Abstract
Haemophilus ducreyi expresses 2 OmpA homologs, designated MOMP and OmpA2, whose genes are arranged in tandem on the chromosome. Northern blot analysis indicated that momp and ompA2 are transcribed independently. Sequences of the momp open reading frame (ORF) lacking the transcriptional start site were amplified by PCR, and an Ω-Km2 cassette was ligated into the ORF. A plasmid containing this construction was electroporated into H. ducreyi 35000HP, and an isogenic MOMP-deficient mutant (35000HP-SMS2) was generated by allele exchange. In Southern blotting, 35000HP-SMS2 contained one copy of the Ω-Km2 cassette in momp. 35000HP and 35000HP-SMS2 had similar outer membrane protein (OMP) and lipooligosaccharide profiles and growth rates except for up-regulation of a putative porin protein in the mutant. Five subjects were inoculated with three doses of live 35000HP-SMS2 on one arm and two doses of live 35000HP and one dose of a heat-killed control on the other arm in a double-blind escalating dose-response trial. Pustules developed at 7 of 10 sites inoculated with 35000HP and at 6 of 15 sites inoculated with 35000HP-SMS2 (P = 0.14). 35000HP and 35000HP-SMS2 were recovered at similar rates from daily surface cultures and semiquantitative cultures. The data suggest that expression of MOMP is not required for pustule formation by H. ducreyi in the human model of infection.
Haemophilus ducreyi is the etiological agent of chancroid, a sexually transmitted genital ulcerative disease that facilitates the transmission of the human immunodeficiency virus (11, 34). H. ducreyi expresses 2 OmpA homologs, designated MOMP (major outer membrane protein) and OmpA2. Depending on gel conditions, both MOMP and OmpA2 may migrate as three bands in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with apparent molecular masses of 37 to 39 kDa and a heat-modifiable 43-kDa species (17, 30). MOMP is estimated to be four to five times more abundant in the outer membrane than OmpA2. The genes encoding MOMP and OmpA2 are separated by 285 bp on the chromosome and share 74% identity (17, 30). Their tandem arrangement on the chromosome may be due to a gene duplication event. Only one other bacterium, Aeromonas salmonicida, has been reported to express two OmpA-like proteins (8).
OmpA proteins are highly conserved in many gram-negative bacteria. OmpA proteins are thought to associate directly with peptidoglycan via their C termini and maintain cell wall integrity and normal cell shape (10, 18, 29). Other known functions of OmpA proteins include serving as phage and colicin receptors (6, 9). The OmpA of Escherichia coli K1 contributes to serum resistance and interacts with glycoproteins expressed on bovine brain microvascular endothelial cells (23, 35). Loss of expression of OmpA decreases E. coli K1 invasiveness 25- to 50-fold (24). E. coli K1 OmpA-deficient mutants are attenuated in the embryonic-chicken and neonatal-rat models of E. coli K1 infection (35). The OmpA homolog of Actinobacillus actinomycetemcomitans, Omp34, is an Fc-γ receptor and may serve to interfere with immunoglobulin-mediated complement fixation or opsonization (19). The PIII protein of Neisseria gonorrhoeae has sequence homology with the E. coli OmpA protein (12). PIII binds blocking antibodies that interfere with the bactericidal activity of antibodies directed to other surface antigens (25). The P5 outer membrane protein of nontypeable Haemophilus influenzae, another OmpA homolog, plays a role in the adherence of nontypeable H. influenzae to mucus and respiratory epithelial cells (28). Immunization with synthetic peptides derived from P5 sequences enhances clearance of H. influenzae in a chinchilla model of otitis media (4). Thus, OmpA proteins may play a role in pathogenesis.
Our laboratories had constructed an H. ducreyi MOMP-deficient mutant (35000.60) by insertion of a chloramphenicol acetyltransferase gene (cat) cassette into the momp open reading frame (ORF) in the same orientation as the momp and ompA2 promoters (17). In Western blotting, monoclonal antibody (MAb) 2C7, which binds to both MOMP and OmpA2, reacts strongly with the 43-kDa band in 35000.60. The increased expression of the 43-kDa band could have been the result of a compensatory increase in OmpA2 expression due to the loss of MOMP or could have resulted from increased expression of OmpA2 due to the presence of the cat cassette in the momp gene. Alternatively, the 43-kDa protein may represent a third protein that binds MAb 2C7. 35000.60 is also more sensitive to the bactericidal effects of normal human serum than the parent, 35000 (15).
In an effort to understand the early events of H. ducreyi pathogenesis, our laboratory developed a human challenge model of H. ducreyi infection (32). Human volunteers are inoculated on the skin of the upper arm via puncture wounds made by an allergy-testing device. The subjects are observed until they achieve a clinical endpoint, defined as the resolution of disease at all sites, development of a painful pustule that is likely to ulcerate, or an infection for 14 days. In the model, inoculation of an estimated delivered dose (EDD) of approximately 27 (95% confidence interval [CI], 14 to 40) CFU results in a pustule formation rate of approximately 50%. Inoculation of approximately 55 (95% CI, 34.9 to 75.3) and 100 (95% CI, 55 to 144) CFU results in pustule formation rates of approximately 70 and 90%, respectively (2). Previous studies have shown that disease outcome (papule, pustule, or resolution) at each site in an individual subject inoculated at multiple sites with identical suspensions of bacteria is independent (3, 31). Therefore, in parent-mutant comparisons, subjects are inoculated with multiple doses of the parent and mutant, and site rather than subject is used as the unit of analysis.
