Summary
The loss of mitochondria in oxymonad protists has been associated with the redirection of the essential Fe-S cluster assembly to the cytosol. Yet as our knowledge of diverse free-living protists broadens, the list of functions of their mitochondrial-related organelles (MROs) expands. We revealed another such function in the closest oxymonad relative, Paratrimastix pyriformis, after we solved the proteome of its MRO with high accuracy, using localization of organelle proteins by isotope tagging (LOPIT). The newly assigned enzymes connect to the glycine cleavage system (GCS) and produce folate derivatives with one-carbon units and formate. These are likely to be used by the cytosolic methionine cycle involved in S-adenosyl methionine recycling. The data provide consistency with the presence of the GCS in MROs of free-living species and its absence in most endobionts, which typically lose the methionine cycle and, in the case of oxymonads, the mitochondria.
Keywords: mitochondrion-related organelle, proteome, LOPIT, spatial proteomics, one-carbon metabolism, methionine cycle, glycine cleavage system, Paratrimastix
Graphical abstract

Highlights
-
•
The proteome of reduced mitochondrion of P. pyriformis was cataloged using LOPIT-DC
-
•
This organelle is involved in neither extended glycolysis nor Fe-S cluster assembly
-
•
The organelle makes folate species and formate essential for the methionine cycle
Zítek et al. report a spatial-proteomic analysis of the free-living metamonad flagellate P. pyriformis. This approach enables distinguishing proteins localized in its reduced mitochondria with high confidence. Metabolic reconstruction suggests that the sole function of this mitochondrion is to provide substrates for the cytosolic methionine cycle.
Introduction
It is well established that the mitochondrion has been acquired prior to the last eukaryotic common ancestor (LECA) and is present in all eukaryotes, with the single known exception of the oxymonad Monocercomonoides exilis.1,2 Eukaryotes thriving in low-oxygen environments usually harbor mitochondrion-related organelles (MROs), the best-studied examples of which are hydrogenosomes and mitosomes.3 The diversity of MROs in free-living protists has been revealed recently,4 but functional methodologies are hindered by their xenic cultivation methods. Their organelles are characterized using transcriptomic and/or genomic data, targeting predictions, and homology searches that use established sets of MRO proteins as baits. Even though this methodology has proven very powerful, it suffers from serious biases. The error rates of predictors are typically unknown; homology-based methods cannot recover proteins that are not already in the researcher’s crosshairs, and the assumption that orthology translates into identical cellular localization is not entirely correct. Finally, experimental verifications using heterologous localization in tractable models suffer the hardship of inaccuracy, and their validity roughly corresponds to the similarity between the tested organelle and its heterologous model.
Consequently, experimental, self-standing de novo identifications of MRO proteomes by proteomic methods are direly required but scarce. The few published proteomes of reduced mitochondria belong to only three lineages of parasitic or endocommensal protists, namely the hydrogenosomes of trichomonads, the hydrogenosomes or mitosomes of diplomonads, and the mitosome of Entamoeba histolytica.5,6,7,8,9,10,11
To the best of our knowledge, the oxymonad flagellate M. exilis (Preaxostyla, Metamonada) is completely devoid of mitochondria and therefore represents a notable exception among eukaryotes. Evidence suggests that in the common ancestor of Preaxostyla, the mitochondrial pathway for iron-sulfur cluster assembly (ISC) was replaced by the sulfur mobilization system (SUF), acquired by lateral gene transfer from unknown bacteria.1,12 This replacement stripped the MRO of one of its essential functions,13 hence pre-adapting the lineage to its subsequent loss. Regardless, not all members of Preaxostyla lack mitochondrion, thus the correlation between the presence of the SUF pathway and the absence of mitochondrion is not rigorous, and MROs likely perform other essential function(s) in Preaxostyla species that retained them. Paratrimastix pyriformis (P. pyriformis), a free-living freshwater bacterivorous flagellate, is one such case and its MRO has been partially characterized.14,15,16 A precise determination of its protein composition and metabolic roles is thus key to understanding the process of MRO reduction and loss in this group.
The only experimentally localized pathway in this organelle is the glycine cleavage system (GCS) composed of four subunits, H, L, P, and T, involved in the degradation of glycine.15 Other biochemical functions remain unknown, namely the localization of Fe-S cluster assembly and extended glycolysis enzymes. The latter comprises a repertoire of proteins present in anaerobic and microaerophilic protists forming acetyl-CoA, with concomitant reduction of ferredoxin17 exemplified by pyruvate:ferredoxin oxidoreductases (PFOs) and [FeFe]hydrogenases (HydA). In the transcriptome of P. pyriformis, several genes encoding HydA and PFO are present, as well as three maturases (HydE, HydF, and HydG),14,15 which are known to be involved in the maturation of the HydA active site in other organisms.15,18
Here, we reconstructed the functions of P. pyriformis MRO using spatial-proteomic data. The established MRO proteome contains 31 proteins including a complete pathway of folate-mediated one-carbon (1C) metabolism, a route connected to GCS and capable of producing formate. We propose that the production of formate and methylated folate species remains an unnoticed yet essential function of MROs in P. pyriformis and potentially other free-living MRO-containing species.
Results
Applying LOPIT-DC robustly assigned more than a thousand proteins into compartments of P. pyriformis cell
We applied localization of organelle proteins by isotope tagging after differential ultracentrifugation (LOPIT-DC) to determine protein localization within P. pyriformis cells.19 This method relies on the assumption that proteins localized in the same organelle co-fractionate and will display similar distribution profiles to known organellar markers.20 Cell lysates were divided by differential centrifugation into 10 fractions (Figure 1A). Inspection of fractions 1–6 using transmission electron microscopy (TEM) confirmed conspicuous changes in fraction content with MROs present mainly in fractions #2 and #3 (Figure S1).
Figure 1.
Application of LOPIT-DC on P. pyriformis cells
(A) Schematic representation of the fractionation workflow.
(B) Venn diagram displaying common and unique proteins identified in four replicates. The total number of quantified proteins for each replicate is shown in brackets.
(C) Distribution profile of marker proteins across ten labeled fractions.
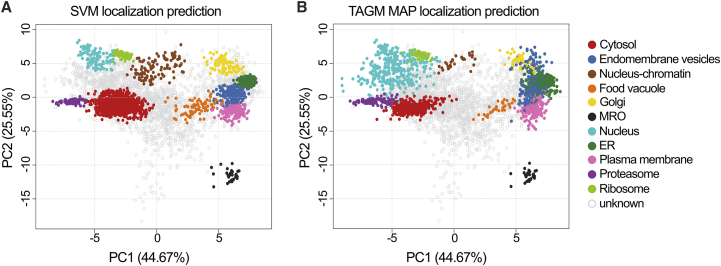
(D) Principal-component analysis (PCA) plot mapping 226 marker proteins for 11 compartments.
See also Figures S1 and S2.
Four biological replicates of 10 fractions were prepared, labeled with tandem mass tag (TMT) 10-plex reagent, and quantified by mass spectrometry for peptide abundances. More than 5,000 proteins were identified across all 10 fractions in each biological replicate, with 4,541 proteins common for all (Figure 1B; Data S1A). This common subset was further analyzed in R21 using pRoloc package.22
Classification using supervised machine learning methods requires a manually curated set of marker proteins with well-known localizations. For this purpose, we compiled a set of 226 markers for 11 compartments (Data S1B) based on up-to-date manually curated annotations, data from previous studies,15 and unsupervised or semi-supervised approaches (see STAR Methods for details). The distribution profiles of marker proteins displayed high similarities between replicates, suggesting acceptable reproducibility (Figures 1C and S2A). The lack of a distribution profile for nuclear proteins indicates that either the nuclei were removed together with unbroken cells or disrupted by sonication. The MRO exhibited the most distinguished profile among all compartments, and it was enriched mainly in fractions #2 and #3, corresponding to EM observations (Figure S1).
Visualization of protein distribution in a 2D plot was achieved by principal-component analysis (PCA; Figure 1D), and among well-separated compartments we could clearly distinguish the MRO. Qsep score23 provided ratios of between-cluster to within-cluster average distances (Figures S2B and S2C) and confirmed that the MRO is well separated.
The localization of the remaining 4,315 proteins was predicted using support vector machine (SVM) and Bayesian approaches, based on t-augmented Gaussian mixture model with maximum a posteriori prediction (TAGM-MAP) (Figure 2). In this respect, 2,503 proteins were classified into one of the 11 compartments by at least one algorithm, and in 1,012 cases both algorithms agreed; 1,786 proteins remained without classification (Data S1B). The MRO and ribosome compartments displayed the most consistent results between algorithms. Altogether, 31 proteins (including 16 markers) were assigned to the MRO (Table 1), and of the 15 proteins classified de novo, both classification methods agreed in 13 cases. The protein composition of the MRO was thus resolved with high confidence.
Figure 2.
PCA plots with proteins assigned to compartments labeled
(A) PCA projection after SVM classification and (B) PCA projection after TAGM MAP classification.
See also Figure S3.
Table 1.
P. pyriformis MRO proteome established by LOPIT-DC analysis
| Accession | Description | Markers | SVM scores | SVM pred | TAGM.MAP. probability | TAGM.MAP. outlier | TAGM.MAP. allocation pred | DeepLoc 2.0 pred |
|---|---|---|---|---|---|---|---|---|
| PaPyr10343 | mitochondrial carrier family protein | MRO | 1 | MRO | 1 | 0 | MRO | yes |
| PaPyr10635 | mitochondrial chaperonin 60 (Cpn60) | MRO | 1 | MRO | 1 | 0 | MRO | yes |
| PaPyr11848 | glycine cleavage system T-protein (GCST) | MRO | 1 | MRO | 1 | 0 | MRO | yes |
| PaPyr1229 | mitochondrial chaperonin 10 (Cpn10) | MRO | 1 | MRO | 1 | 0 | MRO | yes |
| PaPyr1721 | mitochondrial processing protease β subunit (MPPb) | MRO | 1 | MRO | 1 | 0 | MRO | yes |
| PaPyr2518 | Tom40 | MRO | 1 | MRO | 1 | 0 | MRO | yes |
| PaPyr3126 | glycine cleavage system P1-protein (GCSP1) | MRO | 1 | MRO | 1 | 0 | MRO | yes |
| PaPyr3574 | mitochondrial carrier family protein | MRO | 1 | MRO | 1 | 0 | MRO | yes |
| PaPyr4058 | mitochondrial carrier family protein | MRO | 1 | MRO | 1 | 0 | MRO | yes |
| PaPyr5544 | glycine cleavage system L-protein (GSCL) | MRO | 1 | MRO | 1 | 0 | MRO | no |
| PaPyr640 | glycine cleavage system H-protein (GSCH) | MRO | 1 | MRO | 1 | 0 | MRO | yes |
| PaPyr705 | Sam50 | MRO | 1 | MRO | 1 | 0 | MRO | yes |
| PaPyr7885 | mitochondrial processing protease α subunit (MPPa) | MRO | 1 | MRO | 1 | 0 | MRO | yes |
| PaPyr8085 | glycine cleavage system P2-protein (GCSP2) | MRO | 1 | MRO | 1 | 0 | MRO | no |
| PaPyr9 | Tim22 | MRO | 1 | MRO | 1 | 0 | MRO | yes |
| PaPyr9238 | mitochondrial carrier family protein | MRO | 1 | MRO | 1 | 0 | MRO | yes |
| PaPyr5119 | formate-tetrahydrofolate ligase (MTHFD) | unknown | 0.77 | MRO | 1 | 5.98E−24 | MRO | yes |
| PaPyr4383 | hydrogenase maturase HydE | unknown | 0.764 | MRO | 1 | 1.51E−24 | MRO | yes |
| PaPyr2959 | chaperone protein DnaK | unknown | 0.763 | MRO | 1 | 2.07E−23 | MRO | no |
| PaPyr3253 | bifunctional protein FolD | unknown | 0.759 | MRO | 1 | 5.79E−26 | MRO | yes |
| PaPyr4362 | lipoate ligase (LplA) | unknown | 0.755 | MRO | 1 | 9.08E−23 | MRO | yes |
| PaPyr5495 | peroxin-3 | unknown | 0.751 | MRO | 1 | 1.05E−21 | MRO | yes |
| PaPyr9283 | hydrogenase maturase HydF | unknown | 0.746 | MRO | 1 | 8.17E−24 | MRO | yes |
| PaPyr1430 | serine hydroxymethyltransferase (SHMT) | unknown | 0.715 | MRO | 1 | 3.47E−24 | MRO | yes |
| PaPyr1304 | D-serine dehydratase (DSD) | unknown | 0.714 | MRO | 1 | 2.79E−20 | MRO | no |
| PaPyr804 | TRIC | unknown | 0.68 | MRO | 1 | 9.20E−23 | MRO | yes |
| PaPyr4978 | Tom22 | unknown | 0.655 | MRO | 1 | 8.71E−20 | MRO | no |
| PaPyr2234 | VDAC | unknown | 0.646 | MRO | 1 | 1.97E−23 | MRO | no |
| PaPyr2826 | mitochondrial Rho GTPase 1 | unknown | 0.592 | MRO | 1 | 6.18E−17 | MRO | no |
| PaPyr577 | hypothetical protein | unknown | 0.572 | MRO | 2.06E−14 | 1 | unknown | yes |
| PaPyr1418 | Tim23 | unknown | 0.557 | MRO | 3.99E−09 | 1 | unknown | yes |
N termini of MRO proteins have a low positive charge and are often extended compared with prokaryotic homologs
The translation of most proteins destined to mitochondria takes place in the cytosol, and their delivery is ensured by N-terminal or internal targeting sequences.24 N-terminal targeting signals (NTSs) tend to display an amphipathic α-helical structure composed of hydrophobic and positively charged amino acids and are cleaved upon translocation by mitochondrial processing peptidases (MPPs) within an arginine-containing motif.25,26 Bioinformatic tools that predict mitochondrial localization largely focus on these features, often divergent in MROs. Meanwhile, internal localization signals are rarely predicted.27 We took the advantage of the experimentally determined set of MRO proteins of P. pyriformis to calculate the average net charge of their N termini, assess the length of N-terminal extensions relative to their prokaryotic homologs, and test the performance of targeting predictors.
