598) (H. Li); the Excellent Young Scholars of NSFC (82022012) (H. Li); the Two Hundred Program from Shanghai Jiao Tong University School of Medicine (20191830) (H. Li); the Innovative Research Team of High-level Local Universities in Shanghai (SHSMU-ZDCX20212700) (H. Li); the Assistance Publique Hôpitaux de Paris (APHP) DGOS for the Programme hospitalier de Recherche clinique (microbaria) predictive value in identifying individuals with a high risk of developing NAFLD and EU transatlantic project Leducq and JPI-Microdiet (K.C.); the NCATS (5UL1TR001442) (R.L.); the NIDDK (U01DK061734, U01DK130190, R01DK106419, R01DK121378, R01DK124318, and P30DK120515) (R.L.); the NHLBI (P01HL147835) (R.L.); and the NIAAA (U01AA029019) (R.L.).
Footnotes
Competing interests: R.L. serves as a consultant to Aardvark Therapeutics, Altimmune, Anylam/Regeneron, Amgen, Arrowhead Pharmaceuticals, AstraZeneca, Bristol-Myer Squibb, CohBar, Eli Lilly, Galmed, Gilead, Glympse Bio, Hightide, Inipharma, Intercept, Inventiva, Ionis, Janssen Inc., Madrigal, Metacrine Inc., NGM Biopharmaceuticals, Novartis, Novo Nordisk, Merck, Pfizer, Sagimet, Theratechnologies, 89 Bio, Terns Pharmaceuticals, and Viking Therapeutics. In addition, his institutions received research grants from Arrowhead Pharmaceuticals, AstraZeneca, Boehringer-Ingelheim, Bristol-Myers Squibb, Eli Lilly, Galectin Therapeutics, Galmed Pharmaceuticals, Gilead, Intercept, Hanmi, Intercept, Inventiva, Ionis, Janssen, Madrigal Pharmaceuticals, Merck, NGM Biopharmaceuticals, Novo Nordisk, Merck, Pfizer, Sonic Incytes, and Terns Pharmaceuticals. R.L. is a co-founder of LipoNexus Inc. K.H. is the founder of the company Afekta Technologies providing metabolomic services. All other authors declare that they have no competing interests.
SUPPLEMENTARY MATERIALS
www.science.org/doi/10.1126/scitranslmed.abk0855
Data and materials availability:
All data associated with this study are in the paper or the Supplementary Materials. Raw metagenomic sequencing data for all samples have been deposited in NCBI Sequencing Read Archive under BioProject IDs PRJNA728908 and PRJNA686835 (only SRR13279648, SRR13279753, SRR13279664, and SRR13279666). Metabolite profiles have been deposited in MetaboLights (www.ebi.ac.uk/metabolights/) with accession MTBLS2615. The data used to generate main and supplementary figures are in data file S1. Further information and requests for resources and reagents should be directed to G.P. (gianni.panagiotou@leibniz-hki.de).
REFERENCES AND NOTES
- 1.Zheng Y, Ley SH, Hu FB, Global aetiology and epidemiology of type 2 diabetes mellitus and its complications. Nat. Rev. Endocrinol. 14, 88–98 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Younossi ZM, Koenig AB, Abdelatif D, Fazel Y, Henry L, Wymer M, Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 64, 73–84 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Seuring T, Archangelidi O, Suhrcke M, The economic costs of type 2 diabetes: A global systematic review. Pharmacoeconomics 33, 811–831 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Health Organization, “Diet, nutrition and the prevention of chronic diseases” (Technical Report series 916, World Health Organization, 2003). [PubMed] [Google Scholar]
- 5.Adams LA, Anstee QM, Tilg H, Targher G, Non-alcoholic fatty liver disease and its relationship with cardiovascular disease and other extrahepatic diseases. Gut 66, 1138–1153 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Loomba R, Friedman SL, Shulman GI, Mechanisms and disease consequences of nonalcoholic fatty liver disease. Cell 184, 2537–2564 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simon TG, Roelstraete B, Khalili H, Hagström H, Ludvigsson JF, Mortality in biopsy-confirmed nonalcoholic fatty liver disease: Results from a nationwide cohort. Gut 70, 1375–1382 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tilg H, Targher G, NAFLD-related mortality: Simple hepatic steatosis is not as ‘beESnign’ as thought. Gut 70, 1212–1213 (2021). [DOI] [PubMed] [Google Scholar]