In this study, we examined whether momp and ompA2 were independently transcribed. Also, we report the construction and characterization of a new isogenic MOMP-deficient mutant (35000HP-SMS2) in a human-passaged variant of H. ducreyi 35000 (35000HP). We evaluated the role of MOMP in infection by comparing the isogenic MOMP-deficient mutant and 35000HP in the human model of H. ducreyi infection. To our knowledge, this is the first study examining the role of an OmpA protein during infection of human subjects.
MATERIALS AND METHODS
Bacteria and culture conditions.
H. ducreyi 35000HP is a human-passaged variant of 35000 obtained after 13 days of experimental infection with 35000 (3, 31). All H. ducreyi strains were grown on chocolate agar (CA) plates supplemented with 1% IsoVitaleX and incubated at 35°C with 5% CO2 or in broth consisting of proteose peptone, 50 μg of hemin per ml, 1% IsoVitaleX, and 5% heat-inactivated fetal calf serum. Where appropriate, H. ducreyi was grown on CA plates containing kanamycin (20 μg/ml) or vancomycin (3 μg/ml).
PCR.
Genomic DNA isolated from H. ducreyi 35000HP was used as a template for PCR. Synthetic primers were used to amplify the coding sequences. The primers MOMP-For-1 (5′-AGCGGGATCCGGAACAAGAGTGTGTTGTTATTATG-3′) and MOMP-Rev-1 (5′-ACACGAATTCAATGACCGATTACATTGTTACTTCT-3′) were used to amplify momp, and OMPA2-For-4 (5′-GAGGTACCGCGCCACAAGCGGATACTTTTTAT-3′) and OMPA2-Rev-4 (5′-GCTTAAGCGTGGTTTATCTCTTACATTCGCTACA-3′) were used to amplify ompA2. The resulting products were 1.4 and 1.2 kb, respectively. Target DNA was amplified in reactions with a final concentration of the following reagents: 2.5 U of Taq 2000 polymerase (Stratagene, La Jolla, Calif.), MgCl2 (2.25 mM), deoxynucleoside triphosphates (25 μM), 1× reaction buffer (Perkin-Elmer, Branchburg, N.J.), and the appropriate primers (10 pM). Reaction mixtures were held at 94°C for 4 min and then cycled 30 times (94°C for 1 min, 60°C for 1 min, 72°C for 2 min) and held at 4°C using a thermal cycler (Perkin-Elmer PCR System 2400). All PCR products were analyzed by electrophoresis on 0.8% agarose gels and by ethidium bromide staining.
Plasmids.
The plasmid pMOMP1.2, which contains a promoterless momp ORF, was described previously (17). Robert S. Munson, Jr. (Ohio State University, Columbus), kindly provided pRSM1515, which contains the Ω-Km2 cassette. The Ω-Km2 cassette, which encodes kanamycin resistance, was excised with BamHI and ligated into a BglII site located 520 bp downstream of the start codon of the momp ORF in pMOMP1.2 to make the plasmid pMOMP1.3.
MAbs.
MAb 3F12 binds specifically to MOMP and was described previously (17). MAb 2C7 binds to epitopes present on both MOMP and OmpA2 and has been described previously (30).
Northern blot analysis.
35000HP or 35000HP-SMS2 was grown in 10 ml of broth to mid-logarithmic growth (optical density at a wavelength of 660 nm, 0.2). The broth culture was centrifuged at 1,000 × g for 10 min to pellet the bacteria. RNA was extracted from the pellet in 2 ml of Tri-Reagent (Molecular Research Center, Cincinnati, Ohio) according to the manufacturer's instructions. Approximately 15 μg of total RNA was analyzed by electrophoresis on a 1.2% agarose gel with formaldehyde. RNA was transferred overnight to a nylon membrane by a standard capillary method (26).
momp and ompA2 probes were made by amplifying the ORF of each gene by PCR using the primers MOMP-For-1 and MOMP-Rev-1 and OmpA2-For-4 and OmpA2-Rev-4, respectively. DNA probes were labeled by incorporation of [γ-32P]dCTP by random priming with a High Prime DNA labeling kit (Boehringer Mannheim, Indianapolis, Ind.) according to the manufacturer's protocol. Labeled probe was purified from unincorporated [γ-32P]dCTP by NucTrap probe purification columns (Stratagene).
The membrane was incubated at 42°C for 2 to 3 h in prehybridization solution (5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]–5× Denhardts solution–salmon sperm DNA [500 μg/ml]–50% formamide). Hybridization of labeled probe was carried out at 42°C for 18 h in hybridization solution (5× SSC–1× Denhardts solution–salmon sperm DNA [100 μg/ml]–50% formamide). The membrane was washed twice with 2× SSC–0.1% SDS at 25°C, twice with 0.1× SSC–0.1% SDS at 25°C, and once with 0.1× SSC–0.1% SDS at 42°C. Labeled probe was detected by autoradiography.
LOS, whole-cell lysates, OMPs, and Western blotting.
Lipooligosaccharide (LOS), whole-cell lysates, and outer membrane proteins (OMPs) were prepared and subjected to analysis by SDS-PAGE in either 10 or 12.5% acrylamide gels as described previously (5, 17). LOS was stained with silver stain (Bio-Rad Laboratories, Hercules, Calif.). Proteins were stained with Coomassie brilliant blue or transferred to nitrocellulose as described previously (17). Western blots were probed with the MAb followed by either peroxidase-labeled protein A (Kirkegaard and Perry Laboratories, Gaithersburg, Md.) or peroxidase-labeled goat anti-mouse immunoglobulin G plus immunoglobulin M (Kirkegaard and Perry Laboratories) and horseradish peroxidase color developer (Bio-Rad Laboratories) as described previously (17). Densitometry was performed using Quantity One software (Bio-Rad Laboratories) and was performed on three independent OMP preparations.