The average net charge of the first 10, 20, and 30 amino acids of all 31 MRO proteins was calculated using Isoelectric Point Calculator 2.0 (http://ipc2.mimuw.edu.pl/index.html; Data S1C). At neutral pH, the net charge at most N termini was calculated weakly positive close to 1.0, similarly to hydrogenosomal proteins of T. vaginalis but lower than those in yeast, human, or Anaeramoeba, where they fall in the range 3.0–4.0.28,29,30 This suggests that the charge of N termini does not provide strong traction to the mitochondrial matrix as in typical aerobic mitochondria. Also, 20 MRO proteins displayed discernible N-terminal extensions (>10 amino acids), compared with their bacterial homologs, or showed a short extension relative to the beginning of their conserved domain (Data S1C), suggesting that the N-terminal extensions play a role in translocation signaling in these MRO proteins.
The performance of 8 tools for mitochondrial targeting prediction (Cello,31 iPSORT,32 DeepLoc 2.033, MitoFates,34 MultiLoc2,35 NommPred,36 Pprowler,37 and TargetP 2.038) was tested using the set of MRO proteins and 10 negative control sets of the same sizes (n = 30) robustly assigned to 10 other compartments using LOPIT-DC (Data S1D). Specificity and sensitivity were the highest for DeepLoc 2.0 (0.96 and 0.77, respectively). iMLP27 predicted internal signals not only for most MRO proteins but also for negative controls from other compartments and displayed a false positive rate too high to be informative (Data S1D). We took advantage of DeepLoc 2.0’s reasonably high performance and ran the MRO targeting prediction on the full proteome. Altogether, 310 proteins were predicted to the MRO (Data S1E), 24 of which had been robustly assigned to the MRO (Table 1) and 17 to other compartments based on LOPIT-DC data. Of the remaining 269 proteins, 9 were detected by proteomics but remained unclassified based on profile; only 17 have functional annotations, five of which are relevant to mitochondria: Tim14 (PaPyr2934), Tim17 (PaPyr5590), ferredoxin (PaPyr451), Tim44 (PaPyr7077), and folylpolyglutamate synthase (FPGS) (PaPyr1465). As the latter two were detected by proteomics, manual inspection of their profiles (Figure S3B) suggested that they indeed are at least partially localized in MRO, strengthening the notion that those five proteins represent an addition to the MRO proteome.
The mitochondrion-related organelle likely provides one-carbon units to cytosolic pathways
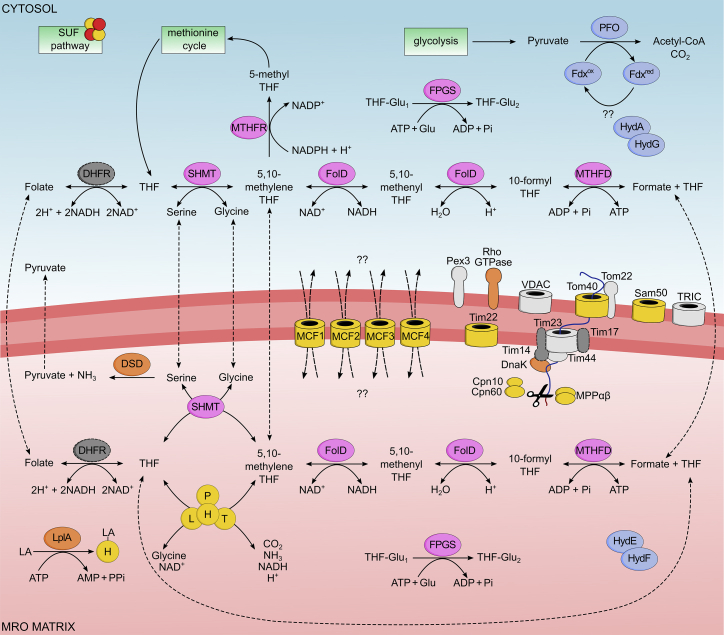
The map of organelle functions built on predicted protein content is shown in Figure 3. Members of the mitochondrial carrier family (MCF), proteins involved in the transport and maturation of MRO proteins, and components of the GCS were used as markers. Among the de novo assigned MRO proteins were bifunctional methylenetetrahydrofolate dehydrogenase/methenyl tetrahydrofolate cyclohydrolase (FolD; PaPyr3253), formate-tetrahydrofolate ligase (MTHFD; PaPyr5119), and serine hydroxymethyltransferase (SHMT; PaPyr1430). These three proteins play a role in folate-mediated 1C metabolism where formate is generated. The first reaction catalyzed by SHMT (PaPyr1430) brings a 1C unit into the folate pool by transferring a methyl group from serine to tetrahydrofolate (THF). Further interconversion of THF derivates into formate is mediated by FolD (PaPyr3253) and MTHFD (PaPyr5119), with concurrent production of ATP (Figure 3). Finally, FPGS (PaPyr1465), involved in the retention and compartmentalization of folate species,39 seems to be dually localized in both the MRO and cytosol (Figure S3B). In mammalian cells, the complete 1C pathway is localized both in cytosol and mitochondria.40 This situation was observed also in P. pyriformis, where the second paralog for each—MTHFD (PaPyr5307), FolD (PaPyr2026), and SHMT (PaPyr5925)—were classified as cytosolic (Figure 3; Data S1B), together with methylenetetrahydrofolate reductase (MTHFR; PaPyr5097). MTHFR catalyzes the irreversible conversion of 5,10-methylene-THF into 5-methyl-THF, the substrate of the methionine cycle.41 Considering all this, we hypothesise that formate or THF species produced within the MRO are exported to the cytosol, where they are utilized to produce 5-methyl-THF supplementing the methionine cycle. All enzymes of the methionine cycle were detected and classified as cytosolic (Data S1B).
Figure 3.
Metabolic map of P. pyriformis MRO based on LOPIT-DC analysis
Proteins used as markers for classification are colored yellow, proteins involved in folate-mediated 1-carbon metabolism are colored pink, proteins involved in extended glycolysis and hydrogen production are colored blue, previously unknown-function proteins are colored light gray, and proteins present in the genome but missing in LOPIT dataset are colored dark gray with a dashed outline. H, L, P, and T, components of glycine cleavage system; SHMT, serine hydroxymethyltransferase; FolD, methylenetetrahydrofolate dehydrogenase/methenyl tetrahydrofolate cyclohydrolase; MTHFD, formate-tetrahydrofolate ligase; FPGS, folylpolyglutamate synthase; MTHFR, methylenetetrahydrofolate reductase; DHFR, dihydrofolate reductase; LplA, lipoate ligase; DSD, D-serine dehydratase; PFO, pyruvate:ferredoxin oxidoreductase; Fdx, ferredoxin; HydA, [FeFe]hydrogenase; HydE, HydF, and HydG, hydrogenase maturases; MPPαβ, mitochondrial processing peptidase subunits α and β; VDAC, voltage-dependent anion channel; TRIC, trimeric intracellular ion channel; Pex3, peroxisomal biogenesis factor 3; Cpn10, mitochondrial chaperonin 10; Cpn60, mitochondrial chaperonin 60; DnaK, chaperone protein; MCF1, MCF2, MCF3, and MCF4, mitochondrial carrier family proteins; THF, tetrahydrofolate; LA, lipoic acid.
Four other MRO-classified proteins are the following. Lipoate ligase (LplA; PaPyr4362) likely mediates attachment of exogenous lipoic acid,42,43 acting as an essential cofactor, to GCS-H.44 Rho GTPase (PaPyr2826), known as Miro, typically localizes in the outer membrane and contains two Ca2+-binding EF-hands domains flanked by GTPase domains on both sides.45 In P. pyriformis, the C-terminal GTPase domain is missing. D-serine dehydratase (PaPyr1304) is a pyridoxal 5′-phosphate (PLP)-dependent enzyme converting serine into pyruvate and ammonia.46 Chaperone protein DnaK (PaPyr2959) is a proteobacterial ortholog of mitochondrial Hsp70, a pivotal player in mitochondrial protein import.47
The dark proteome of the MRO likely consists of membrane proteins involved in its biogenesis
Seven proteins (PaPyr577, PaPyr804, PaPyr1418, PaPyr2234, PaPyr4978, PaPyr5495, and PaPyr7077) assigned to the MRO by LOPIT or DeepLoc 2.0 lacked close hits in NCBI Blast. We searched for conserved motifs and transmembrane domains (HHpred,48 HMMER v3.3,49 and SWISS-MODEL50) to find putative homologs in other organisms.
Using homology modeling with SWISS-MODEL we determined that PaPyr2234 is structurally similar to the porin3 superfamily (PF01459), potentially representing a voltage-dependent anion channel (VDAC). VDACs localize in the outer membrane of mitochondria and perform passive diffusion of ions and small metabolites, including ATP/ADP, NAD/NADH, pyruvate, glutamate, or malate.51,52 Phylogenetic reconstruction (Data S2A) and HHpred search suggest that PaPyr804 represents a trimeric intracellular ion channel (TRIC). In eukaryotes, these monovalent cation channels are predominantly present in the membrane of the endoplasmic reticulum (ER), balancing the charge during Ca2+ uptake or release.53
PaPyr4978, PaPyr7077, and PaPyr1418 were recognized as members of the TOM/TIM translocase machinery Tom22, Tim44, and Tim17/22/23, respectively. In yeast, it has been shown that three different proteins (Tom20, Tom70, and Tom22) recognize proteins to be transported into mitochondria.54 In the genome of P. pyriformis, homologs of Tom20 and Tom70 failed to be identified, hence PaPyr4978 is putatively the one to recognize proteins targeted to the MRO. PaPyr7077 represents a putative homolog of Tim44, a component of the PAM motor complex.55 The presence of Tim44 has been reported in other metamonads,4,6,7,11,29,56 and our phylogenetic reconstructions showed a weak relationship of the long branch-forming PaPyr7077 to Tim44 of other metamonads (Data S2B). Members of the Tim17/22/23 protein family have been identified in all eukaryotes as components of protein translocation complexes in the mitochondrial inner membrane; TIM23 complex facilitates protein import into the matrix, while TIM22 mediates protein insertion into the mitochondrial inner membrane.57,58,59 In the genome of P. pyriformis, two homologs of Tim17/22/23 proteins were already identified (PaPyr9 and PaPyr5590). Both contain the conserved hallmark glycine-zipper (GX3GX3G) and PRAT ((G/A)X2(F/Y)X10RX3DX6(G/A/S)GX3G) motifs. In the last homolog, PaPyr1418, only the glycine-zipper motif was identified. Phylogenetic affiliations of P. pyriformis Tim17/22/23 proteins were determined using the dataset of Žárský and Doležal,58 which is enriched by sequences from diverse eukaryotic lineages not available in the Uniprot database,60 assigning PaPyr5590, PaPyr9, and PaPyr1418 with high support as orthologs of Tim17, Tim22, and Tim23, respectively (Figure 4).
Figure 4.
Schematic phylogeny of the Tim17/22/23 family with the positions of P. pyriformis homologs
Employing a previously compiled dataset,58 the ML tree was inferred by IQ-TREE using the posterior mean site frequency empirical model with the Shimodaira-Hasegawa approximate likelihood ratio test/ultrafast bootstrapping strategy both supporting their phylogenetic position.
See also STAR Methods.
Lastly, a putative function was assigned to PaPyr5495, a protein sharing similarities with peroxisomal biogenesis factor 3 (Pex3) (Data S2C). In human cells, Pex3 is responsible for docking of receptor Pex19 on the peroxisomal membrane during the import of PTS1 C-terminal signal-containing proteins.61 Importantly, Pex19 homolog in P. pyriformis (PaPyr8690) was identified using HMM profiles and shown to be phylogenetically related to other sequences from Preaxostyla (Data S2C). All Preaxostyla Pex19 homologs clearly possess a conserved Pex19 Pfam motif (PF04614) and an N-terminal sequence similar to the Pex3-interaction motif known from plants, fungi, and animals (Data S2C). Since no peroxisomes are present in Preaxostyla,1,62,63 we speculate that the Pex3/Pex19 system may have been recruited to MRO for other functions.
