Summary
Background:
There are limited data on the diagnostic accuracy of gut microbial signatures for predicting hepatic decompensation in patients with cirrhosis.
Aims:
To determine whether a stool metagenome-derived signature accurately detects hepatic decompensation and mortality risk in cirrhosis secondary to non-alcoholic fatty liver disease (NAFLD)
Methods:
Shotgun metagenomic sequencing was performed on faecal samples collected at study entry from a prospective cohort of adults with NAFLD-related cirrhosis. A Random Forest machine learning algorithm was utilised to identify a metagenomic signature of decompensated cirrhosis (defined by ascites, hepatic encephalopathy or variceal haemorrhage) and subsequently validated in an external cohort. A Cox proportional hazards regression model was used to examine predictors of all-cause mortality.
Results:
In all, 25 adults with NAFLD-related cirrhosis (training cohort) were included. Among the 16 participants with decompensated cirrhosis, 33% had ascites, 56% had hepatic encephalopathy and 22% had experienced a variceal haemorrhage (not mutually exclusive). We identified a stool metagenomic signature comprising 13 discriminatory species that reliably distinguished decompensated NAFLD-related cirrhosis (diagnostic accuracy, 0.97, 95% confidence interval [CI] 0.96–0.99). Diagnostic accuracy of the 13-species signature remained high after adjustment for lactulose (area under the curve [AUC] 0.99) and rifaximin use (AUC 0.93). The discriminative ability of 13-species metagenomic signature was robust in an independent test cohort (AUC 0.95, 95% CI 0.81–1.00). The 13-species metagenomic signature (hazard ratio [HR] 1.54, 95% CI 1.10–2.15, p = 0.01) was a stronger predictor of mortality than the Model for End-Stage Liver Disease score (HR 1.25, 95% CI 1.03–1.53, p = 0.03).
Conclusions:
This study provides evidence for a gut metagenome-derived signature with high diagnostic accuracy for hepatic decompensation that predicts risk of mortality in NAFLD-related cirrhosis.
1 |. INTRODUCTION
Non-alcoholic fatty liver disease (NAFLD) is the leading cause of chronic liver disease worldwide, affecting approximately 25% of the global population.1 NAFLD cirrhosis represents the most severe stage of the disease, portending risks including hepatocellular carcinoma and liver-related mortality.2,3 More accurate prognostic scoring systems are needed to risk stratify persons with cirrhosis at highest risk of complications and mortality.
The gut–liver axis has emerged as a major contributor to pathogenesis of chronic metabolic diseases, including NAFLD.4 Cirrhosis of the liver is associated with alterations of the gut–liver axis that are hypothesised to play a role in disease pathogenesis through mechanisms such as perturbations in gut epithelial function and endotoxemia. As such, the gut microbiota represents a potential source of non-invasive biomarkers for liver disease staging and prognosis as well as therapeutic intervention.5 We have previously demonstrated that a microbial signature is highly accurate in predicting both NAFLD-related advanced fibrosis6 and cirrhosis.7,8 Limited data exist concerning the diagnostic accuracy of gut microbiome-derived signatures for detecting stages of NAFLD cirrhosis. Moreover, it is not known whether gut microbiome-derived signatures can predict risk for mortality in adults with NAFLD-related cirrhosis.
To address these gaps in knowledge, we aimed to compare the stool metagenome and as well as the stool and serum metabolome between compensated and decompensated NAFLD cirrhosis to determine whether a microbiome-derived signature accurately detects cirrhosis stage and predicts risk of all-cause mortality.
2 |. MATERIALS AND METHODS
2.1 |. Patient cohorts and clinical staging of NAFLD cirrhosis
We included a training cohort of 25 adults with NAFLD-related cirrhosis prospectively recruited in the outpatient setting between August 2014 and November 2017 at the University of California San Diego (UCSD) NAFLD Research Center and followed clinically thereafter. An independent test cohort included nine adults with NAFLD-related cirrhosis prospectively recruited in the outpatient setting between March 2013 and November 2014 at the UCSD NAFLD Research Center for a separate cohort study. NAFLD was defined according to American Association for the Study of Liver Study Practice Guidelines.9 Participants met criteria for NAFLD-related cirrhosis if they had NAFLD and biopsy-proven cirrhosis (stage 4 fibrosis on histologic review) or a diagnosis of cirrhosis from a board-certified gastroenterologist based on imaging and clinical parameters. Exclusion criteria included any of the following: history of significant alcohol intake (≥7 drinks per week [females] or ≥14 drinks/week [males]); clinical or biochemical evidence of liver disease other than NAFLD including viral hepatitis, autoimmune hepatitis, Wilson’s disease, hemochromatosis, alpha-1-antitrypsin deficiency; use of medications known to cause hepatic steatosis (amiodarone, glucocorticoids, methotrexate and valproic acid) within the last 12 months; history of bariatric surgery; HIV positivity; and females who were pregnant or nursing at the time of the study.
All participants (in training and independent test cohorts) underwent a baseline standardised clinical research visit that included a detailed medical history, physical examination including anthropometric measurements performed by a trained clinical investigator and fasting laboratory tests. A detailed history of medications was obtained. All prior endoscopic procedure results were obtained and reviewed. All study participants provided faecal samples at baseline research visit which were snap frozen at −80°C upon receipt. Participants were counselled to submit their first bowel movement of the day, to minimise postprandial variation. Serum samples were also collected from participants at baseline.
The severity of cirrhosis at the baseline research visit was categorised as compensated or decompensated. Decompensation was defined by any of the following: ascites requiring intervention (diuretics, paracentesis and/or transjugular intrahepatic portosystemic shunt), upper gastrointestinal haemorrhage secondary to gastroesophageal varices or hepatic encephalopathy (grade ≥ 2 based on West Haven Criteria10). Hepatic decompensation was adjudicated and subsequently verified by individual chart review performed by a board-certified transplant hepatologist (S.S.) blinded to microbiome analysis. Demographic, clinical and liver disease characteristics were summarised. Categorical variables were shown as counts and percentages, and associations were tested using the Kruskal–Wallis test or Fisher’s exact test. Normally distributed continuous variables were shown as mean (±standard deviation).
All participants provided a written informed consent, and the study protocol was approved by the UCSD Institutional Review Board (UCSD IRB #140084 and #111282). All authors had access to the study data and reviewed and approved the final manuscript.
2.2 |. Faecal shotgun metagenomic sequencing and metabolomics
DNA extraction from human stool samples and shotgun sequencing were performed by the Center for Microbiome Innovation (CMI) at UCSD. DNA sequencing libraries were prepared using Nextera Library Prep Kits (Illumina). Shotgun DNA sequencing was performed on the Illumina HiSeq4000 platform.
