ABSTRACT
During the first quarter of 2020, a considerable increase in reports of symptomatic hepatitis A cases was noted in Yantai, a coastal city in eastern China. This study aimed to characterize the epidemic and identify the probable source. Serum samples from cases with onsets from 1 January to 31 March 2020 and suspected bivalve mollusk samples from the local seafood market were screened for hepatitis A virus (HAV) RNA by PCR amplification and sequencing of the VP1/2A region. We also analyzed the characteristics and risk exposures of these cases. In total, 110 confirmed cases were notified during the epidemic. Among the 103 cases investigated, the median age was 41 years (range: 25–70 years), and 74 (71.8%) were male. Eighty-eight cases (85.4%) reported having eaten shellfish and 72 (69.9%) specifically oysters. HAV RNA was detected and sequenced successfully in 80.2% (69/86) of the cases, as well as in one oyster out of 20 shellfish samples. Phylogenetic analysis revealed that all isolates belonged to a single genotype IA but presented the co-circulation of five distinct genomic sub-lineages. The oyster-derived HAV strain shared over 98.2% nucleotide identity with all clinical strains obtained during the epidemic, particularly 100% homology with the strains of seven cases. These data indicated that contaminated oyster consumption was probably a common source of this epidemic, although multiple HAV strains were involved. We recommend strengthening shellfish surveillance, changing dietary habits in seafood consumption, and encouraging vaccination for target adults in coastal areas with a high prevalence of hepatitis A.
KEYWORDS: Hepatitis A virus, epidemic, risk factors, phylogenetic analysis, genotype, shellfish
Introduction
Hepatitis A, caused by the hepatitis A virus (HAV), is a common cause of acute hepatitis infections worldwide. Hepatitis A occurs both sporadically and in the form of outbreaks with an average incubation period of 28–30 days (range 15–50 days).1 HAV is primarily transmitted through the fecal-oral route by either person-to-person contact or consuming contaminated food or water.2 Due to the long incubation period of HAV, it is challenging to determine an association between the source of infection and illness, particularly in sporadic cases.3 Outbreaks of hepatitis A commonly involve the consumption of contaminated water and food in both developing and developed countries.4 According to previous reports, the main foods contaminated with HAV corresponded to berry fruits, pomegranate arils, leafy vegetables, green onions, and especially bivalve shellfish.5–10
In China, hepatitis A was a serious public health concern long ago. In 1988, a large epidemic of hepatitis A linked to the consumption of raw clams occurred in Shanghai, resulting in more than 310,000 cases infected and over 8000 patients hospitalized.11 Benefiting from the improvements in hygiene conditions and the introduction of hepatitis A vaccine (HepA) over the past few decades, the incidence of reported cases of hepatitis A in China has declined dramatically from 56 cases per 100 000 population in 1991 to 1.38 cases per 100 000 population in 2019.12,13 Nevertheless, local clusters or outbreaks of hepatitis A have occasionally been reported in recent years, especially in remote rural areas.14
Hepatitis A is a notifiable disease in China, with national coverage of clinical and laboratory surveillance. A total of 110 cases of hepatitis A with onsets from 1 January to 31 March 2020 were reported by the Chinese National Notifiable Disease Reporting System (NNDRS) in Yantai, a coastal city in Shandong province, eastern China. The number of hepatitis A cases was distinctly higher than that in the same period for the preceding 4 years. An investigation was carried out to characterize the epidemic, identify the probable contamination source, and outline the targeted control measures. The paper describes the findings of the investigation.
Materials and methods
Epidemiological investigation
A hepatitis A case was identified on the basis of the National Diagnostic Criteria for Viral Hepatitis A (WS 298-2008, China). A confirmed case was characterized by the positivity for anti-HAV IgM antibodies in serum. Cases were routinely reported by the hospitals to the NNDRS within 24 hours after diagnosis. Given that reported cases began to increase on 1 January and peaked in February, then declined to background levels in early April of 2020, all confirmed cases living in Yantai with onsets from 1 January to 31 March 2020 were included in the study. The geographic distribution map of the reported cases of hepatitis A in 13 counties (districts) of Yantai during this period was created using ArcGIS V.10.8 software.
The cases were interviewed in a face-to-face meeting or by telephone using a standard questionnaire by the staff of the local Center for Disease Control and Prevention (CDC) at the county level. The questionnaire included demographics, clinical characteristics, and possible exposures of infection 30 days before the onset of symptoms (drinking water, vegetable and fruit consumption, seafood consumption, and travel history). This study was approved by the Ethics Committee of the National Institute for Viral Disease Control and Prevention, Chinese Center for Disease Control and Prevention.