The MRO is not involved in extended glycolysis and Fe-S cluster assembly
In the hydrogenosomes of trichomonads, pyruvate undergoes oxidative decarboxylation into acetyl-CoA and CO2 by PFO, while ferredoxins reduced in this reaction are subsequently reoxidized by HydA with concomitant production of H2.64 Five genes encoding for PFO, two for ferredoxins, nine for HydA, and one gene for each of the three HydA maturases (HydE, -F, and -G) are present in the P. pyriformis genome.
Three out of four PFOs (except PFO_3, PaPyr7655) were identified by LOPIT-DC and assigned to the cytosol by at least one classification method. Two genes annotated as ferredoxin (PaPyr13543 and PaPyr13544) showed a distribution profile similar to cytosolic markers but were not classified as cytosolic by algorithms (Figure S3A). Five out of nine HydA enzymes were detected in all four LOPIT-DC replicates, but none of them exhibited a distribution profile suggesting localization into the MRO (Figure S3A). HydA_4 (PaPyr6388) was assigned to the cytosol by both classification tools, whereas HydA_1 (PaPyr1254), HydA_2 (PaPyr1278), and HydA_6 (PaPyr8158) were assigned to the cytosol only by SVM. The last hydrogenase in the dataset, HydA_8 (PaPyr8532_PaPyr8533), was not classified to any compartment. Of hydrogenase maturases, two (HydE; PaPyr4383 and HydF; PaPyr9283) were robustly assigned to the MRO while the third (HydG; PaPyr1252) was clearly cytosolic (Figure S3A). Altogether, these results support a cytosolic localization of extended glycolysis in P. pyriformis but raise an interesting question regarding the role of hydrogenase maturases.
The common function for virtually all mitochondria and MROs is the synthesis of Fe-S clusters, which in Preaxostyla is performed by the SUF. This pathway consists of the ATPase SufC (PaPyr9323), a scaffold protein SufB (PaPyr903), and the cysteine desulfurase fusion protein SufDSU (PaPyr904).12 All three proteins were detected in all four LOPIT-DC replicates and enriched in the cytosolic fraction (Figure S3A). Both classification methods assigned SufC and SufDSU to the cytosol; SufB was classified as cytosolic only by the SVM algorithm, but with high confidence.
Localization by heterologous expression sometimes contradicts the results of LOPIT-DC
A popular experimental approach for assessing protein localization of organisms that cannot be genetically transformed is the expression in heterologous systems. We chose related T. vaginalis possessing hydrogenosomes morphologically, similarly to P. pyriformis MRO, to attest the localization of 14 non-marker proteins classified robustly to the MRO or the cytosol. C-terminally HA-tagged proteins were expressed in T. vaginalis, and their localization was visualized by immunofluorescence assay (IFA) microscopy using an anti-malic enzyme antibody as hydrogenosomal marker (Figure S4A).65 HydE (PaPyr4383), putative VDAC (PaPyr2234), and putative Pex3 (PaPyr5495) displayed hydrogenosomal localization; and 6 proteins displayed cytosolic localization: HydA_2 (PaPyr1278), HydG (PaPyr1252), PFO_2 (PaPyr12211), PFO_4 (PaPyr7937_PaPyr7938), FolD_1 (PaPyr3253), and SufC (PaPyr9323). The last five proteins, HydF (PaPyr9283), HydA_4 (PaPyr6388), HydA_6 (PaPyr8156), MTHFD (PaPyr5119), and putative Tom22 (PaPyr4978), exhibited unclear distribution patterns. Subcellular fractionation and protein protection assay on transfected T. vaginalis revealed that all these proteins were only partially present in the hydrogenosomes and could be found also in lysosomal and microsomal fractions (Figure S4B).
The results of heterologous expression clearly contradicted the outcomes of LOPIT-DC proteomics in the case of FolD_1, which showed a cytosolic pattern in T. vaginalis. Given the fact that FolD_1 exhibited N-terminal extension and was predicted to MRO by Deeploc 2.0, we favor MRO localization in P. pyriformis. In five cases (HydF, HydA_4, HydA_6, MTHFD, and Tom22), the heterologous system provided ambiguous answers. Our results indicate that heterologous expression, at least in this pair of species (P. pyriformis and T. vaginalis), should be carefully interpreted owing to some degree of ambiguity and potential artifactuality. A possible reason could stem from a phenotype affecting the morphology of hydrogenosomes, or a toxic effect created by the ectopic expression.
Discussion
The hyperplexed LOPIT (hyperLOPIT) approach was shown to be a powerful method to map protein distribution within cells,66 and its recent application to Toxoplasma gondii tachyzoites proved successful also for protists.67 Yet hyperLOPIT is not suitable for all cell types owing to the large amount of starting material required to perform subcellular fractionation by density gradient.68 For polyxenically grown P. pyriformis low-cell-density cultures, we employed LOPIT-DC, which produces comparable results with less starting material.19 Here, 4,541 P. pyriformis proteins showed measurable profiles across all four biological replicates, comparable to the number of proteins detected in human U-2 OS cells (6,837 proteins), considering the total size of predicted proteome. Finally, the median distance between clusters, calculated by Qsep in our experiment, was similar to that of Geladaki et al.,19 suggesting that we have reached a satisfactory level of clusters separation.
To analyze LOPIT data by machine learning methods, it is recommended to use at least 13 markers per compartment.69 For model organisms, e.g., Homo sapiens, a marker set of 872 proteins for 12 compartments is available in the pRoloc package.69 Due to the absence of experimental data, our marker set for 11 compartments comprised only 226 proteins. Hence, we focused on the proteome of the MRO, for which we had 16 reliable marker proteins and which displayed the highest separation from other compartments. Furthermore, 31 proteins assigned to the MRO is a relatively small number compared with the proteome of T. vaginalis hydrogenosome that was proposed to contain as many as 600 proteins,6,7,11 and so we expect that some proteins may have escaped detection due to sensitivity limit or were not expressed under experimental conditions. Regardless, LOPIT data provided us with the most reliable picture of protein localization and are less prone to contaminations than classical proteomics based on organelle purification.10
The LOPIT experiments revealed the presence of a complete folate-mediated 1C metabolism in the P. pyriformis MRO, extending the previously experimentally localized GCS. The GCS and the folate pathway in eukaryotes are intrinsically interconnected to deliver 1C units for methylation, methionine regeneration, and de novo formation of purines and thymidine through various folate species serving as 1C carriers.70 P. pyriformis lacks most routes that require different forms of THF, hence 1C metabolism likely only plays a role in the synthesis of 5-methyl-THF, essential for the methionine cycle. In this cycle, methylation of homocysteine regenerates methionine, which is further converted to S-adenosyl methionine (SAM), an essential cofactor for a great number of reactions.41
The partial compartmentation of the folate pathway in the MRO of P. pyriformis possibly provides means to control its flux through a few metabolites (NAD, serine, glycine, and formate) based on metabolic demands. Our model assumes that the pathway is primed by the entry of folate into the MRO, followed by the oxidation of 2 molecules of NADH by the activity of dihydrofolate reductase (DHFR). In the absence of oxidative phosphorylation, this may be the only way to replenish NAD+ used by later enzymatic steps. The genome of P. pyriformis encodes two DHFRs (PaPyr1795 and PaPyr350), although neither was detected by LOPIT-DC nor localized. Next, THF is charged with a 1C unit by the activity of SHMT (from serine) or GCS (from glycine), the latter requiring NAD+. In the last phase, the 1C unit is metabolized into free formate by FolD and MTHFD, requiring another NAD+ and producing ATP. Following its release, formate is exported and THF can be recycled with another 1C unit. As two steps of the pathway require NAD+, the equilibrium toward formate production can be disrupted when the NAD+/NADH ratio decreases. At this point, the NAD+/NADH balance can be restored by 5,10-methylene-THF release into the cytosol for reoxidation (Figure 3). This partial release of the folate-species pool allows for the entry of the corresponding molar amount of folate into MRO and maintains the activity of DHFR.
ATP production by MTHFD may provide energy required for GCS-H lipoylation and folate retention through polyglutamylation.39 Indeed, FPGS (PaPyr1465) is encoded by the P. pyriformis genome and is likely dually localized between MRO and cytosol. The transfer of folate and its reduced derivatives between the mitochondrial and the cytosolic compartments has been thoroughly assessed in a plethora of other cell types and tissues by isotope labeling experiments, using inhibitors and labeled substrates, under aerobiosis and anaerobiosis.71,72,73,74,75 Many of these results support the notion that certain THF species may be capable of crossing from one cellular compartment to another in a carrier-mediated fashion, and the difference in the length of the polyglutamate chain seems to be the defining component of this behavior.
From an evolutionary standpoint, the presence of 1C metabolism known from aerobic mitochondria may not be limited to the MRO of P. pyriformis. Leger et al.4 predicted GCS and SHMT in the MROs of all free-living fornicates and noted that glycine degradation correlates with their free-living lifestyle for an unclear reason. Moreover, GCS and SHMT were predicted to be localized in the MRO of other free-living Metamonada, such as Barthelona sp., Anaeramoeba flamelloides, and A. Ignava29,76 and a number of other free-living, but also endobiotic, anaerobic or microaerophilic protists from the lineages of Archamoebae, breviates, heteroloboseans, ciliates, gregarines, Brevimastigomonas motovehiculus, and Blastocystis sp.77,78,79,80,81,82,83,84,85,86 Our data provide a potential reason for maintaining GCS and SHMT in these organelles, namely the production of 1C-charged folate species to supply cytosolic 1C metabolism, and particularly the reaction in the methionine cycle catalyzed by methionine synthase. In species without methionine synthase, typically parasites,87 this MRO function is not required. Folate pathway enzymes have not been predicted in the other MROs probably due to the limitations of in silico methods. Indeed, P. pyriformis FolD (PaPyr3253) and MTHFD (PaPyr5119), clearly classified as MRO proteins by LOPIT, were predicted as mitochondrial only by DeepLoc 2.0 and Cello (the latter also by NommPred and MultiLoc2-fungal), whereas all other tools and settings considered them cytosolic. In summary, we hypothesize that 1C metabolism remains an overlooked function of some MROs, which cannot be easily revealed by in silico predictions but rather by proteomic studies.
Several MRO proteins with weak homology to database representatives were present in the LOPIT dataset, and most of them enrich the inventory of membrane proteins responsible for organelle biogenesis. The protein translocation machinery of P. pyriformis appears to be reduced, compared with Trichomonas vaginalis and anaeramoebids7,29,88; however, most of the core components were detected in the transcriptome or genome (Tom40, Sam50, Tim17, Tim22, Tim14, Cpn10, Cpn60, and MPPαβ),15 and in our study, we further identified distant homologs of Tom22 (PaPyr4978), Tim44 (PaPyr7077), a Tim23 protein with unusual sequence features (PaPyr1418), and DnaK (PaPyr2959). The presence of a tail-anchored (TA) Rho GTPase Miro should be noted with particular interest. Various studies suggest that Miro proteins are important for mitochondrial transport within the cell, maintenance of their morphology and homeostasis, transport along microtubules, and interactions with the ER.45,89,90 In Dictyostelium discoideum the knockout of Miro does not compromise the transport and distribution of mitochondria yet affects their metabolism.91 So far, the presence of Miro proteins in organisms with reduced mitochondria has been limited to Stygiella incarcerata and Blastocystis,86,92 so its distribution could be more widespread in MROs than previously considered. Finally, P. pyriformis apparently recruited the Pex3-Pex19 system, known to mediate sorting and insertion of peroxisomal membrane proteins, to import TA proteins to the MRO membrane. It was shown that Miro proteins can interact with Pex19 for its membrane insertion.93 In Saccharomyces cerevisiae, Pex19 mediates the import of TA proteins into mitochondria,94 and Pex3 shows mitochondrial localization at the early stage of de novo peroxisomal biogenesis in human fibroblasts.95 Our finding regarding the MRO localization of Pex3 in the absence of peroxisomes adds to the slowly growing body of evidence on the versatility of this system.