Metabolite extraction from human stool and serum samples and the subsequent liquid chromatography with tandem mass spectrometry (LC–MS/MS) was performed by the CMI at UCSD. Dual extraction procedures of serum metabolome were as follows: A 50 μl serum sample was mixed with 300 μl ice-cold methanol in a 1.5 ml Eppendorf vial and vortex for 2 min. The solution was kept at −20°C for 4 h to precipitate proteins. After that, 1000 μl methyl tert-butyl ether was added to extract lipids. After 5 min shake, 350 μl H2O was added to induce the phase separation. The solution was vortex for 10 s and rested at room temperature for 10 min, followed by centrifugation at 14,000 rpm at 4°C for 15 min, for complete phase separation. The clear lower layer was separated into a new vial and dried in SpeedVac at 20°C for 4 h. The dried extract was then reconstituted in 150 μl acetonitrile and water (1:1, v:v) mixed solvent for LC–MS metabolomics analysis. The method blank was also prepared following the same protocol but without adding serum. A 5 μl aliquot from each individual sample was pooled together to make a quality control sample.
LC–MS analysis was performed on an ultra-high-resolution Qq-time-of-flight (UHR-QqTOF) mass spectrometer Impact II (Bruker Daltonics) coupled with a 1290 Infinity II UHPLC system (Agilent Technologies). Hydrophilic interaction liquid chromatography (HILIC) separation was performed using a SeQuant ZIC-pHILIC Column (5 μm, 2.1 mm × 150 mm). For positive mode HILIC-MS analysis, mobile phase A was water with 10 mM ammonium acetate with pH adjusted by acetic acid to 4.8, and mobile phase B was pure ACN. The flow rate was 0.15 ml/min. The LC gradient was as follows: 0 min, 95% B; 20 min, 5% B; 25 min, 5% B; 25.01 min, 95% B; 30 min, 95% B; 35 min, 95% B. The column temperature was set at 30°C, and the injection volume of plasma samples was 3 μl. For the MS, capillary voltage, 4.5 kV; nebuliser gas, 1.6 bar; dry gas, 7 L/min; dry gas temperature, 220 °C; mass scan range, 70–1000 (m/z); spectra rate, 8.00 Hz; and cycle time, 3.0 s. For centroid spectra calculation, the peak summation width was 3 pts. The mass spectrometer was calibrated using NaFA (250 mM) before analysis.
2.3 |. Statistical analyses
2.3.1 |. Development and validation of a model utilising metagenome-derived signatures
We initially performed a cross-sectional analysis to evaluate for a signature of decompensated NAFLD cirrhosis derived from the faecal metagenome. Taxonomy abundance including class, family, genus and species was extracted and used for post-analysis.7 Briefly, the analysis included as follows: (i) quality control of raw reads, (ii) taxa profiling with MetaPhlAn2 (version 2.7.7), (iii) functional pathway profiling with HUMAnN2 (version 0.11.2) with ChocoPhlAn-MetaCyc (iv) and translation search with DIAMOND-UniRef90. The renorm_table command of the HUMAnN2 package was also used to normalise abundance outputs. The curatedMetagenomicData package with the ExperimentHub platform was used to convert the metagenomic output into the R object. For post-analysis including alpha-, beta-diversity and composition, the phyloseq package was utilised. To compute alpha-diversity, the estimate_richness function was utilised using Shannon or InvSimpson methods. For beta-diversity analysis, the ordinate command of the phyloseq was used with PCoA, unweighted-unifrac calculation. To compute permutational analysis of variance (PERMANOVA) significance, the adonis function from the vegan R package was used. For composition analysis, the phyloseq commands and custom script including ggplot functions were utilised.
To develop models capable of distinguishing decompensated cirrhosis samples from compensated cirrhosis samples, we utilised the Random Forest (RF) supervised learning algorithm available in the Classification And Regression Training (caret) package in R to independently identify signatures from metagenomic and metabolomic datasets. A priori relative abundance taxonomic tables were pre-processed using preProcess and predict functions with the zv, scale and centre methods. Briefly, each model was trained using the caret parameter as follows; 10-fold cross-validation, smote sampling mode and 10 repeats in trainControl. To search the best hyperparameters, mtry value for the number of variables randomly sampled as candidates at each split was set at the range (−1, 0 and +1 of the square root of the feature number) on grid tuning. The ntree setting for the number of trees to grow was 501 with the ROC metric option. For feature selection, features (species or metabolites) from the feature importance scores of the RF outcome were examined to find a set of features that trains a forest with the highest overall accuracy of sample classification. To do so, importance scores of features were extracted using varImp function and tested iteratively by ordering from highest to lowest Mean Decrease in Gini index. Then different combinations of features from the top rank were examined by calculating area under the curve (AUC). Feature selection was used to optimise the numbers of the signature sets, with the top-performing models utilising 13 microbiome species. The features used to train a forest with the best-performing in AUC were kept for the further analyses. The trained RF model with final features were also retained for the validation studies. For our training set, total 25 stool samples with 16 of compensated cirrhotic samples and 9 decompensated cirrhotic samples were used. Samples in this study were not split into discovery or validation sets and only used for training process. To calculate AUC, RF outcome was matched to disease status using auc function of the pROC package. For comparison of accuracy/AUC between 13-species model and the Model for End-Stage Liver Disease (MELD) score, the training model was established with MELD scores.
RF models were trained with 13 top-discriminatory taxa abundances. Sequentially, we tested the best performing model of discriminatory signatures with the independent test cohort (Loomba 2017). To do so, we extracted 13 species from the taxa abundance table. We then tested models in the validation dataset using the predict function with the default setting. The predict function of caret was used with the prob type option. To obtain AUC scores, roc function was utilised using the setting of boot.n; 100 and ci.alpha; 0.9. For visualisation of AUC outcome, plot function was utilised in R.
2.3.2 |. Stool and serum metabolomics
Metabolic feature extraction was performed using MS-DIAL platform (version 4.70). Metabolite annotation was carried out by searching against a combined MS/MS reference library provided by MS-DIAL (Version 14). A total score considering the formula and structure matching was utilised for ranking candidates, with isotope ratio tolerance of 20% and MS1 and MS2 tolerance of 0.01 Da. The candidate with the highest total score was chosen as a feature annotation result. Further confirmation was performed using MS-FINDER (Version 3.52) by searching against an in-silico MS/MS spectra database.
2.3.3 |. Evaluating predictors of mortality in NAFLD cirrhosis
We subsequently evaluated whether baseline patient characteristics, including the 13-species metagenomic signature, were associated with longitudinal risk of mortality in NAFLD cirrhosis. A Cox proportional hazards model was used to examine predictors of all-cause mortality in the training cohort. Time to death was defined as time from baseline research visit (stool sample collection) to time of death as documented in the electronic medical record or U.S. Social Security Death Index. If there was no confirmed date of death, patients were censored at the last documented clinical visit or on the date of liver transplantation.
3 |. RESULTS
3.1 |. Patient characteristics: Training and independent test cohorts
This study included 25 adults with NAFLD cirrhosis (training cohort), among which 16 had compensated cirrhosis and 9 had decompensated cirrhosis. Baseline patient characteristics, at the time of stool sample collection, are shown in Table 1. Mean age was 63 and 68 years in those with compensated and decompensated cirrhosis (p = 0.44), respectively. There were no significant differences in race/ethnicity, body mass index and type 2 diabetes in those with and without hepatic decompensation. The median MELD score was 7 in those with compensated cirrhosis and 16 in those with decompensated cirrhosis (p < 0.01). Among those with decompensated cirrhosis, 33% had ascites, 56% had hepatic encephalopathy and 22% had experienced a prior variceal haemorrhage (not mutually exclusive). Not surprisingly, the use of rifaximin and lactulose was significantly higher in the decompensated cirrhosis cohort (p < 0.01 for each). No patients with decompensated cirrhosis were receiving antimicrobial prophylaxis for spontaneous bacterial peritonitis (SBP). Patient characteristics of the independent test cohort, comprised of 9 adults with NAFLD cirrhosis (5 compensated and 4 decompensated), are listed in Table S1.