Virological analysis of clinical samples
Serum samples from 86 cases with hepatitis A during the epidemic were collected in the hospitals and then transferred to the provincial CDC for HAV sequencing. HAV RNA was extracted from 140 μl serum with QIAamp viral RNA extraction Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions. The VP1/2A junction region of HAV was amplified by a nested reverse-transcription polymerase chain reaction (RT-PCR) as previously described.15,16 All necessary precautions were strictly followed to prevent cross-contamination, and negative controls were included in RNA extraction and PCR amplification
Food investigation
The consumption of contaminated shellfish is generally recognized as the most relevant risk factor of epidemic hepatitis A in coastal regions. Yantai is located on the Jiaodong Peninsula of China, where seafood products are abundant. During the period November to April, a variety of fresh seafood is consumed by the residents. Moreover, most people, especially those living in the northern area of Yantai (such as Penglai, Changdao, Longkou, and Kaifaqu), have the habit of eating raw/undercooked shellfish (mainly oysters). Trace-back investigation of the study also focused on shellfish consumption. In order to identify the origin of the suspected shellfish contamination, a total of 20 samples including three species of bivalve mollusks were collected from ten local seafood markets in three counties (Changdao, Laishanqu and Penglai). These selected markets were where some of the HAV cases had purchased shellfish before the illness onset, or where shellfish products were widely sold before the epidemic. Of these, 14 were oysters, 3 were mussels and 3 were scallops. Randomly selected samples were packaged individually and dispatched to the laboratory under chilled conditions. Processing of shellfish was carried out essentially by the methods described in ISO 15216-1: 2017.17
Total RNA was extracted from 280 μl of shellfish homogenate with QIAamp viral RNA extraction Kit (QIAGEN, Hilden, Germany). HAV RNA was tested by using a TaqMan Real-time Reverse Transcriptional Polymerase Chain Reaction (RT-qPCR) kit (Jinhao Biological, Beijing, China) in a Real-Time PCR System (ABI 7500, USA). In parallel, a nested RT-PCR of the VP1-2A junction was also amplified. The primers and thermal profile were the same as molecular detection of clinical samples conducted in this study.
Nucleotide sequencing and phylogenetic analysis
Sequencing of positive RT-PCR products was completed in both directions (Sango Biotech, Shanghai, China). Sequences obtained in this study were first aligned using Mega X software,18 together with 19 reference sequences available from GenBank. A 336 nt region (positions 2926-3261 in the HM-175 reference sequence, GenBank accession no. M14707) of the VP1/2A junction was chosen for genotyping and phylogenetic analysis. The maximum likelihood algorithm with 1000 bootstrap replicates implemented in Mega X was used to construct a phylogenetic tree. In addition, sequences were blasted in GenBank to search for strains with the highest similarity. All sequences obtained in this study have been deposited in the National Center for Biotechnology Information GenBank database (accession nos. ON131030-ON131070).
Results
Epidemiological investigation
A total of 110 confirmed cases of HAV were reported through NNDRS in Yantai, with onsets ranging from 1 January to 31 March 2020. The cases came from 12 counties (districts) of Yantai, of which 37.3% (41/110) resided in Penglai, followed by Longkou (13.6%,15/110), Kaifaqu (10.0%,11/110) and Changdao (8.2%, 9/110). The remaining 34 cases came from other 8 counties (districts) (Figure 1). Compared with the regional historical notifications of the same period for the preceding four years, the number of HAV cases increased steeply (Figure 2).
Figure 1.
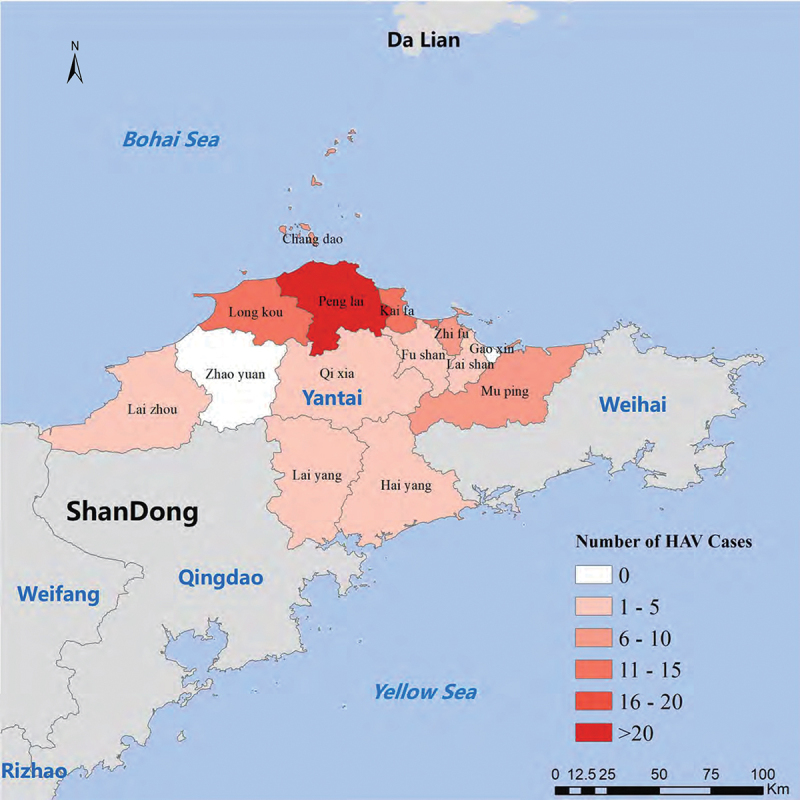
Geographic distribution map of the reported cases of hepatitis A epidemic by county (district) in Yantai, Shandong province, China, onsets of 1 January to 31 March 2020.