N-terminal amino acid sequence analysis.
The N-terminal amino acid sequence of the 45-kDa protein was determined from Sarkosyl-extracted OMPs that were resolved by SDS-PAGE and transferred to a polyvinylidene difluoride membrane as described previously (17).
Colony blotting.
Isolated colonies were picked onto nitrocellulose or lifted directly from CA plates with nitrocellulose. The membrane was dried at 37°C for 1 h, and the blots were probed with MAb 3F12. Antigen-antibody complexes were detected exactly as described for Western blots.
Southern blot analyses.
Chromosomal DNA was digested with the restriction enzyme AvaI and electrophoresed on a 0.8% agarose gel. DNA was transferred to a nylon membrane by a standard capillary method as described previously. The momp probe used in Southern blot analysis was made as described above. The Ω-Km2 cassette probe was obtained by digestion of pRSM1515 with BamHI and was gel purified. We used the Renaissance random-primer fluorescein labeling kit (NEN Life Science Products, Boston, Mass.) to label the probes used in Southern blot analysis. Prehybridization, hybridization, and detection of hybridized probe were performed according to the manufacturer's instructions.
Nucleotide sequence analysis.
Nucleotide sequence analysis was performed with a model 373 automated DNA sequencer (Applied Biosystems, Foster City, Calif.) as described previously (17). Regions of DNA to be sequenced were amplified by PCR. Nucleotide sequences were analyzed using MacVector DNA analysis software (version 6.0; Oxford Molecular Group, Campbell, Calif.).
Human volunteers.
Six healthy adult females (one black, two Asian, and three white; age range, 22 to 56; mean age ± standard deviation, 35.6 ± 12.7 years) volunteered for the study. Informed consent was obtained from the subjects for participation and for human immunodeficiency virus serology, in accordance with the human experimentation guidelines of the U.S. Department of Health and Human Services and the Institutional Review Board of Indiana University-Purdue University at Indianapolis. Enrollment procedures and exclusion criteria are described elsewhere in detail (3, 32).
Human challenge protocol.
The human challenge protocol, inoculation, clinical observations, surface cultures, and study design are described in detail elsewhere (3, 31, 32). The subjects were inoculated at three sites on one arm with suspensions containing the mutant. On the other arm, two sites were inoculated with identical suspensions of 35000HP and one site was inoculated with the highest dose of the heat-killed mutant. The number of CFU in the suspension was determined prior to and after inoculating each group of subjects and was averaged. The bacteria were delivered into the skin with a Multi-Test applicator (Lincoln Diagnostics, Decatur, Ill.), which delivers approximately 1/1,000 of a solution of antigens or bacterial suspensions loaded onto its tines into the epidermis and dermis (16, 27, 31). Although we did not experimentally determine the delivered dose, the EDD was calculated to be 1,000-fold less than the average number of CFU loaded on the tines.
An escalating dose response study was used to compare the parent and mutant (1, 21, 36). In the first iteration, three subjects were enrolled. The target EDD of the parent was 50 CFU, and the target EDDs for the mutant were 25, 50, and 100 CFU. If we observed similar pustule formation rates at sites infected with both the mutant and the parent, we would challenge three more subjects in a similar manner. If the results were confirmed, we would conclude that there was no major difference in the virulence of the mutant and the parent and terminate the trial. If pustules did not develop at sites inoculated with the mutant, and one or more of the sites inoculated with the parent per subject formed pustules, we would inoculate the subjects with 50 CFU of the parent while escalating the dose of the mutant. For example, the next three subjects would be inoculated with target EDDs of the mutant of 100, 200, and 400 CFU. If the mutant did not cause pustules at these doses, we would infect groups of three subjects until the EDD of the mutant was 10- to 100-fold higher than that of the parent. Thus, the mutant-parent comparison would be accomplished with 6 to 12 subjects.
Biopsies.
Skin specimens obtained by the punch biopsy technique were fixed, sectioned, and processed for immunohistochemical staining or semiquantitative cultures as described previously (31, 32). After biopsy, the subjects were treated with ciprofloxacin as described previously (32).
Phenotype characterization of recovered colonies.
Bacteria from the inocula and colonies that were recovered from surface cultures and semiquantitative cultures were tested for antibiotic susceptibility and reactivity with MAb 3F12. If available, at least 30 bacteria from each culture or inoculum were picked and frozen in freezing medium. The bacteria were grown on CA plates with and without kanamycin to determine antibiotic susceptibility. Colony blots were probed with MAb 3F12 to confirm the presence or absence of MOMP.
RESULTS
momp and ompA2 are transcribed as monocistronic messages.