TABLE 1.
Patient characteristics of the non-alcoholic fatty liver disease (NAFLD) cirrhosis cohort.
| Characteristics | Compensated NAFLD cirrhosis (n = 16) | Decompensated NAFLD cirrhosis (n = 9) | p-value |
|---|---|---|---|
| Demographics and co-morbidities | |||
| Age, years | 63 (11) | 68 (8) | 0.44 |
| Female (%) | 82 | 89 | 1.0 |
| Hispanic (%) | 50 | 67 | 0.68 |
| Caucasian (%) | 38 | 22 | 0.66 |
| Type II diabetes (%) | 82 | 89 | 1.0 |
| Body mass index (kg/m2) | 35.3 (11.4) | 29.9 (6.9) | 0.22 |
| Tobacco use (%) | 0 | 0 | 1.0 |
| Laboratory results | |||
| AST (IU/L) | 42 (19) | 58 (28) | 0.17 |
| ALT (IU/L) | 45 (36) | 48 (42) | 0.46 |
| Total bilirubin (mg/dl) | 0.71 (0.45) | 5.1 (11.27) | 0.01 |
| Platelet count (109/L) | 183 (63) | 97 (61) | 0.01 |
| Albumin (g/dl) | 4.3 (0.3) | 3.6 (0.8) | 0.01 |
| INR | 1.1 (0.1) | 1.4 (0.5) | <0.01 |
| Fasting insulin (mIU/L) | 39.1 (30.8) | 40.6 (24.6) | 0.89 |
| Glycated haemoglobin (%) | 7.5 (1.7) | 6.3 (2.1) | 0.07 |
| Triglycerides (mg/dl) | 131 (62) | 179 (213) | 0.83 |
| Low-density lipoprotein (mg/dl) | 83 (33) | 71 (34) | 0.38 |
| High-density lipoprotein (mg/dl) | 52 (15) | 42 (21) | 0.24 |
| Medication use, n (%) | |||
| Proton pump inhibitor | 5 (31%) | 1 (11%) | 0.37 |
| Lactulose | 0 (0%) | 6 (67%) | <0.01 |
| Rifaximin | 0 (0%) | 4 (44%) | <0.01 |
| Metformin | 10 (63%) | 3 (33%) | 0.23 |
| Antibiotic prophylaxis for SBP | – | 0 (0%) | – |
| Liver disease severity | |||
| MELD score | 7.2(1.2) | 15.6 (8.9) | <0.01 |
| Ascites, n (%) | 0 (0%) | 3 (33.33%) | 0.04 |
| Hepatic encephalopathy, n (%) | 0 (0%) | 5 (55.56%) | <0.01 |
| Variceal bleeding, n (%) | 0 (0%) | 2 (22.22%) | 0.12 |
| Prior TIPS, n (%) | 0 (0%) | 1 (11.11%) | 0.36 |
Note: Mean (standard deviation).
Abbreviations: AST, aspartate aminotransferase; ALT, alanine aminotransferase; INR; international normalised ratio; SBP, spontaneous bacterial peritonitis; MELD, Model For End Stage Liver Disease; NAFLD, Non-alcoholic fatty liver disease; TIPS, transjugular intrahepatic portosystemic shunt.
3.2 |. Diagnostic accuracy and validation of a stool metagenomic signature for decompensated NAFLD-related cirrhosis
We initially performed a cross-sectional comparison of the stool metagenome among those with decompensated and compensated cirrhosis at the baseline to evaluate for a stool metagenomic signature of decompensated NAFLD cirrhosis. Using the Shannon diversity index and Inverse Simpson index to measure microbial richness, alpha-diversity was decreased in decompensated cirrhosis compared with compensated cirrhosis (p = 0.022; Figure 1A; Figure S1). Principal coordinates analysis coupled with weighted UniFrac to determine beta-diversity revealed a significant separation between decompensated and compensated NAFLD cirrhosis (Figure 1B; PERMANOVA, p = 0.001). Shifts in taxonomic composition at the class level were noted when comparing decompensated and compensated cirrhosis, including enrichment in Negativicutes, Actinobacteria and Bacteroidia in decompensated cirrhosis (Figure 1C). Significant differences in gut microbial composition of decompensated NAFLD cirrhosis versus compensated NAFLD cirrhosis were also observed at phylum level (Figure S1).
FIGURE 1.
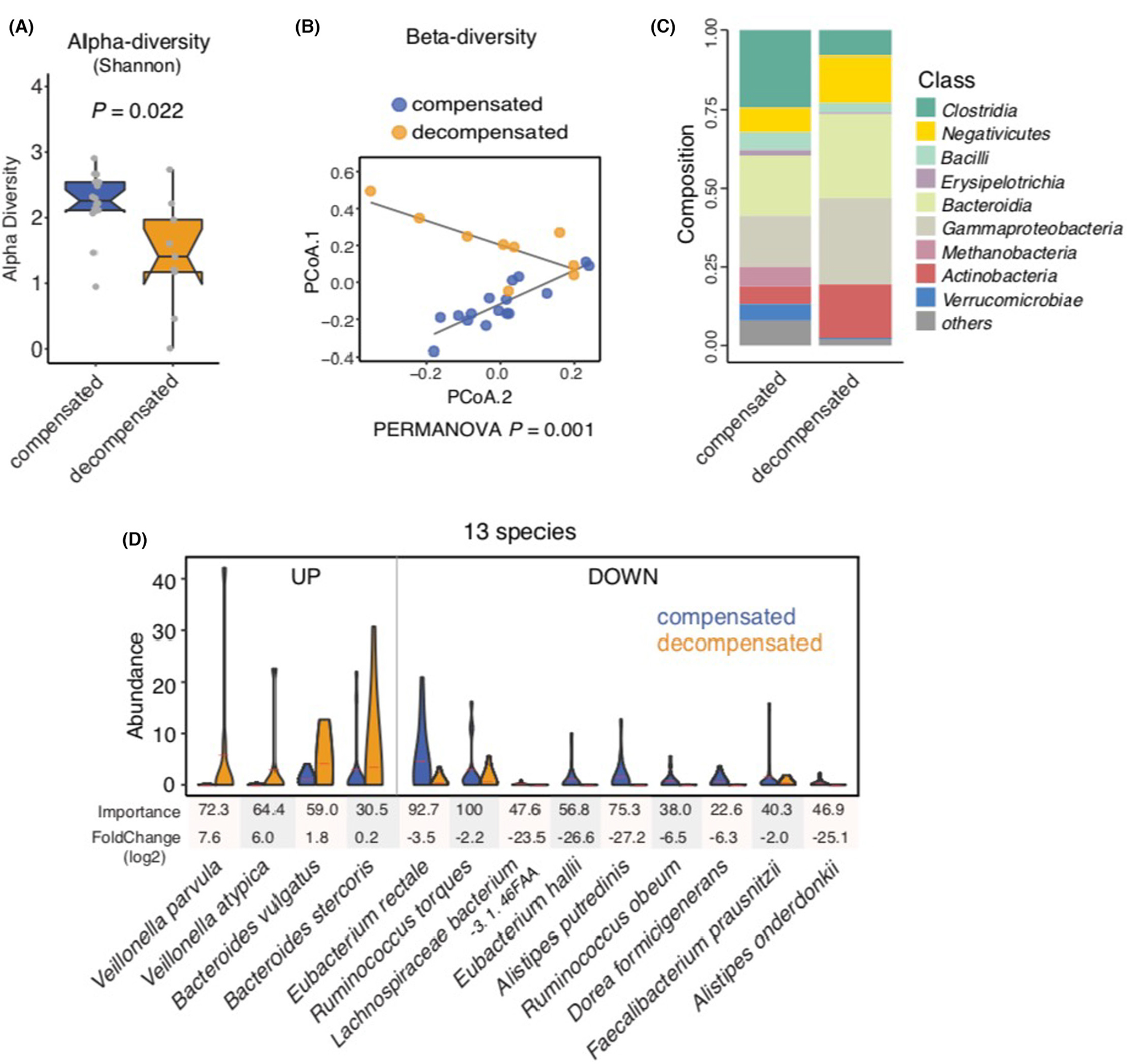