Figure 2.
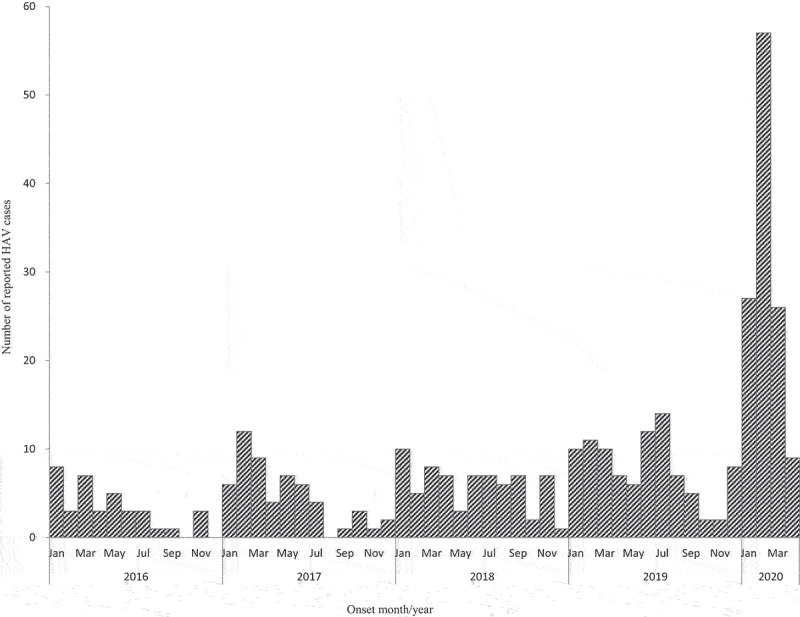
Number of reported cases of hepatitis A by month of onset in Yantai, China, January 2016-April 2020. Bars indicate the number of reported hepatitis A cases.
In total, 103 patients involving 74 males and 29 females were interviewed. The median age of interviewed cases was 41 years (range: 25–70 years). The main clinical signs and symptoms were fever (68.9%), nausea (66.0%), jaundice (41.7%), anorexia (36.9%), and vomiting (20.4%). Eighty-three (82.5%) cases were hospitalized and no death was recorded.
Information about risk exposure was documented in all 103 cases for the 30 days before the onset of symptoms. Ninety-nine (96.1%) cases reported having eaten seafood. Among them, 88 (85.4%) cases had a history of shellfish consumption, and 72 (69.9%) cases specifically having a history of oyster consumption. The consumption of raw vegetable or fruit was documented for 30 cases (29.1%). Only one (1.0%) case reported consuming nonboiled water, and four (3.9%) cases reported traveling to other provinces in China. The demographic, clinical and risk factors of the cases are summarized in Table 1.
Table 1.
Demographic, clinical, and risk factors of hepatitis A cases with onsets from 1 January to 31 March 2020 in Yantai, China (n = 103).
| Variable | |
|---|---|
| Gender, n (%) | |
| Male | 74 (71.8) |
| Female | 29 (28.2) |
| Age (median; range in years) | 41;25-70 |
| Presenting symptom, n (%) | |
| Fever | 71 (68.9) |
| Nausea | 68 (66.0) |
| Vomiting | 21 (20.4) |
| Jaundice | 43 (41.7) |
| Anorexia | 38 (36.9) |
| Biochemical, mean ± SD | |
| ALT (IU/L, n.v. 5–40) | 1891 ± 1715 |
| Exposuresa, n (%) | |
| Seafood consumption | 99 (96.1) |
| Shellfish consumption | 88 (85.4) |
| Oyster consumption | 72 (69.9) |
| Raw vegetable or fruit consumption | 30 (29.1) |
| Nonboiled water | 1 (1.0) |
| Traveling | 4 (3.9) |
SD, Standard Deviation; ALT, alanine aminotransferase; n.v.: normal values.
aExposures within 30 days before the onset of symptoms were documented.
HAV detection in clinical samples and shellfish
Of the 110 notified hepatitis A cases, serum samples from 86 cases (78.2%) were collected for genotyping/subtyping and 69 (80.2%) were successfully sequenced. To reduce the number of sequences in the final dataset to be analyzed, each of two groups of 10 and 21 identical sequences from Penglai was represented by a single sequence (sequences 2A/PL-YT/2020, 1A/PL-YT/2020 in Figure 3).
Figure 3.