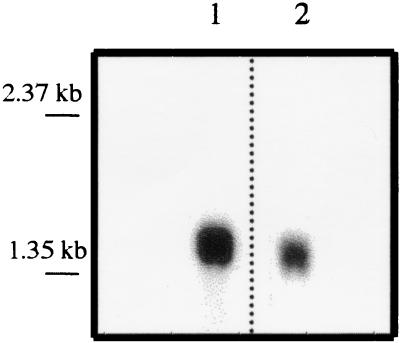
The 1.5-kb momp ORF and the 1.3-kb ompA2 ORF are each flanked by putative promoter regions, ribosomal binding sequences, and transcriptional terminators (17). However, momp and ompA2 are arranged in tandem on the chromosome and may be transcribed as a polycistronic message. To test whether momp and ompA2 are transcribed independently of each other, we performed Northern blot analysis of 35000HP RNA using labeled momp and ompA2 ORFs as probes. When 35000HP total cellular RNA was probed with either the momp or ompA2 probe, a single band migrating with an approximate size of 1.5 or 1.4 kb, respectively, was detected (Fig. 1). A 2.9-kb message was not detected with either probe. The results indicated that each gene was transcribed as a monocistronic message. Thus, the insertion of an Ω-Km2 cassette, which contains transcriptional terminators at both ends of the cassette, into the momp ORF should not have any direct effect on the transcription of ompA2.
FIG. 1.
Northern blot analysis of 35000HP RNA. Lane 1 was probed with the momp ORF, and lane 2 was probed with the ompA2 ORF. The apparent migration of RNA standards is indicated on the left.
Construction and characterization of momp mutant.
An Ω-Km2 cassette was ligated into the momp ORF on pMOMP1.2, resulting in the plasmid pMOMP1.3. H. ducreyi 35000HP was transformed by electroporation with pMOMP1.3, and the momp ORF was insertionally inactivated by allelic exchange. One transformant, 35000HP-SMS2, lacked reactivity with MAb 3F12 by colony blotting and was selected for further characterization.
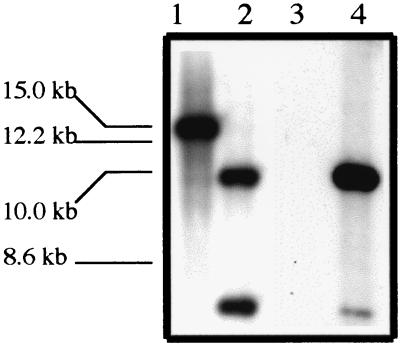
Primers specific to momp were used in PCR to amplify the momp ORF from 35000HP and 35000HP-SMS2 genomic DNA. In agarose gel electrophoresis, the PCR products from the parent and mutant migrated with apparent sizes of 1.4 and 3.7 kb, respectively (data not shown). The difference between the sizes of the amplicons is consistent with the insertion of the Ω-Km2 cassette, which is 2.3 kb in size, into the momp ORF in 35000HP-SMS2. In Southern blotting, genomic DNAs from 35000HP and 35000HP-SMS2 were probed with the momp ORF and Ω-Km2 cassette. The momp probe bound to a 14- to 15-kb DNA fragment in the parent and 6.4- and 10.8-kb DNA fragments in the mutant. The Ω-Km2 cassette did not bind to 35000HP DNA but did bind to two bands measuring 6.4 and 10.8 kb in 35000HP-SMS2 DNA (Fig. 2). We sequenced the area 5′ and 3′ to the insertion of the Ω-Km2 cassette in 35000HP-SMS2 to examine whether other mutations occurred during the allele exchange. Only one nucleotide change (A-to-T transversion) at bp 858 was observed in 35000HP-SMS2 upstream of the Ω-Km2 cassette insertion in comparison to 35000HP. The mutation did not result in an alteration of the amino acid sequence. The nucleotide sequence in 35000HP-SMS2 downstream of the insertion of the Ω-Km2 cassette continuing into the ompA2 ORF exactly matched that of 35000HP (data not shown). Thus, allele exchange occurred within the momp ORF and did not result in linked secondary mutations that should affect ompA2 transcription.
FIG. 2.
Southern blot analysis of genomic DNA from 35000HP (lanes 1 and 3) and 35000HP-SMS2 (lanes 2 and 4). Lanes 1 and 2 were probed with the momp ORF, and lanes 3 and 4 were probed with the Ω-Km2 cassette. The apparent migration of DNA standards is indicated on the left.
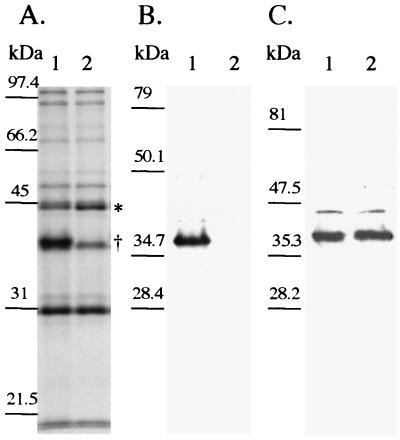
To confirm that 35000HP-SMS2 no longer expressed MOMP and to determine the relative level of OmpA2 expressed by 35000HP-SMS2, Western blots of whole-cell lysates and Sarkosyl-insoluble OMP of 35000HP-SMS2 were probed with either MAb 3F12 or MAb 2C7. MAb 3F12 did not react with the mutant whole-cell lysate or Sarkosyl-insoluble OMP. The amount of OmpA2 expressed by the mutant was not increased compared to the parent, as determined by densitometry of MAb 2C7 reactivity in Western blotting (Fig. 3 and data not shown).
FIG. 3.
(A) SDS–10% PAGE and Coomassie blue staining of OMP prepared from 35000HP (lane 1) and 35000HP-SMS2 (lane 2). Note the location of the 45-kDa protein (∗) and the absence of MOMP (†). (B and C) Western blot analysis of 35000HP (lane 1) and 35000HP-SMS2 (lane 2). Panel B was probed with MAb 3F12, and panel C was probed with MAb 2C7. Molecular mass standards are shown to the left of each panel.