Metagenomic profiling in non-alcoholic fatty liver disease (NAFLD)-related cirrhosis, comparing compensated and decompensated cirrhosis. (A) Shannon α-diversity scores highlighted significant decreases in the richness of gut microbiota in the decompensated NAFLD cirrhosis group (n = 9) compared with the compensated NAFLD cirrhosis group (n = 16; p = 0.022). Grey dots represent values for individual participants. Boxes represent the interquartile range (IQR) between the first and third quartiles. Median values are represented by horizontal lines within the boxes. Notches represent 95% confidence intervals for the medians. Whiskers indicate the range from minimum (first quartile −1.5 × IQR) to maximum (third quartiles +1.5 × IQR). (B) Principal coordinate analysis of stool samples from decompensated and compensated NAFLD cirrhosis patients using weighted-UniFrac distances (PERMANOVA = 0.001). (C) Stacked bar plots depicting class-level differences in gut microbiome composition between decompensated NAFLD cirrhosis and compensated NAFLD cirrhosis. The ‘others’ subcategory includes rare species (<1%). (D) A metagenomic signature of 13 discriminatory species in decompensated NAFLD cirrhosis compared to compensated NAFLD cirrhosis. Species were chosen from the highest scores of Mean Decrease in Gini using Random Forest (RF) feature selection. Importance and log2FoldChange were presented by each species.
Utilising a RF machine learning model, we identified a stool metagenomic signature comprised of 13 discriminatory species that accurately detected decompensated NAFLD cirrhosis (Figure 1D). The identified disease signature included increases in the levels of Veillonella parvula, Veillonella atypica, Bacteroides vulgatus and Bacteroides stercoris. Conversely, there were decreases in the abundance of Eubacterium rectale, Eubacterium hallii, Faecalibacterium prausnitzii, Alistipes onderdonkii, Alistipes putredinis, Dorea formicigenerans, Ruminococcus obeum and Lachnospiraceae bacterium in decompensated NAFLD cirrhosis, as compared to compensated NAFLD cirrhosis.
Next, we evaluated the diagnostic accuracy of the 13-species metagenomic-derived microbial signature, as compared to the MELD score, for distinguishing decompensated and compensated NAFLD cirrhosis. Both the MELD score (area under the receiver operating characteristics [AUROC] 0.84, 95% confidence interval [CI] 0.78–0.90, Figure 2A) and the 13-species signature (AUROC 0.97, 95% CI 0.96–0.99, Figure 2B) were able to reliably distinguish decompensated and compensated NAFLD cirrhosis. However, the 13-species microbial signature had superior discrimination when compared to the MELD score for decompensated cirrhosis (Figure 2C, p < 0.001).
FIGURE 2.
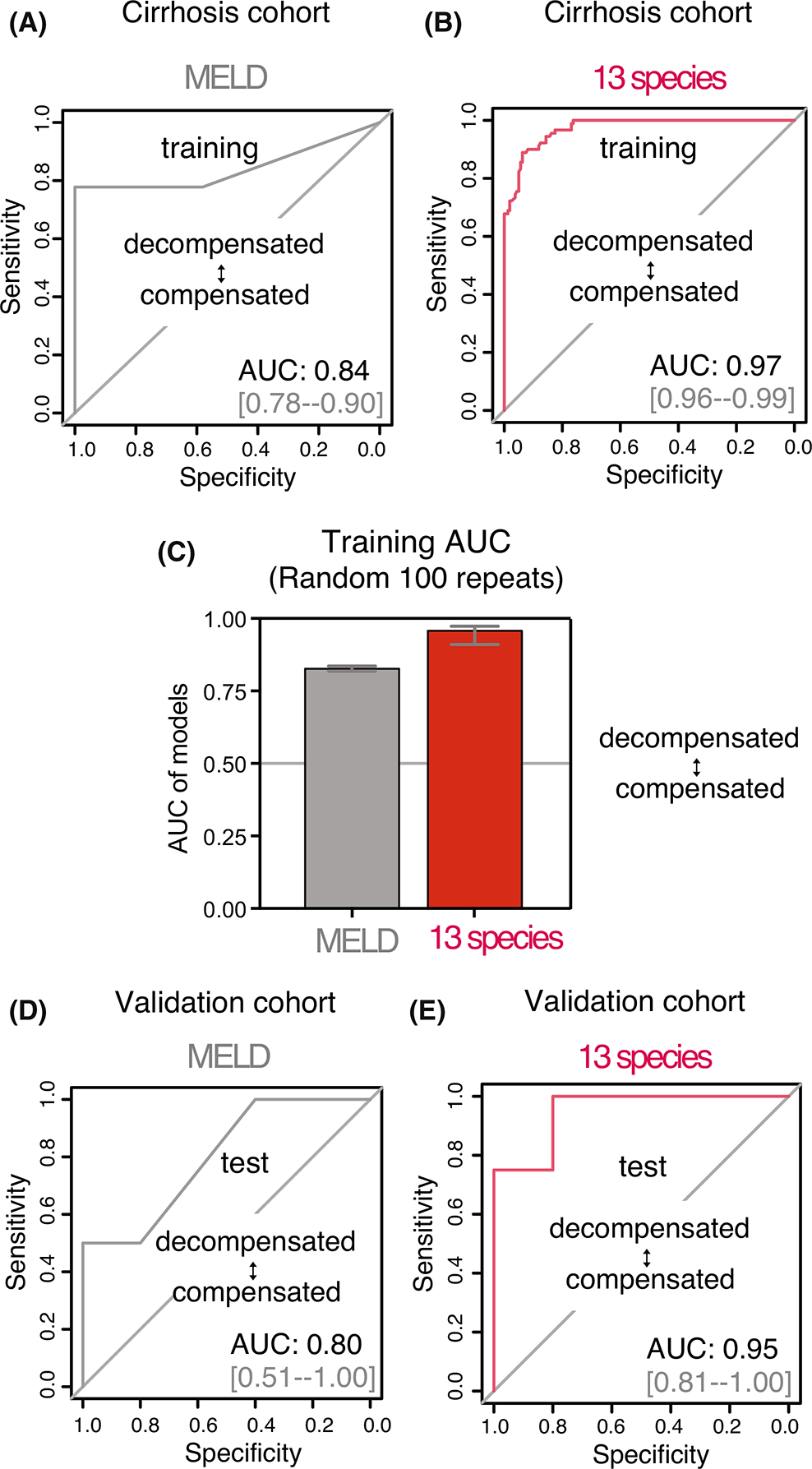
Diagnostic accuracy of faecal metagenome-derived signature for the detection of decompensated non-alcoholic fatty liver disease (NAFLD) cirrhosis. (A) Receiver operating characteristics (ROC) curve using Model for End-Stage Liver Disease (MELD) score for the prediction of decompensated NAFLD cirrhosis in the training cohort. (B) ROC curve of the Random Forest (RF) model using 13 discriminatory species for the prediction of decompensated NAFLD cirrhosis in the training cohort. (C) Comparison of area under the curve (AUC) values from 100 models of repeated RF outcome for decompensated NAFLD cirrhosis in the training cohort. (D) Validation of the RF model using MELD score in an external cohort. ROC curve for prediction of decompensated NAFLD cirrhosis. (E) Validation of the RF model using 13 discriminatory species in an external cohort. Tests demonstrated that the microbial signature had robust accuracy in independent cohorts (AUC 0.97 and AUC 0.95 in training and validation cohorts, respectively).