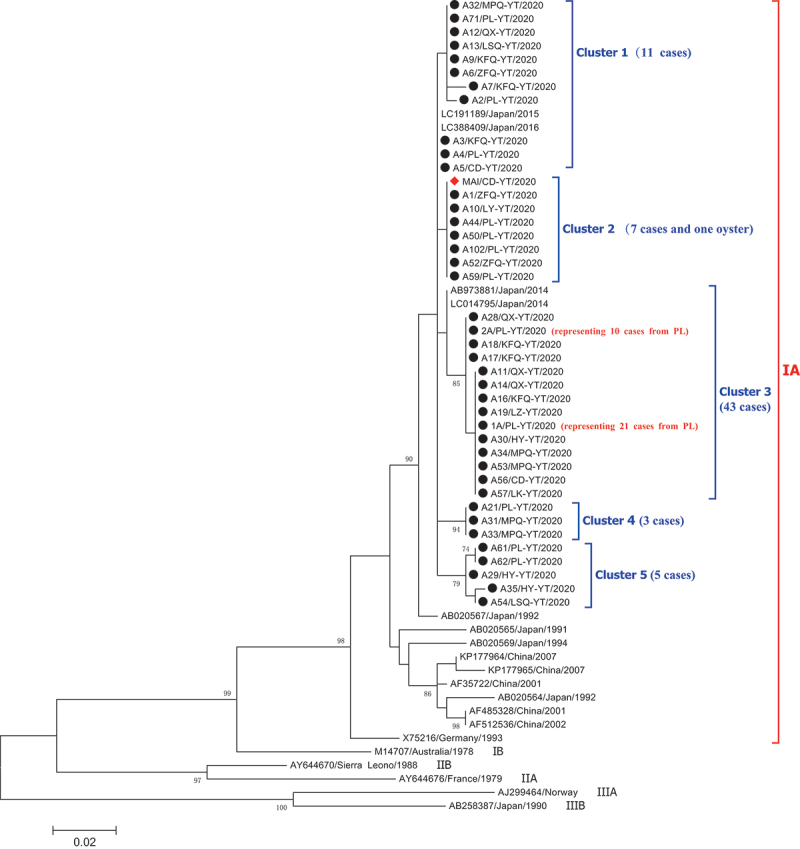
Phylogenetic analysis of the sequences for Hepatitis A virus based on the 336-nt of the VP1/2A junction (positions 2926–3261 of reference sequence M14707), representing 70 sequences from 69 cases and one sequence from an oyster in Yantai. HAV sequences from cases in this work are indicated with black circles. One sequence from an oyster is labeled a red triangle. For each sequence, the county or district (CD: Changdao; HY: Haiyang; KFQ: Kaifaqu; LK: Longkou; LSQ: Laishan; LZ: Laizhou; MPQ: Mupingqu; QX: Qixia; PL: Penglai; ZFQ: Zhifuqu), the city (YT: Yantai), and the isolation year (2020) are reported in the sequence ID. The tree was constructed with MEGA X software (http://www.megasoftware.net) by using the maximum-likelihood algorithm and the Jukes-Cantor model with 1,000 bootstrap replicates. Bootstrap values (%) >70 are indicated at branch nodes.
HAV RNA was successfully detected in 1(5.0%) of 20 shellfish samples by both real-time RT-qPCR and nested RT-PCR methods. The positive sample was derived from an oyster of 5 March 2020, which was taken from a seafood market in Changdao county of Yantai.
Phylogenetic analysis of HAV genome sequences in cases and shellfish
Phylogenetic analysis revealed that all sequences from 69 cases and one oyster belonged to a single genotype IA. However, these sequences showed some variability. Five distinct clusters (named ‘Cluster 1 to 5’) of the IA branch involving as many as 12 different HAV strains were observed after multiple sequences alignment (Figure 3).
Cluster 1 included 11 closely related sequences from the reported cases in 7 counties (districts). Intra-cluster heterogeneities (up to three nucleotide differences) were observed. Among them, six sequences from six different counties (districts) and three sequences from three counties (districts) were identical with 100% homology, respectively. It is generally believed that cases with 100% sequence identity may be considered to share a common source of infection. Further investigation revealed that although cases with 100% homology in each group had separate residences, they all had a history of eating oysters before the illness onset.
Cluster 2 included seven subtype IA sequences isolated from seven cases (4 occurred in Penglai, 2 in Zhifuqu, and 1 in Laiyang) and one sequence from an oyster in Changdao. All the seven sequences from these cases were identical, and they also showed 100% identity with the HAV RNA sequence from implicated oysters. Further investigation revealed that all seven cases (aged: 26–48 years) had consumed oysters from the local seafood market before the onset, although they resided in different places. Our study indicated that some oysters in this area had been contaminated with HAV at the time of our investigation, which likely contributed to this epidemic.
Cluster 3 was the biggest cluster and included 14 subtype IA sequences isolated from 43 cases. Most of them (n = 31) were from Penglai. Two major strains, differing for just a single nucleotide, were observed in 13 cases (living in separate residences) and 30 cases (3, 3, and 2 cases residing in the same village or community, respectively, and the remaining cases living in separate residences), respectively. Moreover, 13 sequences of the first major strain were identical, as well as the 30 sequences of the other strain, with 100% homology. Further analysis found that 11 (84.6%) of the 13 cases with the same strain and 26 (86.7%) of the other 30 cases with an identical strain also had a history of eating bivalve shellfish, especially oysters (9 of 13 cases and 22 of 30 cases, respectively). For a small number of cases with no history of shellfish consumption or living in proximity to one another, the source and chain of infection could not be identified, thus highlighting the importance of collecting more detailed epidemiological records of the cases.