Sarkosyl-insoluble OMPs from 35000HP-SMS2 and 35000HP were compared by SDS-PAGE analysis. The OMP profiles were similar except that a protein migrating with an apparent molecular mass of 45 kDa appeared to be increased in the mutant. The N-terminal amino acid sequence (VTLYEAEGTKIDLDGSIRLV) of the 45-kDa protein indicated that the protein had homology with known or putative classical porins of Actinobacillus pleuropneumoniae, Haemophilus parasuis, and Pasteurella multocida (14). By densitometry, 35000HP-SMS2 contained (1.8 ± 0.42)-fold higher levels of the 45-kDa protein than did 35000HP, as determined in three independent experiments.
35000HP and 35000HP-SMS2 had identical rates of growth in broth (data not shown). Analysis of LOSs isolated from 35000HP and 35000HP-SMS2 by electrophoresis showed similar profiles (data not shown).
Experimental human challenge.
A modified escalating dose-response study was used to compare the virulence of 35000HP-SMS2 and 35000HP (1, 21, 36). In the first iteration, we attempted to inoculate three subjects at three sites with EDDs of 25, 50, and 100 CFU of the mutant and at two sites with an EDD of 50 CFU of the parent. A sixth site was inoculated with the highest dose of the mutant that was heat killed. The actual EDDs were 17, 35, and 70 CFU of the mutant and 120 CFU of the parent. No lesions developed at sites inoculated with the heat-killed control. Papules developed at six of six sites for the parent and nine of nine sites for the mutant. Pustules developed at five of six sites for 35000HP and four of nine sites for 35000HP-SMS2.
The result of the first iteration suggested that the mutant formed pustules at a rate similar to that of the parent. We attempted to infect three more subjects to confirm these results; however, one subject withdrew on the day of inoculation. In the second iteration, two subjects were infected with EDDs of 27, 55, and 110 CFU of 35000HP-SMS2 and 76 CFU of 35000HP. No lesions developed at sites inoculated with heat-killed bacteria. Papules developed at all sites inoculated with live bacteria. Pustules developed at two of four sites inoculated with the parent and two of six sites inoculated with the mutant.
The cumulative results for both iterations showed that papules formed at all sites inoculated with either live 35000HP or 35000HP-SMS2 (Table 1). Pustules developed at 7 of 10 (70%) (95% CI, 34.8 to 93.3%) sites for the parent and 6 of 15 (40%) (95% CI, 16.3 to 67.7%) sites for the mutant (P = 0.14; Fisher's exact test). Thus, expression of MOMP by H. ducreyi was not required for pustule formation in human volunteers.
TABLE 1.
Response to inoculation with live H. ducreyi strainsa
| Subject and isolate | No. of initial papules | Final outcome of initial papules
|
|||
|---|---|---|---|---|---|
| No. of papules | No. of pustules | No. resolved | Days of observation | ||
| 111 | 7 | ||||
| 35000HP | 2 | 2 | |||
| 35000HP-SMS2 | 3 | 3 | |||
| 112 | 14 | ||||
| 35000HP | 2 | 1 | 1 | ||
| 35000HP-SMS2 | 3 | 1 | 2 | ||
| 114 | 8 | ||||
| 35000HP | 2 | 2 | |||
| 35000HP-SMS2 | 3 | 1 | 2 | ||
| 115 | 9 | ||||
| 35000HP | 2 | 2 | |||
| 35000HP-SMS2 | 3 | 1 | 2 | ||
| 116 | 12 | ||||
| 35000HP | 2 | 2 | |||
| 35000HP-SMS2 | 3 | 1 | 2 | ||
Each volunteer was inoculated at two sites with 35000HP and at three sites with 35000HP-SMS2. Volunteers 111, 112, and 114 were inoculated in the first iteration, and 115 and 116 were inoculated in the second iteration.
Cellular infiltrate of lesions.
Two parent and three mutant biopsy specimens were available for histological examination. Similar inflammatory reaction patterns were seen in all specimens. Micropustules with polymorphonuclear leukocytes were present in the epidermis. The dermal infiltrate was composed of perivascular mononuclear cells. Most of these cells were CD3 positive (data not shown).
Recovery of bacteria from lesions.
The recovery rate of H. ducreyi from daily surface cultures of sites inoculated with live bacteria where active disease was present was 9.9% (n = 71) for 35000HP and 5.9% (n = 85) for 35000HP-SMS2 (P = 0.35). All biopsy specimens were semiquantitatively cultured. Bacteria were recovered from two of two parent biopsy specimens and two of three mutant biopsy specimens. The yield ranged from 2.5 × 104 to 5.2 × 104 (geometric mean = 3.83 × 104) CFU/g of tissue from the parent biopsies and 0 to 4.6 × 105 (geometric mean = 2.43 × 105) CFU/g of tissue for the mutant. Thus, 35000HP and 35000HP-SMS2 were recovered at similar rates from experimental lesions.
Confirmation of the phenotypes of the recovered bacteria.
To confirm that the incoula were of the correct phenotype and that no phenotypic changes occurred during infection, individual colonies from each of the broth cultures used to prepare the inocula, from surface cultures, and from biopsy specimens were scored for kanamycin susceptibility and reactivity with MAb 3F12. For the broth cultures used to prepare the inocula, all 72 parent and 72 mutant colonies tested were phenotypically correct (35000HP, Kms and MAb 3F12+; 35000HP-SMS2, Kmr and MAb 3F12−). Forty-nine colonies obtained from surface cultures of parent sites and 42 colonies from mutant sites had the correct phenotypes. Of biopsy specimens that were culture positive, all 57 parent colonies and 72 mutant colonies had the expected phenotypes.