It is widely accepted that Fe-S cluster synthesis is a common function of mitochondria in most eukaryotes.5,13,96 Replacement of the mitochondrial ISC pathway by the bacterial SUF system in Preaxostyla was likely a pre-adaptation for the loss of MRO in Monocercomonoides exilis.1 The localization of the SUF system in P. pyriformis had not been previously addressed experimentally, and here we provide clear evidence that it is indeed localized in the cytosol and does not contribute to the essential role of the MRO. Interestingly, two proteins assigned to the MRO proteome, hydrogenase maturases HydE and HydF, known to bear Fe-S clusters, raise the question of how these proteins are maturated. The presence of these proteins in the MRO is remarkable also in terms of their function, which involves the maturation of the H-cluster of HydA, alongside maturase HydG.97 The third maturase HydG was robustly classified as cytosolic together with four paralogs of HydA; one HydA remained unclassified and four were not detected by LOPIT. It is unclear how the maturation of cytosolic HydA is achieved with maturases distributed across two compartments, yet a similar situation has been reported already in T. vaginalis and G. intestinalis.98,99 The lack of evidence for the MRO localization of HydA is consistent with the apparent lack of PFOs and ferredoxins in this compartment, which further confirms cytosolic localization of extended glycolysis in P. pyriformis.
In conclusion, we demonstrate the feasibility of LOPIT-DC for non-model, non-axenic organisms and identified several new components of P. pyriformis MRO, including a pathway that provides 1C units to the cytosol to supply the methionine cycle. This argues for folate metabolism as another basic function in MROs and in P. pyriformis probably the only reason why the cell maintains the organelle even after Fe-S cluster synthesis relocation to the cytosol.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-HA rat monoclonal antibody | Roche | Cat. # 11867423001; RRID: AB_390918 |
| anti-malic enzyme (hydrogenosomal) antibody | Laboratory of Jan Tachezy | N/A |
| Alexa488 Goat anti-Rat | Invitrogen | Cat. # A32731; RRID: AB_2633280 |
| Alexa594 Goat anti-Rabbit | Invitrogen | Cat. # A-11012; RRID: AB_2534079 |
| Bacterial and virus strains | ||
| E. coli TOP10 chemically competent cells | Invitrogen | Cat. # C404010 |
| Chemicals, peptides, and recombinant proteins | ||
| TMT10plex Isobaric Label Reagent Set | Thermo Fisher Scientific | Cat. # 90110 |
| cOmplete EDTA-free protease inhibitors | Roche | Cat. # 4693132001 |
| Critical commercial assays | ||
| BCA Protein Assay Kit | Sigma | Cat. # 71285-3 |
| Dynabeads mRNA Purification Kit | Invitrogen | Cat. # 61006 |
| SMARTer PCR cDNA synthesis Kit | Takara Bio | Cat. # 634925 |
| Deposited data | ||
| Paratrimastix pyriformis genome | Zenodo | https://doi.org/10.5281/zenodo.6405158 |
| Raw mass-spectrometry-based proteomics data | This paper and PRIDE Archive | PRIDE ID: PXD033021 |
| Alignments used to construct phylogenetic trees in Figure S4 | OSF | https://osf.io/a6drs/ |
| Experimental models: Organisms/strains | ||
| Paratrimastix pyriformis, strain RCP-MX | ATCC | ATCC: 50935 |
| Trichomonas vaginalis, strain T1 | Laboratory of Jan Tachezy | N/A |
| Oligonucleotides | ||
| Primers used in this study are listed in Data S1F | N/A | N/A |
| Recombinant DNA | ||
| TagVag2 vector | Laboratory of Jan Tachezy | Hrdy et al.17 |
| pJET1.2/blunt Cloning Vector | Thermo Fisher Scientific | Cat. # K1231 |
| Software and algorithms | ||
| Proteome Discoverer v2.4 | Thermo Fisher Scientific | https://www.thermofisher.com/cz/en/home/industrial/mass-spectrometry/liquid-chromatography-mass-spectrometry-lc-ms/lc-ms-software/multi-omics-data-analysis/proteome-discoverer-software.html |
| R v4.0.4 | R Core Team21 | https://www.r-project.org/ |
| R Studio v1.4.1106 | RStudio Team100 | https://rstudio.com/ |
| pRolog package v1.30.0 | Dawson et al.101 | https://www.bioconductor.org/packages/release/bioc/html/pRoloc.html |
| Fiji v1.53f51 | Kelley et al.102 | https://fiji.sc/ |
| Huygens v21.10 | Huygens Professional | https://svi.nl/Huygens-Visualization |
| EukProt | Richter et al.60 | https://figshare.com/articles/dataset/EukProt_a_database_of_genome-scale_predicted_proteins_across_the_diversity_of_eukaryotic_life/12417881 |
| HDBSCAN | Gatto and Christoforou103 | https://hdbscan.readthedocs.io/en/latest/how_hdbscan_works.html |
| InterProScan v5.52-86.0 | Breckels et al.104 | https://www.ebi.ac.uk/interpro/search/sequence/ |
| Geneious Prime v2020.1.2 | Crook et al. 105 | https://www.geneious.com/prime/ |
| SWISS-MODEL | Waterhouse et al.50 | https://swissmodel.expasy.org/ |
| PHYRE2 | Wang et al.106 | http://www.sbg.bio.ic.ac.uk/∼phyre2/html/page.cgi?id=index |
| Cello v2.5 | Yu et al.31 | http://cello.life.nctu.edu.tw/ |
| iPSORT | Bannai et al.32 | https://ipsort.hgc.jp/ |
| MitoFates v1.1 | Fukasawa et al.34 | http://mitf.cbrc.jp/MitoFates/ |
| MultiLoc2 | Blum e al.35 | https://abi-services.informatik.uni-tuebingen.de/multiloc2/webloc.cgi |
| NommPred v0.2 | Kume et al.36 | https://gitlab.com/kkei/NommPred |
| DeepLoc v2.0 | Thumuluri et al.33 | https://services.healthtech.dtu.dk/service.php?DeepLoc-2.0 |
| TargetP v2.0 | Almagro Armenteros et al.38 | https://services.healthtech.dtu.dk/service.php?TargetP-2.0 |
| iMLP | Schneider et al.27 | http://imlp.bio.uni-kl.de/ |
| IQ-TREE v1.6.12 | Kearse et al.107 and Katoh and Standley108 | http://www.iqtree.org |
| MAFFT v7.453 | Jones et al.109 | https://mafft.cbrc.jp/alignment/server/ |
| Hhpred | Söding48 | https://toolkit.tuebingen.mpg.de/tools/hhpred |
| HMMER v3.3 | Eddy49 | www.hmmer.org |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Vladimír Hampl (vlada@natur.cuni.cz).
Materials availability
Plasmids and T. vaginalis cell lines generated in this study are available from the lead contact upon request.
Experimental Model and Subject Details
Paratrimastix pyriformis (strain RCP-MX, ATCC 50935) was grown at room temperature in a polyxenic culture in Sonneborn's Paramecium medium ATCC 802110 supplemented with 1% (v/v) of Luria-Bertani (LB) broth.111
Trichomonas vaginalis (strain T1) was maintained at 37°C in tryptone-yeast extract-maltose medium (TYM, pH 6.3) supplemented with 10% (v/v) heat-inactivated horse serum.112 T. vaginalis cells were used as heterologous system to localized P. pyriformis proteins. For this purpose, selected genes of P. pyriformis were amplified using PrimeSTAR MAX DNA Polymerase (Takara Bio) from cDNA and cloned into the TagVag2 vector17 via restriction endonucleases or Gibson assembly. Sequences of primers used for cloning are summarized in Data S1F. Transfection of Trichomonas vaginalis was performed as described by Novák et al.113 Briefly, cells in exponential growth phase were electroporated with 30 μg of circular plasmid. Four hours after electroporation, G418 (200 μg/mL) was added to medium. After 3-6 passages expression of P. pyriformis genes in T. vaginalis cells was examined by immunoblot (not shown) and immunofluorescence microscopy.
Method details
Subcellular fractionation by differential centrifugation
Prior to fractionation, 6-7 L of P. pyriformis culture on exponential phase of growth was filtered through filter paper (80 g/m2) to remove bacterial aggregates and subsequently sieved through a 3 μm polycarbonate filter (Nuclepore Track-Etched Membrane, Whatman) followed by two washing steps with 10% (v/v) LB. P. pyriformis cells were collected into a 50 mL tube and harvested by centrifugation at 1000 × g for 10 min at room temperature. The cell pellet (approximately 2 × 108 cells) was resuspended in 2 mL of pre-cooled lysis buffer (10 mM HEPES pH 7.4, 25 mM sucrose, 2 mM magnesium acetate, 2 mM EDTA) supplemented with cOmplete EDTA-free protease inhibitors (Roche). Subcellular fractionation by differential centrifugation was performed as described in Geladaki et al.19 with slight modifications. Briefly, cells were disrupted by three rounds of sonication of 10 s with a pulse of 1 s ON / 1 s OFF at an amplitude of 20%, using a standard 1/8” diameter probe (Qsonica). After each round, cells were examined under the microscope. Three rounds were sufficient to break 80-90% of cells. The sonicate was centrifuged twice at 200 × g at 4 °C for 10 min to remove unbroken cells. Nine fractions pelleted at different centrifugation speeds were collected and the final supernatant after 120,000 × g represents the cytosol-enriched fraction. Collected pellets were resuspended in 50-100 μL of membrane solubilising buffer [50 mM HEPES pH 8.5, 8 M urea, 0.2 % (w/v) SDS]. Protein concentration was measured by BCA Protein Assay Kit (Sigma-Aldrich) according to the manufacturer’s instructions.
Protein digestion and TMT labeling
A total of 60 μg of protein from each fraction was bound to magnetic HILIC beads according to the SP3 protocol published by Hughes et al.114 After washing out the buffer with 80% (v/v) EtOH, the protein-bead mixture was resuspended in 100 mM triethylammonium bicarbonate (TEAB), disulphide bridges were reduced with 10 mM Tris(2-carboxyethyl)phosphine (TCEP) and cysteines were modified with 50 mM chloroacetamide (one step reaction 30 min at 60 °C). To digest proteins, 2 μg of trypsin was added to the reaction and incubated overnight at 37 °C. The next day, samples were centrifugated and the collected supernatants were acidified with trifluoroacetic acid (TFA) to a final concentration of 1% (v/v). At this step, samples were divided into technical triplicates. 20 μg of peptides were desalted on a homemade desalting column,115 reconstituted in 17μL 100mM TEAB, and labeled with TMT 10plex isobaric tagging reagent (Thermo Fisher Scientific). 0.8 mg of TMT 10plex reagents were resuspended in 42 μL of anhydrous acetonitrile (ACN). Then 6.4 μL of the TMT reagent was added to each fraction sample and incubated for one hour at room temperature. The reaction was stopped by the addition of 1.5 μL of 5% (v/v) hydroxylamine. Subsequently, the respectively indicated fractions were mixed. After evaporation, the sample was desalted on an Opti-trap cartridge C18 column (Optimize Technologies).
The resulting mixture was fractionated on a reverse phase at high pH, flow 2 μL/min, YMC column (300 mm, 0.3 mm, 1.9 μm), linear 60 min gradient [buffer A – 20 mM ammonium formate, 2% (v/v) ACN; buffer B – 20 mM ammonium formate, 80% (v/v) ACN] from 1% B to 60% B.116 Fractions were collected for 64 minutes, and these were combined into 8 resulting pooled fractions by the farthest neighbour method described in Wang et al.117
The fractionated peptides were resuspended in 1% (v/v) TFA + 2% ACN and roughly 1.5 μg were injected onto nanoHPLC Dionex UltiMate 3000RS (Thermo Fisher Scientific) in combination with Thermo Orbitrap Fusion. Thermo PepMap Trap column (pn: 160454) was used for peptides preconcentration and main separation was done on 50 cm EASY Spray Column Thermo (pn: ES903). Each fraction was separated on a 180 min gradient from 2% A to 35% B [A – 0.1% (v/v) formic acid, B – 99.9% ACN, 0.1% (v/v) formic acid]. Cycle time was set to 4 s. The MS2 spectra for identification were measured in an ion trap with CID fragmentation, 60 ms maximum injection time, and the quantification MS3 spectra were measured in an Orbitrap with HCD fragmentation using the SPS function, 118 ms maximum injection time, 10 precursors for synchronous precursors selection feature.118
Electron microscopy
Approximately 3 L of P. pyriformis culture was filtered and fractionated as described above. Pellets of the first six collected fractions were resuspended in 1 mL of fixation buffer containing 200 mM sodium cacodylate (pH 7.4) and 2.5% (v/v) glutaraldehyde. After 3 h of incubation at room temperature, samples were washed twice by 100 mM sodium cacodylate. Next, post-fixation with 2% (v/v) OsO4 in 100 mM sodium cacodylate (pH 7.4) buffer was carried out on ice for 1 hour. Dehydration of fixed subcellular fractions was performed through an ethanol series (35, 50, 70, 80, 95, 100%) and then ethanol was replaced with 100% acetone. Infiltration with a mixture of Epon resin and 100% acetone at a ratio 1:2, 1:1, 2:1 (one hour each) was followed by overnight incubation with absolute Epon resin. After polymerization of the resin (65 °C, 48 h) ultrathin sections were prepared using a Reichert-Jung Ultracut E ultramicrotome and collected onto carbon-and-Formvar coated copper grids. Ultrathin sections were post-contrasted with 4% (w/v) uranyl acetate and 2% (w/v) lead citrate. Prepared sections were examined JEOL 1011 transmission electron microscope.