Cluster 4 was a small cluster and included three subtype IA sequences. All three sequences were identical (the three cases all reported eating oysters). Cluster 5 included five subtype IA sequences. The identical sequences were found in two cases from Penglai, both of whom had also consumed oysters.
Although the 336-nt of the VP1/2A junction in 12 distinct HAV strains showed diversity with 1–18 (0.4–5.3%) nucleotide differences (Additional file 1), all nucleotide variations did not translate to amino acid substitutions, representing that all these nucleotide variations were synonymous. The oyster-derived HAV strain shared 98.2 100% nucleotide similarity with all 69 clinical strains. Based on a BLAST search, cluster 1 and cluster 2 were similar to the sequences isolated from two Japanese patients (GenBank accession nos. LC191189 and LC388409), with an identity of 99.7–100% and 99.7%, respectively. Cluster 3 shared 99.1–99.4% nucleotide similarity between two sequences (GenBank accession nos. AB973881, LC014795) from Japanese patients.
Discussions
This study addressed a community-wide epidemic of hepatitis A in Yantai, eastern China, focusing on epidemiologic characteristics, risk exposure information, and molecular characteristics of isolated sequences among clinical cases and implicated shellfish. Comparative analysis based on sequencing is becoming an increasingly important approach for the investigation of food-borne hepatitis A.19–22 In this study, the epidemiologic investigation and molecular sequencing confirmed a very strong association between this epidemic and the consumption of contaminated oysters, although only one sequence of oysters could be analyzed. HAV testing from food is not part of the routine analysis in most countries, primarily because the ability to identify HAV in foods is hampered by the disease’s long-term incubation period. Hence, it is difficult to recover related foods for HAV testing. To the best of our knowledge, this is the first to identify the occurrence of an identical HAV sequence from both clinical patients and implicated oysters in China. Over 85% of reported cases were from the northern part of Yantai, which may be explained by the fact that the habit of eating raw/undercooked shellfish (especially oysters) is more common in this region. In addition, all confirmed cases of hepatitis A were over 20 years old, and none were in children under 15 years. The reason might be attributable primarily to the beneficial impact of universal childhood vaccination policies against hepatitis A in China since 2008 and the fact that HAV illness in children under 6 years of age is generally asymptomatic.23
A specificity analysis of the VP1/2A genomic region of HAV, commonly used for molecular genotyping and outbreak investigations,24 confirmed that all isolates in this study belonged to a unique genotype IA. The finding was consistent with the commonly detected sub-genotype in China.16,25,26 Also, the strains identified in this study shared high sequence similarities with several HAV IA strains circulating in Japan. In general, HAV is considered to be a virus with low antigenic variability. Most hepatitis A outbreaks are characterized by a common point origin, generally sharing 100% homology for all the sequences.25 However, five distinct phylogenetic clusters, and as many as 12 different HAV strains, were observed simultaneously during this epidemic, likely reflecting the co-circulation of several different HAV genotype IA strains in this region. It should be noted that all these nucleotide variations of HAV were synonymous. The present study revealed that the oyster-derived HAV strain shared over 98.2% nucleotide identity with all clinical strains, particularly 100% homology with the strains of approximately 10% of cases. On the other hand, as many as seven distinct groups in five clusters showed 100% nucleotide homology. Of these, all cases in five groups (21 cases in total) and more than 70% cases in the other two groups (43 cases in total) reported having a history of oyster consumption before the onset of illness. These results indicate that oysters in this region were probably contaminated by multiple HAV strains during the epidemic. Due to the fact that the sampling time was late concerning the date of the possible shellfish contamination, the sample size was small, and the detection rate of HAV in shellfish was relatively low. Unfortunately, only one HAV strain from implicated oysters was available, which might not represent the most dominant circulating strains during the epidemic. In addition, the potential ability of HAV to exist in the form of quasispecies within the viral populations and thus induce genetic mutants should not be overlooked.
Outbreaks of hepatitis A attributed to the consumption of contaminated shellfish, and oysters in particular, have been well documented in many countries or regions.27–31 Shellfish have special abilities to concentrate any pollutants in their tissues. If pathogenic microorganisms appear in the harvesting water, shellfish can accumulate these pathogens considerably. A study on the bioaccumulation of viral indicators demonstrated that oysters could concentrate viruses up to 99-fold compared to the surrounding waters.32 However, the precise cause of HAV contamination in oysters in this region remains under investigation. According to previous reports, the most likely source of viral contamination is generally attributed to the illegal discharge of various wastes into harvest waters or the improper storage of oysters in the guest harbor.27,31
Of note, as seen in Yantai, the incidence of hepatitis A in Dalian city of Liaoning province, China, which is facing Yantai across the Bohai Sea, had also increased rapidly during the same period.33 The Bohai Sea is a nearly closed inland sea and belongs to the category of inland water. These findings indicated that the products of shellfish, especially oysters growing in the part of Bohai off-shore region, were likely to have been contaminated to a certain extent at the same time.