DISCUSSION
H. ducreyi expresses two OmpA-like proteins, MOMP and OmpA2 (17, 30). OmpA proteins have been proposed to be virulence factors for several gram-negative bacteria (19, 23–25, 28, 35), and a previously described MOMP-deficient mutant of H. ducreyi, 35000.60, was more sensitive to 50% normal human serum than its parent (15). In this study, we constructed an isogenic MOMP-deficient mutant of H. ducreyi 35000HP. The MOMP-deficient mutant formed pustules at a rate similar to that of the parent in the human model of H. ducreyi infection.
The original MOMP-deficient mutant, 35000.60, was constructed by insertionally inactivating momp with a cat cassette (17). Prior to reconstruction of a MOMP-deficient mutant in the 35000HP background, we determined that momp and ompA2 were transcribed independently. Nonetheless, we used an Ω-Km2 cassette that contains transcriptional terminators to insertionally inactivate momp and prevent any direct effects on the downstream ompA2 gene. An isogenic MOMP-deficient mutant, 35000HP-SMS2, was generated by allelic exchange. Phenotypic and genetic characterization of 35000HP-SMS2 confirmed that MOMP was not expressed by the mutant, the Ω-Km2 cassette was located in momp, and no secondary mutations had occurred in the intergenic region between momp and ompA2.
MOMP's contribution to the early stages of pathogenesis of chancroid was assessed in the human model of H. ducreyi infection. H. ducreyi clumps in vitro, and it would be technically difficult to perform a mutant-parent comparison using a single dose of bacteria. Therefore, we evaluated the virulence of the mutant using a modified escalating dose-response study, as described previously (1, 21, 36). Within an EDD range of 17 to 120 CFU, where the parent generally forms pustules (2), the parent strain formed pustules at 70% of sites and the mutant formed pustules at 40% of sites (P = 0.14). To achieve 80% power to conclude that the observed difference between the pustule formation rates of the parent (70%) and the mutant (40%) was significant, we would have needed to infect 14 subjects at 70 sites. However, the recovery rates of H. ducreyi from surface cultures and biopsy specimens and from the cellular infiltrate in the lesions caused by the parent and mutant were similar. Taken together, the data indicate that MOMP is not required for pustule formation in human volunteers, but we cannot exclude the possibility that expression of MOMP has a contributing role in pustule formation.
Analysis of Sarkosyl-insoluble OMPs from 35000HP and 35000HP-SMS2 indicated that there was no increase in the amount of OmpA2 in the mutant. This finding suggested that the increased amount of the 43-kDa protein in the previously described MOMP-deficient mutant, 35000.60, may have been due to increased expression of OmpA2 caused by the presence of the cat cassette, which lacks transcriptional terminators (7), in the momp gene. However, 35000HP-SMS2 did contain (1.8 ± 0.42)-fold more of a 45-kDa OMP. The 45-kDa protein shares homology with the OMPs of A. pleuropneumoniae, H. parasuis, and P. multocida, which have antigenic and sequence homology to classical porins of gram-negative bacteria (14). Whether OmpA proteins form nonspecific pores is controversial (22, 33). Previous studies comparing MOMP to classical porins of gram-negative bacteria in planar lipid membranes indicated that MOMP does not form ion-permeable channels (30). The reason for the increase in the 45-kDa protein in 35000HP-SMS2 is unknown; however, the increased amount of the 45-kDa protein may be a compensatory mechanism due to the loss of MOMP. We cannot exclude the possibility that increased expression of the 45-kDa protein influenced pustule formation.
Humans are the only known reservoir for H. ducreyi, which primarily infects the mucosal epithelium of the foreskin and labia during natural infection. However, H. ducreyi also infects stratified squamous epithelium, such as the outer surface of the labia, the shaft of the penis, the buttocks, and the thighs (13, 34). Thus, experimental infection of stratified squamous epithelium is likely to be relevant to natural disease. Although the clinical course and histopathology of experimental infection resembles that of natural disease (3, 20, 31), a limitation of the model is the artificial route of inoculation. Puncture wounds are required to initiate experimental infection (32), but the depth of inoculation required for experimental or natural infection is unknown. The applicator penetrates the skin to a depth of 1.9 mm and delivers bacteria to the epidermis, upper dermis, and deep dermis (unpublished observations). This method of inoculation may mask the role of some virulence determinants required for natural infection. Infection in the model is limited to the pustular stage of disease, and we also cannot evaluate whether virulence factors contribute to ulcer formation. However, an HgbA-deficient mutant and a PAL-deficient mutant were attenuated in the ability to form pustules relative to the parent (1; unpublished results). Thus, the model can discriminate between strains of H. ducreyi based on their abilities to form pustules.
In summary, we have determined that momp and ompA2 are transcribed independently and have constructed an isogenic MOMP-deficient mutant. The mutant formed pustules and was recovered from lesions and daily swabs at a rate similar to that of its parent. Furthermore, the histopathology of sites inoculated with the mutant was indistinguishable from that of the sites inoculated with the parent. Future studies will be directed toward constructing an OmpA2-deficient mutant and a MOMP and OmpA2 double mutant and determining their abilities to form pustules in the human infection model.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grants AI27863, AI31494, AI32011, and MO1RR00750. The clinical trial was also supported by the Sexually Transmitted Diseases Clinical Trials Unit through contract N01-AI75329 from the NIAID.