- 9.Younossi Z, Tacke F, Arrese M, Chander Sharma B, Mostafa I, Bugianesi E, Wai-Sun Wong V, Yilmaz Y, George J, Fan J, Vos MB, Global perspectives on nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Hepatology 69, 2672–2682 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Cleveland ER, Ning H, Vos MB, Lewis CE, Rinella ME, Carr JJ, Lloyd-Jones DM, VanWagner LB, Low awareness of nonalcoholic fatty liver disease in a population-based cohort sample: The CARDIA study. J. Gen. Intern. Med. 34, 2772–2778 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spengler EK, Loomba R, Recommendations for diagnosis, referral for liver biopsy, and treatment of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Mayo Clin. Proc. 90, 1233–1246 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loomba R, Role of imaging-based biomarkers in NAFLD: Recent advances in clinical application and future research directions. J. Hepatol. 68, 296–304 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chalasani N, Younossi Z, Lavine JE, Charlton M, Cusi K, Rinella M, Harrison SA, Brunt EM, Sanyal AJ, The diagnosis and management of nonalcoholic fatty liver disease: Practice guidance from the american association for the study of liver diseases. Hepatology 67, 328–357 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Stefan N, Haring HU, Cusi K, Non-alcoholic fatty liver disease: Causes, diagnosis, cardiometabolic consequences, and treatment strategies. Lancet Diabetes Endocrinol. 7, 313–324 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Zhang HJ, He J, Pan LL, Ma ZM, Han CK, Chen CS, Chen Z, Han HW, Chen S, Sun Q, Zhang JF, Li ZB, Yang SY, Li XJ, Li XY, Effects of moderate and vigorous exercise on nonalcoholic fatty liver disease: A randomized clinical trial. JAMA Intern. Med. 176, 1074–1082 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Mardinoglu A, Wu H, Bjornson E, Zhang C, Hakkarainen A, Rasanen SM, Lee S, Mancina RM, Bergentall M, Pietilainen KH, Soderlund S, Matikainen N, Stahlman M, Bergh PO, Adiels M, Piening BD, Graner M, Lundbom N, Williams KJ, Romeo S, Nielsen J, Snyder M, Uhlen M, Bergstrom G, Perkins R, Marschall HU, Backhed F, Taskinen MR, Boren J, An integrated understanding of the rapid metabolic benefits of acarbohydrate-restricted diet on hepatic steatosis in humans. Cell Metab. 27, 559–571. e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aron-Wisnewsky J, Warmbrunn MV, Nieuwdorp M, Clement K, Nonalcoholic fatty liver disease: Modulating gut microbiota to improve severity? Gastroenterology 158, 1881–1898 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Schattenberg JM, Lazarus JV, Newsome PN, Serfaty L, Aghemo A, Augustin S, Tsochatzis E, de Ledinghen V, Bugianesi E, Romero-Gomez M, Bantel H, Ryder SD, Boursier J, Leroy V, Crespo J, Castera L, Floros L, Atella V, Mestre-Ferrandiz J, Elliott R, Kautz A, Morgan A, Hartmanis S, Vasudevan S, Pezzullo L, Trylesinski A, Cure S, Higgins V, Ratziu V, Disease burden and economic impact of diagnosed non-alcoholic steatohepatitis in five European countries in 2018: A cost-of-illness analysis. Liver Int. 41, 1227–1242 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H, Dong K, Fang Q, Hou X, Zhou M, Bao Y, Xiang K, Xu A, Jia W, High serum level of fibroblast growth factor 21 is an independent predictor of non-alcoholic fatty liver disease: A 3-year prospective study in China. J. Hepatol. 58, 557–563 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Motamed N, Faraji AH, Khonsari MR, Maadi M, Tameshkel FS, Keyvani H, Ajdarkosh H, Karbalaie Niya MH, Rezaie N, Zamani F, Fatty liver index (FLI) and prediction of new cases of non-alcoholic fatty liver disease: A population-based study of northern Iran. Clin. Nutr. 39, 468–474 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Zheng R, Du Z, Wang M, Mao Y, Mao W, A longitudinal epidemiological study on the triglyceride and glucose index and the incident nonalcoholic fatty liver disease. Lipids Health Dis. 17, 262 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lonardo A, Nascimbeni F, Maurantonio M, Marrazzo A, Rinaldi L, Adinolfi LE, Nonalcoholic fatty liver disease: Evolving paradigms. World J. Gastroenterol. 23, 6571–6592 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eslam M, Sanyal AJ, George J; International Consensus Panel MAFLD: A consensus-driven proposed nomenclature for metabolic associated fatty liver disease. Gastroenterology 158, 1999–2014.e1 (2020). [DOI] [PubMed] [Google Scholar]