HAV infection is vaccine-preventable. HepA has been available since 1992 in China and was included in the National Childhood Expand Program on Immunization in 2008. The HepA coverage was consistently greater than 98% among the target children since 2008.34 Live and inactivated HepA are both offered in China. The wider use of HepA in children is primarily responsible for the around 80% decline in the incidence of HAV compared to the time prior to the implementation of program.34 Compared with the reported HAV incidence in inland areas of Shandong province, the incidence of Yantai has been at a relatively high level, with 2.16 per 100,000 population in 2017 and 0.53–1.32 per 100,000 population from 2008 to 2019. Moreover, the reported HAV cases after 2008 largely occurred in individuals over the age of 20. A previous study conducted in Shandong province showed that the seroprevalence of anti-HAV among young adults in 2014 was relatively low, so they would be susceptible to HAV infection.35 In addition, hepatitis A cases in adults generally present symptomatic and are prone to cause acute hepatic damage. Currently, HepA is not routinely recommended for adults living in high-incidence coastal areas in China. Accordingly, HepA for target adults in such special regions becomes important and should be encouraged.
Some limitations of our study should also be taken into consideration. First, a routine method of case–control study was not conducted in the field investigation because of the COVID-19 epidemic during the study period, which may limit the inference of the exact source of infection from an epidemiological perspective. Second, the quality of information collected by the cases, especially the precise details of the food consumption, may be impacted by recall bias due to the long incubation period of the disease. Third, only one HAV strain from oysters was obtained for analysis in this study, which may not be representative of the dominant circulating strains. Despite these problems, the presence of HAV RNA in oysters provided definitive evidence that contaminated oysters were most likely the vehicle for hepatitis A transmission in this epidemic. Ongoing surveillance of HAV contamination in shellfish products in this region is necessary to alert the public promptly.
In conclusion, our findings highlight the fact that contaminated oyster consumption is the most likely common source of this epidemic, although multiple HAV strains are involved. These data could help to improve risk assessment of shellfish-borne diseases and recommend establishing a comprehensive surveillance system to reduce the risk of HAV transmission. In addition, educational campaigns aimed at changing dietary habits in seafood consumption and encouraging HAV vaccination to target adults in coastal areas with a high prevalence of hepatitis A should also be recommended. These measures were adopted in Yantai in late February and March 2020, and then the epidemic was controlled.
Acknowledgments
We are grateful to all the relevant public health workers from CDCs of 12 counties (districts) of Yantai for their assistance with this work.
Funding Statement
This work was supported by grants from Taishan Scholar Program of Shandong Province [No. ts201511105], Shandong Medical Health Science and Technology Development Program [No. 2018WS308], and Science&Technology Innovation and Development Plan of Yantai City [No. 2022MSGY069].
Disclosure statement
No potential conflict of interest was reported by the author(s).
Author contributions
Li Zhang and Aiqiang Xu contributed to the study concept and design. Bingyu Yan, Peng Chen, Jingjing Lu, Xin Meng, and Qing Xu contributed to data acquisition. Peng Chen contributed to the sample collection. Bingyu Yan and Yi Feng were responsible for the hepatitis A experiment. Bingyu Yan and Peng Chen contributed to the data analysis and initial drafting of the manuscript. Li Zhang contributed to the critical review and revision of the article. All authors contributed to the interpretation of data and critical revision of the manuscript.