We subsequently tested the diagnostic accuracy of MELD and the 13-species microbial signature in the independent test cohort. The discriminative ability of both the MELD score (AUROC 0.80, 95% CI 0.51–1.00, Figure 2D) and 13-species metagenomic signature (AUROC 0.95, 95% CI 0.81–1.00, Figure 2E) for decompensated NAFLD cirrhosis remained high, but the microbial signature had superior discrimination in the independent test cohort as well (p < 0.001).
Finally, we conducted sensitivity analyses to examine the diagnostic accuracy of the 13-species metagenomic signature after adjustment for medication exposure, given this differed in decompensated and compensated participants. Diagnostic accuracy of the model remained high after adjustment for rifaximin (AUROC 0.92, Figure S2), lactulose (AUROC 0.99, Figure S2), metformin (AUROC 0.92, Figure S2) and proton pump inhibitor use (AUROC 0.92, Figure S2). As noted previously, no patients with decompensated NAFLD cirrhosis were receiving antibiotic prophylaxis for SBP.
3.3 |. Stool and serum metabolomics
To further refine our gut microbiome signature, we characterised differences in levels of stool and serum metabolites in those with decompensated versus compensated NAFLD-related cirrhosis. Metabolite analysis was performed on 23 matched serum and stool samples (Figure 3A). We examined stool and serum metabolites that were correlated with the 13 discriminatory species. In doing so, we found a set of distinct serum and stool metabolites with significant correlation to the 13 discriminatory species. In serum, we identified that total 32 metabolites positively or negatively associated with a single or multiple species of the metagenomic signature (Figure 3B) including enrichment in 3-indoleacetic acid (correlation with Veillonella parvula) as well as reduction in indoline, tryptophan and phenylalanine (correlation with Ruminococcus obeum). In addition, stool sample analysis demonstrated that 15 metabolites are correlated with discriminatory species (Figure 3C), and similar reductions in indoline, tryptophan and phenylalanine were noted.
FIGURE 3.
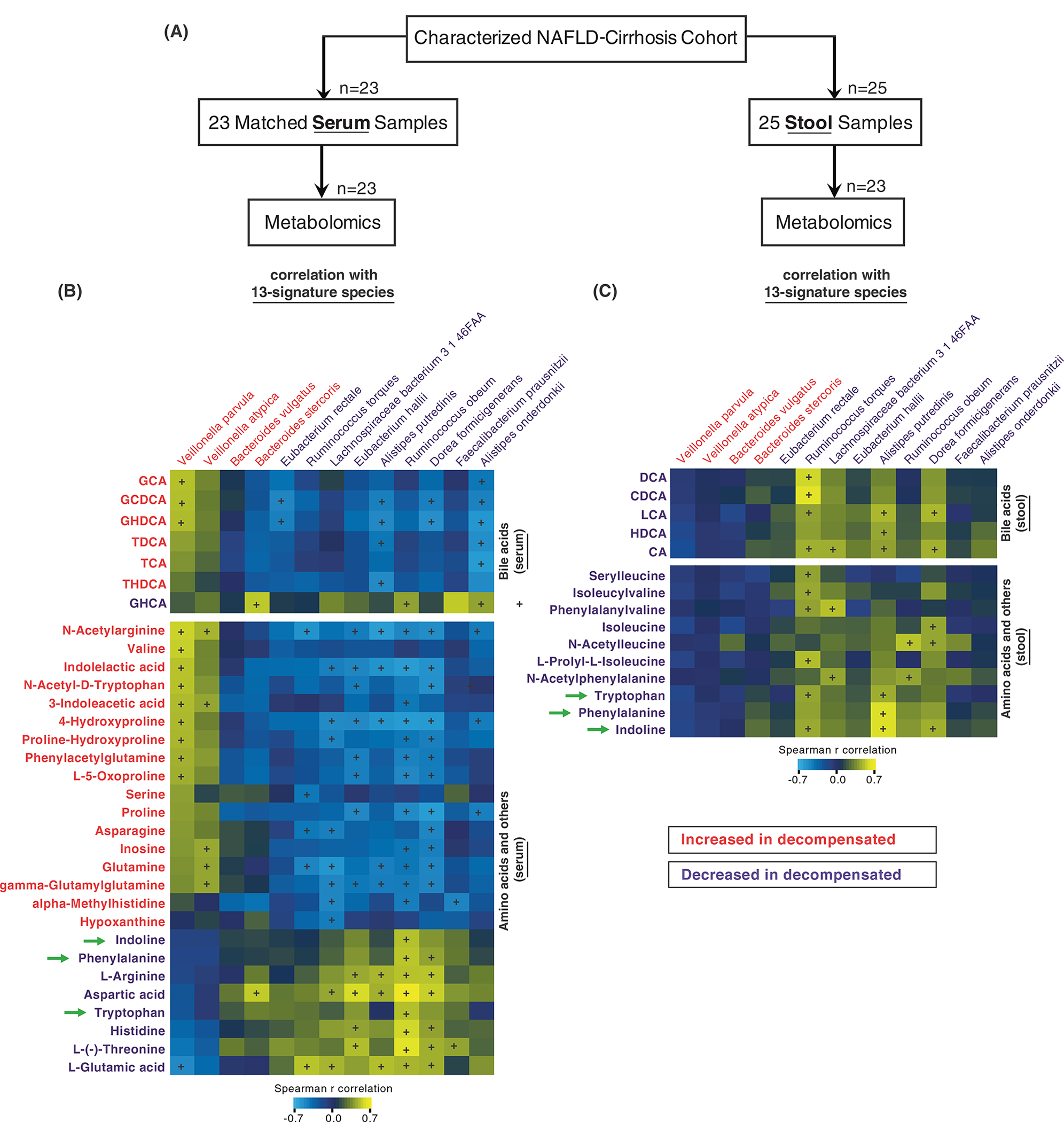
Correlation analysis of metagenomic and metabolomic features for decompensated non-alcoholic fatty liver disease (NAFLD) cirrhosis. (A) A flow chart shows study overview of correlating microbes and metabolites. Metabolite analysis with 23 matched serum or stool samples were correlated top 13-discriminatory species. (B) Correlation analysis identified significant association between serum metabolites and 13-species. The intensity values of bile acids, amino acids and other metabolites were correlated with 13-species abundance levels using Spearman’s correlation analysis. The colour range shows positive (yellow) or negative (light blue) correlation. The + symbol represents the significance of p-value (p < 0.05). (C) Using stool metabolites and 13-species, Spearman’s correlation analysis was performed. The colour range shows positive (yellow) or negative (light blue) correlation. The + symbol represents the significance of p-value (p < 0.05).