RNA extraction and cDNA synthesis
Total RNA was extracted from 2 L of P. pyriformis culture using TRI Reagent (Sigma-Aldrich) Prior RNA extraction cells were filtered and washed as described above. From 75 μg of total RNA, mRNA was isolated using Dynabeads mRNA Purification Kit (Invitrogen). Obtained mRNA was further used to prepare cDNA using SMARTer PCR cDNA synthesis Kit (Takara Bio) through 18 cycles of amplification. Synthesized cDNA was stored at -20°C. All steps of preparing cDNA were done according to the manufacturer's instructions of used kits and reagents.
Immunofluorescence microscopy
Slides for immunostaining were prepared as described previously in Dawson et al.101 with slight modifications. T. vaginalis cells were fixed with 4% (v/v) formaldehyde for 30 min at room temperature. Fixed cells were spread on coverslips coated with 0.1% (v/v) poly-L-lysine solution then left 30 min to adhere and air-dried.
T. vaginalis cells were stained with anti-HA rat monoclonal antibody (Roche) at dilution 1:500 to examine subcellular localization of overexpressed P. pyriformis genes. Hydrogenosomes were stained with rabbit anti-malic enzyme antibody (kindly provided by prof. Jan Tachezy) at a dilution of 1:500. Secondary antibodies Alexa488 Goat anti-Rat (Invitrogen) and Alexa594 Goat anti-Rabbit (Invitrogen) were used at dilution 1:1000. All antibodies were diluted in PEMBALG buffer [100 mM PIPES, pH 6.9, 1 mM EGTA, 0.1 mM MgSO4, 100 mM lysine, and 0.5% (w/v) cold water fish skin gelatine]. Coverslips were mounted to the slides using VECTASHIELD Mounting Medium with DAPI (Vector Laboratories).
Images of prepared slides were captured using a Leica SP8 confocal microscope. Deconvolution of images were performed using Huygens Professional (version 21.10). Deconvolved images were further processed using Fiji software (version 1.53f51).119
Crude fractionation of T. vaginalis
Localization of 5 selected P. pyriformis proteins expressed in Trichomonas vaginalis was examined by subcellular fractionation and obtained samples were further analysed by immunoblotting. For each fractionation 0.5 L of culture was harvested by centrifugation at 2000 × g at 4 °C for 10 min, the pellet was washed once with 20 ml of PBS and centrifugate again at 1000 × g at 4 °C for 10 min. Pelleted cells were resuspended in 20 ml of cold ST buffer (20 mM Tris-HCl, pH 7.2, 250 mM sucrose, 1 mM KCl) supplemented with proteinase inhibitors (50 μg/ml TLCK and 10 μg/ml leupeptin). Cells were homogenised by two rounds of sonication of 30 s with a pulse of 1 s ON / 1 s OFF at an amplitude of 40%, using a standard 1/8” diameter probe (Qsonica). To remove unbroken cells and nuclei sonicate was centrifuged 800 × g at 4 °C for 10 min. Cell lysate was further centrifuged at 17,000 × g at 4 °C for 20 min to separated large granules from cytosol. The crude cytosolic fraction was centrifuged at 100,000 × g at 4 °C for 20 min and the supernatant represent final cytosolic-enriched fraction. The obtained pellet was also kept. The fraction containing large granules was washed once with ST buffer to remove remaining cytosol and pelleted again. The upper white layer enriched in lysosomes was carefully separated from the lower brown layer enriched in hydrogenosomes and transferred into new tube. Both fractions were washed with 1 ml of ST buffer and sedimented by centrifugation at 17,000 × g at 4 °C for 15 min.
Protein protection assay
The protein protection assay was performed on freshly isolated hydrogenosomes, the total volume of the reaction was 400 μl. 50 μg of hydrogenosomes resuspended in ST buffer was treated with 50 μg of proteinase K for 20 min on ice. After this time, the reaction was stopped by adding PMSF to final concentration 2 mM, samples were further centrifuged at 20,000 × g at 4 °C for 10 min. The supernatant was removed, and the pellet was resuspended in 50 μl of 1x SB. To the samples additionally treated with detergents, TritonX-100 and SDS were added to a final concentration of 1% and 0.5%, respectively.
Immunoblotting
Protein concentration in each fraction was measured by BCA Protein Assay Kit (Sigma-Aldrich) and 10 μg of each sample were resolved by SDS-PAGE using 12% (v/v) polyacrylamide gels. Samples were electrophoretically transferred onto PVDF membranes (Amersham). Non-specific binding was blocked by incubation of the membranes in 5% (w/v) non-fat milk in TBST [Tris-buffered saline, 0.1% (v/v) Tween 20]. Membranes were incubated overnight at 4°C with primary antibody (anti-HA, anti-ME), washed 3×10 min with TBST and incubated with a secondary antibody conjugated to HRP. After 1 h of incubation, membranes were washed 3×10 min with TBST. To visualise protein bands via chemiluminescence, Clarity ECL Western Blotting Substrate (Bio-Rad) was applied to the PVDF membrane and detected on an Amersham Imager 600.
Quantification and statistical analysis
Raw LC-MS data processing
Raw data from each biological replicate were searched with Proteome Discoverer 2.4 with Sequest search engine. Method was based on predefined workflow for SPS MS3 isobaric quantification and was used with batch specific correction parameters for TMT 10plex label. Database of Paratrimastix pyriformis (13,533 entries) was used to search along with common contaminants database and databases of selected bacterial taxa known to be co-cultivated with P. pyriformis (ASM2556v1 Enterobacter cloacae, ASM63591v2 Brevundimonas naejangsanensis, ASM64851v1 Citrobaster freundii, ASM76115v1 Pseudomonas rhizosphaerae, ASM383324v1 Paenibacillus xylanexedens, ASM385183v1 Pseudomonas chlororaphis and GCF_900105615.1 Clostridium sphenoides). Dynamic modifications were set as follows: Oxidation / +15.995 Da (M). Protein N-Terminal: Acetyl / +42.011 Da, Met-loss / -131.040 Da (M), Met-loss+Acetyl / -89.030 Da (M). Static Modifications: Carbamidomethyl / +57.021 Da (C), TMT6plex / +229.163 Da (K), Peptide N-Terminus: TMT6plex / +229.163 Da. To assess the false discovery rate (FDR), a percolator was used. For reporter ions detection 20 pmm tolerance was set with the most confident centroid peak integration.
LOPIT-DC data processing
Data were processed in R (version 4.0.4)21 using pRoloc package (version 1.30.0)103 according to the published Bioconductor workflows.69,120 Briefly, experimental replicates were pre-processed to allow decontamination and normalization. Bacterial and other contaminants were removed. Then, average reporter intensity was calculated for each feature based on three technical replicates of the LC-MS measurement. All proteins that contained one or more missing values across ten labeled fractions were removed from the dataset by filterNA function. Normalization of protein intensity-interval was performed by the sum method (each reporter intensity was divided by the sum of the reporter ions intensities). After normalization, four independent LOPIT-DC experiments were merged into one dataset containing 4,541 proteins common for all four replicates.
To search for the subcellular compartments, we used the HDBSCAN unsupervised clustering121 available in Python, and semi-supervised phenotype discovery algorithm (phenoDisco)104 available in the pRoloc package of R. The following parameters were applied for HDBSCAN: min_cluster_size=10, min_samples=8, cluster_selection_method=leaf'. Phenotype discovery was run several times with the group size (GS, minimum number of proteins per group) set to 10, 20 or 30. The content of newly found clusters by either HDBSCAN or phenoDisco were carefully evaluated. Finally, by using these two methods as well as gene annotation, conserved domains, previous studies, and literature, a list of 226 manually curated proteins defining 11 cell compartments (cytosol, proteasome, ribosome, nucleus, nucleus-chromatin, ER, Golgi, endomembrane vesicles, plasma membrane, MRO, food vacuole) was prepared. Different populations of endosomes were merged to form a single class, and the same was performed for 40S/60S ribosomal subunits.
To assign proteins with unknown localization into subcellular clusters two supervised machine learning approaches were used: support vector machine (SVM) and t-augmented Gaussian mixture (TAGM) model105 both available in pRoloc package. In the case of SVM the parameters were optimised as described.69 The best sigma and cost parameters were chosen (sigma = 0.1, cost = 8) based on F1 score. For most clusters, median was used to set up the cut-off scores. Threshold scores were as follows: Cytosol = 0.55, Proteasome = 0.53, Ribosome = 0.63, Nucleus = 0.5, Nucleus-chromatin = 0.42, ER = 0.63, Golgi = 0.4, Endomembrane vesicles = 0.45, Plasma membrane = 0.53, MRO = 0.55, Food vacuole = 0.3. Any prediction of protein localization with a score below these thresholds was labeled as unknown. TAGM MAP, the other used classification method, assigns proteins into compartments based on a probabilistic approach.105 Uniform localization threshold set to 99% probability was applied for this method. To reduce dimensions and visualise the data, principal component analysis (PCA) was used.
Phylogenetic and in silico structural analyses
To clarify the identity of the hypothetical proteins of the P. pyriformis MRO, we employed a combination of phylogenetic reconstruction and in silico structure prediction. For three proteins (PaPyr5495, PaPyr7077 and PaPyr804) we could retrieve multiple BLAST hits, based on which we created HMMER v3.3 (www.hmmer.org)49 profiles for a more comprehensive search of homologues in the NCBI nonredundant protein (nr) database and EukProt.60 Regions flanking the domains of interest as determined by InterProScan v5.52-86.0109 were manually removed in Geneious Prime v2020.1.2107 to allow more reliable sequence alignments. The resulting datasets were aligned by MAFFT v7.453108 using the L-INS-i refinement and a maximum of 1,000 iterations, followed by manual trimming; alignments are attached in Data S1F. ML trees (Figure 4; Data S2) were inferred by IQ-TREE v 1.6.12 using the Posterior Mean Site Frequency (PMSF) empirical model with a LG+C20+F+Γ guide tree, tested by the ultrafast bootstrapping strategy (1,000 replicates) and approximate likelihood ratio test (SH-aLRT 1,000 replicates).106,122 Obvious contaminants were manually pruned based on intermediary phylogenies as well as functional domain predictions by InterProScan. For PaPyr577, a single homologue was found in Trimastix marina (AC: Trimastix_PCT|3971) and the resulting HMMer profile did not recover more homologues in EukProt. To find Pex19 (PaPyr8690), and the members of Tim17/22/23 protein family (PaPyr5590, PaPyr9, PaPyr1418), we used previously constructed alignments kindly provided by Kristína Záhonová and Vojtěch Žárský.58 In addition, we analysed the phylogenetic relationships of enzymes of the cytosolic and MRO folate pathways. Their phylogenetic analysis was performed as above.
For the hypothetical proteins lacking any sequence homologues and thus phylogenetic support, SWISS-MODEL50 and PHYRE2102 structure-based searches were performed (last online access in October 2021) to identify putative functions.
Testing of MRO targeting prediction
For performance comparison of targeting predictors, 30 complete, reliably predicted proteins from the MRO were selected along with 30 complete, randomly chosen, and reliably predicted proteins from each of the remaining compartments. These were analysed by the following algorithms: Cello v2.5 in eukaryote mode (last accessed 5 February 2022),31 iPSORT in non-plant mode,32 MitoFates v1.1 in Metazoa mode,34 MultiLoc2 in animal and fungal modes,35 NommPred v0.2 in MRO mode,36 DeepLoc v2.0 in accurate mode33 and TargetP v2.0 in non-plant mode.38 iMLP predictions in non-plant mode27 (REF) were performed on a smaller subset due to a high false positive rate. From tabulated results, sensitivity and specificity were calculated using formulae:
| (Equation 1) |
| (Equation 2) |
Other than default thresholds were also considered when optimising these statistics but, although ambiguous in a few cases, the decisions made by DeepLoc 2.0 were generally most accurate.
Acknowledgments
This project has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement no. 771592), Grant Agency of Charles University (project no. 1162320), and the Ministry of Education, Youth and Sports (MEYS) of the Czech Republic (CR) in the Centgre for Research of Pathogenicity and Virulence of Parasites (project no. CZ.02.1.01/0.0/0.0/16_019/0000759). We would like to thank Jan Tachezy for providing an anti-malic enzyme antibody, Kristina Záhonová for providing Pex proteins alignments, Konstantin Barylyuk for useful comments regarding data processing, and Jan Pyrih for discussions on 1C metabolism. We acknowledge Imaging Methods Core Facility at BIOCEV, an institution supported by the MEYS CR (Large RI project LM2018129 Czech-BioImaging), and ERDF (project no. CZ.02.1.01/0.0/0.0/18_046/0016045) for their support with obtaining imaging data presented in this paper. Computational resources were supplied by the project “e-Infrastruktura CZ” (e-INFRA CZ LM2018140) supported by the MEYS CR.