References
- 1.Castrodale L, Beller M, Middaugh J, McCumber B, Mynes-Spink K, Bernth G.. Use of liver function tests to predict the magnitude of an outbreak of Hepatitis A. Clin Infect Dis. 2002;35(1):1–8. doi: 10.1086/340864. [DOI] [PubMed] [Google Scholar]
- 2.Lemon SM. Type a viral hepatitis. New developments in an old disease. N Engl J Med. 1985;313(17):1059–1067. doi: 10.1056/NEJM198510243131706. [DOI] [PubMed] [Google Scholar]
- 3.Di Cola G, Fantilli AC, Pisano MB, Ré VE. Foodborne transmission of hepatitis A and hepatitis E viruses: a literature review. Int J Food Microbiol. 2021;338:108986. doi: 10.1016/j.ijfoodmicro.2020.108986. [DOI] [PubMed] [Google Scholar]
- 4.Abutaleb A, Kottilil S. Hepatitis A: epidemiology, natural history, unusual clinical manifestations, and prevention. Gastroenterol Clin North Am. 2020;49(2):191–199. doi: 10.1016/j.gtc.2020.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tavoschi L, Severi E, Fau-Niskanen T, Niskanen T, Fau-Boelaert F, Boelaert F, Fau -Rizzi V, Rizzi V, Fau-Liebana E, Liebana E, et al. Food-borne diseases associated with frozen berries consumption: a historical perspective, European Union, 1983 to 2013. Euro Surveill. 2015;20(29):21193. doi: 10.2807/1560-7917.ES2015.20.29.21193. [DOI] [PubMed] [Google Scholar]
- 6.Ruscher C, Faber M, Werber D, Stark K, Bitzegeio J, Michaelis K, Sagebiel D, Wenzel JJ, Enkelmann J. Resurgence of an international hepatitis A outbreak linked to imported frozen strawberries, Germany, 2018 to 2020. Euro Surveill. 2020;25(37). doi: 10.2807/1560-7917.ES.2020.25.37.1900670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collier MG, Khudyakov YE, Selvage D, Adams-Cameron M, Epson E, Cronquist A, Jervis RH, Lamba K, Kimura AC, Sowadsky R, et al. Outbreak of hepatitis A in the USA associated with frozen pomegranate arils imported from Turkey: an epidemiological case study. Lancet Infect Dis. 2014;14(10):976–981. doi: 10.1016/S1473-3099(14)70883-7. [DOI] [PubMed] [Google Scholar]
- 8.Rosenblum LS, Mirkin IR, Allen DT, Safford S, Hadler SC. A multifocal outbreak of hepatitis A traced to commercially distributed lettuce. Am J Public Health. 1990;80(9):1075–1079. doi: 10.2105/AJPH.80.9.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wheeler C, Vogt TM, Armstrong GL, Vaughan G, Weltman A, Nainan OV, Dato V, Xia G, Waller K, Amon J, et al. An outbreak of hepatitis A associated with green onions. N Engl J Med. 2005;353(9):890–897. doi: 10.1056/NEJMoa050855. [DOI] [PubMed] [Google Scholar]
- 10.Pintó RM, Costafreda MI, Bosch A. Risk assessment in shellfish-borne outbreaks of hepatitis A. Appl Environ Microbiol. 2009;75(23):7350–7355. doi: 10.1128/AEM.01177-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooksley WG. What did we learn from the Shanghai hepatitis A epidemic? J Viral Hepat. 2000;7(Suppl 1):1–3. doi: 10.1046/j.1365-2893.2000.00021.x. [DOI] [PubMed] [Google Scholar]
- 12.Wang F, Sun X, Wang F, Zheng H, Jia Z, Zhang G, Bi S, Miao N, Zhang S, Cui F, et al. Changing epidemiology of hepatitis A in China: evidence from three national serological surveys and the national notifiable disease reporting system. Hepatology. 2021;73(4):1251–1260. doi: 10.1002/hep.31429. [DOI] [PubMed] [Google Scholar]
- 13.Miao Z. Incidence trend and spatial correlation of hepatitis A in Chinese mainland, 2002-2019 [in Chinese]. Henan J Prev Med. 2021;32:325–330. [Google Scholar]
- 14.Yu P, Huang L, Li H, Liu M, Zong J, Li C, Chen F. Epidemiological investigation of an outbreak of hepatitis A in rural China. Int J Infect Dis. 2015;33:191–195. doi: 10.1016/j.ijid.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 15.Tallo T, Norder H, Tefanova V, Ott K, Ustina V, Prukk T, Solomonova O, Schmidt J, Zilmer K, Priimägi L, et al. Sequential changes in hepatitis A virus genotype distribution in Estonia during 1994 to 2001. J Med Virol. 2003;70(2):187–193. doi: 10.1002/jmv.10377. [DOI] [PubMed] [Google Scholar]
- 16.Cao J, Wang Y, Song H, Meng Q, Sheng L, Bian T, Mahemuti W, Yierhali A, Omata M, Bi S, et al. Hepatitis A outbreaks in China during 2006: application of molecular epidemiology. Hepatol Int. 2009;3(2):356–363. doi: 10.1007/s12072-008-9116-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Standardization IOf . Microbiology of the food chain-horizontal method for determination of hepatitis A virus and norovirus using real-time RT-PCR-part 1: method for quantification. 2017. https://www.iso.org/standard/65681.html.