- 24.Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI, The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. U.S.A. 101, 15718–15723 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ley RE, Turnbaugh PJ, Klein S, Gordon JI, Human gut microbes associated with obesity. Nature 444, 1022–1023 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, Almeida M, Arumugam M, Batto J-M, Kennedy S, Leonard P, Li J, Burgdorf K, Grarup N, Jørgensen T, Brandslund I, Nielsen HB, Juncker AS, Bertalan M, Levenez F, Pons N, Rasmussen S, Sunagawa S, Tap J, Tims S, Zoetendal EG, Brunak S, Clément K, Doré J, Kleerebezem M, Kristiansen K, Renault P, Sicheritz-Ponten T, de Vos WM, Zucker J-D, Raes J, Hansen T; MetaHIT consortium P Bork, J. Wang, S. D. Ehrlich, O. Pedersen, Richness of human gut microbiome correlates with metabolic markers. Nature 500, 541–546 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Almeida A, Nayfach S, Boland M, Strozzi F, Beracochea M, Shi ZJ, Pollard KS, Sakharova E, Parks DH, Hugenholtz P, Segata N, Kyrpides NC, Finn RD, A unified catalog of 204,938 reference genomes from the human gut microbiome. Nat. Biotechnol. 39, 105–114 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wahlström A, Sayin SI, Marschall H-U, Bäckhed F, Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab. 24, 41–50 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Sonnenburg ED, Smits SA, Tikhonov M, Higginbottom SK, Wingreen NS, Sonnenburg JL, Diet-induced extinctions in the gut microbiota compound over generations. Nature 529, 212–215 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cani PD, Osto M, Geurts L, Everard A, Involvement of gut microbiota in the development of low-grade inflammation and type 2 diabetes associated with obesity. Gut Microbes 3, 279–288 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Canfora EE, Meex RCR, Venema K, Blaak EE, Gut microbial metabolites in obesity, NAFLD and T2DM. Nat. Rev. Endocrinol. 15, 261–273 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Zhu L, Baker SS, Gill C, Liu W, Alkhouri R, Baker RD, Gill SR, Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: A connection between endogenous alcohol and NASH. Hepatology 57, 601–609 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Wang B, Jiang X, Cao M, Ge J, Bao Q, Tang L, Chen Y, Li L, Altered fecal microbiota correlates with liver biochemistry in nonobese patients with non-alcoholic fatty liver disease. Sci. Rep. 6, 32002 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mouzaki M, Comelli EM, Arendt BM, Bonengel J, Fung SK, Fischer SE, McGilvray ID, Allard JP, Intestinal microbiota in patients with nonalcoholic fatty liver disease. Hepatology 58, 120–127 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Da Silva HE, Teterina A, Comelli EM, Taibi A, Arendt BM, Fischer SE, Lou W, Allard JP, Nonalcoholic fatty liver disease is associated with dysbiosis independent of body mass index and insulin resistance. Sci. Rep. 8, 1466 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.European Association for the Study of the Liver (EASL); European Association for the Study of Diabetes (EASD); European Association for the Study of Obesity (EASO), EASL-EASD-EASO Clinical Practice Guidelines for the management of non-alcoholic fatty liver disease. J. Hepatol. 64, 1388–1402 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Chitturi S, Farrell GC, Hashimoto E, Saibara T, Lau GK, Sollano JD; Asia-Pacific Working Party on NAFLD, Non-alcoholic fatty liver disease in the Asia-Pacific region: Definitions and overview of proposed guidelines. J. Gastroenterol. Hepatol. 22, 778–787 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Gallois Y, Vol S, Caces E, Balkau B; DESIR Study Group, Distribution of fasting serum insulin measured by enzyme immunoassay in an unselected population of 4,032 individuals . Reference values according to age and sex. Diabetes Metab. 22, 427–431 (1996). [PubMed] [Google Scholar]
- 39.Muniyappa R, Lee S, Chen H, Quon MJ, Current approaches for assessing insulin sensitivity and resistance in vivo: Advantages, limitations, and appropriate usage. Am. J. Physiol. Endocrinol. Metab. 294, E15–E26 (2008). [DOI] [PubMed] [Google Scholar]
- 40.Catapano AL, Graham I, De Backer G, Wiklund O, Chapman MJ, Drexel H, Hoes AW, Jennings CS, Landmesser U, Pedersen TR, Reiner Z, Riccardi G, Taskinen MR, Tokgozoglu L, Verschuren WMM, Vlachopoulos C, Wood DA, Zamorano JL, Cooney MT; ESC Scientific Document Group, 2016 ESC/EAS guidelines for the management of dyslipidaemias. Eur. Heart J. 37, 2999–3058 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Pearson TA, Mensah GA, Alexander RW, Anderson JL, Cannon III RO, Criqui M, Fadl YY, Fortmann SP, Hong Y, Myers GL, Rifai N, Smith SC Jr., K. Taubert, R. P. Tracy, F. Vinicor; Centers for Disease Control and Prevention; American Heart Association, Markers of inflammation and cardiovascular disease: Application to clinical and public health practice: A statement for healthcare professionals from the centers for disease control and prevention and the american heart association. Circulation 107, 499–511 (2003). [DOI] [PubMed] [Google Scholar]