Author contributions
V.H. conceived the study and P.P.-D. and V.H. supervised the project. J.Z. performed DC-LOPIT and analyzed data with S.C.T. K.H. performed mass spectrometry analyses; Z.F. performed phylogenetic analyses and evaluation of targeting predictors; and Z.V., D.Z., P.P.-D., and J.Z. performed experiments with the heterologous expression system. Z.F. and J.Z. prepared the figures. J.Z., V.H., Z.F., and P.P.-D. wrote the manuscript. All authors read and approved the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: November 7, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.cub.2022.10.028.
Supplemental information
A) Non-normalised abundance of TMT-labelled peptides and proteins metadata. B) Normalised abundance of TMT-labelled peptides and outcomes of classification methods. C) Calculated net charge of the first 10, 20 and 30 amino acids of MRO proteins. D) Evaluation of targeting predictors. E) Proteins predicted to localise in MRO by DeepLoc 2.0. F) Sequences of primers used for heterologous expression system, Related to STAR Methods.
A) Phylogenetic position of TRIC ion channel (PaPyr804) protein. Maximum likelihood phylogenetic tree constructed using the IQ-TREE’s Posterior Mean Site Frequency model with 1,000 ultra-fast bootstrap replicates and tested with 1,000 SH-aLRT replicates (see STAR Methods). Support values are shown where at least one of UFBoot or SH-aRLT were >50. The P. pyriformis protein PaPyr804 is highlighted by a black background. Albeit the positions of P. pyriformis sequence is not well-supported, its homology with the TRIC protein family is unequivocal. B) Phylogenetic position of Tim44 (PaPyr7077) protein. Maximum likelihood phylogenetic tree constructed and edited as in A. Albeit the positions of P. pyriformis sequence is not well-supported, its homology with the Tim44 protein family is unequivocal. C) Phylogenetic position of peroxin-3 (PaPyr5495) protein. Maximum likelihood phylogenetic tree constructed and edited as in A. Along with peroxin-3, the phylogeny of its interacting partner peroxin-19 is shown. In the lower panel, the alignment of selected Pex19 homologs is shown with the Pex3-interacting domain highlighted in red. Although this is not recognized by the alignment algorithm, this domain is characterised by a sequence of hydrophobic and charged amino acids. The core peroxin-19 domain is located downstream. In the case of PaPyr8690, this domain is not identified by HHpred or Pfam and was inferred by homology with the sequence from Trimastix marina (shown in opaque colors).
Data and code availability
-
•
The predicted proteome of Paratrimastix pyriformis has been deposited at Zenodo and is publicly available as of the date of publication. The mass-spectrometry-based proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository. Alignments used to construct phylogenetic trees are deposited at OSF. All accession numbers are listed in the key resources table. Original western blots and microscopy data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Karnkowska A., Vacek V., Zubáčová Z., Treitli S.C., Petrželková R., Eme L., Novák L., Žárský V., Barlow L.D., Herman E.K., et al. A eukaryote without a mitochondrial organelle. Curr. Biol. 2016;26:1274–1284. doi: 10.1016/j.cub.2016.03.053. [DOI] [PubMed] [Google Scholar]
- 2.Roger A.J., Muñoz-Gómez S.A., Kamikawa R. The origin and diversification of mitochondria. Curr. Biol. 2017;27:R1177–R1192. doi: 10.1016/j.cub.2017.09.015. [DOI] [PubMed] [Google Scholar]
- 3.Santos H.J., Makiuchi T., Nozaki T. Reinventing an organelle: the reduced mitochondrion in parasitic protists. Trends Parasitol. 2018;34:1038–1055. doi: 10.1016/j.pt.2018.08.008. [DOI] [PubMed] [Google Scholar]
- 4.Leger M.M., Kolisko M., Kamikawa R., Stairs C.W., Kume K., Čepička I., Silberman J.D., Andersson J.O., Xu F., Yabuki A., et al. Organelles that illuminate the origins of Trichomonas hydrogenosomes and Giardia mitosomes. Nat. Ecol. Evol. 2017;1:0092. doi: 10.1038/s41559-017-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mi-ichi F., Abu Yousuf M.A., Nakada-Tsukui K., Nozaki T. Mitosomes in Entamoeba histolytica contain a sulfate activation pathway. Proc. Natl. Acad. Sci. USA. 2009;106:21731–21736. doi: 10.1073/pnas.0907106106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beltrán N.C., Horváthová L., Jedelský P.L., Šedinová M., Rada P., Marcinčiková M., Hrdý I., Tachezy J. Iron-induced changes in the proteome of Trichomonas vaginalis hydrogenosomes. PLoS One. 2013;8:e65148. doi: 10.1371/journal.pone.0065148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rada P., Doležal P., Jedelský P.L., Bursac D., Perry A.J., Šedinová M., Smíšková K., Novotný M., Beltrán N.C., Hrdý I., et al. The core components of organelle biogenesis and membrane transport in the hydrogenosomes of Trichomonas vaginalis. PLoS One. 2011;6:e24428. doi: 10.1371/journal.pone.0024428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jedelský P.L., Doležal P., Rada P., Pyrih J., Šmíd O., Hrdý I., Šedinová M., Marcinčiková M., Voleman L., Perry A.J., et al. The minimal proteome in the reduced mitochondrion of the parasitic protist Giardia intestinalis. PLoS One. 2011;6:e17285. doi: 10.1371/journal.pone.0017285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jerlström-Hultqvist J., Einarsson E., Xu F., Hjort K., Ek B., Steinhauf D., Hultenby K., Bergquist J., Andersson J.O., Svärd S.G. Hydrogenosomes in the diplomonad Spironucleus salmonicida. Nat. Commun. 2013;4:2493. doi: 10.1038/ncomms3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang Y.K., Chien K.Y., Huang K.Y., Cheng W.H., Ku F.M., Lin R., Chen T.W., Huang P.J., Chiu C.H., Tang P. Responding to a zoonotic emergency with multi-omics research: Pentatrichomonas hominis hydrogenosomal protein characterization with use of RNA sequencing and proteomics. Omics. 2016;20:662–669. doi: 10.1089/omi.2016.0111. [DOI] [PubMed] [Google Scholar]
- 11.Schneider R.E., Brown M.T., Shiflett A.M., Dyall S.D., Hayes R.D., Xie Y., Loo J.A., Johnson P.J. The Trichomonas vaginalis hydrogenosome proteome is highly reduced relative to mitochondria, yet complex compared with mitosomes. Int. J. Parasitol. 2011;41:1421–1434. doi: 10.1016/j.ijpara.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vacek V., Novák L.V.F., Treitli S.C., Táborský P., Cepicka I., Kolısko M., Keeling P.J., Hampl V. Fe–S cluster assembly in oxymonads and related protists. Mol. Biol. Evol. 2018;35:2712–2718. doi: 10.1093/molbev/msy168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Braymer J.J., Freibert S.A., Rakwalska-Bange M., Lill R. Mechanistic concepts of iron-sulfur protein biogenesis in biology. Biochim. Biophys. Acta Mol. Cell Res. 2021;1868:118863. doi: 10.1016/j.bbamcr.2020.118863. [DOI] [PubMed] [Google Scholar]
- 14.Hampl V., Silberman J.D., Stechmann A., Diaz-Triviño S., Johnson P.J., Roger A.J. Genetic evidence for a mitochondriate ancestry in the “amitochondriate” flagellate Trimastix pyriformis. PLoS One. 2008;3:e1383. doi: 10.1371/journal.pone.0001383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zubáčová Z., Novák L., Bublíková J., Vacek V., Fousek J., Rídl J., Tachezy J., Doležal P., Vlček C., Hampl V. The mitochondrion-like organelle of Trimastix pyriformis contains the complete glycine cleavage system. PLoS One. 2013;8:e55417. doi: 10.1371/journal.pone.0055417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Kelly C.J., Farmer M.A., Nerad T.A. Ultrastructure of Trimastix pyriformis (Klebs) Bernard et al.: similarities of Trimastix species with retortamonad and jakobid flagellates. Protist. 1999;150:149–162. doi: 10.1016/S1434-4610(99)70018-0. [DOI] [PubMed] [Google Scholar]
- 17.Hrdy I., Hirt R.P., Dolezal P., Bardonová L., Foster P.G., Tachezy J., Embley T.M. Trichomonas hydrogenosomes contain the NADH dehydrogenase module of mitochondrial complex I. Nature. 2004;432:618–622. doi: 10.1038/nature03149. [DOI] [PubMed] [Google Scholar]
- 18.McGlynn S.E., Ruebush S.S., Naumov A., Nagy L.E., Dubini A., King P.W., Broderick J.B., Posewitz M.C., Peters J.W. In vitro activation of [FeFe] hydrogenase: new insights into hydrogenase maturation. J. Biol. Inorg. Chem. 2007;12:443–447. doi: 10.1007/s00775-007-0224-z. [DOI] [PubMed] [Google Scholar]
- 19.Geladaki A., Kočevar Britovšek N., Breckels L.M., Smith T.S., Vennard O.L., Mulvey C.M., Crook O.M., Gatto L., Lilley K.S. Combining Lopit with differential ultracentrifugation for high-resolution spatial proteomics. Nat. Commun. 2019;10:331. doi: 10.1038/s41467-018-08191-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sadowski P.G., Dunkley T.P.J., Shadforth I.P., Dupree P., Bessant C., Griffin J.L., Lilley K.S. Quantitative proteomic approach to study subcellular localization of membrane proteins. Nat. Protoc. 2006;1:1778–1789. doi: 10.1038/nprot.2006.254. [DOI] [PubMed] [Google Scholar]
- 21.R Development Core Team . R: A language and environment for statistical computing. R Foundation for Statistical Computing; 2020. [Google Scholar]
- 22.Gatto L., Breckels L.M., Wieczorek S., Burger T., Lilley K.S. Mass-spectrometry-based spatial proteomics data analysis using pRoloc and pRolocdata. Bioinformatics. 2014;30:1322–1324. doi: 10.1093/bioinformatics/btu013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gatto L., Breckels L.M., Lilley K.S. Assessing sub-cellular resolution in spatial proteomics experiments. Curr. Opin. Chem. Biol. 2019;48:123–149. doi: 10.1016/j.cbpa.2018.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wiedemann N., Pfanner N. Mitochondrial machineries for protein import and assembly. Annu. Rev. Biochem. 2017;86:685–714. doi: 10.1146/annurev-biochem-060815-014352. [DOI] [PubMed] [Google Scholar]
- 25.Roise D., Horvath S.J., Tomich J.M., Richards J.H., Schatz G. A chemically synthesized pre-sequence of an imported mitochondrial protein can form an amphiphilic helix and perturb natural and artificial phospholipid bilayers. EMBO J. 1986;5:1327–1334. doi: 10.1002/j.1460-2075.1986.tb04363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kunze M., Berger J. The similarity between N-terminal targeting signals for protein import into different organelles and its evolutionary relevance. Front. Physiol. 2015;6:259. doi: 10.3389/fphys.2015.00259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schneider K., Zimmer D., Nielsen H., Herrmann J.M., Mühlhaus T. iMLP, a predictor for internal matrix targeting-like sequences in mitochondrial proteins. Biol. Chem. 2021;402:937–943. doi: 10.1515/hsz-2021-0185. [DOI] [PubMed] [Google Scholar]
- 28.Garg S., Stölting J., Zimorski V., Rada P., Tachezy J., Martin W.F., Gould S.B. Conservation of transit peptide-independent protein import into the mitochondrial and hydrogenosomal matrix. Genome Biol. Evol. 2015;7:2716–2726. doi: 10.1093/gbe/evv175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stairs C.W., Táborský P., Salomaki E.D., Kolisko M., Pánek T., Eme L., Hradilová M., Vlček Č., Jerlström-Hultqvist J., Roger A.J., Čepička I. Anaeramoebae are a divergent lineage of eukaryotes that shed light on the transition from anaerobic mitochondria to hydrogenosomes. Curr. Biol. 2021;31:5605–5612.e5. doi: 10.1016/j.cub.2021.10.010. [DOI] [PubMed] [Google Scholar]
- 30.Garg S.G., Gould S.B. The role of charge in protein targeting evolution. Trends Cell Biol. 2016;26:894–905. doi: 10.1016/j.tcb.2016.07.001. [DOI] [PubMed] [Google Scholar]
- 31.Yu C.S., Chen Y.C., Lu C.H., Hwang J.K. Prediction of protein subcellular localization. Proteins. 2006;64:643–651. doi: 10.1002/prot.21018. [DOI] [PubMed] [Google Scholar]
- 32.Bannai H., Tamada Y., Maruyama O., Nakai K., Miyano S. Extensive feature detection of N-terminal protein sorting signals. Bioinformatics. 2002;18:298–305. doi: 10.1093/bioinformatics/18.2.298. [DOI] [PubMed] [Google Scholar]