We thank Stacy L. Nelson, Mike Klemsz, and Margaret Bauer for advice and assistance with the manuscript.
REFERENCES
- 1.Al-Tawfiq, J. A., K. R. Fortney, B. P. Katz, C. Elkins, and S. M. Spinola. 1999. An isogenic hemoglobin receptor-deficient mutant of Haemophilus ducreyi is attenuated in the human model of experimental infection. J. Infect. Dis., in press. [DOI] [PubMed]
- 2.Al-Tawfiq J A, Harezlak J, Katz B P, Spinola S M. Cumulative experience with Haemophilus ducreyi 35000 in the human model of experimental infection. Sex Transm Dis. 2000;27:111–114. doi: 10.1097/00007435-200002000-00009. [DOI] [PubMed] [Google Scholar]
- 3.Al-Tawfiq J A, Thornton A C, Katz B P, Fortney K R, Todd K D, Hood A F, Spinola S M. Standardization of the experimental model of Haemophilus ducreyi infection in human subjects. J Infect Dis. 1998;178:1684–1687. doi: 10.1086/314483. [DOI] [PubMed] [Google Scholar]
- 4.Bakaletz L O, Kennedy B, Novotny L A, Duquesne G, Cohen J, Lobet Y. Protection against development of otitis media induced by nontypeable Haemophilus influenzae by both active and passive immunization in a chinchilla model of virus-bacterium superinfection. Infect Immun. 1999;67:2746–2762. doi: 10.1128/iai.67.6.2746-2762.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campagnari A A, Wild L M, Griffiths G E, Karalus R J, Wirth M A, Spinola S M. Role of lipooligosaccharides in experimental dermal lesions caused by Haemophilus ducreyi. Infect Immun. 1991;59:2601–2608. doi: 10.1128/iai.59.8.2601-2608.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chai T-J, Goulds J. Two bacteriophages which utilize a new Escherichia coli major outer membrane protein as part of their receptor. J Bacteriol. 1978;135:164–170. doi: 10.1128/jb.135.1.164-170.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Close T J, Christmann J L, Rodriguez R L. M13 bacteriophage and pUC plasmids containing DNA inserts but still capable of beta-galactosidase alpha-complementation. Gene. 1983;23:131–136. doi: 10.1016/0378-1119(83)90044-6. [DOI] [PubMed] [Google Scholar]
- 8.Costello G, Vipond R, MacIntyre S. Aeromonas salmonicida possesses two genes encoding homologs of the major outer membrane protein, OmpA. J Bacteriol. 1996;178:1623–1630. doi: 10.1128/jb.178.6.1623-1630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Datta D B, Arden B, Henning U. Major proteins of the Escherichia coli outer cell envelope membrane as bacteriophage receptors. J Bacteriol. 1977;131:821–829. doi: 10.1128/jb.131.3.821-829.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Mot R, Vanderleyden J. The C-terminal sequence conservation between OmpA-related outer membrane proteins and MotB suggests a common function in both Gram-positive and Gram-negative bacteria, possibly in the interaction of these domains with peptidoglycan. Mol Microbiol. 1994;12:333–334. doi: 10.1111/j.1365-2958.1994.tb01021.x. [DOI] [PubMed] [Google Scholar]
- 11.Fleming D T, Wasserheit J N. From epidemiological synergy to public health policy and practice: the contribution of other sexually transmitted diseases to sexual transmission of HIV infection. Sex Transm Infect. 1999;75:3–17. doi: 10.1136/sti.75.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gotschlich E C, Seiff M, Blake M S. The DNA sequence of the structural gene of gonococcal protein III and the flanking region containing a repetitive sequence. Homology of protein III with enterobacterial OmpA proteins. J Exp Med. 1987;165:471–482. doi: 10.1084/jem.165.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hammond G W, Slutchuk M, Scatliff J, Sherman E, Wilt J C, Ronald A R. Epidemiologic, clinical, laboratory, and therapeutic features of an urban outbreak of chancroid in North America. Rev Infect Dis. 1980;2:867–879. doi: 10.1093/clinids/2.6.867. [DOI] [PubMed] [Google Scholar]
- 14.Hartmann L, Schroder W, Lubke A. Isolation of the Major Outer-membrane Protein of Actinobacillus pleuropneumoniae and Haemophilus parasuis. J Vet Med B. 1995;42:59–63. doi: 10.1111/j.1439-0450.1995.tb00682.x. [DOI] [PubMed] [Google Scholar]
- 15.Hiltke T J, Bauer M E, Klesney-Tait J, Hansen E J, Munson R S, Jr, Spinola S M. Effect of normal and immune sera on Haemophilus ducreyi 35000HP and its isogenic MOMP and LOS mutants. Microb Pathog. 1999;26:93–102. doi: 10.1006/mpat.1998.0250. [DOI] [PubMed] [Google Scholar]
- 16.Hobbs M M, San Mateo L R, Orndorff P E, Almond G, Kawula T H. Swine model of Haemophilus ducreyi infection. Infect Immun. 1995;63:3094–3100. doi: 10.1128/iai.63.8.3094-3100.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klesney-Tait J, Hiltke T J, Maciver I, Spinola S M, Radolf J D, Hansen E J. The major outer membrane of Haemophilus ducreyi consists of two OmpA homologs. J Bacteriol. 1997;179:1764–1773. doi: 10.1128/jb.179.5.1764-1773.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koebnik R. Proposal for a peptidoglycan-associating alpha-helical motif in the C-terminal regions of some bacterial cell surface proteins. Mol Microbiol. 1995;16:1269–1270. doi: 10.1111/j.1365-2958.1995.tb02348.x. [DOI] [PubMed] [Google Scholar]