- 42.Truong DT, Franzosa EA, Tickle TL, Scholz M, Weingart G, Pasolli E, Tett A, Huttenhower C, Segata N, MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat. Methods 12, 902–903 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Nielsen HB, Almeida M, Juncker AS, Rasmussen S, Li J, Sunagawa S, Plichta DR, Gautier L, Pedersen AG, Le Chatelier E, Pelletier E, Bonde I, Nielsen T, Manichanh C, Arumugam M, Batto J-M, Santos MBQD, Blom N, Borruel N, Burgdorf KS, Boumezbeur F, Casellas F, Doré J, Dworzynski P, Guarner F, Hansen T, Hildebrand F, Kaas RS, Kennedy S, Kristiansen K, Kultima JR, Léonard P, Levenez F, Lund O, Moumen B, Le Paslier D, Pons N, Pedersen O, Prifti E, Qin J, Raes J, Sørensen S, Tap J, Tims S, Ussery DW, Yamada T, Meta HITC, Renault P, Sicheritz-Ponten T, Bork P, Wang J, Brunak S, Ehrlich SD; MetaHIT Consortium, Identification and assembly of genomes and genetic elements in complex metagenomic samples without using reference genomes. Nat. Biotechnol. 32, 822–828 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Million M, Maraninchi M, Henry M, Armougom F, Richet H, Carrieri P, Valero R, Raccah D, Vialettes B, Raoult D, Obesity-associated gut microbiota is enriched in Lactobacillus reuteri and depleted in Bifidobacterium animalis and Methanobrevibacter smithii. Int. J. Obes. (Lond) 36, 817–825 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 45.Muñiz Pedrogo DA, Jensen MD, Van Dyke CT, Murray JA, Woods JA, Chen J, Kashyap PC, Nehra V, Gut microbial carbohydrate metabolism hinders weight loss in overweight adults undergoing lifestyle intervention with a volumetric diet. Mayo Clin. Proc. 93, 1104–1110 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwimmer JB, Johnson JS, Angeles JE, Behling C, Belt PH, Borecki I, Bross C, Durelle J, Goyal NP, Hamilton G, Holtz ML, Lavine JE, Mitreva M, Newton KP, Pan A, Simpson PM, Sirlin CB, Sodergren E, Tyagi R, Yates KP, Weinstock GM, Salzman NH, Microbiome signatures associated with steatohepatitis and moderate to severe fibrosis in children with nonalcoholic fatty liver disease. Gastroenterology 157, 1109–1122 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brahe LK, Le Chatelier E, Prifti E, Pons N, Kennedy S, Hansen T, Pedersen O, Astrup A, Ehrlich SD, Larsen LH, Specific gut microbiota features and metabolic markers in postmenopausal women with obesity. Nutr. Diabetes 5, e159 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Franzosa EA, McIver LJ, Rahnavard G, Thompson LR, Schirmer M, Weingart G, Lipson KS, Knight R, Caporaso JG, Segata N, Huttenhower C, Species-level functional profiling of metagenomes and metatranscriptomes. Nat. Methods 15, 962–968 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oh TG, Kim SM, Caussy C, Fu T, Guo J, Bassirian S, Singh S, Madamba EV, Bettencourt R, Richards L, Yu RT, Atkins AR, Huan T, Brenner DA, Sirlin CB, Downes M, Evans RM, Loomba R, A universal gut-microbiome-derived signature predicts cirrhosis. Cell Metab. 32, 901 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu J, Jiang S, Zhao Y, Sun Q, Zhang J, Shen D, Wu J, Shen N, Fu X, Sun X, Yu D, Chen J, He J, Shi T, Ding Y, Fang L, Xue B, Li C, Geranylgeranyl diphosphate synthase (GGPPS) regulates non-alcoholic fatty liver disease (NAFLD)-fibrosis progression by determining hepatic glucose/fatty acid preference under high-fat diet conditions. J. Pathol. 246, 277–288 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Gabbia D, Roverso M, Guido M, Sacchi D, Scaffidi M, Carrara M, Orso G, Russo FP, Floreani A, Bogialli S, De Martin S, Western diet-induced metabolic alterations affect circulating markers of liver function before the development of steatosis. Nutrients 11, 1602 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aron-Wisnewsky J, Prifti E, Belda E, Ichou F, Kayser BD, Dao MC, Verger EO, Hedjazi L, Bouillot JL, Chevallier JM, Pons N, Le Chatelier E, Levenez F, Ehrlich SD, Dore J, Zucker JD, Clement K, Major microbiota dysbiosis in severe obesity: Fate after bariatric surgery. Gut 68, 70–82 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mardinoglu A, Agren R, Kampf C, Asplund A, Uhlen M, Nielsen J, Genome-scale metabolic modelling of hepatocytes reveals serine deficiency in patients with non-alcoholic fatty liver disease. Nat. Commun. 5, 3083 (2014). [DOI] [PubMed] [Google Scholar]