- 33.Thumuluri V., Almagro Armenteros J.J., Johansen A.R., Nielsen H., Winther O. DeepLoc 2.0: multi-label subcellular localization prediction using protein language models. Nucleic Acids Res. 2022;50:W228–W234. doi: 10.1093/nar/gkac278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fukasawa Y., Tsuji J., Fu S.C., Tomii K., Horton P., Imai K. MitoFates: improved prediction of mitochondrial targeting sequences and their cleavage sites. Mol. Cell. Proteomics. 2015;14:1113–1126. doi: 10.1074/mcp.M114.043083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blum T., Briesemeister S., Kohlbacher O. MultiLoc2: integrating phylogeny and Gene Ontology terms improves subcellular protein localization prediction. BMC Bioinformatics. 2009;10:274. doi: 10.1186/1471-2105-10-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kume K., Amagasa T., Hashimoto T., Kitagawa H. NommPred: prediction of mitochondrial and mitochondrion-related organelle proteins of nonmodel organisms. Evol. Bioinform. Online. 2018;14 doi: 10.1177/1176934318819835. 1176934318819835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hawkins J., Bodén M. Detecting and sorting targeting peptides with neural networks and support vector machines. J. Bioinform. Comput. Biol. 2006;4:1–18. doi: 10.1142/s0219720006001771. [DOI] [PubMed] [Google Scholar]
- 38.Almagro Armenteros J.J.A., Salvatore M., Emanuelsson O., Winther O., von Heijne G., Elofsson A., Nielsen H. Detecting sequence signals in targeting peptides using deep learning. Life Sci. Alliance. 2019;2 doi: 10.26508/lsa.201900429. e201900429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lawrence S.A., Titus S.A., Ferguson J., Heineman A.L., Taylor S.M., Moran R.G. Mammalian mitochondrial and cytosolic folylpolyglutamate synthetase maintain the subcellular compartmentalization of folates. J. Biol. Chem. 2014;289:29386–29396. doi: 10.1074/jbc.M114.593244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tibbetts A.S., Appling D.R. Compartmentalization of mammalian folate-mediated one-carbon metabolism. Annu. Rev. Nutr. 2010;30:57–81. doi: 10.1146/annurev.nutr.012809.104810. [DOI] [PubMed] [Google Scholar]
- 41.Mudd S.H., Brosnan J.T., Brosnan M.E., Jacobs R.L., Stabler S.P., Allen R.H., Vance D.E., Wagner C. Methyl balance and transmethylation fluxes in humans. Am. J. Clin. Nutr. 2007;85:19–25. doi: 10.1093/ajcn/85.1.19. [DOI] [PubMed] [Google Scholar]
- 42.Solmonson A., DeBerardinis R.J. Lipoic acid metabolism and mitochondrial redox regulation. J. Biol. Chem. 2018;293:7522–7530. doi: 10.1074/jbc.TM117.000259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Green D.E., Morris T.W., Green J., Cronan J.E., Guest J.R. Purification and properties of the lipoate protein ligase of Escherichia coli. Biochem. J. 1995;309:853–862. doi: 10.1042/bj3090853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rowland E.A., Snowden C.K., Cristea I.M. Protein lipoylation: an evolutionarily conserved metabolic regulator of health and disease. Curr. Opin. Chem. Biol. 2018;42:76–85. doi: 10.1016/j.cbpa.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fransson A., Ruusala A., Aspenström P. Atypical Rho GTPases have roles in mitochondrial homeostasis and apoptosis. J. Biol. Chem. 2003;278:6495–6502. doi: 10.1074/jbc.M208609200. [DOI] [PubMed] [Google Scholar]
- 46.Dowhan W., Snell E.E. D-serine dehydratase from Escherichia coli. II. Analytical studies and subunit structure. J. Biol. Chem. 1970;245:4618–4628. [PubMed] [Google Scholar]
- 47.Boorstein W.R., Ziegelhoffer T., Craig E.A. Molecular evolution of the HSP70 multigene family. J. Mol. Evol. 1994;38:1–17. doi: 10.1007/BF00175490. [DOI] [PubMed] [Google Scholar]
- 48.Söding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 49.Eddy S.R. Accelerated profile HMM Searches. PLoS Comput. Biol. 2011;7 doi: 10.1371/journal.pcbi.1002195. e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L., et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gincel D., Shoshan-Barmatz V. Glutamate interacts with VDAC and modulates opening of the mitochondrial permeability transition pore. J. Bioenerg. Biomembr. 2004;36:179–186. doi: 10.1023/b:jobb.0000023621.72873.9e. [DOI] [PubMed] [Google Scholar]
- 52.Rostovtseva T., Colombini M. VDAC channels mediate and gate the flow of ATP: implications for the regulation of mitochondrial function. Biophys. J. 1997;72:1954–1962. doi: 10.1016/S0006-3495(97)78841-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yazawa M., Ferrante C., Feng J., Mio K., Ogura T., Zhang M., Lin P.H., Pan Z., Komazaki S., Kato K., et al. TRIC channels are essential for Ca2+ handling in intracellular stores. Nature. 2007;448:78–82. doi: 10.1038/nature05928. [DOI] [PubMed] [Google Scholar]
- 54.Brix J., Dietmeier K., Pfanner N. Differential recognition of preproteins by the purified cytosolic domains of the mitochondrial import receptors Tom20, Tom22, and Tom70. J. Biol. Chem. 1997;272:20730–20735. doi: 10.1074/jbc.272.33.20730. [DOI] [PubMed] [Google Scholar]
- 55.Neupert W., Brunner M. The protein import motor of mitochondria. Nat. Rev. Mol. Cell Biol. 2002;3:555–565. doi: 10.1038/nrm878. [DOI] [PubMed] [Google Scholar]
- 56.Pyrihová E., Motyčková A., Voleman L., Wandyszewska N., Fišer R., Seydlová G., Roger A., Kolísko M., Doležal P. A single Tim translocase in the mitosomes of Giardia intestinalis illustrates convergence of protein import machines in anaerobic eukaryotes. Genome Biol. Evol. 2018;10:2813–2822. doi: 10.1093/gbe/evy215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rassow J., Dekker P.J.T., van Wilpe S., Meijer M., Soll J. The preprotein translocase of the mitochondrial inner membrane: function and evolution. J. Mol. Biol. 1999;286:105–120. doi: 10.1006/jmbi.1998.2455. [DOI] [PubMed] [Google Scholar]
- 58.Žárský V., Doležal P. Evolution of the Tim17 protein family. Biol. Direct. 2016;11:54. doi: 10.1186/s13062-016-0157-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chaudhuri M., Darden C., Soto Gonzalez F.S., Singha U.K., Quinones L., Tripathi A. Tim17 updates: a comprehensive review of an ancient mitochondrial protein translocator. Biomolecules. 2020;10:1–20. doi: 10.3390/biom10121643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Richter D.J., Berney C., Strassert J.F.H., Burki F., de Vargas C. EukProt: a database of genome-scale predicted proteins across the diversity of eukaryotic life. Peer Community J. 2020;2:e56. [Google Scholar]
- 61.Fang Y., Morrell J.C., Jones J.M., Gould S.J. PEX3 functions as a PEX19 docking factor in the import of class I peroxisomal membrane proteins. J. Cell Biol. 2004;164:863–875. doi: 10.1083/jcb.200311131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hampl, V. (2017). Preaxostyla. Handbook of the Protists, Second Edition, pp. 1139–1174.
- 63.Karnkowska A., Treitli S.C., Brzoň O., Novák L., Vacek V., Soukal P., Barlow L.D., Herman E.K., Pipaliya S.V., Pánek T., et al. The oxymonad genome displays canonical eukaryotic complexity in the absence of a mitochondrion. Mol. Biol. Evol. 2019;36:2292–2312. doi: 10.1093/molbev/msz147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lindmark D.G., Muller M., Shio H. Hydrogenosomes in Trichomonas vaginalis. J. Parasitol. 1975;61:552–554. [Google Scholar]
- 65.Doležal P., Vaňáčová S., Tachezy J., Hrdý I. Malic enzymes of Trichomonas vaginalis: two enzyme families, two distinct origins. Gene. 2004;329:81–92. doi: 10.1016/j.gene.2003.12.022. [DOI] [PubMed] [Google Scholar]
- 66.Christoforou A., Mulvey C.M., Breckels L.M., Geladaki A., Hurrell T., Hayward P.C., Naake T., Gatto L., Viner R., Martinez Arias A.M., et al. A draft map of the mouse pluripotent stem cell spatial proteome. Nat. Commun. 2016;7:8992. doi: 10.1038/ncomms9992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Barylyuk K., Koreny L., Ke H., Butterworth S., Crook O.M., Lassadi I., Gupta V., Tromer E., Mourier T., Stevens T.J., et al. A comprehensive subcellular atlas of the Toxoplasma proteome via hyperLOPIT provides spatial context for protein functions. Cell Host Microbe. 2020;28:752–766.e9. doi: 10.1016/j.chom.2020.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mulvey C.M., Breckels L.M., Geladaki A., Britovšek N.K., Nightingale D.J.H., Christoforou A., Elzek M., Deery M.J., Gatto L., Lilley K.S. Using hyperLOPIT to perform high-resolution mapping of the spatial proteome. Nat. Protoc. 2017;12:1110–1135. doi: 10.1038/nprot.2017.026. [DOI] [PubMed] [Google Scholar]
- 69.Breckels L.M., Mulvey C.M., Lilley K.S., Gatto L., Foster L.J., Stekhoven D.J., Zurich E. A bioconductor workflow for processing and analysing spatial proteomics data. F1000Res. 2018;5:2926. doi: 10.12688/f1000research.10411.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ducker G.S., Rabinowitz J.D. One-carbon metabolism in health and disease. Cell Metab. 2017;25:27–42. doi: 10.1016/j.cmet.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tan Y.L., Sou N.L., Tang F.Y., Ko H.A., Yeh W.T., Peng J.H., Chiang E.I. Tracing metabolic fate of mitochondrial glycine cleavage system derived formate in vitro and in vivo. Int. J. Mol. Sci. 2020;21:8808. doi: 10.3390/ijms21228808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Horne D.W., Holloway R.S., Said H.M. Uptake of 5-formyltetrahydrofolate in isolated rat liver mitochondria is carrier-mediated. J. Nutr. 1992;122:2204–2209. doi: 10.1093/jn/122.11.2204. [DOI] [PubMed] [Google Scholar]
- 73.Pasternack L.B., Laude D.A., Appling D.R. Carbon-13 NMR detection of folate-mediated serine and glycine synthesis in vivo in Saccharomyces cerevisiae. Biochemistry. 2002;31:8713–8719. doi: 10.1021/BI00152A005. [DOI] [PubMed] [Google Scholar]
- 74.Cybulski R.L., Fisher R.R. Uptake of oxidized folates by rat liver mitochondria. Biochim. Biophys. Acta. 1981;646:329–333. doi: 10.1016/0005-2736(81)90339-4. [DOI] [PubMed] [Google Scholar]
- 75.Lichtenstein N.S., Oliverio V.T., Goldman I.D. Characteristics of folic acid transport in the L1210 leukemia cell. Biochim. Biophys. Acta. 1969;193:456–467. doi: 10.1016/0005-2736(69)90204-1. [DOI] [PubMed] [Google Scholar]
- 76.Yazaki E., Kume K., Shiratori T., Eglit Y., Tanifuji G., Harada R., Simpson A.G.B., Ishida K.I., Hashimoto T., Inagaki Y. Barthelonids represent a deep-branching metamonad clade with mitochondrion-related organelles predicted to generate no ATP. Proc. Biol. Sci. 2020;287 doi: 10.1098/rspb.2020.1538. 20201538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nývltová E., Stairs C.W., Hrdý I., Rídl J., Mach J., Pa J., Roger A.J., Tachezy J.ɥes. Lateral gene transfer and gene duplication played a key role in the evolution of Mastigamoeba balamuthi hydrogenosomes. Mol. Biol. Evol. 2015;32:1039–1055. doi: 10.1093/molbev/msu408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Záhonová K., Treitli S.C., Le T., Škodová-Sveráková I., Hanousková P., Čepička I., Tachezy J., Hampl V. Anaerobic derivates of mitochondria and peroxisomes in the free-living amoeba Pelomyxa schiedti revealed by single-cell genomics. BMC Biol. 2022;20:56. doi: 10.1186/s12915-022-01247-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rotterová J., Salomaki E., Pánek T., Bourland W., Žihala D., Táborský P., Edgcomb V.P., Beinart R.A., Kolísko M., Čepička I. Genomics of new ciliate lineages provides insight into the evolution of obligate anaerobiosis. Curr. Biol. 2020;30:2037–2050.e6. doi: 10.1016/j.cub.2020.03.064. [DOI] [PubMed] [Google Scholar]
- 80.Gawryluk R.M.R., Kamikawa R., Stairs C.W., Silberman J.D., Brown M.W., Roger A.J. The earliest stages of mitochondrial adaptation to low oxygen revealed in a novel rhizarian. Curr. Biol. 2016;26:2729–2738. doi: 10.1016/j.cub.2016.08.025. [DOI] [PubMed] [Google Scholar]