3.4 |. Predicting risk of all-cause mortality in adults with NAFLD-related cirrhosis
As expected, survival was significantly decreased in adults with decompensated cirrhosis, as compared to those with compensated cirrhosis in the training cohort (log rank p = 0.028; Figure S3). We utilised a Cox proportional hazards model to examine predictors of mortality in the overall cohort of adults with NAFLD cirrhosis (Table 2). Median follow-up was 3.5 years. Serum albumin (hazard ratio [HR] 0.30, 95% CI 0.11–0.82, p = 0.02) and MELD score (HR 1.25, 95% CI 1.03–1.53, p = 0.03) were significantly associated with all-cause mortality in univariate analyses. However, the stool metagenomic signature was the strongest predictor of all-cause mortality (HR 1.54, 95% CI 1.10–2.15, p = 0.01) in our cohort of adults with NAFLD cirrhosis. The association between the stool metagenomic signature and mortality remained significant in multivariate analyses with adjustment for medication exposures including rifaximin (HR 1.49, 95% CI 1.05–2.11, p = 0.02), lactulose (HR 1.54, 95% CI 1.09–2.16, p = 0.01), metformin (HR 1.55, 95% CI 1.09–2.20, p = 0.02) and proton pump inhibitor (HR 1.54, 95% CI 1.10–2.15, p = 0.01; Table S2).
TABLE 2.
Predictors of all-cause mortality in adults with non-alcoholic fatty liver disease (NAFLD) cirrhosis.
| Predictor | HR* | 95% CI | p-value |
|---|---|---|---|
| Age (years) | 1.06 | 0.96–1.17 | 0.22 |
| Body mass index (kg/m2, 1 unit increase) | 1.02 | 0.95–1.10 | 0.59 |
| Female sex | 0.28 | 0.05–1.48 | 0.14 |
| Albumin (g/dl, 1 unit increase) | 0.30 | 0.11–0.82 | 0.02 |
| MELD score | 1.25 | 1.03–1.53 | 0.03 |
| 13-species microbial signature | 1.54 | 1.10–2.15 | 0.01 |
Abbreviations: CI, confidence interval; HR, hazard ratio; MELD, Model for End-Stage Liver Disease.
All results shown represent univariable analyses.
4 |. DISCUSSION
Whether the gut microbiome can serve as a non-invasive biomarker for detection and prognostication in NAFLD-related cirrhosis is not known. Using faecal samples from a deeply phenotyped prospective cohort of adults with NAFLD-related cirrhosis in the ambulatory setting, we provide proof-of-concept data demonstrating that a metagenome-derived signature provides diagnostic and prognostic capabilities. We identified a metagenome-derived signature comprised of 13 microbial species that accurately discriminates decompensated from compensated NAFLD-related cirrhosis, with diagnostic accuracy ranging from AUC 0.96 to 0.99. Diagnostic accuracy remained high in sensitivity analyses adjusted for medication exposures, and validation of this metagenomic signature in an independent test cohort confirmed robust accuracy (AUC 0.81–1.00). Moreover, the 13-species metagenomic signature was more strongly associated with risk for all-cause mortality when compared to the MELD score, a well-validated assessment of chronic liver disease severity. Metabolomic analyses revealed a number of correlations between stool and circulating serum metabolites and bacterial species comprising the metagenomic signature, suggesting that the identified discriminatory species may play important roles in progression from compensated to decompensated cirrhosis. Altogether, our findings suggest a novel microbial biomarker for diagnosis and prognostication in adults with NAFLD-related cirrhosis and provide us with potential points of intervention to prevent hepatic decompensation and its associated mortality.
Prior studies have shown that perturbation of the gut microbiota develops prior to development of cirrhosis and during progression of chronic liver disease.11 Our team has previously described gut microbial signatures with robust diagnostic accuracy for advanced fibrosis and cirrhosis in NAFLD.6–8 Gut microbial features have also been shown to be closely associated with liver fibrosis in a population-based cohort of Hispanics, a patient population with high prevalence of NAFLD.12 A number of the bacterial taxa that comprise our 13-species microbial signature of decompensated NAFLD-related cirrhosis overlap with those taxa previously associated with histologic phenotypes of NAFLD and/or cirrhosis.6,7,13–15 Veillonellaceae is enriched in cirrhosis and associated with fibrosis severity in non-obese adults with non-alcoholic steatohepatitis (NASH),16,17 and we found Veillonella parvula and Veillonella atypica to be enriched in those with decompensated NAFLD cirrhosis, as compared to those with compensated cirrhosis, in our cohort. A shift in Veillonella abundance has been shown to be indicative of histologic improvement in NASH with longitudinal treatment with an analogue of the gut hormone fibroblast growth factor 19, a hormone that affects bile acid synthesis.18 Moreover, Veillonellaceae were previously noted to be increased in those with overt hepatic encephalopathy, as compared to those without hepatic encephalopathy, in a prior study.19 Bacteroides vulgatus, a microbial species in our 13-species signature, was previously noted to be predominant in NASH-related advanced fibrosis which suggests progressive dysbiosis occurs even after development of NAFLD-related cirrhosis.6 Bacteroides vulgatus has also been associated with differences in metabolic pathways between obese youths with and without NAFLD.15 Several taxa were decreased in those with hepatic decompensation, including Alistipes which has been shown to correlate with administration of a glycine-related compound which facilitated NASH improvement in a murine model.20
Emerging data support the role of the gut microbiome as a prognostic biomarker in liver diseases other than NAFLD. Adults with severe acute alcoholic hepatitis and a specific cytolysin-producing Enterococcus strain are at significantly higher risk of mortality compared with those without this microbial strain.21 Serum metabolites linked to the gut microbiota predict inpatient and 30-day mortality in hospitalised adults with decompensated cirrhosis.22,23 The gut microbiome, specifically a measure of gene richness, predicts 90-day survival in hospitalised adults with decompensated cirrhosis (all aetiologies) and acute on chronic liver failure with an AUC 0.708, although accuracy was less robust than the MELD score.24 In our study, we utilised a Cox proportional hazards model to examine predictors of all-cause mortality and found the 13-species metagenomic signature to be a significant predictor of all-cause mortality in NAFLD-related cirrhosis, even after adjustment for a number of clinical factors including the MELD score, serum albumin and medication use. In fact, our 13-species metagenomic signature was a stronger predictor of all-cause mortality in our cohort when compared to the MELD score.