- 54.Hoyles L, Fernández-Real J-M, Federici M, Serino M, Abbott J, Charpentier J, Heymes C, Luque JL, Anthony E, Barton RH, Chilloux J, Myridakis A, Martinez-Gili L, Moreno-Navarrete JM, Benhamed F, Azalbert V, Blasco-Baque V, Puig J, Xifra G, Ricart W, Tomlinson C, Woodbridge M, Cardellini M, Davato F, Cardolini I, Porzio O, Gentileschi P, Lopez F, Foufelle F, Butcher SA, Holmes E, Nicholson JK, Postic C, Burcelin R, Dumas M-E, Molecular phenomics and metagenomics of hepatic steatosis in non-diabetic obese women. Nat. Med. 24, 1070–1080 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gaggini M, Carli F, Rosso C, Buzzigoli E, Marietti M, Della Latta V, Ciociaro D, Abate ML, Gambino R, Cassader M, Bugianesi E, Gastaldelli A, Altered amino acid concentrations in NAFLD: Impact of obesity and insulin resistance. Hepatology 67, 145–158 (2018). [DOI] [PubMed] [Google Scholar]
- 56.Koop AC, Thiele ND, Steins D, Michaëlsson E, Wehmeyer M, Scheja L, Steglich B, Huber S, Schulze Zur Wiesch J, Lohse AW, Heeren J, Kluwe J, Therapeutic targeting of myeloperoxidase attenuates NASH in mice. Hepatol. Commun. 4, 1441–1458 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sztolsztener K, Chabowski A, Harasim-Symbor E, Bielawiec P, Konstantynowicz-Nowicka K, Arachidonic acid as an early indicator of inflammation during non-alcoholic fatty liver disease development. Biomolecules 10, 1133 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rodríguez-Gallego E, Guirro M, Riera-Borrull M, Hernández-Aguilera A, Mariné-Casadó R, Fernández-Arroyo S, Beltrán-Debón R, Sabench F, Hernández M, del Castillo D, Menendez JA, Camps J, Ras R, Arola L, Joven J, Mapping of the circulating metabolome reveals α-ketoglutarate as a predictor of morbid obesity-associated non-alcoholic fatty liver disease. Int. J. Obes. 39, 279–287 (2014). [DOI] [PubMed] [Google Scholar]
- 59.Kindt A, Liebisch G, Clavel T, Haller D, Hörmannsperger G, Yoon H, Kolmeder D, Sigruener A, Krautbauer S, Seeliger C, Ganzha A, Schweizer S, Morisset R, Strowig T, Daniel H, Helm D, Küster B, Krumsiek J, Ecker J, The gut microbiota promotes hepatic fatty acid desaturation and elongation in mice. Nat. Commun. 9, 1–15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tsurutani Y, Inoue K, Sugisawa C, Saito J, Omura M, Nishikawa T, Increased serum dihomo-γ-linolenic acid levels are associated with obesity, body fat accumulation, and insulin resistance in japanese patients with type 2 diabetes. Intern. Med. 57, 2929–2935 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Beloborodova N, Bairamov I, Olenin A, Shubina V, Teplova V, Fedotcheva N, Effect of phenolic acids of microbial origin on production of reactive oxygen species in mitochondria and neutrophils. J. Biomed. Sci. 19, 89 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Takada S, Matsubara T, Fujii H, Sato-Matsubara M, Daikoku A, Odagiri N, Amano-Teranishi Y, Kawada N, Ikeda K, Stress can attenuate hepatic lipid accumulation via elevation of hepatic β-muricholic acid levels in mice with nonalcoholic steatohepatitis. Lab. Invest. 101, 193–203 (2021). [DOI] [PubMed] [Google Scholar]
- 63.Chashmniam S, Ghafourpour M, Rezaei Farimani A, Gholami A, Ghoochani BFNM, Metabolomic biomarkers in the diagnosis of non-alcoholic fatty liver disease. Hepat. Mon. 19, e92244 (2019). [Google Scholar]
- 64.Hodson L, Bhatia L, Scorletti E, Smith DE, Jackson NC, Shojaee-Moradie F, Umpleby M, Calder PC, Byrne CD, Docosahexaenoic acid enrichment in NAFLD is associated with improvements in hepatic metabolism and hepatic insulin sensitivity: A pilot study. Eur. J. Clin. Nutr. 71, 1251 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lundberg S, Lee S-I, A unified approach to interpreting model predictions. arXiv:1705.07874 (2017). [Google Scholar]
- 66.Menni C, Zhu J, Le Roy CI, Mompeo O, Young K, Rebholz CM, Selvin E, North KE, Mohney RP, Bell JT, Boerwinkle E, Spector TD, Mangino M, Yu B, Valdes AM, Serum metabolites reflecting gut microbiome alpha diversity predict type 2 diabetes. Gut Microbes 11, 1632–1642 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grundy SM, Cleeman JI, Daniels SR, Donato KA, Eckel RH, Franklin BA, Gordon DJ, Krauss RM, Savage PJ, Smith SC Jr., Spertus JA, Costa F; American Heart Association; National Heart, Lung, and Blood Institute, Diagnosis and management of the metabolic syndrome: An American Heart Association/National Heart, Lung, and Blood Institute Scientific Statement. Circulation 112, 2735–2752 (2005). [DOI] [PubMed] [Google Scholar]