- 81.Lewis W.H., Lind A.E., Sendra K.M., Onsbring H., Williams T.A., Esteban G.F., Hirt R.P., Ettema T.J.G., Embley T.M. Convergent evolution of hydrogenosomes from mitochondria by gene transfer and loss. Mol. Biol. Evol. 2020;37:524–539. doi: 10.1093/molbev/msz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Salomaki E.D., Terpis K.X., Rueckert S., Kotyk M., Varadínová Z.K., Čepička I., Lane C.E., Kolisko M. Gregarine single-cell transcriptomics reveals differential mitochondrial remodeling and adaptation in apicomplexans. BMC Biol. 2021;19:77. doi: 10.1186/s12915-021-01007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mathur V., Kolísko M., Hehenberger E., Irwin N.A.T., Leander B.S., Kristmundsson Á., Freeman M.A., Keeling P.J. Multiple independent origins of apicomplexan-like parasites. Curr. Biol. 2019;29:2936–2941.e5. doi: 10.1016/j.cub.2019.07.019. [DOI] [PubMed] [Google Scholar]
- 84.Stairs C.W., Eme L., Brown M.W., Mutsaers C., Susko E., Dellaire G., Soanes D.M., van der Giezen M., Roger A.J. A SUF Fe-S cluster biogenesis system in the mitochondrion-related organelles of the anaerobic protist Pygsuia. Curr. Biol. 2014;24:1176–1186. doi: 10.1016/j.cub.2014.04.033. [DOI] [PubMed] [Google Scholar]
- 85.Barberà M.J., Ruiz-Trillo I., Tufts J.Y.A., Bery A., Silberman J.D., Roger A.J. Sawyeria marylandensis (Heterolobosea) has a hydrogenosome with novel metabolic properties. Eukaryot. Cell. 2010;9:1913–1924. doi: 10.1128/EC.00122-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Leger M.M., Eme L., Hug L.A., Roger A.J. Novel hydrogenosomes in the microaerophilic jakobid Stygiella incarcerata. Mol. Biol. Evol. 2016;33:2318–2336. doi: 10.1093/molbev/msw103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nozaki T., Ali V., Tokoro M. Sulfur-containing amino acid metabolism in parasitic protozoa. Adv. Parasitol. 2005;60:1–99. doi: 10.1016/S0065-308X(05)60001-2. [DOI] [PubMed] [Google Scholar]
- 88.Makki A., Rada P., Žárský V., Kereïche S., Kováčik L., Novotný M., Jores T., Rapaport D., Tachezy J. Triplet-pore structure of a highly divergent TOM complex of hydrogenosomes in Trichomonas vaginalis. PLoS Biol. 2019;17 doi: 10.1371/journal.pbio.3000098. e3000098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Saotome M., Safiulina D., Szabadkai G., Das S., Fransson A., Aspenstrom P., Rizzuto R., Hajnóczky G. Bidirectional Ca2+-dependent control of mitochondrial dynamics by the Miro GTPase. Proc. Natl. Acad. Sci. USA. 2008;105:20728–20733. doi: 10.1073/pnas.0808953105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yamaoka S., Hara-Nishimura I. The mitochondrial ras-related GTPase miro: views from inside and outside the metazoan kingdom. Front. Plant Sci. 2014;5:350. doi: 10.3389/fpls.2014.00350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vlahou G., Eliáš M., von Kleist-Retzow J.C., Wiesner R.J., Rivero F. The Ras related GTPase Miro is not required for mitochondrial transport in Dictyostelium discoideum. Eur. J. Cell Biol. 2011;90:342–355. doi: 10.1016/j.ejcb.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 92.Gentekaki E., Curtis B.A., Stairs C.W., Klimeš V., Eliáš M., Salas-Leiva D.E., Herman E.K., Eme L., Arias M.C., Henrissat B., et al. Extreme genome diversity in the hyper-prevalent parasitic eukaryote blastocystis. PLoS Biol. 2017;15 doi: 10.1371/journal.pbio.2003769. e2003769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Covill-Cooke C., Toncheva V.S., Drew J., Birsa N., López-Doménech G., Kittler J.T. Peroxisomal fission is modulated by the mitochondrial Rho-GTPases, Miro1 and Miro2. EMBO Rep. 2020;21 doi: 10.15252/embr.201949865. e49865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cichocki B.A., Krumpe K., Vitali D.G., Rapaport D. Pex19 is involved in importing dually targeted tail-anchored proteins to both mitochondria and peroxisomes. Traffic. 2018;19:770–785. doi: 10.1111/tra.12604. [DOI] [PubMed] [Google Scholar]
- 95.Sugiura A., Mattie S., Prudent J., Mcbride H.M. Newly born peroxisomes are a hybrid of mitochondrial and ER-derived pre-peroxisomes. Nature. 2017;542:251–254. doi: 10.1038/nature21375. [DOI] [PubMed] [Google Scholar]
- 96.Maralikova B., Ali V., Nakada-Tsukui K., Nozaki T., van der Giezen M., Henze K., Tovar J. Bacterial-type oxygen detoxification and iron-sulfur cluster assembly in amoebal relict mitochondria. Cell. Microbiol. 2010;12:331–342. doi: 10.1111/j.1462-5822.2009.01397.x. [DOI] [PubMed] [Google Scholar]
- 97.Britt R.D., Rao G., Tao L. Biosynthesis of the catalytic H-cluster of [FeFe] hydrogenase: the roles of the Fe–S maturase proteins HydE, HydF, and HydG. Chem. Sci. 2020;11:10313–10323. doi: 10.1039/d0sc04216a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lloyd D., Ralphs J.R., Harris J.C. Giardia intestinalis, a eukaryote without hydrogenosomes, produces hydrogen. Microbiol. (Reading) 2002;148:727–733. doi: 10.1099/00221287-148-3-727. [DOI] [PubMed] [Google Scholar]
- 99.Smutná T., Dohnálková A., Sutak R., Narayanasamy R.K., Tachezy J., Hrdý I. A cytosolic ferredoxin-independent hydrogenase possibly mediates hydrogen uptake in Trichomonas vaginalis. Curr. Biol. 2022;32:124–135.e5. doi: 10.1016/j.cub.2021.10.050. [DOI] [PubMed] [Google Scholar]
- 100.RStudio Team (2020). RStudio: integrated development environment for R (RStudio).
- 101.Dawson S.C., Pham J.K., House S.A., Slawson E.E., Cronembold D., Cande W.Z. Stable transformation of an episomal protein-tagging shuttle vector in the piscine diplomonad Spironucleus vortens. BMC Microbiol. 2008;8:71. doi: 10.1186/1471-2180-8-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gatto L., Christoforou A. Using R and bioconductor for proteomics data analysis. Biochim. Biophys. Acta. 2014;1844:42–51. doi: 10.1016/j.bbapap.2013.04.032. [DOI] [PubMed] [Google Scholar]
- 104.Breckels L.M., Gatto L., Christoforou A., Groen A.J., Lilley K.S., Trotter M.W.B. The effect of organelle discovery upon sub-cellular protein localisation. J. Proteomics. 2013;88:129–140. doi: 10.1016/j.jprot.2013.02.019. [DOI] [PubMed] [Google Scholar]
- 105.Crook O.M., Mulvey C.M., Kirk P.D.W., Lilley K.S., Gatto L. A Bayesian mixture modelling approach for spatial proteomics. PLoS Comput. Biol. 2018;14 doi: 10.1371/journal.pcbi.1006516. e1006516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang H.C., Susko E., Roger A.J. The relative importance of modeling site pattern heterogeneity versus partition-wise heterotachy in phylogenomic inference. Syst. Biol. 2019;68:1003–1019. doi: 10.1093/sysbio/syz021. [DOI] [PubMed] [Google Scholar]
- 107.Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C., et al. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jones P., Binns D., Chang H.Y., Fraser M., Li W., McAnulla C., McWilliam H., Maslen J., Mitchell A., Nuka G., et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sonneborn T.M. Methods in the general biology and genetics of Paramecium aurelia. J. Exp. Zool. 1950;113:87–147. [Google Scholar]
- 111.Bertani G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 1951;62:293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Diamond L.S. The establishment of various trichomonads of animals and man in axenic cultures. J. Parasitol. 1957;43:488–490. [PubMed] [Google Scholar]
- 113.Novák L., Zubáčová Z., Karnkowska A., Kolisko M., Hroudová M., Stairs C.W., Simpson A.G.B., Keeling P.J., Roger A.J., Čepička I., Hampl V. Arginine deiminase pathway enzymes: evolutionary history in metamonads and other eukaryotes. BMC Evol. Biol. 2016;16:197. doi: 10.1186/s12862-016-0771-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hughes C.S., Moggridge S., Müller T., Sorensen P.H., Morin G.B., Krijgsveld J. Single-pot, solid-phase-enhanced sample preparation for proteomics experiments. Nat. Protoc. 2019;14:68–85. doi: 10.1038/s41596-018-0082-x. [DOI] [PubMed] [Google Scholar]
- 115.Kulak N.A., Pichler G., Paron I., Nagaraj N., Mann M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat. Methods. 2014;11:319–324. doi: 10.1038/nmeth.2834. [DOI] [PubMed] [Google Scholar]
- 116.Kulak N.A., Geyer P.E., Mann M. Loss-less nano-fractionator for high sensitivity, high coverage proteomics. Mol. Cell. Proteomics. 2017;16:694–705. doi: 10.1074/mcp.O116.065136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang Y., Yang F., Gritsenko M.A., Wang Y., Clauss T., Liu T., Shen Y., Monroe M.E., Lopez-Ferrer D., Reno T., et al. Reversed-phase chromatography with multiple fraction concatenation strategy for proteome profiling of human MCF10A cells. Proteomics. 2011;11:2019–2026. doi: 10.1002/pmic.201000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Le T., Žárský V., Nývltová E., Rada P., Harant K., Vancová M., Verner Z., Hrdý I., Tachezy J. Anaerobic peroxisomes in Mastigamoeba balamuthi. Proc. Natl. Acad. Sci. USA. 2020;117:2065–2075. doi: 10.1073/pnas.1909755117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Crook O.M., Breckels L.M., Lilley K.S., Kirk P.D.W., Gatto L. A bioconductor workflow for the Bayesian analysis of spatial proteomics. F1000Res. 2019;8:446. doi: 10.12688/f1000research.18636.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Campello R.J.G.B., Moulavi D., Sander J. Springer; 2013. Density-based clustering based on hierarchical density estimates. Advances in Knowledge Discovery and Data; pp. 160–172. [Google Scholar]
- 122.Wang H.C., Minh B.Q., Susko E., Roger A.J. Modeling site heterogeneity with posterior mean site frequency profiles accelerates accurate phylogenomic estimation. Syst. Biol. 2018;67:216–235. doi: 10.1093/sysbio/syx068. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A) Non-normalised abundance of TMT-labelled peptides and proteins metadata. B) Normalised abundance of TMT-labelled peptides and outcomes of classification methods. C) Calculated net charge of the first 10, 20 and 30 amino acids of MRO proteins. D) Evaluation of targeting predictors. E) Proteins predicted to localise in MRO by DeepLoc 2.0. F) Sequences of primers used for heterologous expression system, Related to STAR Methods.
A) Phylogenetic position of TRIC ion channel (PaPyr804) protein. Maximum likelihood phylogenetic tree constructed using the IQ-TREE’s Posterior Mean Site Frequency model with 1,000 ultra-fast bootstrap replicates and tested with 1,000 SH-aLRT replicates (see STAR Methods). Support values are shown where at least one of UFBoot or SH-aRLT were >50. The P. pyriformis protein PaPyr804 is highlighted by a black background. Albeit the positions of P. pyriformis sequence is not well-supported, its homology with the TRIC protein family is unequivocal. B) Phylogenetic position of Tim44 (PaPyr7077) protein. Maximum likelihood phylogenetic tree constructed and edited as in A. Albeit the positions of P. pyriformis sequence is not well-supported, its homology with the Tim44 protein family is unequivocal. C) Phylogenetic position of peroxin-3 (PaPyr5495) protein. Maximum likelihood phylogenetic tree constructed and edited as in A. Along with peroxin-3, the phylogeny of its interacting partner peroxin-19 is shown. In the lower panel, the alignment of selected Pex19 homologs is shown with the Pex3-interacting domain highlighted in red. Although this is not recognized by the alignment algorithm, this domain is characterised by a sequence of hydrophobic and charged amino acids. The core peroxin-19 domain is located downstream. In the case of PaPyr8690, this domain is not identified by HHpred or Pfam and was inferred by homology with the sequence from Trimastix marina (shown in opaque colors).
Data Availability Statement
-
•
The predicted proteome of Paratrimastix pyriformis has been deposited at Zenodo and is publicly available as of the date of publication. The mass-spectrometry-based proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository. Alignments used to construct phylogenetic trees are deposited at OSF. All accession numbers are listed in the key resources table. Original western blots and microscopy data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.