We paired stool and serum metabolomics with metagenomic analyses to establish an integral view of the gut microbiome, including microbial gene expression and metabolism, and to evaluate associations with hepatic decompensation and mortality risk. We found a number of stool and serum metabolites that were differentially abundant in those patients with decompensated NAFLD cirrhosis. 3-indoleacetic acid, a metabolite previously noted to be gut microbiota-derived from metabolism of dietary tryptophan,25 was decreased in decompensated disease. Subsequent metabolomic analyses in paired serum samples revealed a significant decrease in 3-indoleacetic acid in patients with decompensated disease which correlated with Veillonella atypica, Veillonella parvula and Ruminococcus obeum. This finding is bolstered by recent evidence linking 3-indoleacetic acid with NAFLD. A recent study demonstrated that 3-indoleacetic acid alleviates high-fat diet-induced hepatotoxicity in mice and is associated with the amelioration of insulin resistance and oxidative stress.26 In humans, serum levels of 3-indoleacetic acid increased after sleeve gastrectomy and negatively correlated with liver fat attenuation.27 Faecal levels of 3-indoleacetic acid have also been found to be lower in adults with severe acute alcoholic hepatitis compared with healthy controls.28 Several other microbial species in the 13-species signature (e.g., Ruminococcus torques) were highly correlated with key metabolites derived from methylamine and branched chain amino acid metabolism and carbohydrate fermentation. This finding is strengthened by pre-clinical data revealing that microbial-derived products of amino acid metabolism and targeting glutaminolysis modulate NAFLD in animal models.20,29–31 We also found a number of bile acid metabolites that were highly correlated with microbial species in the metagenomic signature, which was expected given the gut microbiota regulates the bile acid pool32 and supraphysiologic bile acid concentrations in NAFLD may be precipitated by the gut microbiota.33
This study is innovative because it addresses a clinically important question in a uniquely well-phenotyped group of adults with NAFLD-related cirrhosis in the ambulatory setting, the findings are replicated in an independent test cohort, and the metagenomic signature is cross-validated with serum and stool metabolites that are linked to the key bacterial species, thus yielding new avenues for identifying novel targets for therapy as well as microbial biomarkers of hepatic decompensation. Our cohort was deeply phenotyped; for example, hepatic decompensation was not only prospectively collected, but also subsequently verified by a board-certified transplant hepatologist. However, our study has some limitations. We note that the sample size is small, although we were able to validate our 13-species metagenomic signature in an independent test cohort with high diagnostic accuracy. Multi-centre studies are needed to further validate these findings. Although we cannot account for all confounding factors that may affect the gut microbiome such as dietary intake, we adjusted for key medication exposures that may differ in patients with and without hepatic decompensation. Moreover, prior studies have shown that lactulose has no significant effect on the stool microbiota composition.19 Our cohort study only included adults with NAFLD-related cirrhosis, so whether our microbial signature associates with hepatic decompensation in other aetiologies of cirrhosis is not known. However, a recent study found no gut microbial signal to differentiate among underlying aetiologies of cirrhosis.34 Finally, we recognise that any associations noted between microbial species and metabolites do not imply a causal relationship and therefore follow-up studies are needed.
In summary, our study is novel in that we have identified and validated a metagenomic signature of hepatic decompensation in NAFLD-related cirrhosis that is more closely associated with the risk of all-cause mortality than the MELD score. Our findings suggest that microbial gene markers quantifiable in stool predict hepatic decompensation and risk of mortality in NAFLD-related cirrhosis, thus offering a potential, non-invasive, diagnostic test for risk stratification.
Supplementary Material
ACKNOWLEDGEMENTS
All persons who meet authorship criteria are listed as authors, and all authors certify that they have participated sufficiently in the work to take responsibility for the content, including participation in the concept, design, analysis, writing or revision of the manuscript. All authors approved the final version of the manuscript.
FUNDING INFORMATION
SS received funding support from the AASLD Foundation. RL received funding support from NIEHS (5P42ES010337), NCATS (5UL1TR001442), DOD PRCRP (W81XWH-18-2-0026), NIDDK (U01DK061734, R01DK106419, R01DK121378, R01DK124318, P30DK120515), NHLBI (P01HL147835) and NIAAA (U01AA029019). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
CONFLICTS OF INTEREST
SS has no conflicts of interest. TO has no conflicts of interest. EM has no conflicts of interest. CW has no conflicts of interest. RY has no conflicts of interest. AA has no conflicts of interest. TH has no conflicts of interest. MD has no conflicts of interest. RE has no conflicts of interest. RL serves as a consultant for Anylam/Regeneron, Amgen, Arrowhead Pharmaceuticals, AstraZeneca, Bristol-Myer Squibb, CohBar, Eli Lilly, Galmed, Gilead, Glympse bio, Inipharm, Intercept, Ionis, Janssen Inc., Madrigal, Metacrine, Inc., NGM Biopharmaceuticals, Novartis, Novo Nordisk, Pfizer, Sagimet, 89 bio and Viking Therapeutics. In addition, his institution has received grant support from Allergan, Astrazeneca, Boehringer-Ingelheim, Bristol-Myers Squibb, Eli Lilly, Galectin Therapeutics, Galmed Pharmaceuticals, Genfit, Gilead, Intercept, Inventiva, Janssen, Madrigal Pharmaceuticals, Merck, NGM Biopharmaceuticals, Pfizer and Siemens. RL is also co-founder of Liponexus, Inc.
SUPPORTING INFORMATION
Additional supporting information will be found online in the Supporting Information section.
The Handling Editor for this article was Professor Vincent Wong, and it was accepted for publication after full peer-review.