- 68.Loomba R, Seguritan V, Li W, Long T, Klitgord N, Bhatt A, Dulai PS, Caussy C, Bettencourt R, Highlander SK, Jones MB, Sirlin CB, Schnabl B, Brinkac L, Schork N, Chen C-H, Brenner DA, Biggs W, Yooseph S, Venter JC, Nelson KE, Gut microbiome-based metagenomic signature for non-invasive detection of advanced fibrosis in human nonalcoholic fatty liver disease. Cell Metab. 25, 1054–1062.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee HW, Wong VW-S, Changing NAFLD epidemiology in China. Hepatology 70, 1095–1098 (2019). [DOI] [PubMed] [Google Scholar]
- 70.Estes C, Anstee QM, Arias-Loste MT, Bantel H, Bellentani S, Caballeria J, Colombo M, Craxi A, Crespo J, Day CP, Eguchi Y, Geier A, Kondili LA, Kroy DC, Lazarus JV, Loomba R, Manns MP, Marchesini G, Nakajima A, Negro F, Petta S, Ratziu V, Romero-Gomez M, Sanyal A, Schattenberg JM, Tacke F, Tanaka J, Trautwein C, Wei L, Zeuzem S, Razavi H, Modeling NAFLD disease burden in China, France, Germany, Italy, Japan, Spain, United Kingdom, and United States for the period 2016–2030. J. Hepatol. 69, 896–904 (2018). [DOI] [PubMed] [Google Scholar]
- 71.Albillos A, de Gottardi A, Rescigno M, The gut-liver axis in liver disease: Pathophysiological basis for therapy. J. Hepatol. 72, 558–577 (2020). [DOI] [PubMed] [Google Scholar]
- 72.Aron-Wisnewsky J, Vigliotti C, Witjes J, Le P, Holleboom AG, Verheij J, Nieuwdorp M, Clément K, Gut microbiota and human NAFLD: Disentangling microbial signatures from metabolic disorders. Nat. Rev. Gastroenterol. Hepatol. 17, 279–297 (2020). [DOI] [PubMed] [Google Scholar]
- 73.Hoyles L, Fernandez-Real JM, Federici M, Serino M, Abbott J, Charpentier J, Heymes C, Luque JL, Anthony E, Barton RH, Chilloux J, Myridakis A, Martinez-Gili L, Moreno-Navarrete JM, Benhamed F, Azalbert V, Blasco-Baque V, Puig J, Xifra G, Ricart W, Tomlinson C, Woodbridge M, Cardellini M, Davato F, Cardolini I, Porzio O, Gentileschi P, Lopez F, Foufelle F, Butcher SA, Holmes E, Nicholson JK, Postic C, Burcelin R, Dumas ME, Molecular phenomics and metagenomics of hepatic steatosis in non-diabetic obese women. Nat. Med. 24, 1070–1080 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fromentin S, Forslund SK, Chechi K, Aron-Wisnewsky J, Chakaroun R, Nielsen T, Tremaroli V, Ji B, Prifti E, Myridakis A, Chilloux J, Andrikopoulos P, Fan Y, Olanipekun MT, Alves R, Adiouch S, Bar N, Talmor-Barkan Y, Belda E, Caesar R, Coelho LP, Falony G, Fellahi S, Galan P, Galleron N, Helft G, Hoyles L, Isnard R, Le Chatelier E, Julienne H, Olsson L, Pedersen HK, Pons N, Quinquis B, Rouault C, Roume H, Salem JE, Schmidt TSB, Vieira-Silva S, Li P, Zimmermann-Kogadeeva M, Lewinter C, Sondertoft NB, Hansen TH, Gauguier D, Gotze JP, Kober L, Kornowski R, Vestergaard H, Hansen T, Zucker JD, Hercberg S, Letunic I, Backhed F, Oppert JM, Nielsen J, Raes J, Bork P, Stumvoll M, Segal E, Clement K, Dumas ME, Ehrlich SD, Pedersen O, Microbiome and metabolome features of the cardiometabolic disease spectrum. Nat. Med. 28, 303–314 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Talmor-Barkan Y, Bar N, Shaul AA, Shahaf N, Godneva A, Bussi Y, Lotan-Pompan M, Weinberger A, Shechter A, Chezar-Azerrad C, Arow Z, Hammer Y, Chechi K, Forslund SK, Fromentin S, Dumas ME, Ehrlich SD, Pedersen O, Kornowski R, Segal E, Metabolomic and microbiome profiling reveals personalized risk factors for coronary artery disease. Nat. Med. 28, 295–302 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ruuskanen MO, Erawijantari PP, Havulinna AS, Liu Y, Meric G, Tuomilehto J, Inouye M, Jousilahti P, Salomaa V, Jain M, Knight R, Lahti L, Niiranen TJ, Gut microbiome composition is predictive of incident type 2 diabetes in a population cohort of 5,572 finnish adults. Diabetes Care 45, 811–818 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tas E, Bai S, Ou X, Mercer K, Lin H, Mansfield K, Buchmann R, Diaz EC, Oden J, Børsheim E, Adams SH, Dranoff J, Fibroblast growth factor-21 to adiponectin ratio: A potential biomarker to monitor liver fat in children with obesity. Front. Endocrinol. 11, 654 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li H, Fang Q, Gao F, Fan J, Zhou J, Wang X, Zhang H, Pan X, Bao Y, Xiang K, Xu A, Jia W, Fibroblast growth factor 21 levels are increased in nonalcoholic fatty liver disease patients and are correlated with hepatic triglyceride. J. Hepatol. 53, 934–940 (2010). [DOI] [PubMed] [Google Scholar]