- 19.Mintz K P, Fives-Taylor P M. Identification of an immunoglobulin Fc receptor of Actinobacillus actinomycetemcomitans. Infect Immun. 1994;62:4500–4505. doi: 10.1128/iai.62.10.4500-4505.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palmer K L, Schnizlein-Bick C T, Orazi A, John K, Chen C-Y, Hood A F, Spinola S M. The immune response to Haemophilus ducreyi resembles a delayed-type hypersensitivity reaction throughout experimental infection of human subjects. J Infect Dis. 1998;178:1688–1697. doi: 10.1086/314489. [DOI] [PubMed] [Google Scholar]
- 21.Palmer K L, Thornton A C, Fortney K R, Hood A F, Munson R S, Jr, Spinola S M. Evaluation of an isogenic hemolysin-deficient mutant in the human model of Haemophilus ducreyi infection. J Infect Dis. 1998;178:191–199. doi: 10.1086/515617. [DOI] [PubMed] [Google Scholar]
- 22.Pautsch A, Schulz G E. Structure of the outer membrane protein A transmembrane domain. Nat Struct Biol. 1998;5:1013–1017. doi: 10.1038/2983. [DOI] [PubMed] [Google Scholar]
- 23.Prasadarao N V, Wass C A, Kim K S. Endothelial cell GlcNAcb1-4GlcNAc epitopes for outer membrane protein A enhance traversal of Escherichia coli across the blood-brain barrier. Infect Immun. 1996;64:154–160. doi: 10.1128/iai.64.1.154-160.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prasadarao N V, Wass C A, Weiser J N, Stins M F, Huang S, Kim K S. Outer membrane protein A of Escherichia coli contributes to invasion of brain microvascular endothelial cells. Infect Immun. 1996;64:146–153. doi: 10.1128/iai.64.1.146-153.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rice P A, Vayo H E, Tam M R, Blake M S. Immunoglobulin G antibodies directed against protein III block killing of serum-resistant Neisseria gonorrhoeae by immune serum. J Exp Med. 1986;164:1735–1748. doi: 10.1084/jem.164.5.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 27.San Mateo L R, Toffer K L, Orndorff P E, Kawula T H. Neutropenia restores virulence to an attenuated Cu,Zn superoxide dismutase-deficient Haemophilus ducreyi strain in the swine model of chancroid. Infect Immun. 1999;67:5345–5351. doi: 10.1128/iai.67.10.5345-5351.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sirakova T, Kolattukudy P E, Murwin D, Billy J, Leake E, Lim D, DeMaria T, Bakaletz L. Role of fimbriae expressed by nontypeable Haemophilus influenzae in pathogenesis of and protection against otitis media and relatedness of the fimbrin subunit to outer membrane protein A. Infect Immun. 1994;62:2002–2020. doi: 10.1128/iai.62.5.2002-2020.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sonntag I, Schwartz Y, Hirota Y, Henning U. Cell envelope and shape of Escherichia coli: multiple mutants missing the outer membrane lipoprotein and other major outer membrane proteins. J Bacteriol. 1978;136:280–285. doi: 10.1128/jb.136.1.280-285.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spinola S M, Griffiths G E, Shanks K L, Blake M S. The major outer membrane protein of Haemophilus ducreyi is a member of the OmpA family of proteins. Infect Immun. 1993;61:1346–1351. doi: 10.1128/iai.61.4.1346-1351.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spinola S M, Orazi A, Arno J N, Fortney K, Kotylo P, Chen C-Y, Campagnari A A, Hood A F. Haemophilus ducreyi elicits a cutaneous infiltrate of CD4 cells during experimental human infection. J Infect Dis. 1996;173:394–402. doi: 10.1093/infdis/173.2.394. [DOI] [PubMed] [Google Scholar]
- 32.Spinola S M, Wild L M, Apicella M A, Gaspari A A, Campagnari A A. Experimental human infection with Haemophilus ducreyi. J Infect Dis. 1994;169:1146–1150. doi: 10.1093/infdis/169.5.1146. [DOI] [PubMed] [Google Scholar]
- 33.Sugawara E, Nikaido H. OmpA protein of Escherichia coli outer membrane occurs in open and closed channel forms. J Biol Chem. 1994;269:17981–17987. [PubMed] [Google Scholar]
- 34.Trees D L, Morse S A. Chancroid and Haemophilus ducreyi: an update. Clin Microbiol Rev. 1995;8:357–375. doi: 10.1128/cmr.8.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weiser J N, Gotschlich E C. Outer membrane protein A (OmpA) contributes to serum resistance and pathogenicity of Escherichia coli K-1. Infect Immun. 1991;59:2252–2258. doi: 10.1128/iai.59.7.2252-2258.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Young R S, Fortney K, Haley J C, Hood A F, Campagnari A A, Wang J, Bozue J A, Munson R S, Jr, Spinola S M. Expression of sialylated or paragloboside-like lipooligosaccharides are not required for pustule formation by Haemophilus ducreyi in human volunteers. Infect Immun. 1999;67:6335–6340. doi: 10.1128/iai.67.12.6335-6340.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]