REFERENCES
- 1.Younossi Z, Anstee QM, Marietti M, Hardy T, Henry L, Eslam M, et al. Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol. 2018;15(1):11–20. [DOI] [PubMed] [Google Scholar]
- 2.Hagström H, Nasr P, Ekstedt M, Hammar U, Stål P, Hultcrantz R, et al. Fibrosis stage but not NASH predicts mortality and time to development of severe liver disease in biopsy-proven NAFLD. J Hepatol. 2017;67(6):1265–73. [DOI] [PubMed] [Google Scholar]
- 3.Dulai PS, Singh S, Patel J, Soni M, Prokop LJ, Younossi Z, et al. Increased risk of mortality by fibrosis stage in nonalcoholic fatty liver disease: systematic review and meta-analysis. Hepatology. 2017;65(5):1557–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharpton SR, Ajmera V, Loomba R. Emerging role of the gut microbiome in nonalcoholic fatty liver disease: from composition to function. Clin Gastroenterol Hepatol. 2019;17(2):296–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharpton SR, Schnabl B, Knight R, Loomba R. Current concepts, opportunities, and challenges of gut microbiome-based personalized medicine in nonalcoholic fatty liver disease. Cell Metab. 2021;33(1):21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loomba R, Seguritan V, Li W, Long T, Klitgord N, Bhatt A, et al. Gut microbiome-based metagenomic signature for non-invasive detection of advanced fibrosis in human nonalcoholic fatty liver disease. Cell Metab. 2017;25(5):1054–1062.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oh TG, Kim SM, Caussy C, Fu T, Guo J, Bassirian S, et al. A universal gut-microbiome-derived signature predicts cirrhosis. Cell Metab. 2020;32(5):878–888.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caussy C, Tripathi A, Humphrey G, Bassirian S, Singh S, Faulkner C, et al. A gut microbiome signature for cirrhosis due to nonalcoholic fatty liver disease. Nat Comm. 2019;10(1):1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chalasani N, Younossi ZA-OX, Lavine JE, Charlton M, Cusi K, Rinella M, et al. The diagnosis and management of nonalcoholic fatty liver disease: practice guidance from the American Association for the Study of Liver Diseases. Hepatology. 2018;67(1):328–57. [DOI] [PubMed] [Google Scholar]
- 10.Vilstrup H, Amodio P, Bajaj J, Cordoba J, Ferenci P, Mullen KD, et al. Hepatic encephalopathy in chronic liver disease: 2014 practice guideline by the American Association for the Study of Liver Diseases and the European Association for the Study of the liver. Hepatology. 2014;60(2):715–35. [DOI] [PubMed] [Google Scholar]
- 11.Acharya C, Bajaj JS. Chronic liver diseases and the microbiome-translating our knowledge of gut microbiota to Management of Chronic Liver Disease. Gastroenterology. 2021;160(2):556–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kwan SY, Jiao J, Joon A, Wei P, Petty LE, Below JE, et al. Gut microbiome features associated with liver fibrosis in Hispanics, a population at high risk for fatty liver disease. Hepatology. 2022;75(4):955–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caussy C, Hsu C, Lo M-T, Liu A, Bettencourt R, Ajmera VH, et al. Novel link between gut-microbiome derived metabolite and shared gene-effects with hepatic steatosis and fibrosis in NAFLD. Hepatology (Baltimore, Md). 2018;68:S29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Acharya C, Bajaj JS. Altered microbiome in patients with cirrhosis and complications. Clinical Gastroenterol Hepatol. 2019;17(2):307–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Testerman T, Li Z, Galuppo B, Graf J, Santoro N. Insights from shotgun metagenomics into bacterial species and metabolic pathways associated with NAFLD in obese youth. Hepatol Commun. 2022;6(8):1962–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Y, Yang F, Lu H, Wang B, Chen Y, Lei D, et al. Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology. 2011;54(2):562–72. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Z, Zhai H, Geng J, Yu R, Ren H, Fan H, et al. Large-scale survey of gut microbiota associated with MHE via 16S rRNA-based pyrosequencing. Am J Gastroenterol. 2013;108(10):1601–11. [DOI] [PubMed] [Google Scholar]
- 18.Loomba R, Ling L, Dinh DM, AM DP, Lieu HD, Harrison SA, et al. The commensal microbe Veillonella as a marker for response to an FGF19 analog in nonalcoholic steatohepatitis. Hepatology. 2020;73(1):126–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bajaj JS, Ridlon JM, Hylemon PB, Thacker LR, Heuman DM, Smith S, et al. Linkage of gut microbiome with cognition in hepatic encephalopathy. Am J Physiol Gastrointest Liver Physiol. 2012;302:G168–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rom O, Liu Y, Liu Z, Zhao Y, Wu J, Ghrayeb A, et al. Glycine-based treatment ameliorates NAFLD by modulating fatty acid oxidation, glutathione synthesis, and the gut microbiome. Sci Transl Med. 2020;12(572):eaaz2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duan Y, Llorente C, Lang S, Brandl K, Chu H, Jiang L, et al. Bacteriophage targeting of gut bacterium attenuates alcoholic liver disease. Nat Commun. 2019;575(7783):505–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bajaj JS, Betrapally NS, Hylemon PB, Thacker LR, Daita K, Kang DJ, et al. Gut microbiota alterations can predict hospitalizations in cirrhosis independent of diabetes mellitus. Sci Rep. 2015;5:18559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bajaj JS, Vargas HE, Reddy KR, Lai JC, O’Leary JG, Tandon P, et al. Association between intestinal microbiota collected at hospital admission and outcomes of patients with cirrhosis. Clin Gastroenterol Hepatol. 2019;17(4):756–765.e3. [DOI] [PubMed] [Google Scholar]
- 24.Solé C, Guilly S, da Silva K, Llopis M, le-Chatelier E, Huelin P, et al. Alterations in gut microbiome in cirrhosis as assessed by quantitative metagenomics: relationship with acute-on-chronic liver failure and prognosis. Gastroenterology. 2021;160(1):206–218.e13. [DOI] [PubMed] [Google Scholar]
- 25.Krishnan S, Ding Y, Saedi N, Choi M, Sridharan GV, Sherr DH, et al. Gut microbiota-derived tryptophan metabolites modulate inflammatory response in hepatocytes and macrophages. Cell Rep. 2018;23(4):1099–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ji Y, Gao Y, Chen H, Yin Y, Zhang WA-O. Indole-3-acetic acid alleviates nonalcoholic fatty liver disease in mice via attenuation of hepatic lipogenesis, and oxidative and inflammatory stress. Nutrients. 2019;11(9):2062. 10.3390/nu11092062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Wang G, Bai J, Zhao N, Wang Q, Zhou R, et al. Role of Indole-3-acetic acid in NAFLD amelioration after sleeve gastrectomy. Obes Surg. 2021;31(7):3040–52. [DOI] [PubMed] [Google Scholar]
- 28.Hendrikx T, Duan Y, Wang Y, Oh JH, Alexander LM, Huang W, et al. Bacteria engineered to produce IL-22 in intestine induce expression of REG3G to reduce ethanol-induced liver disease in mice. Gut. 2019;68(8):1504–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao ZH, Xin FZ, Xue Y, Hu Z, Han Y, Ma F, et al. Indole-3-propionic acid inhibits gut dysbiosis and endotoxin leakage to attenuate steatohepatitis in rats. Exp Mol Med. 2019;51(9):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoyles L, Fernández-Real JM, Federici M, Serino M, Abbott J, Charpentier J, et al. Molecular phenomics and metagenomics of hepatic steatosis in non-diabetic obese women. Nat Med. 2018;24(7):1070–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.du K, Chitneni SK, Suzuki A, Wang Y, Henao R, Hyun J, et al. Increased Glutaminolysis Marks active scarring in nonalcoholic steatohepatitis progression. Cell Mol Gastroenterol Hepatol. 2020;10(1):1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ridlon JM, Harris SC, Bhowmik S, Kang DJ, Hylemon PB. Consequences of bile salt biotransformations by intestinal bacteria. Gut microbes. 2016;7(1):22–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yao L, Seaton SC, Ndousse-Fetter S, Adhikari AA, DiBenedetto N, Mina AI, et al. A selective gut bacterial bile salt hydrolase alters host metabolism. Elife. 2018;7:e37182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Naseri M, Houri H, Yadegar A, Asadzadeh Aghdaei H, Zahiri J. Investigation of etiology-specific alterations in the gut microbiota in liver cirrhosis. Expert Rev Gastroenterol Hepatol. 2021;15(12):1435–41. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