- 79.Vilar-Gomez E, Chalasani N, Non-invasive assessment of non-alcoholic fatty liver disease: Clinical prediction rules and blood-based biomarkers. J. Hepatol. 68, 305–315 (2018). [DOI] [PubMed] [Google Scholar]
- 80.Alferink LJ, Kiefte-de Jong JC, Erler NS, Veldt BJ, Schoufour JD, de Knegt RJ, Ikram MA, Metselaar HJ, Janssen H, Franco OH, Darwish Murad S, Association of dietary macronutrient composition and non-alcoholic fatty liver disease in an ageing population: The Rotterdam Study. Gut 68, 1088–1098 (2019). [DOI] [PubMed] [Google Scholar]
- 81.Hashemian M, Merat S, Poustchi H, Jafari E, Radmard AR, Kamangar F, Freedman N, Hekmatdoost A, Sheikh M, Boffetta P, Sinha R, Dawsey SM, Abnet CC, Malekzadeh R, Etemadi A, Red meat consumption and risk of nonalcoholic fatty liver disease in a population with low meat consumption: The golestan cohort study. Am. J. Gastroenterol. 116, 1667–1675 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jung HS, Chang Y, Kwon MJ, Sung E, Yun KE, Cho YK, Shin H, Ryu S, Smoking and the risk of non-alcoholic fatty liver disease: A cohort study. Am. J. Gastroenterol. 114, 453–463 (2019). [DOI] [PubMed] [Google Scholar]
- 83.Wong VW, Adams LA, de Ledinghen V, Wong GL, Sookoian S, Noninvasive biomarkers in NAFLD and NASH - Current progress and future promise. Nat. Rev. Gastroenterol. Hepatol. 15, 461–478 (2018). [DOI] [PubMed] [Google Scholar]
- 84.Jung JY, Shim J-J, Park SK, Ryoo J-H, Choi J-M, Oh I-H, Jung K-W, Cho H, Ki M, Won Y-J, Oh C-M, Serum ferritin level is associated with liver steatosis and fibrosis in Korean general population. Hepatol. Int. 13, 222–233 (2019). [DOI] [PubMed] [Google Scholar]
- 85.Mayneris-Perxachs J, Cardellini M, Hoyles L, Latorre J, Davato F, Moreno-Navarrete JM, Arnoriaga-Rodriguez M, Serino M, Abbott J, Barton RH, Puig J, Fernandez-Real X, Ricart W, Tomlinson C, Woodbridge M, Gentileschi P, Butcher SA, Holmes E, Nicholson JK, Perez-Brocal V, Moya A, Clain DM, Burcelin R, Dumas ME, Federici M, Fernandez-Real JM, Iron status influences non-alcoholic fatty liver disease in obesity through the gut microbiome. Microbiome 9, 104 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Heshiki Y, Vazquez-Uribe R, Li J, Ni Y, Quainoo S, Imamovic L, Li J, Sørensen M, Chow BKC, Weiss GJ, Xu A, Sommer MOA, Panagiotou G, Predictable modulation of cancer treatment outcomes by the gut microbiota. Microbiome 8, 28 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zeevi D, Korem T, Zmora N, Israeli D, Rothschild D, Weinberger A, Ben-Yacov O, Lador D, Avnit-Sagi T, Lotan-Pompan M, Suez J, Mahdi JA, Matot E, Malka G, Kosower N, Rein M, Zilberman-Schapira G, Dohnalová L, Pevsner-Fischer M, Bikovsky R, Halpern Z, Elinav E, Segal E, Personalized nutrition by prediction of glycemic responses. Cell 163, 1079–1094 (2015). [DOI] [PubMed] [Google Scholar]
- 88.Gharaibeh RZ, Jobin C, Microbiota and cancer immunotherapy: In search of microbial signals. Gut 68, 385–388 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liu Y, Wang Y, Ni Y, Cheung CKY, Lam KSL, Wang Y, Xia Z, Ye D, Guo J, Tse MA, Panagiotou G, Xu A, Gut microbiome fermentation determines the efficacy of exercise for diabetes prevention. Cell Metab. 31, 77–91.e75 (2020). [DOI] [PubMed] [Google Scholar]
- 90.Bajaj JS, Betrapally NS, Hylemon PB, Thacker LR, Daita K, Kang DJ, White MB, Unser AB, Fagan A, Gavis EA, Sikaroodi M, Dalmet S, Heuman DM, Gillevet PM, Gut microbiota alterations can predict hospitalizations in cirrhosis independent of diabetes mellitus. Sci. Rep. 5, 18559 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wong T, Wong RJ, Gish RG, Diagnostic and treatment implications of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Gastroenterol. Hepatol. 15, 83–89 (2019). [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang J-Z, Cai J-J, Yu Y, She Z-G, Li H, Nonalcoholic fatty liver disease: An update on the diagnosis. Gene Expr. 19, 187–198 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Duscha A, Gisevius B, Hirschberg S, Yissachar N, Stangl GI, Eilers E, Bader V, Haase S, Kaisler J, David C, Schneider R, Troisi R, Zent D, Hegelmaier T, Dokalis N, Gerstein S, Del Mare-Roumani S, Amidror S, Staszewski O, Poschmann G, Stuhler K, Hirche F, Balogh A, Kempa S, Trager P, Zaiss MM, Holm JB, Massa MG, Nielsen HB, Faissner A, Lukas C, Gatermann SG, Scholz M, Przuntek H, Prinz M, Forslund SK, Winklhofer KF, Muller DN, Linker RA, Gold R, Haghikia A, Propionic acid shapes the multiple sclerosis disease course by an immunomodulatory mechanism. Cell 180, 1067–1080.e1016 (2020). [DOI] [PubMed] [Google Scholar]