- 18.Kumar S, Stecher G, Li M, Knyaz C, Tamura KMX, Battistuzzi FU. Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haddix M, Civen R, Hacker JK, Probert W, New S, Green N, Hemarajata P, Gounder P. Use of molecular epidemiology to inform response to a hepatitis A outbreak — Los Angeles County, California, October 2018–April 2019. MMWR Morb Mortal Wkly Rep. 2020;69(26):820–824. doi: 10.15585/mmwr.mm6926a3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu X, Zhou W, Zhang L, Lv J, Yan B, Zhou Y, Hu W, Dong Y, Chen B, Liu M, et al. Implementing sequencing-based surveillance in developing countries: findings from a pilot rollout for hepatitis A in China. Ann Transl Med. 2021;9(14):1119. doi: 10.21037/atm-21-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nainan OV, Xia G, Vaughan G, Margolis HS. Diagnosis of hepatitis A virus infection: a molecular approach. Clin Microbiol Rev. 2006;19(1):63–79. doi: 10.1128/CMR.19.1.63-79.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Riess M, Enkirch T, Sundqvist L, Lundberg Ederth J. High impact of molecular surveillance on hepatitis A outbreak case detection in Sweden: a retrospective study, 2009 to 2018. Euro Surveill. 2021;26(9):1900763. doi: 10.2807/1560-7917.ES.2021.26.9.1900763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guzman-Holst A, Luna-Casas G, Burguete Garcia A, Madrid-Marina V, Cervantes-Apolinar MY, Andani A, Huerta-Garcia G, Sánchez-González G. Burden of disease and associated complications of hepatitis A in children and adults in Mexico: a retrospective database study. PLoS One. 2022;17(5):e0268469. doi: 10.1371/journal.pone.0268469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robertson BH, Jansen RW, Khanna B, Totsuka A, Nainan OV, Siegl G, Widell A, Margolis HS, Isomura S, Ito K, et al. Genetic relatedness of hepatitis A virus strains recovered from different geographical regions. J Gen Virol. 1992;73(6):1365–1377. doi: 10.1099/0022-1317-73-6-1365. [DOI] [PubMed] [Google Scholar]
- 25.Wang H, Chen W, Zhou W, Qiu F, Yin W, Cao J, Gao P, Yuan Q, Lv M, Bai S, et al. Exploration of a new hepatitis A surveillance system in Beijing, China: based on molecular epidemiology. BMC Infect Dis. 2022;22(1):22. doi: 10.1186/s12879-021-06872-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang H, Wang XY, Zheng HH, Cao JY, Zhou WT, Bi SL. Evolution and genetic characterization of hepatitis A virus isolates in China. Int J Infect Dis. 2015;33:156–158. doi: 10.1016/j.ijid.2015.01.022. [DOI] [PubMed] [Google Scholar]
- 27.Bialek SR, George PA, Xia GL, Glatzer MB, Motes ML, Veazey JE, Hammond RM, Jones T, Shieh YC, Wamnes J, et al. Use of molecular epidemiology to confirm a multistate outbreak of hepatitis A caused by consumption of oysters. Clin Infect Dis. 2007;44(6):838–840. doi: 10.1086/511874. [DOI] [PubMed] [Google Scholar]
- 28.Bellou M, Kokkinos P, Vantarakis A. Shellfish-borne viral outbreaks: a systematic review. Food Environ Virol. 2013;5(1):13–23. doi: 10.1007/s12560-012-9097-6. [DOI] [PubMed] [Google Scholar]
- 29.Shieh YC, Khudyakov YE, Xia G, Ganova-Raeva LM, Khambaty FM, Woods JW, Veazey JE, Motes ML, Glatzer MB, Bialek SR, et al. Molecular confirmation of oysters as the vector for hepatitis A in a 2005 multistate outbreak. J Food Prot. 2007;70(1):145–150. doi: 10.4315/0362-028X-70.1.145. [DOI] [PubMed] [Google Scholar]
- 30.Conaty S, Bird P, Bell G, Kraa E, Grohmann G, McAnulty JM. Hepatitis A in New South Wales, Australia, from consumption of oysters: the first reported outbreak. Epidemiol Infect. 2000;124(1):121–130. doi: 10.1017/S0950268899003386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guillois-Bécel Y, Couturier E, Le Saux JC, Roque-Afonso AM, Le Guyader FS, Le Goas A, Pernès J, Le Bechec S, Briand A, Robert C, et al. An oyster-associated hepatitis A outbreak in France in 2007. Euro Surveill. 2009;14(10):19144. doi: 10.2807/ese.14.10.19144-en. [DOI] [PubMed] [Google Scholar]
- 32.Westrell T, Dusch V, Ethelberg S, Harris J, Hjertqvist M, Jourdan-da Silva N, Koller A, Lenglet A, Lisby M, Vold L, et al. Norovirus outbreaks linked to oyster consumption in the United Kingdom, Norway, France, Sweden and Denmark, 2010. Euro Surveill. 2010;15(12). doi: 10.2807/ese.15.12.19524-en. [DOI] [PubMed] [Google Scholar]
- 33.Jing Sun YL, Fang X, Wang Y, Han Y, Liu Y, Cao J, Zhou W, Zheng H, Yao W. An epidemic of hepatitis A- Liaoning Province, 2020. China CDC Weekly;2020;2:570–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun X, Wang F, Zheng H, Miao N, Yuan Q, Cui F, Yin Z, Zhang G, Levine H. The impact of expanded program on immunization with live attenuated and inactivated hepatitis A vaccines in China, 2004–2016. Vaccine. 2018;36(10):1279–1284. doi: 10.1016/j.vaccine.2018.01.043. [DOI] [PubMed] [Google Scholar]
- 35.Yan BY, Lv JJ, Liu JY, Feng Y, Wu WL, Xu AQ, Zhang L. Changes in seroprevalence of hepatitis A after the implementation of universal childhood vaccination in Shandong Province, China: a comparison between 2006 and 2014. Int J Infect Dis. 2019;82:129–134. doi: 10.1016/j.ijid.2019.03.005. [DOI] [PubMed] [Google Scholar]