- 94.Chen P, Hou X, Hu G, Wei L, Jiao L, Wang H, Chen S, Wu J, Bao Y, Jia W, Abdominal subcutaneous adipose tissue: A favorable adipose depot for diabetes? Cardiovasc. Diabetol. 17, 93 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hou X, Chen P, Hu G, Wei L, Jiao L, Wang H, Liang Y, Bao Y, Jia W, Abdominal subcutaneous fat: A favorable or nonfunctional fat depot for glucose metabolism in chinese adults? Obesity 26, 1078–1087 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Paulson JN, Stine OC, Bravo HC, Pop M, Differential abundance analysis for microbial marker-gene surveys. Nat. Methods 10, 1200–1202 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez J-C, Müller M, pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 12, 77 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhao L, Ni Y, Su M, Li H, Dong F, Chen W, Wei R, Zhang L, Guiraud SP, Martin FP, Rajani C, Xie G, Jia W, High throughput and quantitative measurement of microbial metabolome by gas chromatography/mass spectrometry using automated alkyl chloroformate derivatization. Anal. Chem. 89, 5565–5577 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li J, Sung CYJ, Lee N, Ni Y, Pihlajamäki J, Panagiotou G, El-Nezami H, Probiotics modulated gut microbiota suppresses hepatocellular carcinoma growth in mice. Proc. Natl. Acad. Sci. U.S.A. 113, E1306–E1315 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li H, Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997 [q-bio.GN] (13 March 2013). [Google Scholar]
- 101.Dixon P, VEGAN A package of r functions for community ecology. J. Veg. Sci. 14, 927–930 (2003). [Google Scholar]
- 102.Vavrek MJ, Fossil: Palaeoecological and palaeogeographical analysis tools. Palaeontol. Electronica 14, 16 (2011). [Google Scholar]
- 103.Kuhn M, Building predictive models inRUsing thecaretPackage. J. Stat. Softw. 28, 1–26 (2008).27774042 [Google Scholar]
- 104.Wright MN, Ziegler A, Ranger: A fast implementation of random forests for high dimensional data in C++ and R. J. Stat. Softw. 77, 1–17 (2017). [Google Scholar]
- 105.John CR, MLeval: Machine Learning Model Evaluation. R package version 0.3, (2020); https://CRAN.R-project.org/package=MLeval. [Google Scholar]
- 106.Lundberg SM, Nair B, Vavilala MS, Horibe M, Eisses MJ, Adams T, Liston DE, Low DK-W, Newman S-F, Kim J, Lee S-I, Explainable machine-learning predictions for the prevention of hypoxaemia during surgery. Nat. Biomed. Eng. 2, 749–760 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Artzi NS, Shilo S, Hadar E, Rossman H, Barbash-Hazan S, Ben-Haroush A, Balicer RD, Feldman B, Wiznitzer A, Segal E, Prediction of gestational diabetes based on nationwide electronic health records. Nat. Med. 26, 71–76 (2020). [DOI] [PubMed] [Google Scholar]
- 108.Greenwell B, fastshap: Fast Approximate Shapley Values. R package version 0.0.7, (2021); https://CRAN.R-project.org/package=fastshap. [Google Scholar]
- 109.Kotronen A, Peltonen M, Hakkarainen A, Sevastianova K, Bergholm R, Johansson LM, Lundbom N, Rissanen A, Ridderstrale M, Groop L, Orho-Melander M, Yki-Jarvinen H, Prediction of non-alcoholic fatty liver disease and liver fat using metabolic and genetic factors. Gastroenterology 137, 865–872 (2009). [DOI] [PubMed] [Google Scholar]
- 110.Sterling RK, Lissen E, Clumeck N, Sola R, Correa MC, Montaner J, Sulkowski MS, Torriani FJ, Dieterich DT, Thomas DL, Messinger D, Nelson M; APRICOT Clinical Investigators, Development of a simple noninvasive index to predict significant fibrosis in patients with HIV/HCV coinfection. Hepatology 43, 1317–1325 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data associated with this study are in the paper or the Supplementary Materials. Raw metagenomic sequencing data for all samples have been deposited in NCBI Sequencing Read Archive under BioProject IDs PRJNA728908 and PRJNA686835 (only SRR13279648, SRR13279753, SRR13279664, and SRR13279666). Metabolite profiles have been deposited in MetaboLights (www.ebi.ac.uk/metabolights/) with accession MTBLS2615. The data used to generate main and supplementary figures are in data file S1. Further information and requests for resources and reagents should be directed to G.P. (gianni.panagiotou@leibniz-hki.de).


